Abstract
Background
The development of rational combination therapies is key to overcome inherent treatment resistance of glioblastoma (GBM). We aim at identifying new druggable targets by disturbing GBM cells with inhibitors of bromodomain and extra-terminal motif (BET) proteins to reveal cancer-relevant vulnerabilities that may sensitize to a second drug. BET proteins are epigenetic modulators and have been associated with proto-oncogene overexpression in cancer.
Methods
A GBM-derived sphere-line was treated with the BET inhibitor (BETi) JQ1 over a time-course of 48 hours, followed by RNA-sequencing. Four chromatin marks were investigated by chromatin immunoprecipitation followed by sequencing (ChIP-seq). Signatures of interest were functionally validated in vitro and in orthotopic xenografts. Combination therapies were evaluated for synergistic effects.
Results
Cancer-relevant pathways significantly modulated by JQ1 comprised interferon alpha (IFN-α) response genes and response signatures to histone deacetylase inhibitors (HDACi). The IFN-signature was reminiscent of a GBM-derived IFN-signature comprising CD274 (PD-L1). Functional pathway analysis suggested that JQ1 was acting directly on the transcriptional level of IFN-response genes and not via the canonical JAK/STAT pathway. This was in line with JQ1 modulated expression and BRD4 and Pol II occupancy at IFN-signature genes, supporting a direct mechanistic interaction. Finally, we showed that combining HDACi with JQ1 acts synergistically in reducing cell viability of GS-lines.
Conclusions
Our approach identified BETi-induced vulnerabilities in cancer-relevant pathways, potentially amenable to synergistic combinatorial therapy, such as combination with HDACi. The direct inhibitory effect of BETi on IFN-responsive genes in GBM cells, including CD274, indicates modulation of the tumor immune landscape and warrants further studies.
Keywords: BET inhibitors, epigenetics, glioblastoma, interferon-stimulated genes, synergistic combination therapy
Key Points.
BRD4 regulates IFN-stimulated gene signature.
Combination of BETi and HDACi is synergistic.
Importance of the Study.
Glioblastoma is one of the most difficult tumors to treat and is notorious for its resistance to treatment, despite recent insights on the evolution of the disease gained from multidimensional omics. The failures of single agents tested in clinical trials have dramatically shown that combination therapies are necessary to improve the outcome for patients affected with these treatment-resistant tumors. However, the rational choice for successful combination therapy is the big challenge. Here, we present our approach to harness epigenetic changes that may be targeted by a new class of drugs, bromodomain and extra-terminal (BET) inhibitors. BET proteins are epigenetic regulators that have been associated with overexpression of proto-oncogenes in a variety of cancers and seem to be promising cancer-relevant targets. We disturb the system with a small molecule BET inhibitor, which elucidates emerging pathway vulnerabilities that may become targetable with a second drug.
Glioblastoma (GBM) is the most common and most malignant primary brain cancer in adults with a devastating prognosis. Even under the best care including all approved treatment modalities, median overall survival is just 19 months. Current treatment comprises maximal safe resection followed by radio-chemotherapy with temozolomide and tumor-treating fields.1 Over a dozen clinical trials adding targeted drugs to the standard of care failed to improve overall survival.2 Multiple intrinsic resistance mechanisms leading to treatment evasion have been described. Alterations conferring resistance may be preexisting or brought upon by treatment through acquisition or selection of combinations of epigenetic and genetic alterations.3 Hence, it has become clear that a single targeted drug is not likely to be curative; hence synergistic combinatorial strategies need to be explored. The epigenome has been shown to play a multifaceted role in GBM pathogenesis,4 and to carry cell-of-origin and cell fate information,5,6 but in addition to displaying changes associated with cancer development and progression. Hence, the epigenome informs on the tumor subtype, can be harnessed to identify novel target opportunities and uncover treatment resistance, and thereby serves tumor classification and biomarker development for patient stratification and treatment.7–9 In this study, we aim at disturbing the GBM epigenome using epigenetic drugs to uncover pathway vulnerabilities that may be exploited for combinatorial treatment strategies.
Bromodomain and extra-terminal motif (BET) proteins belong to a class of epigenetic proteins called “chromatin readers” since their function is to recognize acetylated lysine on histone tails and promote the signal downstream.10 BET proteins are bound to promoter regions and active enhancer elements, and recruit mediator complexes to promote target gene transcription. Inhibitors of BET (BETis) are particularly attractive since GBM expresses an arsenal of oncogenes,11 which are unfeasible to inhibit simultaneously with targeted agents due to expected overt toxicity. BETis as single agents showed promising results in preclinical GBM models12–14 and are in clinical testing for progressive grade II and III gliomas, recurrent GBM, and newly diagnosed GBM, respectively (NCT04047303, NCT04324840, and reviewed in Ref.15).
In the current work, we show that the BETi JQ1, a tool drug with excellent penetration of the blood-brain barrier (BBB),16 suppresses the expression of an interferon-stimulated gene (ISG) signature in GBM models, associated with loss of BRD4 binding across ISGs. Moreover, expression changes induced by JQ1 treatment comprise signatures similar to those reported for histone deacetylase inhibitors (HDACi). With the combination of BETi and HDACi, we show a synergistic effect in reducing GBM cell viability.
Methods
Cell Culture
Human GBM-derived sphere (GS)-lines LN-2207GS, LN-2683GS, LN-2540GS, LN-2669GS, LN-3704GS, LN-3708GS, and adherent GBM cell lines LN-18 and LN-428 were established and molecularly characterized17–19 in our laboratory, with ethics approval (CER-VD, PB_2017-00240/F-25/99). U87MG was obtained from ATCC and BS-153 from Adrian Merlo.20 All lines were authenticated as described19 and tested mycoplasma-free (MycoAlert Kit, Lonza). Adherent lines were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco), 5% fetal calf serum (Hyclone, Thermo Fisher Scientific). Experiments with GS-lines were performed under stem cell conditions in DMEM/F12 (Invitrogen) supplemented with B27 (Gibco) (1/50) and growth factors as previously described.21 Spheres were separated into single-cell suspension using Accutase (Gibco). GS-lines were primed with 1000 units/mL interferon alpha (IFN-α) (Peprotech) or 20 ng/mL IFN-γ (GenWay Biotech). (+)-JQ1, its inactive enantiomer (−)-JQ1 (ApexBio Technology LLC) and trichostatin A (TSA, Sigma) were dissolved in dimethyl sulfoxide (DMSO) at 10 mM and 5 mM, respectively.
Cell Assays
The senescence-associated β-galactosidase assay on adherent GBM cells was performed according to the manufacturer’s instructions (BioVision). Immunofluorescence of sphere lines used anti-TUJ1 (Cell Signalling, 1:400) and fluorochrome-conjugated anti-rabbit Alexa647 (Abcam, 1:500). The BrdU incorporation assay was performed using FITC (fluorescein isothiocyanate)-labeled BrdU antibody and 7-AAD (7-aminoactinomycin D, Pharmingen™ BrdU Flow Kit, BD Biosciences) and was analyzed by fluorescence-activated cell sorting (FACS, Calibur). Experimental details are available in the Supplementary Methods.
Cell Viability Assays
For adherent GBM cell lines (+)-JQ1, or (−)-JQ1 was added in a 10-point serial dilution of 1:3, starting from 30 µM. GS-lines were treated using a 10-point serial 1:2 dilution, starting with 30 µM of (+)-JQ1 or (−)-JQ1 (see details in Supplementary Methods). After 72 hours, cell viability was measured using the CellTiter-Blue or CellTiter-Glo Assay (Promega). Lethal concentrations of actinomycin D (1 µg/mL) or DMSO were used as 0% and 100% viability controls. A curve-fitting algorithm with variable Hill slope was applied to derive IC50 values. Experiments were repeated 3 times.
Drug Combination Assay
GS cells were treated using a 7-point serial 1:2 dilution starting with 20 µM, employing a chessboard pattern (8 × 8), and every concentration of JQ1 was tested with every concentration of TSA. DMSO served as control. Cell viability was measured after 5 days using the CellTiter-Blue Assay. For the 10-day assay, JQ1 and TSA were added at a serial dilution starting from 2 µM. After 5 days, drugs were renewed as detailed in the Supplementary Methods. The combination score22,23 was determined using the R package synergyfinder (combination of Lowe and Bliss combination indexes).24 Experiments were repeated 3 times.
Neurosphere Formation Assay
Adapting a protocol by Cheng et al,13 single cells were seeded into a 48-well plate in 100 µL F12/B27 medium (containing EGF [epidermal growth factor]/FGF [fibroblast growth factor]) with 100 (LN-2207GS, LN-2540GS, LN-2683GS) or 1000 cells/well (LN-2669GS). JQ1, diluted in 100 µL F12/B27 to a final concentration of 0, 0.25, 0.5, 1, 2, or 4 µM, was added, with 4 replicates per condition. EGF/FGF were replenished on days 8 and 15 (20 ng/mL). The spheres with a diameter >50 µm were counted after 14 and 21 days. The experiment was repeated 3 times.
RNA Extraction, qRT-PCR
Total RNA isolation and qRT-PCR were performed as described previously21 using primers compiled in Supplementary Table S1. The expression levels were normalized to GAPDH and to the geometrical mean of hGAPDH and C1orf43 for cDNA derived from xenografts.
Western Blot and Nuclear Fractionation
Cells were collected by centrifugation for GS-lines and by scraping for adherent cells. Westerns were done as described21,25 and probed with respective antibodies (Supplementary Methods). Nuclear and cytoplasmic fractionation was performed using the NE-PER Kit (Thermo Scientific) with modifications detailed in the Supplementary Methods.
RNA Sequencing and Differential Expression Analysis
LN-2683GS cells were treated with (+)-JQ1 (1 µM) or DMSO and harvested at 0, 4, 12, 24, and 48 hours. RNA-sequencing was accomplished at the Genomic Technology Facilities (University of Lausanne). Data were preprocessed following standard pipeline and recommendations from bcbio-nextgen (version 0.8, http://bcbio-nextgen.readthedocs.org/en/latest/). Differential expression analysis used a model with full interaction between treatment and time (edgeR package, version 3.12.0). Genes with Bonferroni adjusted P values <.001 were considered differentially expressed. Gene-set enrichment analysis (GSEA) was performed (PGSEA package, version 1.44.0; GAGE package, version 2.20.1) using The Molecular Signatures Database (MSigDB v5, Broad Institute). Gene sets with FDR (false discovery rate)-adjusted P values <.05 were considered significant. Due to patient privacy protection, the raw sequencing data will be made available upon request. See the Supplementary Methods for details.
Chromatin Immunoprecipitation (ChIP)
LN-2683GS cells were treated with 1 µM JQ1, DMSO, and 1000 units/mL IFN-α for 2 hours. U87MG were treated with 1 µM JQ1 or DMSO for 24 hours. Cells were cross-linked and ChIP was performed using iDeal ChIP-seq kit for Transcription Factors (Diagenode) as detailed in the Supplementary Methods, using the following antibodies: anti-Pol II (Cell Signaling, D8L4Y, 1:50), anti-BRD4 (Bethyl Laboratories, A301, 1:60), anti-H3K4me3 (Diagenode, C15410003, 1:300), anti-H3K27ac (Abcam, ab4729, 1:300 or 1:600), control IgG (1:600, Diagenode).
ChIP Sequencing Data Analysis
The preprocessing of ChIP-seq data was performed following the pipeline suggested by project bcbio-nextgen (version 1.1.6, http://bcbio-nextgen.readthedocs.org/). The binding affinity matrix was built by peaks identified in at least 2 samples. The raw read counts were normalized and batch corrected to process ChIP-seq data prior to further analysis. The global effect of JQ1 treatment, IFN stimulation, and their interaction with the IFN-signature for the 4 chromatin marks was tested using Permutational Multivariate Analysis of Variance based on Euclidean distance matrices (ADONIS with 999 permutations26). Corresponding RNA-seq data, normalized and batch corrected was used in the heatmap and to compute pairwise Spearman correlation between Pol II marks and expression of the related genes. The ChIP-seq normalized datasets were weighted by their total inertia for the simultaneous heatmap representation as used in MFA (multiple factor analysis).27 See details in the Supplementary Methods.
Orthotopic Xenografts
Orthotopic xenografts were obtained with 105 U87MG cells transplanted into the striatum of 6-NOD-SCID-gamma knockout mice as previously described.21 Mice were injected i.p., with JQ1 (100 mg/kg) 4 hours before sacrifice. The protocol was approved by the local authorities (VD-3266). See the Supplementary Methods for details.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7 Software and R-3.2.2.28 Bioconductor29 packages for specific tasks are listed in the relevant sections. The Student t test was used to compare the variables between 2 groups, and 2-way ANOVA for more complex experimental design. P values <.05 were considered statistically significant: significance is indicated with asterisks: *P < .05, **P < .01, ***P < .001, and ****P < .0001. Data are presented as mean values, error bars represent standard deviation unless indicated otherwise. Similarity between distance matrices was evaluated by RV (vectorial correlation) coefficient (values between 0 and 1),30 using R package ade431 for pairwise RV coefficient permutation tests.
Results
JQ1 Treatment Reduces Sphere Formation and Viability of GBM Cells In Vitro
First, we evaluated the effects of JQ1 on cell viability of GS-lines (Figure 1A). They appeared more resistant to JQ1 at 72 hours as compared to adherent GBM cell lines, with IC50 ranging between 5.8-18.6 µM and 1.4-11.6 µM, respectively (Supplementary Table S2). A proportion of cells remained viable at high JQ1 concentrations that were associated with a senescence-like phenotype, marked by an increased proportion of β-galactosidase-positive cells (Supplementary Figure S1), similar to other cancer types.32
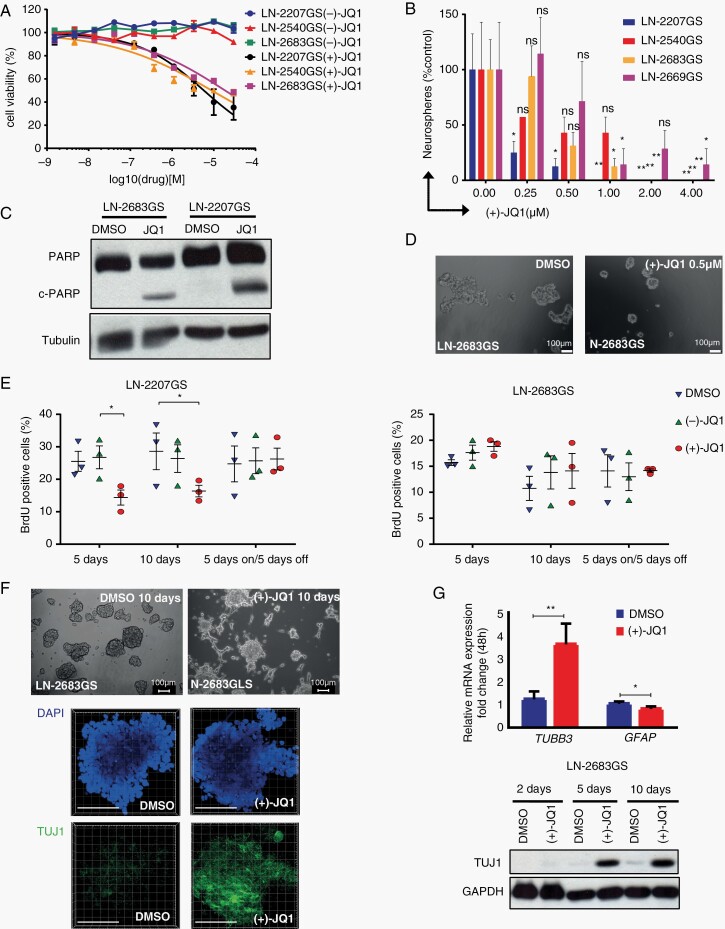
Fig. 1.
Effects of JQ1 on glioma sphere lines. (A) Cell viability of LN-2207GS, LN-2540GS, and LN-2683GS was determined after 72 hours’ incubation with 3-fold serial dilutions of (+)-JQ1 and (−)-JQ1, normalized to DMSO control. Data represent a mean of 3 independent experiments; error bars are SEM. (B) Quantification of neurosphere formation assay of LN-2207GS, LN-2540GS, LN-2683GS, and LN-2669GS. The spheres >50 µm in diameter were counted 21 days after (+)-JQ1 addition. Data represent mean of 4 wells; error bars are SD; representative of at least 3 independent experiments. P values, 2-way ANOVA, adjusted for multiple testing, statistical hypothesis Dunnett test. (C) Western blot for total PARP, cleaved PARP (c-PARP), and tubulin (loading control) of LN-2683GS and LN-2207GS, 10 days after treatment with 1 µM (+)-JQ1 or DMSO. (D) Representative image of neurospheres taken after 21-day incubation of LN-2683GS with 0.5 µM (+)-JQ1 and DMSO. (E) BrdU incorporation assay of LN-2207GS and LN-2683GS treated with 1 µM (+)-JQ1, (−)-JQ1, or DMSO for 5, 10, or 5 days on/5 days off. Data represent summary of 3 independent experiments (*P value < .05, paired t test). (F) Representative microphotographs of LN-2683GS treated 10 days with 1 µM (+)-JQ1 or DMSO. DAPI (blue), TUJ1 (green), scale bars 100 µm. (G) qRT-PCR of TUBB3 (TUJ1) and GFAP after 48 hours’ treatment with 1 µM (+)-JQ1 or DMSO. Data represent mean of 4 independent experiments, error bars are SD, paired t test. Western blot for TUJ1, GAPDH of LN-2683GS treated with 1 µM (+)-JQ1 for 2, 5, and 10 days. Abbreviations: ANOVA, analysis of variance; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; PARP, poly(ADP-ribose) polymerase; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SD, standard deviation; SEM, standard error of the mean.
The treatment of GS-lines with JQ1 (0.25-1 µM) reduced their capacity of sphere formation by 50% over 21 days (Figure 1B and D). Subsequent cell cycle analysis (BrdU incorporation) revealed a decreased proportion of cells in S-phase for LN-2207GS cells treated with 1 µM JQ1 for 5 days (Figure 1E), which was fully reversible upon 5-day drug withdrawal. In contrast, no difference in cell cycle distribution was observed for LN-2683GS. Importantly, 5-day JQ1 treatment induced apoptosis in LN-2207GS and LN-2683GS, based on cleaved poly(ADP-ribose) polymerase (c-PARP) (Figure 1C). Of note, we observed that after short exposure (24 hours) to 1 µM JQ1, LN-2683GS tended to become adherent and formed cell protrusions (Figure 1F). Suspecting JQ1 mediated cell differentiation of LN-2683GS, we assessed markers of neuronal (TUJ1, neuronal-specific class III β-tubulin) and astrocytic (GFAP, glial fibrillary acidic protein) differentiation. TUBB3 (encoding TUJ1) expression was almost 4 times upregulated after 24 hours’ treatment with JQ1, confirmed on the protein level 5 and 10 days after JQ1 addition, suggesting a neuronal differentiation-like phenotype (Figure 1F). MYC was repressed by JQ1 in GS-lines, (Supplementary Figure S2) confirming previous studies.13,33,34
BET Protein Inhibition Causes Extensive Changes in the Transcriptome
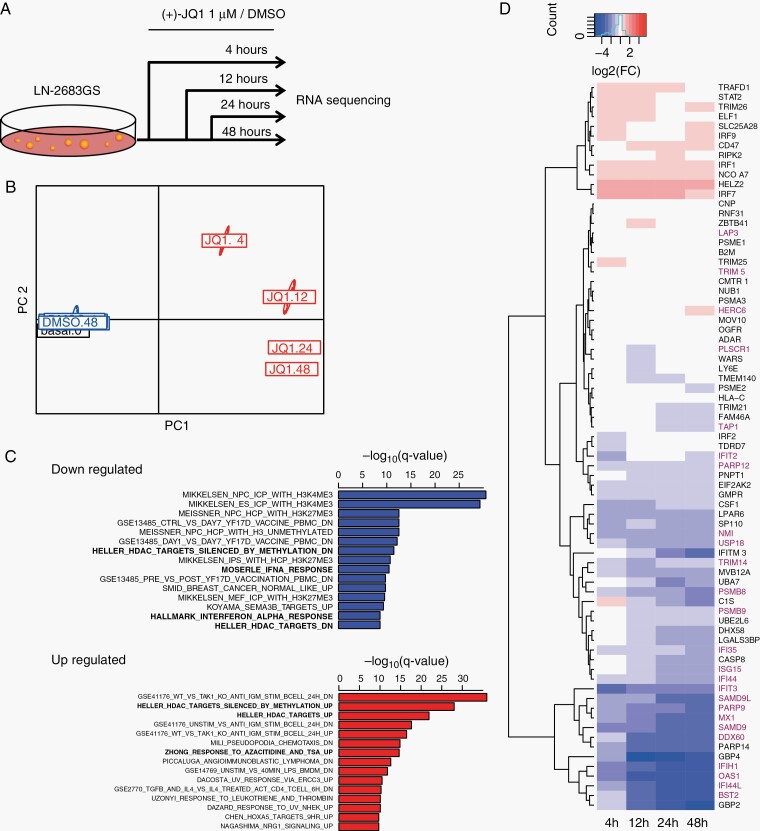
To gain insights into biological changes we performed differential gene expression profiling by RNA-sequencing of LN-2683GS treated with 1 µM JQ1 over a time course of 4, 12, 24, and 48 hours (Figure 2A). This yielded 2709 differentially expressed genes (1278 down, 598 up) using an additive model with interaction between time and treatment (log-likelihood ratio test, Bonferroni, P < .001; fold change >I2I for at least 1 time point) (Supplementary Table S3). Pathway analysis of differentially expressed genes revealed significant modulation of gene sets, such as those associated with particular epigenetic features of neural progenitor cells (NPCs) and embryonic stem (ES) cells, gene sets modulated by epigenetic interference, and inflammation-related signatures (Figure 2C). In JQ1-treated LN-2683GS cells, these comprised downregulation of interferon-stimulated genes (ISG) enriched for Hallmark interferon alpha response (HIAR) signature genes (Figure 2D). Furthermore, we detected enrichment in gene sets associated with cellular response to HDACi (Figure 2C).
Fig. 2.
Transcriptome analysis of JQ1 effects. (A) Experimental setup of differential gene expression analysis by RNA-seq. LN-2683GS cells were treated with 1 µM (+)-JQ1 or DMSO for 4, 12, 24, and 48 hours, the experiment was repeated 3 times, followed by RNA-seq. (B) Principal coordinate analysis of the raw read counts of RNA-seq data. PC1, PC2: principal coordinates 1 and 2. (C) Top 15 downregulated (blue) and upregulated (red) gene sets based on GSEA (q-value, Bonferroni corrected for multiple testing). Gene sets in bold were selected for further analysis. (D) Heatmap of log2FC (FC: fold change) of expression of IFN-α response gene set (HIAR). Genes marked in red overlap with our GBM IFN-signature (G12, Supplementary Figure S3).35 Abbreviations: DMSO, dimethyl sulfoxide; GBM, glioblastoma; GSEA, gene-set enrichment analysis; HIAR, hallmark interferon alpha response; IFN-α, interferon alpha.
Interestingly, we found that the JQ1-modulated HIAR gene set overlapped largely (25 genes) with our previously reported ISGs expression signature (G12) identified in a human GBM dataset35 (Figure 2D). This GBM-derived IFN-signature (G12) shows high similarity of co-expression among publically available GBM datasets (RV values > 0.8; P value = .001; Supplementary Figure S1A and B) and comprises in addition genes associated with IFN-γ response, such as CD274, encoding PD-L1 (programmed death-ligand 1), known for its importance in tumor immune evasion.
Expression of ISG Signature Is Repressed Upon JQ1 Treatment
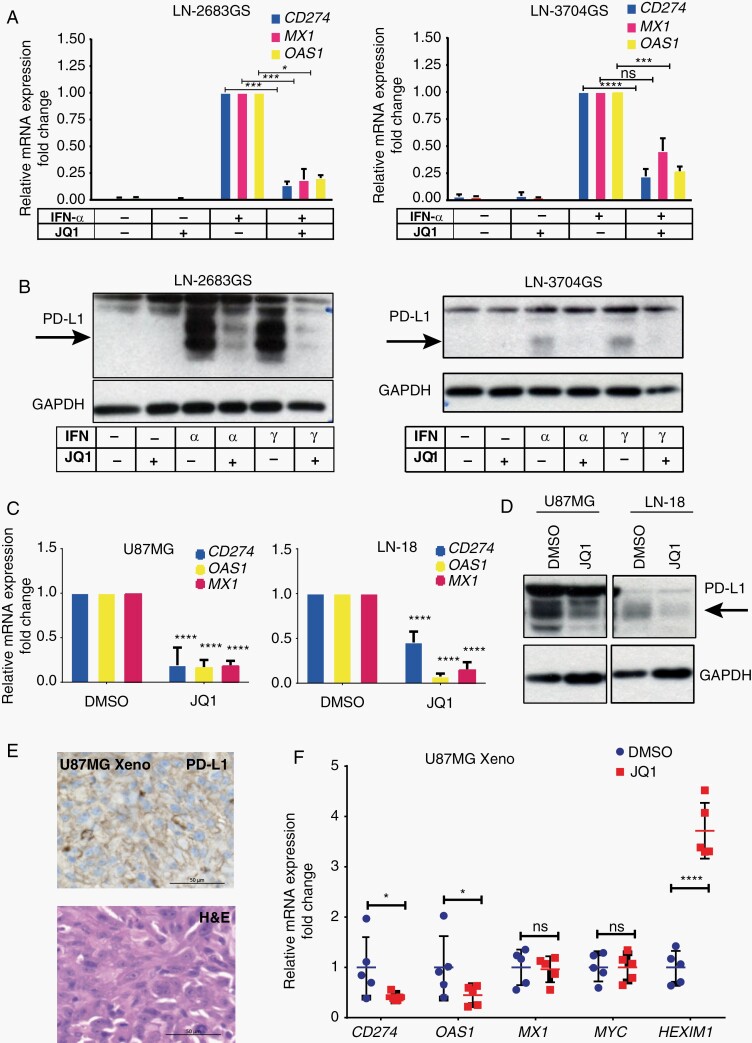
To validate the downregulation of ISGs upon JQ1 treatment, we boosted IFN signaling by priming GS-lines with IFN-α for 4 hours, followed by 4 hours of JQ1 exposure. We chose OAS1 and MX1 as markers of IFN-response (Figure 3A), both carry an interferon-sensitive response element (ISRE) and also belong to the G12 signature (Supplementary Figure S3A). Furthermore, we included CD274 (PD-L1) in the analysis for its importance in anti-tumor immunity. We detected a strong reduction of expression of the ISGs OAS1, MX1, and CD274 in JQ1 - treated GS-lines as compared to DMSO (Figure 3A). Similar results were obtained with IFN-γ priming, confirming repression of CD274 by JQ1 treatment in the GS-lines on the RNA and protein level (Figure 3B).
Fig. 3.
Inhibition of interferon-stimulated genes upon JQ1 treatment in vitro and in vivo. Expression of CD274, MX1, and OAS1 (qRT-PCR) in LN-2683GS and LN3704GS primed 4 hours with IFN-α, followed by 4 hours’ JQ1 (1 µM) treatment (A). Mean of 3 independent experiments, SD; 2-way ANOVA, multiple testing correction, Sidak test. (B) Western blot for PD-L1 and GAPDH for LN-2683GS and LN-3704GS treated as in A, representative of 3 independent experiments. Arrows indicate specific band for PD-L1. (C) Expression of endogenous ISGs, CD274, MX1, and OAS1 (qRT-PCR) in adherent GBM cell lines U87MG and LN-18 after 48 hours’ treatment with 1 µM JQ1 (C) (2-way ANOVA, multiple testing correction, Sidak test). (D) Western blot for PD-L1 in U87MG and LN-18 treated as in (C). (E) U87MG derived orthotopic xenograft immunostained for PD-L1 and H&E, scale bar 50 µm. (F) qRT-PCR analysis of CD274, MX1, OAS1, and c-Myc expression in orthotopic U87MG xenografts. Mice were injected i.p. with 100 mg/kg JQ1 or vehicle control 4 hours before sacrifice (n = 5 mice per group pulled from 3 independent experiments, 2-way ANOVA; adjusted for multiple testing, FDR, 2-stage linear step-up procedure36). Abbreviations: ANOVA, analysis of variance; FDR, false discovery rate; H&E, hematoxylin and eosin; IFN-α, interferon alpha; ISGs, interferon-stimulated genes; PD-L1, programmed death-ligand 1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SD, standard deviation.
To exclude possible effects of JQ1 on JAK/STAT signaling per se, we examined pSTAT1 levels in the nucleus of GS-lines after priming with IFN-α or IFN-γ. While total STAT1 and pSTAT1 were increased in the nucleus after IFN-priming, we did not observe a change upon JQ1 treatment in the GS-lines as compared to DMSO (Supplementary Figure S4).
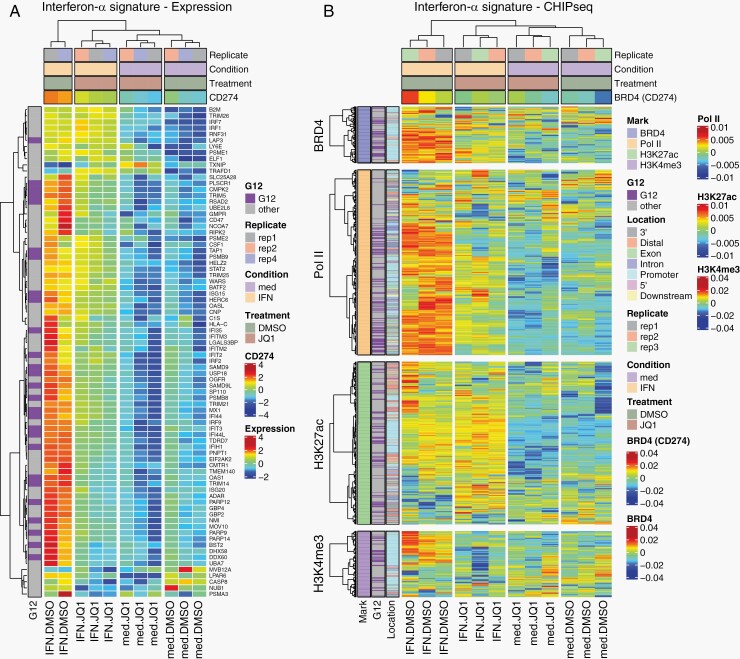
IFN-α Stimulation Enhances BRD4 Occupancy at ISGs
To evaluate whether the ISGs are regulated through BRD4-mediated transcription, we performed ChIP-seq for BRD4 in LN-2683GS cells, with or without 2 hours’ treatment with IFN-α, or JQ1, or their combination. Modulation of BRD4 coverage was observed at our marker genes CD274, MX1, and OAS1 under the 4 experimental conditions, with increased binding upon IFN-α stimulation, and decrease under JQ1 treatment. Acetylation of H3K27 was increased by IFN-α stimulation, along with Pol II recruitment at the active promoters of target genes, while JQ1 treatment did not seem to alter H3K27ac levels, but reduced Pol II occupancy at the active promoters defined by H3K4me3 peaks. The tracks of the consensus peaks for BRD4, Pol II, and the histone marks H3K27ac and H3K4me3 are illustrated for the area within 20 kb of the transcription start sites (TSSs) (Supplementary Figure S5). Next, we explored whether the whole ISG gene signature was modulated by BRD4-binding under the 4 experimental conditions and performed GSEA of BRD4 occupancy using consensus peaks. The results from genome-wide differential binding analysis were significant for the expression of the ISG signature (additive effect), for treatment with JQ1 (adj. P value = .009), for priming with IFN-α (adj. P value = .0018), and interaction between JQ1 treatment and IFN-α stimulation (adj. P value = .0021). In order to illustrate that BRD4 occupancy is similarly modulated across the IFN-signature genes under the 4 experimental conditions, a heatmap of the BRD4 consensus peaks associated with these ISG signature genes is shown, complemented with respective heatmaps for RNA-seq and ChIP-seq for Pol II, H3K27ac, and H3K4me3 (Figure 4). Similar to the 3 marker genes, CD274, MX1, and OAS1, most of the IFN-signature genes revealed increased BRD4 occupancy with IFN-α stimulation that was repressed upon JQ1 treatment, which was also reflected on the respective RNA expression levels. Furthermore, IFN-α stimulation triggered acetylation of H3K27 along with Pol II binding, whereas JQ1 treatment did not significantly alter H3K27ac levels, but significantly reduced Pol II occupancy. The respective multivariate analysis of the effects on the IFN-signature by experimental modality is summarized in Supplementary Table S4 for the 4 chromatin marks and RNA expression.
Fig. 4.
JQ1 impairs the transcriptional program activated by IFN-α. (A) Heatmap representation of RNA-seq for ISGs from LN-2683GS under 4 experimental conditions: LN-2683GS cells were treated for 2 hours with IFN-α, 1 µM JQ1, or the combination thereof. The samples are annotated for treatment (JQ1, DMSO), condition (IFN-α, media [med]), and replicate. The normalized CD274 expression was added as a supplementary variable. Genes overlapping with the G12 GBM IFN-signature are annotated in a separate column (G12, purple, nonG12, gray). (B) Heatmap shows consensus peak intensities (ChIP-seq) of BRD4, Pol II, H3K27ac, and H3K4me3 at ISGs in LN-2683GS under 4 experimental conditions as in (A). The sample dendrogram is based on the BRD4 ChIP-seq dataset. The BRD4 consensus peak of CD274 was added as a supplementary variable. The consensus peak locations (promoter, intergenic, exon, intron, etc.) of BRD4, Pol II, H3K27ac, and H3K4me3 are annotated in a color code as indicated. The peaks overlapping with G12 genes are annotated (G12, purple; nonG12, gray). Abbreviations: DMSO, dimethyl sulfoxide; GBM, glioblastoma; IFN-α, interferon alpha; ISGs, interferon-stimulated genes.
No super-enhancers associated with the modulation of the expression of the ISGs were identified using H3K27ac marks and previously published methodology.37,38 The Spearman correlation between mRNA expression and Pol II binding in the promoter region ranged from 0.7 to over 0.9 for our marker genes OAS1, MX1, and CD274. Together, these results support the notion that ISGs may be direct targets of BRD4, and therefore are subjected to transcriptional repression by JQ1 in GS.
Endogenous Expression of ISGs Is Repressed Upon JQ1 Treatment In Vitro and In Vivo
To investigate whether JQ1 represses endogenous expression of ISGs, we treated U87MG and LN-18 cell lines, which express CD274 (PD-L1) endogenously, with 1 µM JQ1 for 48 hours. As a result, CD274, OAS1, and MX1 expression were largely reduced, and PD-L1 protein was diminished (Figure 3C and D). In accordance, ChIP-seq analysis of U87MG, visualized for CD274 (Supplemental Figure S6), indicates binding of BRD4, H3K27ac, Pol II, and H3K4me3 at the TSS before JQ1 treatment (IFN-untreated).
To determine the effects of JQ1 on ISGs expression in vivo, we administered JQ1 (100 mg/kg, i.p.) or vehicle control to mice bearing orthotopic U87MG-derived xenografts for 4 hours before their sacrifice. Using human-specific primers, a significant decrease in CD274 and OAS1 expression was detected in the tumor xenografts upon JQ1 treatment, whereas no significant difference was observed for MX1 and c-Myc (Figure 3F). Together with the strong JQ1-induced upregulation (4-fold) of HEXIM1, a pharmacodynamic marker for BETi exposure,39 the data suggested that a pharmacologically relevant concentration of JQ1 was reached in the orthotopic tumor xenografts.
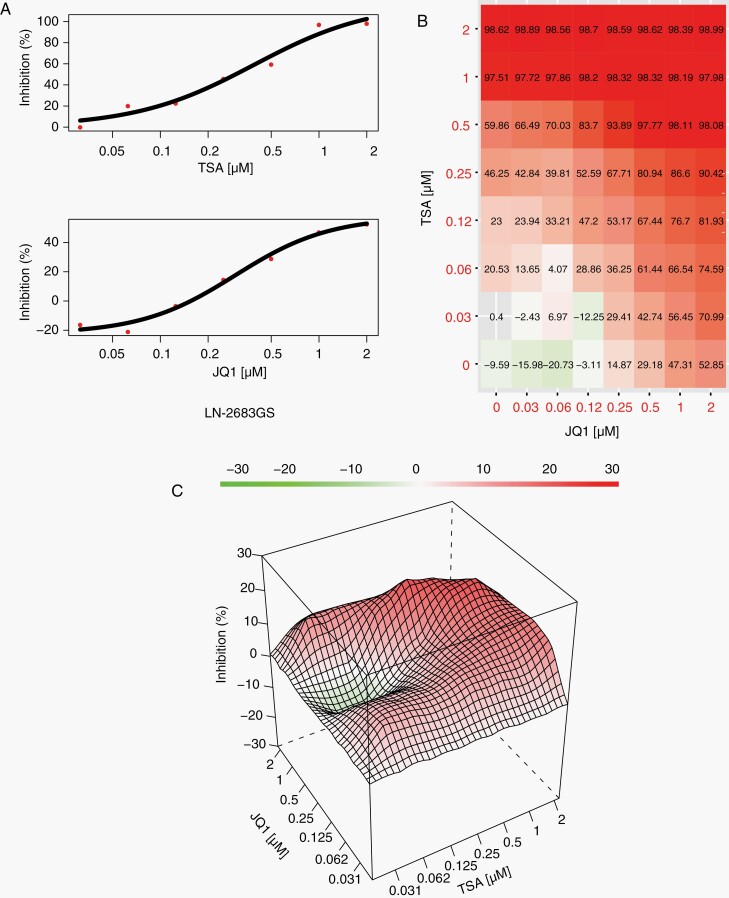
JQ1 and TSA Act Synergistically to Reduce Cell Viability of GS-Lines
The JQ1-induced modulation of gene sets related to HDACi-associated cellular response signatures40 motivated us to test whether JQ1 sensitizes GS-lines to HDACi treatment. We therefore tested TSA, a small molecule pan-HDAC inhibitor in combination with JQ1 in the GS-lines LN-2683GS and LN-3708GS. A scheme of 64 possible combinations of JQ1 and TSA was employed, ranging from 0 µM to 10 µM for a 3-day assay and from 0 µM to 2 µM for a 10-day assay (Figure 5A–C, Supplementary Figure S7). The dose-response matrix (Figure 5B) was analyzed to determine the range of concentrations with synergistic drug interactions (Figure 5C). Based on the performed analysis, synergistic drug interactions explained 5% of cell deaths in LN-2683GS after 3 days, and 9% after 10 days, and for LN-3708GS 15% and 4% after 3 and 10 days, respectively (Supplementary Figure S7).
Fig. 5.
Combination of JQ1 with TSA shows synergy in reducing cell viability of GS-lines. (A) Single-drug cell viability curves for LN-2683GS treated with serial 2-fold dilutions of TSA or JQ1, respectively, starting with 2 µM for 10 days. Data represent mean of 3 plates (technical replicates). (B) Combination matrix of cell inhibition by JQ1 and TSA. 100% is inhibition of LN-2683GS cells treated with the lethal dose of 1 µg/mL actinomycin D, 0% is inhibition of LN-2683GS exposed to DMSO. Data represent mean of 3 wells (technical replicates). (C) Combination landscape, synergistic range of TSA and JQ1 concentrations in red, additive range in green, representative of 3 independent experiments. The synergy score is estimated by the zero interaction potency (ZIP) model, where the expected response corresponds to an additive effect as if the 2 drugs do not affect the potency of each other.22,23 The ZIP score has a unit of percentage inhibition and can be interpreted as the proportion of cellular responses that can be attributed to the drug interactions; ZIP score = 0 no interaction, ZIP score >0 synergistic, and <0 for antagonistic effect. Abbreviations: DMSO, dimethyl sulfoxide; GS, glioblastoma-derived sphere; TSA, trichostatin A.
Discussion
BETis have demonstrated efficacy in NUT-midline carcinoma that is driven by the oncogenic BRD4-NUTM1 translocation. BETis are currently studied in other solid tumors including glioma, where they are tested as single agents or in combination with standard radio-chemotherapy. Hence novel strategies for rational combination strategies are warranted.41,42
In GBM models, the BETis JQ1, I-BET151, OTX015, and the BET-degrader, dBET6, have displayed some positive preclinical efficacy.12–14,43,44 Our data support the anti-proliferative activities of JQ1 in both adherent cell lines and patient-derived GS-lines, showed apoptotic cell death in patient-derived GS-lines, and uncovered a differentiation-like phenotype in some GS-lines.
In this study, we focus on predicting rational combination strategies by identifying pathway vulnerabilities induced by the BETi JQ1. Pathway analysis of genes disturbed by JQ1 treatment uncovered an IFN-response signature among the top downregulated gene sets, suggesting that BETi suppresses transcription of a significant proportion of IFN-response genes in GBM. This is of interest, as around 50% of GBM display enhanced expression of a reminiscent IFN-signature, as we have reported previously (Supplementary Figure S3).35 Indeed, based on the integrated analysis of ChIP-seq and RNA-seq data, a large fraction of IFN-response genes are activated through acetylation of H3K27 and BRD4 recruitment upon IFN stimulation. Thus, transcription of ISGs, including CD274, is directly repressed by JQ1-induced release of BRD4 from the chromatin. Of note IFN-α induced acetylation of H3K27 at IFN-signature genes was not significantly affected by JQ1 treatment, in accordance with a recent report suggesting that BETi blunts transcription, but retains enhancer-promoter contact.45 JQ1 impaired expression of endogenous, as well as interferon-induced ISGs. This suggests that BRD4 chromatin occupancy of ISGs is a key regulator of response to signals from the GBM microenvironment, and may regulate intrinsic ISGs expression. Previously, we have shown that PD-L1 is detectable by immunohistochemistry in the majority of human GBMs, while higher CD274 expression is enriched in the mesenchymal GBM subtype.46 Of note, the mesenchymal GBM subtype has a more prominent component of brain-resident microglia and tumor-infiltrating immune cells.47 Interestingly, Hogg et al48 reported a stronger tumor response to BETi in immune-competent tumor-bearing mice as compared to immunocompromised mice, which incited the authors to combine BETi with immune checkpoint inhibition targeting PD-1 that further improved outcome. Along similar lines BETi has been suggested to modulate T cells, thereby improving the effectiveness of checkpoint inhibitors in a melanoma model.49 On the other hand, conservation of an IFN-γ signature in melanoma was associated with better response to immune checkpoint blocking therapy in patient cohorts.50 Hence, the effects of BETis on immune cells in the GBM microenvironment will need to be addressed, particularly as immune checkpoint inhibiting antibodies are currently tested in multiple GBM trials (NCT02617589; NCT02667587).
In our study, a data-driven approach based on differential gene expression analysis was used to predict a therapeutic potential of combining BETi and HDACi. In fact, a synergy between HDACi and BETi was previously reported from models of several tumor types.33,51 In our experiments, the combination of JQ1 and the pan-HDAC inhibitor TSA showed synergism in reducing cell viability. In line, a combination of BETi and HDACi treatment has been reported to modulate the expression of pro- and anti-apoptotic genes as compared to single-agent treatments.33 Of note, this study demonstrated inefficacy of the combination of the HDACi Panobinostat and BETi OTX015, when compared to OTX015 alone in orthotopic GS-line-derived xenografts, while the same combination reduced subcutaneous tumor growth more efficiently than the single agents, indicating limited penetrance of the BBB.33 The poor BBB penetrance of HDACi may explain in part why the pan-HDACi Vorinostat, FDA approved for the treatment of lymphoma, failed in trials for recurrent GBM patients as a monotherapy, or in combinations in newly diagnosed GBM.52,53
Collectively, our study defines BETi target genes and provides evidence that BRD4 is involved in the transcriptional regulation of a GBM interferon response signature. We have also shown that the combination of an HDACi with a BETi, in therapeutically relevant concentrations, can synergistically reduce the viability of GS-lines. However, the prospective translation to the clinic may only be possible once both BETi and HDACi are optimized for penetrance of the BBB.
Given that epigenetic drugs modulate the inherent, individual expression profile of tumors (and the tumor environment), specific combination therapies will require respective patient selection based on biomarker signatures. This has also been suggested by the authors of a phase II trial for HDACi in newly diagnosed GBM. Although their study was negative, a post hoc subgroup analysis searching expression signatures as predictive factors identified subgroups of patients that may have benefitted from treatment.42,52
Supplementary Material
Acknowledgments
We thank the patients and their families for allowing the use of tumor tissue for research. We thank Tham Thi Nguyen for assisting with intracranial injections, Janine Horlbeck, Raphael Genolet, Corinne Peter, and Orbicia Riccio for technical assistance. The collaborators at the Animal-, Cellular Imaging Facility (CIF), Flow Cytometry, Mouse Pathology, and the Genomic Technology Facility (GTF) of the University of Lausanne are gratefully acknowledged for their support. This study has used data generated by The Cancer Genome Atlas. Information about TCGA and the investigators and institutions constituting the TCGA research network can be found at “http://cancergenome.nih.gov.”
Funding
This work was supported by the Swiss National Science Foundation (SNSF 163297, 182821), the Swiss Cancer Research Foundation (KFS-4461-02-2018), and the Summer Undergraduate Research Programme at the University of Lausanne (to Y.L. and J.P.).
Conflict of interest statement. M.E.H. and O.G. received a research grant from Orion Pharma.
Authorship statement. O.G. and M.E.H. designed the study. O.G. performed the experiments, analyzed and interpreted the data, and wrote the manuscript. M.D.T.P., Y.L., and J.P. performed the experiments and analyzed the data. P.B. performed data analysis (RNA-seq, ChIP-seq) interpreted the bio-statistical analyses. R.T.D. provided study material. M.D. interpreted the bio-statistical analysis. M.E.H. directed the study, interpreted the data, and wrote the manuscript. All authors contributed to manuscript writing.
References
- 1.Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Weller M, Lee EQ, et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22(8):1073–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyler CE, Matsunaga H, Hovestadt V, Vantine SJ, van Galen P, Bernstein BE. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol. 2020;21(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips RE, Soshnev AA, Allis CD. Epigenomic reprogramming as a driver of malignant glioma. Cancer Cell. 2020;38(5):647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bady P, Delorenzi M, Hegi ME. Sensitivity analysis of the MGMT-STP27 model and impact of genetic and epigenetic context to predict the MGMT methylation status in gliomas and other tumors. J Mol Diagn. 2016;18(3):350–361. [DOI] [PubMed] [Google Scholar]
- 9.Hegi ME, Genbrugge E, Gorlia T, et al. MGMT promoter methylation cutoff with safety margin for selecting glioblastoma patients into trials omitting temozolomide: a pooled analysis of four clinical trials. Clin Cancer Res. 2019;25(6):1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: from structures to applications. Epigenetics. 2017;12(5):323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berenguer-Daizé C, Astorgues-Xerri L, Odore E, et al. OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int J Cancer. 2016;139(9):2047–2055. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Z, Gong Y, Ma Y, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19(7):1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastori C, Daniel M, Penas C, et al. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics. 2014;9(4):611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58. [DOI] [PubMed] [Google Scholar]
- 16.Matzuk MM, McKeown MR, Filippakopoulos P, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150(4):673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sciuscio D, Diserens AC, van Dommelen K, et al. Extent and patterns of MGMT promoter methylation in glioblastoma- and respective glioblastoma-derived spheres. Clin Cancer Res. 2011;17(2):255–266. [DOI] [PubMed] [Google Scholar]
- 18.Kurscheid S, Bady P, Sciuscio D, et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bady P, Diserens AC, Castella V, et al. DNA fingerprinting of glioma cell lines and considerations on similarity measurements. Neuro Oncol. 2012;14(6):701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones G, Machado J Jr, Merlo A. Loss of focal adhesion kinase (FAK) inhibits epidermal growth factor receptor-dependent migration and induces aggregation of nh(2)-terminal FAK in the nuclei of apoptotic glioblastoma cells. Cancer Res. 2001;61(13):4978–4981. [PubMed] [Google Scholar]
- 21.Vassallo I, Zinn P, Lai M, Rajakannu P, Hamou MF, Hegi ME. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2016;35(1):12–21. [DOI] [PubMed] [Google Scholar]
- 22.Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J. 2015;13:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav B, Wennerberg K, Aittokallio T, Tang J. Corrigendum to “Searching for drug synergy in complex dose-response landscapes using an interaction potency model” [Comput. Struct. Biotechnol. J. 13 (2015) 504–513]. Comput Struct Biotechnol J. 2017;15:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, Kulesskiy E, Saarela J, et al. Methods for high-throughput drug combination screening and synergy scoring. Methods Mol Biol. 2018;1711:351–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenain C, Gusyatiner O, Douma S, van den Broek B, Peeper DS. Autophagy-mediated degradation of nuclear envelope proteins during oncogene-induced senescence. Carcinogenesis. 2015;36(11):1263–1274. [DOI] [PubMed] [Google Scholar]
- 26.McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82(1):290–297. [Google Scholar]
- 27.Escofier B, Pagès J. Multiple factor analysis (AFMULT package). Comput Stat Data Anal. 1994;18(1):121–140. [Google Scholar]
- 28.R Core Team. A language and environment for statistical computing; 2015. http://www.R-project.org [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escoufier Y. Le traitement des variables vectorielles. Biometrics. 1973;29:750–760. [Google Scholar]
- 31.Chessel D, Dufour A-B, Thioulouse J. The ade4 package - I: One-table methods. R News 2004;4(1):5–10. [Google Scholar]
- 32.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Ishida CT, Ishida W, et al. Combined HDAC and bromodomain protein inhibition reprograms tumor cell metabolism and elicits synthetic lethality in glioblastoma. Clin Cancer Res. 2018;24(16):3941–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen N, Guo B, Zheng H, et al. Bromodomain inhibitor jq1 induces cell cycle arrest and apoptosis of glioma stem cells through the VEGF/PI3K/AKT signaling pathway. Int J Oncol. 2019;55(4):879–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93(3):491–507. [Google Scholar]
- 37.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47(1):8–12. [DOI] [PubMed] [Google Scholar]
- 38.Lovén J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin X, Huang X, Uziel T, et al. HEXIM1 as a robust pharmacodynamic marker for monitoring target engagement of BET family bromodomain inhibitors in tumors and surrogate tissues. Mol Cancer Ther. 2017;16(2):388–396. [DOI] [PubMed] [Google Scholar]
- 40.Heller G, Schmidt WM, Ziegler B, et al. Genome-wide transcriptional response to 5-aza-2′-deoxycytidine and trichostatin A in multiple myeloma cells. Cancer Res. 2008;68(1):44–54. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Romano P, Baldini C, Postel-Vinay S. How much can we bet on activity of BET inhibitors beyond NUT-midline carcinoma? JNCI Cancer Spectr. 2020;4(2):pkz092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morel D, Jeffery D, Aspeslagh S, Almouzni G, Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol. 2020;17(2):91–107. [DOI] [PubMed] [Google Scholar]
- 43.Xu L, Chen Y, Mayakonda A, et al. Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc Natl Acad Sci USA. 2018;115(22):E5086–E5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lam FC, Morton SW, Wyckoff J, et al. Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat Commun. 2018;9(1):1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crump NT, Ballabio E, Godfrey L, et al. BET inhibition disrupts transcription but retains enhancer-promoter contact. Nat Commun. 2021;12(1):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berghoff AS, Kiesel B, Widhalm G, et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogg SJ, Vervoort SJ, Deswal S, et al. BET-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand PD-L1. Cell Rep. 2017;18(9):2162–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikbakht N, Tiago M, Erkes DA, Chervoneva I, Aplin AE. BET inhibition modifies melanoma infiltrating T cells and enhances response to PD-L1 Blockade. J Invest Dermatol. 2019;139(7):1612–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grasso CS, Tsoi J, Onyshchenko M, et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020;38(4):500–515.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazur PK, Herner A, Mello SS, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21(10):1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galanis E, Anderson SK, Miller CR, et al. ; Alliance for Clinical Trials in Oncology and ABTC . Phase I/II trial of vorinostat combined with temozolomide and radiation therapy for newly diagnosed glioblastoma: results of Alliance N0874/ABTC 02. Neuro Oncol. 2018;20(4):546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friday BB, Anderson SK, Buckner J, et al. Phase II trial of vorinostat in combination with bortezomib in recurrent glioblastoma: a north central cancer treatment group study. Neuro Oncol. 2012;14(2):215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.