Abstract
Background and aims
Acinetobacter baumannii is among the most concerning cause of nosocomial infections due to its high level of antibiotic resistance and high mortality. The aim of this study was to determine the role of efflux pumps in resistance of A. baumannii strains to three disinfectants, including MICROZED ID‐MAX, NANOSIL D2, and OPIDEX OPA.
Methods
Twenty‐eight environmental and clinical isolates of A. baumannii were collected from selected hospitals of central Iran. The minimum inhibitory concentrations of the disinfectants were determined and real time reverse transcriptase‐PCR was performed to investigate the expression level of qacEΔ1, amvA, abeM, and adeB efflux pump genes.
Results
Considering both clinical and environmental isolates, there was a significant difference in the mean expression level of qacEΔ1 gene between susceptible and resistant strains to MICROZED ID‐MAX disinfectant, of amvA and abeM genes between susceptible and resistant strains to NANOSIL D2 disinfectant and of abeM gene in susceptible and resistant strains to OPIDEX OPA disinfectant (all P ˂ .05). The expression levels of abeM and amvA genes were higher in the environmental isolates that were resistant to NANOSIL D2 disinfectant compared to those that were susceptible (P ˂ .05).
Conclusions
This study provided evidence for the role of abeM and amvA genes in the resistance of environmental isolates to disinfectants, particularly hydrogen peroxide derivatives.
Keywords: Acinetobacter baumannii, biocides, disinfectants, efflux pumps
1. INTRODUCTION
Health‐care associated infections or nosocomial infections are the leading cause of mortality in hospitalized patients and are regarded as a concern for both patients and medical staff.1 These infections are usually caused by resistant microbial strains such as Acinetobacter baumannii, an emerging pathogen associated with various outbreaks in hospitals all over the world.2
A. baumannii are nonfermentative gram‐negative coccobacilli that colonize various organs of hospitalized patients and could survive for long time on both humid and dry environmental surfaces.3, 4 There are increasing reports of carbapenem‐resistant and multidrug resistant A. baumannii and several evidence show dissemination of common clones of drug resistant A. baumannii strains among patients within or between hospitals. A. baumannii can tolerate desiccation and survive in the inanimate healthcare environment including surfaces and equipment for long period of time. A combination of these properties and the enhanced resistance of A. baumannii to antibiotic and biocide, make it a great challenge in healthcare settings. A. baumannii causes pneumonia, bacteremia, secondary meningitis, burn and wound infection, urinary tract infection, and soft tissue infection.5, 6, 7
Disinfectants are chemicals that inactivate pathogenic microorganisms on contaminated equipment and surfaces by different mechanisms. Various commercially available disinfectants are currently used to reduce or completely eliminate the microbial burden of health care facilities8 and their activity depends on several factors such as temperature, concentration, chemical nature, and pH.5
One of the key mechanisms of low susceptibility/resistance of bacteria to biocides is the function of efflux transport systems. Efflux pumps are proteins which are localized in plasma membrane of bacteria9 and efflux function allows the microorganisms to regulate their internal environment by removing toxic substances, including antimicrobial agents, metabolites, and biocides. According to their substrates, composition, number of membrane spanning segments, and energy sources, bacterial efflux pumps are classified into seven families including the resistance‐nodulation‐cell division superfamily (RND), the small multidrug resistance family (SMR), the major facilitator superfamily (MFS), the ATP‐binding cassette superfamily (ABC), the multidrug and toxic compound extrusion protein family (MATE), the p‐aminobezoyl‐glutamate transporter, and the proteobacterial antimicrobial compound efflux.10, 11, 12
A. baumannii have been isolated from surfaces, equipment, solutions, monitors, and beds in hospitals and most of the A. baumannii infections are directly associated with the length of hospital stay, especially in the intensive care units. To control the emergence and outbreaks of multiple drug resistance (MDR) A. baumannii in health care settings, there is a need to focus simultaneously on environmental isolates in order to disrupt transmission of A. baumannii clones between environment and hospitalized patients and/or healthcare‐workers.13 In this study, we have analyzed the transcription levels of four efflux pumps genes from four different families including: qacEΔ1 (SMR), adeB (RND), amvA (MFS), and abeM (MATE) in the environmental and clinical isolates of A. baumannii. The association of activity of efflux pumps with susceptibility/resistance to common commercially available disinfectants was also evaluated.
2. MATERIALS AND METHODS
2.1. Disinfectants
Three commonly known disinfectants, including MICROZED ID‐MAX (Atrineh Saziba, Iran), NANOSIL D2 (Kimiafaam Pharmaceutical, Iran), and OPIDEX OPA (Sung Kwang Pharm, Korea), which are routinely used in hospitals of Iran to disinfect surfaces, equipment, and medical devices, were selected. MICROZED ID‐MAX is a concentrated disinfectant solution classified as quaternary ammonium compounds. NANOSIL D2 is a ready to use disinfectant for surfaces and might be used in a wide range of ambient temperatures (0°C‐95°C) composed of hydrogen peroxide and silver ions. OPIDEX OPA is a ready to use solution composed of 0.55% ortho‐phthalaldehyde (Table 1).
TABLE 1.
Properties of disinfectants
| Disinfectants | |||
|---|---|---|---|
| MICROZED ID‐MAX | NANOSIL D2 | OPIDEX OPA | |
| Composition |
‐ Didecylmethylpoly(oxyethyl) ammonium propionate ‐ Polyethylene glycol ‐ N,N‐Dimethyldodecylamine ‐ Anti‐corrosion agent ‐ Complexing agent |
Hydrogen peroxide and silver ion | Ortho‐phthalaldehyde 0.55 g/100 mL |
| Properties |
‐ Yellowish clear liquid ‐ Medium disinfectant solution |
‐ Ready to use disinfectant ‐ Usable in a wide range of thermal ranges (0°C‐95°C) |
‐ Ready to use disinfectant ‐ Transparent light blue liquid pH: 7.5 |
| Mechanism of action | ‐ Inactivation of the cellular enzyme system and protein degradation and cell membrane disruption |
‐ Binding of silver ions to cellular enzymes and inhibition of bacterial cell metabolic activities ‐ Damage to the cell wall by oxygen radicals released from hydrogen peroxide |
‐ Protein degradation of microorganisms ‐ Prevent of spore germination |
2.2. Settings and sampling
Samples were collected from environmental surfaces, equipment, and hospitalized patients of intensive care unit (ICU) and neonatal intensive care unit wards of four hospitals located at central Iran including Qom and the capital Tehran provinces. For environmental sampling, the sterile swab was moistened with sterile saline and rubbed by swab over a 10 cm square surface of environment. In the case of liquids, 1 mL of the solution was used as sample for culture.14
Clinical MDR strains were isolated from skin ulcers, trachea, and bronchoalveolar specimens of the hospitalized patients. Also, environmental strains were isolated from bed sheets, sinks, faucets, carrying tables, and medical devices such as suction tubes. The clonal distribution of the isolates was previously determined which belongs to nine different clones.14
2.3. Identification of A. baumannii strains
The swabs were directly cultured on blood agar medium and then were subcultured on MacConkey agar medium (Merck, Germany) and incubated at 37°C for 48 hours. The resulting single colonies were used further for culture and biochemical tests. MacConkey agar, OF (oxidative fermentative basal agar), TSI (triple sugar iron agar) (all from Merck, Germany), oxidase (Padtan Teb Co, Iran), and growth test at 44°C were used to identify A. baumannii.14 Nonfermentative Gram‐negative bacilli which identified as A. baumannii were further confirmed by PCR targeting bla OXA‐51 gene. For this purpose, the strains were cultured on trypticase soy agar (TSA) medium and incubated at 37°C for 18 to 24 hours. DNAs were extracted by alkaline lysis method,15 then PCR of the target gene was performed using specific primers (F: 5′‐TAATGCTTTGATCGGCCTTG‐3′ and R: 5′‐TGGATTGCACTTCATCTTGG‐3′). The reaction consisted of 15.5 μL of distilled water, 2 μL of 10× buffer, 0.6 μL of MgCl2 (50 mM), 0.4 μL of dNTPs, 0.5 μL of each primer, 0.1 μL of Taq DNA polymerase, and 0.5 μL of DNA. The temperature steps include initial denaturation at 95°C for 5 minutes, then 35 cycles of denaturation at 95°C for 45 seconds, annealing at 58°C for 45 seconds, extension at 72°C for 45 seconds, followed by final extension at 72°C for 10 minutes. The presence of bla OXA‐51 gene was confirmed by the visualization of 353 bp band on 1% agarose gel after electrophoresis. Confirmed isolates were kept at −20°C in trypticase soy broth (Merk, Germany) solution until further use.
2.4. MIC and MBC determination
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of disinfectants were determined by microdilution broth method.
2.4.1. Preparation of disinfectants
A 2‐fold serially diluted concentrations of disinfectants were prepared in triplicate with distilled water in 96‐well microtiter plates. The concentrations prepared for MICROZED ID‐MAX disinfectants were from 2.5% to 0.019%, for NANOSIL D2 disinfectant were from 5% to 0.039%, and for OPIDEX OPA were from 100% to 0.78%. Although these disinfectants are ready‐to‐use solutions, a broader range around the concentrations recommended by the manufacturers were used to determine the MICs. In other words, concentrations recommended by manufacturers, along with one or two concentrations higher or lower, were applied to determine MICs. Numerous experiments were performed to obtain the optimum concentrations.
The disinfectants were kept in the standard conditions recommended by the manufacturers and their stability was evaluated by testing the MIC against a standard strain, A. baumannii ATCC 19606, in each round of experiment. One hundred microliter of each of eight concentrations of disinfectants were pipetted into each well of microtiter plates.
2.4.2. Preparation of microbial suspensions
A suspension of 0.5 McFarland was prepared for each of the isolates using normal saline, then each microbial suspension was diluted 1:100 in Mueller Hinton Broth (MHB) medium (Merck, Germany) and 100 μL of bacterial suspensions were added to each well of plates containing disinfectant. The positive control wells contained 100 μL of 1% microbial solution without disinfectant and the negative control wells contained 100 μL of MHB solution without microbial suspension. A. baumannii strain ATCC 19606 was used as the standard strain. The experiments were performed in triplicate. The microtiter plates were incubated at 37°C for 18 to 24 hours. MIC is defined as the lowest concentration of a disinfectant that inhibited the visible growth of the bacteria after overnight incubation, and MBC is defined as the lowest concentration of a disinfectant that prevented the growth of the bacteria. MIC50 (concentration of disinfectant in which 50% growth of strains are inhibited) and MIC90 (concentration of disinfectant in which 90% growth of strains are inhibited) were determined as criteria for determining the susceptibility of the strains to each of the disinfectants. The isolates which had MIC concentrations ≤MIC50 were considered susceptible and isolates with MIC concentrations ≥MIC90 were considered resistant to each disinfectant.16 Overall, the isolates that were resistant to at least one of the three disinfectants were considered resistant, and isolates that were susceptible to all three disinfectants were considered susceptible.
2.4.3. Minimum bactericidal concentration
After MIC determination, the well containing MIC and the well after that were cultured on TSA medium and incubated at 37°C for 18 to 24 hours. Then, plates without colony growth with the lowest disinfectant concentration were designated as MBC.
2.5. Real time reverse transcriptase‐PCR
RNA extraction of the samples was performed using Trizol reagents (Sigma), according to the manufacturer's instruction. RNA was kept at −70°C until further use. Measurement of absorption at wavelengths of 260 and 280 nm was performed to determine the concentration and purity of extracted RNA.
cDNA synthesis was performed by reverse transcription using M‐MLV enzyme (Takara, Japan). Briefly, 10 to 100 ng of RNA was mixed with 1 μL random hexamer and 2.5 μL diethyl pyrocarbonate (DEPC) treated water and incubated at 70°C for 5 minutes. After chilling on ice, 4 μL 5× buffer, 1 μL dNTPs, 0.5 μL RNasin, and 1 μL M‐MLV (10 000 U) were added to the tube and incubated for 1 hour at 37°C and 5 minutes at 70°C and used as cDNA samples.17
For real time PCR, in a 25 μL reaction mixture 9.5 μL of distilled water, 0.5 μL of each primer pairs (Table 2), 12.5 μL SYBR Green (Takara, Japan), and 2 μL cDNA were prepared. The cycling program were set on Corbett machine (Corbett Rotorgene 6000, Qiagen, UK) as: initial denaturation at 95°C for 15 minutes, 40 cycles of replication including denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and elongation at 72°C for 30 seconds. Target genes were qacEΔ1 (SMR), adeB (RND), amvA (MFS), and abeM (MATE).
TABLE 2.
Sequence of primers of efflux genes
| Gene | Primernucleotide sequence (5′ → 3′) | Expected size (bp) | Reference | |
|---|---|---|---|---|
| adeB | F | GAATAAGGCACCGCAACAAT | 124 | 38 |
| R | TTTCGCAATCAGTTGTTCCA | |||
| amvA | F | GCCGCTCAATTATTTTGCCA | 137 | 32 |
| R | TTGCTGCGCCACTACAACTA | |||
| qacEΔ1 | F | ATCGCAATAGTTGGCGAAGT | 226 | 30 |
| R | CAAGCTTTTGCCCATGAAGC | |||
| abeM | F | AGGGACGTATTATGGCGAAA | 165 | 39 |
| R | CTGCTGTGCTTAGACCAATTTTT | |||
| 16S rRNA | F | CAGCTCGTGTCGTGAGATGT | 150 | 30 |
| R | CGTAAGGGCCATGATGACTT |
The housekeeping gene 16S rRNA was used as the internal standard for normalization. qPCR results were calculated based on Pfaffl method using C t value in 2−ΔΔCt formula.17 The relative expression of each gene was calculated as the normalized ratio of gene expression of a given strain to gene expression in reference strain. A. baumannii strain ATCC19606 was used as the standard strain.
2.6. Statistical analysis
Data were analyzed using SPSS software ver. 20.0 (SPSS Inc., Chicago, Illinois). Parameters such as mean and SD were calculated as descriptive results. Pearson correlation coefficient and nonparametric tests of Mann‐Whitney U test and Kruskal‐Wallis were used to compare between groups. The correlation intensity is graded as follows: from 0.1 to 0.29 = weak, from 0.3 to 0.49 = average, from 0.5 to 1 = strong. Significance level less than .05 was considered significant.
3. RESULTS
3.1. Bacterial strains
During the 6 months of sampling, a total of 122 samples including 88 environmental and 34 clinical were collected from ICUs of four referral hospitals in central Iran. Of them, 31 were identified as Acinetobacter spp. using phenotypic tests (including OF fermentation, oxidase, and growth at 44°C). A total of 28 strains, including 10 environmental isolates and 18 clinical isolates were definitively identified as A. baumannii by amplification of bla OXA‐51.
3.2. MICs of three disinfectants
MIC, MIC50, and MIC90 were determined for all three disinfectants and are shown in Table 3. Also, the mean of MICs and MBCs in all 28 strains was calculated and compared. The mean ± SD MIC of MICROZED ID‐MAX disinfectant was 0.09 ± 0.026, NANOSIL D2 was 0.3 ± 0.16, and OPIDEX OPA was 16.62 ± 6.6. The mean MBC of MICROZED ID‐MAX disinfectant was 0.09 ± 0.026, NANOSIL D2 was 0.41 ± 0.29, and OPIDEX OPA was 16.62 ± 6.6 (Figure 1). Based on the Mann‐Whitney U test, the difference between the mean MICs of NANOSIL and other disinfectants (P < .05), and between OPA and other disinfectants (P < .001) were significant. Also, the difference between the mean MBCs of NANOSIL and other disinfectants (P < .05), and between OPA and other disinfectants (P < .05) were significant. Then, susceptible and resistant strains were determined based on MIC50 and MIC90. The MIC50 and MIC90 in MICROZED ID‐MAX disinfectant were 0.07% and 0.15%, in NANOSIL were 0.31% and 0.62%, and in OPIDEX OPA were 12.5% and 25%, respectively. According to MIC50 and MIC90, the highest percentage of strains was susceptible to MICROZED ID‐MAX disinfectant (24 strains, 86%), while NANOSIL D2 (23 strains, 82%) and OPIDEX OPA (18 strains, 64%) disinfectants ranked second and third, respectively. Therefore, the strains had the highest resistance to OPIDEX OPA disinfectant (Table 4).
TABLE 3.
Serial dilutions of disinfectants for MIC determination
| Dilution number | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Disinfectants | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | MIC50 (%) | MIC90 (%) | |
| MICROZED ID‐MAX | Serial concentrations of disinfectants (%) | 2.5 | 1.25 | 0.62 | 0.31 | 0.15 | 0.07 | 0.03 | 0.01 | 0.07 | 0.15 |
| No. of strains containing specified MICs | 0 | 0 | 0 | 0 | 4 | 24 | 0 | 0 | |||
| NANOSIL D2 | Serial concentrations of disinfectants (%) | 5 | 2.5 | 1.25 | 0.62 | 0.31 | 0.15 | 0.07 | 0.03 | 0.31 | 0.62 |
| No. of strains containing specified MICs | 0 | 0 | 0 | 5 | 13 | 10 | 0 | 0 | |||
| OPIDEX OPA | Serial concentrations of disinfectants (%) | 100 | 50 | 25 | 12.5 | 6.25 | 3.12 | 1.56 | 0.78 | 12.5 | 25 |
| No. of strains containing specified MICs | 0 | 0 | 10 | 17 | 0 | 1 | 0 | 0 | |||
Abbreviation: MIC, minimum inhibitory concentration.
FIGURE 1.
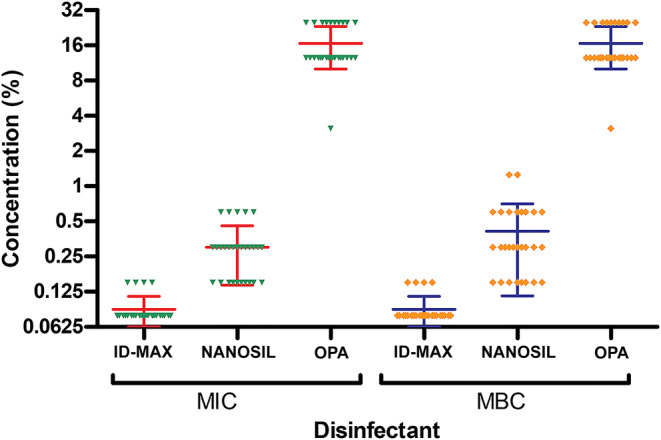
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values of three disinfectants in Acinetobacter baumannii isolates. Mean of MIC and MBC values of three disinfectants has been shown in A. baumannii isolates. Mean and distribution of MICs and MBCs are shown. To determine MIC, different concentrations of disinfectants were applied to the microbial suspension by microdilution method. After MIC determination, part of their contents were cultured in trypticase soy agar medium 18 to 24 hours and wells without colony growth at the lowest concentration of disinfectant designated as MBC
TABLE 4.
Susceptibility ranges of Acinetobacter baumannii isolates to three disinfectants
| Disinfectants | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MICROZED ID‐MAX | NANOSIL D2 | OPIDEX OPA | |||||||||
| Strains | MIC (%) | MBC (%) | Susceptible/resistant | MIC (%) | MBC (%) | Susceptible/resistant | MIC (%) | MBC (%) | Susceptible/resistant | Overall sensitivity | |
| 1 | Clinical | 0.078 | 0.078 | Susceptible | 0.15 | 0.6 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible |
| 2 | 0.078 | 0.078 | Susceptible | 0.3 | 0.6 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 3 | 0.078 | 0.078 | Susceptible | 0.15 | 0.15 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 4 | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 5 | 0.15 | 0.15 | Resistant | 0.15 | 0.15 | Susceptible | 25 | 25 | Resistant | Resistant | |
| 6 | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 25 | 25 | Resistant | Resistant | |
| 7 | 0.078 | 0.078 | Susceptible | 0.6 | 1.25 | Resistant | 12.5 | 12.5 | Susceptible | Resistant | |
| 8 | 0.078 | 0.078 | Susceptible | 0.6 | 0.6 | Resistant | 12.5 | 12.5 | Susceptible | Resistant | |
| 9 | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 25 | 25 | Resistant | Resistant | |
| 10 | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 11 | 0.078 | 0.078 | Susceptible | 0.15 | 0.15 | Susceptible | 25 | 25 | Resistant | Resistant | |
| 12 | 0.078 | 0.078 | Susceptible | 0.3 | 0.6 | Susceptible | 25 | 25 | Resistant | Resistant | |
| 13 | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 14 | 0.078 | 0.078 | Susceptible | 0.15 | 0.15 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 15 | 0.078 | 0.078 | Susceptible | 0.15 | 0.15 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 16 | 0.078 | 0.078 | Susceptible | 0.3 | 0.6 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 17 | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 25 | 25 | Resistant | Resistant | |
| 18 | 0.078 | 0.078 | susceptible | 0.6 | 0.6 | Resistant | 3.12 | 3.12 | Susceptible | Resistant | |
| 19 | Environmental | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 25 | 25 | Resistant | Resistant |
| 20 | 0.078 | 0.078 | Susceptible | 0.6 | 1.25 | Resistant | 25 | 25 | Resistant | Resistant | |
| 21 | 0.15 | 0.15 | Resistant | 0.15 | 0.3 | susceptible | 25 | 25 | Resistant | Resistant | |
| 22 | 0.078 | 0.078 | Susceptible | 0.3 | 0.3 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 23 | 0.078 | 0.078 | Susceptible | 0.15 | 0.15 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 24 | 0.078 | 0.078 | Susceptible | 0.3 | 0.6 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 25 | 0.15 | 0.15 | Resistant | 0.15 | 0.15 | Susceptible | 12.5 | 12.5 | Susceptible | Resistant | |
| 26 | 0.15 | 0.15 | Resistant | 0.3 | 0.3 | Susceptible | 25 | 25 | Resistant | Resistant | |
| 27 | 0.078 | 0.078 | Susceptible | 0.15 | 0.15 | Susceptible | 12.5 | 12.5 | Susceptible | Susceptible | |
| 28 | 0.078 | 0.078 | Susceptible | 0.6 | 0.6 | Resistant | 12.5 | 12.5 | Susceptible | Resistant | |
Abbreviations: MBC, minimum bactericidal concentration (the lowest antibacterial concentration that leads to bacterial death); MIC, minimum inhibitory concentration (the lowest concentration of antibacterial agent that prevents visible bacterial growth).
3.3. The overall frequency of susceptible/resistant strains among clinical and environmental isolates of A. baumannii
The frequency of resistant/susceptible strains to different disinfectants among environmental and clinical strains is shown in Figure 2. Of the total 28 strains, 10 were environmental (35.7%), of which 14.3% were susceptible and 21.4% were resistant. Also, 18 strains (64.3%) were clinical, of which 9 strains (32.1%) were susceptible and the same numbers were resistant. Thus, out of the total isolates, 13 strains (46.4%) were susceptible and 15 strains (53.6%) were resistant. According to these data, the frequency of resistance to all three disinfectants in environmental strains (n = 6 of 10) was significantly higher than that of clinical strains (n = 9 of 18) (P < .05) (Figure 2).
FIGURE 2.
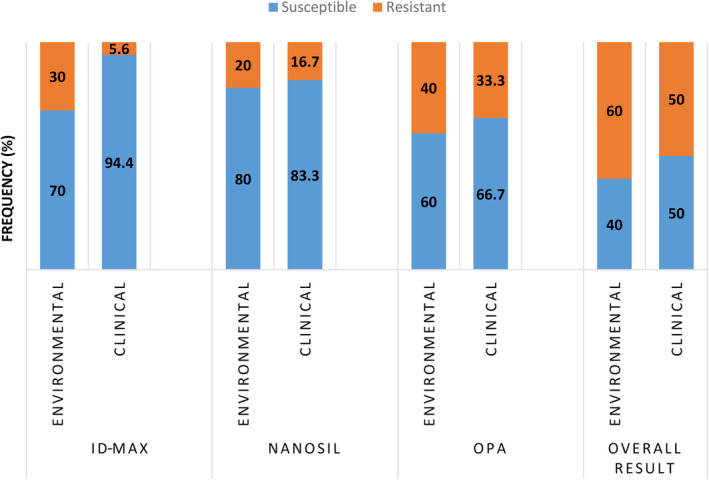
The frequency of susceptible and resistant strains of Acinetobacter baumannii to each disinfectant, in terms of environmental/clinical sources. Resistance of bacterial isolates to different disinfectants was calculated based on the minimum inhibitory concentrations. Overall, 60% of environmental and 50% of clinical isolates were designated as resistant strain
3.4. Distribution of efflux genes expression in susceptible and resistant strains
When comparing all strains in general (both environmental and clinical isolates), there was a significant difference in the mean expression level of qacEΔ1 and abeM genes between susceptible (qacEΔ1 = 1.19 ± 0.08; abeM = 7.0 ± 14.91) and resistant (qacEΔ1 = 2.12 ± 2.07; abeM = 24.04 ± 39.89) strains to MICROZED ID‐MAX disinfectant (P ˂ .05). Also, there was a significant difference in the mean expression level of amvA (P < .05) and abeM (P ˂ .05) genes between susceptible (amvA = 1.57 ± 0.14; abeM = 2.45 ± 3.67) and resistant (amvA = 3.16 ± 2.09; abeM = 10.95 ± 21.88) strains to NANOSIL D2 disinfectant. Similarly, there was a significant difference in the mean expression level of abeM gene in susceptible (2.36 ± 3.45) and resistant (13.36 ± 24.25) strains to OPIDEX OPA disinfectant (P ˂ .05). The mean expression levels of abeM and amvA genes were significantly higher in environmental isolates which were susceptible (amvA = 1.82 ± 0.37; abeM = 3.02 ± 2.14) compared to those that were resistant (amvA = 4.09 ± 2.02; abeM = 23.86 ± 31.59) to NANOSIL D2 disinfectant (P ˂ .05). However, there was no significant difference between the mean expression of abeM, qacEΔ1, amvA, and adeB genes in resistance vs susceptible clinical isolates (Figure 3).
FIGURE 3.
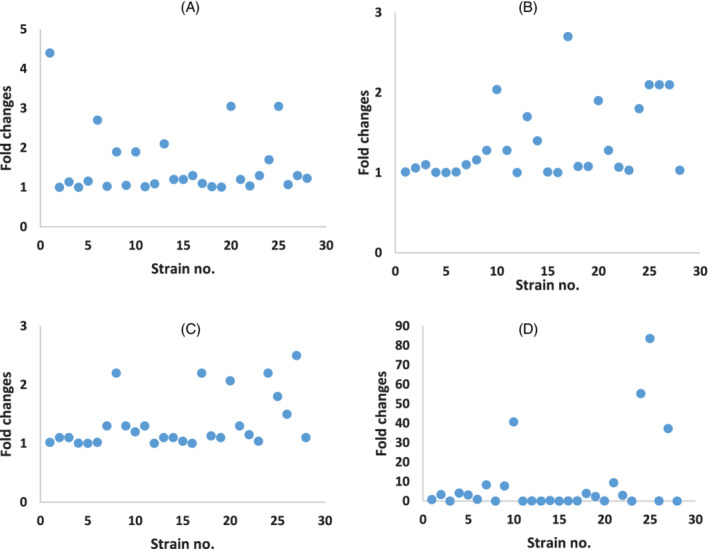
(A‐D). Relative expression of efflux genes in Acinetobacter baumannii strains isolated from clinical and environmental settings. Total RNA was extracted from each strain and reverse transcription to cDNA was performed using M‐MLV enzyme. Real‐time PCR was set on cDNA samples using SYBR Green I system and specific primer pairs. Threshold cycles (C ts) of each amplicon was used for further analysis. The relative quantities of the target genes were normalized against the 16 seconds rRNA gene. Fold‐expression changes of (A) qacEΔ1, (B) adeB, (C) amvA, (D) abeM genes were calculated in each strain compared to reference strain and represented
3.5. The correlation between the rates of expression of the studied genes
The correlation between the expression of four studied genes was investigated, and the results showed that the relationship between gene expression of amvA and adeB was positive, strong, and significant (P < .001), between abeM and adeB (P < .05) and between abeM and amvA (P < .05) was also significant.
4. DISCUSSION
In this study, the resistance of A. baumannii isolates to disinfectants (MICROZED ID‐MAX, NANOSIL D2, and OPIDEX OPA) was measured based on MIC. Then, the association between efflux gene expression rate and MIC changes was determined. The serial dilutions of the three disinfectants used in this study were prepared considering the recommended concentrations of the manufacturers. However, the recommended concentration, which is also applied in hospitals, was higher than enough to inhibit the growth and the isolates were inhibited or killed with lower dilutions. The maximum concentration of OPIDEX OPA disinfectant that inhabited the growth was a solution of 25% while the manufacturer recommended the concentration of 100%, and for NANOSIL D2 it was 0.6% while the recommended concentration was 100%. Similarly, in several other studies, the MIC of the biocides used was less than that of recommended by manufacturers.18, 19 For instance, in the study of the effect of hospital disinfectants including quaternary ammonium compounds (similar to MICROZED ID‐MAX) on Staphylococcus epidermidis, it was suggested that the MIC was from 6 to 8 times lower than the concentrations recommended by the manufacturer.20 However, there are studies that reported that all A. baumannii isolates were resistant to recommended concentrations of disinfectants.21
The use of concentrations above the effective level may lead to the emergence and spread of more resistant strains due to selective pressure and becomes a great challenge in nosocomial infection control.19, 21, 22, 23, 24 It was noted that the high concentrations of biocides are toxic to humans and the environment, in addition to imposing cost to health system.22 It seems that the difference in the effective concentration of disinfectants depends on the type of disinfectant or the bacteria tested.2, 16, 25 However, it can be assumed that the concentrations recommended by the manufacturers probably target all common infectious agents, not just a particular species such as A. baumannii.
In this study, MIC and MBC of MICROZED ID‐MAX disinfectant were equal in all isolates. Similarly in the OPIDEX OPA disinfectant, MIC and MBC were the same for all isolates. This point to the concentrations of these disinfectants applied here, which do not inhibit the growth of the isolates but kill them. In another study on two species of Enterococcus, didecyldimethyl ammonium chloride disinfectant had a similar pattern as its MIC and MBC were the same against the tested strains; Enterococcus faecalis and Enterococcus faecium.26
According to the results, the rate of resistance to OPIDEX OPA was significantly higher than that of the other two disinfectants. The high resistance of A. baumannii to OPIDEX OPA disinfectant may justify the use of high concentrations of this disinfectant in hospitals, according to the manufacturer's recommendation. It should be noted that the variation in response of A. baumannii's to these three disinfectants is not unexpected owing to the difference in structure and mechanisms of disinfectant tested here. In another study, a significant association between the MICs of benzalkonium chloride and benzetonium chloride was attributed to the similarity of their chemical structure.16
According to this study, the percentage of resistant strains in environmental isolates (60%) was higher than that of resistant strains in clinical isolates (50%). This may be due to the fact that environmental strains are more exposed to disinfectants than clinical strains. In contrast to the current study, a study showed that clinical strains of A. baumannii were more resistant to disinfectants than environmental strains.27 The reason for the difference can be related to different testing conditions such as geographical location, type of disinfectants, and to the method of using disinfectants. The effectiveness of a disinfection method depends on the contact time, temperature, concentration of the active substance and the persistence of disinfectants,2 and the type of growth of the isolates in the environment (biofilm or plankton).28
In this study, the level of efflux gene expression was compared in environmental and clinical strains and showed that the mean expression of amvA and adeB genes was significantly higher in the environmental than clinical isolates of A. baumannii. There is further evidence for the role of adeB and amvA gene expression in resistance to biocides. In one study, strains of A. baumannii exposed to chlorhexidine digluconate had increased adeB gene expression.29 Also, the expression of the adeB gene in A. baumannii, which was resistant to triclosan, was significantly higher than the susceptible strains.30, 31 In MDR A. baumannii, which were exposed to methyl viologen and ethidium bromide, amvA expression levels increased compared to the susceptible strains and inhibiting the amvA pump confirmed the role of this gene in resistance.32 The most probable reason for the high resistance of environmental isolates is that they are more exposed to biocides than clinical isolates.
Based on the current result, the difference in the expression of qacEΔ1 gene was significant between susceptible and resistant strains to MICROZED ID‐MAX disinfectant. Previously, a relationship between the presence of qac gene and increased MIC in A. baumannii isolates was reported.33 MICs were suggested to be significantly high in strains of Salmonella spp., Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus containing the qacEΔ1 gene.25, 34 In contrast, there are other studies that do not suggest a significant difference between the presence or absence of qacE gene and susceptible/resistant strains of A. baumannii.18, 23
This study showed that among clinical isolates, there was no significant difference in amvA gene expression between susceptible and resistant ones. There was also no significant difference in abeM gene expression between susceptible and resistant clinical isolates. In contrast, there was a significant difference in efflux gene expression between resistant and susceptible isolates of environmental source.
A study by Rajamohan et al suggested that by inactivation of these genes, susceptibility to multiple antibiotics and disinfectants was increased.32 While we did not find a role for adeB efflux gene function in disinfectant resistance, in other studies the role of efflux genes including the adeB gene in antiseptic‐resistant isolates of Acinetobacter spp. was demonstrated.35, 36
Based on the current results, there was a positive significant correlation between gene expression of abeM and amvA, abeM and adeB, and amvA and adeB in all samples, both clinical and environmental, and resistant and susceptible. This significant association among the expressions of efflux genes indicates that efflux's mechanism of biocides occurs through multiple genes function.
The results showed that the effective concentrations of disinfectants in inhibiting the growth of A. baumannii strains are less than the amounts recommended by manufacturing companies. Therefore, to prevent selective pressure and consequently the spread of biocide‐resistant strains, it is advisable to keep the concentration of the disinfectants according to their calculated MICs and bacterial type. EU GMP and US FDA recommend the use of disinfectants by rotations to prevent microbial resistance.37
5. CONCLUSIONS
The significant difference in the expression of efflux pumps genes studied in this study between the susceptible and resistant strains of A. baumannii shows the association of efflux function in resistance to disinfectants. Particularly abeM and amvA genes contribute to resistance of environmental isolates of A. baumannii to hydrogen peroxide derivatives. However, the contribution of efflux pumps mechanism in reducing the susceptibility of the clinical strains to disinfectants was not as evident as in the environmental isolates.
FUNDING
This study was funded by Tehran University of Medical Sciences, Tehran, Iran. Tehran University of Medical Sciences had no role in the study design; collection, analysis, and interpretation of data; writing of the report; and the decision to submit the report for publication.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceptualization: Mahmoud Nateghi Rostami
Data Curation: Mojtaba Ranjbar
Formal Analysis: Tahereh Rostami, Mahmoud Nateghi Rostami
Methodology: Tahereh Rostami, Mojtaba Ranjbar, Sedighe Ghourchian, Fatemeh Darzi
Supervision: Masoumeh Douraghi
Writing—Original Draft Preparation: Tahereh Rostami
Writing—Review and Editing: Masoumeh Douraghi, Mahmoud Nateghi Rostami
All authors have read and approved the final version of the manuscript.
Mahmoud Nateghi Rostami had full access to all of the data in this study and takes complete responsibility for the integrity of the data and the accuracy of the data analysis.
TRANSPARENCY STATEMENT
Mahmoud Nateghi Rostami affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
ACKNOWLEDGMENTS
The authors would like to thank staff of laboratory at Department of Parasitology, Pasteur Institute of Iran, for their kind help during the molecular experiment.
Rostami T, Ranjbar M, Ghourchian S, Darzi F, Douraghi M, Nateghi‐Rostami M. Upregulation of abeM, amvA, and qacEΔ1 efflux pump genes associated with resistance of Acinetobacter baumannii strains to disinfectants. Health Sci Rep. 2021;4:e395. doi: 10.1002/hsr2.395
Funding information Amol University of Special Modern Technologies; Tehran University of Medical Sciences and Health Services
Contributor Information
Masoumeh Douraghi, Email: mdouraghi@tums.ac.ir.
Mahmoud Nateghi‐Rostami, Email: m_nateghi@pasteur.ac.ir.
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article.
REFERENCES
- 1.Mehta Y, Gupta A, Todi S, et al. Guidelines for prevention of hospital acquired infections. Indian J Crit Care Med. 2014;18(3):149‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montagna MT, Triggiano F, Barbuti G, et al. Study on the in vitro activity of five disinfectants against nosocomial bacteria. Int J Environ Res Public Health. 2019;16(11):1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martí S, Rodríguez‐Baño J, Catel‐Ferreira M, et al. Biofilm formation at the solid‐liquid and air‐liquid interfaces by Acinetobacter species. BMC Res Notes. 2011;4(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawamura‐Sato K, Wachino J‐I, Kondo T, Ito H, Arakawa Y. Reduction of disinfectant bactericidal activities in clinically isolated Acinetobacter species in the presence of organic material. J Antimicrob Chemother. 2008;61(3):568‐576. [DOI] [PubMed] [Google Scholar]
- 5.Russell AD. Bacterial resistance to disinfectants: present knowledge and future problems. J Hosp Infect. 1999;43:S57‐S68. [DOI] [PubMed] [Google Scholar]
- 6.Coyne S, Courvalin P, Périchon B. Efflux‐mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55(3):947‐953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bialvaei AZ, Kouhsari E, Salehi‐Abargouei A, et al. Epidemiology of multidrug‐resistant Acinetobacter baumannii strains in Iran: a systematic review and meta‐analysis. J Chemother. 2017;29(6):327‐337. [DOI] [PubMed] [Google Scholar]
- 8.Bridier A, Briandet R, Thomas V, Dubois‐Brissonnet F. Resistance of bacterial biofilms to disinfectants: a review. Biofouling. 2011;27(9):1017‐1032. [DOI] [PubMed] [Google Scholar]
- 9.McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12(1):147‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Housseini B, Issa K, Phan G, Broutin I. Functional mechanism of the efflux pumps transcription regulators from Pseudomonas aeruginosa based on 3D structures. Front Mol Biosci. 2018;5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du D, van Veen HW, Murakami S, Pos KM, Luisi BF. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr Opin Struct Biol. 2015;33:76‐91. [DOI] [PubMed] [Google Scholar]
- 12.Hassan KA, Liu Q, Elbourne LDH, et al. Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram‐negative pathogens. Res Microbiol. 2018;169(7):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manchanda V, Sanchaita S, Singh N. Multidrug resistant Acinetobacter . J Glob Infect Dis. 2010;2(3):291‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nateghi Rostami M, Mehrban F, Ghourchian S. Genetic diversity of OXA producing carbapenem‐resistant Acinetobacter baumannii from environment of tertiary hospitals in central Iran. Arch Clin Infect Dis. 2020;15(1):e95602. [Google Scholar]
- 15.Ciccolini LAS, Ayazi Shamlou P, Titchener‐Hooker NJ, Ward JM, Dunnill P. Rheological properties of chromosomal and plasmid DNA during alkaline lysis reaction. Bioprocess Eng. 1999;21(3):231‐237. [Google Scholar]
- 16.Kawamura‐Sato K, Wachino J‐I, Kondo T, Ito H, Arakawa Y. Correlation between reduced susceptibility to disinfectants and multidrug resistance among clinical isolates of Acinetobacter species. J Antimicrob Chemother. 2010;65(9):1975‐1983. [DOI] [PubMed] [Google Scholar]
- 17.Nateghi Rostami M, Keshavarz H, Edalat R, et al. CD8+ T cells as a source of IFN‐γ production in human cutaneous leishmaniasis. PLoS Negl Trop Dis. 2010;4(10):e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babaei MR, Sulong A, Hamat RA, Nordin SA, Neela VK. Extremely high prevalence of antiseptic resistant quaternary ammonium compound E gene among clinical isolates of multiple drug resistant Acinetobacter baumannii in Malaysia. Ann Clin Microbiol Antimicrob. 2015;14(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi M, Kawamura K, Matsui M, et al. Reduction in chlorhexidine efficacy against multi‐drug‐resistant Acinetobacter baumannii international clone II. J Hosp Infect. 2017;95(3):318‐323. [DOI] [PubMed] [Google Scholar]
- 20.Asadollahi P, Jabalameli F, Beigverdi R, Emaneini M. Assessment of disinfectant and antibiotic susceptibility patterns and multi‐locus variable number tandem repeat analysis of Staphylococcus epidermidis isolated from blood cultures. Iran J Microbiol. 2018;10(2):90‐97. [PMC free article] [PubMed] [Google Scholar]
- 21.Nowroozi J, Akhavan Sepahi A, Tahmasebinejad Kamarposhti L, Razavipour R, Mazhar F. Evaluation of ciprofloxacin (gyrA, parC genes) and tetracycline (tetB gene) resistance in nosocomial Acinetobacter baumannii infections. Jundishapur J Microbiol. 2014;7(2):e8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas D, Tiwari M, Tiwari V. Comparative mechanism based study on disinfectants against multidrug‐resistant Acinetobacter baumannii . J Cell Biochem. 2018;119(12):10314‐10326. [DOI] [PubMed] [Google Scholar]
- 23.Romao C, Miranda CA, Silva J, Mandetta Clementino M, de Filippis I, Asensi M. Presence of qacEΔ1 gene and susceptibility to a hospital biocide in clinical isolates of Pseudomonas aeruginosa resistant to antibiotics. Curr Microbiol. 2011;63(1):16‐21. [DOI] [PubMed] [Google Scholar]
- 24.Liu WJ, Fu L, Huang M, et al. Frequency of antiseptic resistance genes and reduced susceptibility to biocides in carbapenem‐resistant Acinetobacter baumannii . J Med Microbiol. 2017;66(1):13‐17. [DOI] [PubMed] [Google Scholar]
- 25.Wu G, Yang Q, Long M, et al. Evaluation of agar dilution and broth microdilution methods to determine the disinfectant susceptibility. J Antibiot (Tokyo). 2015;68(11):661‐665. [DOI] [PubMed] [Google Scholar]
- 26.Schwaiger K, Harms K, Bischoff M, et al. Insusceptibility to disinfectants in bacteria from animals, food and humans—is there a link to antimicrobial resistance? Front Microbiol. 2014;5:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanjri S, Uwingabiye J, Frikh M, et al. In vitro evaluation of the susceptibility of Acinetobacter baumannii isolates to antiseptics and disinfectants: comparison between clinical and environmental isolates. Antimicrob Res Infect Cont. 2017;6(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanković T, Goić‐Barišić I, Hrenović J. Reduced susceptibility to disinfectants of Acinetobacter baumannii biofilms on glass and ceramic. Arh Hig Rada Toksikol. 2017;68(2):99‐108. [DOI] [PubMed] [Google Scholar]
- 29.Fernández‐Cuenca F, Tomás M, Caballero‐Moyano F‐J, et al. Reduced susceptibility to biocides in Acinetobacter baumannii: association with resistance to antimicrobials, epidemiological behaviour, biological cost and effect on the expression of genes encoding porins and efflux pumps. J Antimicrob Chem. 2015;70(12):3222‐3229. [DOI] [PubMed] [Google Scholar]
- 30.Lin F, Xu Y, Chang Y, Liu C, Jia X, Ling B. Molecular characterization of reduced susceptibility to biocides in clinical isolates of Acinetobacter baumannii . Front Microbiol. 2017;8:1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu K, Zhang Y, Xu W, et al. Hyper‐expression of the efflux pump gene adeB was found in Acinetobacter baumannii with decreased triclosan susceptibility. J Glob Antimicrob Resist. 2020;22:367‐373. [DOI] [PubMed] [Google Scholar]
- 32.Rajamohan G, Srinivasan VB, Gebreyes WA. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii . J Antimicrob Chemother. 2010;65(9):1919‐1925. [DOI] [PubMed] [Google Scholar]
- 33.Gomaa FAM, Helal ZH, Khan MI. High prevalence of blaNDM‐1, blaVIM, qacE, and qacEΔ1 genes and their association with decreased susceptibility to antibiotics and common hospital biocides in clinical isolates of Acinetobacter baumannii . Microorganisms. 2017;5(2):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafaati M, Boroumand M, Nowroozi J, Amiri P, Kazemian H. Correlation between qacE and qacE∆1 efflux pump genes, antibiotic and disinfectant resistant among clinical isolates of E.coli. Recent Pat Antiinfect Drug Discov. 2016;11(2):189‐195. [DOI] [PubMed] [Google Scholar]
- 35.Dostvandi S, Abiri R, Mohajeri P, Alvandi A. Genotypic and phenotypic characteristics of efflux pumps in Acinetobacter spp. isolated from inpatients. J Mazandaran Uni Med Sci. 2015;25(126):1‐10. [Google Scholar]
- 36.Rajamohan G, Srinivasan VB, Gebreyes WA. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J Antimicrob Chemother. 2009;65(2):228‐232. [DOI] [PubMed] [Google Scholar]
- 37.Vijayakumar R, Sandle T. A review on biocide reduced susceptibility due to plasmid‐borne antiseptic‐resistant genes—special notes on pharmaceutical environmental isolates. J Appl Microbiol. 2019;126(4):1011‐1022. [DOI] [PubMed] [Google Scholar]
- 38.Nowak J, Seifert H, Higgins PG. Prevalence of eight resistance‐nodulation‐division efflux pump genes in epidemiologically characterized Acinetobacter baumannii of worldwide origin. J Med Microbiol. 2015;64(6):630‐635. [DOI] [PubMed] [Google Scholar]
- 39.Rumbo C, Gato E, López M, et al. Contribution of efflux pumps, porins, and β‐lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii . Antimicrob Agents Chemother. 2013;57(11):5247‐5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


