Abstract
Inhibitor of β-catenin and T-cell factor (ICAT) was first found as a polypeptide that blocks β-catenin–TCF interaction. Abundant evidence has shown that ICAT has different functions in diverse cancers’ progression. Nevertheless, the roles it plays in colorectal cancer (CRC) have not been described. Here, we documented that ICAT expression was higher in CRC tissue than in the adjacent normal tissue and that prognosis was better in high-ICAT expression patients. The overexpression of ICAT inhibited CRC cell proliferation both in vitro and in vivo. Wnt pathway transcriptional activity was suppressed in the CRC cells with ICAT overexpression, where the CCND1 and MYC expression, which occurs downstream of the Wnt signaling pathway, was inhibited. Co-immunoprecipitation experiments showed that ICAT bound with β-catenin in stable overexpression cell lines; immunofluorescence showed the co-localization of ICAT and β-catenin in the cytoplasm. Overall, our study reveals that ICAT inhibits CRC cell proliferation by binding to cytoplasm-located β-catenin, and prevents its translocation, which results in Wnt signaling pathway inactivation. It may provide a scientific foundation for focusing on ICAT in treatments for CRC.
Keywords: ICAT, colorectal cancer, proliferation, Wnt signaling pathway, β-catenin
Introduction
According to the most recent statistics, colorectal cancer (CRC) is ranked third among the most frequent malignancies worldwide.1 Better diagnostic methods and treatments have improved the survival of CRC patients during the last few years.2 However, regional and distal progression of CRC persists as the major event leading to treatment failure.3 Therefore, investigation of the molecular mechanism that underlies CRC development may facilitate early intervention for patients with CRC.
The Wnt signaling pathway is a fundamental growth control pathway in many physiological and pathological processes. It plays roles in cancer initiation, development, differentiation, and metastasis.4–6 β-Catenin is a transcriptional coactivator in the Wnt signaling pathway.7 The Tcf/Lef family is bound by stabilized β-catenin, activating Wnt target gene nuclear transcription.8
ICAT is a protein encoded by the catenin beta-interacting protein 1 (CTNNBIP1) gene.9,10 ICAT was once regarded as an inhibitor of β-catenin-mediated transcription and an important target in Wnt-dependent cancer therapy.11 Sekiya et al.12 found that, in CRC cells that had APC mutations, proliferation was inhibited by ICAT. Another study confirmed that ICAT can inhibit glioma cell proliferation by inhibiting Wnt signaling pathway activation.13 Lung and gastric cancer studies have discovered a similar cancer suppressor role of ICAT.14,15 Further researches on ICAT have yielded conflicting results revealing that ICAT can promote cancer invasion and metastasis in some cancers,16–18 suggesting that it may play different roles in different cancers.
Only a few studies published previously focused on ICAT function in CRC. ICAT induced G2 arrest and cell death in CRC cells.12 Another study revealed that ICAT is not a putative cancer suppressor by analyzing human CRC specimens.19 Therefore, the function of ICAT in CRC progression remains elusive.
In the present study, we evaluated ICAT expression in CRC samples and explored the potential mechanisms. CRC tissue had higher ICAT expression than normal tissue. Patients with CRC with high-ICAT expression had a better prognosis. ICAT overexpression inhibited the proliferation of CRC cells by stabilizing cytoplasmic β-catenin and inhibiting its translocation to the nucleus. This resulted in inactivation of the Wnt signaling pathway. Our data highlight ICAT as a tumor suppressor in CRC progression. It may provide a scientific basis for targeting ICAT in CRC interventions.
Materials and Methods
Database Search
We analyzed ICAT expression levels in different tumors with the Gene Expression Profiling Interactive Analysis (GEPIA) online database (http://gepia.cancer-pku.cn), which is based on The Cancer Genome Atlas (TCGA) data. The association between the prognosis of patients with CRC and their levels of ICAT expression was evaluated with GEPIA using the Kaplan–Meier plotter. The correlation analysis between ICAT and Wnt target genes was performed with LinkedOMimcs (http://www.linkedomics.org) from TCGA data.
Cell Culture and Transfection
The CRC cell lines SW480 and DLD1 were purchased from American Type Culture Collection. We cultured the CRC cells in RPMI 1640 medium (Gibco) that contained 10% fetal bovine serum and 100 U/mL penicillin–streptomycin. The cells were all cultured at 37 °C in 5% CO2. ICAT overexpression vector (pLVX-IRES-Puro-ICAT) and vector control (pLVX-IRES-Puro) were transfected by lentivirus into the SW480 and DLD1 cells and then selected by puromycin for a stably overexpression cell lines.
Western Blotting
After harvesting, the cells were lysed in lysis buffer for radioimmunoprecipitation assay (RIPA) (Meiluncio). Protein concentrations were quantified using bicinchoninic acid (BCA) analysis. Subsequently, the samples were transferred onto a polyvinylidene fluoride membrane (Millipore) after separation by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. After 1 h blocking with 5% defatted milk, the membrane was incubated at 4 °C overnight with primary antibodies against ICAT (A7122, dilution 1:2000, ABclonal), MYC (1472-1, dilution 1:1000, Epitomics), CCND1 (2978S, dilution 1:1000, Cell Signaling Technology), β-catenin (8480S, dilution 1:5000, Cell Signaling Technology), ACTB (6609-1-Ig, dilution 1:5000, Proteintech), and GAPDH (10494-AP, dilution 1:5000, Proteintech). All primary antibodies used in this article are anti-human species. Then, we incubated the membranes with secondary antibodies that had been conjugated with horseradish peroxidase. We detected the signals using a chemiluminescence system (Bio-Rad). We analyzed the data using Image Lab Software (Bio-Rad).
Real-Time Quantitative Polymerase Chain Reaction
We extracted total RNA using an RNA-Quick Purification Kit (ESscience Biotech). PrimeScript RT Master Mix (Takara Biomedical Technology) was used to perform reverse transcription. SYBR GREEN Premix Ex Taq II (Takara Biomedical Technology) was used for detecting the gene expression levels. The comparative threshold cycle (2−ΔΔCt) method was used to analyze the results. The sequences of the primers used (purchased from Sango Biotech) were as follows:
CTNNBIP1: forward (F): 5′-CTATGCAGGGGTGGTCAACA-3′, reverse (R): 5′-CTGGAAAACGCCATCACCAC-3′;
MYC: F: 5′-GGCTCCTGGCAAAAGGTCA-3′, R: 5′-CTGCGTAGTTGTGCTGATGT-3′;
CCND1: F: 5′-GCTGCGAAGTGGAAACCATC-3′, R: 5′-CCTCCTTCTGCACACATTTGAA-3′.
Cell Counting Kit-8 Assay
We seeded stable ICAT overexpression cells and vector control (2000 cells per well) in a 96-well plate, and the cells were cultured at 37 °C in 5% CO2. We detected the cell viability every 24 h. First, we added reagent from Cell Counting Kit-8 (CCK8) (10 μL, Biosharp) to each well and mixed it gently. After 3h incubation, we measured the optical density at 450 nm with an enzyme-labeled instrument (Varioskan Flash, Thermo Scientific).
Immunohistochemistry
The tissues were embedded in paraffin and sectioned by Servicebio. We baked the fixed tissue sections for 1 h at 65 °C. Then, we deparaffinized the sections in xylene, followed by graded alcohol rehydration, and incubation at room temperature with 0.3% H2O2 for 10 min to destroy the endogenous peroxidase activity. The slides, in EDTA solution, were heated in a microwave oven at 100 °C for 3 min at high heat, and then at medium–low heat for 25 min. The slides were blocked with goat serum for 1 h. Then, the slides were incubated overnight with primary antibodies against ICAT (LS-B16681, dilution 1:500, LSBio) and Ki67 (12202, dilution 1:500, Cell Signaling Technology). The primary antibody against ICAT is anti-human species,but the antibody against Ki67 is anti-mouse species. Subsequently, we incubated the slides for 1 h with secondary antibody, and then stained with DAKO and hematoxylin. The immunohistochemistry (IHC) results were evaluated by two doctors. ICAT expression was evaluated by an intensity score of 1 to 4 (Figure 1E). The percentage of positive cells was also scored 1 to 4. The scores were determined by the intensity score multiplied by the percentage of positive cells score. The final score of each tissue spot was the average score of the two evaluators. Based on the median score, ICAT expression level was divided into high and low expression groups.
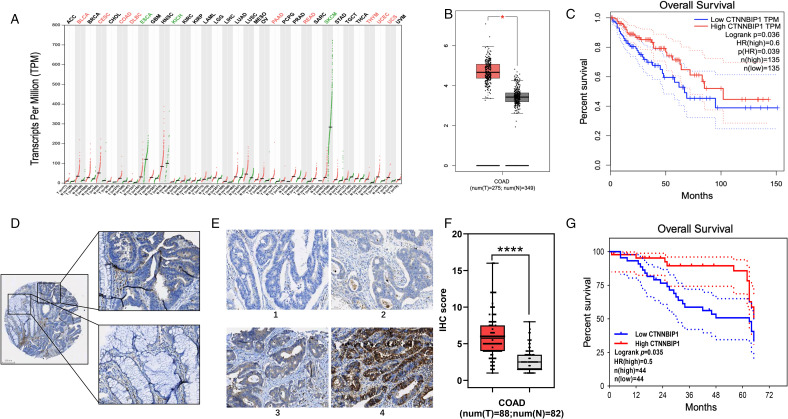
Figure 1.
ICAT is upregulated in CRC. (A) ICAT expression in various cancers was analyzed by the GEPIA online database. (B) The expression of ICAT in COAD and normal tissues of online data was analyzed. (C) Comparison of the overall survival of low- and high-ICAT expression patients in TCGA database. (D) ICAT expression in normal tissue and cancer in single slice sample. (E) The standard of IHC scores in cancer tissue. (F) Tissue chip analysis showing higher ICAT expression in COAD than in adjacent normal tissues. (G) Comparison of the overall survival of low- and high-ICAT expression patients in the tissue chip. *P < 0.05, ****P < 0.0001.
Abbreviations: ICAT, inhibitor of β-catenin and T-cell factor; CRC, colorectal cancer; GEPIA, gene expression profiling interactive analysis; COAD, colon adenocarcinoma; TCGA, The Cancer Genome Atlas; IHC, immunohistochemistry.
Tissue Chip Microarray
The tissue chip (HColA180Su12) was purchased from Shanghai Outdo Biotech. The chip included 87 pairs of colon cancer tissue and the adjacent normal tissues, 6 colon cancer tissues without paired adjacent normal tissues. The patients’ clinical-pathological characteristics and survival details were recorded. The details of the process are described in the Immunohistochemistry section. After IHC staining, 88 colon cancer tissues and 82 adjacent normal tissues were included in the analysis. We were unable to evaluate 5 colon cancer tissues and 6 adjacent normal tissues due to incomplete or destroyed tissue in the tissue chip.
Co-Immunoprecipitation (Co-IP)
Stable ICAT overexpression cells and the vector control were seeded 2 × 106 in 100-mm plates. When the cells had filled the plate, they were lysed in IP lysis buffer (Beyotime). We kept the samples on ice for approximately 20 min until flocculent turbidity appeared. After 10 min centrifugation at 14 000 rpm, the supernatant was carefully removed and divided into two portions: input, and one portion to be mixed with Anti-Flag Affinity Gel (Bimake) that had been resuspended in IP lysis buffer. The samples were placed on a shaker at 4 °C overnight. Then, the samples were centrifuged at 7000 rpm for 2 min, and the supernatant was discarded carefully. The Anti-Flag Affinity Gel was gently washed with IP lysis buffer three times. Subsequently, 50 μL 1 × loading buffer was added to the Anti-Flag Affinity Gel, vortexed, and heated at 100 °C for 4 min. The protein samples were analyzed by western blotting.
Immunofluorescence
Cells (1 × 106 per well) were seeded in a 24-well plate with a climbing sheet on the bottom for 24 h, and fixed in methanol for 15 min. We used 10% goat serum to block the climbing sheets, then incubated them at 4 °C overnight with the following primary antibodies: anti-DYKDDDDK tag (66008-3-Ig, dilution 1:500, Proteintech) and anti–β-catenin (8480S, dilution 1:200, Cell Signaling Technology). All primary antibodies used in immunofluorescence are anti-human species. Subsequently, we incubated the climbing sheets at room temperature for 1 h with Alexa Fluor Plus 594 or Plus 488 sary antibody against rabbit or mouse (A11034; A11037; A11029; A11005, dilution 1:200, Thermo Scientific). We observed the climbing sheets using laser scanning confocal microscopy (Leica TCS-SP8).
Luciferase Reporter Assay
Stable ICAT overexpression cells and negative control (5 × 106 per well) were seeded in a 6-well plate. After 24-h culture, cells were transfected for 24 h with pGL4.49/Wnt reporter plasmid (Promega), with pRT/Renilla luciferase as the internal reference (Promega). The firefly and Renilla luciferase were detected by a luciferase assay kit (Promega). The luminescence was normalized against Renilla luciferase.
Animal Experiments
The animal experiments (IACUC-2020050602) were approved by the Committee on the Ethics of Animal Experiments of the Sixth Affiliated Hospital, Sun Yat-sen University. We obtained male BALB/c nude mice (age, 4-5 weeks) from the Model Animal Research Center of Nanjing University. Matrigel was added in equivalent amounts of phosphate-buffered saline to resuspend the cells. The right sides of the mice were injected subcutaneously with stable ICAT overexpression SW480 cells and negative cells (5 × 106 cells in 300 μL suspension). A week after injection, we measured the subcutaneous tumor volumes every 2 days. At 31 days following the injection, the mice were fed. Subsequently, the mice were killed and we removed the subcutaneous tumors. After they had been weighed, the tumors were sent for paraffin-embedding and sectioning.
Statistical Analysis
The data in this study are presented as the mean ± SD and were obtained from triplicate assays. All data were analyzed with GraphPad Prism 8.0 (GraphPad Software). Differences were analyzed with Student's t-test and Chi-square test. P-value of <0.05 was considered statistically significant.
Results
ICAT Is Upregulated in CRC
We first analyzed ICAT transcript expression in a TCGA pan-cancer dataset obtained from the GEPIA online database, and identified that ICAT is abnormally expressed in many cancers (Figure 1A). In colon adenocarcinoma (COAD), ICAT expression was significantly higher than in normal tissues (Figure 1B). We performed IHC staining on tumor tissues and corresponding adjacent normal tissues to determine whether the protein expression level of ICAT is correlated with the clinical progression of CRC. We confirmed that ICAT expression was significantly higher in cancer as compared with that in the normal tissues (Figure 1D–F). The clinical-pathological characteristics of the patients of tissue chip HColA180Su12 were shown in Table 1. Although there was no association between ICAT expression with the pathological stage of CRC, survival analysis showed that the patients with higher ICAT expression had a better prognosis in both the TCGA database and the tissue chip (Figure 1C and G).
Table 1.
The Clinical-Pathological Characteristics of the Patients of Tissue Chip HColA180Su12.
| ICAT expression | P-value | |||
|---|---|---|---|---|
| Parameters | Number of patients | Low (<median) |
High (>median) |
|
| Number | 88 | 44 | 44 | |
| Age (years) | ||||
| ≥60 | 61 | 30 | 31 | 0.817 |
| <60 | 27 | 14 | 13 | |
| Gender | ||||
| Male | 49 | 26 | 23 | 0.520 |
| Female | 39 | 18 | 21 | |
| Lymph node involvementa | ||||
| Absent (pN0) | 50 | 29 | 21 | 0.165 |
| Present (pN+) | 33 | 14 | 19 | |
| Distal metastasis | ||||
| Absent (M0) | 84 | 41 | 43 | 0.616 |
| Present (M1) | 4 | 3 | 1 | |
| TNM stageb | ||||
| I | 3 | 1 | 2 | 0.188 |
| II | 41 | 25 | 16 | |
| III | 31 | 12 | 19 | |
| IV | 4 | 3 | 1 | |
| Differentiation | ||||
| Grade 1 | 2 | 2 | 0 | 0.358 |
| Grade 2 | 70 | 34 | 36 | |
| Grade 3 | 16 | 8 | 8 | |
| Location | ||||
| Proximal colon | 37 | 18 | 19 | 0.829 |
| Distal colon | 51 | 26 | 25 | |
The information of lymph node involvement was absent in 5 patients.
The information of TNM stage was absent in 9 patients.
Overexpression of ICAT Inhibits Cell Proliferation In Vitro
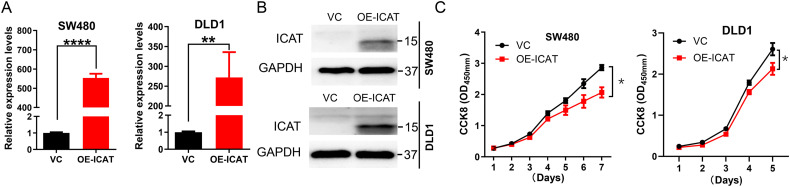
To study the potential biological roles of ICAT in CRC progression, SW480 and DLD1 cells were infected with lentivirus to establish the stably transfected cell lines. Stable ICAT overexpression was confirmed by RT-PCR and western blotting (Figure 2A and B). CCK8 assay showed that overexpression of ICAT inhibited cell proliferation in both the SW480 and DLD1 cell lines (Figure 2C).
Figure 2.
ICAT overexpression inhibits CRC cell proliferation. (A and B) Western blotting and RT-PCR analysis of ICAT expression after transfection and selection in SW480 and DLD1 cells. (C) CCK8 assay measurement of cell proliferative ability. The proliferative abilities of ICAT overexpression cells and the control were compared in the SW480 and DLD1 cells. *P < 0.05, **P < 0.01, ****P < 0.0001.
Abbreviations: ICAT, inhibitor of β-catenin and T-cell factor; CRC, colorectal cancer; RT-PCR, real-time quantitative polymerase chain reaction; CCK8, Cell Counting Kit-8.
Overexpression of ICAT Inhibits CRC Proliferation In Vivo
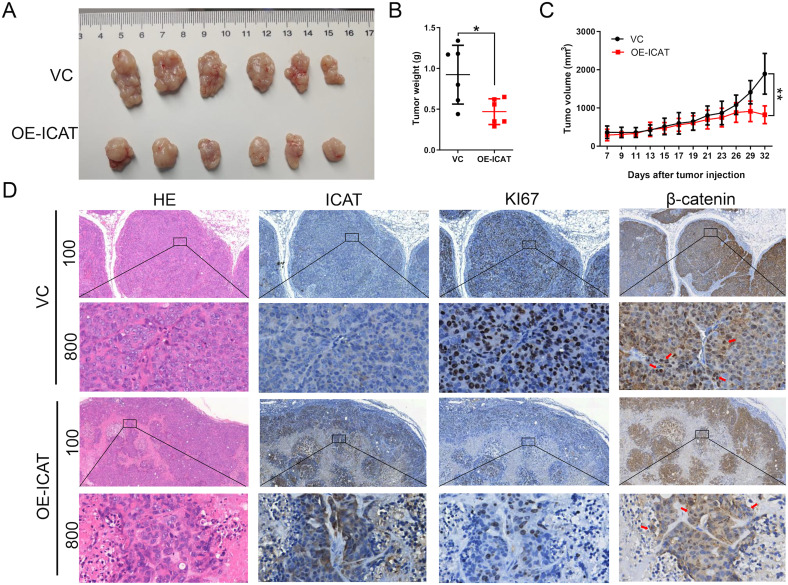
The effect of ICAT overexpression in vivo was examined with a nude mouse xenograft model. Compared with the vector control, ICAT overexpression significantly reduced tumor volume and weight (Figure 3A-C), suggesting that ICAT overexpression restrained CRC xenograft growth significantly. Ki67 expression is considered an indication of activated cell proliferation.20 Here, IHC studies of the subcutaneous tumors revealed that decreased expression of Ki67 in the ICAT overexpression group (Figure 3D). Both the in vivo and in vitro experiments indicated that ICAT overexpression can inhibit CRC cell proliferation.
Figure 3.
ICAT overexpression inhibits CRC cell growth in the xenograft model. (A) Subcutaneous tumor specimen. (B) The weight of the subcutaneous tumors of each group. (C) The tumor volume of each group was measured every 2 days. (D) HE staining and IHC of ICAT, Ki67 and β-catenin in subcutaneous tumor slides. *P < 0.05, **P < 0.01.
Abbreviations: ICAT, inhibitor of β-catenin and T-cell factor; CRC, colorectal cancer; HE, hematoxylin and eosin; IHC, immunohistochemistry.
ICAT Inhibits the Wnt Signaling Pathway in CRC
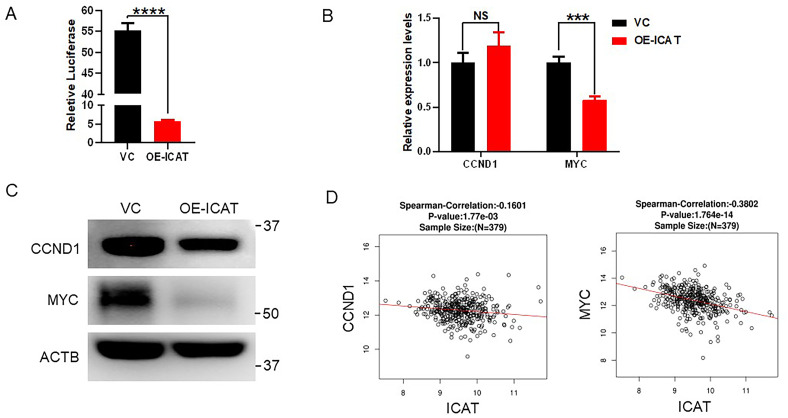
As mentioned above, ICAT was identified previously as a β-catenin interaction protein resulting in the inactivation of the Wnt signaling pathway. The Wnt signaling pathway report system was used to analyze the effect of ICAT in Wnt signaling pathway. We transferred pGL4.49 and pRT into the stable ICAT overexpression cell line and the vector control. The ICAT-overexpressing cells had lower Wnt signaling pathway transcriptional activity than the vector control (Figure 4A). Next, we detected the Wnt downstream target CCND1 and MYC genes and protein products of the Wnt signaling pathway with RT-PCR and western blotting (Figure 4B and C). Although the CCND1 mRNA levels in the ICAT overexpression cells were a little higher than that of the control (P > 0.05), western blotting showed that ICAT overexpression inhibited the expression of CCND1. Both RT-PCR and western blotting showed that ICAT overexpression inhibited the expression of MYC. Finally, we also used the TCGA data to analyze a link between ICAT and Wnt classical target genes. Consistently with our results, ICAT is correlated negatively with the Wnt classical target genes, ie, CCND1 and MYC (Figure 4D). Taken together, ICAT inhibits the Wnt signaling pathway in CRC cell lines.
Figure 4.
ICAT inhibits the Wnt signaling pathway in CRC. (A) The Wnt pathway transcriptional activity in the vector control and ICAT overexpression SW480 cells. (B) RT-PCR results for CCND1 and MYC in the vector control and ICAT overexpression cells. (C) Western blotting results for CCND1 and MYC in the vector control and ICAT overexpression cells. (D) TCGA database analysis of the relationship between ICAT and the Wnt target genes CCND1 andMYC. ***P < 0.001, ****P < 0.0001.
Abbreviations: ICAT, inhibitor of β-catenin and T-cell factor; TCGA, The Cancer Genome Atlas.
ICAT May Inhibit the Wnt Signaling Pathway by Binding to β-Catenin in the Cytoplasm
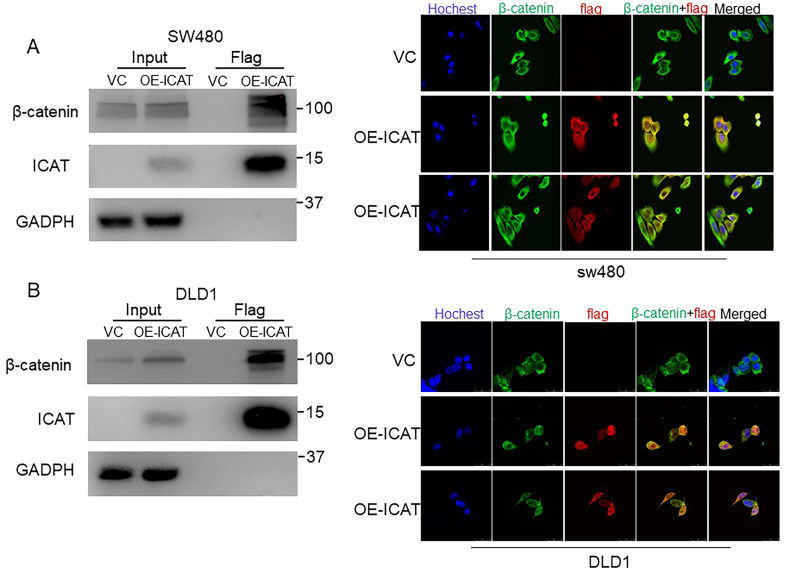
Since ICAT was reported to both inhibit or increase the functions of β-catenin by binding the β-catenin.9,10,18 Thus, the co-immunoprecipitation (co-IP) and IF experiments were performed to investigate the interaction between ICAT and β-catenin. As shown in Figure 5A and B, ICAT bound with β-catenin in the stable overexpression cell lines. Immunofluorescence results showed that ICAT and β-catenin were co-localization in the cytoplasm in stable ICAT overexpression cells whithout β-catenin translocated to the nucleus (Figure 5A and B). IHC of ICAT and β-catenin in subcutaneous tumor slides showed the co-localization trend in animal experiment (Figure 3D). These findings suggest ICAT can combine with β-catenin in the cytoplasm inhibting β-catenin translocation to the nucleus to exercise its inhibitory function. Our results would explain the observation that overexpression of ICAT induces stabilization of β-catenin, but inhibit the Wnt signaling activity.
Figure 5.
ICAT may inhibit Wnt signaling pathway by binding to β-catenin in CRC. (A) Co-IP and immunofluorescence detection of ICAT and β-catenin binding in stable overexpression SW480 cells. (B) Co-IP and immunofluorescence detection of ICAT and β-catenin binding in stable overexpression DLD1 cells.
Abbreviations: ICAT, inhibitor of β-catenin and T-cell factor; CRC, colorectal cancer; co-IP, co-immunoprecipitation.
Discussion
The canonical Wnt signaling pathway is important in tumorigenesis and cancer development.4–6 Several studies have considered ICAT as a Wnt–β-catenin pathway suppressor with anti-cancer function in several cancers.12–15 On the other hand, increasing evidencing has demonstrated that ICAT could also promote cancer progression. As early as 2002, Koyama et al.19 reported that the mRNA expression level of ICAT was overexpressed in almost 50% of CRC cases, but the mRNA expression level of ICAT was not a prognostic factor, which may be due to the sample size limitation. In the present study, we analyzed ICAT expression in TCGA database and revealed its abnormal expression in several cancers. In patients with CRC, there was a higher expression of ICAT in cancer tissue than in normal tissue. Although no association between ICAT expression and cancer stage was found, ICAT-high expression patients had better overall survival than patients with low ICAT expression. Furthermore, we also used IHC to analyze ICAT protein expression level in patients with colon cancer. Consistently, ICAT expression in cancer tissue was higher than that in the adjacent normal tissue. The overall survival in ICAT-high expression patients was better than that of patients with low ICAT expression. These results are consistent with the results of TCGA database analysis. Based on these results, ICAT may serve as an anti-cancer factor in CRC.
To examine the exact role played by ICAT in CRC, we constructed CRC cell lines stably overexpressing ICAT. ICAT overexpression inhibited the proliferation of CRC cells. In the animal experiments, ICAT overexpression inhibited subcutaneous tumor proliferation. In a glioblastoma xenograft model, Zhang et al.13 found that ICAT could slow down cancer growth by inhibiting glioma cell proliferation. Wang et al.21 silenced the CTNNBIP1 gene in psoriasis, revealing disrupted keratinocyte proliferation. ICAT also inhibited the proliferation of CRC cells with APC mutation,12 which was similar to our results. With the above results, it is suggested that ICAT may promote a good prognosis by inhibiting the proliferation of CRC cells.
CCND1 and MYC are downstream targets in the canonical Wnt signaling pathway.22 Wnt signaling pathway mutations are frequently observed in CRC. The APC gene is mutated in familial adenomatous polyposis,23,24 while the loss of both APC alleles has been observed in most sporadic CRC.25 Abnormal APC function results in inappropriate stabilization of β-catenin and β-catenin–Tcf-4 complex formation.26 In the present study, ICAT overexpression inhibited the expression of the downstream genes of the Wnt signaling pathway in CRC cells. TCGA data analysis showed that ICAT correlated negatively with downstream genes in the Wnt signaling pathway, suggesting that ICAT inhibits the Wnt signaling pathway in CRC cell lines. In the present study, we reported that MYC was decreased in both mRNA and protein levels in SW480 cells overexpressing ICAT. However, the mRNA levels of CCND1 did not decrease. Meanwhile, TCGA data analysis showed that ICAT correlated negatively with both MYC and CCND1 in CRC. These results suggest that ICAT inhibits the Wnt signaling pathway in CRC, but the ICAT influences the final output in a context-dependent manner in cell lines. The decrease observed in protein expression of CCND1 in ICAT-overexpressing SW480 cells could be a result of phosphorylation by other proteins, which trigger the degradation of CCND1 protein in the cytoplasm.27,28 Further studies are needed to understand this precise mechanism of ICAT in different cell types.
ICAT was reported selectively inhibiting the binding of Tcf to β-catenin and β-catenin-mediated transcription.9 ICAT has been considered a Wnt pathway suppressor. Structurally, an N-terminal helical domain and an extended C-terminal region in ICAT binds to repeats 10 to 12 and repeats 5 to 9 of β-catenin, respectively.10,11 In the present study, co-IP showed that ICAT bound to β-catenin in the stable overexpression cell lines. In canonical Wnt signaling pathway activation, accumulated β-catenin translocates to the nucleus and interacts with Tcf/Lef family transcription factors.29 The translocation of β-catenin is seen as the main molecular event in the formation of CRC.30 Interestingly, in the present study, ICAT overexpression led to the upregulation of β-catenin. Ji et al.18 have demonstrated that the β-catenin destruction complex is inhibited by ICAT, and that β-catenin is stabilized by ICAT blocking APC–β-catenin interaction. Here, immunofluorescence showed that the overexpressed ICAT and β-catenin co-localized in the cytoplasm without β-catenin translocated to the nucleus. We assumed that although ICAT stabilizes β-catenin by binding to it, it does not induce β-catenin nuclear translocation, a key molecular event in the activated Wnt signaling pathway. ICAT is believed to inhibit Wnt signaling pathway activation by selectively inhibiting Tcf binding to β-catenin.9–11 However, another study revealed that ICAT stabilized β-catenin in DLD1 cells.18 In the present study, ICAT overexpression led to the upregulation of β-catenin without β-catenin translocating to the nucleus, which resulted in Wnt signaling pathway inactivation. The present results can explain the conflicting phenomena reported previously.
In summary, we comprehensively analyzed the role of ICAT in CRC from the aspects of tissue, cell, and animal experiments. Our study shows the potential tumor suppressor role of ICAT in CRC. ICAT overexpression inhibited CRC cell proliferation by stabilizing β-catenin but inhibiting the translocation of β-catenin, resulting in Wnt signaling pathway inactivation. We will explore the possibility of targeting ICAT in CRC interventions in further studies.
Abbreviations
- 2−ΔΔCt
comparative threshold cycle
- co-IP
co-immunoprecipitation
- CCK8
Cell Counting Kit-8
- COAD
colon adenocarcinoma
- CRC
colorectal cancer
- GEPIA
gene expression profiling interactive analysis
- ICAT
inhibitor of β-catenin and T-cell factor
- IHC
immunohistochemistry
- RT-PCR
real-time quantitative polymerase chain reaction
- TCGA
The Cancer Genome Atlas
Footnotes
Authors’ Contributions: All authors contributed to final approval of manuscript. H L and X Y contributed to conception and design. J H contributed to collection and assembly of data, data analysis and interpretation. J H and Z W contributed to manuscript writing. Z W and J C contributed to experiment operation. J H and Z Y contributed to IHC scoring. J Z, W L and M L contributed to collection and assembly of data. No patient consent is needed for this study. All data in this study are available upon request.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This study was supported by National Key Clinical Discipline, Project to Attract High Level Foreign Experts (G20190019023), and Medical Science and Technology Foundation of Guangdong Province of China (A2019487).
Ethics Statement: The animal experiments (IACUC-2020050602) were approved by the Committee on the Ethics of Animal Experiments of the Sixth Affiliated Hospital, Sun Yat-sen University.
ORCID iDs: Jiancong Hu https://orcid.org/0000-0003-1116-4177
Huanliang Liu https://orcid.org/0000-0002-1006-6666
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. 2020. Cancer statistics. CA Cancer J Clin. 2020;70(1):7-30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Goding Sauer A, et al. 2020. Colorectal cancer statistics. CA Cancer J Clin. 2020;70(3):145-164. doi: 10.3322/caac.21601 [DOI] [PubMed] [Google Scholar]
- 4.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9-26. doi: 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985-999. doi: 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 6.Gammons M, Bienz M. Multiprotein complexes governing Wnt signal transduction. Curr Opin Cell Biol. 2018;51:42-49. doi: 10.1016/j.ceb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 7.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103(2):311-320. [DOI] [PubMed] [Google Scholar]
- 8.Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21(10):2303-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tago KI, Nakamura T, Nishita M, et al. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev. 2000;14(14):1741-1749. doi: 10.1101/gad.14.14.1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels DL, Weis WI. ICAT Inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol Cell. 2002;10(3):573-584. [DOI] [PubMed] [Google Scholar]
- 11.Graham TA, Clements WK, Kimelman D, Xu W. The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT. Mol Cell. 2002;10(3):563-571. [DOI] [PubMed] [Google Scholar]
- 12.Sekiya T, Nakamura T, Kazuki Y, et al. Overexpression of Icat induces G(2) arrest and cell death in tumor cell mutants for adenomatous polyposis coli, beta-catenin, or axin. Cancer Res. 2002;62(11):3322-3326. [PubMed] [Google Scholar]
- 13.Zhang K, Zhu S, Liu Y, et al. ICAT inhibits glioblastoma cell proliferation by suppressing Wnt/β-catenin activity. Cancer Lett. 2015;357(1):404-411. doi: 10.1016/j.canlet.2014.11.047 [DOI] [PubMed] [Google Scholar]
- 14.Chang J-M, Tsai AC-D, Huang W-R, Tseng R-C. The alteration of CTNNBIP1 in lung cancer. Int J Mol Sci. 2019;20(22):5684. doi: 10.3390/ijms20225684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosari-Monfared M, Nikbakhsh N, Fattahi S, et al. CTNNBIP1 downregulation is associated with tumor grade and viral infections in gastric adenocarcinoma. J Cell Physiol. 2019;234(3):2895-2904. doi: 10.1002/jcp.27106 [DOI] [PubMed] [Google Scholar]
- 16.Domingues MJ, Rambow F, Job B, et al. β-catenin inhibitor ICAT modulates the invasive motility of melanoma cells. Cancer Res. 2014;74(7):1983-1995. doi: 10.1158/0008-5472.CAN-13-0920 [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, Ren W, Wang W, et al. Inhibitor of β-catenin and TCF (ICAT) promotes cervical cancer growth and metastasis by disrupting E-cadherin/β-catenin complex. Oncol Rep. 2017;38(5):2597-2606. doi: 10.3892/or.2017.5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji L, Lu B, Wang Z, et al. Identification of ICAT as an APC inhibitor, revealing Wnt-dependent inhibition of APC-axin interaction. Mol Cell. 2018;72(1):37-47. e4. doi: 10.1016/j.molcel.2018.07.040 [DOI] [PubMed] [Google Scholar]
- 19.Koyama T, Tago K-I, Nakamura T, et al. Mutation and expression of the beta-catenin-interacting protein ICAT in human colorectal tumors. Jpn J Clin Oncol. 2002;32(9):358-362. [DOI] [PubMed] [Google Scholar]
- 20.Lopes MBS. The 2017 World Health Organization classification of tumors of the pituitary gland: a summary. Acta Neuropathol. 2017;134(4):521-535. doi: 10.1007/s00401-017-1769-8 [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Wang H, Peng Y, et al. CTNNBIP1 modulates keratinocyte proliferation through promoting the transcription of β-catenin/TCF complex downstream genes. J Eur Acad Dermatol Venereol. 2020;35(2):368-379. doi: 10.1111/jdv.16725 [DOI] [PubMed] [Google Scholar]
- 22.Zhang K, Zhang J, Han L, Pu P, Kang C. Wnt/beta-catenin signaling in glioma. J Neuroimmune Pharmacol. 2012;7(4):740-749. doi: 10.1007/s11481-012-9359-y [DOI] [PubMed] [Google Scholar]
- 23.Kinzler KW, Nilbert MC, Su LK, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253(5020):661-665. [DOI] [PubMed] [Google Scholar]
- 24.Nishisho I, Nakamura Y, Miyoshi Y, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253(5020):665-669. [DOI] [PubMed] [Google Scholar]
- 25.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108-1113. [DOI] [PubMed] [Google Scholar]
- 26.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275(5307):1784-1787. [DOI] [PubMed] [Google Scholar]
- 27.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alao JP, Stavropoulou AV, Lam EWF, Coombes RC, Vigushin DM. Histone deacetylase inhibitor, trichostatin A induces ubiquitin-dependent cyclin D1 degradation in MCF-7 breast cancer cells. Mol Cancer. 2006;5:8. doi: 10.1186/1476-4598-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brabletz T, Jung A, Hermann K, Günther K, Hohenberger W, Kirchner T. Nuclear overexpression of the oncoprotein beta-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol Res Pract. 1998;194(10):701-704. [DOI] [PubMed] [Google Scholar]
- 30.Lin P-L, Wu D-W, Huang C-C, et al. MicroRNA-21 promotes tumour malignancy via increased nuclear translocation of β-catenin and predicts poor outcome in APC-mutated but not in APC-wild-type colorectal cancer. Carcinogenesis. 2014;35(10):2175-2182. doi: 10.1093/carcin/bgu110 [DOI] [PubMed] [Google Scholar]