Abstract
Objective
We develop natural language processing (NLP) methods capable of accurately classifying tumor attributes from pathology reports given minimal labeled examples. Our hierarchical cancer to cancer transfer (HCTC) and zero-shot string similarity (ZSS) methods are designed to exploit shared information between cancers and auxiliary class features, respectively, to boost performance using enriched annotations which give both location-based information and document level labels for each pathology report.
Materials and Methods
Our data consists of 250 pathology reports each for kidney, colon, and lung cancer from 2002 to 2019 from a single institution (UCSF). For each report, we classified 5 attributes: procedure, tumor location, histology, grade, and presence of lymphovascular invasion. We develop novel NLP techniques involving transfer learning and string similarity trained on enriched annotations. We compare HCTC and ZSS methods to the state-of-the-art including conventional machine learning methods as well as deep learning methods.
Results
For our HCTC method, we see an improvement of up to 0.1 micro-F1 score and 0.04 macro-F1 averaged across cancer and applicable attributes. For our ZSS method, we see an improvement of up to 0.26 micro-F1 and 0.23 macro-F1 averaged across cancer and applicable attributes. These comparisons are made after adjusting training data sizes to correct for the 20% increase in annotation time for enriched annotations compared to ordinary annotations.
Conclusions
Methods based on transfer learning across cancers and augmenting information methods with string similarity priors can significantly reduce the amount of labeled data needed for accurate information extraction from pathology reports.
Keywords: natural language processing, cancer, pathology
INTRODUCTION
Personalized healthcare depends on detailed and accurate patient data. The massive amounts of unstructured medical text in electronic health records are a primary source of this data, and the ability to reliably extract clinical information is a crucial enabling technology. As a result, there has been much interest in natural language processing (NLP) and information extraction methods to tackle healthcare text data1–4 which have been used in health informatics, precision medicine, and clinical research.5,6
Implementing such extraction systems in practice remains challenging, as many systems rely on large amounts of annotated textual data to perform well. However, annotating healthcare text is a largely manual and time-consuming process that requires training and medical knowledge. Combined with privacy considerations that limit sharing of corpora, it can be difficult to obtain sufficient amounts of annotated data across many clinical domains. While deep learning has been shown to be extremely powerful in NLP, it can underperform in biomedical applications due to smaller training sets. Therefore, it is of considerable practical importance to develop methods in biomedical NLP that perform well with small amounts of labeled data.
Despite a long history of approaches to biomedical information extraction which include rules-based methods,7–9 classical machine learning methods,10–12 and deep learning methods,13–15 few works have focused on sample efficient learning. Yala et al10 carried out a performance analysis of boosting tree extraction models and found that approximately 400 training examples were required to obtain an accuracy of 0.9 for 20 breast cancer attributes, though they only considered tumor attributes that take on present or absent values. We previously showed non-deep learning methods largely outperformed deep learning methods with data sizes below 256 for prostate cancer reports.16 We also previously developed a novel supervised line attention (SLA) approach using more fine-grained, location-based annotations and showed the fully supervised location-based approach outperformed the state-of-the-art methods using training data sizes below 186 for colon and kidney cancer reports.17 However, these methods still require hundreds of labeled examples.
Transfer learning has been shown as a promising approach to improve information extraction in medical text. Qiu et al13 found that jointly training a convolutional neural network (CNN) on both lung and breast cancer pathology reports for predicting the tumor site was more effective than training on each cancer individually. Using a corpus size of 942, their transfer approach led to improvements of up to 0.04 in micro-F1 and 0.09 in macro-F1 over single cancer training. Alawad et al14 show improvements of multi-task CNNs trained across cancer registers over single registry models. They achieve up to 0.17 improvement in the macro-F1 score for the primary tumor site and topography with a corpus size of 71 223.
Zero-shot learning is also a promising avenue for achieving better sample efficiency in limited data settings.18–20 It is a setting where the model learns to classify test instances with labels not previously seen in the training set. Typically, a zero-shot learning approach learns to make a prediction by using the original features of an instance and auxiliary information of classes, which are related to the feature space. For example, for a document classification task, the features could include the document text, while the class name and description could be used as the class auxiliary information. The learned relational information between auxiliary information and features allows the model to generalize to new classes when auxiliary information is available.
OBJECTIVE
In this work, we extend the existing SLA approach based on enriched annotations using transfer learning and zero-shot learning. For tumor attributes with labels that are shared across colon, kidney, and lung cancers, we develop a cancer-to-cancer transfer learning procedure to leverage cancers with many labeled examples for cancers with few labeled examples. Transfer learning is applicable here, since much of the language is shared when reporting an individual attribute that is shared across different cancers. For tumor attributes with labels that are unique to a specific cancer type, we develop zero-shot string similarity (ZSS) methods to augment our SLA approach. ZSS finds the predicted label by calculating string similarity scores between the label and text. Note that character-based similarities can be calculated for unseen labels as long as the label name is available at prediction time. Since ZSS only requires a string similarity score to make a prediction, ZSS can generalize to labels never seen during training.
MATERIALS AND METHODS
Data sources
We randomly sampled 250 pathology reports each across colon, lung, and kidney cancers from the University of California, San Francisco from 2002 to 2019. For each cancer type, we sampled and annotated 250 reports to create 10 random cancer-specific, train-validation-test splits. Each split for a cancer consists of the same 250 annotated reports overall, but the individual training, validation, and test sets differ due to randomness. The full train, validation, and test sets consist of 40, 20, and 190 annotated reports, respectively. We chose to place a majority of the data in the test set and limit the number of reports in the training and validation splits, since we are interested in performance in the low data regime (10–40 examples). Each experiment is run separately on each of the 10 splits. We obtain confidence intervals for the evaluation metric scores computed on the test set of each of the 10 splits. Further details on the reports and text preprocessing are included in the Supplementary Material.
Tumor attributes
Tumor attributes of interest are histologic grade and the presence of lymphovascular invasion for transfer learning on shared labels across cancers and tumor site, histologic type, and the surgical procedure carried out on a patient for ZSS on cancer-specific labels, details in Supplementary Tables S1 and S2.
Enriched annotations
We annotated the pathology reports with document-level labels and highlighted text throughout the report relevant to the label, as in prior work.17 These fine-grained annotations provide specific lines in the report that determine the value of a tumor attribute and were generated using the Multi-document Annotation Environment software.21 Similar to prior work, we use a “reduced annotation set,” which consists of the minimal set of annotations containing the line of a given tumor attribute’s value in the synoptic comment. If the report does not contain a synoptic comment, the first line that contains the relevant information is used.17 These synoptic comments are typically common in more recent pathology reports and are a brief standardized portion of the text where relevant cancer attributes are reported. Location-specific annotations take 20% longer to generate on average than typical annotations.17
SUPERVISED LINE ATTENTION
Our models are based on the SLA framework, previously reported by our group.17 The goal is to predict the lines in the report that contain information on a specific tumor attribute and then use the predicted lines to make the final class prediction for an attribute. There are 2 separate classifiers for the line prediction task and the class prediction task trained using location-based and label annotations. As in previous work, separate XGBoost models are used for the line prediction task and the class prediction task.
Tumor attributes are divided into 2 distinct categories. The first category contains tumor attributes with shared labels across cancers, such as the histologic grade. Most cancers are graded on a numeric or ordinal scale, and while the underlying biology and clinical significance of the grades differ, the labels are similar. The second category contains tumor attributes whose labels are not shared across cancers. An example is the procedure; for each organ system or cancer type, there are a different set of surgical procedures for resecting tumors. The first group is a natural candidate for a transfer learning approach, whereas transfer learning is less applicable for the second group, since the labels are not shared across cancer types. We propose 2 methods to perform extraction depending on whether the labels are shared across cancers.
Shared labels
When labels are the same across cancers, knowledge can be transferred from one cancer type to another. For example, for the presence of lymphovascular invasion, identifying the relevant lines in a report is domain-independent because lymphovascular invasion is a relevant attribute for many cancers. Furthermore, identifying the correct label is again domain-independent because the categories (present and not identified) are the same across cancers. Shared knowledge is important because reports from other domains can be used to improve performance through data augmentation.
We create a transfer learning technique to learn data extraction using the shared information across cancer types for the relevant tumor attributes. We build off SLA by training both the line classifier and the final classifier on reports from all domains, which we refer to as hierarchical cancer to cancer transfer learning (HCTC). We apply cancer-to-cancer transfer learning hierarchically at both stages of SLA: predicting the relevant lines in the report and predicting the final classification of a report. As a sensitivity analysis, we report results with ablations to HCTC where we only share information for the line classifier (HCTC-line) or the final classifier (HCTC-final).
Unique labels
Unlike the shared labels case, we opt against a transfer learning approach as the applicability is uncertain due to a different label space for each cancer. Furthermore, some attributes have a large number of labels (there are 32 possible labels for kidney histologic type). It is highly likely that we will encounter labels at test time that were not present in the training data. Typical machine learning models need a sufficient number of examples for each possible label to learn classification tasks and generalize to new data. As seen by the large set of possible labels for our attributes (Supplementary Table S2), it is possible to see few or no examples of a class during training. Consequently, a technique that can handle a large set of labels is essential here and in particular, a method capable of zero-shot learning is necessary.
We develop a novel method that enables a more sample efficient method capable of zero-shot learning, referred to as ZSS. At a high level, ZSS first predicts the relevant lines for the label using the line classifier as in Altieri et al,17 then calculates the string similarity score between each possible label and the concatenated text of the top 3 lines output from the line classifier using a subroutine we call the fuzzy jaccard score (Algorithm 1). Finally, we take the label with the highest fuzzy jaccard score as the final prediction. The possible labels we use are defined in the College of American Pathology reporting guidelines.22
Algorithm 1.
Fuzzy jaccard subroutine for the Zero-shot similarity algorithm
Input: Predicted line in a report and a candidate label for a tumor attribute
Output: Similarity score between the predicted line and candidate label
For each unique word in the candidate label, calculate pairwise string similarity scores with each unique word in the predicted line using the Ratcliff-Obershelp contiguous matching subsequence algorithm.
From the pairwise string similarity scores from step 1, find the maximum score for each unique word in the label.
Sum up the max similarity scores from step 2 across unique words in the candidate label
Scale the resulting sum from step 3 by the number of unique words in the label. This score represents the similarity score between the candidate label and the predicted line
ZSS involves calculating pairwise character-based similarity scores between a predicted line of a report and each possible label using the fuzzy jaccard score as a subroutine (Algorithm 1). We use the line in the report with the highest probability computed using the line classifier as the predicted line. The similarity between the predicted line and a candidate label is computed with the Ratcliff-Obershelp algorithm.23 We evaluated several character-level string similarity algorithms, such as the Jaro–Winkler similarity, Levenshtein similarity, and Hamming distance but found that the Ratcliff–Obershelp approach performed best according to the mean F1-micro score using the training set averaged across the all splits for lung, colon, and kidney cancers for each data size. The label with the highest Ratcliff–Obershelp score is used as the final prediction. The full routine is described in Algorithm 2.
Algorithm 2.
Zero-shot similarity
Input: Predicted line in a report, the probability for whether a given line is relevant for a tumor attribute taken from the line classifier, a set of labels for a given attribute (the possible values that a tumor attribute can take), and a learned cutoff parameter used to predict “NA” or not reported
Output: final predicted label of a report for the tumor attribute in question
For each candidate label, calculate its fuzzy jaccard score (Algorithm 1) with the predicted line
Take the label with the highest fuzzy jaccard score. If there is a tie between multiple labels, take the label with the most characters as the prediction
If the fuzzy jaccard score is less than 0.5, then replace predicted label with “other”
If the line probability is less than the cutoff, then replace predicted label with “NA”
Ensembling string similarity with the SLA approach
While we found ZSS to be effective on its own, we identified a few weaknesses of the approach. In particular, we found that the “other” class was particularly challenging, as it consists of all possible values the attribute can take outside of the defined label set in the CAP protocols. For example, if the attribute is “procedure,” then the “other” class corresponds to all other possible procedures not listed in the CAP protocols, which will all have low string similarity to the label “other.” Additionally, ZSS may underperform when synonyms of the class name are used in the actual text which can have low string similarity with the actual class name (e.g. central vs middle pole for kidney tumor site). Therefore, we aim to get the best of both ZSS and the SLA approach by developing a hybrid approach to the problem called ZSS-thresholding. If the final string similarity score is above a learned threshold, then we output the ZSS prediction; otherwise, we output the SLA prediction.
We include an oracle method that chooses the SLA prediction if it is equal to the ground truth and the ZSS prediction otherwise. This oracle method serves as an upper bound on the performance of ensembling ZSS and SLA. Our final method is ZSS-doc which is using ZSS on the entire text of the report instead of the lines output from the line classifier. This allows us to gauge how necessary the location targeting approach is for the ZSS methods.
BASELINE METHODS
Shared labels
Our first set of baselines includes document-level classification methods, such as logistic regression, XGBoost, random forest, and support vector machines. These methods take as input all the tokens in a given report and predict the class value of a particular tumor attribute. A bag-of-ngrams approach is used to vectorize the text in each document. This approach only uses the final document-level labels and is trained on the cancer of interest as well as the out-domain cancers.
Our next baseline is the hierarchical attention network (HAN) for document classification.24 In particular, we study HAN in terms of transfer learning. We pretrain the model on out-domain reports for a shared attribute and then fine-tune the model on in-domain reports.
We also use the SLA approach in Altieri et al17 as another baseline. XGBoost models are used for line prediction and final label prediction. Location-based annotations are used to train the line prediction model, while document-level label annotations are used to train the final classifier.
Unique labels
The baselines in the unique labels scenario include the ordinary document-level classifiers and SLA like the previous case. All the methods are trained on a single cancer domain.
RESULTS
We run 2 sets of experiments across lung, colon, and kidney cancers: one for the shared labels case and another for the unique labels case. Each method reliant on the SLA method is trained on training data of 8, 17, and 33 in-domain reports, while all other methods are trained on 10, 20, and 40 in-domain reports. The validation set size for each trial is half of the corresponding training set size. The difference in the training set sizes and validation set sizes between SLA methods and all others is to account for the difference in annotation times between ordinary annotations and location-based annotations. This difference is based on our prior work, noting that location-based annotations took 20% longer than ordinary annotations when highlighting the first line containing the relevant information.17 Additionally, for the shared labels case, 372 out-domain reports are additionally used to augment the training set. The test set for all experiments consists of 186 held out reports from the domain in question.
Each experiment is run 10 times where the training, validation, and test splits are randomly formed. We compare across methods using the mean micro-F1 and macro-F1 scores and obtain uncertainty bounds around the means. Details on hyperparameter tuning can be found in the Supplementary Notes.
Shared labels
Our experiments show that HCTC and HCTC-final consistently outperforms all other methods (Table 1). Compared to boosting, which performs the best among baselines on macro-F1, HCTC achieves performance gains by 0.03–0.04 in macro-F1 across data sizes. Compared to SLA, which performs the best among baselines on micro-F1, HCTC requires half the data to perform better in both macro-F1 and micro-F1. Additionally, we find that for data sizes 17 and 33, HCTC-final outperforms HCTC, suggesting the main benefit of transfer learning comes from the final classifier and not the line classifier.
Table 1.
Average micro-f1 and macro-f1 performance as a function of 10, 20, and 40 labeled examples on colon, kidney, and lung cancer pathology reports
| Macro-F1 | Micro-f1 | |||||
|---|---|---|---|---|---|---|
| In-domain training sizes | 10 | 20 | 40 | 10 | 20 | 40 |
| Hierarchical attention network | 0.298 | 0.287 | 0.355 | 0.580 | 0.574 | 0.718 |
| Logistic | 0.344 (0.055) | 0.441 (0.059) | 0.467 (0.073) | 0.634 (0.039) | 0.676 (0.037) | 0.708 (0.047) |
| Random forest | 0.276 (0.025) | 0.307 (0.034) | 0.340 (0.044) | 0.586 (0.039) | 0.614 (0.030) | 0.641 (0.036) |
| SVM | 0.221 (0.048) | 0.269 (0.034) | 0.310 (0.034) | 0.519 (0.102) | 0.560 (0.051) | 0.570 (0.052) |
| Boost | 0.436 (0.036) | 0.468 (0.044) | 0.548 (0.052) | 0.704 (0.049) | 0.732 (0.037) | 0.789 (0.038) |
| SLA* | 0.211 (0.024) | 0.338 (0.037) | 0.466 (0.043) | 0.579 (0.031) | 0.700 (0.029) | 0.790 (0.026) |
| HCTC* | 0.461 (0.038) | 0.508 (0.034) | 0.544 (0.028) | 0.797 (0.023) | 0.832 (0.022) | 0.858 (0.018) |
| HCTC-final* | 0.421 (0.034) | 0.502 (0.047) | 0.584 (0.048) | 0.776 (0.027) | 0.842 (0.030) | 0.882 (0.024) |
| HCTC-line* | 0.205 (0.013) | 0.341 (0.035) | 0.473 (0.040) | 0.579 (0.044) | 0.700 (0.041) | 0.800 (0.025) |
Note: The results presented include the mean performance and standard deviation across 10 random splits of the data for the shared labels case.
Methods marked with are trained on 8, 17, and 33 reports to adjust for annotation time. Note due to computational reasons we only run HAN once for all experiments.
Unique labels
ZSS-thresholding also requires approximately half the data to perform similarly or better than the baseline methods for the unique labels setting (Table 2). ZSS-thresholding with 8 points achieves an increase of 0.14 in micro-F1 and 0.16 in macro-F1 over boosting trained on 20 data points. Furthermore, ZSS-thresholding trained on 17 data points achieves an increase of 0.05 in micro-F1 and an increase of 0.06 in macro-F1 compared to boosting trained on 40 data points. A similar trend holds when computing differences in extraction quality between ZSS-thresholding and SLA which suggests that the string similarity approach enhances models trained on small data.
Table 2.
Average micro-f1 and macro-f1 performance across a function of 10, 20, and 40 labeled examples on colon, kidney, and lung cancer pathology reports
| Macro-f1 | Micro-f1 | |||||
|---|---|---|---|---|---|---|
| In-domain training sizes | 10 | 20 | 40 | 10 | 20 | 40 |
| Hierarchical attention network | 0.051 | 0.026 | 0.079 | 0.255 | 0.118 | 0.264 |
| Logistic | 0.208 (0.029) | 0.277 (0.047) | 0.354 (0.048) | 0.473 (0.033) | 0.578 (0.053) | 0.651 (0.033) |
| Random forest | 0.177 (0.045) | 0.223 (0.024) | 0.323 (0.043) | 0.438 (0.044) | 0.516 (0.042) | 0.618 (0.028) |
| SVM | 0.152 (0.029) | 0.172 (0.031) | 0.239 (0.030) | 0.387 (0.066) | 0.425 (0.036) | 0.517 (0.044) |
| Boost | 0.155 (0.021) | 0.288 (0.040) | 0.382 (0.034) | 0.421 (0.029) | 0.608 (0.051) | 0.715 (0.028) |
| SLA* | 0.095 (0.015) | 0.178 (0.012) | 0.219 (0.016) | 0.472 (0.036) | 0.651 (0.023) | 0.736 (0.016) |
| ZSS* | 0.442 (0.024) | 0.436 (0.017) | 0.428 (0.028) | 0.743 (0.016) | 0.737 (0.011) | 0.742 (0.009) |
| ZSS-doc* | 0.359 (0.023) | 0.356 (0.022) | 0.341 (0.019) | 0.546 (0.024) | 0.540 (0.021) | 0.528 (0.007) |
| ZSS-thresholding* | 0.441 (0.024) | 0.447 (0.022) | 0.449 (0.029) | 0.739 (0.017) | 0.765 (0.018) | 0.780 (0.015) |
| Oracle* | 0.454 (0.031) | 0.501 (0.029) | 0.529 (0.024) | 0.775 (0.019) | 0.829 (0.017) | 0.862 (0.011) |
Note: The results presented include the mean performance and standard deviation across 10 random splits of the data for the unique labels case.
Methods marked with are trained on 8, 17, and 33 reports to adjust for annotation time. Note due to computational reasons we only run HAN once for all experiments.
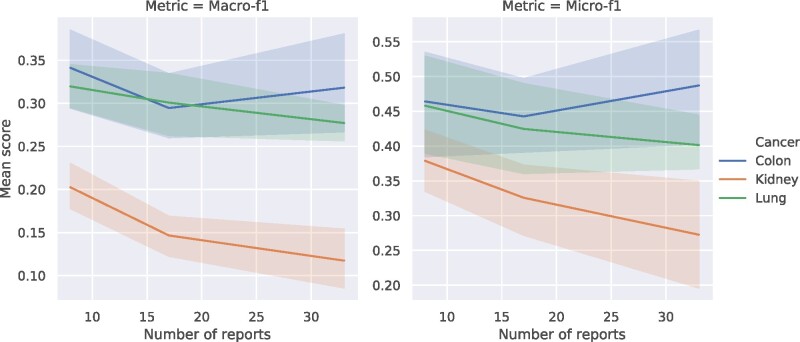
For ZSS and ZSS-thresholding, we additionally include micro-F1 and macro-F1 scores computed on test instances which have labels never seen during training across colon, kidney, and lung cancers (Figures 1 and 2). For colon cancer, the zero-shot performances are near or above 0.3 macro-F1 and 0.4 micro-F1 for both methods. For lung cancer, the performances are consistently near or above 0.25 macro-F1 and 0.4 micro-F1. For kidney cancer, the performance we see benefits of up to 0.1 macro-F1 and 0.25 micro-F1. These metrics show ZSS is a viable zero-shot approach for this application and is able to learn to predict classes never observed in the training set.
Figure 1.
Average macro-f1 (A) and micro-f1 (B) performance for test instances where the label is not seen during training as a function of 10, 20, and 40 labeled examples on colon, kidney, and lung cancer pathology reports. The results presented include the mean performance using ZSS across 10 random splits of the data and 95% confidence intervals for the unique labels case. Note that the number of zero-shot test instances decreases as the number of training instances increase.
Figure 2.
Average macro-f1 (A) and micro-f1 (B) performance for test instances where the label is not seen during training as a function of 10, 20, and 40 labeled examples on colon, kidney, and lung cancer pathology reports. The results presented include the mean performance using ZSS-thresholding across 10 random splits of the data and 95% confidence intervals for the unique labels case. Note that the number of zero-shot test instances decreases as the number of training instances increase.
DISCUSSION
We have developed 2 ways to improve the performance of learning-based extraction systems when the amount of annotated reports is limited. For attributes where the tumor attribute and labels are shared across domains, it is natural to aggregate annotations across domains to augment the data used to train the models. Our experiments with enhancing the SLA method show that the gain in performance is consistent across data sizes; there is a 0.09 increase in micro-F1 and 0.02 increase in macro-F1 for data size 8 and 0.09 increase in micro-F1 and 0.04 increase in macro-F1 for data size 33 over the state-of-the-art averaged across the 3 cancers. We note that, to the authors’ best knowledge, this is the first work to investigate transfer learning techniques across more than 2 cancers in NLP.
In the case of attributes where the labels differ across domains, we opt for a string similarity enhancement instead of a transfer approach. Because the categories for these attributes are unique for each domain, there is less room for improvement via transfer learning due to cancer-unique labels. String similarity is a more viable approach because typically in this case the text will contain strings close to the label names. Our experiments show that interpolating learning-based solutions with string similarity prediction can lead to a significant increase in performance—up to 0.26 micro-F1 and 0.23 macro-F1 for data size 8 and 0.04 micro-F1 and 0.06 macro-F1 for data size 33 over the state-of-the-art averaged across the 3 cancers. In terms of zero-shot performance of ZSS, the results vary across cancers and tops at 0.55 micro-F1 and 0.34 macro-F1 for a specific cancer.
Zero-shot learning has previously been studied in the context of medical information extraction, specifically on the public MIMIC II and MIMIC III datasets.25,26 Rios and Kavululu18 used natural language descriptors of labels and label space structure as auxiliary information to achieve zero-shot learning. Their approach matches textual summaries of reports obtained from attention-based CNNs to feature vectors of labels obtained from graphical neural networks and achieves recall-at-10 scores of up to 0.362 on MIMIC II and 0.495 on MIMIC III for zero-shot labels. Lu et al19 similarly use pre-defined label relations, label descriptions, and pre-trained word embeddings as auxiliary information. Information from multiple graphs built on label descriptions, label taxonomy, and label co-occurrences obtained from graph convolutional networks are matched with document embeddings obtained from CNN encoders. They achieve recall-at-10 scores of up to 0.462 on MIMIC II and 0.553 on MIMIC III. While deep learning has been shown to be extremely effective in the presence of a large amount of labeled data, it can often struggle on smaller datasets. We note that MIMIC II and III contain 18 822 and 37 016 patients, respectively, orders of magnitude larger than our dataset size of 40.
Our findings motivate future directions for information extraction with small data regimes. While in preliminary experiments, we found that using pre-trained word embeddings to measure similarity performed worse than our string-based method, we believe one promising direction is taking advantage of models pre-trained using large corpuses of text on language modeling tasks. Recent work in NLP has shown fine-tuning such models on specific tasks with small amounts of data lead to improvements in performance for a given task. Combining such models, such as BERT27 with the SLA framework can potentially improve upon ZSS-based methods especially for cases when synonyms of class names are used in the report in lieu of the class name. Furthermore, we did not study how much transfer learning benefits learning across different attributes for a particular cancer. Though most attributes have different label sets for a given cancer, there are instances where knowledge can be transferred. One such case is when a pathologist denotes that a particular attribute is not reported in the text which is applicable to many tumor attributes. Hence a fully unified extraction model may perform better than a model trained on a specific tumor attribute. In practice, it is also easier to maintain one model over maintaining many individual models.
Another promising direction is improving the ensemble approach between machine learning methods and rules-based or string similarity methods for the unique labels case. Our results on the oracle ensembling model shows that there is still much room for improvement when combining predictions. For example, the oracle model has up to 0.08 improvement in macro-F1 and 0.08 improvement in micro-F1 over our thresholding method based on the learned similarity score cutoff. Potential approaches include basing the decision-making process on the uncertainties of each algorithm or combining model probabilities and string similarity scores for each label.
CONCLUSION
Large datasets in medical contexts are expensive to generate, limiting the generalizability of many NLP systems. We develop a novel cancer-to-cancer transfer learning approach and a ZSS approach that can halve the amount of labeled data required, which potentially opens doors to more widespread implementation of these systems in the real world.
FUNDING
Partial support is gratefully acknowledged from ARO grant W911NF1710005, NSF grants DMS-1613002 and IIS 1741340, the Center for Science of Information (CSoI), a US NSF Science and Technology Center, under grant agreement CCF-0939370, and Adobe Research Grant.
AUTHOR CONTRIBUTIONS
BP and NA implemented, tested, and validated the experiments. All authors were involved in designing and developing the study and writing the paper.
ETHICS APPROVAL
This study was approved by the Institutional Review Board (#18-25222).
SUPPLEMENTARY MATERIAL
Supplementary material is available at JAMIA Open online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The code for the SLA, ZSS, and ZSS-thresholding algorithms is available in the Zenodo Repository at https://zenodo.org/record/5514358#.YU5_vdNKJOR.
Supplementary Material
REFERENCES
- 1.Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burger G, Abu-Hanna A, de Keizer N, et al. Natural language processing in pathology: a scoping review. J Clin Pathol 2016; jclinpath-2016-203872. [DOI] [PubMed] [Google Scholar]
- 3.Martinez D, Yue L. Information extraction from pathology reports in a hospital setting. In: Proceedings of the 20th ACM International Conference on Information and Knowledge Management. 2011: 1877–82; Glasgow Scotland, UK.
- 4.Napolitano G, Fox C, Middleton R, et al. Pattern-based information extraction from pathology reports for cancer registration. Cancer Causes Control 2010; 21 (11): 1887–94. [DOI] [PubMed] [Google Scholar]
- 5.Schroeck FR, Patterson OV, Alba PR, et al. Development of a natural language processing engine to generate bladder cancer pathology data for health services research. Urology 2017; 110: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yim WW, Yetisgen M, Harris WP, et al. Natural language processing in oncology: a review. JAMA Oncol 2016; 2 (6): 797–804. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen AN, Lawley MJ, Hansen DP, et al. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc 2010; 17 (4): 440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weegar R, Hercules D. Creating a rule based system for text mining of Norwegian Breast Cancer Pathology reports. In: Proceedings of the Sixth International Workshop on Health Text Mining and Information Analysis2015. Stroudsburg, PA, USA: Association for Computational Linguistics.
- 9.Mykowiecka A, Marciniak M, Kupść A.. Rule-based information extraction from patients’ clinical data. J Biomed Inform 2009; 42 (5): 923–36. [DOI] [PubMed] [Google Scholar]
- 10.Yala A, Barzilay R, Salama L, et al. Using machine learning to parse breast pathology reports. Breast Cancer Res Treat 2017; 161 (2): 203–11. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Martinez D.. Information extraction of multiple categories from pathology reports. In: 2010 Australasian Language Technology Workshop (ALTW 2010); 2010; Melbourne, Australia.
- 12.Zhou G, Zhang J, Su J, et al. Recognizing names in biomedical texts: a machine learning approach. Bioinformatics 2004; 20 (7): 1178–90. [DOI] [PubMed] [Google Scholar]
- 13.Qiu JX, Yoon H-J, Fearn PA, et al. Deep learning for automated extraction of primary sites from cancer pathology reports. IEEE J Biomed Health Inform 2018; 22 (1): 244–51. [PMC][28475069] [DOI] [PubMed] [Google Scholar]
- 14.Alawad M, Gao S, Qiu J, et al. Deep transfer learning across cancer registries for information extraction from pathology reports. In: IEEE EMBS International Conference on Biomedical & Health Informatics. 2019: 1–4; Chicago, IL. [DOI] [PMC free article] [PubMed]
- 15.Alawad M, Gao S, Qiu J, et al. Automatic extraction of cancer registry reportable information from free-text pathology reports using multitask convolutional neural networks. J Am Med Inform Assoc 2020; 27 (1): 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odisho A, Park B, Altieri N, et al. Natural language processing systems for pathology parsing in limited data environments with uncertainty estimation. JAMIA Open 2020; 3 (3): 431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altieri N, Park B, Olson M, et al. Supervised line attention for tumor attribute classification from pathology reports: Higher performance with less data. J Biomed Inform 2021; 122:103872. [DOI] [PubMed]
- 18.Rios A, Kavululu R.. Few-shot and zero-shot multi-label learning for structured label spaces. In: Proceedings of the Conference on Empirical Methods in Natural Language Processing. Conference on Empirical Methods in Natural Language Processing. Vol. 2018. 2018: 3132–42; Brussels, Belgium. [PMC free article] [PubMed]
- 19.Lu J, Du L, Liu M, et al. Multi-label few/zero-shot learning with knowledge aggregated from multiple label graphs. In: Conference on Empirical Methods in Natural Language Processing; 2020; Punta Cana, Dominican Republic. [Google Scholar]
- 20.Xia Y, Lampert CH, Schiele B, et al. Zero-shot learning—a comprehensive evaluation of the good, the bad and the ugly. IEEE Trans Pattern Anal Mach Intell 2019; 41 (9): 2251–65. [DOI] [PubMed] [Google Scholar]
- 21.Stubbs A.MAE and MAI: lightweight annotation and adjudication tools. In: Proceedings of the 5th Linguistic Annotation Workshop. 2011: 129–33; Portland, OR.
- 22.Cancer Protocol Templates. College of American Pathologists. https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/cancer-protocol-templates (Accessed 6 May 2020).
- 23.Ratcliff JW, Metzener D. Pattern matching: the gestalt approach. Dr. Dobb's Journal 1988; 46.
- 24.Gao S, Young MT, Qiu JX, et al. Hierarchical attention networks for information extraction from cancer pathology reports. J Am Med Inform Assoc 2018; 25 (3): 321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson A, Pollard T, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Scott D, Villarroel M, et al. Open-access MIMIC-II database for intensive care research. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. 2011: 8315–8; Boston, MA. [DOI] [PMC free article] [PubMed]
- 27.Devlin J, Chang MW, Lee K, et al. BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Vol. 1; 2019; Minneapolis, MN. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for the SLA, ZSS, and ZSS-thresholding algorithms is available in the Zenodo Repository at https://zenodo.org/record/5514358#.YU5_vdNKJOR.