Summary
CCNE1-amplified ovarian cancers (OVCAs) and endometrial cancers (EMCAs) are associated with platinum resistance and poor survival, representing a clinically unmet need. We hypothesized that dysregulated cell-cycle progression promoted by CCNE1 overexpression would lead to increased sensitivity to low-dose WEE1 inhibition and ataxia telangiectasia and Rad3-related (ATR) inhibition (WEE1i-ATRi), thereby optimizing efficacy and tolerability. The addition of ATRi to WEE1i is required to block feedback activation of ATR signaling mediated by WEE1i. Low-dose WEE1i-ATRi synergistically decreases viability and colony formation and increases replication fork collapse and double-strand breaks (DSBs) in a CCNE1 copy number (CN)-dependent manner. Only upon CCNE1 induction does WEE1i perturb DNA synthesis at S-phase entry, and addition of ATRi increases DSBs during DNA synthesis. Inherent resistance to WEE1i is overcome with WEE1i-ATRi, with notable durable tumor regressions and improved survival in patient-derived xenograft (PDX) models in a CCNE1-level-dependent manner. These studies demonstrate that CCNE1 CN is a clinically tractable biomarker predicting responsiveness to low-dose WEE1i-ATRi for aggressive subsets of OVCAs/EMCAs.
Keywords: CCNE1 copy number, WEE1, ATR, biomarker, ovarian and endometrial cancer
Graphical abstract

Highlights
CCNE1 induction increases ATR signaling and sensitivity to WEE1i-ATRi treatment
WEE1i-ATRi increases tumor regression in a CCNE1-level-dependent manner in PDXs
Differential molecular effects of WEE1i and ATRi promote replication fork collapse
CCNE1 amplification is a reliable biomarker predictive of response to WEE1i-ATRi
CCNE1 amplification is associated with platinum resistance and poor survival in ovarian and endometrial cancers. Xu et al. identify CCNE1 overexpression as a sensitizer to combination WEE1 and ATR inhibition that allows lower-dosing strategies to decrease toxicity, thus identifying a new treatment for patients with limited options.
Introduction
Gynecological cancers can be heterogeneous in anatomic location, histology, and genomics. High-grade serous ovarian cancer (HGSOC) is the most common and deadly subtype of ovarian cancer.1 Although aggressive histological subtypes of endometrial cancer (EMCA), including uterine serous cancer and uterine carcinosarcomas, account for only 10%–20% of EMCAs, they are responsible for 40% of deaths from this disease.2 Most patients with advanced HGSOC (>80%) and aggressive subtypes of EMCA (50%–60%) experience recurrence and ultimately succumb to their disease.2,3 Survival from these aggressive gynecological cancers has not significantly improved over the past decade.4
CCNE1, an oncogenic driver in many cancers including HGSOC and EMCA, is associated with poor outcome and platinum resistance.5,6 In HGSOC, CCNE1 amplification (CCNE1Amp; copy number [CN] > 5) occurs in 22% of cases, and another 34% exhibit CCNE1 gain (CCNE1Gain; CN 2–5).7 In uterine cancers, CCNE1 is amplified in 50% of serous EMCAs,8 45% of uterine carcinosarcomas,9 and 8% of endometrioid EMCAs.10 Cyclin E1 protein complexes with CDK2 to promote cell-cycle progression from G1 to S phase.7 Overexpression of cyclin E1 promotes premature entry into S phase, resulting in increased stress at replication forks and double-strand DNA breaks (DSBs) that are repaired by the high-fidelity homologous recombination pathway.7,11 TP53 mutations, which are ubiquitous in HGSOC12 and found in >90% of serous EMCAs13 and uterine carcinosarcomas,9 exacerbate the effects of cyclin E1 overexpression.7 Thus, in CCNE1-overexpressing cells, further loss of control at the G1-S checkpoint by aberrant p53 occurs, increasing dependency on S phase and, more importantly, the G2-M cell-cycle checkpoint for survival.14, 15, 16 Targeted combination regimens that strategically exploit these oncogene-dependent vulnerabilities in these diseases are lacking and are the focus of our study.
Critical S and G2-M cell-cycle checkpoint functions are regulated by ataxia telangiectasia and Rad3-related (ATR) and WEE1. ATR is activated by replication stress, blocks G2-M cell-cycle progression, and prevents premature activation of the SLX1-SLX4, MUS81-EME1, and XPF-ERCC1 (SMX) complex, which promotes DSB reformation at replication forks.17,18 Because of its role in suppressing breaks under replication stress, cells with increased oncogenic stress, TP53 defects, or CCNE1 overexpression are especially sensitive to ATR inhibition (ATRi).19, 20, 21, 22 In contrast, WEE1 limits progression from G1 to S and from S/G2 to M through inhibitory phosphorylation of CDK2 and CDK1, respectively.23, 24, 25 Thus, when WEE1 inhibition (WEE1i) occurs, both G1-S and G2-M checkpoints are abrogated, leading to premature S-phase and M-phase entry.19 Notably, WEE1i exacerbates existing high levels of replication stress associated with CCNE1 overexpression, ultimately causing mitotic catastrophe.14,26,27 We hypothesize replication stress caused by cyclin E1 overexpression can be further increased to toxic levels by dual inhibition of WEE1 and ATR that allows lower-dosing strategies, thus selectively promoting cell death and tumor regression while ameliorating toxicity.
The ATRi, AZD6738, is highly selective and potent and has demonstrated activity and tolerability as monotherapy in phase I clinical trials.28,29 It has entered phase II studies both alone and in combination with other DNA-damaging agents (clinicaltrials.gov). Similarly, the WEE1i, AZD1775, is being evaluated in phase I/II clinical trials as monotherapy and in combination with other therapies. The side effects of myelosuppression and diarrhea associated with WEE1i and to a lesser degree with ATRi will require novel dosing schedules.30,31 ATRi and WEE1i combinations have not been tested in humans (clinicaltrials.gov).
We reasoned that inhibiting both WEE1 and ATR (WEE1i-ATRi) will selectively target CCNE1-overexpressing HGSOC and EMCA cells and allow lower-dosing strategies to mitigate off-target toxicity. Herein, we tested this possibility using an experimental drug development platform designed to rapidly move novel therapies into the clinic. CCNE1 induction increases ATR/CHK1 signaling as previously shown.32 We show that a low-dose WEE1i-ATRi combination is synergistic in decreasing HGSOC and EMCA cell survival and colony formation in a CCNE1-level-dependent manner. Dual inhibition led to increased replication stress, mitotic catastrophe, and cell death in CCNE1Amp EMCA and ovarian cancer (OVCA) lines. In cells with high CCNE1, low-dose WEE1i abrogates CDK2 inhibitory phosphorylation and nucleotide incorporation, leading to defective DNA replication at S-phase entry. Adding ATRi to WEE1i increases γH2AX in S phase, leading to fork collapse and suggesting that defective DNA synthesis caused by WEE1i leads to increased reliance on ATR for fork stability. Indeed, WEE1i-ATRi treatment caused significant tumor regression and improved survival compared with WEE1i alone in a CCNE1-level-dependent manner in OVCA/EMCA patient-derived xenograft (PDX) models. We find that a high CCNE1 CN is a reliable genomic biomarker predictive of response to treatment in these tumor models. We have identified a combination treatment exploiting aggressive CCNE1Amp OVCA/EMCAs addressing a clinically important, unmet need.
Results
CCNE1 induction increases ATR signaling and sensitivity to low-dose WEE1-ATR inhibition
We hypothesized that induction of CCNE1 would increase replication stress and dependency on the G2-M checkpoint as reported.11,33 As expected, CCNE1 induction increased expression of pRb, FOXM1, cyclin B1, pCDK1, pCHK2, pATR, ATM/pATM, and WEE1/pWEE1 by reverse-phase protein array (RPPA) analysis; in aggregate, all are involved in S and G2-M cell-cycle progression and checkpoint responses in CCNE1 inducible FT282 (FT282induc) immortalized fallopian tube cells (Figure 1A).7 An increase in cell-cycle checkpoint markers pWEE1 and pCDK1(Y15), a WEE1 substrate, with CCNE1 induction was confirmed (Figure 1B). Given that CCNE1 induction increased markers for G1-S progression, effects on the replication fork stabilizer and G2-M checkpoint regulator, ATR, were evaluated. CCNE1 induction significantly increased both pCHK1(S345) (Figure 1C) and pATR (Figure S1A), suggesting sensitivity to drugs targeting this pathway.
Figure 1.
Cyclin E induction increases ATR signaling and sensitivity to combination WEE1i-ATRi
(A) Select proteins involved in G1-S and G2-M cell-cycle regulation by RPPA analysis from FT282 cells after cyclin E1 induction (FT282 CCNE1induc) by doxycycline treatment (500 nM) for indicated times. Data are presented as the log2 fold change relative to 0 h (n = 2; mean).
(B and C) Immunoblot of the indicated proteins after FT282 CCNE1induc cells were treated with doxycycline at the indicated time points.
(D) Viability of FT282 CCNE1induc cells pretreated ± doxycycline (24 h) followed by 120 h of WEE1i (black) or ATRi (gray) monotherapy at the indicated dosages. The inlay is the immunoblot for cyclin E1 after 120 h ± doxycycline (n = 6; mean ± SD; ± doxycycline: WEE1i, p < 0.0001, ATRi, p = 0.0002; doses highlighted red).
(E) Immunoblot of the indicated proteins in FT282 CCNE1induc cells ± doxycycline and then 24 h of the indicated drug.
(F) Cell viability for FT282 CCNE1induc cells ± doxycycline and then 120 h of the indicated monotherapy and combination (n = 6; mean ± SD; ± doxycycline: p < 0.0001). Coefficient of drug interaction (CDI) relative to fraction affected (Fa) comparing ± doxycycline (blue versus gray), with selected doses highlighted in red (right) (CDI = 0.8, 1.0).
(G and H) Immunoblot for cyclin E1 in OVKATE (G) and SNU685 (H) CCNE1induc cells ± doxycycline at the indicated time points (left) for up to 5 days (right).
(I and J) Cell viability analysis of OVKATE (I) and SNU685 (J) CCNE1induc cells ± doxycycline at the indicated doses (n = 3–4; mean ± SD; ± doxycycline: p < 0.0001 for select combination doses shown in red). CDI relative to Fa comparing ± doxycycline (blue versus gray), with selected doses highlighted in red (right) (OVKATE: CDI = 0.8, 1.1; SNU685: CDI = 0.4, 0.7).
(K and L) Quantification of colony formation for OVKATE (K) and SNU685 (L) CCNE1induc cells ± doxycycline followed by the indicated treatment for 10 days. Doses for representative images are shown in red (± doxycycline: p < 0.0001; n = 3; representative image shown). CDI relative to the Fa plot comparing ± doxycycline (blue versus gray), with selected doses highlighted in red (right) (OVKATE: CDI = 0.05, 0.5; SNU685: CDI = 0.2, 0.6).
Significance determined by two-way ANOVA followed by Tukey’s multiple comparison test in (D), (F), and (I)–(L). Representative data are shown (B–L) for one of 3 biologically independent experiments. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Given that CCNE1 induction increases ATR signaling, we hypothesized that targeting replication stress and cell-cycle checkpoint regulators would lead to more effective anti-tumor responses. With CCNE1 induction, FT282induc cells were more sensitive to monotherapy with WEE1i (AZD1775) than with ATRi (AZD6738), but neither drug alone markedly decreased viability (Figure 1D). FT282induc cells with doxycycline (Dox) treatment for 5 days retained high cyclin E1 protein (Figure 1D). Using monotherapy doses that were not cytotoxic to FT282 control cells (Figure 1D), WEE1i (200 nM) treatment alone, however, significantly increased pCHK1(S345) (Figure 1E), likely due to inhibition of DNA synthesis and consequential ATR activation.34 The addition of ATRi to WEE1i blocked the WEE1i-induced increase of pCHK1 (Figure 1E). WEE1i alone modestly decreased pCDK1(Y15), and the addition of ATRi to WEE1i further decreased pCDK1(Y15), implying loss of the S/G2 to M checkpoint (Figure 1E). Finally, there was a dramatic increase in phospho-histone H2AX (γH2AX), a marker for DNA DSBs,35 with the WEE1i-ATRi combination compared with WEE1i or ATRi alone (Figure 1E). With CCNE1 induction, we found low-dose WEE1i-ATRi significantly decreased viability and was more synergistic across multiple doses tested compared with non-induced cells (±Dox, red, p < 0.0001; Figure 1F). Dosing was optimized for further studies using a low dose of WEE1i (200 nM) or ATRi (1 μM) monotherapy that had maximal effect on CCNE1 inducible fallopian cells and minimal effect on cells without CCNE1 induction (highlighted red in Figures 1D and 1F).
Given that the FT282 cell line consists of immortalized fallopian tube cells, we next explored whether CCNE1 induction in HGSOC and EMCA cells with low baseline cyclin E1 levels would increase sensitivity to WEE1i-ATRi. OVKATE and SNU685 (both CCNE1 CN neutral/low [CCNE1Low], CN < 2, with low cyclin E1 protein) were transfected with inducible CCNE1 (CCNE1induc). Upon CCNE1 induction with doxycycline (Figures 1G and 1H), both OVKATE CCNE1induc and SNU685 CCNE1induc cells were more sensitive to WEE1i or ATRi monotherapy compared with non-induced controls (OVKATE: ATRi, p = 0.0002, WEE1i, p = 0.0009; SNU685: ATRi, p = 0.0057, WEE1i, p = 0.0087; Figures 1I and 1J). WEE1i-ATRi was synergistic and had significantly decreased cell viability (OVATE: p < 0.0001, red highlighting, coefficient of drug interaction [CDI] = 0.8, 1.1; SNU685: p < 0.0001, red highlighting, CDI = 0.4, 0.7, red dots; Figures 1I and 1J), and colony formation ability (Figures 1K and 1L) in cells subjected to CCNE1 induction compared with non-induced controls (Figures 1I–1L). We did not perform knockdown studies given previous reports that CCNE1 knockdown in CCNE1Amp HGSOC cells is not viable.36 Thus, CCNE1 induction increases sensitivity to low-dose WEE1i-ATRi, which is more effective than monotherapy and not toxic to normal fallopian tube cells.
Combination low-dose WEE1i-ATRi decreases viability and colony formation in CCNE1Amp compared with CCNE1Low cells
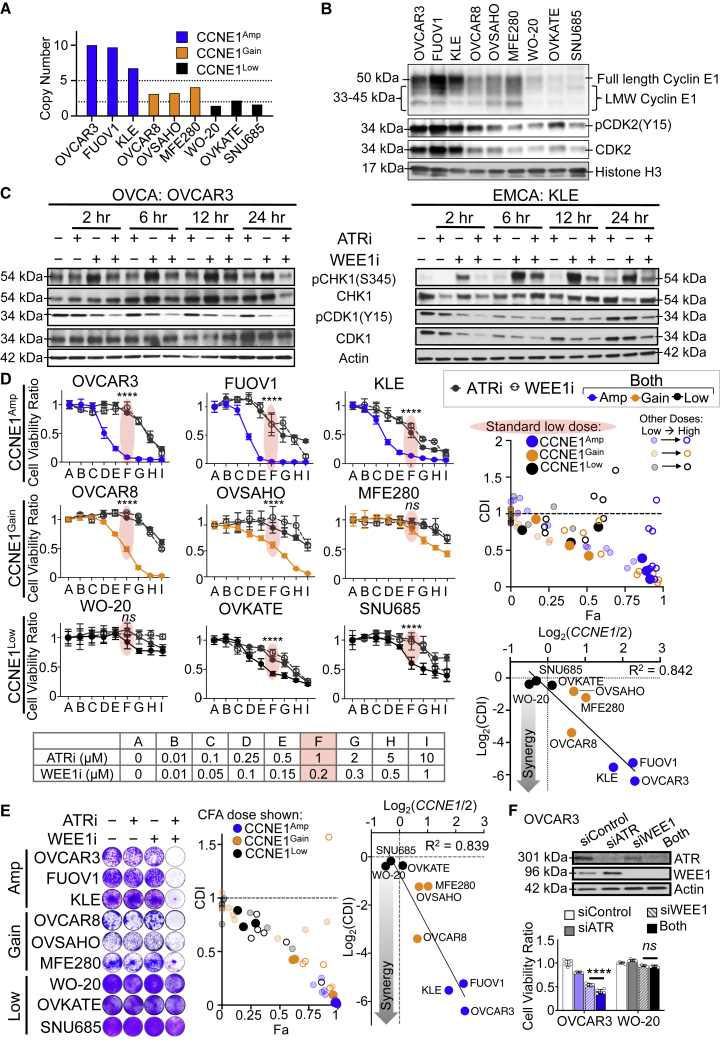
To test whether WEE1i-ATRi treatment effects depend on CCNE1 in established cell lines, HGSOC and EMCA cell models with varying CCNE1 CN and cyclin E1 protein expression levels were studied (Figures 2A and 2B). CCNE1 gene CN levels correlated with cyclin E protein expression in all nine models evaluated (Figures 2A and 2B; Figure S1B).
Figure 2.
Combination WEE1i-ATRi decreases viability and colony formation in CCNE1Amp compared with CCNE1Low cells
(A) Copy number analysis in CCNE1Amp (blue), CCNE1Gain (orange), and CCNE1Low (black) cells based on the Cancer Cell Line Encyclopedia (CCLE) data (https://sites.broadinstitute.org/ccle).
(B) Immunoblot of the indicated proteins, including full-length and low molecular weight (LMW) cyclin E1, in cell lines normalized by cell number. Histone H3 was nuclear loading control.
(C) Immunoblot for indicated proteins in CCNE1Amp OVCAR3 and KLE cells after treatment (200 nM WEE1i, 1 μM ATRi, or both) at the indicated times.
(D) Cell viability of the indicated cell lines after 5 days of treatment at the indicated doses (left). Selected dose is defined as the "standard low dose" for comparison, highlighted in red (n = 3–5; mean ± SD). CDI relative to the Fa plot comparing CCNE1Amp (blue), CCNE1Gain (orange), and CCNE1Low (black) cell lines (right upper). The standard low dose is the bold dot, the higher doses are open dots, and lower doses are faded dots. Synergy is represented by log2(CDI) plotted against CCNE1 CN, displayed as log2(CN/2) for standard low doses (right lower). The trend line with correlation coefficient R2 is shown.
(E) Colony formation (CF) after all lines were treated with 0.25 μM ATRi, 0.05 μM WEE1i, or both for 10 days (n = 3; representative image shown). CF quantified with ImageJ (Figure S1) and mean CF were used to calculate CDI. CDI relative to the Fa plot as in (D) (middle). Log2(CDI) versus CCNE1 CN as in (D) (right).
(F) Knockdown efficiency of WEE1 siRNA (2 nM) and ATR siRNA (10 nM) in OVCAR3 cells measured by immunoblot 48 h post-transfection (top). Viability for combination WEE1 and ATR siRNA treatment in CCNE1Amp OVCAR3 cells and CCNE1Low WO-20 cells at 48 h (n = 5; mean ± SD; CDI = 0.9).
Significance determined by one-way ANOVA followed by Tukey’s multiple comparison test for (D) and (F). Representative data are shown for one of 3 biologically independent experiments. ∗∗∗∗p < 0.0001; ns, not significant.
Treatment with low-dose WEE1i monotherapy increased pCHK1(S345) and modestly decreased pCDK1(Y15) in CCNE1Amp HGSOC (OVCAR3) and EMCA (KLE) cell lines in a time-dependent manner (Figure 2C). The addition of ATRi to WEE1i abrogates the WEE1i-induced increase in pCHK1 by 24 h and decreased pCDK1 more so than with WEE1i alone (Figure 2C). Given that the WEE1i-ATRi combination abrogates critical S and G2-M checkpoint regulators more efficiently than either alone, cell viability was assayed using multiple doses across cell lines (Figure 2D). Low-dose WEE1i-ATRi (200 nM and 1 μM, red highlighting) demonstrated the greatest synergy scores (CDI) and cell fraction affected (Fa), most notably in high cyclin E1 protein, CCNE1Amp cells (solid blue); with intermediate scores in CCNE1Gain cells (solid orange); and minimally in low cyclin E1 protein, neutral CCNE1 CN (CCNE1Low) cells (solid black; Figure 2D). Stronger drug synergy scores correlated with an increasing CCNE1 CN (R2 = 0.842; Figure 2D) and increasing cyclin E1 protein (R2 = 0.561; Figure S1C). WEE1i-ATRi treatment at similar low doses across all cell lines significantly inhibited colony formation the most in CCNE1Amp cells, followed by CCNE1Gain cells, and the least in CCNE1Low cells (Figure 2E; Figure S1D shows the full dose range tested). CDI and fraction affected were the highest in CCNE1Amp cells (solid blue dot), followed by gain models (solid orange dot), and the least in low models (solid black dot; Figure 2E). In addition, a stronger synergy score correlated with an increasing CCNE1 CN (R2 = 0.839; Figure 2E, right) and increasing cyclin E1 protein levels (R2 = 0.529). Higher doses were required to decrease colony formation in CCNE1Low cells (Figure S1D). Furthermore, the combination of WEE1 and ATR knockdown with small interfering RNA (siRNAs) significantly decreased survival in CCNE1Amp cells, but not CCNE1Low cells (OVCAR3; p < 0.0001; WO-20; p = 0.7264; Figure 2F), demonstrating drug effects were not off target. WEE1i-ATRi is more effective than monotherapy, allowing lower-dosing strategies that should mitigate off-target toxicity. High CN amplification provides a genomic biomarker predictive of sufficient CCNE1 protein levels for reliable sensitivity to WEE1i-ATRi. These data suggest that WEE1i-ATRi sensitivity correlates with CCNE1 CN and protein in OVCA/EMCA cell lines.
CCNE1 expression is a biomarker predictive of response to combination WEE1i-ATRi in PDX models
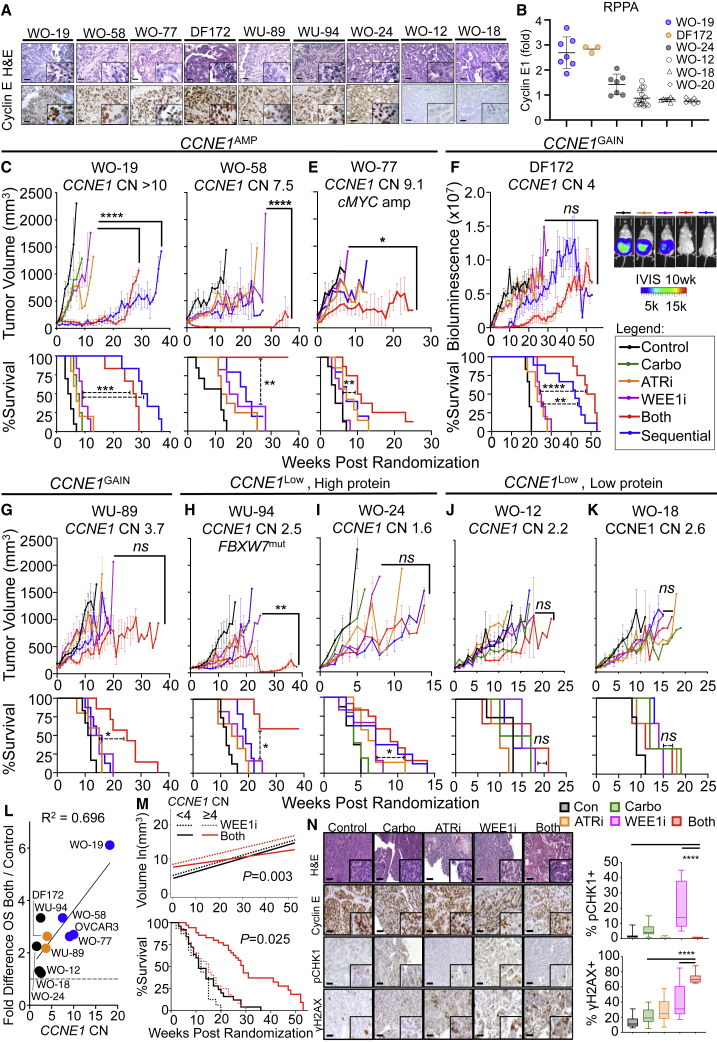
We established a preclinical drug development platform composed of PDX models to identify and optimize drug schedules that exploit genetic vulnerabilities with the goal of bringing scientifically rational therapies into clinical trials (Figure S2A).37, 38, 39 A reliable biomarker for CCNE1 overexpression will be critical for clinical implementation. We thus tested how CCNE1 CN correlates with cyclin E1 protein assessed by immunohistochemistry (IHC) and RPPA analysis in PDX models with a diverse range of CCNE1 CN levels. Despite a broad range of CNs in PDX models (CN > 10 to 1.6; Figures 3C–3K), cyclin E1 protein staining by IHC was less quantitative than RPPA. CCNE1 CN appears to correlate better with cyclin E1 protein by RPPA rather than IHC in the models tested (Figures 3A and 3B; Figure S2B).
Figure 3.
CCNE1 expression is a biomarker predictive of response to combination WEE1i-ATRi in PDX models
(A) Representative H&E and cyclin E1 protein by IHC in PDX models (scale bar, 50 μm; 40× magnification, 100× inlay).
(B) Quantification of cyclin E1 protein by RPPA analysis (n = 4–9; mean ± SD). WO-20 has a CCNE1 CN of 2 and low cyclin E1 protein serving as control.
(C–K) Tumor volume growth curve (upper) and survival by Kaplan-Meier analysis (lower) for CCNE1Amp OVCA PDXs: (C) WO-19, (D) WO-58, and (E) WO-77; CCNE1Gain PDXs: (F) DF-172 OVCA ascites PDX (inlay is IVIS [in vivo imaging system] imaging of treatment groups at 10 weeks) and (G) WU-89 EMCA; CCNE1Low with high cyclin E1 protein PDXs: (H) WU-94 FBXW7MUT EMCA and (I) WO-24 OVCA; and CCNE1Low with low cyclin E protein OVCA PDXs: (J) WO-12 and (K) WO-18 (Table S1). Mice were randomized to treatment groups: (1) control (n = 6–10), (2) carboplatin at 30 mg/kg intraperitoneally (i.p.) weekly (n = 4–5), (3) ATRi at 40 mg/kg/day oral gavage (OG) 5 days weekly (n = 5–8), (4) WEE1i at 60 mg/kg/day OG 5 days weekly (n = 4–6), (5) WEE1i + ATRi 5 days weekly (n = 5–12), and (6) sequential WEE1i at 90 mg/kg/day 7 days weekly during week 1 + ATRi at 50 mg/kg/day 7 days weekly during week 2, and repeat (n = 5–9). For WO-12 and WO-18, n = 3–4 mice per group. For WU-94, WEE1i and ATRi were dosed at 30 mg/kg/day. Treatment continued until progression (tumor volume > 1,000 mm3 or ascites score of 5). Both significantly resulted in tumor regression compared with WEE1i alone in WO-19 (p < 0.0001 for both and sequential), WO-58 (p < 0.0001), WO-77 (p = 0.03), and WO-94 (p = 0.001). Overall survival (OS) was improved in both versus WEE1i in WO-19 (p = 0.0005), WO-58 (p = 0.005), WO-77 (p = 0.001), DF-172 (p < 0.0001), WO-24 (p = 0.04), WU-89 (p = 0.03), and WU-94 (p = 0.01). For exact p values for all comparisons, see Table S2.
(L) Median overall survival for combination relative to control plotted against CCNE1 CN for each PDX model and trend line with correlation coefficient R2 shown.
(M) Higher CCNE1 CN dichotomized at the median value (≥4) is predictive of response (growth rates, log scale; top) and improved OS (bottom) with WEE1i-ATRi combination compared with WEE1i alone in PDX models. Test for interaction: p = 0.003 for differential growth rates using a nested model and p = 0.025 for differential OS. WU-94 was excluded from the analysis because of the FBXW7 mutation.
(N) Representative H&E staining and IHC detection of the indicated proteins (H&E 10× and IHC 20×, with 40× insets) in WO-19 PDX tumors in treatment groups collected at 2–3 weeks on treatment (left) and quantification (right). For boxplots, bound boxes show interquartile range, whiskers show maximum and minimum, and center lines indicate median (n = 3 mice except combination for 2 mice; 9 high power field [HPF]
per tumor; scale bar, 50 μm).
Tumor growth shown is mean ± SEM. Longitudinal tumor growth was analyzed by linear mixed effects modeling with type II ANOVA and pairwise comparisons across groups. Data were analyzed for overall survival using the Mantel-Cox log-rank test. Data analysis for IHC was determined by one-way ANOVA followed by Tukey’s multiple comparison test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
We next tested drug effects in CCNE1Amp (WO-19, WO-58, WO-77, and OVCAR3), CCNE1Gain (DF-172 and WU-89), CCNE1Low with high cyclin E1 protein (WO-24 and WU-94), and CCNE1Low with low cyclin E1 protein (WO-12 and WO-18) PDX models (Figures 3C–3K; representative gross tumor and ultrasound images of PDXs; Figures S2C–S2E). Although carboplatin or WEE1i monotherapy had only modest effects, even at the maximum tolerated doses (MTDs) (Figure S2F), low-dose WEE1i-ATRi resulted in near-complete tumor regression in the CCNE1Amp HGSOC PDX models WO-19 (CN > 10) and WO-58 (CN = 7). Notably, there was an overall response rate of 68%, with complete and partial responses seen in 36% (n = 8/22) and 32% (n = 7/22), respectively, with the drug combination (including concomitant and sequential dosing) in CCNE1Amp PDXs (WO-19 and WO-58). There was significant tumor regression with concomitant WEE1i-ATRi relative to monotherapy groups (p < 0.0001) and significant improvement in survival (both versus WEE1i: p < 0.001 for WO-19, p = 0.005 for WO-58) in these CCNE1Amp PDX models (Figures 3C and 3D; Figure S3, Table S2). Combination treatments were also synergistic by Bliss (synergy index of 0.9 for WO-19 and 0.9 for WO-58; Table S2).40 In WO-19, sequential dosing was superior to concomitant dosing of WEE1i-ATRi in terms of overall survival (p = 0.02; Figure 3C; Figure S3; Table S2), whereas concomitant dosing was better in the WO-58 model (Figure 3D; Figure S3; Table S2). However, both dosing schedules were equally tolerable, as demonstrated by stable mice weights (Figure S4). Two additional CCNE1Amp xenograft models, WO-77 (CN = 9.1; c-MYC amplified) and metastatic OVCAR3 (CN = 10), were also tested. Concomitant WEE1-ATRi synergistically suppressed WO-77 tumor growth (p = 0.02; Figure 3E; Figure S3; Table S2) and OVCAR3 xenograft tumor burden and ascites generation (p = 0.009; Figures S2C, S3, and S4J; Table S2), and improved overall survival (p = 0.001 for WO-77, p < 0.0001 for OVCAR3) relative to WEE1i monotherapy. Response to combination therapy was synergistic relative to monotherapies by Bliss (synergy index of 0.78; Table S2) for WO-77.
The WEE1i-ATRi combination also caused tumor regressions in CCNE1Gain PDX models, including the DF-172 HGSOC (CN = 4) metastatic ascites model and WU-89 EMCA (CN = 3.7), but was less robust than in the CCNE1Amp models (Figures 3F and 3G; Figure S3; Table S2).41 Treatment response was synergistic when using combination treatment compared with monotherapies (synergy index of 0.40 for DF-172 and 0.82 for WU-89), and combination treatment improved overall survival (p < 0.0001 for DF-172 and p = 0.0257 for WU-89; Figures 3F and 3G; Figure S3; Table S2) and suppressed ascites generation (Figure S4K). Sequential dosing showed survival improvement similar to that of concomitant treatment in DF-172 (p = 0.07), but not WU-89 (p = 0.002).
We next evaluated this combination in CCNE1Low PDX models with high cyclin E protein (WU-94: CCNE1 CN = 2.5 and FBXW7 mutation, known to result in increased cyclin E1 protein;42 WO-24: CCNE1 CN = 1.6 and ARID1A mutation) and low cyclin E protein (WO-12: CCNE1 CN = 2.2; WO-18: CCNE1 CN = 2.6; Figures 3A, 3B, and 3H–3K). WU-94 EMCA PDX demonstrated a robust and synergistic response with concurrent WEE1i-ATRi relative to WEE1i monotherapy (p = 0.001; synergy index of 0.75) and improved overall survival (p = 0.01; Figure 3H; Figure S3; Table S2). Although there was a significant improvement in overall survival relative to WEE1i monotherapy (p = 0.039) in WO-24, this difference was small (median overall survival of 9 versus 7 weeks; Figure 3J; Figure S3; Table S2), and there was only a partial response in 8.3% (1/12) of mice with the combination compared with a 36% complete response (CR) rate in CCNE1Amp models (for concurrent and sequential dosing, versus 55% CR for concurrent dosing in WO-19 and WO-58; Figure S3). Finally, the CCNE1Low and cyclin E1 protein low models WO-12 and WO-18 were tested, and no significant difference in tumor response or overall survival compared with WEE1i monotherapy was found (p > 0.05; Figures 3J and 3K; Figure S3; Table S2).
Correlation analysis confirmed that PDXs with higher CCNE1 CNs showed increased overall survival benefit with WEE1i-ATRi combination treatment compared with PDX models with lower CCNE1 CNs (R2 = 0.696; Figure 3L). In addition, a CCNE1 CN of 4 and higher is predictive of better treatment response and overall survival in PDX models in comparing WEE1i-ATRi combination with WEE1i alone (test for interaction for differential tumor growth rate, p = 0.003). WEE1i-ATRi combination is associated with a statistically significantly survival benefit with a hazards ratio (HR) of 0.23 (95% confidence interval [CI] = 0.13, 0.42; p < 0.0001) compared with WEE1i alone. Thus, WEE1i-ATRi reduces the hazard of death of 77% compared with WEE1i. There was no significant survival difference in PDX models with a CCNE1 CN < 4 (test for interaction, p = 0.025; Figure 3M).
Further evaluation of drug effects by pCHK1 and γH2AX IHC in CCNE1Amp PDX (WO-19) revealed that addition of ATRi reversed increased pCHK1 resulting from WEE1i monotherapy treatment and WEE1i-ATRi increased γH2AX protein more than monotherapies (Figure 3N). These PDX data suggest that CCNE1 CN is a reliable biomarker that predicts drug response and improved outcome with WEE1i-ATRi treatment compared with WEEi monotherapy.
WEE1i-ATRi combination increases M-phase entry, replication fork instability, and apoptosis in CCNE1Amp cancer cells
Given that ATRi added to WEE1i blocks WEE1i-induced pCHK1 and further decreases pCDK1, key regulators of the S and G2-M checkpoints, we evaluated drug effects on the cell cycle in CCNE1Amp and CCNE1induc HGSOC and EMCA cells (Figures S5A–S5D). As expected, WEE1i monotherapy significantly decreased G1. More pronounced was the time-dependent increase in G2-M (OVCAR3, p < 0.0001; KLE, p = 0.02) and decrease in S phase with combination WEE1i-ATRi compared with WEE1i monotherapy in both CCNE1Amp cells (OVCAR3 and KLE, p < 0.0001), as well as with CCNE1 induction (OVKATE and SNU685 cells + Dox, p = 0.0001). Furthermore, treatment caused minimal effect in CCNE1Low cells (p > 0.05). Combination WEE1i-ATRi significantly upregulated pHH3, a mitosis marker, compared with WEE1i monotherapy in CCNE1Amp cells (Figures S5E and S5F). Altogether, these results suggest that WEE1i-ATRi leads to loss of G1-S and S/G2-M cell-cycle control with cell buildup in M phase.
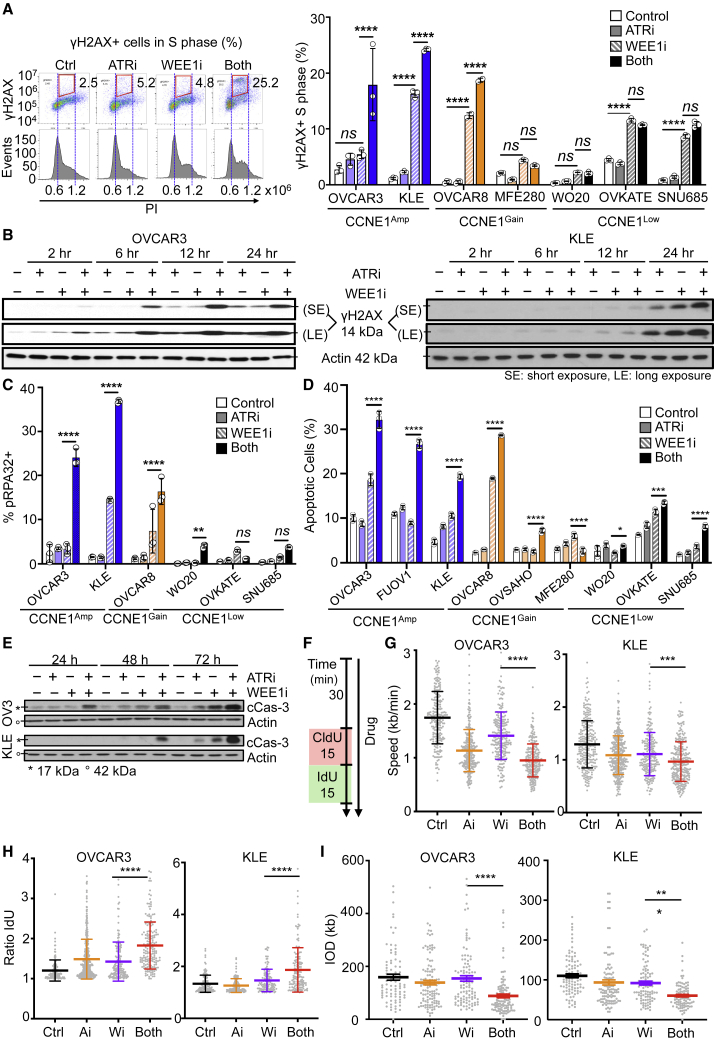
Drug effects on DNA DSBs, replication stress, and apoptosis were next studied in CCNE1Amp versus CCNE1Low cells. To assess DNA damage that occurred in S phase, γH2AX+ cells were gated by DNA content (Figure 4A, left). Treatment with WEE1i alone increased γH2AX induction in most cell lines by 8 h (Figure 4A, right), and the addition of ATRi to WEE1i led to a significant increase in γH2AX in the CCNE1Amp and CCNE1Gain cells compared with CCNE1Low cells. Similarly, combination WEE1i-ATRi significantly induced DNA damage in whole cells by γH2AX at 24 h compared with monotherapies (Figure 4B).
Figure 4.
WEE1i-ATRi combination increases M-phase entry, replication fork instability, and apoptosis in CCNE1Amp cancer cells
(A) Detection of γH2AX-positive cells in S phase of WEE1i-ATRi-treated CCNE1Amp, CCNE1Gain, and CCNE1Low cells. Cells were treated with DMSO (control) or 200 nM WEE1i, 1 μM ATRi, or both for 8 h and then fixed and stained with γH2AX and PI for flow cytometry. γH2AX-positive cells in S phase were quantified. Representative images of OVCAR3 (left) and quantified data (right) are shown (both versus WEE1i monotherapy: OVCAR3, p < 0.0001; KLE, p < 0.0001; OVCAR8, p < 0.0001; MFE280, p = 0.8151; WO-20, p = 0.9997; OVKATE, p = 0.8630; SNU685, p = 0.2171; n = 3; mean ± SD).
(B) Representative immunoblots for γH2AX in OVCAR3 (left) and KLE (right) cells after they were treated as in (A) for the indicated time. Actin is loading control.
(C) Quantification of pRPA32(S33) by flow cytometry of the indicated cells after treatment similar to (A) after 24 h (n = 3; mean ± SD; WEE1i versus both shown by an asterisk).
(D) Flow cytometry quantification of apoptotic cells by Annexin V-APC (allophycocyanin) and propidium iodide (PI) staining of the indicated cells after treatment similar to (A) for 48 h (n = 3; mean ± SD; WEE1i versus both: MFE280, p = 0.0022; OVKATE, p = 0.0084; OVCAR3, p = 0.0005; OVSAHO, p = 0.0001; WO-20, p = 0.0002; SNU685, p < 0.0001).
(E) Representative immunoblots for cleaved caspase-3 in OVCAR3 (upper) and KLE (lower) treated for the indicated time as in (A).
(F) Experimental design for replication fork analysis. OVCAR3 and KLE cells were pretreated with a drug as in (A) for 30 min and pulse labeled with 5-chloro-2′-deoxyuridine (CIdU) (red) followed by 5-iodo-2′-deoxyuridine (IdU) (green) for 15 min each in the continuous presence of inhibitors.
(G) Quantification of replication fork speed (length of track/duration of both pulses). At least 200 intact, unidirectional tracks were counted for each condition (mean ± SEM; WEE1i versus both shown by an asterisk).
(H) Quantification of fork asymmetry as calculated by long green length/short green length replication initiation tracks. At least 130 intact initiation tracks were counted for each condition (mean ± SEM; WEE1i versus both shown by an asterisk).
(I) Quantification of inter-origin distance (IOD) for firing calculated by the distance between two nearby origins on the same fiber. At least 100 intact tracks were counted per condition (mean ± SEM; WEE1i versus both shown by an asterisk).
Individual samples are presented as data points, and data were analyzed using one-way ANOVA followed by Tukey’s multiple comparison test. Representative data are shown (A–E) for one of 3 biologically independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
Given the increase in DSBs with WEE1i-ATRi, we next assessed ATR/ataxia telangiectasia mutated (ATM)/DNA-dependent protein kinase (DNA-PK) activation near single-strand DNA by pRPA32 (a marker for replication stress). The WEE1i-ATRi combination dramatically increased pRPA32 in CCNE1Amp cells (Figure 4C). Effects on pRPA32 were less robust although still significant in CCNE1Gain models but minimal in CCNE1Low models. Further studies revealed that combination WEE1i-ATRi significantly increased apoptosis in CCNE1Amp cells, as shown by increased Annexin V staining and elevated cleaved caspase-3; gave a modest increase in CCNE1Gain cells; and had a smaller increase in the CCNE1Low cells at the same ATRi-WEE1i doses (Figures 4D and 4E). Thus, WEE1i-ATRi increases genomic instability by an increase in γH2AX in S-phase, pRPA, leading to apoptosis in CCNE1Amp cells compared with CCNE1Low cells.
Because DSBs can occur as a consequence of defects in replication fork progression, we examined the effect of WEE1i-ATRi on DNA synthesis rates by more in-depth replication fork studies (e.g., DNA combing) in CCNE1Amp cells, given the increased genomic instability seen (OVCAR3 and KLE;Figures 4F–4I). Both ATRi and WEE1i monotherapy significantly reduced fork speed (Figure 4G). WEE1i-ATRi significantly reduced fork speed relative to control (p < 0.0001), as well as ATRi (OVCAR3, p < 0.0001; KLE, p = 0.004) and WEE1i (OVCAR3, p < 0.0001; KLE, p = 0.0003) monotherapies.
Because a reduced DNA polymerization rate and an increased frequency of fork stalling or collapse can impair replication, we next interrogated replication fork asymmetry (Figure 4H). A decrease in polymerization speed would affect two forks emanating in opposite directions from the same origin equally (ratio close to 1), whereas fork stalling would affect them independently, resulting in fork asymmetry.43 ATRi or WEE1i exposure alone caused a significant increase in fork asymmetry in OVCAR3. More importantly, WEE1-ATRi further increased asymmetric fork ratios relative to ATRi or WEE1i in both OVCAR3 cells (ATRi versus both, p < 0.0001; WEE1i versus both, p = 0.01) and KLE cells (ATRi versus both, p < 0.0001; WEE1i versus both, p < 0.0001).
Replication origins were mapped in the middle of individual replication tracks corresponding to replicons, and inter-origin distances (IODs) were estimated by the distances between midpoints of two adjacent replication tracks. The WEE1i-ATRi combination decreased IOD more than monotherapies in both OVCAR3 and KLE (both versus WEE1i: p < 0.0001 for OVCAR3, p = 0.02 for KLE; Figure 4I). Altogether, these results suggest that WEE1i-ATRi caused replication fork stalling, increased origin firing, and reduced inter-origin distances, likely leading to fork collapse in CCNE1Amp cells to a level that exceeds that by ATRi or WEE1i monotherapy. Collectively, these data suggest that WEE1i-ATRi increased replication fork instability and DSBs, leading to apoptosis in CCNE1Amp HGSOC and EMCA cells.
Differential effects of WEE1i and ATRi upon CCNE1 induction result in fork collapse in early S phase
Next, we sought to determine the mechanism by which CCNE1 induction increases response to WEE1i-ATRi. FT282 immortalized fallopian tube cells were used because they lack mutations characteristic of cancer cells that may impact the effect of these drugs in unpredictable ways. FT282 CCNE1induc cells were arrested in G0 by serum starvation (0.1% fetal bovine serum [FBS]), followed by release (10% FBS) for up to 16 h. CCNE1 induction accelerated entry into S phase (12–16 h without CCNE1induc versus 8 h with CCNE1induc; p < 0.0001; Figure 5A). Notably, while CCNE1induc increased S-phase entry, it also increased the levels and inhibitory phosphorylation of CDK2(Y15), a key regulator of DNA replication origin firing. This phosphorylation was inhibited by low-dose WEE1i (200 nM) alone, but not ATRi treatment (Figure 5B). Premature entry into S phase has been proposed to cause slowed DNA synthesis rates because of the suboptimal synthesis of replication factors and deoxyribonucleotides before origin firing.34 Consistent with these findings, an accumulation of cells in early S phase was observed with WEE1i alone with CCNE1 induction beginning at 8 h (control [Ctrl] versus WEE1i, p < 0.0001; Figure 5C). Moreover, the nucleotide incorporation rate (ratio of mean 5-ethynyl-2'-deoxyuridine (EdU) intensity; S/G0-G1) was significantly decreased by WEE1i monotherapy following CCNE1 induction (Ctrl versus WEE1i, p < 0.0001), suggesting WEE1i leads to defective DNA synthesis (Figure 5D). Accentuated effects on nucleotide incorporation were observed with higher doses of WEE1i (1 μM known to be toxic to normal cells [Figure 1] and show off-target effects), but again, these effects were not observed with ATRi (Figure S6).
Figure 5.
Differential effects of WEE1i and ATRi upon CCNE1 induction results in fork collapse in early S phase
(A) Representation of cell-cycle distribution for FT282 CCNE1induc cells ± doxycycline. Cells were synchronized by 24 h FBS deprivation (0.1%), released with 10% FBS, and collected at the indicated time points (± doxycycline). Cell-cycle distribution was measured with EdU and 7-AAD. Representative images (left) and quantification (right) are shown (n = 3; mean ± SD; p < 0.0001 for S phase ± doxycycline at 12 and 16 h, shown by an asterisk).
(B) Detection of the indicated proteins after FT282 cells were arrested, released as in (A), and treated with DMSO (control) or 1 μM ATRi, 200 nM WEE1i, or both for 8 h (± doxycycline).
(C and D) Cell-cycle distribution after drug treatment as in (B). Representative images (C, left), quantification of early, mid-, and late S phase (C, right) are shown (n = 3; mean ± SD; control versus WEE1i, statistics shown for early S phase, ∗∗∗∗p < 0.0001). Nucleotide incorporation by mean EdU intensity (S/G0-G1) (D) is shown (control versus WEE1i, p < 0.0001).
(E) Detection of S-phase entry and nucleotide incorporation by PCNA and EdU immunofluorescences after cells were treated as in (B) and fixed at 8 h. Percentages of PCNA+, PCNA+EdU+, and PCNA+EdU− cells were quantified by ImageJ. Representative images of cells with CCNE1 induction (bottom) are shown (scale bar, 20 μM; n = 3; mean ± SD; control versus WEE1i, p = 0.012).
(F) Measurement of γH2AX and EdU after treatment as in (B). Representative images (top) and quantification of γH2AX+EdU−, γH2AX+EdU+, and γH2AX+EdU+/EdU+ (bottom) ± doxycycline are presented in CCNE1induc cells. With CCNE1induc, addition of ATRi to WEE1i affected γH2AX+EdU− (WEE1i versus both, p < 0.0001; γH2AX+EdU−, p < 0.0001; γH2AX+EdU+/EdU+, p < 0.0001) at 16 h.
Data were analyzed using one-way ANOVA followed by Tukey’s multiple comparison test (C–F). Representative data are shown for one of 3 biologically independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
Because WEE1i potentiates CDK2 activation, and thereby origin firing, we set out to determine whether WEE1i causes premature entry into S phase. Given that FT282 CCNE1induc cells begin to enter S phase at 8 h with CCNE1 induction, cells were released, treated with drugs for 8 h, and evaluated by immunofluorescence staining of proliferating cell nuclear antigen (PCNA) and EdU (Figure 5E). Surprisingly, we observed no changes of PCNA positivity (PCNA+) in FT282 CCNE1induc cells after WEE1i with CCNE1 induction, suggesting cells enter S phase at the same rate. However, WEE1i decreased PCNA+EdU+ and increased PCNA+EdU− populations (p = 0.012), indicating that WEE1i-treated cells are entering S phase but unable to incorporate nucleotides normally (Figure 5E). We observed a similar trend with combination WEE1i-ATRi treatment. However, ATRi did not cause an observable decrease in nucleotide incorporation in PCNA+ cells (p = 0.26). Altogether, these data indicate that WEE1i has a greater inhibitory effect on nucleotide incorporation than ATRi following S-phase entry. Thus, ATRi and WEE1i have distinct effects on DNA synthesis in early S phase.
Replication stalling is known to cause increased reliance on ATR to prevent replication fork collapse.44 By extension, we hypothesized that WEE1i-mediated defects in DNA replication generate a similar dependence on ATR. Consistent with this model, the addition of ATRi to WEE1i significantly increased DNA DSBs, as evaluated by γH2AX.45,46 This increase was particularly apparent after CCNE1 induction, and it occurred in both cells with highly defective DNA synthesis (γH2AX+EdU−) and those with nucleotide incorporation in S phase (γH2AX+EdU+) (Figure 5F). Although increases in γH2AX-positive cells were also observed with WEE1i alone and ATRi alone, these effects were far less impactful than the WEE1i-ATRi combination (Ctrl versus WEE1i, p = 0.9927; Ctrl versus ATRi, p = 0.0312; Ctrl versus both, p < 0.0001; Figure 5D).
Similar trends in γH2AX were observed in HGSOC and EMCA cells with treatment upon CCNE1 induction (Figures 6A and 6B). In addition, pRPA32 and cleaved caspase-3 double-positive cells significantly increased when using WEE1i-ATRi treatment compared with monotherapy upon CCNE1 induction compared with non-induced cancer models (p < 0.0001; Figures 6C and 6D). This suggests that upon CCNE1 induction and after WEE1i-ATRi treatment, cells demonstrating increased replication stress are undergoing apoptosis. Furthermore, apoptosis detection with Annexin V confirms that WEE1i-ATRi increased cell apoptosis in both HGSOC and EMCA cells upon CCNE1 induction (all p < 0.0001; Figures 6E and 6F). These findings indicate that low-dose WEE1i causes defects in early DNA replication selectively in CCNE1-overexpressing cells and that the addition of ATRi promotes replication fork collapse into DNA DSBs in this context, suggesting distinct mechanisms of action (Figure 7). Thus, CCNE1 overexpression fosters a particular sensitivity to combination WEE1i-ATRi that is associated with distinct effects on DNA synthesis and replication fork stability.
Figure 6.
Combination WEE1i-ATRi leads to replication stress, double-strand DNA breaks, and apoptosis with CCNE1 induction in OVCA and EMCA cell lines
(A and B) OVKATE (A) and SNU685 (B) CCNE1induc cells were pretreated 24 h ± doxycycline; treated with DMSO (control) or 200 nM WEE1i, 1 μM ATRi, or both for 8 h; and evaluated for γH2AX and PI by flow cytometry. γH2AX in OVKATE and SNU685 with CCNE1induc (both versus WEE1i, p < 0.0001; n = 3; mean ± SD).
(C and D) OVKATE (C) and SNU685 (D) CCNE1induc cells were treated as in (A) for 24 h and evaluated for pRPA32 and cleaved caspase-3 by flow cytometry. pRPA32-positive and cleaved caspase-3-positive cells in OVKATE and SNU685 cells with CCNE1induc compared with non-induced (both versus WEE1i, p < 0.0001; n = 3; mean ± SD).
(E and F) OVKATE (E) and SNU685 (F) CCNE1induc cells were treated drug as in (A) for 48 h and evaluated for Annexin V staining by flow cytometry. Annexin V-positive populations in OVKATE and SNU685 cells with CCNE1induc compared with non-induced (both versus WEE1i, p < 0.0001; n = 3; mean ± SD).
Data were analyzed using one-way ANOVA followed by Tukey’s multiple comparison test (C–F). Representative data are shown for one of 3 biologically independent experiments. ∗∗∗∗p < 0.0001; ns, not significant.
Figure 7.
Distinct mechanism of actions for WEE1i and ATRi with CCNE1 overexpression
(A) Normal cell showing the G1-S and G2 -M cell-cycle progression.
(B) When cyclin E1 is overexpressed, there is premature S-phase entry, increased replication initiation, and perturbed replication fork progression, leading to a prolonged S phase.
(C) Treatment with a WEE1i in cyclin E1-overexpressing cells leads to an increase in early S phase and defective nucleotide incorporation. This leads to activation of the ATR/CHK1 pathway to protect replication forks and stop progression through G2-M to allow DNA repair.
(D) Addition of ATRi to WEE1i leads to increased DNA double-strand breaks and replication fork collapse. Because ATR also plays a role in G2-M cell-cycle checkpoint control, damaged DNA can now progress through G2-M unchecked, leading to mitotic catastrophe and cell death.
Discussion
CCNE1 is a commonly amplified oncogene in HGSOC and EMCAs, often conferring resistance to standard-of-care platinum chemotherapy and consequent poor overall survival.5,47 Given the lack of effective treatment options for these aggressive cancers, we sought to address a clinically unmet need by identifying a treatment strategy that targets critical survival pathways for CCNE1-dependent cancers and to identify biomarkers predictive of response. Because the emergence of resistance to monotherapy for oncogene-addicted cancers is essentially universal,48 a combination strategy was investigated. A combination therapy that exploits genetic liabilities such as CCNE1 amplification provides an opportunity to interdict drug resistance and potentially permit utilization of lower drug concentrations, thereby decreasing toxicity.49 We hypothesized that combination WEE1i-ATRi would increase replication fork instability and complete tumor regression more so than monotherapy in CCNE1-overexpressing HGSOC and EMCAs in a CCNE1-level-dependent manner.
We identified a means to exploit CCNE1 oncogene addiction vulnerabilities with low-dose WEE1 and ATR inhibition, a dosing strategy that should be tolerable and feasible in the clinic. We show the induction of CCNE1 activates replication fork stabilizers ATR/CHK1/WEE1, sensitizing cells to ATRi and even more to WEE1i monotherapy. However, monotherapy is less effective or requires high doses (WEE1i > 200 nM) that are toxic to normal cells and associated with potential off-target effects (Figure 1).50 Treatment with WEE1i alone also significantly increased ATR-CHK1 signaling (pCHK1) but the addition of ATRi to WEE1i blocked WEE1i-mediated induction of the feedback loop that may represent an escape pathway, suggesting dual blockade of WEE1 and ATR will be required for optimal activity (Figure 1E). In addition, treatment of CCNE1Amp HGSOC PDX tumors with WEE1i monotherapy initially resulted in tumor regression, but resistance rapidly emerged by 4–5 weeks at the MTD, suggesting a need for combinatorial therapy to induce durable responses (Figure S2F). We show that low-dose WEE1 and ATR inhibition, when combined, is synergistic, decreasing viability and colony formation ability, and that sensitivity increases with higher CCNE1 CN (Figures 1 and 2). With induction of CCNE1, lower doses of the combination are required to achieve therapeutic effects; therefore, this is expected to minimize toxicity to normal cells (Figures 1, 2, 4, and 5). By exploiting CCNE1 overexpression with low-dose WEE1i-ATRi, we can balance the threshold that kills cancer cells, but not normal cells, generating a therapeutic index that is critical in moving combinations into the clinic.
A high CCNE1 CN is a promising genomic biomarker of sensitivity to combination WEE1i-ATRi. Our data show that response to low-dose WEE1i-ATRi is dependent on CCNE1 levels in many diverse established and inducible cell lines and PDX models (Figures 1, 2, 4, 5, and 6). Higher responses to WEE1i-ATRi were demonstrated in CCNE1Amp (CN > 5), followed by CCNE1 high copy gain (CN 2–5) and CCNE1Low HGSOC and EMCA models (Figure 2). Similarly, we demonstrate that increased durable tumor regression and overall survival benefit when using combination WEE1i-ATRi compared with WEE1i monotherapy was most significant in high CCNE1-expressing HGSOC and EMCA PDX models (Figure 3). A sequential and concomitant dosing schedule was evaluated, with the concern that the concomitant dosing regimen could be toxic in the clinic. Both dosing schedules were tolerable by similar body weight and condition scores and were active in the murine models, although the sequential schedule (7 days on/off) appeared slightly less active overall than concomitant dosing (Figure S4). Sensitivity to combination correlated most with CCNE1 CN and cyclin E1 protein by RPPA analysis rather than protein measured by IHC (Figure 3). In addition, our results suggest that CCNE1 copy number is a biomarker predictive of improved overall survival for WEE1i-ATRi compared with WEE1i in PDX models (HR = 0.23; 95% CI = 0.13, 0.42; p < 0.0001; Figure 3). Somatic tumor testing for CCNE1 CN (amplified versus not) is currently available for patients as a validated Clinical Laboratory Improvement Amendments (CLIA)-certified assay and is thus a clinically readily available biomarker. Validation of CCNE1 protein by quantitative detection methods for CCNE1-amplified tumors in the clinic should be considered, because not all amplified tumors result in increased CCNE1 protein expression.7 Although CCNE1 is rarely amplified in breast cancer, increased sensitivity to WEE1i has been demonstrated in CCNE1-amplified breast cancers, supporting our findings.32 This WEE1i-ATRi combination has also been reported to have significant activity in blood malignancies and colon cancers, but not preselected by genetic context.51, 52, 53, 54 In summary, a high CCNE1 CN is a genomic biomarker indicating sufficient levels of cyclin E1 protein overexpression that may be used to predict reliable sensitivity to combination WEE1i-ATRi.
WEE1i-ATRi drug synergy results from differential mechanisms of action for WEE1i and ATRi in CCNE1-amplified cells. As expected, we observed that WEE1i-ATRi leads to loss of G2 arrest, increased γH2AX, and mitotic catastrophe in CCNE1Amp ovarian and endometrial cancer cells.51, 52, 53, 54 We also show that combination WEE1i-ATRi increased replication fork instability, as assessed by decreasing fork speed and increasing fork asymmetry, in CCNE1Amp ovarian and endometrial cancer models (Figures 4F and 4G). Finally, inter-origin distance between replication sites decreased when using combination treatment compared with WEE1i alone (Figure 4H), consistent with the expected promotion of origin firing by WEE1i-mediated deregulation of CDK2 activity (Figure 5B). In this context, the failure of WEE1i to cause premature S-phase entry as determined by PCNA staining (Figure 5E) may result from the steady-state loss of PCNA from collapsed replication forks, which is accentuated by CCNE1 overexpression (Figure 5E).
In our study, we investigated how induction of CCNE1 increases sensitivity to the WEE1i-ATRi combination. Importantly, we show that this sensitivity of CCNE1-overexpressing cells to WEE1i-ATRi results from the differential effects from each drug. CCNE1 induction accelerates S-phase entry, and WEE1i treatment exacerbates this aberrant state by causing defective DNA synthesis after replisome assembly. This conclusion is evidenced by reduced nucleotide incorporation in PCNA+ cells (Figure 5). ATRi treatment did not yield the same outcome, indicating that defective nucleotide incorporation is more strongly associated with WEE1i than ATRi treatment. Instead, the addition of ATRi to WEE1i treatment increases CDK1 activation (Figure 2), thus exacerbating fork collapse through premature M-phase entry and activation of the SMX complex.18,55,56 This conclusion is supported by increased DNA double-strand breaks (γH2AX+EdU+) upon addition of ATRi to WEE1i in CCNE1-overexpressing cells, which is not seen to the same degree with ATRi or WEE1i alone (Figures 5 and 6). Collectively, these data indicate that the combinatorial effects of these drugs in CCNE1-overexpressing cells hinge on the combination of two distinct mechanisms: (1) hyperactivation of CCNE1-CDK2 and perturbation of DNA synthesis by WEE1i treatment and (2) further inhibition of the G2-M checkpoint by the addition of ATRi; together, these amplify replication fork collapse (Figure 7).
In summary, we have identified a treatment option for an aggressive subset of OVCA and EMCA patients who have limited treatment options. By exploiting oncogene-addicted cell-cycle checkpoints and DNA repair mechanisms with combination WEE1i-ATRi, low-dosing strategies are possible. CCNE1 CN is a practical biomarker that predicts sensitivity to WEE1i-ATRi combination therapy and should be included in future clinical trials addressing this patient population.
Limitations of the study
There are limitations with regards to extrapolating cell culture and xenograft study results to the clinic. Although long-term WEE1i-ATRi treatment was well tolerated in mice, this may not be predictive in the clinic, where patients have medical comorbidities and have exhausted multiple prior lines of treatment. Although we tested multiple cell lines and PDXs with a range of CCNE1 CN levels and cyclin E protein levels, a larger sample size would be required for biomarker validation. Even though the models used represent genetic profiles we commonly see in the clinic, other genetic alterations—in addition to CCNE1 overexpression—present in established cancer lines and PDX models may have contributed to the response to the combination.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti- phospho-CHK1(Ser345) | Cell Signaling Technology | cat. # 2348, Lot# 18, RRID: AB_331212 |
| Rabbit anti- CHK1 | Santa Cruz Biotechnology | cat. # sc8408, Lot# I2515, RRID: AB_627257 |
| Rabbit anti- phospho-ATR | EMD Millipore | cat. # ABE462, Lot# Q2475126 |
| Goat anti- ATR | Santa Cruz Biotechnology | cat. # sc1887, Lot# A1515, RRID: AB_630893 |
| Mouse anti- Cyclin E1 | Cell Signaling Technology | cat. # 4129, Lot# 11, RRID: AB_2071200 |
| Rabbit anti- Cyclin E1 | Cell Signaling Technology | cat. # 20808, RRID: AB_2783554 |
| Rabbit anti- pWEE1(Ser642) | Cell Signaling Technology | cat. # 4910, Lot# 3, RRID: AB_2215870 |
| Rabbit anti- WEE1 | Cell Signaling Technology | cat. # 13084, Lot# 1, RRID: AB_2713924 |
| Rabbit anti- pCDK1(Y15) | Cell Signaling Technology | cat. # 9111, Lot# 1, RRID: AB_331460 |
| Mouse anti- CDK1 | Cell Signaling Technology | cat. # 9116, Lot# 2, RRID: AB_2074795 |
| Rabbit anti- pRPA32 (S33) | Bethyl Laboratories | cat. # A300-246A, Lot# 8, RRID: AB_2180847 |
| Rabbit anti- Phospho-Histone H3 (Ser10) | Cell Signaling Technology | cat. # 53348, Lot# 1, RRID: AB_2799431 |
| Rabbit anti- γH2AX | Cell Signaling Technology | cat. # 9718, Lot # 17, RRID: AB_2118009 |
| Mouse anti- β-Actin | Cell Signaling Technology | cat. # 3700, Lot# 15, RRID: AB_2242334 |
| Rabbit anti- Caspase3 | Cell Signaling Technology | cat. # 9664, Clone 5A1E, Lot# 21, RRID: AB_2070042 |
| Rabbit anti- Cleaved Caspase 3 (Alexa Fluor® 488 Conjugate) | Cell Signaling Technology | cat. # 9603, RRID: AB_11179205 |
| Rabbit anti- PAX8 | Proteintech Group | cat. # 10336-1-AP, Lot# 00019427, RRID: AB_2236705 |
| Mouse anti- CK7 | Thermo Fisher Scientific | cat. # 14-9005-82, Lot# E13276-102, RRID: AB_10669584 |
| Rabbit anti- Phospho-CDK2(Y15) | Thermo Fisher Scientific | cat. # MA5-33128, RRID: AB_2811944 |
| Rabbit anti- CDK2 | Cell Signaling Technology | cat. # 2546, RRID: AB_2276129 |
| Mouse anti- PCNA | Santa Cruz Biotechnology | cat. # sc-56, RRID: AB_628110 |
| anti-Rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | catalog 7074, Lot# 28, RRID: AB_2099233 |
| anti-Mouse IgG, HRP-linked Antibody | Cell Signaling Technology | catalog 7076, Lot# 32, RRID: AB_330924 |
| Secondary antibody goat anti-Rabbit IgG (H+L), Alexa Fluor® 647 | Thermo Fisher Scientific | cat. # A-21246, Lot# 2051068, RRID: AB_2535814 |
| Bacterial and virus strains | ||
| FUW-Luc-mCherry-puro lentivirus (FmC) | Liu et al., 201741 | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5332350/ |
| Biological samples | ||
| Human: WO-19 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WO-58 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WO-77 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WO-24 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WO-20 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WO-12 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WO-18 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WU-89 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: WU-94 Patient-derived xenografts (PDXs) | Hospital of University of Pennsylvania | https://www.med.upenn.edu/OCRCBioTrust/ |
| Human: DF-172 Patient-derived xenografts (PDXs) | Liu et al., 201741 | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5332350/ |
| Chemicals, peptides, and recombinant proteins | ||
| Cholera Toxin | Sigma-Aldrich | cat. # 227036 |
| OCMI-E media | Live Tumor Culture Core at Sylvester Comprehensive Cancer Center, Miller School of Medicine | https://www.nature.com/articles/ncomms8419 |
| Carboplatin | Hospira | NDC 61703-339-56 |
| WEE1i | AstraZeneca | AZD1775 |
| ATRi | AstraZeneca | AZD6738 |
| D-Luciferin, Potassium Salt | Gold Biotechnology | cat. # LUCK-100 |
| Puromycin dihydrochloride | Sigma-Aldrich | cat. # P9620 |
| Polybrene Infection / Transfection Reagent | Sigma-Aldrich | cat. # TR-1003-G |
| MTT | Sigma-Aldrich | cat. # M2003 |
| Crystal violet | Sigma-Aldrich | cat. # V5265 |
| 5-chloro-2′-deoxyuridine (CldU) | Sigma-Aldrich | cat. # C6891 |
| 5-iodo-2′-deoxyuridine (IdU) | Sigma-Aldrich | cat. # I7125 |
| Nocodazole | Sigma-Aldrich | cat. # M1404 |
| Critical commercial assays | ||
| eBioscience Annexin V Apoptosis Detection Kit APC | Thermo Fisher Scientific | cat. # 88-8007-74 |
| FITC- BrdU Staining Kit from | BD Biosciences | cat. # 559619 |
| Click-iT Plus EdU Alexa Fluor 488 Flow Cytometry Assay Kit | Thermo Fisher Scientific | cat. # C10633 |
| Genomic Vision FiberPrep® kit | Genomic Vision | cat. # EXTR-001 |
| Click-iT EdU Alexa Fluor 555 Imaging Kit | Thermo Fisher Scientific | cat. # C10338 |
| NEBNext® Ultra™ DNA Library Prep Kit for Illumina® | New England BioLabs | cat. # E7530L |
| Lipofectamine 3000 | Thermo Fisher Scientific | cat. # L3000008 |
| ATR siRNA Assay ID 82 | Thermo Fisher Scientific | AM51331 (ID 82) |
| WEE1 siRNA Assay ID 404 | Thermo Fisher Scientific | AM51331 (ID 404) |
| Experimental models: Cell lines | ||
| Human: OVCAR3 ovarian cancer cells | American Tissue Type Collection (ATCC) | cat. # HTB-161 |
| Human: FUOV1 ovarian cancer cells | Leibniz Institute DSMZ | cat. # ACC-444 |
| Human: KLE endometrial cancer cells | ATCC | cat. # CRL-1622 |
| Human: OVCAR8 ovarian cancer cells | NCI-DTP | cat. # OVCAR-8, RRID: CVCL_1629 |
| Human: OVSAHO ovarian cancer cells | Japanese Collection of Research Bio resources Cell Bank (JCRB) | cat. # JCRB1046 |
| Human: MFE280 endometrial cancer cells | Sigma-Aldrich | cat. #98050131 |
| Human: OVKATE ovarian cancer cells | Japanese Collection of Research Bio resources Cell Bank (JCRB) | cat. # JCRB1044 |
| Human: SNU685 endometrial cancer cells | AcceGen Biotech | cat. # ABC-TC1104 |
| Human: WO-20 patient derived primary cells | University of Pennsylvania | This paper |
| Human: WO-24 patient derived primary cells | University of Pennsylvania | This paper |
| Human: FT282 fallopian tube epithelium | Karst et al., 20147 | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4517944/ |
| Deposited Data | ||
| WO-12 patient derived xenograft | SRA database; Mendeley | PRJNA750496; DOI 10.17632/2xxxzsg6hz.1 |
| WO-18 patient derived xenograft | SRA database; Mendeley | PRJNA750496; DOI 10.17632/2xxxzsg6hz.1 |
| WO-20 patient derived xenograft | SRA database; Mendeley | PRJNA750496; DOI 10.17632/2xxxzsg6hz.1 |
| WO-24 patient derived xenograft | SRA database; Mendeley | PRJNA750496; DOI 10.17632/2xxxzsg6hz.1 |
| WO-77 patient derived xenograft | SRA database; Mendeley | PRJNA750496; DOI 10.17632/2xxxzsg6hz.1 |
| WU-89 patient derived xenograft | SRA database; Mendeley | PRJNA750496; DOI 10.17632/2xxxzsg6hz.1 |
| WU-94 patient derived xenograft | SRA database; Mendeley | PRJNA750496; DOI 10.17632/2xxxzsg6hz.1 |
| WO-19 patient derived xenograft | GEO database; Mendeley | PRJNA626436; DOI 10.17632/2xxxzsg6hz.1 |
| WO-58 patient derived xenograft | GEO database; Mendeley | PRJNA626436; DOI 10.17632/2xxxzsg6hz.1 |
| Experimental models: Organisms/strains | ||
| NSG mice (NOD/SCID IL2Rγ−/−) | Stem Cell and Xenograft Core (SCXC) at the University of Pennsylvania | https://www.med.upenn.edu/scxc/ |
| Recombinant DNA | ||
| CCNE1 | Harvard PlasmID Repository HSCD00326535, | RefSeq NM_001322261 |
| pCW57-MCS1-2A-MCS2 | Addgene | cat. #71782 |
| Software and algorithms | ||
| ImageJ | National Institutes of Health | 1.50i; Fiji |
| FlowJo | Tree Star | version VX |
| FiberStudio | Genomic Vision | version 2.0 |
| Tumor Manager software | Biopticon | version 3.3.4 |
| TumGrowth web tool | Enot et al., 201857 | https://kroemerlab.shinyapps.io/TumGrowth/ |
| GraphPad Prism | Graphpad Software | version 8.4.2 |
| Broad Institute Cancer Cell Line Encyclopedia (CCLE) | Barretina et al., 201258 | https://sites.broadinstitute.org/ccle |
| MuTect2 | Benjamin et al., 201959 | https://www.biorxiv.org/content/10.1101/861054v1v |
| ANNOVAR | Chang and Wang, 201260 | https://pubmed.ncbi.nlm.nih.gov/22717648/ |
| Genome Analysis Toolkit (GATK) | McKenna et al., 201061 | https://pubmed.ncbi.nlm.nih.gov/20644199/ |
| CODEX v2 | Jiang et al., 201862 | https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1578-y |
| Microsoft Excel | Microsoft | version 2016 |
| MTT calorimetric assay software Gen5 ELISA | BioTek | v 1.04.5 |
| Other | ||
| SonoSite Edge II Ultrasound System | Sonosite | Edge II |
| Leica TCS SP8 WLL Confocal with STED 3X | Leica | TCS SP8 STED 3X |
| Nikon Eclipse 80i microscope | Nikon | Eclipse 80i |
| FiberVision scanner | Genomic Vision | SCN-002 |
| MTT calorimetric assay plate reader | BioTek | EL800 |
| HiSeq 4000 | Illumina | HiSeq 4000 |
| Perkin Elmer IVIS Spectrum | Perkin Elmer | IVIS Spectrum |
| BD LSR II | Becton Dickinson | LSR II |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fiona Simpkins (fiona.simpkins@pennmedicine.upenn.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. There are restrictions to the availability of PDX tumors due to the lack of an external centralized repository for its distribution and our need to maintain the early passage stocks. We are glad to share with reasonable compensation by requestor for processing and shipping.
Experimental model and subject details
Cell lines and primary cells
OVCAR3, KLE cell lines were purchased from ATCC (Manassas, Virginia); FUOV1 was obtained from Leibniz Institute DSMZ; OVCAR8 was obtained from NCI-DTP; OVSAHO and OVKATE obtained from the Japanese Collection of Research Bioresources Cell Bank (JCRB). MFE280 was purchased from Sigma-Aldrich and SNU685 from AcceGen Biotech (Fairview, NJ). CCNE1 DNA copy number data were derived from the Affymetrix SNP 6.0 array and downloaded from the CCLE portal https://sites.broadinstitute.org/ccle.58 CCNE1Amp lines were: OVCAR3, FUOV1, KLE; CCNE1Gain: OVCAR8, OVSAHO, MFE280; CCNE1 copy neutral (CCNE1Low): OVKATE, SNU685. Ovarian cancer cell lines included: OVCAR3, FUOV1, OVCAR8, OVSAHO, OVKATE. Endometrial cancer cell lines included: KLE, MFE280, SNU685.
OVCAR3, OVCAR8, OVSAHO, OVKATE and SNU685 cells were maintained in RPMI 1640 media with 10% fetal bovine Serum (FBS; Thermo Fisher) and 1% penicillin/streptomycin (P/S; Thermo Fisher). MFE280 was grown in MEM media (Thermo Fisher, Rockford, IL) with 10% FBS and 1% P/S. FUOV1 and KLE cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 media with 10% FBS and 1% P/S.
FT282 cells, human fallopian tube epithelium was immortalized with hTERT, and transduced with TP53R175H, a common conformational TP53 mutants identified in HGSOC as described7. FT282 cells were cultured in DMEM/F12 media supplemented with 10% FBS and 1% P/S.
The WO-20 and WO-24 primary ovarian cancer tumor cultures were generated in our laboratory. Fresh tumor obtained at the time of ovarian cancer debulking surgery was minced, digested, and grown in OCMI-E media (Live Tumor Culture Core at Sylvester Comprehensive Cancer Center, Miller School of Medicine, Miami, FL) with the addition of 30 ng/mL Cholera Toxin (Sigma-Aldrich, St Louis, MO) as described39 and characterized by immunofluorescence shown to confirm ovarian cancer in origin. Cell lines were authenticated by short tandem repeat (STR) analysis at the Oncogenomics Core at Wistar Institute and confirmed mycoplasma negative by end-point PCR at the Cell Center Service at the University of Pennsylvania.
Mouse Models
NOD-SCID IL2Rγ−/− (NSG) mice were purchased from the Stem Cell and Xenograft Core (SCXC) at the University of Pennsylvania (UPENN, Philadelphia, PA). All mice experiments were performed in adherence to the policies of NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee (IACUC).
Five to eight week old female mice were used for orthotopic tumor transplantation or intraperitoneal (IP) injection as previously described.41,63 The WO-19, WO-58, WO-77, WO-24, WO-20, WO-12, WO-18, WU-89, and WU-94 PDX models were developed by orthotopic transplantation of patient tumor to the ovaries/fallopian tubes of mice for high grade serous ovarian cancer (HGSOC) and uterine horn for endometrial cancer (EMCA) PDX models using methods previously described.63 After tumors were palpable, tumor volume was measured weekly by ultrasound (SonoSite Edge II Ultrasound System) by a trained sonographer blinded by treatment arm. For the DF-172 PDX and OVCAR3 models, cells expressing firefly luciferase were injected IP to generate ascites models. The intraperitoneal model DF-172 PDX model utilized HGSOC ascites cells after transduced with FUW-Luc-mCherry-puro lentivirus (FmC) as previously described.41 For the OVCAR3 intraperitoneal model developed, OVCAR3 cells were transduced with lentivirus FmC in medium containing polybrene (8 μg/ml). The cells were selected and maintained with puromycin (5 μg/ml) 2 days after virus infection. Selected cells were then confirmed to be expressing RFP by fluorescent microscopy and luciferase activity detection by in vitro luminescence assay with 150 ug/ml D-luciferin. Approximately, 2x106 DF-172 or 1x106 OVCAR3-Luc cells were suspended in PBS and injected into NSG mice. Intraperitoneal luciferized tumors were measured by IVIS Lumina II (PerkinElmer, Waltham, MA) weekly by intraperitoneal injection of 10 μl/g D-luciferin at 15mg/ml (Gold Biotechnology, St. Louis, MO). The ascites, body weights, and condition scores of mice were monitored and recorded weekly. Tumor volume criteria for randomization to treatment arms was 70-100 mm3 forthe orthotopic PDX studies and 1.5-2.5 × 105 Luminescence for intraperitoneal PDX studies.
For all preclinical studies, mice were randomized into 6 treatment groups: vehicle (2-hydroxylpropyl-β-cyclodextrin); carboplatin (30 mg/kg IP weekly; Hospira); WEE1i (AZD1775; 60 mg/kg/day 1-5 weekly by oral gavage; AstraZeneca); ATRi (AZD6738; 40 mg/kg daily on day 1-5 weekly by oral gavage; AstraZeneca); combination WEE1i + ATRi (AZD1775 60 mg/kg/day + AZD6738 40 mg/kg day 1-5 weekly by oral gavage; drugs were dosed WEE1i followed by ATRi 1-2 h apart); and sequential WEE1i + ATRi (AZD1775 90 mg/kg daily on days 1-7 + AZD6738 50 mg/kg daily on days 8-14 by oral gavage for a 2 week cycle and repeated), except WU-94 (WEE1i and ATRi at 30 mg/kg/day with same treatment schedule as other PDXs). In all the models, percentage change in body weight during treatment was used as a marker for toxicity and dose level adjustments. Significant treatment toxicity was defined as a 15% drop in body weight and the mice require treatment reduction at 25% dose and supplements supportive. For mice with 20% drop in body weight, treatment was stopped and supportive measures (i.e., food supplement and subcutaneous fluid) were provided. Body weight was rechecked every 3-4 days. Once improved, treatment was restarted with a 25% dose reduction. If body weight was not regained after one week, PDX was sacrificed in accordance with the Institutional Animal Care and Use Committee (IACUC) protocols. Trial endpoints were defined as tumor volume > 1000 mm3 for orthotopic PDX model or significant ascites (defined as score of 5 using an ascites range of 1-5) for intraperitoneal PDX models or poor condition score (defined as score of 1 on a 1-5-point scale). Mice were euthanized according to Institutional Animal Care and use Committee guidelines. Tumors were collected and snap frozen for protein and genomic analysis and fixed in formalin for IHC.
Method details
Cell line transduction and knock-down
Human fallopian tube epithelial cells were immortalized with hTERT, and transduced with TP53R175H, a common conformational TP53 mutant identified in HGSOC (FT282) as described.7 Cyclin E was then overexpressed in FT282 cells in a doxycycline-inducible manner (FT282 CCNE1 induc) as described.7 To generated CCNE1 inducible OVKATE and SNU685 cells, the CCNE1 inducible lentivirus was produced. pCW57-CCNE1 was generated by PCR subcloning CCNE1 (Harvard PlasmID Repository, HSCD00326535, RefSeq NM_001322261) into pCW57-MCS1-2A-MCS2 (Addgene #71782). All cloning was verified by DNA sequencing. Replication-deficient lentivirus was produced by transient transfection of 6.0 μg psPAX2 (Addgene #12260), 2.0 μg pMD2.G (Addgene #12259), and 8.0 μg transfer plasmid into HEK293T cells in a 10 cm dish with Lipofectamine 3000 reagent (Thermo Fisher Scientific, Carlsbad, CA), according to the manufacturer’s instructions. Viral supernatants were collected at 48 h and passed through a 0.2 μm filter. Functional titration was performed by transduction of OVKATE and SNU685 cells with serially diluted virus in the presence of polybrene (4 μg/ml, Sigma-Aldrich) for 6 h followed by puromycin (5 μg/ml, Life Technologies) selection for 48 h post-infection.
To evaluate the off target effect of ATR and WEE1 inhibitors, ATR and WEE1 siRNAs (Thermo Fisher Scientific) were transfected with Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) following the transfection protocol. 2 nM WEE1i, 10 nM ATRi or combination were transfected into OVCAR3 and WO-20 cells. The cell viability and protein expression were detected 48 h post transfection.
In vitro cytotoxicity assays
Cells were seeded into 96-well plates with the cell number normalized based on cell doubling time (Table S3). Cells were treated with control (DMSO), WEE1i (AZD1775), ATRi (AZD6738) or combination at indicated concentrations in triplicate for 5 days. Drugs were clinical grade and obtained from AstraZeneca. At the end of the treatment period, an MTT colorimetric assay was performed to detect the cell viability. Cells were incubated with 10 μL of MTT at 5 mg/ml (Sigma Chemical Co., St Louis, MO) for 4 h at 37°C. The supernatant was removed and 50 μL DMSO (Fisher Scientific, Hampton, NH) was used to dissolve the MTT formazan. Absorbance was measured in a microplate reader at a wavelength of 570 nm. Relative cell viability was calculated, with the non-treatment group as a control.
Colony formation Assay
For colony formation assay, cells were plated onto 24-well plates and cell number normalized based on cell doubling time (Table S3) and cultured overnight in triplicate. They were then treated with DMSO vehicle, WEE1i, ATRi, or combination as indicated every 3 days for a total of 10 days. Cells were then fixed and stained with 0.5% Crystal violet in 20% methanol solution. The plates were washed, air-dried, scanned, and quantified in ImageJ (National Institutes of Health, Bethesda, MD).
Western blot
Cells were treated and collected at indicated time, then washed and incubated with 2 × Laemmli Sample Buffer (4% SDS, 20% Glycerol, 0.12M Tris-HCl at pH 6.8 in distilled water) containing a protease and phosphatase inhibitor cocktail (EMD Millipore, Billerica, MA). With protein concentration determined by BCA kit (BioRad, Hercules, CA), whole cell lysates (15 μg) were separated on reducing 4%–15% SDS-PAGE gels, electrotransferred to PVDF membrane (Bio-Rad, Hercules, CA), blocked with 5% BSA (ThermoFisher) in 1x Tris-buffered saline (ThermoFisher) with 0.1% Tween20 (ThermoFisher) (1x TBST), and immunoblotted with respective primary antibodies. After that, membranes were washed and blotted with species-appropriate horseradish peroxidase conjugated anti-rabbit (catalog 7074, Cell Signaling Tech) or anti-mouse (catalog 7076, Cell Signaling Tech) secondary antibody in 5% BSA in 1x TBST for 1 h, followed by chemiluminescent substrate (Thermo Scientific, Rockford, IL) incubation and film development. Actin was used as loading control for whole cell lysate and total histone H3 was considered as loading control for nucleic proteins.
Flow cytometry detection of apoptosis
Cells were plated, incubated overnight, and treated with DMSO vehicle, 0.2 μM WEE1i (AZD1775), 1 μM ATRi (AZD6738), or combination for 48 h. Apoptosis assay was performed with eBioscience Annexin V Apoptosis Detection Kit APC (ThermoFisher Scientific), according to the manufacturer’s instruction. Annexin V-APC and propidium iodide labeled cells were detected by BD Accuri C6 Cytometer (BD Biosciences, San Jose, CA). The acquired data was analyzed with FlowJo (Tree Star, Inc., Ashland, OR).
Flow cytometry detection of intracellular protein
Cells were seeded in triplicate and then incubated in 0.2 μM WEE1i (AZD1775), 1 μM ATRi (AZD6738) or combination treatments for 8 h or 24 h. Cells were then trypsinized, fixed washed and incubated with blocking buffer. Cells were then stained with the following primary antibodies diluted in blocking buffer at 1:300: γH2AX (catalog 9718, Cell Signaling Technology, Inc), pRPA32 (S33, catalog A300-246A, Bethyl Laboratories, Inc.) or phospho-histone H3 (pHH3, Ser10, catalog 53348, Cell Signaling Technology, Inc). The cells were washed, and incubated with secondary antibody goat anti-Rabbit IgG (H+L), Alexa Fluor® 647 (ThermoFisher Scientific) for 30 min. The cells were then incubated with 50 μg/mL propidium iodide (Sigma-Aldrich) and subjected to flow cytometry acquisition on BD LSRII (BD Biosciences) and data analysis with FlowJo (Tree Star, Inc., Ashland, OR). For Phospho-Histone H3 (Ser10), cells were treated with 0.2 μM WEE1i, 1 μM ATRi or combination for 6 h or 12 h then 500 nM nocodazole for 6 h to prevent cells from exiting mitosis. The supernatant and attached cells were collected and fixed. The staining and detection process was performed same as above.
Cell cycle analysis
For cell cycle detection via BrdU staining, cells were seeded in triplicate and treated with 0.2 μM WEE1i, 1 μM ATRi or combination for 12, 24, and 48 h in OVCAR3, KLE, and 24 h in CCNE1induc OVKATE, CCNE1induc SNU685 cells with or without 0.5 μM doxycycline. Cell cycle was evaluated using the FITC- BrdU Staining Kit from (BD Biosciences, San Jose, CA) per the manufacturer’s instructions. Flow analysis was conducted on a BD LSRII (BD Biosciences) and data analysis with FlowJo (Tree Star, Inc., Ashland, OR).
For cell cycle detection via via EdU incorporation, the CCNE1induc FT282 were serum starved for 24hrs with DMEM/F12 and RPMI 1640 medium containing 0.1% FBS, respectively; then treated and collected as indicated at various times (0, 8, 12, 16 h), in the presence or absence of 0.5 μM doxycycline with 10 μM EdU added 1 h in advance of cell cycle analysis. The cells were costained with γH2AX (Cell Signaling Technology, Cat #9718) and 7-AAD to detect DNA damage and DNA content. Click-iT Plus EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Thermo Fisher, Cat #C10633) was applied to for cell cycle analysis.
DNA combing
OVCAR3 and KLE cells were treated with DMSO, 0.2 μM WEE1i, 1 μM ATRi or combination for 30min, pulse-labeled with 100 μM 5-chloro-2′-deoxyuridine (CldU; cat. # C6891, Sigma-Aldrich, St. Louis, MO) followed by 100 μM 5-iodo-2′-deoxyuridine (IdU; cat. # I7125, Sigma-Aldrich, St. Louis, MO) for 15min each treatment, in the presence of drug. After treatment, cells were chilled, trypsinized and embedded into agarose plugs with the Genomic Vision FiberPrep® kit (Genomic Vision, Bagneux, France) per manufacturer’s protocol. The DNA combing assay was performed following EasyComb. The stained cover slides were scanned with FiberVision® and images were stored and analyzed with Genomic Vision FiberStudio® software. Intact red and green staining, indicating CldU and IdU respectively, on the same fiber were selected for replication fork speed and asymmetry calculation. The distances between midpoints of two adjacent replication origins in the same fiber were counted as inter-origin distance (IODs).
Immunofluorescence staining
For PCNA-EdU staining, the FT282 cells were seed into 24-well plates with cover slides (Thermo Fisher) sterilized by 75% ethanol at 105 cells/well. After 12hrs culture, the cells were washed with PBS twice and incubated with DMEM/F12 containing 0.1% FBS for 24hrs to arrest cells in G0. They were released into DMEM/F12 containing 10% FBS, and then treated with DMSO, 0.2 μM WEE1i, 1 μM ATRi or combination for 8 h, in the presence or absence of 0.5 μg/ml doxycycline. The cells were fixed with 4% PFA, penetrated by 0.5% Triton X-100 in PBS containing 5% FBS for 1 h, and then incubated with PCNA antibody (Santa Cruz, cat #sc-56) overnight at 4 degree. The cover slides with cells were washed with PBS, stained with anti-mouse secondary antibody conjugated with Alexa 488 for 1 h at room temperature. Afterward, the EdU were stained with Click-iT EdU Alexa Fluor 555 Imaging Kit (Thermo Fisher, Cat #C10338), incubated with DAPI, and sealed with VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA). Images were captured on a Confocal Microscope (Leica TCS SP8 WLL Confocal with STED 3X). These images were then analyzed via ImageJ software.
For primary ovarian tumor culture characterization, cells were seeded at 2 × 103 cells/well, fixed, washed, blocked with blocking buffer (1 × PBS containing 2% FBS, 0.01% Tween 20; Thermo Fisher Scientific and 0.01% Triton X-100), for 30 min, followed by anti-PAX8 (Cat. 10336-1-AP, Proteintech Group, Rosemont, IL) or anti-CK7 (Cat. 14-9005-82, ThermoFisher Scientific) incubation, and then Goat anti-Mouse IgG (H+L), Alexa Fluor® 488 (ThermoFisher) and Goat anti-Rabbit IgG (H+L), Alexa Fluor® 555 (ThermoFisher). The slides were then sealed with VECTASHIELD Antifade Mounting Medium with DAPI and imaged under the Nikon Eclipse 80i microscope.
Targeted DNA Sequencing
Massively parallel sequencing was performed on DNA from cell lines and PDX models using a custom designed panel of 157 genes known to be implicated in HGSOC and EMCA tumors.63 500ng of genomic DNA was sheared randomly into 200 base pair fragments with the Covaris™ LE220 Focused-UltraSonicator (Covaris®, Woburn, MA). Sheared DNA was A-tailed and ligated with adaptor-embedded indexes using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England BioLabs, Inc., Ipswich, MA). DNA quality, fragment size, and concentration of library preps were measured using Agilent’s DNA 1000 chips in conjunction with the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). High quality samples were pooled and hybridized to a custom capture library using Agilent SureSelect kits. Hybridization pools were assessed for fragment length and concentration using the Qubit® 2.0 fluorometer (Life Technologies, Carlsbad, CA) and the Bioanalyzer. Sequencing was performed on the HiSeq 4000 (Illumina) at the University of Pennsylvania Next Generation Sequencing Core. All PDX and parent tumors were authenticated using STR analysis (Wistar Genomics Facility).
Targeted DNA Sequencing Analysis, Variant Calling, and Copy Number Profiling
Sequencing reads were aligned to human reference genome NCBI Build 37 using the Burrows-Wheeler Aligner and following Genome Analysis Toolkit (GATK) best practices.61 Disambiguation of sequencing read origin was performed by mapping reads to both the mouse and human genome. Reads that mapped with high accuracy to the mouse genome were removed. Samples were removed due to poor sequencing quality if more than 10% of targets had 0% coverage, or if at least 10x coverage was achieved in less than 50% of targets.
Variant calling was performed using MuTect2, and ANNOVAR was used for variant annotation.60,59 Variant identification and classification was performed using a strict filtering and analysis pipeline that has previously been described and validated.64 Locus-specific databases, ClinVar, dbSNP, and COSMIC were utilized to help identify suspected deleterious variants, and any missense variant calls that could not be confirmed in the literature were considered variants of undetermined significance (VUS) and excluded from analysis. Copy number variation (CNV) was assessed in primary cells and PDX models using CODEX2 on targeted sequencing data.62 Segmentation was restricted to exons for all genes. Visual confirmation of CNV calls was done in Nexus 7.5 (BioDiscovery) software. CNV for DF-172 was performed and reported previously.41 For both MuTect2 and CODEX2, a panel of normal samples (germline DNA from male patients) was used as controls.
Reverse Phase Protein Array
Patients’ tumor tissues, PDX tumor tissues at different mouse passage numbers and primarily cultured cells are collected and frozen for evaluation. Reverse Phase Protein Array (RPPA) was performed by the MD Anderson Center RPPA core facility as described65 and the results were reported as normalized linear as well as normalized Log2. Four hundred and Seventy-two phosphorylated and total proteins were evaluated (https://www.mdanderson.org/research/research-resources/core-facilities/functional-proteomics-rppa-core.html). The relative protein levels were analyzed by GraphPad Prism. Heatmaps were generated using the Morpheus software from Broad Institute (https://software.broadinstitute.org/morpheus).
Immunohistochemistry
The tumors were collected and immunohistochemistry staining was performed in PDX models. Samples were fixed with 10% formalin overnight and maintained in 70% ethanol at 4°C. The tissue samples were dehydrated in graded ethanol, xylene, and embedded in paraffin. The paraffin blocks were cut into 4-6 μm sections and placed onto slides. After deparaffinized with Histo-Clear, rehydrated with degraded ethanol, and antigen retrieval with 1x target retrieval solution, the endogenous hydrogen peroxidase activity was blocked with hydrogen peroxide for 10 min. Slides were individually immunoblotted with primary antibodies pCHK1 (Cat. 2348, Cell Signaling Technology, Inc.), Phospho-Histone H2A.X (Ser139) (Cat. 9718, Cell Signaling Technology, Inc.), CyclinE1 (Cat. 20808, Cell Signaling Technology) at 1:500 titer for 60 min. The slides were incubated with anti-rabbit or anti-mouse horseradish peroxidase polymer for 30 min following primary antibodies wash out. The slides were developed using 3,3′-diaminobenzidine (DAB)+ chromogen for 10 min and washed with water. The slides were then counterstained, dehydrated, and mounted with mounting media. The pictures were taken under Nikon Eclipse 80i microscope. The quantification of immunohistochemistry signal was performed blindly by a pathologist (K.D, L.S.).
Statistical Analysis
In vitro studies were performed using at least three biological replicates per sample and as three independent experiments. Two-tailed unpaired t tests were used when comparing two groups. One-way ANOVA followed by Tukey’s post hoc comparison was performed for multiple group comparisons. p < 0.05 was considered statistically significant. Drug interaction between ATRi and WEE1i was analyzed using the coefficient of drug interaction (CDI).66 CDI = AB/(AxB); AB is the ratio of two-drug combination group to control, and A or B is ratio of a single drug to control. CDI < 1 indicates synergism, CDI < 0.7 indicates significant synergism, CDI = 1 indicates additivity, and CDI > 1 indicates antagonism. GraphPad Prism (Graphpad Software version 8.4.2, San Diego CA) was used for statistical analyses.
For Analysis of PCNA and EdU positivity, an ROI Mask analysis technique was performed with Fiji/ImageJ. Over 100 cells were analyzed from each replicate and Corrected Total Cell Fluorescence (CTCF) was calculated by subtracting the background fluorescence per area for each fluorescent channel. To count foci, we used a prominence threshold of 175 to detect PCNA foci within the DAPI ROI mask regions. We then calculated foci by using a single point analysis of PCNA maxima and dividing the raw integrated density of the single points by the maximum pixels per nucleus. FT282 cells with a CTCF 1000 or greater were considered EdU positive. FT282 cells with 20 or more foci were considered PCNA positive cells.
For statistical power for in vivo studies, we transplanted 12 mice/arm.63,66 After randomization, once pre-specified tumor volume was achieved, there were approximately 10 mice/arm (range 4-12). Randomization was performed using Tumor Manager software (v 3.3.4, Biopticon). Mice that died for unknown reason (low tumor burden, normal weight and condition scores) were excluded from analysis. Weekly ultrasound measurements, weights, and condition scores were obtained in a blinded manner. Longitudinal analysis of tumor growth was carried out by linear mixed-effect modeling with type II ANOVA and pairwise comparisons across groups on log pre-processed tumor sizes using the TumGrowth web tool (https://kroemerlab.shinyapps.io/TumGrowth/).57 Natural log transformed tumor volume was used to better satisfy normal distribution. Survival data was analyzed by Mantel-Cox log rank test. Using the slopes obtained from PRISM for each treatment group as effects measurements, synergy under the Bliss definition of independence was used. The index was defined as the ratio of sums of slopes (Both and Control over ATRi and WEE1i) would indicate synergy, additivity, and antagonism if < 1, 1, > 1 respectively.
To determine whether higher CCNE1 is associated with more reduction in tumor growth rates for the combination over WEE1i monotherapy in the PDX models (i.e., CCNE1 is a predictive biomarker), we tested the differential treatment effects using a linear mixed effects model for the log transformed tumor volumes with an interaction term of treatment assignment, time, and whether CN > = 4 or not. Because the data indicated no treatment effects in PDX models with CN < 4, we refitted the data with a smaller nested linear mixed model assuming treatment effects only appeared in models with CN > = 4 Similarly, CCNE1 as a predictive biomarker for OS was tested using a Cox proportional hazard model with the indicators of the two treatment groups, CN > = 4 versus < 4, and the interaction term of the two indicator terms. A significant interaction implies CN > = 4 is associated with more improvement of OS for the combination over WEE1i monotherapy (i.e., predictive). A two-sided p value is considered statistically significant.
Acknowledgments
The authors thank Mei Zheng for immunohistochemistry staining. This work was supported by the Mary Kay Foundation (017-64 to F.S.), Rivkin Center for Ovarian Cancer (to F.S. and E.G.), Ovarian Cancer Research Alliance (600095 to H.X. and 545152 to G.B.M.), AACR-AstraZeneca Ovarian Cancer Research Fellowship (to E.G.), NIH (5R37CA215436-02 to F.S. and 5R01CA189743 to E.J.B.), Kaleidoscope of Hope (to F.S.), George and Emily McMichael Harrison Grant (to E.G.), 5-K12-HD-001265-20 (to E.G.), 3-K12-HD-000849-34S1 (to E.G.), NCI (CA217685, CA217842, and CA098258 to G.B.M.), and a gift from the Miriam and Sheldon G. Adelson Medical Research Foundation (to G.B.M. and R.D.).
Author contributions
H.X., E.G., and F.S. designed the study, analyzed the data, and drafted the manuscript. H.X. and E.G. performed in vitro experiments. H.X., S.M., E.G., Y.K., B.F., T.M., and H.K. did the animal experiments. J.D.P. performed PCNA experiments. C.J.B. and A.R.K. generated FT282 Dox-inducible cell models. K.M.D. and L.E.S. quantified the immunohistochemistry data. J.B.S., K.D., B.W., and K.L.N. performed the next-generation DNA sequencing and analyzed the data. G.B.M. carried out the proteomic assay. R.D. established the FT282 cells. F.S., E.J.B., G.B.M., R.D., and W.-T.H. interpreted the data and revised the manuscript. H.X. and E.G. are co-first authors based on their equal contributions. All authors read and approved the manuscript.
Declaration of interests
F.S. serves on a scientific advisory board and has received funding for clinical trials from AstraZeneca. G.B.M. receives support or acts as a consultant for AstraZeneca, ImmunoMET, Ionis, Nanostring, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Abbvie, Amphista, Ellipses Pharma, Eli Lilly, Medacorp, Turbine, Zentalis Pharmaceuticals, and Tarveda and has transferred technology to Myriad and Nanostring. E.J.B. serves on the scientific advisory board of Atrin Pharmaceuticals and has been an advisor for Sierra Oncology. R.D. serves on the scientific advisory board of Repare Therapeutics and Siamab Therapeutics and advises Mersana Therapeutics and nVision Medical.
Published: September 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100394.
Supplemental information
Data and code availability
The published article includes all datasets generated or analyzed during this study. All Sequencing data has been deposited in NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/bioproject) with accession number PRJNA750496 and NCBI GEO database (https://www.ncbi.nlm.nih.gov/gds/) with NCBI accession # PRJNA626436. All the data are also available in Mendeley Data with DOI 10.17632/2xxxzsg6hz.1.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Birrer, M.J., and Ceppi, L., eds. (2017). Translational Advances in Gynecologic Cancers (Academic Press).
- 3.Armstrong D.K., Bundy B., Wenzel L., Huang H.Q., Baergen R., Lele S., Copeland L.J., Walker J.L., Burger R.A., Gynecologic Oncology Group Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N. Engl. J. Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 4.Winter W.E., 3rd, Maxwell G.L., Tian C., Carlson J.W., Ozols R.F., Rose P.G., Markman M., Armstrong D.K., Muggia F., McGuire W.P., Gynecologic Oncology Group Study Prognostic factors for stage III epithelial ovarian cancer: a Gynecologic Oncology Group Study. J. Clin. Oncol. 2007;25:3621–3627. doi: 10.1200/JCO.2006.10.2517. [DOI] [PubMed] [Google Scholar]
- 5.Patch A.M., Christie E.L., Etemadmoghadam D., Garsed D.W., George J., Fereday S., Nones K., Cowin P., Alsop K., Bailey P.J., Australian Ovarian Cancer Study Group Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 6.Rosen D.G., Yang G., Deavers M.T., Malpica A., Kavanagh J.J., Mills G.B., Liu J. Cyclin E expression is correlated with tumor progression and predicts a poor prognosis in patients with ovarian carcinoma. Cancer. 2006;106:1925–1932. doi: 10.1002/cncr.21767. [DOI] [PubMed] [Google Scholar]
- 7.Karst A.M., Jones P.M., Vena N., Ligon A.H., Liu J.F., Hirsch M.S., Etemadmoghadam D., Bowtell D.D., Drapkin R. Cyclin E1 deregulation occurs early in secretory cell transformation to promote formation of fallopian tube-derived high-grade serous ovarian cancers. Cancer Res. 2014;74:1141–1152. doi: 10.1158/0008-5472.CAN-13-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao S., Choi M., Overton J.D., Bellone S., Roque D.M., Cocco E., Guzzo F., English D.P., Varughese J., Gasparrini S. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc. Natl. Acad. Sci. USA. 2013;110:2916–2921. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherniack A.D., Shen H., Walter V., Stewart C., Murray B.A., Bowlby R., Hu X., Ling S., Soslow R.A., Broaddus R.R., Cancer Genome Atlas Research Network Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell. 2017;31:411–423. doi: 10.1016/j.ccell.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama K., Rahman M.T., Rahman M., Nakamura K., Ishikawa M., Katagiri H., Sato E., Ishibashi T., Iida K., Ishikawa N., Kyo S. CCNE1 amplification is associated with aggressive potential in endometrioid endometrial carcinomas. Int. J. Oncol. 2016;48:506–516. doi: 10.3892/ijo.2015.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones R.M., Mortusewicz O., Afzal I., Lorvellec M., García P., Helleday T., Petermann E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744–3753. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., Benz C.C., Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobbelstein M., Sørensen C.S. Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 2015;14:405–423. doi: 10.1038/nrd4553. [DOI] [PubMed] [Google Scholar]
- 15.Castedo M., Perfettini J.L., Roumier T., Andreau K., Medema R., Kroemer G. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 16.O’Connor M.J. Targeting the DNA Damage Response in Cancer. Mol. Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 17.Couch F.B., Bansbach C.E., Driscoll R., Luzwick J.W., Glick G.G., Bétous R., Carroll C.M., Jung S.Y., Qin J., Cimprich K.A., Cortez D. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ragland R.L., Patel S., Rivard R.S., Smith K., Peters A.A., Bielinsky A.K., Brown E.J. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013;27:2259–2273. doi: 10.1101/gad.223180.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forment J.V., O’Connor M.J. Targeting the replication stress response in cancer. Pharmacol. Ther. 2018;188:155–167. doi: 10.1016/j.pharmthera.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Reaper P.M., Griffiths M.R., Long J.M., Charrier J.D., Maccormick S., Charlton P.A., Golec J.M., Pollard J.R. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 21.Gilad O., Nabet B.Y., Ragland R.L., Schoppy D.W., Smith K.D., Durham A.C., Brown E.J. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–9702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoppy D.W., Ragland R.L., Gilad O., Shastri N., Peters A.A., Murga M., Fernandez-Capetillo O., Diehl J.A., Brown E.J. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J. Clin. Invest. 2012;122:241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell P., Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 24.De Witt Hamer P.C., Mir S.E., Noske D., Van Noorden C.J., Würdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin. Cancer Res. 2011;17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 25.Geenen J.J.J., Schellens J.H.M. Molecular Pathways: Targeting the Protein Kinase Wee1 in Cancer. Clin. Cancer Res. 2017;23:4540–4544. doi: 10.1158/1078-0432.CCR-17-0520. [DOI] [PubMed] [Google Scholar]
- 26.Lin A.B., McNeely S.C., Beckmann R.P. Achieving Precision Death with Cell-Cycle Inhibitors that Target DNA Replication and Repair. Clin. Cancer Res. 2017;23:3232–3240. doi: 10.1158/1078-0432.CCR-16-0083. [DOI] [PubMed] [Google Scholar]
- 27.Mir S.E., De Witt Hamer P.C., Krawczyk P.M., Balaj L., Claes A., Niers J.M., Van Tilborg A.A., Zwinderman A.H., Geerts D., Kaspers G.J. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18:244–257. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dillon M.T., Boylan Z., Smith D., Guevara J., Mohammed K., Peckitt C., Saunders M., Banerji U., Clack G., Smith S.A. PATRIOT: A phase I study to assess the tolerability, safety and biological effects of a specific ataxia telangiectasia and Rad3-related (ATR) inhibitor (AZD6738) as a single agent and in combination with palliative radiation therapy in patients with solid tumours. Clin. Transl. Radiat. Oncol. 2018;12:16–20. doi: 10.1016/j.ctro.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foote K.M., Blades K., Cronin A., Fillery S., Guichard S.S., Hassall L., Hickson I., Jacq X., Jewsbury P.J., McGuire T.M. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J. Med. Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- 30.Leijen S., van Geel R.M., Pavlick A.C., Tibes R., Rosen L., Razak A.R., Lam R., Demuth T., Rose S., Lee M.A. Phase I Study Evaluating WEE1 Inhibitor AZD1775 As Monotherapy and in Combination With Gemcitabine, Cisplatin, or Carboplatin in Patients With Advanced Solid Tumors. J. Clin. Oncol. 2016;34:4371–4380. doi: 10.1200/JCO.2016.67.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leijen S., van Geel R.M., Sonke G.S., de Jong D., Rosenberg E.H., Marchetti S., Pluim D., van Werkhoven E., Rose S., Lee M.A. Phase II Study of WEE1 Inhibitor AZD1775 Plus Carboplatin in Patients With TP53-Mutated Ovarian Cancer Refractory or Resistant to First-Line Therapy Within 3 Months. J. Clin. Oncol. 2016;34:4354–4361. doi: 10.1200/JCO.2016.67.5942. [DOI] [PubMed] [Google Scholar]
- 32.Chen X., Low K.H., Alexander A., Jiang Y., Karakas C., Hess K.R., Carey J.P.W., Bui T.N., Vijayaraghavan S., Evans K.W. Cyclin E Overexpression Sensitizes Triple-Negative Breast Cancer to Wee1 Kinase Inhibition. Clin. Cancer Res. 2018;24:6594–6610. doi: 10.1158/1078-0432.CCR-18-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang Y., McGrail D.J., Sun C., Labrie M., Chen X., Zhang D., Ju Z., Vellano C.P., Lu Y., Li Y. Sequential Therapy with PARP and WEE1 Inhibitors Minimizes Toxicity while Maintaining Efficacy. Cancer Cell. 2019;35:851–867.e7. doi: 10.1016/j.ccell.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck H., Nähse-Kumpf V., Larsen M.S., O’Hanlon K.A., Patzke S., Holmberg C., Mejlvang J., Groth A., Nielsen O., Syljuåsen R.G., Sørensen C.S. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 2012;32:4226–4236. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinner A., Wu W., Staudt C., Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Au-Yeung G., Lang F., Azar W.J., Mitchell C., Jarman K.E., Lackovic K., Aziz D., Cullinane C., Pearson R.B., Mileshkin L. Selective Targeting of Cyclin E1-Amplified High-Grade Serous Ovarian Cancer by Cyclin-Dependent Kinase 2 and AKT Inhibition. Clin. Cancer Res. 2017;23:1862–1874. doi: 10.1158/1078-0432.CCR-16-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domcke S., Sinha R., Levine D.A., Sander C., Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weroha S.J., Becker M.A., Enderica-Gonzalez S., Harrington S.C., Oberg A.L., Maurer M.J., Perkins S.E., AlHilli M., Butler K.A., McKinstry S. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin. Cancer Res. 2014;20:1288–1297. doi: 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ince T.A., Sousa A.D., Jones M.A., Harrell J.C., Agoston E.S., Krohn M., Selfors L.M., Liu W., Chen K., Yong M. Characterization of twenty-five ovarian tumour cell lines that phenocopy primary tumours. Nat. Commun. 2015;6:7419. doi: 10.1038/ncomms8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demidenko E., Miller T.W. Statistical determination of synergy based on Bliss definition of drugs independence. PLoS ONE. 2019;14:e0224137. doi: 10.1371/journal.pone.0224137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J.F., Palakurthi S., Zeng Q., Zhou S., Ivanova E., Huang W., Zervantonakis I.K., Selfors L.M., Shen Y., Pritchard C.C. Establishment of Patient-Derived Tumor Xenograft Models of Epithelial Ovarian Cancer for Preclinical Evaluation of Novel Therapeutics. Clin. Cancer Res. 2017;23:1263–1273. doi: 10.1158/1078-0432.CCR-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tetzlaff M.T., Yu W., Li M., Zhang P., Finegold M., Mahon K., Harper J.W., Schwartz R.J., Elledge S.J. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. USA. 2004;101:3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nieminuszczy J., Schwab R.A., Niedzwiedz W. The DNA fibre technique—tracking helicases at work. Methods. 2016;108:92–98. doi: 10.1016/j.ymeth.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 44.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown E.J., Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y. GammaH2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Etemadmoghadam D., deFazio A., Beroukhim R., Mermel C., George J., Getz G., Tothill R., Okamoto A., Raeder M.B., Harnett P. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin. Cancer Res. 2009;15:1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim S.Y., Menzies A.M., Rizos H. Mechanisms and strategies to overcome resistance to molecularly targeted therapy for melanoma. Cancer. 2017;123(S11):2118–2129. doi: 10.1002/cncr.30435. [DOI] [PubMed] [Google Scholar]
- 49.Yap T.A., Omlin A., de Bono J.S. Development of therapeutic combinations targeting major cancer signaling pathways. J. Clin. Oncol. 2013;31:1592–1605. doi: 10.1200/JCO.2011.37.6418. [DOI] [PubMed] [Google Scholar]
- 50.Wright G., Golubeva V., Remsing Rix L.L., Berndt N., Luo Y., Ward G.A., Gray J.E., Schonbrunn E., Lawrence H.R., Monteiro A.N.A., Rix U. Dual Targeting of WEE1 and PLK1 by AZD1775 Elicits Single Agent Cellular Anticancer Activity. ACS Chem. Biol. 2017;12:1883–1892. doi: 10.1021/acschembio.7b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young L.A., O’Connor L.O., de Renty C., Veldman-Jones M.H., Dorval T., Wilson Z., Jones D.R., Lawson D., Odedra R., Maya-Mendoza A. Differential Activity of ATR and WEE1 Inhibitors in a Highly Sensitive Subpopulation of DLBCL Linked to Replication Stress. Cancer Res. 2019;79:3762–3775. doi: 10.1158/0008-5472.CAN-18-2480. [DOI] [PubMed] [Google Scholar]
- 52.Qi W., Xu X., Wang M., Li X., Wang C., Sun L., Zhao D., Sun L. Inhibition of Wee1 sensitizes AML cells to ATR inhibitor VE-822-induced DNA damage and apoptosis. Biochem. Pharmacol. 2019;164:273–282. doi: 10.1016/j.bcp.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 53.Bukhari A.B., Lewis C.W., Pearce J.J., Luong D., Chan G.K., Gamper A.M. Inhibiting Wee1 and ATR kinases produces tumor-selective synthetic lethality and suppresses metastasis. J. Clin. Invest. 2019;129:1329–1344. doi: 10.1172/JCI122622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin J., Fang H., Yang F., Ji W., Guan N., Sun Z., Shi Y., Zhou G., Guan X. Combined Inhibition of ATR and WEE1 as a Novel Therapeutic Strategy in Triple-Negative Breast Cancer. Neoplasia. 2018;20:478–488. doi: 10.1016/j.neo.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyatt H.D., Laister R.C., Martin S.R., Arrowsmith C.H., West S.C. The SMX DNA Repair Tri-nuclease. Mol. Cell. 2017;65:848–860.e11. doi: 10.1016/j.molcel.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyatt H.D., Sarbajna S., Matos J., West S.C. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 57.Enot D.P., Vacchelli E., Jacquelot N., Zitvogel L., Kroemer G. TumGrowth: An open-access web tool for the statistical analysis of tumor growth curves. OncoImmunology. 2018;7:e1462431. doi: 10.1080/2162402X.2018.1462431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamin D., Sato T., Cibulskis K., Getz G., Stewart C., Lichtenstein L. Calling Somatic SNVs and Indels with Mutect2. bioRxiv. 2019:861054. doi: 10.1101/861054. [DOI] [Google Scholar]
- 60.Chang X., Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 2012;49:433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Y., Wang R., Urrutia E., Anastopoulos I.N., Nathanson K.L., Zhang N.R. CODEX2: full-spectrum copy number variation detection by high-throughput DNA sequencing. Genome Biol. 2018;19:202. doi: 10.1186/s13059-018-1578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.George E., Kim H., Krepler C., Wenz B., Makvandi M., Tanyi J.L., Brown E., Zhang R., Brafford P., Jean S. A patient-derived-xenograft platform to study BRCA-deficient ovarian cancers. JCI Insight. 2017;2:e89760. doi: 10.1172/jci.insight.89760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fishbein L., Khare S., Wubbenhorst B., DeSloover D., D’Andrea K., Merrill S., Cho N.W., Greenberg R.A., Else T., Montone K. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nat. Commun. 2015;6:6140. doi: 10.1038/ncomms7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tibes R., Qiu Y., Lu Y., Hennessy B., Andreeff M., Mills G.B., Kornblau S.M. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 66.Kim H., George E., Ragland R., Rafial S., Zhang R., Krepler C., Morgan M., Herlyn M., Brown E., Simpkins F. Targeting the ATR/CHK1 Axis with PARP Inhibition Results in Tumor Regression in BRCA-Mutant Ovarian Cancer Models. Clin. Cancer Res. 2017;23:3097–3108. doi: 10.1158/1078-0432.CCR-16-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. All Sequencing data has been deposited in NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/bioproject) with accession number PRJNA750496 and NCBI GEO database (https://www.ncbi.nlm.nih.gov/gds/) with NCBI accession # PRJNA626436. All the data are also available in Mendeley Data with DOI 10.17632/2xxxzsg6hz.1.