Abstract
With applications in object detection, image feature extraction, image classification, and image segmentation, artificial intelligence is facilitating high-throughput analysis of image data in a variety of biomedical imaging disciplines, ranging from radiology and pathology to cancer biology and immunology. Specifically, a growth in research on deep learning has led to the widespread application of computer-visualization techniques for analyzing and mining data from biomedical images. The availability of open-source software packages and the development of novel, trainable deep neural network architectures has led to increased accuracy in cell detection and segmentation algorithms. By automating cell segmentation, it is now possible to mine quantifiable cellular and spatio-cellular features from microscopy images, providing insight into the organization of cells in various pathologies. This mini-review provides an overview of the current state of the art in deep learning– and artificial intelligence–based methods of segmentation and data mining of cells in microscopy images of tissue.
Machine learning has led to rapid advancements in the biomedical sciences, particularly image quantitation. Deep learning, a branch of machine learning focused on artificial neural networks, is driving the growth of research in the imaging sciences, radiomics, and computational pathology. In conjunction with more conventional machine-learning methods, deep learning has facilitated the automation of image-analysis tasks such as cell classification and tissue-type identification, thereby reducing the time and effort needed for analyzing microscopy data. This mini-review discusses the recently developed applications of artificial intelligence (AI) to digital pathology and cell-image analysis, focusing on novel machine-learning and deep-learning methods and associated challenges.
Artificial Intelligence in Digital Pathology
Computer visualization and machine learning have been applied in medical imaging for decades. Early computer-visualization techniques in medical imaging were developed for application in radiography and have since been widely applied in histopathology.1,2 Digital pathology produces a wealth of complex data, making automated, high-throughput analysis desirable. Manual annotation of cell-level features (eg, surface marker expression) and tissue-level features (eg, tumor boundaries) from microscopy data is a resource-intensive task that usually requires time and the experience of a trained pathologist. The extraction of quantitative information from image data sets for research purposes has historically been limited by the availability and participation of these clinical experts. AI provides a means for high-throughput, standardized, quantitative analysis of pathology images.3, 4, 5
The development of early machine-learning applications in medical imaging was facilitated by digitization, which allowed for mathematical operations to be performed on image data.1,4 For lesion grading and assessment in radiology and pathology, imaging specialists worked in tandem with clinicians to identify the qualitative features recognizable to the well-trained eye, then mathematically designed filters to enhance such characteristics, leading to computerized lesion detection and segmentation. Once segmented, a lesion can be quantitatively described using human-engineered features such as size, shape, pixel intensity, and local texture.4,6 Texture analysis and lesion classification are two of the early building blocks of computational and digital pathology and have been applied in clinical tasks such as diagnosis and grading, and in studies of disease pathogenesis.4,6
Deep learning was widely adopted in the imaging sciences after AlexNet, a deep convolutional neural network (DCNN), achieved a 15% error rate (nearly 10% better than its competitors) on the ImageNet Large Scale Visual Recognition Challenge.7 The implications for medical image analysis were immediately apparent. Within a year, multiple studies were published in which DCNNs were applied in medical image analysis, followed by continued growth in the application of deep learning to biomedical image analysis.8,9 Since the success of AlexNet, a variety of DCNN architectures have been developed for specialized medical and biological imaging tasks, ranging from lesion detection in magnetic resonance imaging to cell segmentation in microscopy.4,10 DCNNs recognize patterns in raw image data without the constraints of human-defined equations. These patterns may not be interpretable by human observers but are determined by the network to be the features most useful in making robust and accurate decisions in image classification or segmentation.
Deep Learning in Multiplexed Microscopy
Deep learning facilitates the rapid analysis of high-content, experimental imaging modalities, including multiplexed microscopy. Multiplexed microscopy refers to the use of image data in which the spatial distribution of multiple biological markers, such as cell surface proteins, has been captured via immunohistochemistry analysis or immunofluorescence. Previously, these type of data were either analyzed manually or used only for the purpose of presenting representative examples of biological phenomena. DCNNs have been particularly useful for automating the segmentation and classification of regions, structures, and cells in tissue.10 This automation has facilitated previously intractable quantification of microscopy data.
Simultaneously, multiplexed microscopy has advanced to the point of generating novel computational challenges. Improved microscope designs and staining protocols have greatly increased the number of markers that can be captured in a given sample, allowing for co-localization of upward of 40 markers per individual frame.11, 12, 13 This level of phenotypic resolution was previously accessible only through tissue-destructive methods such as flow cytometry and RNA sequencing. The destructive processing required for these methods does not conserve the spatial arrangement of cells and other tissue structures. In contrast, highly multiplexed imaging produces rich data sets that include detailed information on phenotype and cell spacing. Novel AI solutions have been implemented in the location and segmentation of these phenotype-rich images.11,13
Cell Detection and Segmentation in Tissue Microscopy Images
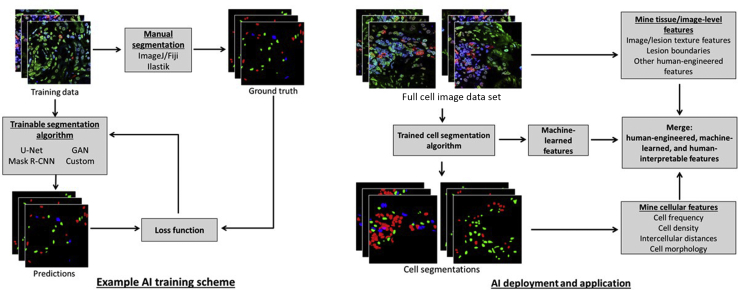
A key application of machine learning in tissue microscopy is in the automation of the identification of structures and individual cells within images. Cell segmentation is achieved through iterative training of AI computer-visualization algorithms (Figure 1). These trained models can then be deployed for predicting cell segmentation in new data, allowing for high-throughput mining of quantitative descriptors of tissue pathology. This section describes deep-learning architectures and open-source programs used for performing cell segmentation in microscopy data.
Figure 1.
Supervised AI computer-vision algorithms are trained using manual ground truth to achieve accurate cell segmentations on a specific domain of image data (left). A trained algorithm can then be deployed to automatically extract quantitative features from microscopy data to answer clinic- and biology-related questions (right). CNN, convolutional neural network; GAN, generative adversarial networks. Fiji, NIH, Bethesda, MD; https://imagej.nih.gov/ij. Ilastik; http://ilastik.org. ImageJ, NIH; http://imagej.nih.gov.ij.
DCNN Architectures for Analyzing Cells in Microscopy Images
The detection and segmentation of cells in ex vivo tissue allow for high-throughput quantification of cell features, including cell frequency, cell morphology, cell-specific signal intensity, and spatial distribution of cells. Biomedical image segmentation is often conducted using U-Net or similarly structured architectures, which provide pixel-level classification, referred to as semantic segmentation.14 However, these methods can underperform in dense regions of cells. Object-detection and semantic-segmentation tasks can be combined to perform instance segmentation, which can be accomplished through object-based detection networks such as Mask R-CNN or by combining a U-Net with a region-proposal network.15,16 Image-translation methods have also shown potential in segmentation of microscopy images.17 These generative methods can also improve image quality by converting the image to a higher resolution or converting between image modalities.18,19
Semantic Segmentation
Encoder–decoder architectures have dominated biomedical image segmentation since the advent of U-Net and its derivatives. These have been applied to semantic segmentation tasks in medical imaging across scales, with excellent performance on full-organ segmentation in computed tomography and nuclear segmentation in high-resolution microscopy.14,20,21 The contracting, or encoder, portion of the architecture captures contextual features within the image, while the expanding, or decoder, portion of the architecture generates precise localization. In pathology images, semantic segmentation has been used to differentiate between tumor and healthy tissue, or segment pathogenic or prepathogenic areas of tissue. Cell and cell-nucleus segmentation can also be performed through semantic segmentation schemes. Quantitative characteristics acquired from these segmentations, such as nucleus-to-cytoplasm ratio, can be indicative of cancer grade.22 However, semantic segmentation of cells can fail in crowded regions or in images with a low signal-to-noise or signal-to-background ratio.
Instance Segmentation
While semantic segmentation alone is not always sufficient for accurate cell counting in images, it can be combined with object-detection methods to generate object-level segmentation of cells rather than image-level segmentation. This combined task, called instance segmentation, generates pixel-level segmentation of individual cells in an image, which allows for the separation of clustered or overlapping cells in an image, resulting in improved cell-frequency data. Region-based CNNs from the Fast R-CNN family perform multi-object, multi-class segmentation with high accuracy in natural images, and have also shown promise in biomedical image analysis tasks, including cell segmentation in a variety of contexts.23,24 Additionally, region-proposal networks have been incorporated into U-Net architectures to perform object-level cell segmentation in biomedical images.15,25 NuSeT, a U-Net + region proposal network architecture, has shown high accuracy in nuclear segmentation in a variety of contexts, including segmentation in images from different modalities and different tissues and pathologies.15
Generative Networks for Segmentation
Generative adversarial networks (GANs) have also been successfully implemented in segmentation in biological images.17 However, GANs take a different approach to image segmentation. A typical GAN architecture consists of two networks, a generator and a discriminator, which are trained competitively to generate a simulated version of the input image as it would present in a different context, for example translating a daytime landscape image to appear as if it were taken at night. GAN segmentation works by converting an input image into the mask domain, in which each class is represented as a specific pixel value. In addition to image segmentation, image conversion can also mitigate generalizability-related problems in deep learning. de Bel et al17 have shown the ability of GANs to normalize staining variability across multiple institutions and multiple tissue types. Additionally, generative architectures have been trained to rectify image artefacts, to de-noise low signal-to-noise ratio images, and to generate isotropic resolution in three dimensions.19 These de-noising and resolution adjustments have led to improved segmentation of cells in three-dimensional images, videos, and image projections. Keikhosravi et al18 have trained generative networks to perform cross-modality synthesis on images of hematoxylin and eosin (H&E) staining to produce second harmonic generation images of collagen organization in biopsy samples. A second harmonic generation system is not feasible to operate in the clinic, but by converting standard H&E images, deep-learning methods can provide more detailed information on samples.
Examples of Current Open-Source Software
Several open-source implementations of machine-learning tools have been designed for scientists who may not have the extensive background in programming required for low-level algorithm control. Many of these programs employ pretrained networks for common tasks such as nuclear segmentation. Some enable the training of machine-learning models from scratch. One of the benefits of these open-source pipelines is that they include graphical user interfaces, making them user-friendly and accessible to a broader audience. However, the user-friendly environment comes at the expense of the user losing low-level control of the algorithm, making task-specific optimization via hyperparameter tuning or transfer learning challenging.
ImageJ
ImageJ (NIH, Bethesda, MD; http://imagej.nih.gov.ij; alias Fiji) is an open-source tool that has been used extensively for many image-analysis tasks. Users frequently build and share plug-ins that can be used to perform complex analyses. To date, several plug-ins have been developed to make machine learning–based segmentation more accessible. For example, the Trainable Weka segmentation plug-in allows users to train segmentation algorithms using the ImageJ software graphical user interface.26 Similarly, the laboratory that invented U-Net developed a U-Net plug-in (Unet-segmentation), which uses the Caffe deep-learning framework (University of California, Berkeley, CA) to implement the architecture in ImageJ.20 DeepImageJ can run pretrained TensorFlow models in ImageJ, and is bundled with a set of models for particular tasks. One of the bundled models is a deep CNN that can virtually stain an unlabeled tissue autofluorescence image to produce an approximation of the corresponding H&E, Masson trichrome, or Jones stain image.27
CellProfiler
CellProfiler is a widely used software that has been cited >9000 times since the publication on its original release in 2006.28 It has a point-and-click graphical user interface that allows users with minimal programing experience to string together several image-analysis modules into an analytical pipeline. CellProfiler comes with >50 modules that allow for standard image-analysis procedures, and further allows users to write their own modules. As of 2018, CellProfiler software version 3.0 featured a pretrained U-Net–based semantic segmentation module (ClassifyPixels-Unet).29 A drawback is that no task-specific training or fine-tuning can be applied, which can limit its utility in tissue microscopy analysis, as tissue background can have a substantial impact on network generalizability. However, the modularity of CellProfiler allows users to integrate trained models from other sources into a large analytical pipeline. For example, Sadanandan et al30 wrote a module that allowed users to run pretrained, Caffe-based models within CellProfiler pipelines.
Ilastik
Ilastik is a deep-learning software package used for performing semantic segmentation tasks by extracting a series of predefined pixel-level features to train random forest classifiers.31 It provides a user interface that enables the generation of sparse training labels, and an interactive mode in which users can provide feedback to the network during training. This flexibility has made it a useful tool in the context of highly multiplexed imaging, in which training-set generation is an expensive process.32,33 Several recent papers have reported on the combination of CellProfiler with Ilastik for segmenting cells in tissue-microscopy images. For example, a CellProfiler pipeline can be generated to perform preprocessing steps, then Ilastik can be used to produce a probability map for semantic segmentations.34,35 This map is then returned to CellProfiler for object detection.
QuPath
QuPath, an open-source platform designed specifically for whole-slide image analysis, can be used for both immunohistochemistry analysis and immunofluorescence imaging.36 Like Ilastik, it employs random forest classifiers that can be trained to segment cell classes based on cell surface marker expression. It is useful for quantifying the distribution of a particular marker across a whole-tissue section.
Downstream Applications of Algorithm Outputs
The output of a given machine-learning or deep-learning algorithm is often not the culmination of a digital pathology analysis. These techniques are widely applied to automate the extraction of quantifiable metrics from cell images to answer biology-related questions. This section addresses how machine-extracted features are used in downstream analyses.
Extracting Human-Interpretable Features from Deep-Learning Algorithms
Beyond automating the laborious parts of image analysis, deep learning provides the advantage of utilizing image features that are not generated or perceived by the user. However, the black-box nature of DCNNs is a common critique of applying deep learning to medical and biological image analysis. DCNNs are excellent for recognizing patterns, but to optimize and apply DCNNs, it needs to be understood whether they are recognizing either patterns specific to the imaging system or protocol, or biological features such as cell density and tissue structure. Minimal insight into how a DCNN arrives at a classification limits the opportunities to strategically improve the algorithm or training data. The ability to explain and interpret how deep-learning algorithms arrive at classification scores has therefore become a topic of active research by AI developers and AI users, respectively. The internal machine-learned features can be extracted to better interpret the rationale of decision making of a trained network. Attention maps allow for visualization of these machine-learned features, highlighting the parts of a given image that were influential in classifying or segmenting the image.37 Faust et al38 have used feature map visualization methods to compare learned features with human-interpretable features of images. Machine-learned features were found to correlate to multiple tissue and cell features, including the presence of epithelial cells, fibrosis, and luminal space.
An understanding of the internal DCNN features and the ways in which they relate to known markers of tissue structure and pathology provides insight into the decision-making capabilities of these AI algorithms. Human-interpretable features can also be generated from DCNN results to better characterize pathologic tissue. U-Nets and their derivatives can be used to segment both cells and tissues within a sample, producing a multiscale map of the cell populations that reside in given regions of tissue. If a network that segments cancerous tissue is combined with a network that segments cells, the resulting segmentations can be mined for human-interpretable features of the spatial distribution of cells, such as lymphocyte density near the cancer boundary and relative density of immune cells and cancer cells within the tumor.39 The method of combining deep automated cell and lesion detection with human-engineered computer-visualization features provides high-throughput computation of interpretable features that previously would have required costly and time-intensive manual annotation of images.
Quantifying Spatial Organization of Cells in Microscopy Images
Automation of annotation facilitates spatial analyses that are more complex than was previously possible. Specifically, determining the interspatial organization of various cell classes unlocks the capacity to determine the ways in which the cell constituents of tissue interact. This spatial data allows for the generation of novel features that can be used to stratify patients and increase the understanding of cell processes in tissue.
This idea has been applied extensively in the study of tumor immunology. Nearchou et al40 examined sequential sections of surgically resected colorectal cancer specimens and developed the spatial immuno-oncology index, composed of patient-level spatial features, including mean CD3+ density, mean number of lymphocytes in proximity to tumor buds, and ratio of CD68+/CD163+ macrophages, and used as a predictor of prognosis. Similarly, Lazarus et al41 examined the abundance and spatial distribution of various subsets of T cells, antigen-presenting cells, and tumor cells in liver metastases of colorectal cancer. The automated detection and segmentation of immune cells facilitated the finding that cytotoxic T lymphocytes were typically positioned further from epithelial cells and antigen-presenting cells that were expressing a programmed cell death checkpoint molecule. Furthermore, they found that high levels of engagement, defined as an intercell distance of <15 μm, between cytotoxic T lymphocytes and epithelial cells, helper T cells, and regulatory T cells was predictive of increased survival. Thus, automated image analysis allows for the extraction of features related to the interactions between tumors and the immune system that can be used to predict patient outcomes. Wang et al42 used a Mask R-CNN architecture to segment lymphocytes and tumors cells in whole-slide H&E images of hepatocellular carcinoma samples. These segmentations were then used to generate 246 image features that included tumor nuclei morphology, density of lymphocytes and tumors, and the spatial relationships between the two cell types. These features were used to perform unsupervised consensus clustering that identified three distinct subgroups within the patient population.
Predicting prognosis is not the final goal of these analyses. Rather, associating specific intercell interactions with positive or negative patient outcomes is the first step in identifying immunologic phenomena that have potential as therapeutic targets. Understanding the cell interactions taking place in tissue, and the ways in which these interactions correlate with outcome, allows for the identification of interactions that should be promoted and those that should be disrupted.
Challenges in Applying AI to Cell Image Analysis
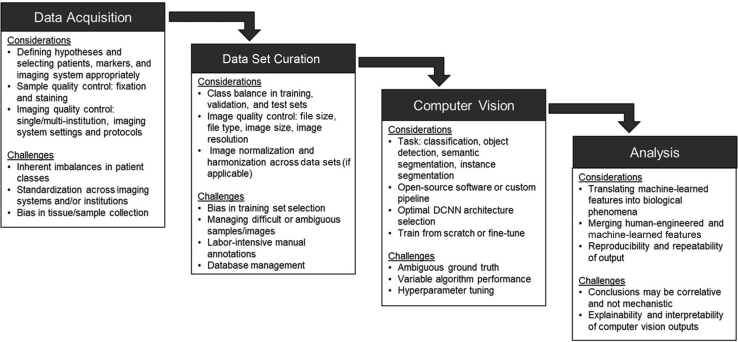
Use of deep learning for image analysis as a standard in the experiment toolkit necessitates considering the ways in which image data acquisition can be optimized for downstream machine-learning applications (Figure 2). In particular, it is important to understand which features of data acquisition can influence the performance of segmentation algorithms, including: i) selection bias, ii) sample processing, and iii) imaging system.43,44
Figure 2.
The application of AI to the microscopy of cells requires careful consideration and planning at all stages of the process, including data acquisition, data set curation, selection and deployment of computer-vision methods, and post–deep-learning analysis. If not properly addressed, the challenges associated with each step may limit the scope of the conclusions. DCNN, deep convolutional neural network.
Selection Bias
Selection bias in AI for cell imaging has multiple components. This section discusses selection bias as it relates to both tissue sampling and building training, validation, and testing sets of data.
Bias in Tissue Collection and Sampling
Selection bias is always a concern with ex vivo imaging, as the full organ or tissue cannot always be excised for ex vivo analysis.45 In sampling a small region of the tissue, ex vivo two-dimensional imaging may or may not contain the most severe region, meaning that at the cellular level, sampling error must be considered for in-depth evaluation of the tissue. While selection bias does affect the capacity of AI to accurately predict patient diagnosis or prognosis, tissue analysis by a pathologist is also subject to selection bias. Further improvements in the specificity of biopsy sample–acquisition methods will be useful in reducing the effects of selection bias on both human and machine decision-making in pathology.
Bias in Training Data
AI methods consist of trainable algorithms that must learn from a predetermined set of samples. If an algorithm is trained using data that is biased toward one population or class, this bias will be inherent in the trained model when it is applied to new samples.46 Certain biases in training data are difficult to address in smaller-scale studies. For example, if a training set is composed of samples from a single institution, or if one institution is more represented than another, the resulting model will likely perform better on samples from this overrepresented institution. The same concept can be applied to patient or sample groups (ie, patient age or sex) present in the training data. This bias can be partially mitigated through training-set normalization or harmonization but should be noted as a limitation of any trained model.
Sample Processing
The fixation method also affects the results of AI analysis of ex vivo tissue samples. A significant difference was found between quantitative features of cell morphology in cells detected by deep CNNs in tissue samples that were fresh-frozen compared to formalin-fixed, paraffin-embedded tissue samples.16 As these cell morphology features can be used as descriptors of disease states, it becomes important to interpret studies on patient diagnosis or disease pathogenesis relative to the way in which the biopsy sample was stored after acquisition. Variability in tissue staining is another concern. Even with the use of the same protocols, findings from H&E, immunohistochemistry, and immunofluorescence staining can vary between technicians, or within samples stained by the same technician on different days. With different institutions and different protocols, stain quality and appearance can vary even further.44 These differences in staining can affect the performance of deep-learning algorithms in diagnosis or lesion/cell segmentation, but they also limit the generalizability of these algorithms. While deep-learning algorithms excel in the recognition of patterns in data sets, changes in the hue, relative intensity, or staining specificity can change these patterns, causing AI to learn institution-specific features and thus reducing algorithm generalizability.
Imaging System
Performance of AI in the analysis of microscopy images is also sensitive to the imaging system on which the training data were acquired, and trained algorithms may not generalize well to new microscopes or slide scanners.47 Many variables associated with the imaging system can affect performance, including light source power and spectrum, magnification, pixel size, and objective numeric aperture. A large training set representative of the diversity of images used in the algorithm should be built, including images from multiple microscopes or slide scanners, if necessary. Additionally, a small fine-tuning data set can be used to adapt a trained network to a new imaging system or imaging protocol. Generalization between imaging systems is often further complicated by proprietary image file formats offered by various vendors, which can be difficult to convert into a standardized format. There is a need for an industry-wide file standard, analogous to the digital imaging and communications in medicine (DICOM) format used in radiologic imaging modalities.
Image Annotation
Many of the methods discussed in this review are considered supervised learning methods, in which a specialist must annotate a large training set of data to establish ground truth. Generating these training sets is a costly and time-intensive task, but algorithm performance hinges on the acquisition of a well-annotated training set. Fully unsupervised algorithms, such as clustering or dimensionality reduction, are machine-learning tools useful in digital pathology.48 Rather than learning the features associated with specific classes, unsupervised methods learn the inherent distribution of the training data. Groups of points that emerge from an unsupervised algorithm will all have similar features, but the groups may or may not have direct relevance to human-interpretable groups. Unsupervised machine-learning algorithms do not require the manual annotation overhead of supervised algorithms. However. they address separate hypotheses, and probe questions such as, How do these data points group together?, compared to supervised questions, such as, What are the similarities between data points in each class?
New developments in AI are pushing toward semi- or self-supervised machine-learning methods. Contrastive learning, a form of self-supervision, has recently been applied to medical imaging tasks to minimize the need for manual annotations. In contrastive learning, a network is trained to learn feature representations of images, ultimately identifying the features that are similar across similar images, without human-defined truth on the class of those images.49 AI in digital and computational pathology is currently dominated by data-hungry algorithms, so the application of contrastive learning methods may be useful in mitigating the need for large sets of training data. Additionally, transfer learning can be implemented to leverage trained networks to apply to new image domains, reducing the minimal amount of manual data needed for achieving high accuracy.
Summary
AI in microscopy image analysis is a rapidly evolving discipline that affects both clinical and basic science research. The automation of image classification and cell detection using computer-visualization techniques facilitates high-throughput analysis of information-dense microscopy images, particularly in the case of multiplexed microscopy. These tools are becoming increasingly mainstream, with the availability of open-source software making them accessible to a broader range of users. Machine learning–enabled feature extraction can allow for novel insights into cell environments associated with disease. However, the use of machine-learning methods introduces a set of experiment-related considerations that must be addressed to generate data sets that produce robust and meaningful insights.
Footnotes
Artificial Intelligence and Deep Learning in Pathology Theme Issue
Supported by National Institute of Allergy and Infectious Diseases (NIH) grants U19 AI082724 (M.R.C.), R01 AR055646 (M.R.C.), R01 AI148705 (M.R.C.), and U01 CA195564 (M.L.G.); and US Department of Defense grant LR180083 (M.R.C.). The content is the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosures: Maryellen L. Giger holds stock options in and receives royalties from Hologic, Inc.; holds equity in and is a co-founder of Quantitative Insights, Inc. (now, Qlarity Imaging); holds stock options in QView Medical, Inc.; and receives royalties from General Electric Company, Median Technologies, Riverain Technologies LLC, Mitsubishi, and Toshiba. It is the University of Chicago's conflict of interest policy that investigators disclose publicly actual or potential significant financial interests that would reasonably appear to be directly and significantly affected by the research activities.
This article is part of a mini-review series on the applications of artificial intelligence and deep learning in advancing research and diagnosis in pathology.
Contributor Information
Madeleine S. Durkee, Email: durkeems@uchicago.edu.
Maryellen L. Giger, Email: m-giger@uchicago.edu.
References
- 1.Bansal G.J. Digital radiography. A comparison with modern conventional imaging. Postgrad Med J. 2006;82:425–428. doi: 10.1136/pgmj.2005.038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giger M.L., Chan H.P., Boone J. Anniversary paper: history and status of CAD and quantitative image analysis: the role of medical physics and AAPM. Med Phys. 2008;35:5799–5820. doi: 10.1118/1.3013555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H., Shang S., Long L., Hu R., Wu Y., Chen N., Zhang S., Cong F., Lin S. Biological image analysis using deep learning-based methods: literature review. Digit Med. 2018;4:157–165. [Google Scholar]
- 4.Gurcan M.N., Boucheron L.E., Can A., Madabhushi A., Rajpoot N.M., Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xing F., Yang L. Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: a comprehensive review. IEEE Rev Biomed Eng. 2016;9:234–263. doi: 10.1109/RBME.2016.2515127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bera K., Schalper K.A., Rimm D.L., Velcheti V., Madabhushi A. Artificial intelligence in digital pathology-new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2014 14091556. [Google Scholar]
- 8.Sahiner B., Pezeshk A., Hadjiiski L.M., Wang X., Drukker K., Cha K.H., Summers R.M., Giger M.L. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46:e1–e36. doi: 10.1002/mp.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tizhoosh H.R., Pantanowitz L. Artificial intelligence and digital pathology: challenges and opportunities. J Pathol Inform. 2018;9:38. doi: 10.4103/jpi.jpi_53_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper L.A.D., Carter A.B., Farris A.B., Wang F., Kong J., Gutman D.A., Widener P., Pan T.C., Cholleti S.R., Sharma A., TKurc T.M., Brat D.J., Saltz J.H. Digital pathology: data-intensive frontier in medical imaging. Proc IEEE Inst Electr Electron Eng. 2012;100:991–1003. doi: 10.1109/JPROC.2011.2182074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radtke A.J., Kandov E., Lowekamp B., Speranza E., Chu C.J., Gola A., Thakur N., Shih R., Yao L., Yaniv Z.R. IBEX: a versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc Natl Acad Sci U S A. 2020;117:33455–33465. doi: 10.1073/pnas.2018488117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsujikawa T., Kumar S., Borkar R.N., Azimi V., Thibault G., Chang Y.H., Balter A., Kawashima R., Choe G., Sauer D. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017;19:203–217. doi: 10.1016/j.celrep.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G.P. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ronneberger O, Fischer P, Brox T: U-Net: Convolutional Networks for Biomedical Image Segmentation. New York: Springer. 234-241.
- 15.Yang L., Ghosh R.P., Franklin J.M., Chen S., You C., Narayan R.R., Melcher M.L., Liphardt J.T. NuSeT: a deep learning tool for reliably separating and analyzing crowded cells. PLoS Comput Biol. 2020;16:e1008193. doi: 10.1371/journal.pcbi.1008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durkee M.S., Abraham R., Ai J., Veselits M., Clark M.R., Giger M.L. Quantifying the effects of biopsy fixation and staining panel design on automatic instance segmentation of immune cells in human lupus nephritis. J Biomed Opt. 2021;26:1–17. doi: 10.1117/1.JBO.26.2.022910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Bel T., Bokhorst J.-M., van der Laak J., Litjens G. Residual CycleGAN for robust domain transformation of histopathological tissue slides. Med Image Anal. 2021;70:102004. doi: 10.1016/j.media.2021.102004. [DOI] [PubMed] [Google Scholar]
- 18.Keikhosravi A., Li B., Liu Y., Conklin M.W., Loeffler A.G., Eliceiri K.W. Non-disruptive collagen characterization in clinical histopathology using cross-modality image synthesis. Commun Biol. 2020;3:414. doi: 10.1038/s42003-020-01151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigert M., Schmidt U., Boothe T., Müller A., Dibrov A., Jain A., Wilhelm B., Schmidt D., Broaddus C., Culley S., Rocha-Martins M., Segovia-Miranda F., Norden C., Henriques R., Zerial M., Solimena M., Rink J., Tomancak P., Royer L., Jug F., Myers E.W. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat Methods. 2018;15:1090–1097. doi: 10.1038/s41592-018-0216-7. [DOI] [PubMed] [Google Scholar]
- 20.Falk T., Mai D., Bensch R., Çiçek Ö., Abdulkadir A., Marrakchi Y., Böhm A., Deubner J., Jäckel Z., Seiwald K., Dovzhenko A., Tietz O., Dal Bosco C., Walsh S., Saltukoglu D., Tay T.L., Prinz M., Palme K., Simons M., Diester I., Brox T., Ronneberger O. U-net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16:67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Z., Rahman Siddiquee M.M., Tajbakhsh N., Liang J. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Stoyanov D., Taylor Z., Carneiro G., Syeda-Mahmood T., Martel A., Maier-Hein L., Tavares J.M.R.S., Bradley A., Papa J.P., Belagiannis V., Nascimento J.C., Lu Z., Conjeti S., Moradi M., Greenspan H., Madabhushi A., editors. Springer International Publishing; Cham: 2018. UNet++: a nested U-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan D.J., Rexhepaj E., O'Brien S.L., McSherry E., O'Connor D.P., Fagan A., Culhane A.C., Higgins D.G., Jirstrom K., Millikan R.C. Altered cytoplasmic-to-nuclear ratio of survivin is a prognostic indicator in breast cancer. Clin Cancer Res. 2008;14:2681–2689. doi: 10.1158/1078-0432.CCR-07-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren S., He K., Girshick R., Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE. 2017;39:1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 24.He K., Gkioxari G., Dollár P., Girshick R. IEEE Computer Society; Venice, Italy: 2017. Mask R-CNN; pp. 2961–2969. 2017 International Conference on Computer Vision (ICCV 2017). Oct 24-27. [Google Scholar]
- 25.Lin T.-Y., Goyal P., Girshick R., He K., Dollár P. IEEE Computer Society; Venice, Italy: 2017. Focal loss for dense object detection; pp. 2980–2988. 2017 International Conference on Computer Vision (ICCV 2017). Oct 24-27. [DOI] [PubMed] [Google Scholar]
- 26.Rodemerk J., Junker A., Chen B., Pierscianek D., Dammann P., Darkwah Oppong M., Radbruch A., Forsting M., Maderwald S., Quick H.H. Pathophysiology of intracranial aneurysms: Cox-2 expression, iron deposition in aneurysm wall, and correlation with magnetic resonance imaging. Stroke. 2020;51:2505–2513. doi: 10.1161/STROKEAHA.120.030590. [DOI] [PubMed] [Google Scholar]
- 27.Rivenson Y., Wang H., Wei Z., de Haan K., Zhang Y., Wu Y., Günaydın H., Zuckerman J.E., Chong T., Sisk A.E. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng. 2019;3:466–477. doi: 10.1038/s41551-019-0362-y. [DOI] [PubMed] [Google Scholar]
- 28.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:1–11. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadanandan S.K., Ranefall P., Le Guyader S., Wählby C. Automated training of deep convolutional neural networks for cell segmentation. Sci Rep. 2017;7:1–7. doi: 10.1038/s41598-017-07599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M. Ilastik: interactive machine learning for (bio) image analysis. Nat Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 32.McMahon N.P., Jones J.A., Kwon S., Chin K., Nederlof M.A., Gray J.W., Gibbs S.L. Oligonucleotide conjugated antibodies permit highly multiplexed immunofluorescence for future use in clinical histopathology. J Biomed Opt. 2020;25:1–18. doi: 10.1117/1.JBO.25.5.056004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rashid R., Gaglia G., Chen Y.-A., Lin J.-R., Du Z., Maliga Z., Schapiro D., Yapp C., Muhlich J., Sokolov A. Highly multiplexed immunofluorescence images and single-cell data of immune markers in tonsil and lung cancer. Sci Data. 2019;6:1–10. doi: 10.1038/s41597-019-0332-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wills J.W., Robertson J., Summers H.D., Miniter M., Barnes C., Hewitt R.E., Keita Å.V., Söderholm J.D., Rees P., Powell J.J. Image-based cell profiling enables quantitative tissue microscopy in gastroenterology. Cytometry A. 2020;97:1222–1237. doi: 10.1002/cyto.a.24042. [DOI] [PubMed] [Google Scholar]
- 35.Schürch C.M., Bhate S.S., Barlow G.L., Phillips D.J., Noti L., Zlobec I., Chu P., Black S., Demeter J., McIlwain D.R. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;182:1341–1359. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:1–7. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. IEEE Computer Society; Venice, Italy: 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. 2017 International Conference on Computer Vision (ICCV 2017). Oct 24-27. [Google Scholar]
- 38.Faust K., Bala S., van Ommeren R., Portante A., Al Qawahmed R., Djuric U., Diamandis P. Intelligent feature engineering and ontological mapping of brain tumour histomorphologies by deep learning. Nat Mach Intell. 2019;1:316–321. [Google Scholar]
- 39.Diao J.A., Chui W.F., Wang J.K., Mitchell R.N., Rao S.K., Resnick M.B., Lahiri A., Maheshwari C., Glass B., Mountain V., Kerner J.K., Montalto M.C., Khosla A., Wapinski I.N., Beck A.H., Taylor-Weiner A., Elliott H.L. Dense, high-resolution mapping of cells and tissues from pathology images for the interpretable prediction of molecular phenotypes in cancer. bioRxiv. 2020 doi: 10.1101/2020.08.02.233197. [Preprint] doi: [DOI] [Google Scholar]
- 40.Nearchou I.P., Gwyther B.M., Georgiakakis E.C.T., Gavriel C.G., Lillard K., Kajiwara Y., Ueno H., Harrison D.J., Caie P.D. Spatial immune profiling of the colorectal tumor microenvironment predicts good outcome in stage II patients. NPJ Digit Med. 2020;3:1–10. doi: 10.1038/s41746-020-0275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazarus J., Maj T., Smith J.J., Lanfranca M.P., Rao A., D'Angelica M.I., Delrosario L., Girgis A., Schukow C., Shia J. Spatial and phenotypic immune profiling of metastatic colon cancer. JCI Insight. 2018;3:e121932. doi: 10.1172/jci.insight.121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Jiang Y., Li B., Cui Y., Li D., Li R. Single-cell spatial analysis of tumor and immune microenvironment on whole-slide image reveals hepatocellular carcinoma subtypes. Cancers. 2020;12:3562. doi: 10.3390/cancers12123562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niethammer M., Borland D., Marron J.S., Woosley J., Thomas N.E. Appearance normalization of histology slides. Mach Learn Med Imaging. 2010;6357:58–66. doi: 10.1007/978-3-642-15948-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bancroft J.D., Gamble M. Elsevier Health Sciences; New York: 2008. Theory and Practice of Histological Techniques. [Google Scholar]
- 45.Hoppin J.A., Tolbert P.E., Taylor J.A., Schroeder J.C., Holly E.A. Potential for selection bias with tumor tissue retrieval in molecular epidemiology studies. Ann Epidemiol. 2002;12:1–6. doi: 10.1016/s1047-2797(01)00250-2. [DOI] [PubMed] [Google Scholar]
- 46.Jiang H., Nachum O. Identifying and correcting label bias in machine learning. Proc Mach Learn Res. 2020;108:702–712. [Google Scholar]
- 47.Weinstein R.S., Graham A.R., Richter L.C., Barker G.P., Krupinski E.A., Lopez A.M., Erps K.A., Bhattacharyya A.K., Yagi Y., Gilbertson J.R. Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future. Hum Pathol. 2009;40:1057–1069. doi: 10.1016/j.humpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 48.Rashidi H.H., Tran N.K., Betts E.V., Howell L.P., Green R. Artificial intelligence and machine learning in pathology: the present landscape of supervised methods. Acad Pathol. 2019;6 doi: 10.1177/2374289519873088. 2374289519873088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen T., Kornblith S., Norouzi M., Hinton G. A simple framework for contrastive learning of visual representations. Proc Mach Learn Res. 2020;119:1597–1607. [Google Scholar]