Addition of a functional copy of the b-globin gene and reactivation of fetal hemoglobin (HbF) are promising therapeutic approaches for b-globinopathies such as sickle cell disease (SCD) and b-thalassemia. Results from ongoing clinical trials for b-globinopathies indicate that a successful outcome is genotype-dependent. In studies that included patients with the b0/b0 genotype or SCD, vectors expressing a single curative gene have failed or have required several integrations per genome to produce enough adult hemoglobin (HbA) levels to correct the phenotype of the patients.1-6 Therefore, to achieve HbA synthesis at therapeutic levels in the most severe of genotypes, and with minimal vector copy number (VCN) per cell, more powerful and versatile vectors are required.
Figure 1.
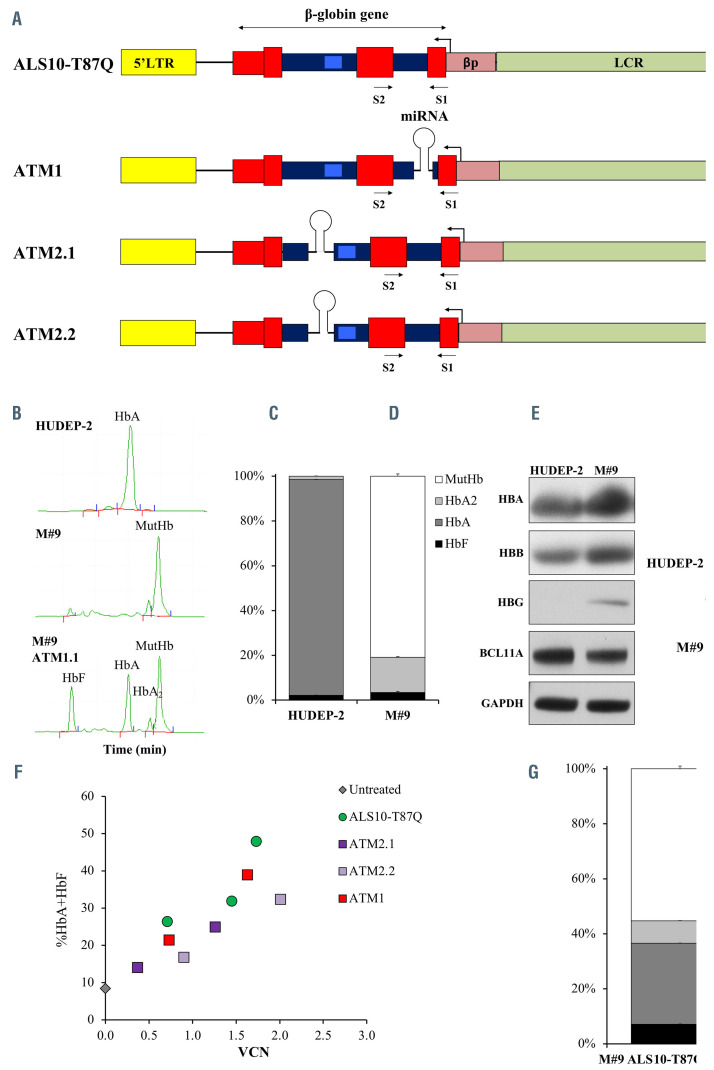
Initial constructs design, Hudep9 (M#9) line development and characterization of constructs efficiency. (A) Initial constructs used in the study: ALS10- T87Q, ATM1.1, ATM2.1, ATM2.2. The miR-E-BCL11A5 sequences were synthesized by Genscript USA (NJ) by combining the passenger and guide sequences of short hairpin RNA (shRNAmiR5) with the flanking miR-E backbone sequences.3,4 The combined miR-E-BCL11A5 sequences were cloned in intron 1 position c.79+36 of ALS10-T87Q (ATM1) and intron 2 positions c.303-163 and c.303-172 (ATM2.1 and ATM2.2, respectively) of the b-globin transgene. Viral production and titration were performed according to methods previously reported.15 All vectors include the human b-globin gene and regulatory elements (β-globin promoter (bp) and portions of the locus control region (LCR) as in our previously described vector AnkT9).8 Some of these vectors present a miRNA as described in the main manuscript. Erythroid specific expression of the b-globin gene and shRNAmiR is achieved by using the b-globin promoter and its locus control region (LCR), in a pol II-promoter driven system. Transgene expression under these regulatory elements will be limited to the latest stages of erythroid development,8 preventing unwanted effects as previously seen in B lymphocytes and hematopoetic stem cellks.10 (B) Representative chromatographic separation (high-performance liquid chromatography [HPLC])2 of hemolysates from the HUDEP-2 (top) and M#9 (middle) erythroblasts (developed using methodology previously described),15 showing fetal hemoglobin (HbF), adult hemoglobin (HbA), HbA2 and mutant HbA (MutHbA) peaks at day 7 of differentiation as described.1,15 At the bottom, a representative chromatographic separation15 of the M#9 cell line after transduction with one of the vectors that simultaneously produced HbAT87Q and induced HbF expression. (C) Relative proportion of HbF, HbA, HbA2 and MutHb in HUDEP-2 and M#9 quantified by HPLC at day 7 of differentiation. (D) Representative western blot showing HBA, HBB, HBG and BCL11A proteins in HUDEP-2 and M#9 at day 7 of differentiation. The evaluation was performed using rabbit anti- HBB (Proteintech, Chicago, Il), rabbit anti-HBA (Santa Cruz Biotechnology), mouse anti-BCL11A (Abcam, Cambridge, MA, USA) rabbit anti-HBG (Santa Cruz Biotechnology, Santa Cruz, CA) and rabbit anti-GAPDH (Proteintech) primary antibodies. Secondary, anti-mouse and anti-rabbit IgG HRP-linked antibodies (Cell Signaling) were used for detection with the Amersham enhanced chemiluminescence (ECL) western blotting analysis system (GE Healthcare, Malborough, MA, USA). (E) Representative fluorescence-activated cell sorting (FACS) plots of HUDEP-2 and M#9 stained for GPA, CD71, c-KIT and CD36 markers adapted from and analyzed as previously described.8 (F) Percentage of total curative hemoglobins (HbAT87Q+HbF) over a range of viral integrations after transducing M#9 cells. (G) Proportion of the different hemoglobins produced in M#9 cells transduced with ALS10-T87Q with vector copy number (VCN)= 2.
Using a previous vector developed by our group indicated as AnkT9W, we generated ALS10-T87Q (Figure 1A), a vector that produces a functional b-globin gene that carries a modification (indicated as T87Q) with improved anti-sickling activity.7-8 Additionally, ALS10- T87Q vector includes the full sequence of the intron 2 of the b-globin gene (Figure 1A). We incorporated into the b-globin coding sequence a short hairpin RNA (shRNAmiR) targeting the transcription factor BCL11A, a known repressor of γ-globin.
The shRNAmiR sequences targeting BCL11A9-10 were flanked by an optimized backbone termed “miR-E” (11) to increase HbF levels through downregulation of BCL11A (Figure 1A). Our goal is to overcome some of the limitations of the vectors presently in clinical trials by simultaneous i) production of transgenic HbA; ii) reactivation of endogenous HbF; and iii) decrease in production of endogenous mutant b-globin mRNA and/or protein.
The erythroid HUDEP-2 immortalized human cord blood cell line was generated by inducible expression of the encoding genes HPV16-E6/E7 derived from the human Papilloma virus 16.12 These cells can be used as a semi-primary adult hematopoietic cell model that can be propagated indefinitely in the presence of tetracycline or differentiated into red cells under appropriate culture conditions. In order to assess the level of HbAT87Q and HbF produced by the new vectors, we mutagenized the first exon of the human b-globin gene (HBB) in erythroid HUDEP-2 cells using the CRISPR/Cas9 system. We established a clonal cell line, named M#9, carrying a homozygous deletion of codon 6, thus generating a mutant HBB gene (Hb-mutant). This line produced a predominant mutant adult HbA (Hb-mutant) distinguishable from the endogenous wild-type (WT) HbA by high-performance liquid chromatography (HPLC) (Figure 1A and B, top and middle panel, and Figure 1C), which was also visible by western blot analysis (Figure 1D). The change did not alter erythroid maturation (Figure 1E). The HbAT87Q generated by ALS10-T87Q and HbF induced by the shRNAmir were also both distinguishable from the Hb-mutant produced by M#9 via chromatographic separation (Figure 1B, bottom panel). We were able to assess the relative production of functional HbAT87Q +HbF after gene transfer and correlate these values to VCN in a dose/effect relationship. Upon transduction in the M#9 cell line, ALS10-T87Q generated 18%, 23% and 44% of HbAT87Q with VCN=0.6, 1.0 and 2.0, respectively (Figure 1F and G). We cloned the miR-E-BCL11A sequence either in the b-globin intron 1 (ATM1), or in two different regions of intron 2 (ATM2.1 and ATM2.2) (Figure 1A). Upon transduction of M#9, all ATM vectors reduced BCL11A level and induced HbF production by western blot (Online Supplementary Figure S1A), confirming functional repression from the miR-E-BCL11A sequence. ATM1, the most effective among the ATM vectors, showed the highest HbF induction (Online Supplementary Figure S1B) and an HbA production equivalent to ~90% of that made by ALS10-T87Q at VCN=1 (Online Supplementary Figure S1C). However, the total production of HbAT87Q +HbF by ATM1 vector was suboptimal, as the combination of the miR-E-BCL11A with the b-globin gene did not lead to an overall increase of therapeutic hemoglobins (HbAT87Q+HbF) compared to ALS10-T87Q (Figure 1F; Online Supplementary Figure S1D to E).
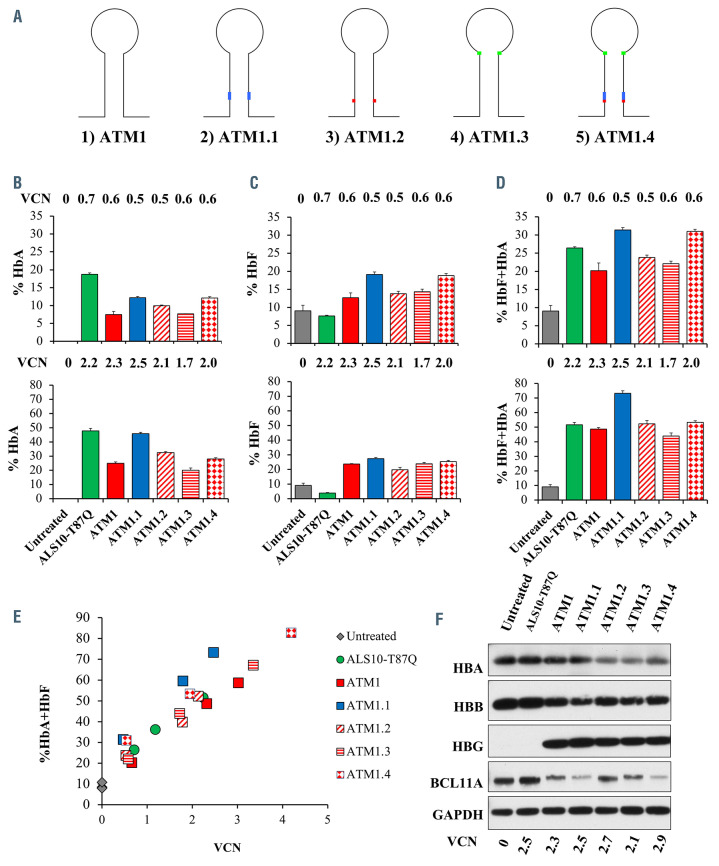
In order to overcome this limitation, we modified the sequence of miR-E-BCL11A within intron 1, generating additional vectors named ATM1.1, ATM1.2, ATM1.3 and ATM1.4 (Figure 2A; Online Supplementary Table S1A). We tested two different modifications of the guide and one modification of the loop of the miR scaffold itself. ATM1.1 differs from ATM1 in the last four bases of the guide to adapt the shift from shRNA to miR-based design, as indicated in the legend of Figure 2A. These changes ensure higher 5’ passenger stability for efficient guide recognition and incorporation into RISC complex as previously shown.9
Among these constructs, ATM1.1 produced the highest HbAT87Q and HbF expression levels (Figure 2B and C, top and bottom panels). ATM1.1 produced between ~25% and ~47% more therapeutic hemoglobins (HbAT87Q+HbF) compared to the HbAT87Q produced by ALS10-T87Q, at low and high VCN, respectively (Figure 2D, top and bottom panels). Using a mixed-effects linear regression model, the increase was quantified in ~33% more HbAT87Q+HbF in ATM1.1 compared to the HbAT87Q made by ALS10-T87Q (looking at similar levels of VCN; Figure 2E; Online Supplementary Table S1B). Western blot analysis confirmed the simultaneous reduction of BCL11A and the increase of γ-globin protein levels in M#9 samples treated with ATM1.1 (Figure 2F).
In order to verify that the mRNA or DNA of the ATM1.1 vector did not recombine due to the inclusion of the miR-E-BCL11A, both during integration and expression of the transcript, we introduced silent mutations in exon1 and exon2 of the b-globin gene (Silb-globin gene) to distinguish, in human cells, DNA and mRNA of the endogenous and the transgenic Silb-globin sequences (Figure 3G). We confirmed that the oligonucleotides (Online Supplementary Table S1D) designed to amplify the Silb-globin gene amplified the transgenic Silb-globin gene exclusively (Online Supplementary Figure S1F). Using a murine erythroleukemia cell line (MEL), we confirmed a linear relationship between VCN, chimeric HbA (mouse α-globin/human b-globin-T87Q tetramer; Online Supplementary Figure S1G), protein levels (Online Supplementary Figure S1H), and human b-globin RNA (Online Supplementary Figure S1I) produced upon integration of ALS10-T87Q, ALS10-T87Q-silent, ATM1.1 and ATM1.1-silent. Furthermore, in a pilot experiment on CD34+-derived SCD erythroblasts treated with ALS10-T87Q, ALS10-T87Q-silent, ATM1.1 and ATM1.1-silent vectors, we were able to discriminate between the SCD and b-globin-T87Q mRNA and that encoded by the silent vectors (Online Supplementary Figure S2A). Amplification of the region encompassing exon 1 and 2 (Figure 1A), performed using oligonucleotides (Online Supplementary Table S1D) that selectively anneal to the silent b-globin mRNA, suggested that the presence of the miR-E-BCL11A in intron 1 did not affect the length and splicing of the portion of the b-globin mRNA amplified in the double vector (Online Supplementary Figure S2B). This indicated no apparent interference between the expression of the transgenic b-globin and the miR-EBCL11A components.
Figure 2.
Optimization of BCL11A miRNA design. (A) Illustration of changes introduced into BCL11A miRNA sequence of ATM1.1 vector. ATM1.2 contains 1 basepair (bp) mismatch between the guide and the passenger creating a ”bulge” in our design, based on evidence from Fellman’s group, which demonstrated that inclusion of such an element can boost the potency of the miRNA.11 ATM1.3 presents a 2 bp modification of the published miR-30 loop to improve performance. 11 The modifications tested in ATM1.1, -1.2 and -1.3 were combined in the final design of ATM1.4. (B) Adult hemoglobin (HbAT87Q) (C) fetal hemoglobin (HbF) and (D) HbAT87Q+HbF levels at low (0.6+/-0.1, top panels) or high (2.0+/-0.3, bottom panels) integrations per cell after transduction of M#9 with ALS10-T87Q, ATM1, and its variants ATM1.1, ATM1.2, ATM1.3, or ATM1.4. (E) Graphic representation of the proportion of curative HbAT87Q+HbF hemoglobin over a range vector copy number (VCN) after transduction of M#9 with ALS10-T87Q, ATM1, ATM1.1, ATM1.2, ATM1.3, or ATM1.4. Data are represented as mean ± standard deviation, n=3. (F) Representative western blot showing side by side the HBA, HBB, HBG and BCL11A expression in control and different vectors transduced M#9 cells at day 7 of differentiation.
Figure 3.
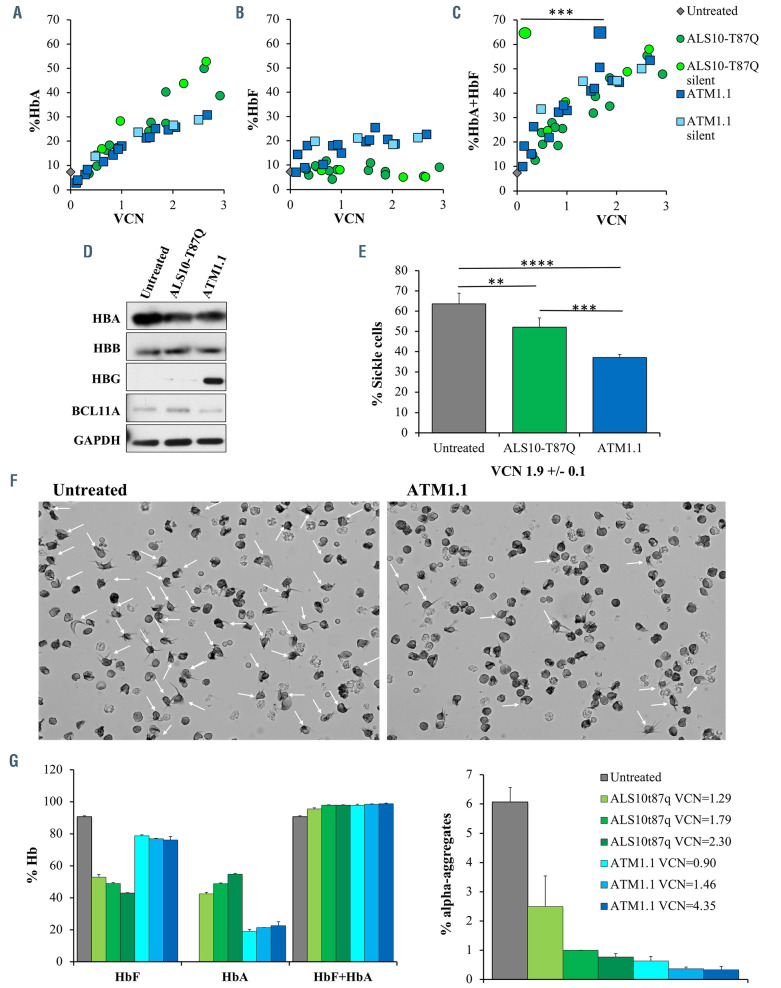
Efficiency of ATM1.1 construct in sickle cell disease and b0/b0 patients’ cells. Dose/response analyses obtained by plotting integration levels (vector copy number [VCN]) against the percentage values of (A) adult hemoglobin (HbAT87Q), (B) fetal hemoglobin (HbF) and (C) HbAT87Q+HbF, respectively, in sickle cell disease (SCD) erythroid cells untreated or treated with ALS10-T87Q, ALS10-T87Q-silent, ATM1.1, or ATM1.1-silent. Data are represented as mean ± standard deviation (SD), n=3. (D) Representative western blot showing the HBA, HBB, HBG and BCL11A protein content in differentiated SCD cells. (E) Percentage of blind counts of sickle-shaped cells exposed to hypoxia, as previously described,5 in four fields from untransduced SCD erythroid cells or after treatment with ALS10- T87Q or ATM1.1. Averages and SD are indicated. (F) Representative image of patient-derived SCD erythroid cultures under hypoxia. Arrows mark cells with sickle- like morphology. (G) Dose/response analyses obtained for HbF, HbAT87Q, and HbAT87Q+HbF, with increasing VCN, in patient-derived b0/b0 thalassemic erythroid cells untreated or treated with ALS10-T87Q, ATM1.1., left panel. Percentage of corresponding alpha aggregates, right panel. Statistical significance was assessed by computing means with one-way Anova test followed by a Tukey test to compare every mean with every other mean. All tests were done using GraphPad Prism software, version 7.0. software package was used for statistical analysis.
In order to evaluate the potential use of this vector in gene therapy for SCD, we transduced CD34+-derived hematopoietic progenitor cells isolated from peripheral blood of three patients with SCD. We compared the production of HbAT87Q and HbF by ATM1.1 with its counterpart ALS10-T87Q. We observed a dose-dependent induction of HbF and HbAT87Q in SCD erythroblasts treated with ATM1.1 (Figure 3A and B). Moreover, our results showed that ATM1.1 outperformed ALS10-T87Q, inducing the highest levels of curative hemoglobins (HbAT87Q+HbF) (Figure 3C; Online Supplementary Table S2C). Western blot analysis confirmed the induction of γ-globin chains in cells transduced with ATM1.1 (Figure 3D). While not completely suppressed, the partial knockdown of BCL11A observed in cells treated with ATM1.1 (Figure 3D) could be advantageous. Reported evidence suggests that disruption of BCL11A leads to an erythroid differentiation defect or survival disadvantage.13-14 Gene expression analysis of SCD cells showed comparable expression of transgenic b-globin, irrespectively of whether they carried the silent-b-globin mRNA or not (Online Supplementary Figure S2C). This also indicated that introduction of the silent mutation did not affect the ability of the vectors to produce the b-globin mRNA (Online Supplementary Figure S2C, top panel). Both ATM1.1 and its silent variant presented a significantly increased level of γ-globin mRNA, tightly correlating with the protein yield (Online Supplementary Figure S2A to C). Moreover, the SCD erythroblasts treated with ATM1.1 and exposed to hypoxic conditions showed lower percentage of sickle-like morphology (58.1%) by comparison to parallel specimens transduced with ALS10-T87Q (81.9%) (Figure 3E to F). We analyzed b0/b0 thalassemic patients’ cells and compared the percentage of total curative hemoglobin produced by ALS10-T87Q and ATM1.1. Untransduced b0/b0 cells showed over 90% of baseline HbF (Figure 3G, left panel). The high level of HbF is in part due to intrinsic features of the culture conditions and lack of b-globin expression in these cells. Upon transduction, we observed that the total content of HbF+HbA was close to 100%, although in different proportions (Figure 3G, left panel). The most informative change was the abundance of a HPLC peak corresponding to α-globin aggregates, which we previously characterized in b0/b0 CD34+ derived erythroid cells.8 ATM1.1-transduced cells showed lower levels of these aggregates when compared to the same cells transduced with ALS10-T87Q at similar VCN (Figure 3G, right panel; Online Supplementary Figure S2D). The higher suppression of free α-chains suggests that, overall, ATM1.1 was able to produce more curative chains (b+γ).
In conclusion, our results show that HbA and HbF can be elevated simultaneously using a single lentiviral construct, as our ATM1.1 vector. The ability of ATM1.1 to induce functional hemoglobin production (HbF+HbA) surpasses that of a vector expressing b-globin alone. While the results in vitro demonstrate proof-of-principle potency of the vectors used, the level of functional hemoglobin produced in vivo may vary. Future studies will evaluate this approach analyzing erythroid cells derived from transduced SCD or thalassemic HSC in vivo. These studies will be performed after cloning the best miR-E-BCL11A into an enhanced vector recently developed in our lab.15
Supplementary Material
Acknowledgments
We would like to thank Vanessa Carrion for her assistance in identifying the SCD specimens.
References
- 1.Thompson AA, Walters MC, Kwiatkowski JL, et al. Northstar-2: updated safety and efficacy analysis of lentiglobin gene therapy in patients with transfusion-dependent b-thalassemia and non-b0/b0 genotypes. Blood. 2019;134(Suppl1):S3543. [Google Scholar]
- 2.Lal A, Locatelli F, Kwiatkowski JL, et al. Northstar-3: interim results from a phase 3 study evaluatinglLentiglobin gene therapy in patients with transfusion-dependent b-thalassemia and either a b0 or IVS-I- 110 mutation at both alleles of the HBB gene. Blood. 2019;134(Suppl 1):S815. [Google Scholar]
- 3.Ribeil JA, Hacein-Bey-Abina S, Payen E, et al. Gene therapy in a patient with sickle cell disease. N Engl J Med. 2017;376(9):848-855. [DOI] [PubMed] [Google Scholar]
- 4.Kanter J, Walters MC, Hsieh MM, et al. Interim results from a phase 1/2 clinical study of lentiglobin gene therapy for severe sickle cell disease. Blood. 2016;128(22):1176. [Google Scholar]
- 5.Kanter J, Tisdale JF, Mapara MY, et al. Resolution of sickle cell disease manifestations in patients treated with lentiglobin gene therapy: updated results from the phase 1/2 Hgb-206 Group C Study. Blood. 2019;134(Suppl 1):S990. [Google Scholar]
- 6.Marktel S, Scaramuzza S, Cicalese MP, et al. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ss-thalassemia. Nat Med. 2019;25(2):234-241. [DOI] [PubMed] [Google Scholar]
- 7.Pawliuk R, Westerman KA, Fabry ME, et al. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294(5550):2368-2371. [DOI] [PubMed] [Google Scholar]
- 8.Breda L, Casu C, Gardenghi S., et al. , Therapeutic hemoglobin levels after gene transfer in beta-thalassemia mice and in hematopoietic cells of beta-thalassemia and sickle cells disease patients. PLoS One. 2012;7(3):32345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guda S, Brendel C, Renella R., et al. , miRNA-embedded shRNAs for lineage-specific BCL11A knockdown and hemoglobin F induction. Mol Ther. 2015;23(9):1465-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brendel C, Guda S, Renella R, et al. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J Clin Invest. 2016;126(10):3868-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellmann C, Hoffmann T, Sridhar V., et al. , An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5(6):1704-1713. [DOI] [PubMed] [Google Scholar]
- 12.Kurita R, Suda N, Sudo K., et al. , Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8(3):59890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luc S, Huang J, McEldoon JL, et al. Bcl11a-deficiency leads to hematopoietic stem cell defects with an aging-like phenotype. Cell Rep. 2016;16(12):3181-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Psatha N, Reik A, Phelps S, et al. Disruption of the BCL11A erythroid enhancer reactivates fetal hemoglobin in erythroid cells of patients with b-thalassemia major. Mol Ther Methods Clin Dev. 2018; 10:313-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breda L, Ghiaccio V, Tanaka N, et al. Lentiviral vector ALS20 yields high hemoglobin levels with low genomic integrations for treatment of beta-globinopathies. Mol Ther. 2021;29(4):1625-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.