Author's summary
Genetic cardiomyopathies are an important cause of sudden cardiac death across all age groups. Genetic testing in heart failure clinics is useful for family screening and providing individual prognostic insight. Obtaining a family history of at least three generations, including the creation of a pedigree, is recommended for all patients with primary cardiomyopathy. Additionally, when appropriate, consultation with a genetic counsellor can aid in the success of a genetic evaluation. Clinical screening should be performed on all first-degree relatives of patients with genetic cardiomyopathy.
Keywords: Cardiomyopathy, Hypertrophic cardiomyopathy, Dilated cardiomyopathy, Genetics, Heart failure
Abstract
Genetics has played an important role in the understanding of different cardiomyopathies, and the field of heart failure (HF) genetics is progressing rapidly. Much research has also focused on distinguishing markers of risk in patients with cardiomyopathy using genetic testing. While these efforts currently remain incomplete, new genomic technologies and analytical strategies provide promising opportunities to further explore the genetic architecture of cardiomyopathies, afford insight into the early manifestations of cardiomyopathy, and help define the molecular pathophysiological basis for cardiac remodeling. Cardiovascular physicians should be fully aware of the utility and potential pitfalls of incorporating genetic test results into pre-emptive treatment strategies for patients in the preliminary stages of HF. Future work will need to be directed towards elucidating the biological mechanisms of both rare and common gene variants and environmental determinants of plasticity in the genotype-phenotype relationship. This future research should aim to further our ability to identify, diagnose, and treat disorders that cause HF and sudden cardiac death in young patients, as well as prioritize improving our ability to stratify the risk for these patients prior to the onset of the more severe consequences of their disease.
INTRODUCTION
Cardiomyopathies constitute a diverse group of diseases that are the leading cause of heart failure (HF) and are defined by structural or functional disorders of myocardium in the absence of secondary causes of HF such as hypertension, valvular heart disease, ischemic heart disease, or congenital heart disease that are sufficient to explain the observed myocardial abnormality.1),2),3) Over the last quarter-century, an incredible amount of progress has been made in elucidating the genetic components of cardiomyopathy. The clinical translation of this research through genetic testing makes it possible to more precisely classify affected patients according to molecular etiology and identify individuals with no symptoms of disease who maybe at increased risk of developing cardiomyopathy. This progress allows insights into the early manifestations of cardiomyopathy and could help define the molecular pathophysiological basis for cardiac remodeling. New genomic tools and analytic methodologies provide unprecedented opportunities to thoroughly examine the genetic architecture of cardiomyopathies, even though these efforts are yet incomplete. Such data suggest that gaining a better understanding of variant gene-specific pathophysiology may result in the identification of novel therapeutic targets and herald the dawn of precision medicine. Cardiomyopathies may present late in their course with advanced disease, including HF, arrhythmias and sudden cardiac death. Therefore, the rationale for identifying genetic risk is compelling, as it allows those who are at risk to undergo interval screening to detect the phenotype's earliest manifestations. The early detection of a phenotype allows for earlier interventions,4) such as lifestyle modifications; medication to slow or stop the progression of the disease or to prevent thromboembolism; and procedures, drugs, or devices to reduce the risk of sudden cardiac death. Individuals at-risk, whether affected but asymptomatic or clinically unaffected, may benefit from genetic counseling and reproductive decision-making if they are identified. Genetic testing is recommended for patients with hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), arrhythmogenic right ventricular cardiomyopathy (ARVC), cardiomyopathy with other extra cardiac manifestations, and left ventricular noncompaction (LVNC).5 ) Substantial genetic overlap has become evident between these cardiomyopathies; both genetic and phenotypic overlap among them has been observed. Careful evaluation of family history, phenotypic screening, and genetic counseling are recommended for relatives of patients with genetically confirmed inherited cardiomyopathies. We intend to provide clinicians with a focused, evidence-based guide to the use of genetic testing to aid the management of inherited cardiomyopathies in this review.
THE NECESSARY TERMINOLOGY FOR CLINICIANS
The terms “mutation” and “polymorphism,” which are widely misused interchangeably, frequently cause confusion due to inaccurate assumptions regarding their pathogenic and benign effects, respectively. Thus, it is recommended that both terms be replaced by the term “variant.” The following modifiers have been described by the American College of Medical Genetics (ACMG) and Genomics and the Association for Molecular Pathology6):
• Pathogenic variant (PV): A genetic alteration with sufficient evidence to contribute to the development of disease based on population frequency, computational and predictive data, functional data, segregation data, de novo data, allelic data, and reputable database. Some PVs may show reduced penetrance.
• Likely pathogenic variant (LPV): A genetic alteration with high likelihood (greater than 90% certainty) to classify as pathogenic or disease-causing.
• Variant of uncertain significance (VUS): A genetic alteration with limited and/or conflicting evidence regarding pathogenicity.
• Likely benign variant (LBV): A genetic alteration with high likelihood (greater than 90% certainty) to classify as benign.
• Benign variant (BV): A genetic alteration with sufficient evidence against pathogenicity.
HYPERTROPHIC CARDIOMYOPATHY
HCM is a genetic disease of the cardiac muscle with substantial variation in morphology, clinical manifestation, genetic etiology, and cardiac outcome.7) HCM has been an important model disease for studying many inherited cardiac diseases because it is also relatively common, with an estimated 1 in 500 people affected.8),9) HCM can be diagnosed clinically in adult patients by imaging, specifically with 2D echocardiography or cardiovascular magnetic resonance (CMR) that demonstrates a maximal end-diastolic wall thickness of ≥15 mm anywhere in the left ventricle, in the absence of a confounding cause of hypertrophy.10) A patient with more limited hypertrophy, only a wall thickness of 13–14 mm, can also be diagnosed with HCM when present in conjunction with a family history of HCM or a positive genetic test. HCM is typically inherited in an autosomal dominant pattern, but there is considerable variation in its expression and penetrance.11),12),13) The offspring of each affected family member has a 50% chance of inheriting the genetic variant. In the case of HCM, the chance of developing the disease is significantly increased among family members who carry a pathogenic variant, but the age at which the disease manifests in an individual is variably distributed. The mechanisms are not clear, but these genetic alterations result in the characteristic pathological (myocyte disarray and fibrosis) and morphological (hypertrophied, nondilated left ventricle) features of HCM. The phenotypic variability is due, in part, to the causal PVs that interact with a variety of other genetic and nongenetic effects. Around 60% of patients with HCM have clearly identifiable familial disease.14) Although autosomal recessive and X-linked inheritance modes have been described, they are uncommon.15) A genetic basis for HCM was first described in 1990, when a PV in a gene encoding a sarcomeric protein was identified in a large family with HCM, sudden death, and HF.16),17) Since then, reports of multiple other PVs in genes encoding distinct sarcomere proteins,18),19) which act as effectors and regulators of contraction and relaxation of the heart, established HCM as a genetically heterogeneous disease.
Hypertrophic cardiomyopathy caused by pathogenic variants in genes encoding sarcomeric proteins
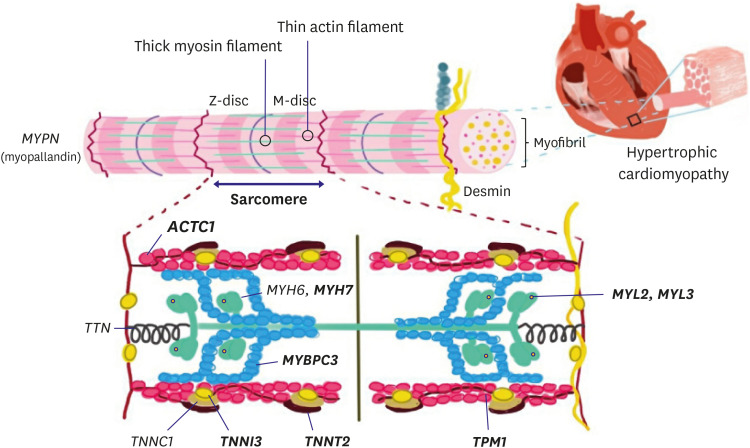
PVs in genes encoding sarcomeric proteins have been implicated in the majority of HCM cases (Figure 1 and Table 1). Among the known causal genes, the β-myosin heavy chain (MYH7) and myosin binding protein C (MYBPC3) are the two most common,20),21),22) followed by PVs in the genes encoding cardiac troponin T (TNNT2), cardiac troponin I (TNNI3), and α-tropomyosin (TPM1).11),23) PVs in the genes encoding cardiac α-actin 1 (ACTC1), myosin light chain 2 (MYL2), myosin light chain 3 (MYL3), and cysteine and glycine rich protein 3 (CSRP3) have also been identified as less frequent causes of HCM.23),24) Combined, MYH7 and MYBPC3 account for up to 50% of all clinically recognized cases of HCM, and constitute at least 75% of probands when a PV is identified, whereas other HCM genes account for <10% of cases.25),26),27) These PVs generally cause single-residue substitutions in proteins that become incorporated into the sarcomere; however, approximately half of the reported PVs in MYBPC3 variant are truncations, and some MYBPC3 missense variants can result in haploinsufficiency, a condition in which the gene product of the wild-type allele is not sufficient to compensate for the decreased or absent protein from the variant allele.28),29)
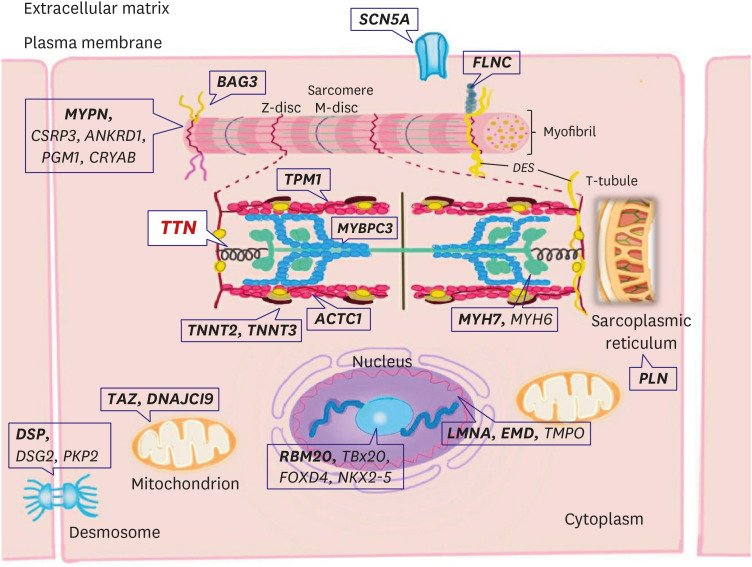
Figure 1. Main structural elements of the cardiac sarcomere and their respective genes involved in the pathogenesis of hypertrophic cardiomyopathy. A schematic of definitive (bold) hypertrophic cardiomyopathy genes in sarcomere.
ACTC1 = cardiac α-actin 1; MYBPC = myosin binding protein C; MYH6 = α-myosin heavy chain; MYH7 = β-myosin heavy chain; MYL= myosin light chain; TNNI = cardiac troponin I; TNNT = cardiac troponin T; TPM1 = α-tropomyosin; TTN = titin.
Table 1. Genes associated with sarcomere hypertrophic cardiomyopathy.
| Gene | Protein | Estimated prevalence in HCM (%) | Notes* | |
|---|---|---|---|---|
| Sarcomere | Force generation/Transmission | |||
| Thick myofilament | ||||
| MYH7 | β-Myosin heavy chain | 25–40% | - Prognosis variable depending on site of genetic variant; homogeneity within families | |
| - Sometimes restrictive cardiomyopathy phenotype | ||||
| MYBPC3 | Myosin-binding protein C | 25–40% | - Prognosis variable depending on site of genetic variant; homogeneity within families | |
| MYL2 | Myosin light chain 2, regulatory | <5% | ||
| MYL3 | Myosin light chain 3, essential | <5% | ||
| Thin myofilament | ||||
| TNNT2 | Cardiac troponin T | 5–10% | - Left ventricular hypertrophy is rare in children with TNNT2 mutations. Particularly among males, SCD-predisposition and minimal hypertrophy (some reports) | |
| TNNI3 | Cardiac troponin I | 5% | - This mutation is associated with sudden death at any age and dilated cardiomyopathy-like features in those aged >40 years | |
| - Sometimes restrictive cardiomyopathy phenotype | ||||
| TNNC1 | Cardiac troponin C | <1% | ||
| ACTC1 | Cardiac α-actine | <1% | - Benign prognosis and apical LV hypertrophy morphology | |
| TPM1 | α-Tropomyosin | 1–5% | ||
HCM = hypertrophic cardiomyopathy; LV = left ventricular; SCD = sudden cardiac death.
*As the field of prognostically relevant mutations expanded, a series of studies directly contradicted, or at least failed to replicate, many of these initial observations.
Pathogenic variants in genes encoding nonsarcomeric proteins
Additionally, the spectrum of HCM-associated genes has expanded to include nonsarcomeric genes, such as those encoding proteins found in the Z-disk, sarcoplasmic reticulum, and plasma membrane. However, these variants are uncommon, and our understanding of their role in disease is limited. Several of these nonsarcomeric genes are strongly associated with genetic evidence, such as disease segregation or in vivo functional data (e.g., CSRP3)30); however, many of these genes' involvement with HCM (e.g., MYH6, MYLK2, and TCAP) is supported only by the presence of variants in affected individuals, absence from controls, and computational assessments. Other genes' clinical significance has not been fully elucidated due to a lack of evidence. Titin (TTN), α-actinin (ACTN1), α-myosin heavy chain (MYH6), CSRP3, telethonin (TCAP), and vinculin (VCL) are among these genes.31),32),33),34) Despite their rarity in isolated cases and small families, variants in myozenin 2 (MYOZ2), ubiquitin E3 ligase tripartite motif protein 63 (TRIM63), and four-and-a-half LIM domains 1 (FHL1) have been described as causes of HCM.35),36)
Coexistence of a restrictive cardiomyopathy phenotype with the genetic basis of hypertrophic cardiomyopathy
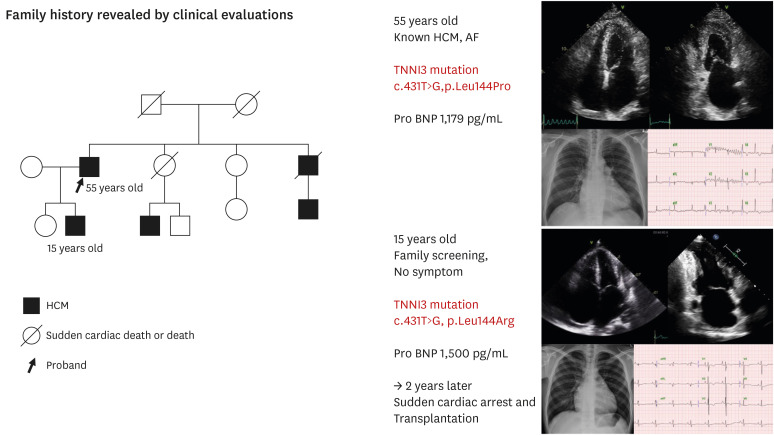
In the gray zone (minimal or no left ventricular hypertrophy [LVH, maximum LV wall thickness ≤13 mm] and severe diastolic dysfunction) between HCM and RCM, diagnosis may be problematic. HCM may share many hallmarks with other cardiomyopathies and progress to a compensatory phase.37),38) Kubo et al.39) showed that typical RCM phenotypes were found in 1.5% of the over 1,200 patients with familial HCM. PVs in in either MYH7 or TNNI3 were found in half of the RCM probands.39) On biopsy, all RCM patients with a PV showed either marked myofibrillar disarray, indicating a mixed phenotype with HCM, or had relatives with a clear HCM diagnosis.39) The restrictive phenotype comprised of increased dyspnea, decreased exercise capacity, and a significantly higher composite rate (56%) of mortality, cardiac transplantation, or implantable cardioverter defibrillator (ICD) discharges.39) The study's limitation is that it focused exclusively on known HCM-causing genes; it is uncertain whether additional genetic modifiers contributed to the RCM phenotype. Mogensen et al.40),41) identified a c.569A>G (p.Asp190Gly) variant of TNNI3 that was associated with marked restrictive physiology and a history of sudden cardiac death (SCD). Variants in TNNI3 now appear to be responsible for the development of RCM in a large proportion of young patients.40) Although the majority of PVs in TNNI3 that have been reported are generally missense, frame shift or splice variants.41) TNNI3-related variant phenotypes manifest with significant myofibrillar disarray even in the absence of typically LVH. A recent report described a genetic investigation of nine unrelated HCM patients with a marked restrictive filling pattern, significant atrial dilation, and normal wall thickness.42) Myocardial histology revealed the distinctive myofibrillar disarray associated with HCM. The majority of these patients had a TTNI3 variant. Hwang et al.43) described a large family in which a single TNNI3 PV resulted in variable phenotypic expression ranging from restrictive cardiomyopathy to HCM to a near-normal phenotype. It has been proposed that different variants exhibit varying degree of Ca2+ sensitivity; strong sensitivity leads to HCM or RCM, whereas low sensitivity results in DCM.44) These discoveries may have an effect on the evolution of the so called “restrictive phenotype.” However, not all patients from the same family develop the restrictive phenotype, suggesting the involvement of additional genetic and/or environmental variables and highlighting the genetic/phenotypic variability of HCM (Figure 2).
Figure 2. Restrictive phenotype of the hypertrophic cardiomyopathy family.
During family screening for hypertrophic cardiomyopathy, we found that a 15-year-old male had restrictive physiology cardiomyopathy. The patient had no symptoms at that time, but the pro-BNP level was elevated and both atria were hugely enlarged. The gene test showed the same variant as his father, and the patient suddenly presented with dyspnea and cardiac arrest. The patient required heart transplantation.
AF = atrial fibrillation; HCM = hypertrophic cardiomyopathy; TNNI = cardiac troponin I.
Genetic testing for diagnosis of hypertrophic cardiomyopathy associated conditions and phenocopies
LVH can also be induced by metabolic or storage disorders such as Danon disease (LAMP2 gene),45) Fabry disease (GLA gene), and metabolic storage disease associated with Wolff-Parkinson-White syndrome (PRKAG2 gene). PVs in the TTR gene are associated with familial transthyretin amyloidosis, a condition marked by the accumulation of amyloid protein in the autonomic and peripheral nervous systems. In families with HCM and Wolff-Parkinson-White syndrome, PVs in genes producing AMP-activated γ2 noncatalytic subunit of protein kinase A (AMPK), a regulator of cell bioenergetics, have been identified.46),47) The phenotype described ranges from that of preexcitation and conduction abnormalities and slight hypertrophy to that of early-onset and severe hypertrophy, with the small number of patients exhibiting preexcitation. LVH is also a common feature in cardiofaciocutaneous syndrome, a rare genetic condition caused by PVs in genes involved in the RAS-mitogen-activated protein kinase (MAPK) signaling pathway.48) HCM phenocopies in LEOPARD, Noonan, and Costello syndromes, collectively referred to as RASopathies, are frequently observed in affected children but may be less noticeable in adulthood; this characteristic may account for some unexplained adult cardiac arrest.49) HCM is also associated with Friedreich's ataxia, an inherited disorder caused by an intrinsic GAA sequence expansion in the frataxin (FXN) gene. The disease typically begins in childhood with peripheral muscle weakness and visual and auditory impairment and advances faster in males.50)
Implications of genetic testing in hypertrophic cardiomyopathy
Guidelines
2020 American College of Cardiology/American Heart Association (ACC/AHA),10) Heart Rhythm Society/European Heart Rhythm Association (HRS/EHRA)*, and 2014 European Society of Cardiology (ESC) guidelines51)
(1) Genetic testing recommended in patients fulfilling diagnostic criteria for or with signs or symptoms suggestive of HCM (class I).
(2) In patients with suspected HCM, comprehensive physical examination and complete medical and three-generation family history are recommended as part of the initial diagnostic assessment (class I).
(3) Pathogenic variant-specific cascade screening is recommended (class I).
(4) In first-degree relatives of patients with HCM, both clinical screening (electrocardiogram [ECG] and echocardiography) and cascade genetic testing should be offered (class I).
(5) Genetic testing should be considered in deceased patients with pathologically confirmed HCM to facilitate cascade screening (class I).
(6) In families affected by HCM, preconception and prenatal reproductive and genetic counseling should be offered (class I).
*Updated in 2018 in a joint publication with the ACMG.52)
Diagnostic value
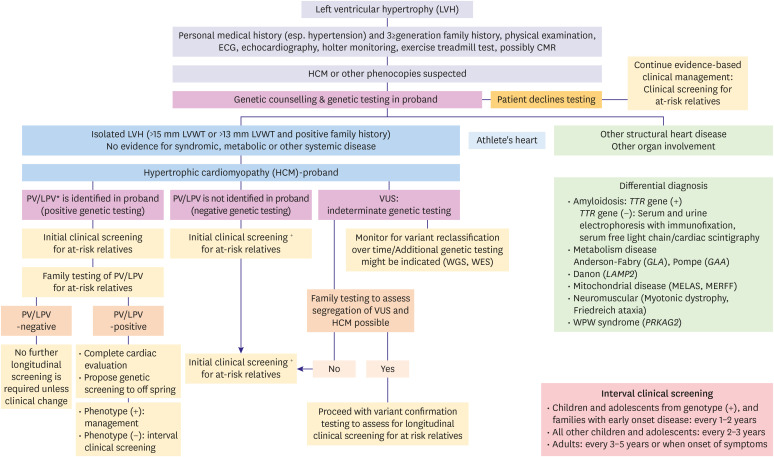
There are a number of clinical benefits to having genetic testing in HCM, including but not limited to, confirmation of diagnosis, detect the presence of preclinical disease, enable cascade genetic testing in the family and informed reproductive decisions.53),54) The “genetic substrate” is typically present in approximately 60–70% of patients with HCM whose family history includes HCM, and it is present in 10–50% of those without a family history of the disease.10),55),56) However, as the genetic architecture of HCM is still being mapped, it would be premature to rule out its genetic nature in patients with negative tests. Because HCM is primarily a sarcomere disease, first-line genetic testing focuses on panel testing for genes with strong evidence for being disease-causing in HCM.57) Numerous technological platforms are available for genetic testing, including gene panels, exome sequencing, and whole genome sequencing.54) MYH7, MYBPC3, TNNI3, TNNT2, TPM1, MYL2, MYL3, and ACTC1 are the eight sarcomere genes that are typically included in the gene panels. Typically, the index case (proband) undergoes initial genetic testing.53) When the targeted gene panel testing does not find a PV, the choice may be to perform exome sequencing on a clinical or research basis, which can provide additional support when used alongside genetic counseling to explain the low yield of the test and the likelihood of finding susceptibility variants associated with disease other than the disorder being studied. About 40% of patients with HCM have no sarcomere disease-causing variant and no family history of the disease.56) Although the discovery of a VUS is not a clinically actionable result, it can be useful for clarifying the pathogenicity of variants by conducting further investigations that include cosegregation analysis in family members, parental DNA testing to see whether the VUS is de novo, and functional studies (Figure 3).
Figure 3. Schematic for employing hypertrophic cardiomyopathy genetic testing in the proband (index patient) and family members.
CMR = cardiovascular magnetic resonance; ECG = electrocardiogram; HCM = hypertrophic cardiomyopathy; LPV = likely pathogenic variant; LVH = left ventricular hypertrophy; LVWT = left ventricular wall thickness; PV = pathogenic variant; VUS = variant of uncertain significance; WES = whole exome sequencing; WGS = whole genome sequencing.
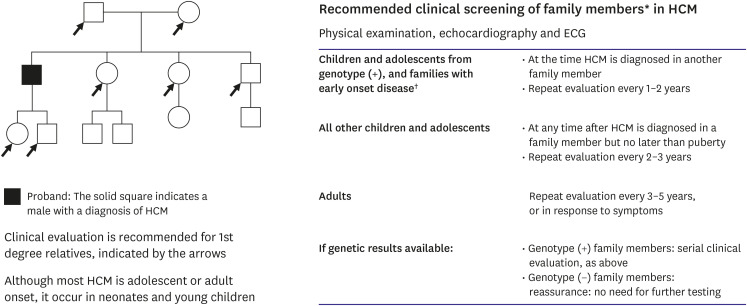
It can help elucidate the diagnosis and provide the opportunity for cascade (predictive) testing of at-risk family members if a clinically actionable result (i.e., likely pathogenic or pathogenic) from genetic testing is identified in the proband.10),58) A higher yield of genetic testing and a more severe phenotype are associated with diagnosis of HCM during early childhood to adolescence.59) The construction and analysis of family pedigrees enables the identification of specific features, such as familial history of SCD, unexplained HF, cardiac transplantation, pacemaker and ICD implants, and evidence for systemic disease, which may aid in diagnosis and help to determine the mode of inheritance in the family. Because PVs in HCM variant are highly penetrant, carriers have a substantial lifetime risk (>95%) of developing the phenotype.10) In general, the penetrance (proportion of variant carriers with clinically detectable disease) of HCM increases with age. However, in 2 large pediatric studies, approximately 9.9% of children screened for HCM were already phenotype-positive at first evaluation. The median age at HCM onset was 8.9–10 (4–13) years with earlier onset in males, those with a family history of SCD, and PVs in MYH7/MYBPC3.60),61) Similarly, the median time interval between the onset of HCM and major cardiac events, such as death, SCD, or cardiac intervention (myectomy, ICD), was 1.5 years. These studies suggest that the prevalence of HCM in children and preadolescents is comparable to that in adolescents owing to the fact that the phenotype of familial HCM in childhood varies and includes severe disease. This finding implies the importance of initiating clinical screening at a younger age. Taken together, these findings support the concept of initiating family screening in childhood and repeating it periodically. Therefore, regular follow-up in genotype-positive/phenotype-negative individuals with serial ECG, transthoracic echocardiography (TTE), and clinical assessment at periodic intervals (every 1–2 years in children and adolescents and every 3–5 years in adults) is recommended for early detection of clinical disease expression and timely initiation of therapy (Figure 4).10)
Figure 4. Recommended clinical screening of family members* in HCM. The screening interval may be modified (e.g., at the onset of new symptoms or in families with a malignant clinical course or late-onset HCM). These screening intervals apply to at-risk family members when genetic testing has not been performed or is uninformative in the proband, or when it has identified a likely pathogenic or pathogenic variant in the at-risk family member.
HCM = hypertrophic cardiomyopathy; ECG = electrocardiogram; LVH = left ventricular hypertrophy.
*Includes all asymptomatic, phenotype-negative, first-degree relatives deemed to be at risk for developing HCM based on family history or genotype status, and may sometimes include more distant relatives based on clinical judgment. †Severe family history of early HCM-related death, sudden cardiac death, early development of LVH, or other adverse complications.
Factors that influence the hypertrophic cardiomyopathy phenotypes
The morphological, histological, and clinical phenotypes of HCM result from complex interactions among a large number of contributing factors, ranging from the effects of causal gene variants to environmental factors. Penetrance of variants increase with age but remains less than 100%. In the majority of patients with HCM, hypertrophy begins throughout adolescence. According to Lafreniere-Roula et al.,60) 9.9% of children evaluated for HCM were already phenotype-positive at initial evaluation and experienced major adverse cardiac events (MACEs) at a median follow-up of 1.5 (0.5–4.1) years from HCM beginning. This indicates that many children by the time of diagnosis have already developed a well-defined phenotype, which also supports the notion that disease prevalence and penetrance in children are comparable to those in adolescents.60) Clinical presentation may differ from asymptomatic to SCD, the latter of which targets younger adults. According to genotype-phenotype correlation studies, certain causal genes are strongly correlated with the phenotypic expression of HCM, such as MYH7, TNNT2, and MYBPC3. These genes appear to influence the severity of cardiac hypertrophy and the likelihood of SCD.62),63),64),65),66) The PVs in MYH7 are generally found in younger patients, and they have more significant hypertrophy, along with a higher risk of SCD.67),68),69) On the other hand, PVs in MYBPC3 are linked to relatively mild hypertrophy and late-onset clinical manifestations.64),70),71) PVs in the MYBPC3 are often associated with low penetrance, mild hypertrophy, and a low incidence of SCD. The phenotype frequently manifests late, and may coincide with the concomitant presence of hypertension. Despite the overall benign nature of PVs in MYBPC3, significant variability exists in the phenotypic expression of HCM; so-called malignant PVs have also been reported in MYBPC3 genes.60),70) TNNT2 PVs may cause premature sudden cardiac death in patients with only mild or no LVH.72) PVs in TPM1 are generally benign and associated with mild LVH. Despite a mild degree of hypertrophy, a high rate of SCD has also been reported.73) Certain studies have suggested that PVs in essential and regulatory myosin light chains (MYL1, MYL2) can lead to mid-cavity obstruction in HCM and skeletal myopathy in some patients,74) while other studies have found no correlation.75) Although PVs are quite rare in TTN 76) and ACTC1,77) they have been detected in a small number of HCM phenotype families.
Although the exact mechanisms by which sarcomere variants cause clinically relevant phenotypes have not been thoroughly elucidated, it can be presumed that these mechanisms may involve contractile elements. In vitro and in vivo research reveal that PVs exhibit a diverse array of functional defects, such as reduced actin-myosin interaction, reduced cross-bridging kinetics, myocyte contractility, ATPase activity of myosin, and altered Ca2+ sensitivity.78),79) Hypertrophy, along with other clinical and pathological phenotypes, is considered compensatory phenotypes that occur as a result of functional defects. Various disease features, including abnormal intramural coronary arteries causing small vessel ischemia, elongated mitral valve leaflets, and congenital anomalies of the sub-mitral valve apparatus, are widely recognized components of the HCM phenotype which appear to have no known direct connection with sarcomere variants.
Is genotype-phenotype correlation in hypertrophic cardiomyopathy really clinically useful in predicting prognosis?
Some studies have noted that although MYH7-p.Val606Met is known “benign” variant, it appears to be associated with increased risk of SCD in a family with HCM.80) A previous study observed that three individuals, including a son of the index case, experienced SCD at a young age with marked septal hypertrophy. Among the surviving four HCM phenotype-positive individuals with the p. Val606Met variant, all were diagnosed before 18 years of age, and three demonstrated either nonsustained ventricular tachycardia (VT) or atrial fibrillation (AF).
After Menon et al.81) identified its association with SCD in 4/9 variant-positive individuals, the “malignant,” the TNNT2-p.Ile89Asn variant also suffers from similar contradictions variant.81) The variant was identified in each of the nine affected family members who were diagnosed with RCM (n=2), DCM (n=2), HCM (n=4), and mixed cardiomyopathy (n=1). Furthermore, although there was a high incidence of atrial tachyarrhythmia, there was no incidence of SCD. The results of these studies question the association between individual genetic variants and patient survival. There are also contradictory reports regarding the role of specific variants influencing other aspects of HCM, such as septal hypertrophy morphology.
Recent advances in the prognostic role of genotyping in hypertrophic cardiomyopathy
There are several ongoing controversies regarding the relative value of variant-specific and genotype-specific risk stratification, but recent evidence suggests that unrelated patients with a positive HCM genetic test have a greater likelihood of developing a more severe phenotype (diastolic and systolic dysfunction and symptoms) than those without genetic variants. According to a Mayo Clinic study, patients who were genotype positive were found to be younger at diagnosis, demonstrated greater hypertrophy, were more likely to have a positive family history of HCM, and were more likely to receive an ICD than negative genotype counterparts.82) Furthermore, a positive genetic test conferred a hazard ratio of 4.3 (95% confidence interval, 1.5–12.5%), which was greater than that of age, degree of outflow tract obstruction, and presence of AF. These differences were also observed in patients with HCM from Italy between those who tested positive and negative for HCM variants. In this study, Olivotto et al.83) found that patients with a positive myofilament genetic test had drastically different outcomes. Of the cases in this cohort of 203 index cases, 62% (126 of 203) of those who had a positive HCM genetic test had an increased combined risk of cardiovascular death, nonfatal stroke, or progression to New York Heart Association class III or IV symptoms, whereas the rest (38%) with a negative genetic test result had lower risk. Additionally, myofilament-positive patients were more likely to have severe LV systolic (EF <50%) and diastolic (restrictive filling pattern) dysfunction. These studies indicate for the first time that a “positive genetic test” may be associated with adverse outcomes in patients with HCM. These observations remain to be translated in a clinically meaningful way. Perhaps clinicians can consider that patients with a negative result for HCM ultimately exhibit milder disease phenotypes and are far less likely to progress. HCM is a complex disease that presents itself with variable hypertrophy and outflow tract obstruction at any age. While knowledge of genetic variants are becoming increasingly understood, HCM susceptibility variant and their roles in disease pathogenesis still present a difficult challenge. This is particularly due to the multiple factors including that the same genetic variant can lead to dramatically different phenotypic variations of HCM in different individuals. Until these factors and the relationship between specific HCM variants and its impact on phenotypes are fully understood, specific isolated variants currently do not have feasible prognostic clinical utility.
DILATED CARDIOMYOPATHY GENETICS
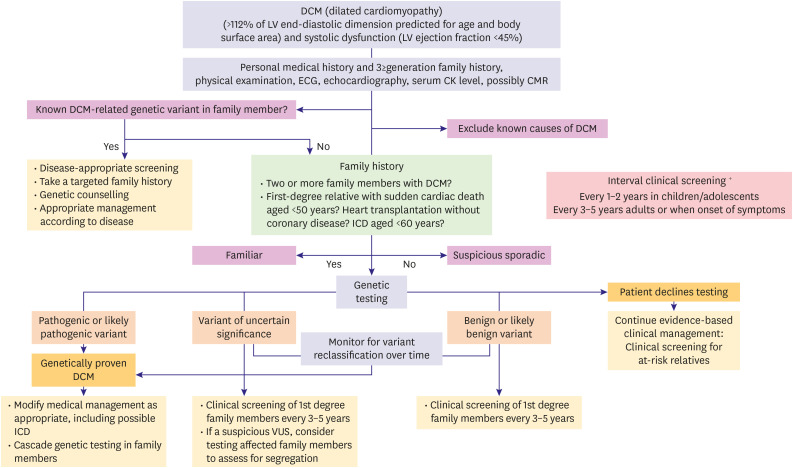
DCM is defined by the presence of LV or biventricular dilatation (>112% of LV end-diastolic dimension predicted for age and body surface area) and systolic dysfunction (LVEF <45%) in the absence of abnormal loading conditions (hypertension, valve disease) or coronary artery disease sufficient to cause global systolic impairment.84) Familial DCM is diagnosed when 2 or more family members meet the criteria for DCM or when the patient of interest (proband) meets the criteria for DCM, and a first-degree relative has experienced sudden cardiac death aged <35 years.85) However, the definitions of these terms are merely arbitrary and, in many cases, can only be documented partially. In general, if family members of the proband have been reported to have HF or premature sudden cardiac death with no identified cause, it is reasonable to consider familial DCM.86)
DCM has a highly diverse genetic basis. Approximately up to 40% of DCM may have a genetic basis, and over 50 distinct genes have been implicated, many of which influence the functionality of diverse protein assemblies.87),88),89) Despite the fact that these genes are implicated in various genetic pathways, the end result is the same: a specific DCM phenotype. The sheer numbers of genes and diversity of variants identified (allelic and locus heterogeneity) with DCM are more extensive than with the other cardiomyopathies.3),90),91) The range of diversity among these individuals is extremely relevant to the practice of clinical medicine because DCM, more so than any other etiology, underlies a large proportion of HF cases and their resulting morbidity and mortality. The majority of genetic DCM is inherited in an autosomal dominant pattern with variable expressivity and penetrance, although specific forms of autosomal recessive, X-linked recessive, and mitochondrial inheritance do infrequently occur.92),93),94) De novo variant can also lead to genetic cardiomyopathy and is defined as the absence of the offspring's variant in parents. Variants that are considered de novo have been described in many different genes, and the existence of a de novo variant can be used to infer the pathogenic status of genetic variants. This is because it is quite rare for de novo variants to appear in the coding exons of each genome. So, these newly developed pathogenic variants, which introduce changes that alter the protein in the cardiomyopathy gene, may be considered pathogenic.95) Although many of these variants may be potential pathogenic variants, a new pattern of oligogenic inheritance is emerging (for example, diseases caused by a small number of variants in more than one gene), which requires genetic counseling and results in predictive testing becoming an important challenge. This may provide one explanation for the variation in disease penetrance seen in some individual families. From a clinical standpoint, it is critical to conduct a thorough phenotypic evaluation of patients and their families in order to correctly interpret genetic findings.87)
Table 2 summarizes the most frequently implicated genes in DCM and their estimated prevalence in non-syndromic DCM.96),97),98) The location of the proteins encoded by these genes in cardiomyocytes is depicted in Figure 5. While some ethnic differences in the proportion of causative genes exist, TTN truncating variants and lamin A/C (LMNA) variants are considered major genetic factors for the development of DCM.
Table 2. Major genes associated with dilated cardiomyopathy.
| Gene | Protein | Estimated prevalence in DCM (%) | Age of onset of DCM (years) | Notes |
|---|---|---|---|---|
| TTN | Titin | 12–25% | DCM: Usually 40–59 | - Usually, isolated DCM |
| - More common in women than in men; variable prognosis but family clustering | ||||
| LMNA | Lamin A/C | 5–10% (up to 30 if conduction disease also present) | 30–49 and penetrance is almost complete at the age of 70; conduction disease usually prominent | - Subset of Emery–Dreifuss muscular dystrophy |
| Nuclear membrane | - Phenotypic expression is characterized by a relatively high incidence of sudden cardiac death or major ventricular arrhythmias, even before the development of systolic left ventricular dysfunction | |||
| - Class IIa for ICD implantation: NSVT during monitoring, EF <45% at first evaluation, male and non-missense mutations96),97) | ||||
| MYH7 | β-Myosin heavy chain | 4–10% | Variable: adolescence to 49 | - Primarily HCM; also, LVNC and RCM |
| Sarcomere | - Prognosis variable depending on site of genetic variant; homogeneity within families | |||
| MYH6 | α-Myosin heavy chain | 4% | Variable: adolescence to 49 | - Primarily HCM |
| Sarcomere | - Prognosis variable depending on site of genetic variant; homogeneity within families | |||
| MYPN | Myopalladin | 3–4% | Variable: adolescence to 59 | - Usually, isolated DCM |
| Sarcomere, Z-disc | - Variable presentation with some early deaths due to heart failure | |||
| DSP | Desmoplakin | 3–4% | Variable: mostly 40–49 | - ARVC |
| Desmosome | - Identified in multiple patients with advanced DCM although natural history uncertain; arrhythmic burden seems to be much lower than in ARVC | |||
| RBM20 | RNA-binding protein 20 | 1–5% | Adolescence to 50; can present with SCD or advanced heart failure | - Usually, isolated DCM |
| Spliceosome | - Rapid progression: SCD and end-stage heart failure are common, with a high incidence of ventricular arrhythmias | |||
| - Severe disease expression in male patients98) | ||||
| TNNT2 | Cardiac muscle troponin T | 2–3% | Variable: adolescence to 49 | - HCM, LVNC, and RCM |
| Sarcomere | - Prognosis variable depending on site of genetic variant; homogeneity within families | |||
| SCN5A | SCN5A (sodium channel protein type 5, α subunit) | 2–3% | Adolescence | - Brugada syndrome and LQTS |
| Ion channel | - Early onset, commonly with atrial fibrillation; not associated with risk of ventricular arrhythmias | |||
| TPM1 | α-Tropomyosin | 0.5–1.0% | Variable: adolescence to 49 | - Other cardiomyopathy: HCM, LVNC, and RCM |
| Sarcomere | - Prognosis variable depending on site of genetic variant; homogeneity within families |
ARVC = arrhythmogenic right ventricular cardiomyopathy; DCM = dilated cardiomyopathy; EF = ejection fraction; HCM = hypertrophic cardiomyopathy; ICD = implantable cardioverter defibrillator; LQTS = long QT syndrome; LVNC = left ventricular noncompaction; NSVT = nonsustained ventricular tachycardia; RCM = restrictive cardiomyopathy; SCD = sudden cardiac death.
Figure 5. A schematic of definitive (bold) and posited dilated cardiomyopathy genes with the subcellular localization of the encoded proteins. Pathogenic genes encode proteins that participate in many diverse biological processes of cardiomyocytes. TTN truncating mutations are the most frequent cause of dilated cardiomyopathy.
ACTC1 = cardiac α-actin 1; BAG3 = BAG cochaperone 3; DES = desmin; DSG2 = desmoglein-2; DSP = desmoplakin; EMD = emerin; FLNC = filamin C; LMNA = lamin A/C; MYBPC = myosin binding protein C; MYH6 = α-myosin heavy chain; MYH7 = β-myosin heavy chain; MYPN = myopalladin; PKP2 = plakophilin-2; PLN = phospholamban; RBM20 = RNA-binding protein 20; SCN5A = sodium voltage-gated channel alpha subunit 5; TMPO = trimethylphosphate; TNNT = cardiac troponin T; TPM1 = α-tropomyosin; TTN = titin.
Genetic variants causing dilated cardiomyopathy
TTN
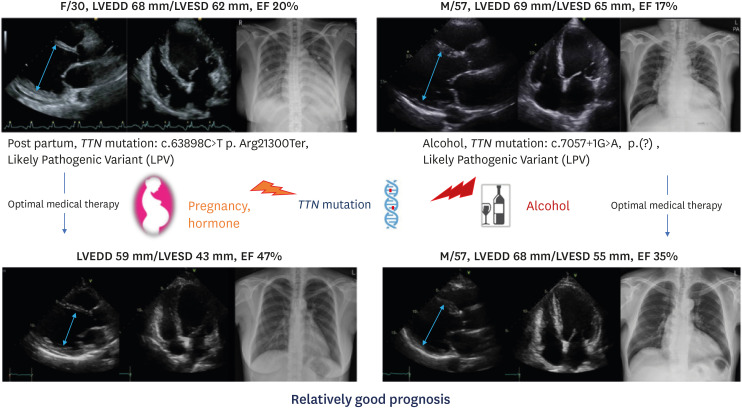
TTN truncating variants are the most common cause of DCM,99) accounting for about 15–25% of DCM cases.88),100) Titin is the giant protein that encodes the TTN gene, the largest known protein expressed in the heart. Titin has 4 major protein domains (the Z-disk, the I-band, the A-band, and the M-band) that are located on either side of the length of the sarcomere. Titin serves as a spring providing both an inner and outer force. Additionally, it regulates sarcomere contraction and signaling.101) The clinical phenotypes of TTN truncating variants include low penetrance, low association with abnormalities in the conduction system, relatively good prognosis, sex differences in prognosis (males have a worse prognosis than females), and a tendency for LV reverse remodeling.88),89),102) Despite the low penetrance of TTN truncating variants, additional cardiac insults such as pregnancy and alcohol abuse may cause cardiac dysfunction.103),104) Patients who have a TTN truncating variant tend to develop severe LV dysfunction at diagnosis, but with optimal medical therapy they tend to have a good outcome, and the patients' cardiac function may often improve significantly (Figure 6).88)
Figure 6. Two cases of TTN-related dilated cardiomyopathy. Although cardiomyopathy patients with TTN truncating variants show severe left ventricular dysfunction at diagnosis, they tend to present with a good response to appropriate medical therapy and their cardiac function improves drastically.
EF = ejection fraction; LPV = likely pathogenic variant; LVEDD = left ventricular end diastolic dimension; LVESD = left ventricular end systolic dimension; TTN = titin.
LMNA
LMNA variants are the second most common cause of DCM,105) accounting for 5–10% of DCM cases.88),89) The single LMNA gene encodes lamins A and C, and differential splicing at the 3′ end results in two proteins that are identical across their first 566 amino acids. PVs in LMNA lead to a constellation of diseases, from premature aging to myopathies and DCM.106) There is compelling evidence for the clinical significance of LMNA variants. LMNA variants have high penetrance, a young -onset, coexistence with an abnormalities in the conduction system, high occurrence of ventricular arrhythmias and SCD, a poor response to medical therapy, and a frequent need for heart transplantation.89),102),107),108) Due to the frequent coexistence of LMNA variants with ventricular arrhythmias, the European Society of Cardiology guidelines recommends the use of an ICD in patients with DCM and a confirmed disease-causing LMNA variant to prevent sudden cardiac death despite not meeting left ventricular ejection fraction criteria.97),109) It is suggested that pathogenic LMNA variants result in a failure in mechano-transduction and the development of laminopathy, which can lead to various clinical symptoms, including skeletal and/or cardiac muscular dystrophy, lipodystrophy, dysplasia, neuropathy, leukodystrophy, and premature aging.109) Overall, out of a selected tertiary referral population of patients with familial DCM, 30% had the LMNA variant while 88% had the DCM/muscular dystrophy syndrome combination, and 25% had the isolated conduction system disease without DCM.110)
Sarcomeric (motor) genes
PVs in genes encoding proteins that form thick and thin sarcomeric filaments have been implicated in DCM. These proteins (myosin-heavy chain alpha and beta [MYH6 and MYH7, respectively], MYBPC3, troponins [TNNT2, TNNI3, TNNC1], tropomyosin 1 [TPM1], cardiac actin 1 [ACTC1], and myopalladin [MYPN]) share catalytic activity and are involved in sarcomeric contraction. Structural properties found in Z-disk genes are also shared by MYPN. Variants that have been discovered to contribute to DCM, have phenotypic heterogeneity, often each variant resulting in a different DCM manifestation. Up to 10% of familial DCM result from genetic variants that encode contractile proteins, with substantial phenotypic variability.111)
RBM20
RBM20 encodes the RNA binding motif 20 protein, a chaperone protein that interacts with mRNA and regulates exon splicing to generate distinct gene isoforms. RBM20 regulates the splicing of TTN, which is one of several cardiac targets.112) In rats, an RBM20 deletion alters TTN splicing and results in a DCM phenotype. With respect to prevalence in DCM families, RBM20 represents a rare genotype, accounting for 1–5% of cases.113),114),115) For this reason, evidence-based genotype-phenotype correlations remain unproven. However, it has been shown that RBM20 variants can be found in those who have a higher risk of developing DCM phenotype characterized by rapid progression of HF, high risk for arrhythmias, and the need for cardiac transplantation, especially in males.98),116) These findings underscore the importance of conducting clinical and genetic investigations to identify patients at risk of developing disease complications to initiate proper and timely treatment.
FLNC (structural cytoskeleton-Z-disk gene)
More recently, PVs in FLNC encoding filamin C, have been described in DCM.117),118) Deletion of FLNC in mice causes skeletal muscle weakness, which suggests a positive interaction between filamin C and the dystrophin complex.119) In humans, truncating variants lead to cardiomyopathy, which is associated with a high rate of ventricular arrhythmias and SCD; these findings suggest that filamin C plays a role in the cardiac conduction system in addition to cardiomyocyte function.120)
SCN5A (ion channel gene)
The major sodium channel expressed in the heart is encoded by SCN5A. In both long QT syndrome and Brugada syndrome, heterozygous autosomal dominant PVs are found in SCN5A. Missense variants in SCN5A have been described in familial DCM, and these PVs carry a higher risk for arrhythmias.121),122) There is significant amount of genetic heterogeneity in the SCN5A gene in the general population, which makes it challenging to accurately interpret pathogenicity of variation in SCN5A.
DSP, PKP2 (Desmosomal genes)
The desmosome is a symmetric myocyte structure in which each part resides in the cytoplasm of one of a pair of adjacent cells, anchoring the intermediate filaments in the cytoskeleton to the cell surface. It is essential that the desmosome serves as a coupling agent between cardiac myocytes and cell gap junctions to ensure both the mechanical and electrical integrity of the heart. There is a traditional correlation between ARVC and variants in genes encoding desmosomal proteins. However, a number of desmosomal genes, usually inherited from an autosomal dominant heritage, have been identified in patients with DCM patterns independent of right ventricular involvement. Elliott et al. discovered a prevalence of 5% of desmosomal protein-coding gene variants among 100 unrelated patients with DCM patients.123) Therefore, the true pathogenesis of ARVC or DCM caused by variants in desmosomal gene products has not been fully characterized.
Implications of the genetic test in dilated cardiomyopathy
Guidelines
Similar to HCM, the identification of the genetic cause of DCM provides significant clinical insight into family screening, management, and prognosis, but the yield of genetic testing is lower as compared to HCM.86) The prevalence of underlying PVs is estimated to be between 15% and 25% in unselected DCM patients and between 20% and 40% in familial DCM patients.52),124),125),126) While modern genetic testing methods enable the evaluation of a greater number of genes, the detection rate has remained relatively constant over the last decade.125),126) The guidelines vary slightly for sporadic DCM (without a family history of the disease), primarily due to the unknown diagnostic yield and clinical utility of genetic screening in this population.
ACC/AHA*,127),128) HRS/EHRA†, and 2010 ESC guidelines129)
(1) Obtaining a family history of at least three generations, including the creation of a pedigree, is recommended for all patients with primary cardiomyopathy (class I).
(2) Clinical (phenotypic) screening for cardiomyopathy in at-risk 1st-degree relatives is recommended (class I).
(3) Variant-specific cascade screening is recommended (class I).
(4) Comprehensive or LMNA/SCN5A-targeted genetic testing is recommended for all patients with DCM and severe cardiac conduction disease or a family history of SCD (class I).
*2013 ACC/AHA Heart Failure Guideline, 2016 AHA Scientific Statement on DCM.
†Updated in 2018 in a joint publication with the ACMG.52)
Diagnostic value
Genetic testing is not utilized to make a diagnosis in patients with DCM but rather to search for an underlying cause. The identification of a PV does not significantly alter clinical care in many patients with overt DCM. However, DCM genetic testing allows the identification of populations at risk of developing DCM and facilitates the close monitoring of DCM cases associated with a high risk of SCD.55) Because these aggressive forms of DCM are often difficult to recognize at early stages, genetic testing appears to be beneficial in young individuals with early symptoms of DCM. The results of genetic testing should be interpreted in conjunction with comprehensive cardiac and systemic evaluations.
In contrast, genetic test results (PV or LPV) in a DCM susceptibility gene are useful for assessing asymptomatic family members, allowing identification of those at risk of future disease and rationalizing follow-up. Echocardiographic screening is recommended for asymptomatic family members. Testing of asymptomatic family members highlights the necessity for echocardiographic screening and is a cost-effective approach. Family members who are both genotype-positive and phenotype-negative require routine echocardiographic evaluation to detect early signs of contractile dysfunction that may manifest even before symptoms develop.5),130) In the past, it was thought that no further cardiac evaluation was necessary if an at-risk family member tested negative for a previously identified pathogenic or likely pathogenic variant in the context of a negative baseline cardiac evaluation, depending on the strength of the association between the variant and the disease.5) However, with growing awareness of the molecular complexity of DCM in many groups, there may be a way to identify DCM-susceptible individuals as being at lower risk of developing the disease rather than no risk, and these individuals could have their periodic clinical visits tailored accordingly. A comprehensive evaluation of index patients with DCM should ideally include family pedigree for at least 3–4 generations to identify the potential familial occurrence of the disease (Figure 7).
Figure 7. Schematic for employing dilated cardiomyopathy genetic testing in the proband (index patient) and family.
CK = creatine kinase; CMR = cardiovascular magnetic resonance; DCM = dilated cardiomyopathy; ECG = electrocardiogram; ICD = implantable cardioverter defibrillator; LV = left ventricular; VUS = variant of uncertain significance.
Complexity and phenotype correlates with definitive and putative dilated cardiomyopathy genes
The treatment of genetic DCM depends on the following factors: 1) the penetrance of the gene variant; 2) the phenotypic presentation; 3) the underlying molecular mechanisms leading to cardiac involvement; and 4) the gene–environmental interactions determining the DCM phenotype.90)
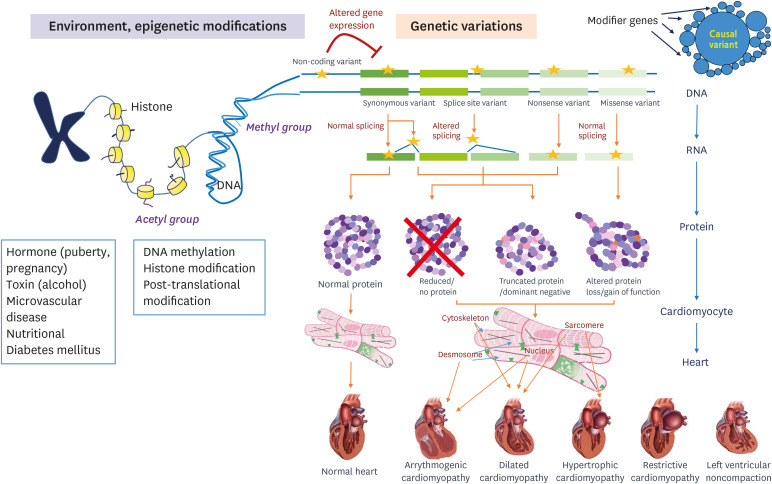
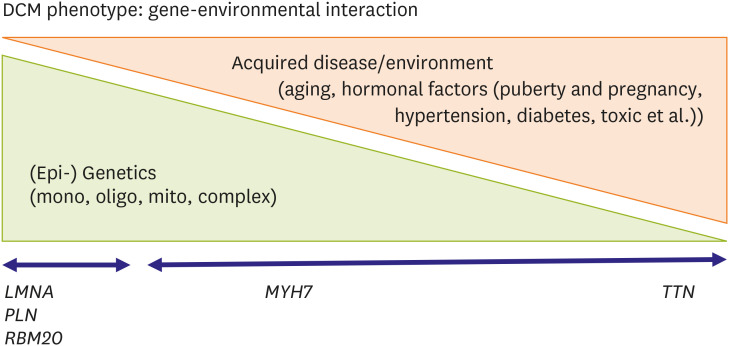
The clinical phenotype of DCM is much more complex than commonly observed. Genetic and nongenetic interactions may influence the severity and outcome of DCM cases, including HF, arrhythmia, stroke, and the need for cardiac transplantation.95) The complicated underlying architecture of the phenotype emerges from many complex and dynamic interactions that arise from multiple genetic and nongenetic factors. This unknown, mysterious influence of epigenetic regulation of gene expression on the clinical phenotype is another example of something we need to study further.131) Additionally, it is estimated that 94% of human genes undergo extensive alternative splicing, but the alternative splicing's effect on the expression of the clinical phenotype remains yet to be determined.132) A variety of posttranslational modifications, including phosphorylation, acylation, glycosylation, ubiquitinylation, and disulfide bridges, modify protein function, and thus they can also affect the phenotype. Additional acquired conditions such as toxic exposure, diabetes, arrhythmia, myocarditis, hormonal imbalance, and pregnancy also contribute to the phenotype and outcome.133) Often, genetic determinants increase susceptibility to DCM or alter risk factors in the presence of an external cause. Thus, the line between a genetic disorder and acquired disease becomes increasingly blurred, complicating patients' assessment but also opening new avenues for understanding and treating this etiologically heterogeneous disease (Figures 8 and 9).90)
Figure 8. The diversity of the cardiomyopathies results from genetic, allelic, epigenetic, and environmental heterogeneity, all of which contribute to the phenotype. Hypertrophic cardiomyopathy caused by mutations in genes encoding sarcomeric proteins. TTN truncating variants and LMNA variants are considered to be major factors for the development of dilated cardiomyopathy. Desmosome is major cause of arrhythmogenic cardiomyopathy. The treatment of a genetic cardiomyopathy will depend on the 1) penetrance of the pathogenic variant, 2) the phenotypic presentation, 3) the underlying molecular mechanisms leading cardiac involvement, and 4) the gene-environmental interactions determining the DCM phenotype.
LMNA = lamin A/C; TTN = titin.
Figure 9. Gene-environmental interaction in dilated cardiomyopathies. DCM is a complex multi-factorial disease related to genetic determinants interfering with environmental factors.
DCM = dilated cardiomyopathy; LMNA = lamin A/C; MYH7 = β-myosin heavy chain; PLN = phospholamban; RBM20 = RNA-binding protein 20; TTN = titin.
Prognostic value
According to emerging evidence, variants in LMMA or SCN5A could put individuals at a high risk of developing HF and cardiac arrhythmias. As a result LMNA-related cardiac disease patients are at risk of experiencing SCD, receiving ICD therapy, or suffering from end-stage HF that necessitates heart transplantation.134) Eventhough not all variants in these genes are necessarily arrhythmogenic or cause advanced HF, each variant should be evaluated independently for its potential pathogenicity. Also, guidelines do recommend as Class II to consider ICD implantation in mildly reduced LVEF patients with LMNA variant under certain conditions.
ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY/ARRYTHMOGENIC CARDIOMYOPATHY GENETICS
The prevalence of ARVC in the general population is approximately 1 in 5,000135); however, due to the difficulty of its diagnosis, the disease is not widely recognized.136) In order to standardize the clinical diagnosis of ARVC, the working group revised the guidelines in 2010 to improve diagnostic sensitivity, mainly for the clinical screening of family members; this goal was achieved by providing quantitative criteria for diagnosing right ventricular abnormalities and adding molecular genetic criteria.136) However, due to the low specificity of electrocardiographic abnormalities, the various causes of right ventricular arrhythmias, and the difficulty of using imaging to assess the structure of the right ventricle, challenges in phenotypic diagnosis increases the difficulty in interpreting genetic testing results.137),138) International expert documents highlight the potential limitations of current criteria to propose solutions for better clinical use and identifying potential areas of improvement, such as the identification of early disease in the pediatric population.138)
ARVC is a progressive myocardial disease, which is defined as the replacement of myocardial cells with fat and fibrosis. The disease is related to abnormalities in the structure and function of the right ventricle (RV).137) The classical form of ARVC is an autosomal dominant genetic disease, which accounts for 30–50% of ARVC cases with a variable penetrance. ARVC also includes Naxos disease (a cardiocutaneous syndrome characterized by palmoplantar keratosis, wooly hair, and heart muscle disease)139) and Carvajal syndrome, which are two specific autosomal recessive inherited diseases. While RV disease defines the condition, there are a number of features that may overlap with DCM, as defined in this statement. In particular, the degree of LV involvement ranges from cardiac magnetic resonance imaging (MRI) scarring to severe LV dilation and systolic deterioration. There is also an overlap in causation; for example, PVs in the desmosomal genes in ARVC are also relatively common in patients with a clinical diagnosis of DCM.140) Although the degree of family coexistence of both DCM and ARVC has not been well described, the presence of RV abnormalities, such as ventricular dilatation and ventricular ectopy of RV origin in relatives of patients with DCM, may be a diagnostic red flag (Table 3) for the presence of familial disease.141) Similarly, it is important to keep in mind that the presence of LV dysfunction in a patient with clearly defined ARVC does not imply another disease. Myocarditis with or without a DCM phenotype may resemble ARVC, and this sometimes requires endomyocardial biopsy for proper differential diagnosis.141)
Table 3. Family history of red flags.
| Any of the following findings in the family history should prompt further investigation: |
|---|
| Sudden cardiac death (aged <50 years) |
| Sudden infant death syndrome |
| Unexplained syncope, particularly if it occurs during physical activity |
| Seizures without objective ictal activity |
| Heart transplantation in the absence of coronary artery disease |
| Presence of a pacemaker and/or implantable cardioverter-defibrillator in a young person (aged <50 years) |
| Muscle weakness or myopathy |
| Individual who died of a supposed heart attack at a young age, in the absence of known coronary risk factors and with no autopsy performed. |
Arrhythmogenic right ventricular cardiomyopathy is often caused by pathogenic variants in the desmosome genes
Patients with ARVC usually have genetic abnormalities in the genes encoding desmosomal proteins. Desmosomes are mechanical bridges that link cardiomyocytes. When these proteins are dysfunctional, the mechanical bonds that hold cardiomyocytes together stop functioning. This defect plays a key role in the pathogenesis of fibrofatty replacement of the myocardium and the development of the disease phenotype.137),142) This separation triggers the process of scarring and fat replacement, which can be increased by high physical activity, which explains why it is common in young athletes.143) Pooled data from major studies on molecular genetic screening for desmosomal gene variants have shown that the overall success rate of genetic testing for patients who meet the diagnostic criteria was about 50%.144) A small number of affected patients may have defective nondesmosomal genes, such as TMEM43 and PLN, in certain groups due to the influence of the founder.145),146) The most common causative variants occur in PKP2 (encoding plakophilin-2, 10–45%), followed by DSP (encoding desmoplakin, 10–15%), DSG2 (encoding desmoglein-2, 7–10%), and DSC2 (encoding desmocollin-2, 2%).147),148),149) PVs in these genes have also been associated with ARVC, RCM, DCM, and SCD.150),151) Affected patients usually suffer from HF, advanced atrioventricular and VT that require pacemaker implantation and/or ICD.
Implications of the genetic test in arrhythmogenic right ventricular cardiomyopathy
Guidelines
HRS/EHRA*
(1) Variant-specific cascade screening is recommended. (class I).
(2) Clinical (phenotypic) screening in at-risk 1st degree relatives is recommended (level of evidence A).
(3) Comprehensive or targeted (DSC2, DSG2, DSP, JUP, PKP2, and TMEM43) arrhythmogenic cardiomyopathy (ACM)/ARVC genetic testing can be useful for patients satisfying task force diagnostic criteria for ACM/ARVC (class IIa).
(4) Genetic testing may be considered for patients with possible ACM/ARVC (1 major or 2 minor criteria) according to the 2010 Task Force Criteria (class IIb).
*Updated in 2018 in a joint publication with the ACMG.52)
†Clinical phenotype screening includes the following:
• Medical history, with special attention to HF symptoms, arrhythmias, presyncope or syncope, and thromboembolism.
• Electrocardiography, holter, and echocardiography.
• Cardiac magnetic resonance is recommended if echocardiography is insufficient to define the phenotype.
Diagnostic value
Unlike all other forms of cardiomyopathy, the diagnostic criteria for ARVC include the presence of a pathogenic variant in genes associated with ARVC as the major diagnostic criteria.152) However, it has not been determined which variants have sufficient evidence to be considered as disease-causing; therefore, it may be challenging to define the pathogenicity to a variant. In addition, a negative genetic test does not rule out the possibility that the phenotype is due to a variant in an unknown gene or that the molecular genetic screening technique cannot detect all disease-causing variants, including large deletions or duplications. According to the general recommendations for molecular genetic testing in inherited cardiomyopathies,153) genetic tests will be performed to detect a PV or LPV in a proband who have already fulfills the diagnostic criteria for ARVC phenotype. When the test is positive in an ARVC proband, cascade screening is recommended for family members. Early detection of asymptomatic at-risk individuals can suggest lifestyle changes (restrict vigorous physical exercise) to delay development of the phenotype.
Genotype-phenotype correlation
Based on the current classifications of other cardiomyopathies such as HCM and DCM and according to the 2019 HRS Expert Consensus Statement on arrhythmogenic cardiomyopathy,154) it is appropriate to propose a disease classification that covers various conditions of different etiologies, including: the RV, the LV, or both and either genetic or non-genetic causes, are collectively referred to as “arrhythmogenic cardiomyopathy (ACM).” The common denominator is obvious nonischemic ventricular myocardial scarring and scar-related ventricular arrhythmias. Since myocardial fibrosis acts as a substrate for malignant ventricular arrhythmias, all disease conditions manifesting with the ACM phenotype are associated with a distinctively significantly increased risk of SCD. Therefore, the existence of significant arrhythmogenic ventricular scarring necessitates ICD implantation for patients affected by either genetic or nongenetic ACM, as their primary prevention regardless of the patient’s systolic ventricular function.
Prognostic value (risk stratification)
Patients with multiple PVs in major desmosomal genes (compound or digenic heterozygosity), ranging from 6%155) to 20%,156),157) has been associated with earlier manifestation of disease, a higher prevalence of arrhythmic events, and more frequent LV dysfunction and risk of HF158) than those with a single mutation. It is noteworthy that multiple desmosomal gene mutations and male gender resulted in independent predictors of life threatening arrhythmic events with hazard ratio of 2.3 and 2.9, respectively.159) Women with desmosomal genetic variants have a lower risk of expressing the disease than men and are therefore more likely to be healthy carriers. It is not fully understood why there are differences in the prevalence of sexual penetrance, but there could be differences due to varying levels of participation in competitive and intensive sports among men, or due to the influence of hormones.160)
Nearly 86% of variant-carrying relatives of ARVC patients were asymptomatic at the time of diagnosis. The presence of phenotypic expression has been shown to be a prerequisite for malignant ventricular arrhythmias and SCD. In a cohort of ARVC desmosomal genetic variant carriers who were prospectively investigated during a long-term follow-up (8.5 years), Merner et al.145) found that major arrhythmic events generally occurred in desmosomal gene mutation carriers who fulfilled morpho-functional criteria and had major risk factors. Patients with desmosomal gene mutation and no apparent ARVC phenotype have a relatively benign prognosis. The electrical and structural abnormalities found on CMR in ARVC genetic variant carriers identified those who are at risk for arrhythmic events and may benefit from ICD implantation.152),161)
RESTRICTIVE CARDIOMYOPATHY GENETICS
Although rare, RCM in part shares a genetic basis with HCM. RCM is usually diagnosed by TTE, that demonstrates the characteristic (yet nonspecific) morphological constellation of a nonhypertrophied, nondilated ventricle with preserved ejection function and dilated atria.162) Doppler techniques reveal restrictive filling dynamics of the LV and RV and an abnormally high E/A ratio, indicating accentuated early filling with diminished late filling.
Guidelines52),55)
(1) Obtaining a family history of at least three generations, including the creation of a pedigree, is recommended for all patients with RCM (level of evidence A).
(2) Clinical (phenotypic) screening for cardiomyopathy in at-risk 1st-degree relatives is recommended (level of evidence A).
The frequency of RCM is extremely low, which has limited understanding its genetic characteristics to assessing a small number of genes in small patient groups. Familial RCM is usually passed on in an autosomal dominant manner, but other possible modes of inheritance include autosomal recessive and compound heterozygous forms. Hereditary forms of RCM may not typically be a distinct genetic cardiomyopathy; but may be part of the broad phenotypic spectrum of HCM that is manifested by limited (or absent) hypertrophy and restrictive physiology. A main observation was that the RCM phenotype coexists with HCM-related gene variants as discussed above. Variants have also been described in other sarcomeric genes, including TTNT2, MYBPC3, MYL2 and MYL3, and ACTC1.163),164) Menon et al.81) have reported a PV in TTNT2 that is inherited in an autosomal dominant manner and may present with three distinct clinical phenotypes: RCM, HCM, or DCM. Linkage analysis followed by sequencing identified a missense variant resulting in p.Ile89Asn substitution in TTNT2. De novo sarcomeric variants appear to be associated with severe disease expression and premature death or heart transplantation in childhood.165) Nonsarcomeric variants have also recently been identified in RCM. PVs in MYPN and TTN may cause myofibrillar disarray and restrictive physiology.166) The mechanism proposed for the progress of RCM is altered myofibrillogenesis. PVs in FLNC have also been recognized in several families and demonstrate autosomal dominant inheritance.167)
In addition, various systemic diseases that distress the heart can mimic the RCM phenotype, including hereditary amyloidosis (mainly caused by PVs in TTR gene, but also in CST3, FGA, LYZ, GSN, APOA2, and APOA1), metabolic disorders such as glycogen storage diseases, Fabry disease (GLA), hemochromatosis (HFE), cystinosis, sarcoidosis, lymphoma, Danon's disease (LAMP2), and PRKAG2-mediated heart disease.168)
It is also important that clinicians be aware that there are forms of familial RCM for which a recognized genetic basis does not exist, and some RCM patients with an identified variant will not have an affected family member due to variable penetrance.
Implications of the genetic test in restrictive cardiomyopathy
The yield of genetic tests in RCM remains undetermined. It is presently unclear whether genetic testing has any value in classifying the risk stratification for patients with isolated RCM, but it can be important when evaluating patients with secondary causes of RCM, such as infiltrative diseases (amyloidosis, sarcoidosis) or storage disorders (Gaucher disease, glycogen storage disease, and Fabry disease).162) In addition, it is crucial to ascertain the genetic status of asymptomatic members to determine their potential risk of developing RCM or other cardiomyopathy phenotypes later in life.
LEFT VENTRICULAR NONCOMPACTION CARDIOMYOPATHY GENETICS
LVNC is cardiomyopathy characterized by prominent trabeculations of the LV with deep intertrabecular recesses and thinning of the compact epicardium.169) Current imaging diagnostic criteria, including the most commonly used echocardiographic Jenni criteria, are based on the ratio between a severely thickened myocardium. This myocardium has a noncompacted layer that is at least twice as thick as the compacted layer, as measured in systole in the short-axis view.170) LVNC is thought to occur in approximately 1 in 7,000 live births and occurs in newborns, young children, and adults. Major adverse events include life-threatening arrhythmias, thromboembolism, and HF. Infants who have conditions such as systemic disease and metabolic derangement have reportedly experienced the worst outcomes. It is estimated that 30–50% of cases of LVNC are genetic, as each year differently sized cohorts of pediatric and adult cases are studied. Autosomal dominant transmission is the most common mode of inheritance, followed by X-linked and maternal modes.171),172),173),174) In addition, sporadic cases are common and are thought to be involved in 60–70% of cases. Although several known LVNC susceptibility genes have been identified, no single susceptibility gene predominates, and systematic evaluations of large populations have not been conducted to date. Variants in more than 15 genes encoding sarcomeric proteins (MYH7, MYBPC3, ACTC1, TNNT2, and TPM1),175),176),177) ion channel proteins (HCN4, SCN5A),178),179) and other proteins have been linked to LVNC. Previous studies have identified that sarcomere-encoding genes are most common in approximately 32–41% of the adult cohorts with LVNC.173),180) These studies, however, used various definitions for LVNC, and many did not account for the co-occurrence of hypertrophic or DCM. Among the LVNC genes, MYH7, MYBPC3, and TTN are the most prevalent and are also frequent causes of HCM and DCM. Previous family studies of LVNC occasionally reported relatives with HCM or DCM, apparently without noncompaction, in familial LVNC. Waning et al. showed that cardiac features and genetic defects in index cases may help to predict the cardiomyopathy phenotype and associated risk for relatives. Familial segregation of isolated LVNC, LVNC with DCM, or LVNC with HCM have been observed, and families in which a range of different LVNC phenotypes occurred have been studied. When the index case had isolated LVNC, relatives were more likely to have similarly isolated LVNC, which was linked to a better prognosis with less RV and LV systolic dysfunction and a low risk for MACEs.173) Almost half of the patients with isolated LVNC had a variant, mainly in the head domain of MYH7. LVNC with DCM was the most common LVNC phenotype and variants in the tail domain of MYH7 and in TTN, which are associated with increased risk for LV systolic dysfunction and MACEs.173) Chances of cardiac symptoms, LV systolic dysfunction, and MACEs increased in the first year of life for children with a variant. In contrast, risk of MACEs was low in children with sporadic LVNC. LV systolic dysfunction is a high risk for MACEs in adults with a variant. Additionally, nuclear proteins LMNA and RBM20 pathogenic variants were associated with worse outcomes.171),172) HCN4 variants may also cause ventricular fibrillation (VF) in patients with LVNC. It is important to keep in mind that while the age of manifestation and the phenotype displayed by those affected by a LVNC-associated variant may differ in families.181) As well, genetic testing is necessary to make an early diagnosis of syndromic forms of LVNC (i.e. Danon disease), allowing for early medical interventions and family planning. Interestingly, some patients in the same family can exhibit different phenotypes with the same variant (variable expression of the disease), varying from LVNC to HCM or DCM.
Implications of the genetic test in left ventricular non-compaction
Diagnostic value
The diagnostic yield of genetic testing in adult proband patients with LVNC is low. In patient with LVNC accompanied by cardiac and syndromic features, genetic testing is most helpful for accurate diagnosis.182) In addition, identification of a definite PV in clinically affected probands should lead to screening of all at-risk individuals (siblings, parents, children), including those with a negative phenotype. One of the important aspects of genetic testing is the ability to confirm PV-positive or PV-negative status in family members of clinically affected, genotype-positive subjects. Due to possible X-linked inheritance in some patients, affected male family members may have a noticeable disease expression, while females may have milder signs of LVNC (or none at all); thus, these females may have a role as transmitters for subsequent generations.182)
Prognostic role of genetic testing for left ventricular non-compaction
Thus far, genotype-phenotype correlations has not been well characterized in LVNC; therefore, prognostic value of genetic testing is unclear.
IMPORTANT INHERITED SYNDROMES ASSOCIATED WITH CARDIAC DISEASE
Table 4 summarizes the most frequently implicated genes and clinical features in inherited syndromes associated with cardiac disease.183),184),185),186)
Table 4. Inherited syndromes associated with cardiac disease.
| Gene | Protein | Disease | Clinical features | Note |
|---|---|---|---|---|
| DMD | Large sarcolemmal protein dystrophin. | Duchenne muscular dystrophy | Early-onset DCM with history of muscular dystrophy | X-linked, recessive |
| Duchenne: 1:4,000 to 1:6,000 live male births182) | ||||
| Becker muscular dystrophy | Milder and later onset age of onset of DCM tends to be later | Becker: 1:18,000 live male births183) | ||
| Mitochondria | Supply and regulation of energy metabolism | MERRF syndrome | Myoclonic epilepsy, septal hypertrophy and DCM | Maternal transmission or autosomal dominant |
| MELAS syndrome | Mitochondrial encephalopathy | Maternal transmission or autosomal dominant | ||
| Lactic acidosis | ||||
| Stroke like episodes | ||||
| Heart failure | ||||
| Symmetrical LVH | ||||
| Kearns-Sayre syndrome | Ophalomoplegia, conduction defects and DCM, heart block | Maternal transmission or autosomal dominant | ||
| FRDA/FXN | Frataxin: a mitochondrial protein of iron homeostasis | Friedreich ataxia | Cerebellar ataxia, dysarthria, HCM, diabetes, neurosensory hearing loss, and visual impairment supraventricular tachycardia | Spinocerebellar ataxia |
| PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2 | PRKAG2-related cardiomyopathy | Symmetrical nonobstructive hypertrophy, diastolic dysfunction184),185) | Skeletal myopathy |
| TTR | Transthyretin | Amyloidosis | ECG: pseudo-infarct pattern, low-voltage QRS, conduction abnormalities | Neuropathy, autonomic dysfunction, Carpal tunnel syndrome, renal failure and proteinuria |
| Echo: LVH+right ventricular hypertrophy | ||||
| MRI: global subendocardial or transmural late enhancement, suboptimal myocardial nulling | ||||
| LAMP2 | Lysosome-associated membrane glycoprotein 2 | Danon disease | ECG: high-voltage QRS, preexcitation pattern, short PR interval | Skeletal myopathy, intellectual disability |
| Echo: LVH | ||||
| PTPN11RAF1 | Noonan syndrome | ECG: left or extreme right axis deviation | Typical facies, short stature, webbed neck, pectus excavatum/carinatum, developmental delay, bleeding disorders | |
| Echo: congenital heart defects (mainly pulmonary stenosis and hypertrophic cardiomyopathy) | ||||
| GLA | Galactosidase Alpha | Fabry disease | ECG: LVH, high-voltage QRS, short PR interval, preexcitation, sinus node dysfunction, conduction abnormalities, atrioventricular block, supraventricular tachyarrhythmia8 | Angiokeratoma, cornea verticillata, neuropathic pain, stroke/transient ischemic attack, vertigo, tinnitus, hearing impairment, renal failure and proteinuria, GI symptoms |
| Echo: LVH |
DCM = dilated cardiomyopathy; ECG = electrocardiography; GI = gastrointestinal; LVH = left ventricular hypertrophy; MELAS = mitochondrial encephalo-myopathy with lactic acidosis and stroke-like episodes; MERRF = myoclonic epilepsy with ragged red fibers; MRI = magnetic resonance imaging.
Neuromuscular disease
Neuromuscular diseases may accompany cardiomyopathies; in some forms of neuromuscular disease, irregular heart rhythm may be the critical feature symptom. Patients with PVs in the LMNA gene may or may not have muscle disease, and it ranges from limb-girdle muscular dystrophy to Emery–Dreifuss muscular dystrophy, which is usually associated with contractures of the elbows and Achilles tendon.187),188) Cardiac involvement includes progressive arrhythmias (brady/tachyarrhythmias, SCD) and often progresses to DCM. The genetic variants in DMD that encode dystrophin, are present in individuals who have an X-linked recessive pattern of inheritance. Dystrophin connects sarcomeric proteins to the plasma membrane via the dystroglycan complex. Duchenne muscular dystrophy is caused by muscle contraction, which results in the loss of cell membrane integrity, ultimately resulting in cell death and the replacement of the affected muscle tissue with fibro-fatty tissue.189) Duchenne muscular dystrophy cardiomyopathy usually manifests in the second decade of life, and screening for cardiomyopathy is crucial and is performed yearly in adult patients.190) Friedreich ataxia is an autosomal recessive inheritance caused by the loss of function variants in the FXN gene. The key clinical manifestations are neurologic dysfunction (e.g., progressive ataxia of limbs), HF and diabetes mellitus. Echocardiographic and electrocardiographic abnormalities are those of morphologically mild asymmetric septal hypertrophy with gradually systolic dysfunction.191),192)
Mitochondrial myopathies
Mitochondrial myopathies have 2 inheritance forms: maternal inheritance and autosomal dominant transmission.193) The final diagnosis is only made after pathological confirmation, including mitochondrial quantity and function or genetic testing.194) Mitochondrial DNA-related diseases constitute approximately 3% of DCM cases. MELAS is the syndrome of mitochondrial encephalo-myopathy with lactic acidosis and stroke-like episodes, and it is a maternally inherited multisystemic disorder caused by PVs in mitochondrial DNA. MELAS frequently manifests in childhood after normal early development.195) Cardiac involvement is occurred in over 1/3 of patients with MELAS syndrome and exhibits a bimodal age-related distribution with different final outcomes. Adult-onset cardiomyopathy appears to be a mild and nonprogressive mid-term manifestation.196)
Amyloid cardiomyopathy
Cardiac disease due to the deposition of amyloid fibrils is seen in familial amyloid cardiomyopathy and hereditary TTR amyloidosis, as well as in nongenetic diseases (e.g., senile TTR amyloidosis). The cardiac manifestations of these diseases have been reviewed in prior articles.197),198)
Fabry disease
Fabry disease is an X-linked genetic multisystem disorder that results in a deficiency of the enzyme alpha galactosidase. Cardiac manifestations may be the predominant clinical features, usually with a phenotype of HCM and may also present in females.199),200),201)
IMPORTANT CONSIDERATIONS IN GENETIC CARDIOMYOPATHIES
The diversity of cardiomyopathies results from the multifactorial involvement of genetic, allelic, epigenetic, and environmental variables, all of which play a role in determining the cardiomyopathy phenotype.
Incomplete and age-related penetrance
Inherited cardiomyopathies typically exhibit significant phenotypic variability, and families can show greater diversity in their manifestations. Penetrance—the percentage of PV carriers with clinically detectable disease—increases with age but remains below 100%. Hypertrophy occurs in the majority of patients with HCM during adolescence, whereas sarcomeric DCM has a bimodal age of onset (with peaks during childhood and mid-adulthood).202) It is infrequent to find multiple family members with clinically apparent ARVC in a single pedigree, suggesting a reduced penetrance. Therefore, it is difficult to identify family members with subtle phenotypes, because it may take decades to develop a reliable and robust phenotype. In addition, there is increasing evidence that, regardless of the phenotype, certain cardiomyopathy-related variants have a high arrhythmia prevalence, which make cascade genetic screening a key tool for identifying high-risk individuals in affected families.203) The risk of developing inherited cardiac disease is affected by multiple genetic and environmental factors, and every case/family must be individually evaluated by a multidisciplinary expert team.
Missing causal genes
It is difficult to identify the remaining causal genes, usually called missing causal genes, which are usually related to cardiomyopathy manifesting as sporadic cases or in small families. The missing causal gene in cardiomyopathy might partly be due to the difficulty in determining the causality of the genetic variants in sporadic cases and small families.204)
Generally, the effect size gradient of genetic variants ranges from a large causal relationship to an insignificant scale.205),206),207) Genetic variants that have influential large effect sizes are considered to be PVs accountable for autosomal dominant single-gene disorders in large families including cardiomyopathy. Through robust genetic techniques, such as cosegregation and linkage analyses, such PVs have been mapped and identified. The MYH7 and MYBPC3 genetic variants are good examples. Clinically small or subtle effects on the genetic level can be categorized as ‘effect sizes’ on the other end of the spectrum. When it is in the functional state, such variants have a moderate effect on the disorders in phenotype, but do not cause monogenic disorders. The center of the spectrum represents a subset of genetic variants with intermediate to large effect sizes and incomplete penetrance; their effects are influenced by other genetic and nongenetic factors. Although it is believed that these variants cause cardiomyopathy, the penetrance of these variants is insufficient or low. There is no clear cosegregation of the genetic variant and the phenotype in this case; thus, it is impossible to establish a strong linkage, even more so when the family size is small and the disease is sporadic. This difficulty is partly due to the large number of variants in each genome. One strategy for identifying missing causal genes is to conduct large-scale association studies followed by rare variant analysis. The evidence for an association between a candidate gene and cardiomyopathy in a discovery population is considered tentative (i.e., hypothesis-generating) and requires replication in a separate population.
Modifier genes
The variability in the phenotype of cardiomyopathy is partly attributed to the influence of modifier genetic variants and environmental factors. Each human nuclear genome contains, on average, approximately 11,000 nonsynonymous variants, 160 premature protein truncation variants, and 500,000 variants in known regulatory regions.208),209) PVs in genes and pathways implicated in regulating cardiac hypertrophy and fibrosis could influence the expression of the cardiomyopathy phenotype and function as modifier genetic variants. Modifier genes are genes other than the causal genes that affect the phenotypic expression of genetic disorders. Modifier genes are the genetic background of affected individuals, which differs because of DNA polymorphisms. Modifier genes are neither sufficient nor necessary to cause cardiomyopathy, but rather affect the severity of cardiomyopathy phenotypes, such as the degree of hypertrophy and the risk of SCD.
Epidemiological and family studies in humans210),211) and experimental data in animals212) have shown that the genetic background has an impact on the degree of cardiac hypertrophy observed in response to the hypertrophic stimulus. Clinical observations show a significant degree of variability in the phenotypic expression of hypertrophy, sudden death, and cardiac failure in patients with cardiomyopathy, a genetic model of the cardiac hypertrophic response; this variability underscores the influence of modifier genes on cardiac phenotypes. A large degree of heterogeneity in the phenotypic expression of the disease is readily apparent when individuals who have the same causative variants or family members who share the same variations all have varied outcomes. The final phenotype is the product of causal variants, modifier genes, and environmental factors. Thus, given the significance of the modifier genes in modulating cardiac phenotypes in cardiomyopathy, their identification and characterization of their role will complement the primary (causal) variant in predicting prognosis and could be essential for designing specific and more comprehensive therapies.
COUNSELING BEFORE GENETIC TESTING
Genetic counseling is recommended for all patients with cardiomyopathy and their family members. It is essential to take a multigenerational (preferably at least three generations) family history of cardiomyopathy and suspected SCD events. All patients with cardiomyopathy should receive counseling before a genetic test, ideally by an experienced physician with knowledge of cardiomyopathy genetics, such as a genetic counselor. Through use of this strategy, patients will understand the rationale for testing, gain empowerment. They will also be better equipped to communicate and deal with the dynamics within their families. Given that patients often communicate the results and implications of genetic testing to at-risk family members, their understanding of this information is imperative. Pretest genetic counseling is informed by the benefits and potential harm (including psychosocial, ethical, and insurability) of finding a genetic cause of the disease. Many patients may face financial stress because of the high cost of optional genetic tests that do not directly affect healthcare. Ensuring that patients have a full understanding of financial and insurance implications of genetic testing is thus of critical importance, as private insurance coverage is not consistent. Theoretically, a genetic counselor is qualified to give financial advice about genetic testing, but most genetic counselors are not trained to provide financial advice on genetic testing, and they are not certified in this area.
CHOICE OF GENETIC TEST
Genetic testing, if positive, will assist the diagnosis in a proband; however, if the test is negative, it will not be ruled out. If a proband with a positive test is identified, cascade screening, or testing for the presence of the variant in family members should be undertaken. A positive screen, that is an identification of a relative carrying the same PV as detected in the proband with cardiomyopathy, should lead to a detailed examination for the presence of the cardiomyopathy phenotype. If the phenotype is manifested, the relative may be diagnosed with cardiomyopathy and treated appropriately. Relatives who are PV carriers but do not have the phenotype should undergo periodic clinical evaluations (history, physical examination, electrocardiogram, and echocardiogram), or more frequently if symptoms develop. Improved genome sequencing approaches have resulted in a remarkable increase in the speed and efficiency of testing and reduced costs, which has transformed our insights into cardiomyopathy variant genomic disease.213),214),215)
Next-generation sequencing (NGS) of cardiomyopathy genes for standard panels has emerged as a rapid testing alternative to Sanger sequencing, providing analytical characteristics for the comprehensive exploration of genetic causes.215) Various commercially available gene panels are routinely used to identify genetic variants in cardiomyopathies. On the other hand, NGS has some shortcomings, including a lack of representation and coverage of cartain exons; this raises the risk of reduced sensitivity and the potential for false negatives with clinically relevant PV detection. As a result, the procedure that first required target enrichment and then NGS for high-throughput genetic testing of disease genes for DCM, HCM, and other cardiomyopathies has now become possible and has technically been validated by the nearly complete coverage and high accuracy of the approach.
Inherent to more extended genetic screening using whole exome sequencing (WES) and whole genome sequencing (WGS) is the detection of more variants of unknown significance and variants in genes that are unlikely to be associated with the phenotype. It is not recommended to report these results back to the patients on a routine basis. Careful variant classification according to the ACMG guidelines5) is mandatory, and genetic counseling plays a central role in validating variants through familial segregation analysis, as well as in discussing the limits of risk stratification for relatives. Genetic counseling should include discussions on potential reanalysis and even retesting after predefined intervals in gene-elusive patients with a high suspicion of genetic disease.
Guided but systematic implementation of genetic testing in clinical practice is expected to allow us to precisely distinguish cardiomyopathies within broad phenotypes, improve the effectiveness of counseling for patients and their relatives, and better inform important clinical decisions, such as defibrillator implantation.
CONCLUSIONS
Although it is now easy to obtain human genome sequence data, the approach is far from perfect; this has encouraged the notion of incorporating genomic data into routine health care in order to advance precision medicine. Increasing numbers of rare genetic variants have been validated, and new common genetic markers for the risk of cardiomyopathy have been identified. The usefulness of the research depends on answering those questions, which in turn are contingent on whether and when other patients and relatives who have detrimental genetic variants should be treated, and how to investigate therapeutic efficacy. Despite these difficulties, a new clinical framework for future evidence-based and personalized HF care in cardiomyopathy is already emerging from the integration of comprehensive sequence analyses with detailed phenotyping. Genotype-phenotype correlations may seem inconsistent because traditional phenotypes (LVH, HF, and SCD) are late manifestations of the disease. Studying the earlier manifestations of a disease allows researchers to establish stronger correlations, discover more mechanistic pathways, and potentially find disease-modifying treatments. It is our hope that future research will allow the use of genetic testing to identify variants and subsequently improve treatment outcomes.
ACKNOWLEDGMENTS
We would like to thank Chang-Seok Ki, MD, PhD and Jin-Sung Lee for their thoughtful comments on the manuscript and for their critical suggestions, discussion, and manuscript review.
Footnotes
Funding: This research was supported from the Korean Cardiac Research Foundation (202003-05).
Conflict of Interest: The authors have no financial conflicts of interest.
Data Sharing Statement: The data generated in this study is available from the corresponding author(s) upon reasonable request.
- Conceptualization: Kim KH, Pereira NL.
- Writing - original draft: Kim KH.
- Writing - review & editing: Pereira NL.
References
- 1.Reichart D, Magnussen C, Zeller T, Blankenberg S. Dilated cardiomyopathy: from epidemiologic to genetic phenotypes: a translational review of current literature. J Intern Med. 2019;286:362–372. doi: 10.1111/joim.12944. [DOI] [PubMed] [Google Scholar]
- 2.Sabater-Molina M, Pérez-Sánchez I, Hernández Del Rincón JP, Gimeno JR. Genetics of hypertrophic cardiomyopathy: a review of current state. Clin Genet. 2018;93:3–14. doi: 10.1111/cge.13027. [DOI] [PubMed] [Google Scholar]
- 3.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 4.Bennette CS, Gallego CJ, Burke W, Jarvik GP, Veenstra DL. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015;17:587–595. doi: 10.1038/gim.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hershberger RE, Givertz MM, Ho CY, et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J Card Fail. 2018;24:281–302. doi: 10.1016/j.cardfail.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242–255. doi: 10.1016/S0140-6736(12)60397-3. [DOI] [PubMed] [Google Scholar]
- 8.Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–1254. doi: 10.1016/j.jacc.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA study. Coronary artery risk development in (Young) adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 10.Ommen SR, Mital S, Burke MA, et al. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2020;76:e159–240. doi: 10.1016/j.jacc.2020.08.045. [DOI] [PubMed] [Google Scholar]
- 11.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 12.Ho CY. Genetics and clinical destiny: improving care in hypertrophic cardiomyopathy. Circulation. 2010;122:2430–2440. doi: 10.1161/CIRCULATIONAHA.110.978924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 14.Marian AJ, Braunwald E. Hypertrophic cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res. 2017;121:749–770. doi: 10.1161/CIRCRESAHA.117.311059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmannova H, Kubanek M, Sramko M, et al. Isolated X-linked hypertrophic cardiomyopathy caused by a novel mutation of the four-and-a-half LIM domain 1 gene. Circ Cardiovasc Genet. 2013;6:543–551. doi: 10.1161/CIRCGENETICS.113.000245. [DOI] [PubMed] [Google Scholar]
- 16.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 17.Seidman CE, Seidman JG. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: a personal history. Circ Res. 2011;108:743–750. doi: 10.1161/CIRCRESAHA.110.223834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita H, Nagai R, Seidman JG, Seidman CE. Sarcomere gene mutations in hypertrophy and heart failure. J Cardiovasc Transl Res. 2010;3:297–303. doi: 10.1007/s12265-010-9188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuusisto J. Genetics of hypertrophic cardiomyopathy: what is the next step? Heart. 2020;106:1291–1292. doi: 10.1136/heartjnl-2020-317043. [DOI] [PubMed] [Google Scholar]
- 20.Kaski JP, Syrris P, Esteban MT, et al. Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2009;2:436–441. doi: 10.1161/CIRCGENETICS.108.821314. [DOI] [PubMed] [Google Scholar]
- 21.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108:751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlossarek S, Mearini G, Carrier L. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: mechanisms and therapeutic opportunities. J Mol Cell Cardiol. 2011;50:613–620. doi: 10.1016/j.yjmcc.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Erdmann J, Daehmlow S, Wischke S, et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin Genet. 2003;64:339–349. doi: 10.1034/j.1399-0004.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 24.Walsh R, Buchan R, Wilk A, et al. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur Heart J. 2017;38:3461–3468. doi: 10.1093/eurheartj/ehw603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Liu W, Hu D, et al. Mutation spectrum in a large cohort of unrelated Chinese patients with hypertrophic cardiomyopathy. Am J Cardiol. 2013;112:585–589. doi: 10.1016/j.amjcard.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 26.Curila K, Benesova L, Penicka M, et al. Spectrum and clinical manifestations of mutations in genes responsible for hypertrophic cardiomyopathy. Acta Cardiol. 2012;67:23–29. doi: 10.1080/ac.67.1.2146562. [DOI] [PubMed] [Google Scholar]
- 27.Andersen PS, Havndrup O, Hougs L, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum Mutat. 2009;30:363–370. doi: 10.1002/humu.20862. [DOI] [PubMed] [Google Scholar]
- 28.Marston S, Copeland O, Jacques A, et al. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 29.van Dijk SJ, Dooijes D, dos Remedios C, et al. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 30.Geier C, Gehmlich K, Ehler E, et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum Mol Genet. 2008;17:2753–2765. doi: 10.1093/hmg/ddn160. [DOI] [PubMed] [Google Scholar]
- 31.Niimura H, Patton KK, McKenna WJ, et al. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 32.Chiu C, Bagnall RD, Ingles J, et al. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J Am Coll Cardiol. 2010;55:1127–1135. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Geier C, Perrot A, Özcelik C, et al. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–1395. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- 34.Bos JM, Poley RN, Ny M, et al. Genotype-phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Mol Genet Metab. 2006;88:78–85. doi: 10.1016/j.ymgme.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedrich FW, Wilding BR, Reischmann S, et al. Evidence for FHL1 as a novel disease gene for isolated hypertrophic cardiomyopathy. Hum Mol Genet. 2012;21:3237–3254. doi: 10.1093/hmg/dds157. [DOI] [PubMed] [Google Scholar]
- 36.Osio A, Tan L, Chen SN, et al. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007;100:766–768. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maron MS, Rowin EJ, Olivotto I, et al. Contemporary natural history and management of nonobstructive hypertrophic cardiomyopathy. J Am Coll Cardiol. 2016;67:1399–1409. doi: 10.1016/j.jacc.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 38.Rowin EJ, Maron BJ, Carrick RT, et al. Outcomes in patients with hypertrophic cardiomyopathy and left ventricular systolic dysfunction. J Am Coll Cardiol. 2020;75:3033–3043. doi: 10.1016/j.jacc.2020.04.045. [DOI] [PubMed] [Google Scholar]
- 39.Kubo T, Gimeno JR, Bahl A, et al. Prevalence, clinical significance, and genetic basis of hypertrophic cardiomyopathy with restrictive phenotype. J Am Coll Cardiol. 2007;49:2419–2426. doi: 10.1016/j.jacc.2007.02.061. [DOI] [PubMed] [Google Scholar]
- 40.Mogensen J, Arbustini E. Restrictive cardiomyopathy. Curr Opin Cardiol. 2009;24:214–220. doi: 10.1097/hco.0b013e32832a1d2e. [DOI] [PubMed] [Google Scholar]
- 41.Mogensen J, Kubo T, Duque M, et al. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111:209–216. doi: 10.1172/JCI16336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostareva A, Gudkova A, Sjöberg G, et al. Deletion in TNNI3 gene is associated with restrictive cardiomyopathy. Int J Cardiol. 2009;131:410–412. doi: 10.1016/j.ijcard.2007.07.108. [DOI] [PubMed] [Google Scholar]
- 43.Hwang JW, Jang MA, Jang SY, et al. Diverse phenotypic expression of cardiomyopathies in a family with TNNI3 p.Arg145Trp mutation. Korean Circ J. 2017;47:270–277. doi: 10.4070/kcj.2016.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD. Mutations in troponin that cause HCM, DCM AND RCM: what can we learn about thin filament function? J Mol Cell Cardiol. 2010;48:882–892. doi: 10.1016/j.yjmcc.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 45.Yang Z, McMahon CJ, Smith LR, et al. Danon disease as an underrecognized cause of hypertrophic cardiomyopathy in children. Circulation. 2005;112:1612–1617. doi: 10.1161/CIRCULATIONAHA.105.546481. [DOI] [PubMed] [Google Scholar]
- 46.Gollob MH, Green MS, Tang AS, et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 47.Blair E, Redwood C, Ashrafian H, et al. Mutations in the γ2 subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet. 2001;10:1215–1220. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 48.Wu X, Simpson J, Hong JH, et al. MEK-ERK pathway modulation ameliorates disease phenotypes in a mouse model of Noonan syndrome associated with the Raf1L613V mutation. J Clin Invest. 2011;121:1009–1025. doi: 10.1172/JCI44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaski JP, Syrris P, Shaw A, et al. Prevalence of sequence variants in the RAS-mitogen activated protein kinase signaling pathway in pre-adolescent children with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2012;5:317–326. doi: 10.1161/CIRCGENETICS.111.960468. [DOI] [PubMed] [Google Scholar]
- 50.Bos JM, Towbin JA, Ackerman MJ. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54:201–211. doi: 10.1016/j.jacc.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 51.Authors/Task Force members. Elliott PM, Anastasakis A, et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur Heart J. 2014;35:2733–2779. doi: 10.1093/eurheartj/ehu284. [DOI] [PubMed] [Google Scholar]
- 52.Hershberger RE, Givertz MM, Ho CY, et al. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2018;20:899–909. doi: 10.1038/s41436-018-0039-z. [DOI] [PubMed] [Google Scholar]
- 53.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 54.Bagnall RD, Ingles J, Dinger ME, et al. Whole genome sequencing improves outcomes of genetic testing in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2018;72:419–429. doi: 10.1016/j.jacc.2018.04.078. [DOI] [PubMed] [Google Scholar]
- 55.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 56.Ingles J, Burns C, Bagnall RD, et al. Nonfamilial hypertrophic cardiomyopathy: prevalence, natural history, and clinical implications. Circ Cardiovasc Genet. 2017;10:e001620. doi: 10.1161/CIRCGENETICS.116.001620. [DOI] [PubMed] [Google Scholar]
- 57.Ingles J, Goldstein J, Thaxton C, et al. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med. 2019;12:e002460. doi: 10.1161/CIRCGEN.119.002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maron BJ. Clinical course and management of hypertrophic cardiomyopathy. N Engl J Med. 2018;379:655–668. doi: 10.1056/NEJMra1710575. [DOI] [PubMed] [Google Scholar]
- 59.Loar RW, Bos JM, Will ML, Ommen SR, Ackerman MJ. Genotype-phenotype correlations of hypertrophic cardiomyopathy when diagnosed in children, adolescents, and young adults. Congenit Heart Dis. 2015;10:529–536. doi: 10.1111/chd.12280. [DOI] [PubMed] [Google Scholar]
- 60.Lafreniere-Roula M, Bolkier Y, Zahavich L, et al. Family screening for hypertrophic cardiomyopathy: is it time to change practice guidelines? Eur Heart J. 2019;40:3672–3681. doi: 10.1093/eurheartj/ehz396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Norrish G, Jager J, Field E, et al. Yield of clinical screening for hypertrophic cardiomyopathy in child first-degree relatives: evidence for a change in paradigm. Circulation. 2019;140:184–192. doi: 10.1161/CIRCULATIONAHA.118.038846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erdmann J, Raible J, Maki-Abadi J, et al. Spectrum of clinical phenotypes and gene variants in cardiac myosin-binding protein C mutation carriers with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2001;38:322–330. doi: 10.1016/s0735-1097(01)01387-0. [DOI] [PubMed] [Google Scholar]
- 63.Epstein ND, Cohn GM, Cyran F, Fananapazir L. Differences in clinical expression of hypertrophic cardiomyopathy associated with two distinct mutations in the beta-myosin heavy chain gene. A 908Leu----Val mutation and a 403Arg----Gln mutation. Circulation. 1992;86:345–352. doi: 10.1161/01.cir.86.2.345. [DOI] [PubMed] [Google Scholar]
- 64.Charron P, Dubourg O, Desnos M, et al. Clinical features and prognostic implications of familial hypertrophic cardiomyopathy related to the cardiac myosin-binding protein C gene. Circulation. 1998;97:2230–2236. doi: 10.1161/01.cir.97.22.2230. [DOI] [PubMed] [Google Scholar]
- 65.Watkins H, Rosenzweig A, Hwang DS, et al. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med. 1992;326:1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 66.Marian AJ, Mares A, Jr, Kelly DP, et al. Sudden cardiac death in hypertrophic cardiomyopathy. Variability in phenotypic expression of beta-myosin heavy chain mutations. Eur Heart J. 1995;16:368–376. doi: 10.1093/oxfordjournals.eurheartj.a060920. [DOI] [PubMed] [Google Scholar]
- 67.Marian AJ, Roberts R. The molecular genetic basis for hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2001;33:655–670. doi: 10.1006/jmcc.2001.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marian A. Molecular genetic basis of hypertrophic cardiomyopathy. Circ Res. 2021;128:1533–1553. doi: 10.1161/CIRCRESAHA.121.318346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anan R, Greve G, Thierfelder L, et al. Prognostic implications of novel beta cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J Clin Invest. 1994;93:280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adalsteinsdottir B, Burke M, Maron BJ, et al. Hypertrophic cardiomyopathy in myosin-binding protein C (MYBPC3) Icelandic founder mutation carriers. Open Heart. 2020;7:e001220. doi: 10.1136/openhrt-2019-001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niimura H, Bachinski LL, Sangwatanaroj S, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 72.Watkins H, McKenna WJ, Thierfelder L, et al. Mutations in the genes for cardiac troponin T and α-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 73.Karibe A, Tobacman LS, Strand J, et al. Hypertrophic cardiomyopathy caused by a novel α-tropomyosin mutation (V95A) is associated with mild cardiac phenotype, abnormal calcium binding to troponin, abnormal myosin cycling, and poor prognosis. Circulation. 2001;103:65–71. doi: 10.1161/01.cir.103.1.65. [DOI] [PubMed] [Google Scholar]
- 74.Poetter K, Jiang H, Hassanzadeh S, et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 75.Flavigny J, Richard P, Isnard R, et al. Identification of two novel mutations in the ventricular regulatory myosin light chain gene (MYL2) associated with familial and classical forms of hypertrophic cardiomyopathy. J Mol Med (Berl) 1998;76:208–214. doi: 10.1007/s001090050210. [DOI] [PubMed] [Google Scholar]
- 76.Satoh M, Takahashi M, Sakamoto T, Hiroe M, Marumo F, Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem Biophys Res Commun. 1999;262:411–417. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 77.Mogensen J, Klausen IC, Pedersen AK, et al. α-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103:R39–R43. doi: 10.1172/JCI6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marian AJ, Salek L, Lutucuta S. Molecular genetics and pathogenesis of hypertrophic cardiomyopathy. Minerva Med. 2001;92:435–451. [PMC free article] [PubMed] [Google Scholar]
- 79.Marian AJ. Pathogenesis of diverse clinical and pathological phenotypes in hypertrophic cardiomyopathy. Lancet. 2000;355:58–60. doi: 10.1016/s0140-6736(99)06187-5. [DOI] [PubMed] [Google Scholar]
- 80.Havndrup O, Bundgaard H, Andersen PS, et al. The Val606Met mutation in the cardiac beta-myosin heavy chain gene in patients with familial hypertrophic cardiomyopathy is associated with a high risk of sudden death at young age. Am J Cardiol. 2001;87:1315–1317. doi: 10.1016/s0002-9149(01)01532-6. [DOI] [PubMed] [Google Scholar]
- 81.Menon SC, Michels VV, Pellikka PA, et al. Cardiac troponin T mutation in familial cardiomyopathy with variable remodeling and restrictive physiology. Clin Genet. 2008;74:445–454. doi: 10.1111/j.1399-0004.2008.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80:739–744. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 83.Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 84.Pinto YM, Elliott PM, Arbustini E, et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37:1850–1858. doi: 10.1093/eurheartj/ehv727. [DOI] [PubMed] [Google Scholar]
- 85.Taylor MR, Carniel E, Mestroni L. Cardiomyopathy, familial dilated. Orphanet J Rare Dis. 2006;1:27. doi: 10.1186/1750-1172-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosenbaum AN, Agre KE, Pereira NL. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol. 2020;17:286–297. doi: 10.1038/s41569-019-0284-0. [DOI] [PubMed] [Google Scholar]
- 87.Haas J, Frese KS, Peil B, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–1135. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 88.Tobita T, Nomura S, Fujita T, et al. Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling. Sci Rep. 2018;8:1998. doi: 10.1038/s41598-018-20114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 90.Bondue A, Arbustini E, Bianco A, et al. Complex roads from genotype to phenotype in dilated cardiomyopathy: scientific update from the Working Group of Myocardial Function of the European Society of Cardiology. Cardiovasc Res. 2018;114:1287–1303. doi: 10.1093/cvr/cvy122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH. Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era. J Mol Diagn. 2013;15:158–170. doi: 10.1016/j.jmoldx.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 92.Lefeber DJ, de Brouwer AP, Morava E, et al. Autosomal recessive dilated cardiomyopathy due to DOLK mutations results from abnormal dystroglycan O-mannosylation. PLoS Genet. 2011;7:e1002427. doi: 10.1371/journal.pgen.1002427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khogali SS, Mayosi BM, Beattie JM, McKenna WJ, Watkins H, Poulton J. A common mitochondrial DNA variant associated with susceptibility to dilated cardiomyopathy in two different populations. Lancet. 2001;357:1265–1267. doi: 10.1016/S0140-6736(00)04422-6. [DOI] [PubMed] [Google Scholar]
- 94.Muntoni F, Cau M, Ganau A, et al. Brief report: deletion of the dystrophin muscle-promoter region associated with X-linked dilated cardiomyopathy. N Engl J Med. 1993;329:921–925. doi: 10.1056/NEJM199309233291304. [DOI] [PubMed] [Google Scholar]
- 95.McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Rijsingen IA, Nannenberg EA, Arbustini E, et al. Gender-specific differences in major cardiac events and mortality in lamin A/C mutation carriers. Eur J Heart Fail. 2013;15:376–384. doi: 10.1093/eurjhf/hfs191. [DOI] [PubMed] [Google Scholar]
- 97.Priori SG, Blomström-Lundqvist C, Mazzanti A, et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC) Eur Heart J. 2015;36:2793–2867. doi: 10.1093/eurheartj/ehv316. [DOI] [PubMed] [Google Scholar]
- 98.Hey TM, Rasmussen TB, Madsen T, et al. Pathogenic RBM20-variants are associated with a severe disease expression in male patients with dilated cardiomyopathy. Circ Heart Fail. 2019;12:e005700. doi: 10.1161/CIRCHEARTFAILURE.118.005700. [DOI] [PubMed] [Google Scholar]
- 99.Herman DS, Lam L, Taylor MR, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schafer S, de Marvao A, Adami E, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet. 2017;49:46–53. doi: 10.1038/ng.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anderson BR, Granzier HL. Titin-based tension in the cardiac sarcomere: molecular origin and physiological adaptations. Prog Biophys Mol Biol. 2012;110:204–217. doi: 10.1016/j.pbiomolbio.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Akhtar MM, Lorenzini M, Cicerchia M, et al. Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circ Heart Fail. 2020;13:e006832. doi: 10.1161/CIRCHEARTFAILURE.119.006832. [DOI] [PubMed] [Google Scholar]
- 103.Ware JS, Amor-Salamanca A, Tayal U, et al. Genetic etiology for alcohol-induced cardiac toxicity. J Am Coll Cardiol. 2018;71:2293–2302. doi: 10.1016/j.jacc.2018.03.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ware JS, Li J, Mazaika E, et al. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N Engl J Med. 2016;374:233–241. doi: 10.1056/NEJMoa1505517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 106.Lu JT, Muchir A, Nagy PL, Worman HJ. LMNA cardiomyopathy: cell biology and genetics meet clinical medicine. Dis Model Mech. 2011;4:562–568. doi: 10.1242/dmm.006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hasselberg NE, Haland TF, Saberniak J, et al. Lamin A/C cardiomyopathy: young onset, high penetrance, and frequent need for heart transplantation. Eur Heart J. 2018;39:853–860. doi: 10.1093/eurheartj/ehx596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gigli M, Merlo M, Graw SL, et al. Genetic risk of arrhythmic phenotypes in patients with dilated cardiomyopathy. J Am Coll Cardiol. 2019;74:1480–1490. doi: 10.1016/j.jacc.2019.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swift J, Ivanovska IL, Buxboim A, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Tintelen JP, Hofstra RM, Katerberg H, et al. High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. Am Heart J. 2007;154:1130–1139. doi: 10.1016/j.ahj.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 111.Kamisago M, Sharma SD, DePalma SR, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 112.Guo W, Schafer S, Greaser ML, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brauch KM, Karst ML, Herron KJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li D, Morales A, Gonzalez-Quintana J, et al. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3:90–97. doi: 10.1111/j.1752-8062.2010.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang XL, Xie J, Lan RF, et al. Genetic basis and genotype–phenotype correlations in Han Chinese patients with idiopathic Dilated cardiomyopathy. Sci Rep. 2020;10:2226. doi: 10.1038/s41598-020-58984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van den Hoogenhof MM, Beqqali A, Amin AS, et al. RBM20 mutations induce an arrhythmogenic dilated cardiomyopathy related to disturbed calcium handling. Circulation. 2018;138:1330–1342. doi: 10.1161/CIRCULATIONAHA.117.031947. [DOI] [PubMed] [Google Scholar]
- 117.Begay RL, Tharp CA, Martin A, et al. FLNC gene splice mutations cause dilated cardiomyopathy. JACC Basic Transl Sci. 2016;1:344–359. doi: 10.1016/j.jacbts.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ortiz-Genga MF, Cuenca S, Dal Ferro M, et al. Truncating FLNC mutations are associated with high-risk dilated and arrhythmogenic cardiomyopathies. J Am Coll Cardiol. 2016;68:2440–2451. doi: 10.1016/j.jacc.2016.09.927. [DOI] [PubMed] [Google Scholar]
- 119.Fujita M, Mitsuhashi H, Isogai S, et al. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev Biol. 2012;361:79–89. doi: 10.1016/j.ydbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 120.Begay RL, Graw SL, Sinagra G, et al. Filamin C truncation mutations are associated with arrhythmogenic dilated cardiomyopathy and changes in the cell–cell adhesion structures. JACC Clin Electrophysiol. 2018;4:504–514. doi: 10.1016/j.jacep.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McNair WP, Sinagra G, Taylor MR, et al. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol. 2011;57:2160–2168. doi: 10.1016/j.jacc.2010.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mann SA, Castro ML, Ohanian M, et al. R222Q SCN5A mutation is associated with reversible ventricular ectopy and dilated cardiomyopathy. J Am Coll Cardiol. 2012;60:1566–1573. doi: 10.1016/j.jacc.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 123.Elliott P, O'Mahony C, Syrris P, et al. Prevalence of desmosomal protein gene mutations in patients with dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:314–322. doi: 10.1161/CIRCGENETICS.110.937805. [DOI] [PubMed] [Google Scholar]
- 124.Ganesh SK, Arnett DK, Assimes TL, et al. Genetics and genomics for the prevention and treatment of cardiovascular disease: update: a scientific statement from the American Heart Association. Circulation. 2013;128:2813–2851. doi: 10.1161/01.cir.0000437913.98912.1d. [DOI] [PubMed] [Google Scholar]
- 125.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:155–161. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Spaendonck-Zwarts KY, van Rijsingen IA, van den Berg MP, et al. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years' experience. Eur J Heart Fail. 2013;15:628–636. doi: 10.1093/eurjhf/hft013. [DOI] [PubMed] [Google Scholar]
- 127.Yancy CW, Jessup M, Bozkurt B, et al. 2013 ACCF/AHA guideline for the management of heart failure: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. Circulation. 2013;128:1810–1852. doi: 10.1161/CIR.0b013e31829e8807. [DOI] [PubMed] [Google Scholar]
- 128.Bozkurt B, Colvin M, Cook J, et al. Current diagnostic and treatment strategies for specific dilated cardiomyopathies: a scientific statement from the American Heart Association. Circulation. 2016;134:e579–646. doi: 10.1161/CIR.0000000000000455. [DOI] [PubMed] [Google Scholar]
- 129.Charron P, Arad M, Arbustini E, et al. Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2010;31:2715–2726. doi: 10.1093/eurheartj/ehq271. [DOI] [PubMed] [Google Scholar]
- 130.Fatkin D members of the CSANZ Cardiac Genetic Diseases Council Writing Group. Guidelines for the diagnosis and management of familial dilated cardiomyopathy. Heart Lung Circ. 2011;20:691–693. doi: 10.1016/j.hlc.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 131.Kaneda R, Takada S, Yamashita Y, et al. Genome-wide histone methylation profile for heart failure. Genes Cells. 2009;14:69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 132.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giudicessi JR, Shrivastava S, Ackerman MJ, Pereira NL. Clinical impact of secondary risk factors in TTN-mediated dilated cardiomyopathy. Circ Genom Precis Med. 2021;14:e003240. doi: 10.1161/CIRCGEN.120.003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kayvanpour E, Sedaghat-Hamedani F, Amr A, et al. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. 2017;106:127–139. doi: 10.1007/s00392-016-1033-6. [DOI] [PubMed] [Google Scholar]
- 135.Norman MW, McKenna WJ. Arrhythmogenic right ventricular cardiomyopathy: perspectives on disease. Z Kardiol. 1999;88:550–554. doi: 10.1007/s003920050324. [DOI] [PubMed] [Google Scholar]
- 136.Marcus FI, McKenna WJ, Sherrill D, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation. 2010;121:1533–1541. doi: 10.1161/CIRCULATIONAHA.108.840827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Corrado D, Link MS, Calkins H. Arrhythmogenic right ventricular cardiomyopathy. N Engl J Med. 2017;376:61–72. doi: 10.1056/NEJMra1509267. [DOI] [PubMed] [Google Scholar]
- 138.Corrado D, van Tintelen PJ, McKenna WJ, et al. Arrhythmogenic right ventricular cardiomyopathy: evaluation of the current diagnostic criteria and differential diagnosis. Eur Heart J. 2020;41:1414–1429. doi: 10.1093/eurheartj/ehz669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McKoy G, Protonotarios N, Crosby A, et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 140.Sen-Chowdhry S, Morgan RD, Chambers JC, McKenna WJ. Arrhythmogenic cardiomyopathy: etiology, diagnosis, and treatment. Annu Rev Med. 2010;61:233–253. doi: 10.1146/annurev.med.052208.130419. [DOI] [PubMed] [Google Scholar]
- 141.Mahon NG, Madden BP, Caforio AL, et al. Immunohistologic evidence of myocardial disease in apparently healthy relatives of patients with dilated cardiomyopathy. J Am Coll Cardiol. 2002;39:455–462. doi: 10.1016/s0735-1097(01)01762-4. [DOI] [PubMed] [Google Scholar]
- 142.Corrado D, Basso C, Judge DP. Arrhythmogenic cardiomyopathy. Circ Res. 2017;121:784–802. doi: 10.1161/CIRCRESAHA.117.309345. [DOI] [PubMed] [Google Scholar]
- 143.Thiene G, Nava A, Corrado D, Rossi L, Pennelli N. Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med. 1988;318:129–133. doi: 10.1056/NEJM198801213180301. [DOI] [PubMed] [Google Scholar]
- 144.Marcus FI, Edson S, Towbin JA. Genetics of arrhythmogenic right ventricular cardiomyopathy: a practical guide for physicians. J Am Coll Cardiol. 2013;61:1945–1948. doi: 10.1016/j.jacc.2013.01.073. [DOI] [PubMed] [Google Scholar]
- 145.Merner ND, Hodgkinson KA, Haywood AF, et al. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet. 2008;82:809–821. doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van Rijsingen IA, van der Zwaag PA, Groeneweg JA, et al. Outcome in phospholamban R14del carriers: results of a large multicentre cohort study. Circ Cardiovasc Genet. 2014;7:455–465. doi: 10.1161/CIRCGENETICS.113.000374. [DOI] [PubMed] [Google Scholar]
- 147.Gerull B, Heuser A, Wichter T, et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36:1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 148.Hoorntje ET, Te Rijdt WP, James CA, et al. Arrhythmogenic cardiomyopathy: pathology, genetics, and concepts in pathogenesis. Cardiovasc Res. 2017;113:1521–1531. doi: 10.1093/cvr/cvx150. [DOI] [PubMed] [Google Scholar]
- 149.Syrris P, Ward D, Evans A, et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in the desmosomal gene desmocollin-2. Am J Hum Genet. 2006;79:978–984. doi: 10.1086/509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brodehl A, Dieding M, Klauke B, et al. The novel desmin mutant p.A120D impairs filament formation, prevents intercalated disk localization, and causes sudden cardiac death. Circ Cardiovasc Genet. 2013;6:615–623. doi: 10.1161/CIRCGENETICS.113.000103. [DOI] [PubMed] [Google Scholar]
- 151.Klauke B, Kossmann S, Gaertner A, et al. De novo desmin-mutation N116S is associated with arrhythmogenic right ventricular cardiomyopathy. Hum Mol Genet. 2010;19:4595–4607. doi: 10.1093/hmg/ddq387. [DOI] [PubMed] [Google Scholar]
- 152.Costa S, Medeiros-Domingo A, Gasperetti A, et al. Impact of genetic variant reassessment on the diagnosis of arrhythmogenic right ventricular cardiomyopathy based on the 2010 task force criteria. Circ Genom Precis Med. 2021;14:e003047. doi: 10.1161/CIRCGEN.120.003047. [DOI] [PubMed] [Google Scholar]
- 153.Nielsen JC, Lin YJ, de Oliveira Figueiredo MJ, et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) expert consensus on risk assessment in cardiac arrhythmias: use the right tool for the right outcome, in the right population. Europace. 2020;22:1147–1148. doi: 10.1093/europace/euaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Towbin JA, McKenna WJ, Abrams DJ, et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm. 2019;16:e301–72. doi: 10.1016/j.hrthm.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 155.Bhonsale A, Groeneweg JA, James CA, et al. Impact of genotype on clinical course in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated mutation carriers. Eur Heart J. 2015;36:847–855. doi: 10.1093/eurheartj/ehu509. [DOI] [PubMed] [Google Scholar]
- 156.Rigato I, Bauce B, Rampazzo A, et al. Compound and digenic heterozygosity predicts lifetime arrhythmic outcome and sudden cardiac death in desmosomal gene-related arrhythmogenic right ventricular cardiomyopathy. Circ Cardiovasc Genet. 2013;6:533–542. doi: 10.1161/CIRCGENETICS.113.000288. [DOI] [PubMed] [Google Scholar]
- 157.Bao J, Wang J, Yao Y, et al. Correlation of ventricular arrhythmias with genotype in arrhythmogenic right ventricular cardiomyopathy. Circ Cardiovasc Genet. 2013;6:552–556. doi: 10.1161/CIRCGENETICS.113.000122. [DOI] [PubMed] [Google Scholar]
- 158.Bauce B, Nava A, Beffagna G, et al. Multiple mutations in desmosomal proteins encoding genes in arrhythmogenic right ventricular cardiomyopathy/dysplasia. Heart Rhythm. 2010;7:22–29. doi: 10.1016/j.hrthm.2009.09.070. [DOI] [PubMed] [Google Scholar]
- 159.Akdis D, Saguner AM, Shah K, et al. Sex hormones affect outcome in arrhythmogenic right ventricular cardiomyopathy/dysplasia: from a stem cell derived cardiomyocyte-based model to clinical biomarkers of disease outcome. Eur Heart J. 2017;38:1498–1508. doi: 10.1093/eurheartj/ehx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.James CA, Bhonsale A, Tichnell C, et al. Exercise increases age-related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J Am Coll Cardiol. 2013;62:1290–1297. doi: 10.1016/j.jacc.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zorzi A, Rigato I, Pilichou K, et al. Phenotypic expression is a prerequisite for malignant arrhythmic events and sudden cardiac death in arrhythmogenic right ventricular cardiomyopathy. Europace. 2016;18:1086–1094. doi: 10.1093/europace/euv205. [DOI] [PubMed] [Google Scholar]
- 162.Muchtar E, Blauwet LA, Gertz MA. Restrictive cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res. 2017;121:819–837. doi: 10.1161/CIRCRESAHA.117.310982. [DOI] [PubMed] [Google Scholar]
- 163.Kaski JP, Syrris P, Burch M, et al. Idiopathic restrictive cardiomyopathy in children is caused by mutations in cardiac sarcomere protein genes. Heart. 2008;94:1478–1484. doi: 10.1136/hrt.2007.134684. [DOI] [PubMed] [Google Scholar]
- 164.Wu W, Lu CX, Wang YN, et al. Novel phenotype-genotype correlations of restrictive cardiomyopathy with myosin-binding protein C (MYBPC3) gene mutations tested by next-generation sequencing. J Am Heart Assoc. 2015;4:e001879. doi: 10.1161/JAHA.115.001879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sen-Chowdhry S, Syrris P, McKenna WJ. Genetics of restrictive cardiomyopathy. Heart Fail Clin. 2010;6:179–186. doi: 10.1016/j.hfc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 166.Purevjav E, Arimura T, Augustin S, et al. Molecular basis for clinical heterogeneity in inherited cardiomyopathies due to myopalladin mutations. Hum Mol Genet. 2012;21:2039–2053. doi: 10.1093/hmg/dds022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schubert J, Tariq M, Geddes G, Kindel S, Miller EM, Ware SM. Novel pathogenic variants in filamin C identified in pediatric restrictive cardiomyopathy. Hum Mutat. 2018;39:2083–2096. doi: 10.1002/humu.23661. [DOI] [PubMed] [Google Scholar]
- 168.Pereira NL, Grogan M, Dec GW. Spectrum of restrictive and infiltrative cardiomyopathies: part 2 of a 2-part series. J Am Coll Cardiol. 2018;71:1149–1166. doi: 10.1016/j.jacc.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 169.Elliott P, Andersson B, Arbustini E, et al. Classification of the cardiomyopathies: a position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 170.Jenni R, Oechslin E, Schneider J, Attenhofer Jost C, Kaufmann PA. Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart. 2001;86:666–671. doi: 10.1136/heart.86.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van Waning JI, Caliskan K, Hoedemaekers YM, et al. Genetics, clinical features, and long-term outcome of noncompaction cardiomyopathy. J Am Coll Cardiol. 2018;71:711–722. doi: 10.1016/j.jacc.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 172.Sedaghat-Hamedani F, Haas J, Zhu F, et al. Clinical genetics and outcome of left ventricular non-compaction cardiomyopathy. Eur Heart J. 2017;38:3449–3460. doi: 10.1093/eurheartj/ehx545. [DOI] [PubMed] [Google Scholar]
- 173.van Waning JI, Caliskan K, Michels M, et al. Cardiac phenotypes, genetics, and risks in familial noncompaction cardiomyopathy. J Am Coll Cardiol. 2019;73:1601–1611. doi: 10.1016/j.jacc.2018.12.085. [DOI] [PubMed] [Google Scholar]
- 174.Xing Y, Ichida F, Matsuoka T, et al. Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol Genet Metab. 2006;88:71–77. doi: 10.1016/j.ymgme.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 175.Chang B, Nishizawa T, Furutani M, et al. Identification of a novel TPM1 mutation in a family with left ventricular noncompaction and sudden death. Mol Genet Metab. 2011;102:200–206. doi: 10.1016/j.ymgme.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 176.Ripoll Vera T, Monserrat Iglesias L, Hermida Prieto M, et al. The R820W mutation in the MYBPC3 gene, associated with hypertrophic cardiomyopathy in cats, causes hypertrophic cardiomyopathy and left ventricular non-compaction in humans. Int J Cardiol. 2010;145:405–407. doi: 10.1016/j.ijcard.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 177.Dellefave LM, Pytel P, Mewborn S, et al. Sarcomere mutations in cardiomyopathy with left ventricular hypertrabeculation. Circ Cardiovasc Genet. 2009;2:442–449. doi: 10.1161/CIRCGENETICS.109.861955. [DOI] [PubMed] [Google Scholar]
- 178.Shan L, Makita N, Xing Y, et al. SCN5A variants in Japanese patients with left ventricular noncompaction and arrhythmia. Mol Genet Metab. 2008;93:468–474. doi: 10.1016/j.ymgme.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 179.Servatius H, Porro A, Pless SA, et al. Phenotypic spectrum of HCN4 mutations: a clinical case. Circ Genom Precis Med. 2018;11:e002033. doi: 10.1161/CIRCGEN.117.002033. [DOI] [PubMed] [Google Scholar]
- 180.Hoedemaekers YM, Caliskan K, Michels M, et al. The importance of genetic counseling, DNA diagnostics, and cardiologic family screening in left ventricular noncompaction cardiomyopathy. Circ Cardiovasc Genet. 2010;3:232–239. doi: 10.1161/CIRCGENETICS.109.903898. [DOI] [PubMed] [Google Scholar]
- 181.Towbin JA, Ballweg J, Johnson J. In: Heart Failure in the Child and Young Adult: from Bench to Bedside. Jefferies JL, Chang AC, Rossano JW, Shaddy RE, Towbin JA, editors. Cambridge (MA): Academic Press; 2018. Chapter 20. Left ventricular noncompaction cardiomyopathy; pp. 269–290. [Google Scholar]
- 182.Ross SB, Singer ES, Driscoll E, et al. Genetic architecture of left ventricular noncompaction in adults. Hum Genome Var. 2020;7:33. doi: 10.1038/s41439-020-00120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bushby K, Finkel R, Birnkrant DJ, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 184.Bushby KM, Thambyayah M, Gardner-Medwin D. Prevalence and incidence of Becker muscular dystrophy. Lancet. 1991;337:1022–1024. doi: 10.1016/0140-6736(91)92671-n. [DOI] [PubMed] [Google Scholar]
- 185.Reetz K, Dogan I, Costa AS, et al. Biological and clinical characteristics of the European Friedreich's Ataxia Consortium for Translational Studies (EFACTS) cohort: a cross-sectional analysis of baseline data. Lancet Neurol. 2015;14:174–182. doi: 10.1016/S1474-4422(14)70321-7. [DOI] [PubMed] [Google Scholar]
- 186.Weidemann F, Rummey C, Bijnens B, et al. The heart in Friedreich ataxia: definition of cardiomyopathy, disease severity, and correlation with neurological symptoms. Circulation. 2012;125:1626–1634. doi: 10.1161/CIRCULATIONAHA.111.059477. [DOI] [PubMed] [Google Scholar]
- 187.Muchir A, Worman HJ. Emery-Dreifuss muscular dystrophy: focal point nuclear envelope. Curr Opin Neurol. 2019;32:728–734. doi: 10.1097/WCO.0000000000000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Baban A, Cicenia M, Magliozzi M, et al. Cardiovascular involvement in pediatric laminopathies. report of six patients and literature revision. Front Pediatr. 2020;8:374. doi: 10.3389/fped.2020.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 190.Kamdar F, Garry DJ. Dystrophin-deficient cardiomyopathy. J Am Coll Cardiol. 2016;67:2533–2546. doi: 10.1016/j.jacc.2016.02.081. [DOI] [PubMed] [Google Scholar]
- 191.Payne RM, Wagner GR. Cardiomyopathy in Friedreich ataxia: clinical findings and research. J Child Neurol. 2012;27:1179–1186. doi: 10.1177/0883073812448535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mejia E, Lynch A, Hearle P, et al. Ectopic burden via Holter monitors in Friedreich ataxia. Pediatr Neurol. 2021;117:29–33. doi: 10.1016/j.pediatrneurol.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Brega A, Narula J, Arbustini E. Functional, structural, and genetic mitochondrial abnormalities in myocardial diseases. J Nucl Cardiol. 2001;8:89–97. doi: 10.1067/mnc.2001.112755. [DOI] [PubMed] [Google Scholar]
- 194.Zaragoza MV, Brandon MC, Diegoli M, Arbustini E, Wallace DC. Mitochondrial cardiomyopathies: how to identify candidate pathogenic mutations by mitochondrial DNA sequencing, MITOMASTER and phylogeny. Eur J Hum Genet. 2011;19:200–207. doi: 10.1038/ejhg.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kaufmann P, Engelstad K, Wei Y, et al. Natural history of MELAS associated with mitochondrial DNA m.3243A>G genotype. Neurology. 2011;77:1965–1971. doi: 10.1212/WNL.0b013e31823a0c7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Brambilla A, Favilli S, Olivotto I, et al. Clinical profile and outcome of cardiac involvement in MELAS syndrome. Int J Cardiol. 2019;276:14–19. doi: 10.1016/j.ijcard.2018.10.051. [DOI] [PubMed] [Google Scholar]
- 197.Garcia-Pavia P, Rapezzi C, Adler Y, et al. Diagnosis and treatment of cardiac amyloidosis. A position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur J Heart Fail. 2021;23:512–526. doi: 10.1002/ejhf.2140. [DOI] [PubMed] [Google Scholar]
- 198.Ruberg FL, Grogan M, Hanna M, Kelly JW, Maurer MS. Transthyretin amyloid cardiomyopathy: JACC state-of-the-art review. J Am Coll Cardiol. 2019;73:2872–2891. doi: 10.1016/j.jacc.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pieroni M, Moon JC, Arbustini E, et al. Cardiac involvement in Fabry disease: JACC review topic of the week. J Am Coll Cardiol. 2021;77:922–936. doi: 10.1016/j.jacc.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 200.Hagège A, Réant P, Habib G, et al. Fabry disease in cardiology practice: literature review and expert point of view. Arch Cardiovasc Dis. 2019;112:278–287. doi: 10.1016/j.acvd.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 201.Luis SA, Maleszewski JJ, Young PM, Schaff HV, Pereira NL. Previously unreported in women Galactosidase Alpha Pro409Ser variant is associated with fabry disease. Circ Cardiovasc Genet. 2017;10:e001661. doi: 10.1161/CIRCGENETICS.116.001661. [DOI] [PubMed] [Google Scholar]
- 202.Lakdawala NK, Dellefave L, Redwood CS, et al. Familial dilated cardiomyopathy caused by an alpha-tropomyosin mutation: the distinctive natural history of sarcomeric dilated cardiomyopathy. J Am Coll Cardiol. 2010;55:320–329. doi: 10.1016/j.jacc.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wordsworth S, Leal J, Blair E, et al. DNA testing for hypertrophic cardiomyopathy: a cost-effectiveness model. Eur Heart J. 2010;31:926–935. doi: 10.1093/eurheartj/ehq067. [DOI] [PubMed] [Google Scholar]
- 204.Marian AJ. The case of “missing causal genes” and the practice of medicine: a Sherlock Holmes approach of deductive reasoning. Circ Res. 2016;119:21–24. doi: 10.1161/CIRCRESAHA.116.308830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Marian AJ. Clinical interpretation and management of genetic variants. JACC Basic Transl Sci. 2020;5:1029–1042. doi: 10.1016/j.jacbts.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Marian AJ. Causality in genetics: the gradient of genetic effects and back to Koch's postulates of causality. Circ Res. 2014;114:e18–21. doi: 10.1161/CIRCRESAHA.114.302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.1000 Genomes Project Consortium. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Saltzman AJ, Mancini-DiNardo D, Li C, et al. Short communication: the cardiac myosin binding protein C Arg502Trp mutation: a common cause of hypertrophic cardiomyopathy. Circ Res. 2010;106:1549–1552. doi: 10.1161/CIRCRESAHA.109.216291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Adams TD, Yanowitz FG, Fisher AG, et al. Heritability of cardiac size: an echocardiographic and electrocardiographic study of monozygotic and dizygotic twins. Circulation. 1985;71:39–44. doi: 10.1161/01.cir.71.1.39. [DOI] [PubMed] [Google Scholar]
- 211.Schunkert H, Bröckel U, Hengstenberg C, et al. Familial predisposition of left ventricular hypertrophy. J Am Coll Cardiol. 1999;33:1685–1691. doi: 10.1016/s0735-1097(99)00050-9. [DOI] [PubMed] [Google Scholar]
- 212.Innes BA, McLaughlin MG, Kapuscinski MK, Jacob HJ, Harrap SB. Independent genetic susceptibility to cardiac hypertrophy in inherited hypertension. Hypertension. 1998;31:741–746. doi: 10.1161/01.hyp.31.3.741. [DOI] [PubMed] [Google Scholar]
- 213.Sturm AC, Hershberger RE. Genetic testing in cardiovascular medicine: current landscape and future horizons. Curr Opin Cardiol. 2013;28:317–325. doi: 10.1097/HCO.0b013e32835fb728. [DOI] [PubMed] [Google Scholar]
- 214.Burke MA, Cook SA, Seidman JG, Seidman CE. Clinical and mechanistic insights into the genetics of cardiomyopathy. J Am Coll Cardiol. 2016;68:2871–2886. doi: 10.1016/j.jacc.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jacoby D, McKenna WJ. Genetics of inherited cardiomyopathy. Eur Heart J. 2012;33:296–304. doi: 10.1093/eurheartj/ehr260. [DOI] [PMC free article] [PubMed] [Google Scholar]