Abstract
Histone acetylation plays an important role in regulating chromatin structure and thus gene expression. Here we describe the functional characterization of HDAC4, a human histone deacetylase whose C-terminal part displays significant sequence similarity to the deacetylase domain of yeast HDA1. HDAC4 is expressed in various adult human tissues, and its gene is located at chromosome band 2q37. HDAC4 possesses histone deacetylase activity intrinsic to its C-terminal domain. When tethered to a promoter, HDAC4 represses transcription through two independent repression domains, with repression domain 1 consisting of the N-terminal 208 residues and repression domain 2 containing the deacetylase domain. Through a small region located at its N-terminal domain, HDAC4 interacts with the MADS-box transcription factor MEF2C. Furthermore, HDAC4 and MEF2C individually upregulate but together downmodulate c-jun promoter activity. These results suggest that HDAC4 interacts with transcription factors such as MEF2C to negatively regulate gene expression.
In eukaryotic cells, genetic information is packaged into chromatin, a highly organized DNA-protein complex which controls gene activities. A central question in studying eukaryotic gene regulation is how the generally repressive chromatin structure is regulated when necessary. In the past several years, three regulatory mechanisms have been recognized: DNA methylation, posttranslational modifications of histones, and ATP-dependent chromatin remodeling (53, 55, 57). The most extensively studied form of posttranslational modifications of histones is acetylation of ɛ-amino groups of lysine residues located at the flexible N-terminal tails of core histones (53, 55). The level of histone acetylation at a given region of chromatin correlates well with its transcriptional activity (39). Mechanistically, histone acetylation affects nucleosome stability and/or internucleosomal interaction (2, 29). The dynamic level of histone acetylation in vivo is maintained through opposing actions of histone acetyltransferases and deacetylases. Several known transcriptional coactivators possess intrinsic histone acetyltransferase activity (14, 27, 49, 57).
The first histone deacetylase, originally called HD1 (histone deacetylase 1) and later renamed HDAC1 (histone deacetylase 1), was cloned from mammalian cells (18, 50). HDAC1 was found to be highly homologous to the known yeast transcriptional coregulator RPD3 (50). Two HDAC1 homologs (HDAC2 and HDAC3) have been cloned from human cDNA libraries (10, 58, 59). Transcriptional repressors recruit RPD3 or HDAC1 to -3 to downregulate transcription (reviewed in references 41 and 56). The deacetylase activity of HDAC1 and RPD3 has been found to be important for transcriptional repression (18, 24), suggesting that histone deacetylation directly leads to transcriptional repression. Consistent with this contention, recruitment of RPD3 by the yeast repressor Ume6 leads to local histone deacetylation and formation of a highly localized domain of repressed chromatin in vivo (25).
Two distinct yeast histone deacetylase complexes have been characterized: one possesses RPD3 as its catalytic subunit, while the other contains the histone deacetylase HDA1 (6, 43). The N-terminal domain of HDA1 shows some sequence similarity to the catalytic domain of the RPD3/HDAC family (amino acid sequence identity, 26%; similarity, 49%), whereas its C-terminal domain exhibits no sequence similarity to known proteins. A great deal of knowledge has been acquired about the function of the RPD3/HDAC family of histone deacetylases in transcriptional regulation (14, 27, 49, 57). In contrast, it is entirely unclear if and how HDA1 plays a role in transcriptional regulation.
In vertebrates, the MEF2 family of transcription factors, also called RSRFs (related to serum response factors), is composed of four isoforms, MEF2A, -B, -C, and -D, all of which contain MADS-box DNA-binding domains at their N termini and adjacent MEF2-specific motifs (4, 36, 42). Although MEF2s were initially identified as myocyte enhancer-binding factors activating muscle-specific genes, their roles in nonmuscle cells have also been demonstrated (7, 15, 16, 26, 44, 63). In nonmuscle cells, MEF2s serve as nuclear targets of several signaling pathways (7, 9, 15, 26, 63). Moreover, it has been suggested that MEF2s are involved in negative transcriptional regulation (40). How this occurs remains largely unexplored.
Here we report that HDAC4, a human histone deacetylase whose C-terminal region is highly related to the N-terminal portion of HDA1, physically and functionally interacts with the transcription factor MEF2C: through the N-terminal domain of HDAC4, MEF2C recruits HDAC4 to repress transcription. Furthermore, MEF2C and HDAC4 individually upregulate but together downmodulate c-jun promoter activity. These results suggest that like RPD3 and HDAC1 to -3, HDAC4 is recruited to promoters by target transcription factors to regulate transcription.
MATERIALS AND METHODS
Molecular cloning.
Plasmid construction and DNA sequencing were performed according to standard procedures. The cDNA clone KIAA0288 (GenBank accession no. AB006626) was kindly provided by T. Nagase (Kazusa DNA Research Institute, Chiba, Japan). This clone was used to construct expression plasmids for HDAC4 and its mutants except that the coding sequence for its N-terminal 221 residues was obtained from a human bone marrow cDNA library (the KIAA0288 clone contains a C-to-T nonsense mutation at nucleotide 1135, as kindly communicated by T. Nagase). This mutation has also been identified by Grozinger et al. (13). The partial clone for HDAC7 was amplified from a human brain cDNA library by PCR with primers based on the sequences of four human bacterial artificial chromosome clones (GenBank accession no. AC002124, AC002088, AC002410, and AC002433). Northern analyses on poly(A) RNA blots (Clontech) were carried out according to the manufacturer’s instructions. The reporter tk-Luc was derived from pGL2 (Promega) by insertion of the thymidine kinase (tk) core promoter (−105 to +52). Gal4-tk-Luc was constructed from tk-Luc by insertion of five copies of the Gal4-binding site upstream from the tk promoter. Gal4-SV40-Luc was constructed from pGL2-Control (Promega) by insertion of the Gal4-binding sites from Gal4-tk-Luc. Gal4-AdML-Luc and Gal4-CD4-Luc have been described elsewhere (34, 62). MEF2-E4-Luc was derived from the 3TP-Lux luciferase reporter (19) by replacement of its NheI/BamHI region with an oligonucleotide duplex consisting of 5′-CTA GCT GGG CTA TTT TTA GG-3′ and 5′-GAT CCC TAA AAA TAG CCC AG-3′ (the MEF2-binding sites are underlined).
FISH.
Fluorescence in situ hybridization (FISH) was performed on human lymphocytes as described elsewhere (21). The probe was a 5.5-kb HDAC4 cDNA fragment biotinylated with dATP by using a BioNick labeling kit (Gibco).
Protein expression and purification.
For expression in 293T cells, 10 μg of plasmid expressing HDAC4 or its mutants were used to transfect 1 × 106 to 1.5 × 106 cells (in a 10-cm diameter dish) with 24 μl of SuperFect transfection reagent (Qiagen). After 48 h, cells were washed twice with phosphate-buffered saline and collected in 1 ml of buffer B (20 mM Tris-HCl [pH 8.0], 10% glycerol, 5 mM MgCl2, 0.1% NP-40, protease inhibitors) containing 0.5 M KCl. The same buffer was used for washing M2-agarose beads immobilized with Flag-HDAC4; for elution, the concentration of KCl was reduced to 0.15 M.
For expression of HDAC4 mutants in Spodoptera frugiperda Sf9 cells, recombinant baculoviruses were generated by the BaculoGold (Pharmingen) or Bac-to-Bac (Gibco) systems. HDAC4 mutants were affinity purified as described above.
Deacetylase assay.
[3H]acetyl-histones were prepared from HeLa cells. Briefly, after incubation for 2 to 6 h in medium containing 50 μCi of [3H]acetate (2.4 Ci/mmol; NEN Life Sciences) per ml and 3 μM trichostatin A (TSA; Wako), HeLa cells were harvested and lysed in buffer N (10 mM Tris-HCl [pH 8.0], 250 mM sucrose, 2 mM MgCl2, 1 mM CaCl2, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitors), and nuclei were isolated as described elsewhere (51). To isolate histones, the nuclei were extracted with 0.4 N H2SO4, and acid-extracted histones were precipitated with 9 volumes of acetone. After at least 1 h on ice, histones were collected by centrifugation; the histone pellet was dissolved in 0.1 ml of 100 mM Tris-HCl (pH 8.0) and precipitated with cold acetone three to four times. Histones were air dried and dissolved in 2 mM HCl. Levels of histone acetylation were verified by using Triton-acetic acid-urea gels (22).
[3H]acetyl-histones were also prepared by in vitro labeling: 50-μg aliquots of histones (Sigma) were incubated with 50 pmol of [3H]acetylcoenzyme A (4.7 Ci/mmol; Amersham) and 0.5 μg of Flag-PCAF (p300/CBP-associated factor) in 100 μl of buffer A (50 mM Tris-HCl [pH 8.0], 10% glycerol, 1 mM dithiothreitol, 0.1 mM EDTA, 1 mM PMSF) at 30°C for 30 min. The expression and purification of Flag-PCAF has been described elsewhere (60). To remove unincorporated [3H]acetyl coenzyme A, histones were precipitated by adding 2 μl of 5 M NaCl, 1 ml of cold acetone, and 65 μg of bovine serum albumin. The tube was left on dry ice for 2 h and subsequently centrifuged at 4°C for 5 min. The resulting pellet was washed with 1 ml of cold acetone, air dried, and dissolved in 100 μl of 2 mM HCl.
Deacetylase activity was determined by analysis of the release of [3H]acetate from [3H]acetyl-histones (20, 23). Assays were carried out in 0.2 ml of buffer H (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 0.1 mM PMSF) containing [3H]acetyl-histones (25,000 dpm). The reaction was allowed to proceed at 37°C for 90 min and stopped by addition of 0.1 ml of 0.1 M HCl–0.16 M acetic acid. Released [3H]acetate was extracted with 0.9 ml of ethyl acetate. After centrifugation, 0.6 ml of the upper organic phase was quantified by liquid scintillation counting.
DNA-binding assay.
A modified filter-binding assay was used (17). Briefly, sheared fish sperm DNA (100 ng; Boehringer Mannheim) was labeled with [α-32P]dCTP in a Ready-To-Go DNA labeling reaction tube (Pharmacia) and separated from free [α-32P]dCTP on a G-25 spin column. Flag-HDAC4 was immobilized on 10 μl of M2-agarose and incubated with 2 ng of 32P-labeled fish sperm DNA fragments. After extensive washing, bound DNA was quantified by liquid scintillation counting.
Protein-protein interaction.
To examine the interaction between HDAC4 and MEF2C in vivo, HDAC4 (Flag tagged) and/or MEF2C expression plasmids were cotransfected into 293T cells, and transfected cells were collected in buffer B–0.15 M KCl as described above. One-third of the extract was used for immunoprecipitation with anti-Flag M2-agarose beads (Sigma). Beads with bound immunocomplexes were washed four times with buffer B–0.15 M KCl, and bound proteins were eluted with the Flag peptide or 0.1 M glycine-HCl (pH 2.5). After separation by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred to nitrocellulose membranes for Western blot analyses with anti-Flag and anti-MEF2C antibodies. Blots were developed with the Supersignal chemiluminescent substrate (Pierce). The same procedure was followed to examine the in vivo interaction between HDAC4 and MEF2D except that endogenous MEF2D was detected due to its reasonable expression level in 293T cells.
For in vitro MBP pull-down assays, the MEF2C fragment M178 was expressed as a fusion with maltose-binding protein (MBP) in Escherichia coli, immobilized on amylose-agarose beads and used to study the interaction with HDAC4 and its mutants, which were synthesized in vitro with the TNT-T7 coupled reticulocyte lysate system (Promega) in the presence of Redivue l-[35S]methionine (Amersham). After rotation for 30 min at 4°C, the complexes bound to agarose beads were washed three times with buffer B–0.15 M KCl and once with buffer B–0.5 M KCl and boiled in 1 × SDS sample buffer prior to separation by SDS-PAGE and autoradiography.
Reporter gene assays.
SuperFect transfection reagent (Qiagen) was used to transiently transfect a luciferase reporter plasmid (50 to 200 ng) and/or mammalian expression plasmids (50 to 200 ng) into NIH 3T3 or 293T cells. pBluescript KSII(+) was used to normalize the total amount of plasmids used in each transfection, and pCMV-β-Gal (50 ng) was cotransfected for normalization of transfection efficiency. After 48 h, cells were lysed in situ, and luciferase reporter activity was determined by using d-(−)-luciferin (Boehringer Mannheim) as the substrate. Galactosidase activity was measured with Galacto-Light Plus (Tropix) as the substrate. The chemiluminescence from activated luciferin or Galacto-Light Plus was measured on a Luminometer plate reader (Dynex). As indicated, transfected cells were exposed to TSA (3 μM) for 16 h prior to reporter assays. Each transfection was performed at least four times.
RESULTS
A family of human histone deacetylases related to yeast HDA1.
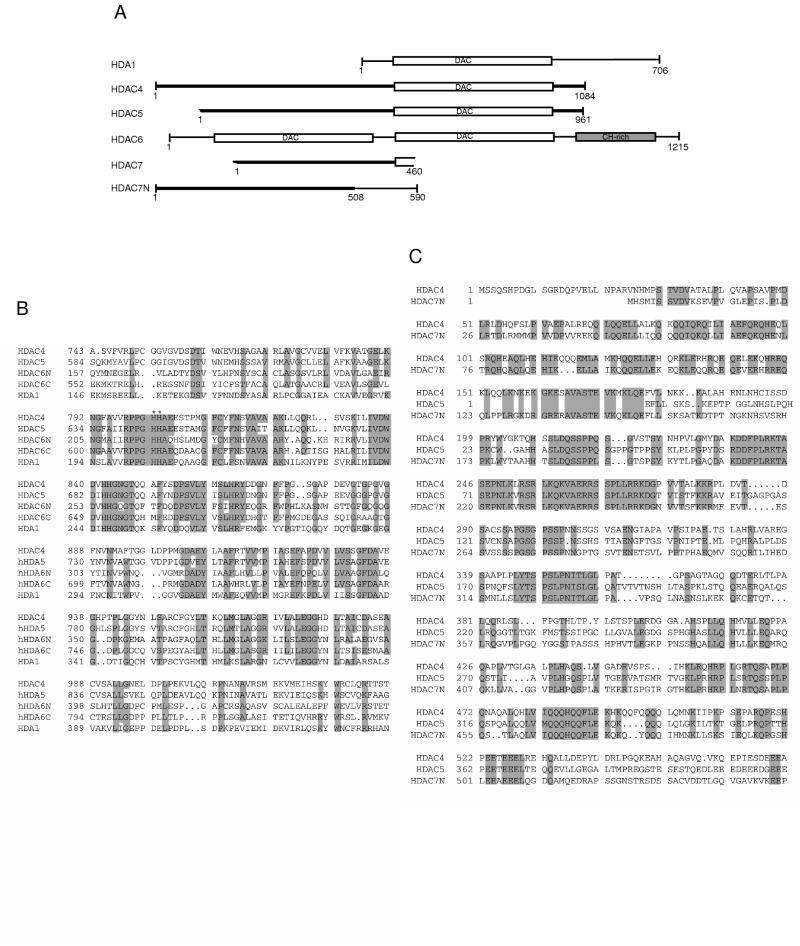
To identify new mammalian histone deacetylases, we performed sequence database searches with BLAST and PSI-BLAST (1). Using the amino acid sequence of yeast HDA1 as the bait, we found several human cDNA and genomic clones encoding polypeptides with significant sequence similarity to the catalytic domain of HDA1. Figure 1A shows the schematic representation of these novel polypeptides. Most of these clones were isolated in DNA sequencing projects, whereas HDAC5 was also isolated as a clone coding a human colon cancer antigen recognized by an autologous antibody (37, 38, 45). Available sequence data indicated that HDAC4, -5, and -7 are homologous, with their C-terminal parts similar to the catalytic domain of HDA1 (Fig. 1A and B). Sequence alignment of the N-terminal domains of HDAC4, -5, and -7N is shown in Fig. 1C. HDAC6 possesses two homologous regions similar to the catalytic domain of HDA1, and a cysteine/histidine-rich domain located at its C-terminal part (Fig. 1A and B). The putative catalytic domains of HDAC4, -5, and -6 are more similar to yeast HDA1 (sequence identity of 35%) than to human HDAC1, -2, and -3 (sequence identity of 26%), suggesting that HDAC4, -5, and -6 and probably HDAC7 constitute a new subfamily of human histone deacetylases, with HDAC4, HDAC5, and probably HDAC7 more similar to each other than to HDAC6. Since HDAC4 was identified first and its full-length cDNA was available, we chose to characterize it further.
FIG. 1.
Comparison of HDAC4-7 with HDA1. (A) Schematic representation of HDA1 and HDAC4 to -7. The N terminus of HDAC5 is incomplete, as are both termini of HDAC7. HDAC7N may be an alternatively spliced variant of HDAC7. The conserved deacetylase domains are boxed and labeled “DAC.” Other domains shared by HDAC4, -5, and -7 and HDAC7N are shown in bold lines. HDAC6 has a cysteine/histidine-rich domain (CH-rich; shaded box) at its C terminus. This diagram was generated based on BLAST search results. Sequences (GenBank accession numbers) referred to are HDA1 (P53973), HDAC4 (AB006626), HDAC5 (AB011172 and AF039691), HDAC6 (AJ011972), HDAC7 (AF124924), and HDAC7N (AB018287). A genomic clone (GenBank accession no. AC004466) contains some coding sequences related to HDAC4, -5, and -7 and may encode HDAC8. (B) Sequence alignment of catalytic domains of HDAC4 to -6 and HDA1. Identical or highly conserved residues (four of five sequences) are shaded. For simplicity, only S/T, R/K, and D/E are considered to be highly conserved. Asterisks denote histidines 802 and 803 of HDAC4, residues that may be important for deacetylase activity. (C) Sequence alignment of the N-terminal domains of HDAC4, -5, and -7N. Identical residues are shaded.
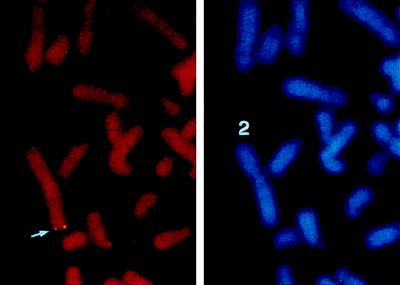
To determine tissue distribution of HDAC4, Northern blot analyses were performed. These analyses indicated that HDAC4 is expressed in skeletal muscle, brain, leukocyte, colon, small intestine, and ovary but not in liver, lung, and placenta (Fig. 2). To map the chromosomal localization of the HDAC4 gene, FISH analyses were performed. These analyses revealed that the HDAC4 gene is located at chromosome band 2q37.2 (Fig. 3). Abnormalities in this region have been implicated in developmental delay and predisposition to certain cancers (8, 33). Moreover, this band has been found to contain a cellular senescence gene (52).
FIG. 2.
Expression of HDAC4 in various adult human tissues. Poly(A) RNA blots (Clontech; 2 μg/lane) were probed with an HDAC4 cDNA fragment derived from the 3′ untranslated region (top). As a loading control, the same blots were reprobed with a β-actin cDNA probe (bottom). Molecular size markers are shown at the right.
FIG. 3.
Chromosomal localization of the HDAC4 gene. Left, FISH signals detected at chromosome band 2q37.2, indicated by an arrow; right, the same mitotic cell stained with DAPI (4′,6-diamidino-2-phenylindole) to identify chromosomes. Human blood lymphocytes were used for FISH; the hybridization efficiency was 81% (i.e., 81 of 100 checked mitotic figures showed the indicated localization).
Histone deacetylase activity of HDAC4.
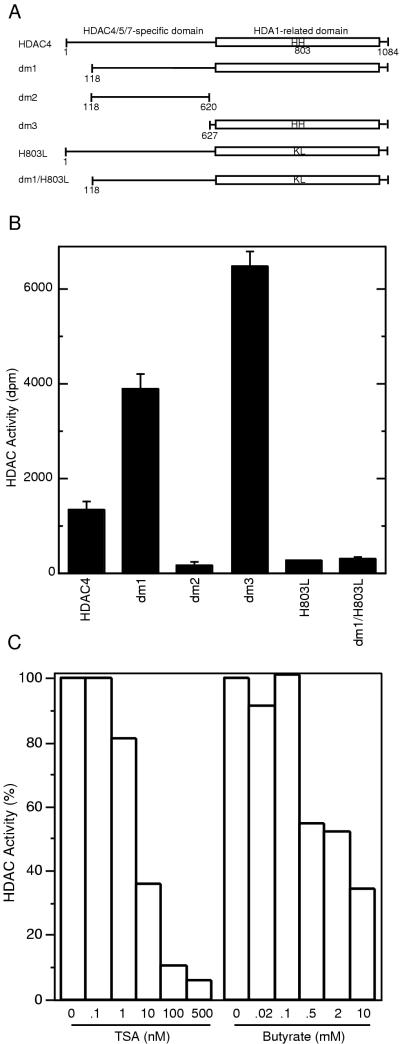
To determine the histone deacetylase activity of HDAC4, Flag-tagged HDAC4 and deletion mutants dm1, -2, and -3 (Fig. 4A) were expressed in 293T cells and subject to histone deacetylase assays. As shown in Fig. 4B, affinity-purified HDAC4 efficiently deacetylated [3H]acetyl-histones. Mutant dm1 had activity 2.9-fold higher than that of full-length HDAC4. Whereas dm2 had minimal activity, dm3 was slightly more active than dm1, suggesting that dm3 contains a deacetylase domain. This is consistent with the observation that the HDA1-related domain of HDAC4 is located at its C-terminal part (Fig. 1A).
FIG. 4.
Characterization of histone deacetylase activity of HDAC4. (A) Schematic representation of HDAC4 and its mutants used for deacetylase assays. The letters “HH” denote histidines 802 and 803, which may be essential for deacetylase activity. (B) Deacetylase activity of HDAC4 and its mutants. Deacetylase activity, measured as disintegrations per minute of [3H]acetate released from [3H]acetyl-histones, was normalized to relative protein concentration determined by Western analyses with an anti-Flag antibody. During purification of Flag-tagged proteins, a buffer containing 0.5 M KCl was used for extensive washing; under such conditions, with untransfected cell extracts, equivalent amounts of M2-agarose beads retained deacetylase activity close to background levels. (C) Effects of TSA and sodium butyrate on deacetylase activity of dm3.
To establish that the observed deacetylase activity is intrinsic to HDAC4 (but not due to any associated proteins), we prepared mutants with histidines 802 and 803 replaced with lysine and leucine, respectively (Fig. 4A, H803L and dm1/H803L). Histidine residues at equivalent positions have been found to be important for the deacetylase activity of HDAC1 and RPD3 (18, 24). Compared with HDAC4 and dm1, both mutants had much lower deacetylase activity (Fig. 4B), suggesting that HDAC4 has intrinsic deacetylase activity and the histidine residues are important for the enzymatic activity.
To examine the effects of deacetylase inhibitors, we determined the deacetylase activity of dm3 in the presence of various concentrations of TSA or sodium butyrate. As shown in Fig. 4C, TSA dramatically inhibited the activity of dm3, with a 50% inhibitory concentration of 5 nM, whereas sodium butyrate (up to 5 mM) had much smaller effects. HDAC1 and HDAC3 are more sensitive to sodium butyrate than HDAC4 (10).
Mutants dm1 and dm3 were also expressed in Sf9 cells, using the baculovirus expression system. Proteins prepared this way had activity inversely proportional to their expression levels. Even the most active preparations possessed much lower activity than those obtained from 293T cells (data not shown), suggesting that an elusive factor(s) required for deacetylase activity may not present in sufficient quantities in insect cells.
Tethered HDAC4 functions as a repressor.
The possession of intrinsic deacetylase activity by HDAC4 suggests that it may be involved in transcriptional regulation. To test this hypothesis, we first investigated if HDAC4 functions as a repressor when artificially tethered to a promoter. For this purpose, a mammalian vector was constructed to express HDAC4 fused to the Gal4 DNA-binding domain and tested by cotransfection assays with the Gal4-tk-Luc reporter (Fig. 5A) in NIH 3T3 cells. As shown in Fig. 5B, while the Gal4 DNA-binding domain itself activated transcription 2-fold, GAL4-HDAC4 repressed transcription 14-fold. To delineate the repression domain(s), mammalian vectors were constructed to express various HDAC4 mutants fused to the Gal4 DNA-binding domain. HDAC4 mutants tested include dm1 to -3 (Fig. 4A), dm4 (residues 1 to 208), and dm5 (residues 1 to 114). As shown in Fig. 5B, similar to Gal4-HDAC4, Gal4-dm1 repressed transcription 11-fold. While Gal4-dm2 had minimal effects (∼2-fold), Gal4-dm3 repressed transcription 83-fold. In contrast, Gal4-dm3 had a much smaller repressive effect on the tk-Luc reporter (1.8-fold [data not shown]). Western analyses with an anti-Gal4 antibody indicated that Gal4-HDAC4 and Gal4-dm1 to -5 were indeed expressed (Fig. 5C). All of these results suggest that dm3 contains an active, strong repression domain. Unexpectedly, Gal4-dm4 repressed transcription 14-fold whereas both Gal4-dm2 and Gal4-dm5 had minimal effects (Fig. 5B), suggesting that residues 1 to 208 of HDAC4 constitute another repression domain.
FIG. 5.
Tethered HDAC4 represses transcription. (A) Schematic representation of the luciferase reporter Gal4-tk-Luc. Upstream from the tk core promoter (−152 to +50) are five copies of the Gal4-binding site. (B) Repression of Gal4-tk-Luc by HDAC4 and its mutants in NIH 3T3 cells. The mutants dm1 to -3 and dm1/H803A are depicted in Fig. 4A; dm4 and dm5 contain the N-terminal 208 and 114 residues of HDAC4, respectively. Mammalian constructs expressing HDAC4 and its mutants fused to the C terminus of Gal4(1-147) were transfected into NIH 3T3 cells with the reporter Gal4-tk-Luc. Luciferase (Luc) activities were normalized to the internal β-galactosidase control; the normalized luciferase activity from the transfection without any effector plasmid was arbitrarily set to 1.0. (C) Expression of Gal4-HDAC4 and its mutants. Extracts (10 μg/lane), prepared from 293T cells transfected with expression plasmids for indicated fusion proteins, were subjected to Western blotting analyses using an anti-Gal4 antibody (RK5C1; Santa Cruz Biotechnology). Molecular size markers are shown at the right. (D) Repression of reporters with different core promoters by Gal4-dm3 in 293T cells. The reporters possess indicated core promoters replacing the tk region of Gal4-tk-Luc (A); 100 and 300 ng of expression plasmids were used as indicated.
The repression observed with dm3 is stronger than that reported for HDAC1, -2, and -3 (59). To assess if the repression by Gal4-dm3 is cell line dependent, we performed similar transfection assays in 293T cells. As shown in Fig. 5D, Gal4-dm3 repressed Gal4-tk-Luc reporter activity in these cells in a dose-dependent manner. Since repression mediated by HDAC1 was found to be promoter dependent (30), we assessed if Gal4-dm3 is able to repress reporters containing other core promoters. For this purpose, transfection assays were performed with TATA-containing (Gal4-AdML-Luc and Gal4-SV40-Luc) as well as TATA-less (Gal4-CD4-Luc) reporters. As shown in Fig. 5D, Gal4-dm3 was able to repress transcription of all of these reporters. Taken together, these results suggest that once tethered to a promoter, the deacetylase domain of HDAC4 functions as a transcriptional repressor.
Requirement of HDAC4 deacetylase activity for repression.
The repression observed with HDAC4 could be due to deacetylation mediated by HDAC4 and/or to association with a repressor(s). This prompted us to examine whether the intrinsic deacetylase activity of HDAC4 is important for the observed repression. Since TSA inhibited deacetylase activity of HDAC4 (Fig. 4C), we determined effects of TSA on HDAC4-mediated repression. TSA only partially relieved repression mediated by Gal4-HDAC4 and Gal4-dm1 (Fig. 5B). TSA had a much more dramatic effect on the repression mediated by Gal4-dm3 (Fig. 5B), suggesting that histone deacetylase activity is important for the repression observed with Gal4-dm3. Substitution of histidines 802 and 803 reduced repression by Gal4-dm1, and TSA had no effects on residual repression observed with Gal4-dm1/H803L (Fig. 5B; compare Gal4-dm1 and Gal4-dm1/H803L). TSA did not relieve repression mediated by Gal4-dm4 (Fig. 5B). Taken together, these results suggest that while the histone deacetylase activity of HDAC4 is important for its repression function, mechanisms independent of deacetylation are also involved.
HDAC4 does not directly bind to DNA.
Since promoter tethering of HDAC4 leads to transcriptional repression, we next asked how HDAC4 is recruited to promoters in vivo. One possibility is that HDAC4 possesses intrinsic DNA-binding ability. Sequence-specific DNA-binding proteins can, although with lower affinity, bind to nonspecific DNA. To address if HDAC4 directly binds to DNA, we performed a DNA-binding assay to determine if HDAC4 could nonspecifically bind to fish sperm DNA (17). This assay revealed that Flag-HDAC4 immobilized on M2-agarose could not retain a significantly higher amount of DNA than M2-agarose itself (data not shown). Therefore, HDAC4 does not have intrinsic DNA-binding ability.
HDAC4 physically interacts with MEF2 transcription factors.
Since HDAC4 does not bind to DNA by itself, we reasoned that other transcription factors might mediate the recruitment of HDAC4 to promoters. To identify such target transcription factors, we tested several active repressors, including human Groucho homolog TLE1 (12, 48), zinc finger oncoprotein Evi1 (3), Polycomb-group protein EZH2 (28), and adenovirus protein E1B (61). Protein-protein interaction studies and reporter gene assays indicated that none of these repressors interact with HDAC4 (data not shown).
A novel Xenopus laevis repressor protein, termed MITR (GenBank accession no. Z97214; reference 47), was identified as an interaction partner for the Xenopus myocyte enhancer-binding factors SL-1 and -2. Xenopus MITR is a homolog of HDAC7N (sequence identity, 59%; similarity, 67%). As illustrated in Fig. 1A, HDAC7N is composed of two regions, the N-terminal part of which shows significant sequence similarity to HDAC4 (sequence identity, 46%; similarity, 58%). In light of these observations, we tested if HDAC4 interacts with human MEF2 transcription factors.
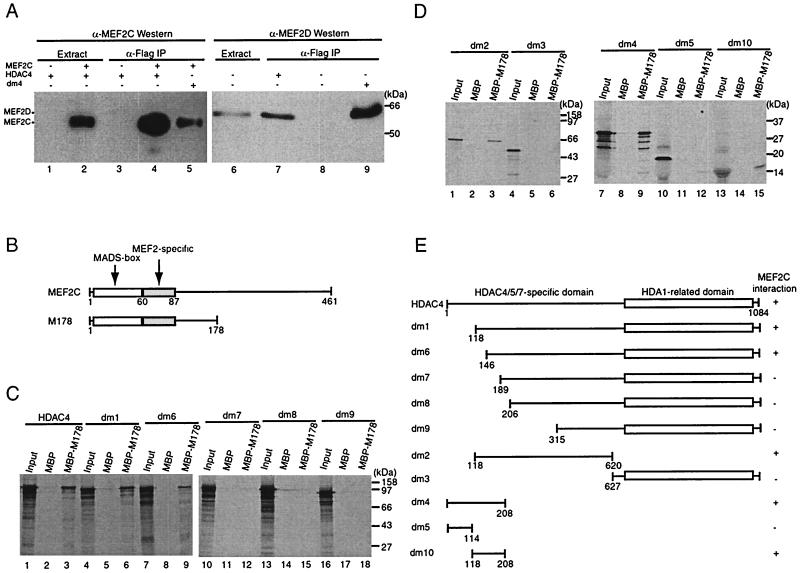
To examine in vivo interaction between HDAC4 and MEF2s, we performed immunoprecipitation experiments in which HDAC4 (Flag tagged) and/or MEF2C expression plasmids were cotransfected into 293T cells, and extracts prepared from the transfected cells were subjected to immunoprecipitation with anti-Flag M2-agarose. Eluted immunocomplexes were subjected to Western blotting analyses with anti-Flag and anti-MEF2C antibodies. As shown in Fig. 6A, MEF2C specifically precipitated with Flag-tagged HDAC4 (lanes 1 to 4). Similar immunoprecipitation experiments revealed that HDAC4 precipitated with endogenous MEF2D (lanes 6 to 8). These results indicate that HDAC4 interacts with MEF2C and MEF2D in vivo.
FIG. 6.
HDAC4 interacts with MEF2 in vivo and in vitro. (A) Immunoprecipitation of HDAC4 with MEF2C (lanes 1 to 5) or MEF2D (lanes 6 to 9). Flag-tagged HDAC4 (lanes 1 to 4 and 7) or dm4 (lanes 5 and 9) was expressed with (lanes 2, 4, and 5) or without (lanes 1, 3, and 6 to 9) MEF2C in 293T cells and immunoprecipitated (IP) with anti-Flag M2-agarose. Extracts (lanes 1, 2, and 6) and immunoprecipitated proteins eluted with Flag peptide (lanes 3 to 5 and 7 to 9) were subjected to Western blotting analyses with an anti-MEF2C (lanes 1 to 5) or anti-MEF2D (lanes 6 to 9) polyclonal antibody. The presence of Flag-tagged HDAC4 and dm4 was confirmed by Western blotting analyses of the same samples with an anti-Flag monoclonal antibody (data not shown). (B) Schematic representation of MEF2C and its mutant M178 (consisting residues 1 to 178). (C and D) Interaction of M178 with HDAC4 and its deletion mutants in vitro. MBP or MBP-M178 was immobilized on amylose-agarose and tested for interaction with HDAC4 or its deletion mutants, synthesized in vitro in the presence of [35S]methionine. Input lanes represent 20% of HDAC4 or its mutants used for interaction. (E) Schematic representation of HDAC4 and its deletion mutants used in the interaction assays (A, C, and D). The + symbol denotes that the protein shown at left interacts with MEF2C.
These immunoprecipitation data also suggest that conserved regions of MEF2C and MEF2D mediate their interaction with HDAC4. Since the N-terminal regions of MEF2C and MEF2D contain the MADS-box and MEF2-specific domains and are the most conserved, we next examined whether the MEF2C mutant M178 could interact with HDAC4 (Fig. 6B). For this, M178 was expressed in E. coli as a fusion with MBP and used for in vitro pull-down assays. As shown in Fig. 6C, M178 specifically interacted with HDAC4 (lanes 1 to 3). To delineate regions of HDAC4 required for such interaction, we used a series of HDAC4 mutants (Fig. 6E). M178 interacted with dm1 (Fig. 6C, lanes 4 to 6) and less strongly with dm6 (lanes 7 to 9). By contrast, M178 did not interact with dm7 to -9 (lanes 10 to 18), suggesting that residues 118 to 188 of HDAC4 are essential for interaction with M178. Consistent with this contention, dm2 but not dm3 interacted with M178 (Fig. 6D, lanes 1 to 6). To further map the MEF2 interaction domain, dm4 and dm5 were tested. Unlike dm5, dm4 interacted with M178 (lanes 7 to 12), suggesting that residues 118 to 208 of HDAC4 are essential for interacting with M178. To determine whether these residues are sufficient, dm10 was used (Fig. 6E). This mutant was found to interact with M178 (Fig. 6D, lanes 13 to 15), confirming that residues 118 to 208 of HDAC4 are sufficient for interaction with MEF2C. Furthermore, in immunoprecipitation experiments, dm4 was found to interact with MEF2C (Fig. 6A, lane 5) or MEF2D (lane 9) in vivo. Taken together, these results indicate that residues 118 to 208 of HDAC4 contain a MEF2 interaction domain (Fig. 6E).
HDAC4 represses MEF2C-dependent transcription.
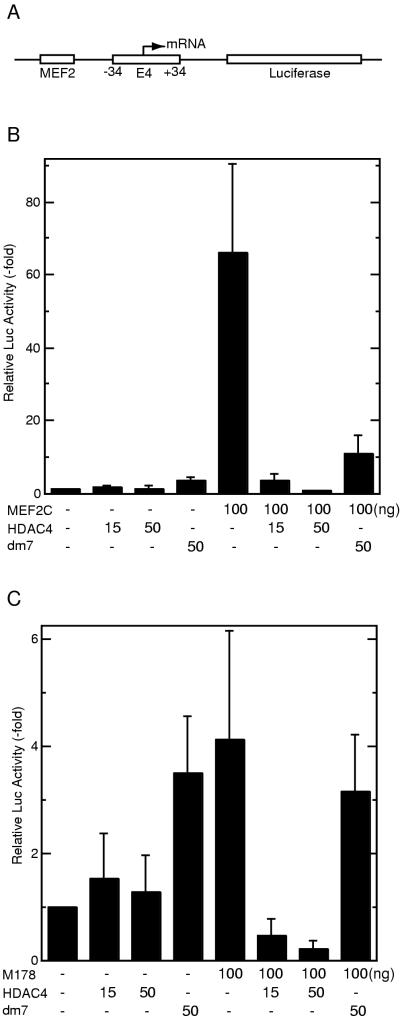
To explore the functional relevance of the observed physical interaction between HDAC4 and MEF2C, we constructed a luciferase reporter containing a MEF2-binding site (MEF2-E4-Luc [Fig. 7A]). This reporter was transfected into NIH 3T3 cells with or without expression plasmids for HDAC4 and/or MEF2C. As expected, MEF2C activated the reporter (Fig. 7B). While HDAC4 itself had minimal effects on the reporter activity in the absence of cotransfected MEF2C, HDAC4 repressed MEF2C-dependent transcription in a dose-dependent manner. The HDAC4 mutant dm7, which lacks a MEF2-binding site, had a much smaller effect. Since recruitment of HDAC4 by MEF2C repressed the reporter activity below the control level, HDAC4 may not be only inhibitory to the activation function of MEF2C. To substantiate this point, the MEF2C mutant M178 was tested. This mutant only weakly stimulated the reporter activity since it lacks the MEF2C activation domain located at its C-terminal part (Fig. 7C). In a dose-dependent manner, HDAC4 repressed the reporter activity below the control level. On the other hand, dm7 had minimal effects. Western blotting analyses revealed that HDAC4 and dm7 were expressed at similar levels (data not shown). Taken together, these results suggest that MEF2C recruits HDAC4 to repress transcription.
FIG. 7.
HDAC4 represses transcription in a MEF2C-dependent manner. (A) Schematic representation of the reporter MEF2-E4-Luc, which contains one copy of the MEF2-binding site upstream from the adenovirus E4 core promoter (−34 to +34) and the luciferase coding sequence. (B) HDAC4 represses MEF2C-dependent transcription. MEF2-E4-Luc was cotransfected into NIH 3T3 cells with the expression plasmids at indicated amounts. Luciferase (Luc) activities were normalized to the internal β-galactosidase control; the normalized luciferase activity from the transfection without any effector plasmid was arbitrarily set to 1.0. (C) Recruitment of HDAC4 by the MEF2C mutant M178 leads to repression. Reporter assays were performed as for panel B except that the expression plasmid for M178 was used instead.
HDAC4 cooperates with MEF2C to inhibit c-jun promoter activity.
Next we wished to examine a native promoter containing a MEF2-binding site. In nonmuscle cells, MEF2C regulates the expression of the proto-oncogene c-jun (15, 26, 63). Therefore, we tested the reporter pJLuc (Fig. 8A), which contains the c-jun promoter upstream from the luciferase gene (Fig. 8A; reference 16).
FIG. 8.
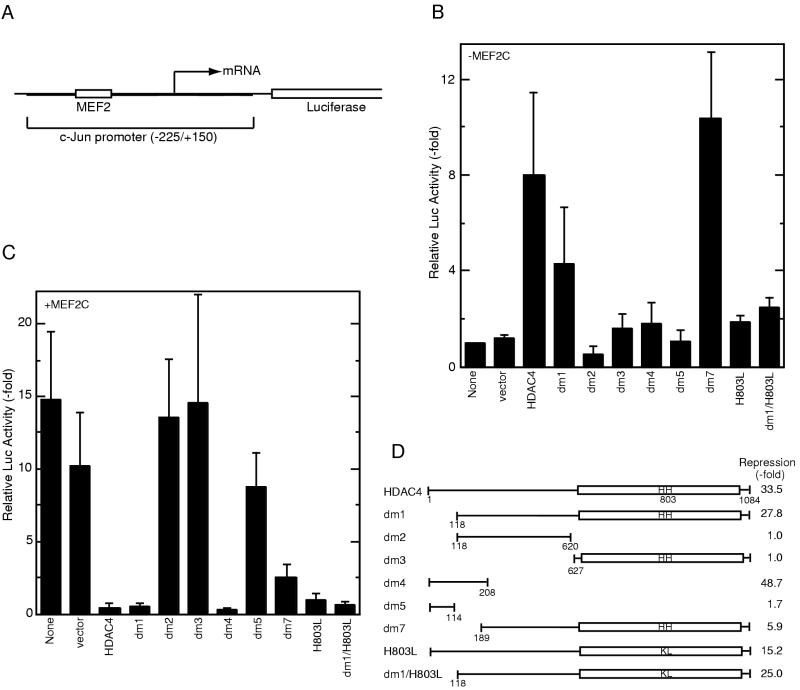
HDAC4 and MEF2C cooperatively regulate c-jun promoter activity. (A) Schematic representation of the reporter pJLuc, which contains positions −225 to +150 of the c-jun promoter upstream of the luciferase coding sequence. (B) HDAC4 activates c-jun promoter activity in a MEF2C-independent manner. The pJLuc reporter and expression plasmids for HDAC4 or its mutants were cotransfected into NIH 3T3 cells. Luciferase (Luc) activities were normalized to the internal β-galactosidase control; the normalized luciferase activity from the transfection without any effector plasmid was arbitrarily set to 1.0. (C) HDAC4 represses MEF2C-dependent transcription. Together with the MEF2C expression plasmid, pJLuc and indicated HDAC4 plasmids were cotransfected into NIH 3T3 cells. Reporter assays were determined and calculated as for panel B. (D) Schematic representation of HDAC4 and its mutants used for panels B and C. Repression of MEF2C-dependent transcription by each construct is shown at the right.
First, the expression plasmid for HDAC4 was cotransfected with this reporter to verify that HDAC4 does not regulate the promoter in the absence of cotransfected MEF2C. Unexpectedly, HDAC4 increased the reporter activity eightfold (Fig. 8B). To localize regions of HDAC4 involved in such activation, several deletion mutants were tested. While mutants dm2 to -5 had minimal effects, dm1 and dm7 activated the reporter 4- and 10-fold, respectively. Since dm7 lacks MEF2C-binding ability (Fig. 6E), HDAC4-mediated activation of pJLuc may be independent of MEF2C. Substitution of histidines 802 and 803 greatly diminished the activation ability of both HDAC4 and dm1 (Fig. 8B; compare the mutants H803L and dm1/H803L with HDAC4 and dm1, respectively), suggesting that the histone deacetylase activity of HDAC4 is important for activation of the c-jun promoter.
We then investigated the effects of MEF2C on the reporter pJLuc. As expected, transfection of MEF2C activated the expression of this reporter 15-fold (Fig. 8C). Cotransfection of HDAC4 repressed the activation mediated by MEF2C below the control level (Fig. 8C), raising an intriguing regulation scheme: transfected HDAC4 and MEF2C individually activate but together repress c-jun promoter activity. To determine which region of HDAC4 is required for this repression, we tested HDAC4 deletion mutants. Mutant dm1 repressed transcription 28-fold, whereas dm2 and dm3 had minimal effects (Fig. 8C and D), suggesting that both the deacetylase domain and residues 118 to 626 are required for dm1 to repress MEF2C-dependent transcription. dm7 repressed the reporter activity less efficiently than dm1 (Fig. 8C and D). Since dm7 lacks the MEF2C-binding domain (Fig. 6E), these results suggest that the MEF2C interaction domain is important for dm1 to repress transcription of the reporter pJLuc.
Mutant dm4 repressed transcription 49-fold, whereas dm5 had minimal effects (Fig. 8C and D). Western blotting analyses revealed that dm4 and dm5 were expressed at similar levels (data not shown). Therefore, HDAC4 represses MEF2C-dependent transcription through two repression domains. This may explain why substitution of histidines 802 and 803 had minimal effects on the ability of HDAC4 to repress MEF2C-dependent transcription (Fig. 8C). Surprisingly, the same mutation also had minimal effects on the ability of dm1 to repress MEF2C-dependent transcription, implying the existence of additional repression mechanisms. Taken together, these results suggest that through a MEF2C interaction domain and at least two repression domains, HDAC4 counteracts MEF2C-dependent activation of the c-jun promoter.
DISCUSSION
HDAC4 has intrinsic histone deacetylase activity.
Numerous studies have established that yeast RPD3 and human HDAC1 to -3 constitute one family of histone deacetylases (10, 50, 58, 59). The plant histone deacetylase HD2 may represent the first member of another family of deacetylases, one which does not display any sequence similarity to RPD3 or HDAC1 to -3 (31). Human HDAC4 to -7 and yeast HDA1 constitute a third family of histone deacetylases, one which displays some sequence similarity to RPD3 and HDAC1 to -3 (Fig. 1). While this paper was under revision, characterization of the histone deacetylase activity of human HDAC4-6 was reported (11, 13). Homologs of HDAC4-6 have been identified in the mouse (54) and other organisms (GenBank accession no. Q20296 and P56523).
HDAC4 possesses intrinsic histone deacetylase activity (Fig. 4; reference 35). HDAC4 mutants dm1 and dm3 were found to be slightly more active than full-length HDAC4 (Fig. 4B). One explanation for this difference is that these proteins had differential posttranslational modifications. Alternatively, the difference may suggest that the deacetylase activity of HDAC4 is subject to negative regulation by its N-terminal domain. If so, this raises the intriguing possibility that other proteins regulate the activity of HDAC4 by counteracting its autoinhibitory function.
HDAC4 possesses at least two transcriptional repression domains.
As implied by its deacetylase activity, HDAC4 repressed transcription when it was artificially tethered to promoters (Fig. 5). Intriguingly, we have found that HDAC4 possesses at least two repression domains, one composed of the N-terminal 208 residues and the other consisting of the HDA1-related deacetylase domain (Fig. 9). In contrast, HDAC1, -2, and -3 do not appear to possess repression domains other than their deacetylase domains (10, 18, 59). The possession of redundant repression domains by HDAC4 reflects similar themes described for the histone acetyltransferases p300 and CBP, both of which possess transcriptional activation domains in addition to their acetyltransferase domains (14, 27, 49, 57).
FIG. 9.
Functional domain organization of HDAC4. HDAC4 possesses at least two repression domains, with repression domain 1 located at the N terminus and repression domain 2 at the C-terminal part including the HDA1-related deacetylase domain. The MEF2C interaction domain resides within the N-terminal domain; the region C-terminal from the MEF2C interaction domain may be involved in activation of the c-jun promoter in the absence of transfected MEF2C.
Unlike its N-terminal repression domain, the deacetylase domain of HDAC4 mediates TSA-sensitive repression. The mutation at histidines 802 and 803 greatly diminished the deacetylase activity of HDAC4 (Fig. 4), but its effects on the transcriptional ability of HDAC4 were somewhat mixed: (i) it reduced the repression function of Gal4-dm1 (Fig. 5B); (ii) it abolished the ability of HDAC4 and dm1 to activate the c-jun promoter (Fig. 8B); and (iii) it had minimal effects on the ability of HDAC4 and dm1 to repress the activation function of MEF2C (Fig. 8C). There are several possible explanations for why the mutation had such varied effects. First, HDAC4 possesses at least one repression domain besides its deacetylase domain. Second, HDAC4 may homodimerize or heterodimerize with other histone deacetylases. This is consistent with the recent finding that HDAC4 interacts with HDAC3 (13). Third, transiently transfected reporters may not possess standard chromatin structure. Further studies of integrated reporters or endogenous c-jun promoter will certainly clarify this. From the present study, we conclude that the deacetylase activity of HDAC4 is important for repression, but additional mechanisms are also involved.
Recruitment of HDAC4 to promoters may lead to local deacetylation and thus transcriptional repression. Since histone acetyltransferases have been found to acetylate transcription factors, HDAC4 may also regulate acetylation levels of transcription factors. Therefore, the repression mediated by HDAC4 could be due either to deacetylation of hyperacetylated chromatin and subsequent formation of repressive chromatin structure or to deacetylation of acetylated transcription factors. Further investigation is needed to elucidate how HDAC4 is involved in transcriptional repression.
HDAC4 physically and functionally interacts with MEF2C.
How is HDAC4 recruited to promoters in vivo? HDAC4 does not have intrinsic DNA-binding ability and therefore must be recruited by interaction with target transcription factors. Compared to HDA1, HDAC4 has a long N-terminal domain (Fig. 1A). By immunoprecipitation experiments and in vitro binding assays, we have demonstrated that HDAC4 interacts with MEF2C and MEF2D and mapped the MEF2 interaction domain to residues 118 to 208 of HDAC4 (Fig. 6). This is consistent with a model in which the N-terminal domain of HDAC4 mediates its interaction with target transcription factors such as MEF2C and MEF2D.
Using the luciferase reporter MEF2-E4-Luc, we have shown that HDAC4 is recruited by MEF2C to repress transcription (Fig. 7). Independently, it has been demonstrated that HDAC4 associates with MEF2A and represses MEF2A-dependent transcription (35). Furthermore, MITR interacts with MEF2 and negatively regulates MEF2-dependent transcription (47).
MEF2s are known transcriptional activators, so it is somewhat unexpected that MEF2s recruit HDAC4 or MITR to repress transcription. However, it has been suggested that MEF2s negatively regulate transcription by associating with a negatively acting accessory factor (40). These findings suggest that HDAC4 or MITR may be such an accessory factor. Interestingly, more and more transcription factors are being found to have dual function. For example, the transcriptional activator E2F binds to the tumor suppressor Rb and recruits HDAC1 to repress transcription (5, 30, 32). Therefore, it is tempting to propose that MEF2s play a dual role in transcriptional regulation.
HDAC4 and MEF2C cooperatively regulate c-jun promoter activity.
The proto-oncogene product c-Jun is one of the immediate-early genes products whose expression is rapidly induced by treatment of cells with serum and many growth factors (reference 7 and reference therein). c-Jun regulates cell cycle progression in a p53-dependent manner (46). When cotransfected with MEF2C, HDAC4 repressed c-jun promoter activity (Fig. 8C). Like HDAC4, both dm1 and dm4 repressed c-jun promoter activity in the presence of transfected MEF2C (Fig. 8C). These results are consistent with a model that in the presence of transfected MEF2C, HDAC4 represses c-jun promoter activity via at least two repression domains (Fig. 9).
Unexpectedly, in the absence of cotransfected MEF2C, HDAC4 activated the c-jun promoter in NIH 3T3 cells (Fig. 8B). The MEF2 interaction domain appears to be dispensable for this activation, suggesting that activation of the c-jun promoter by HDAC4 operates through MEF2C-independent mechanisms. It is possible that HDAC4 activates the c-jun promoter by regulating the function and/or protein level of a required transcription factor(s). We favor the model in which HDAC4 downmodulates the expression of a repressor whose function is required for repression of the c-jun promoter and thus leads to activation. In NIH 3T3 cells, dependent on whether MEF2C is cotransfected, HDAC4 exerts opposing actions on the c-jun promoter. In other types of cells, relative expression levels of HDAC4, MEF2C and the elusive repressor may dictate which action takes place. It is also possible that the actions of HDAC4 are subject to regulation by various signaling pathways. Therefore, we propose that HDAC4 regulates the c-jun promoter in a context-dependent manner.
In summary, we have demonstrated that HDAC4, a human histone deacetylase related to HDA1, is composed of multiple functional domains: its N-terminal part possesses repression domain 1 and a MEF2C interaction region, whereas its C-terminal part constitutes repression domain 2 and functions as the catalytic domain conducting deacetylation (Fig. 9). In NIH 3T3 cells, dependent on the expression level of MEF2C, HDAC4 exerts opposing actions on the c-jun promoter, suggesting that HDAC4 and probably its homologs HDAC5 and HDAC7 cooperate with the MEF2 family of transcription factors to regulate their target genes such as c-jun in a context-dependent manner. It will be interesting to determine if and how the interaction of HDAC4 with MEF2s is regulated to fulfill their roles in various types of cells.
ACKNOWLEDGMENTS
We thank T. Nagase for the cDNA clone KIAA0288; R. Prywes for the pJLuc reporter and anti-MEF2D antibody; E. N. Olson for MEF2 expression plasmids; W. M. Yang and E. Seto for Gal4 expression plasmids; P. Haus-Seuffert and M. Meisterernst for Gal4-CD4-Luc; Y. Zhang and D. Reinberg for Gal4-AdML-Luc; D. Sparrow, T. J. Mohun, and T. Kouzarides for communicating results prior to publication; and A. Nepveu for helpful discussions.
This work was supported by funds from the National Cancer Institute of Canada (to X.-J.Y.) and grants from the Medical Research Council (MRC) of Canada (to X.-J.Y. and J.T.). X.-J. Y. is an MRC scholar.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausio J, van Holde K E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomew C, Kilbey A, Clark A M, Walker M. The Evi-1 proto-oncogene encodes a transcriptional repressor activity associated with transformation. Oncogene. 1997;14:569–577. doi: 10.1038/sj.onc.1200864. [DOI] [PubMed] [Google Scholar]
- 4.Black B L, Olson E N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 5.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 6.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 7.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-Jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad B, Dewald G, Christensen E, Lopez M, Higgins J, Pierpont M E. Clinical phenotype associated with terminal 2q37 deletion. Clin Genet. 1995;48:134–139. doi: 10.1111/j.1399-0004.1995.tb04073.x. [DOI] [PubMed] [Google Scholar]
- 9.Coso O A, Montaner S, Frosman C, Lacal J C, Prywes R, Teramoto H, Gutkind J S. Signaling from G protein-coupled receptors to the c-jun promoter involves the MEF2 transcription factor. J Biol Chem. 1997;272:20691–20697. doi: 10.1074/jbc.272.33.20691. [DOI] [PubMed] [Google Scholar]
- 10.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischle W, Emilian S, Hendzel M J, Nagase T, Nomura N, Voelter W, Verdin E. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 12.Fisher A L, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 13.Grozinger C M, Hassig C A, Schreiber S L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 16.Han T-H, Prywes R. Regulatory role of MEF2D in serum induction of the c-Jun promoter. Mol Cell Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada K, Dufort D, Denis-Larose C, Nepveu A. Conserved Cut repeats in the human Cut homeodomain protein function as DNA binding domains. J Biol Chem. 1994;269:2062–2067. [PubMed] [Google Scholar]
- 18.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata A, Lo R S, Wotton D, Lagna G, Massague J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 20.Hendzel M J, Delcuve G P, Davie J R. Histone deacetylase is a component of the internal nuclear matrix. J Biol Chem. 1991;266:21936–21942. [PubMed] [Google Scholar]
- 21.Heng H H, Squire J, Tsui L-C. High resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc Natl Acad Sci USA. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henzel M J, Nishioka W K, Raymond Y, Bazett-Jones D, Th’ng J P H. Chromatin condensation is not associated with apotosis. J Biol Chem. 1998;273:24470–24478. doi: 10.1074/jbc.273.38.24470. [DOI] [PubMed] [Google Scholar]
- 23.Inoue A, Fujimoto D. Histone deacetylase from calf thymus. Biochim Biophy Acta. 1970;220:307–316. doi: 10.1016/0005-2744(70)90015-x. [DOI] [PubMed] [Google Scholar]
- 24.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J-D. BMK1/ERK5 regulate serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo M-H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 28.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of Zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luger K, Mader A W, Richmond R K, Sarget D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 30.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 31.Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 32.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 33.Magnani I, Larizza L, Doneda L, Weitnauer L, Rizzi R, Di Lernia R. Malformation syndrome with t(2;22) in a cancer family with chromosome instability. Cancer Genet Cytogenet. 1989;38:223–227. doi: 10.1016/0165-4608(89)90663-8. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miska, E., C. Karlsson, E. Langley, S. Nielsen, J. Pines, and T. Kouzarides. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 36.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- 38.Ohara O, Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Nomura N. Construction and characterization of human brain cDNA libraries suitable for analysis of cDNA clones encoding relatively large proteins. DNA Res. 1997;4:53–59. doi: 10.1093/dnares/4.1.53. [DOI] [PubMed] [Google Scholar]
- 39.O’Neill L P, Turner B M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ornatsky O I, McDermott J C. MEF2 protein expression, DNA binding specificity and complex formation, and transcriptional activity in muscle and non-muscle cells. J Biol Chem. 1996;271:24927–24933. doi: 10.1074/jbc.271.40.24927. [DOI] [PubMed] [Google Scholar]
- 41.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 42.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 43.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satyaraj E, Storb U. MEF2 proteins, required for muscle differentiation, bind an essential site in the lg lambda enhancer. J Immunol. 1998;161:4795–4802. [PubMed] [Google Scholar]
- 45.Scanlan M J, Chen Y T, Williamson B, Gure A O, Stockert E, Gordan J D, Tureci O, Sahin U, Pfreundschuh M, Old L J. Characterization of human colon cancer antigens recognized by autologous antibodies. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 46.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner E F. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sparrow, D. B., E. A. Miska, E. Langley, S. Reynaud-Deonauth, S. Kotecha, N. Towers, G. Spohr, T. Kouzarides, and T. J. Mohun. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 48.Stifani S, Blaumueller C M, Redhead N J, Hill R E, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of Split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 49.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 50.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 51.Th’ng J P H, Guo X-W, Swank R A, Crissman H A, Bradbury E M. Inhibition of mitotic histone phosphorylation by staurosporine leads to chromosome decondensation. J Biol Chem. 1994;269:9568–9573. [PubMed] [Google Scholar]
- 52.Uejima H, Shinohara T, Nakayama Y, Kugoh H, Oshimura M. Mapping a novel cellular-senescence gene to human chromosome 2q37 by irradiation microcell-mediated chromosome transfer. Mol Carcinog. 1998;22:34–45. doi: 10.1002/(sici)1098-2744(199805)22:1<34::aid-mc5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 53.van Holde K E. Chromatin. Berlin, Germany: Springer-Verlag; 1989. [Google Scholar]
- 54.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 55.Wolffe A. Chromatin: structure and function. 2nd ed. New York, N.Y: Academic Press, Harcourt Brace & Company; 1995. [Google Scholar]
- 56.Wolffe A P. Transcriptional control: sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 57.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 58.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 60.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 61.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–64. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 63.Zhao M, New L, Kravchenko V V, Kato Y, Gram H, Padova F D, Olson E N, Ulevitch R J, Han J. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1998;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]