Abstract
The B-cell architecture of nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) is complex since it is composed of malignant lymphocyte-predominant cells along with a rich B-cell bystander environment. To gain insight into molecular determinants of disease transformation, we studied B-cell evolutionary trajectories in lymphoma tissue from diagnosis to relapse or transformation to non- Hodgkin lymphoma by next-generation sequencing of immunoglobulin heavy chains. Patients with NLPHL that later transformed were older and showed IgD negativity, absence of the characteristic IGHV3/IGHD3/IGHJ6 lymphocyte-predominant rearrangement and high repertoire clonality. We constructed phylogenetic trees within the compartment of the malignant clone to investigate clonal evolution. In all relapsing cases, the lymphocyte-predominant rearrangement was identical at diagnosis and relapse. NLPHL cases with transformation showed more complex trajectories with strong intraclonal diversification. The dominant founder clone in transformations showed clonal evolution if derived from the same cell of origin, or arose from a different cell of origin. Together, our data point to a significant role of antigenic drive in the transformation of NLHPL and identify high B-cell repertoire clonality with dominant intraclonal lymphocyte-predominant cell diversification as a hallmark of transformation. Sequencing of initial paraffin-embedded tissue may therefore be applied diagnostically to identify NLPHL cases with high risk of transformation.
Introduction
Nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) accounts for approximately 5-16% of all cases of Hodgkin lymphoma and has a male predilection. The monoclonal tumor cells, termed lymphocyte-predominant (LP)1-3 cells, differ from Hodgkin-Reed-Sternberg cells of classical Hodgkin lymphoma in terms of morphology and immunophenotype (sIg+, CD20+, BCL6+, CD79a+, CD45+, CD30–, CD15–, EBER–).4,5 LP cells have been described to express IgG B-cell receptors (BCR),6,7 while in a distinct subgroup of NLPHL, LP cells have been found to be IgDpositive by immunohistochemical staining (~25%). This subgroup has attracted interest recently because of emerging evidence of shared immunoglobulin characteristics and the role of Moraxella bacterial antigens in its pathogenesis. 6,8,9 Histologically, most NLPHL cases show LP cells located within large nodules composed of reactive B cells. A few cases present with variant growth patterns in which reactive T cells and/or histiocytes predominate over the reactive B cells.10 These cases are more often diagnosed in an advanced stage and relapse earlier.3,11 In general, relapses occur in around 15% of patients and can arise even after many years.12 Still, NLPHL - both at diagnosis and at relapse - has a rather indolent clinical behavior with a favorable prognosis and there are effective treatment options. However, an important complication of NLPHL is transformation into a more aggressive B-cell non-Hodgkin lymphoma (NHL) such as diffuse large B-cell lymphoma (DLBCL), which is observed in 10-30% of cases.13,14 Past studies have identified both cases with NHL clones related to the LP-cell clone and cases emerging from different cells of origin.15-17 Cases with disease transformation have a significantly higher mortality and faster progression, thus requiring a different treatment approach. Markers that are able to predict the risk of transformation early would, therefore, be beneficial.
Here we used next-generation immunoglobulin heavy chain sequencing (IGH NGS) to investigate B-cell repertoire metrics and evolutionary B-cell trajectories in NLPHL cases that later relapsed or transformed to high-grade NHL. As the most informative controls, we used nonmalignant lymphoproliferations that can precede or follow NLPHL or Hodgkin lymphoma.18-21 Our major goals were to characterize better the LP-cell clone and its bystander B cells in these NLPHL subsets, to address the question of clonal identity at diagnosis, relapse and transformation, and to find B lineage signatures potentially predictive of subsequent transformation with a technique that can be applied to paraffin-embedded tissue and that is robust enough to be used in routine diagnostics. Our analysis revealed pathogenic clues in NLPHL showing that the majority of NLPHL relapses and some transformations arise from the same cell of origin and that this cell of origin is subject to ongoing antigenic selection in many of these lymphomas. Moreover, the data presented here provide a set of molecular determinants for transformation that may be derived from targeted NGS on paraffin-embedded lymphoma tissue.
Methods
Patients’ characteristics
Eighty-six paraffin-embedded tissues from patients and controls were collected between 1987 and 2020 at the University Medical Centers of Frankfurt, Halle, Tübingen, Kiel, Würzburg, Stuttgart, Lübeck (Germany) and Tampere (Finland). Three of 81 tissues were previously used for LP-cell dissection and immunoglobulin heavy chain (IGH) and light chain (IGL) sequencing as published previously9 (sample denomination case 3 = NLPHL01-initial diagnosis, case 6 = NLPHL02-initial diagnosis, case 8 = NLPHL10-initial NLPHL). Our cohort included five cases of NLPHL that did not relapse or undergo transformation within 5 years after diagnosis (cohort 1), 16 NLPHL cases with paired samples at initial diagnosis and relapse(s) (cohort 2), ten cases with paired samples of initial NLPHL with transformation to DLBCL at relapse (cohort 3) and 28 control cases. These control cases comprised ten cases of nonspecific lymphadenitis, four cases of progressive transformation of germinal centers, ten cases of DLBCL (activated B-cell subtype) and four cases of T-cell/histiocyte- rich large B-cell lymphoma. The clinical characteristics of cohorts 1-3 are listed in Online Supplementary Table S1. NLPHL was diagnosed according to the World Health Organization (WHO) classification 2017.4 All variants were classified by one of the authors (SH) according to the histopathological patterns described by Fan et al.10 Pattern A was considered ‘typical’, all other patterns were termed ‘atypical’ in Figures 1 and 5. Example pictures of histopathological CD20 staining of the most relevant cases are shown in Online Supplementary Figure S1. The study was approved by the local ethics committee of the University of Frankfurt (n. 157/17) and Halle (n. 2017-81) and was performed in accordance with the Declaration of Helsinki of 1975.
IgD staining
IgD immunostaining was performed as previously described.22 In brief, paraffin slides were deparaffinized and heat-pretreated at pH 8. They were subsequently incubated with a polyclonal IgD antibody (Agilent, Santa Clara, CA, USA) for 30 min at room temperature. The Envision-FLEX Kit (Agilent) was used for detection.
Next-generation sequencing of IGH repertoires
IGH repertoires were obtained from bulk lymphoid tissue as described elsewhere.23-28 The rearranged IGH locus was amplified in a multiplex polymerase chain reaction from 250 ng of genomic DNA and BIOMED2-FR1 or -FR3 primer pools.29 NGS and demultiplexing were performed on an Illumina MiSeq sequencer (601-cycle single indexed, paired-end run, V3-chemistry). The MiXCR framework30 was used for data processing with the IMGT library31 as reference for sequence alignment. Only productive reads were used and all repertoires were normalized to 20,000 reads. All analyses and data plotting were performed using RStudio version 3.5.1. and the tcR,32 ade4,33 ggplot2,34 bubbles35 and tidyverse36 packages.
Using FR3 polymerase chain reactions, the CDR3 sequence as well as the IGHD- and IGHJ-gene segments could be reliably identified, while the alignment of the IGHV-gene segment was less robust because of the small size of the amplified segment. To account for this weakness, we decided to indicate only the IGHV1-7 families, but not the individual IGHV genes.
A detailed description of the NGS and data analysis is included in the Online Supplementary Information.
Results
Clinical and pathological characteristics of cases of nodular lymphocyte-predominant Hodgkin lymphoma that did or did not subsequently transform into non-Hodgkin lymphoma
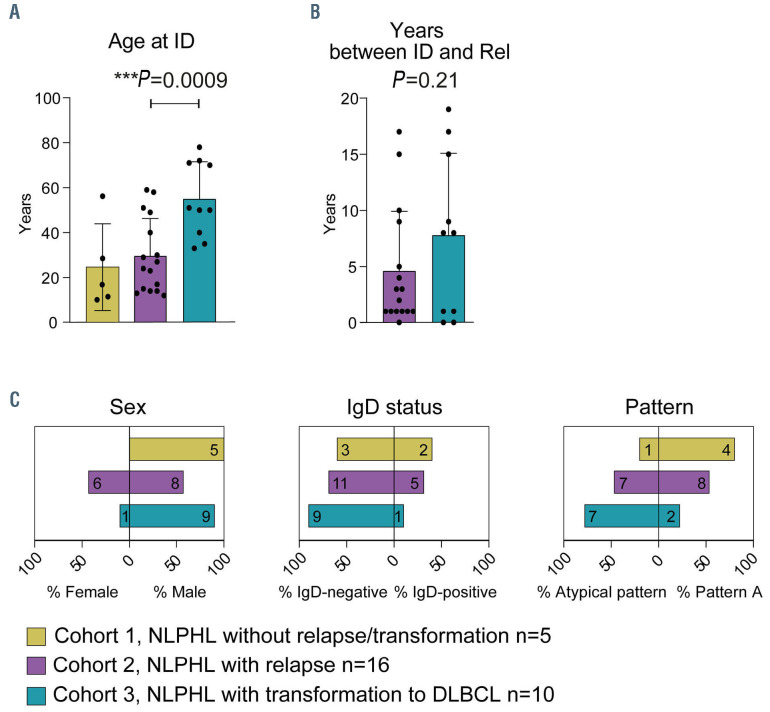
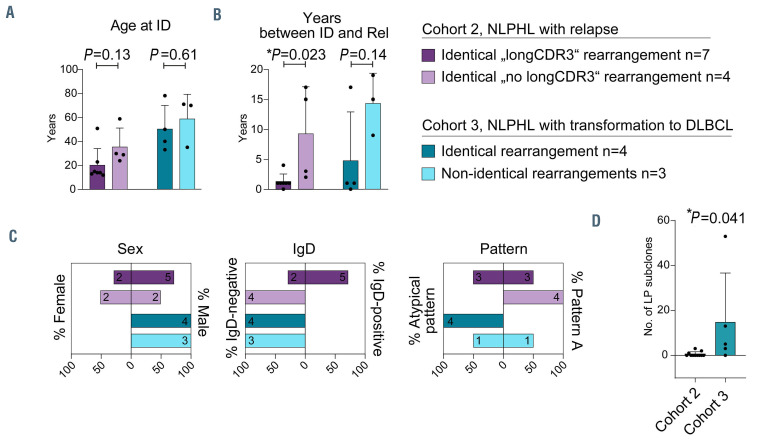
NLPHL has a high rate of transformation into subtypes of NHL such as DLBCL. We investigated three unselected subcohorts to identify determinants of transformation: five NLPHL cases (5 samples) without relapse or transformation with a mean/median follow-up time of 16/17 (6 to 19) years (cohort 1), 16 NLPHL cases (33 samples) with subsequent NLPHL relapses (cohort 2) and ten cases (20 samples) with transformation to NHL at relapse (cohort 3). The clinical characteristics of cohorts 1-3 are listed in Online Supplementary Table S1. Patients in cohort 3 were generally older at first diagnosis than cases in cohorts 1 and 2 (median age: 50.5 vs. 17 years and 25.5 years, respectively) (Figure 1A) suggesting that higher age may be a risk factor for transformation. Time to relapse/transformation was numerically, but not statistically different in cohorts 2 and 3 (Figure 1B). A clear male predominance was observed in all NLPHL cohorts, with relatively more females in cohort 2 (Figure 1C). There was a clear correlation of IgD status and transformation, in that cohort 3 cases were only rarely IgD-positive (Figure 1C). Moreover, this cohort included more cases with variant histological patterns (Figure 1C).
IGH repertoire characteristics of nodular lymphocyte-predominant Hodgkin lymphoma that did or did not subsequently transform into non-Hodgkin lymphoma
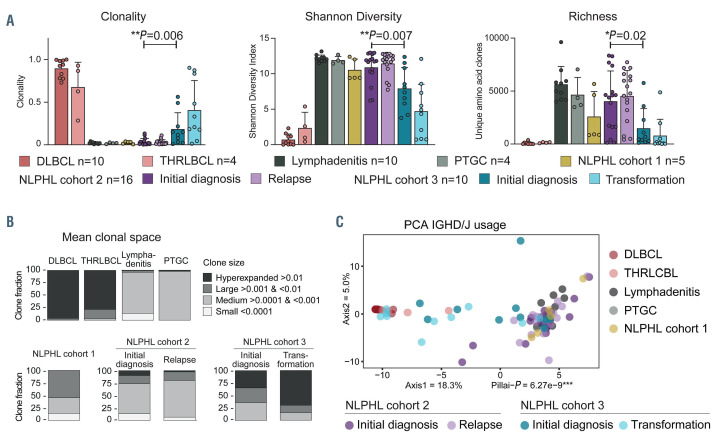
We used IGH NGS on DNA derived from paraffinembedded lymphoma tissue to investigate whether NLPHL cases with subsequent transformation harbored characteristic B lineage patterns predictive of transformation to NHL. In addition to our three cohorts, 28 control samples (DLBCL, T-cell/histiocyte-rich large B-cell lymphoma, lymphadenitis, progressive transformation of germinal centers) were available for this analysis. For repertoire metrics analyses, all IGH sequences including the malignant LP-cell rearrangement as well as those deriving from the bystander B-cell repertoire were used. We found that NLPHL cases that subsequently underwent transformation (cohort 3) showed NHL-typical quantitative repertoire metrics characterized by significantly lower B lineage diversity and richness as well as higher clonality compared to other NLPHL and non-malignant lymphoproliferations (Figure 2A, B). In addition, principal component analysis based on IGHD/J gene usage of IGH repertoires demonstrated a significant segregation of cohort 3 from NLPHL without transformation at both the NLPHL and DLBCL stages of the disease with the transformation cases clustering close to NHL cases already at the pre-transformation stage (Figure 2C). These data suggest that global IGH repertoire metrics (lower diversity, higher clonality) may be predictive of later transformation of NLPHL to NHL.
Characteristics of lymphocyte-predominant-cell IGH rearrangement in nodular lymphocyte-predominant Hodgkin lymphoma cases with subsequent relapse or transformation
We set out to characterize the properties of the malignant clone’s LP-cell IGH rearrangement to be able to compare it between NLPHL cases with relapse and transformation. For this analysis bulk lymphoma tissue from involved lymph nodes or other organs was used without microdissection of LP cells, so the sequenced IGH repertoires contained the LP-cell rearrangement along with the rearrangements of all other B lineage cells present in the B-cell rich bystander environment of NLPHL. Online Supplementary Figure S2 shows example plots reflecting the distribution of IGHV, IGHD and IGHJ genes in the lymphoma tissues as well as in non-malignant controls. As expected, the B lineage pattern of NLPHL resembled that of reactive lymphoproliferations, while NHL samples (DLBCL or T-cell/histiocyte-rich large B-cell lymphoma) showed much more pronounced clonal dominance in the B-cell lineage, compatible with the repertoire metrics reported above.
Figure 1.
Clinical and pathological characteristics of cohorts 1-3 with nodular lymphocyte-predominant Hodgkin lymphoma. (A) Age at initial diagnosis. (B) Years between initial diagnosis and relapse/transformation. (C) Sex, IgD status and histological variant pattern of patients with nodular lymphocyte-predominant Hodgkin lymphoma in cohort 1 (no relapse/transformation), cohort 2 (relapsing cases) and cohort 3 (transforming cases). Bars correspond to the mean + standard deviation. Statistical test: unpaired, two-tailed t-test. ID: initial diagnosis; Rel: relapse/transformation; NLPHL: nodular lymphocyte-predominant Hodgkin lymphoma; DLBCL: diffuse large B-cell lymphoma.
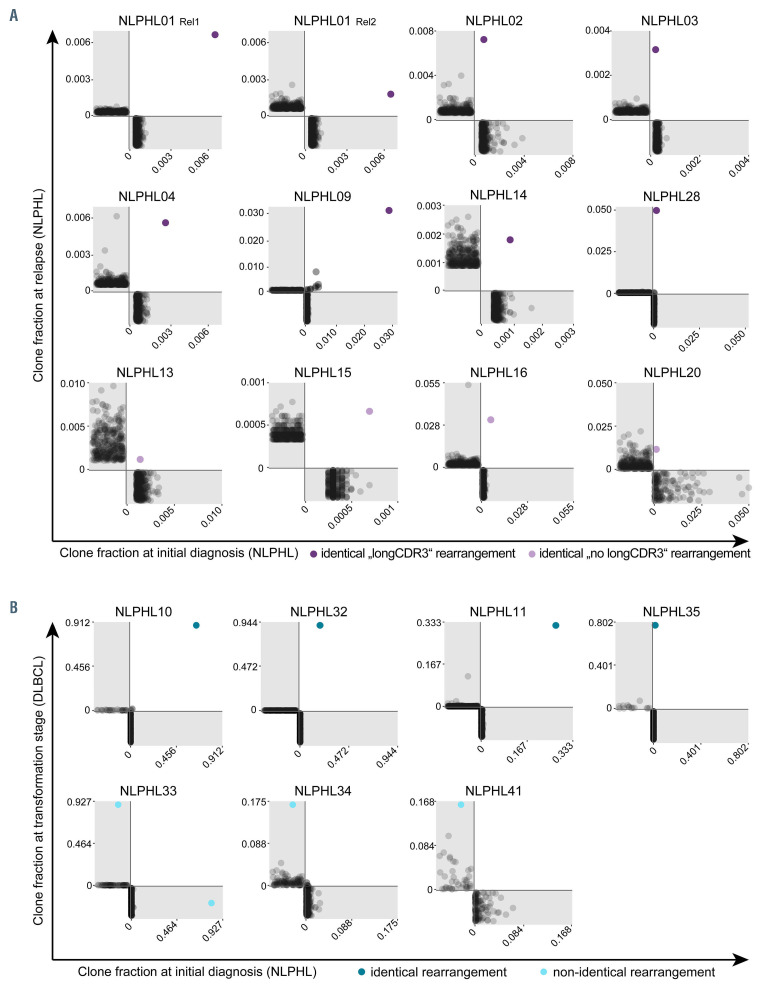
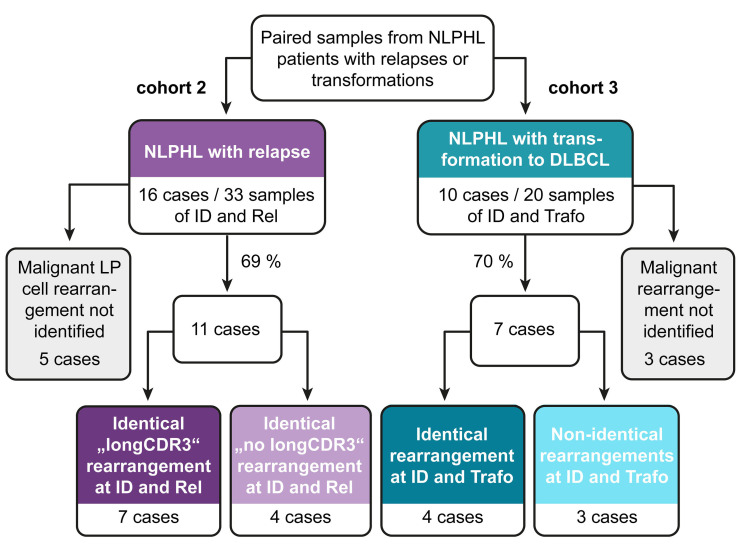
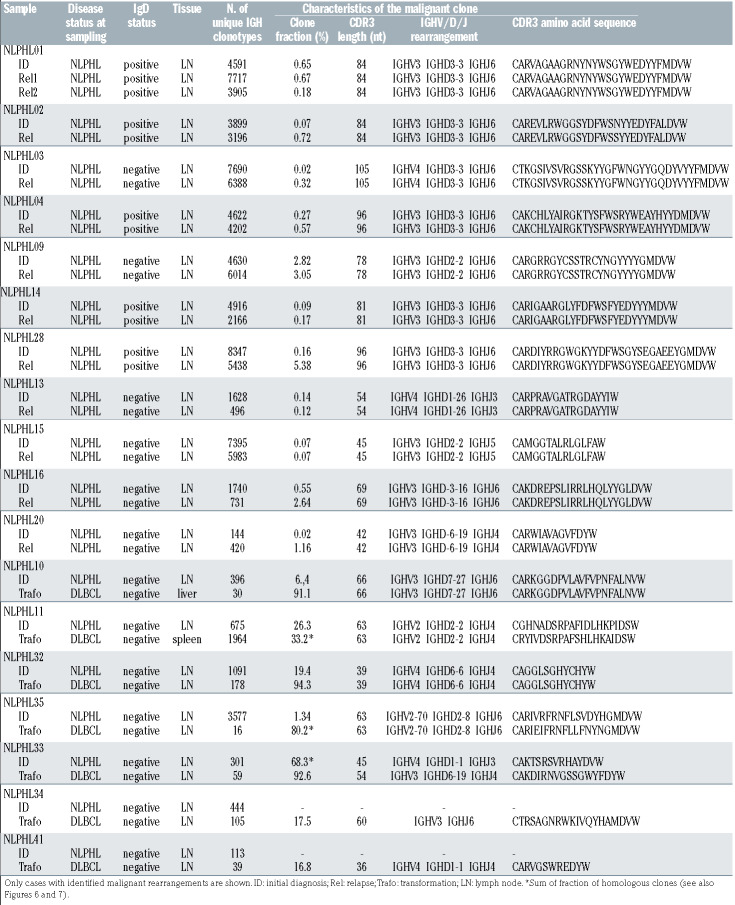
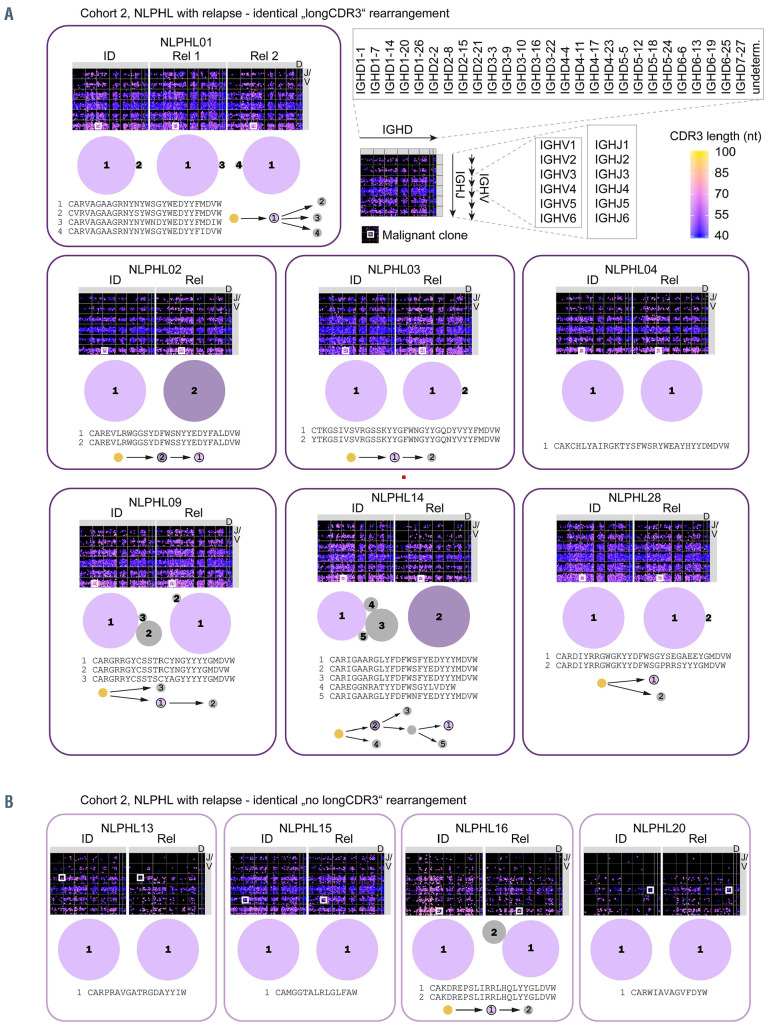
We used two possible criteria to define a malignant clone. We defined a dominant clone with >10% of the repertoire as a malignant clone. A dominant clone was present exclusively in the cases with transformation of cohort 3. In cohorts 1 and 2, the clone fraction of the most frequent clone was never >10% and usually <1% because of pronounced B-cell-rich bystander environment. Our strategy to identify the malignant clone in cases without clonal dominance (all of cohort 2) was to look at the overlap in paired samples taken at initial diagnosis and relapse. To determine the specificity of an identical rearrangement in lymph node material from the same patient, we analyzed non-malignant lymphadenitis of NLPHL01 and NLPHL02. There was no overlap between the repertoires of lymph nodes affected by lymphadenitis and NLPHL of the same patients (Online Supplementary Figure S3). In ten of 16 cases of cohort 2, there was a single identical rearrangement in the overlap of the paired samples (Figure 3). We defined these clones as malignant clones. One case (NLPHL09) had more than one overlapping clone; however, one of those was far more frequent and additional clones had similar sequences and could have been subclones of the malignant clone which had arisen from ongoing mutation.
Overall, we deduced the malignant clone’s rearrangement in paired samples from the same patient in about 70% of cases (Figures 3 and 4, Table 1). This is in line with the reported frequency of 66% detection of IGH rearrangements using FR3 primer sets.37 To confirm correct identification by our NGS strategy, we included two cases in our analysis that had been previously studied by Thurner et al.9 using LP-cell microdissection and IGH sequencing (denominated NLPHL01 and NLPHL02 in our cohort and cases 3 and 6 in the cohort described by Thurner et al.9). Indeed, our sequences were identical to the sequences derived from LP-cell microdissection. The characteristics of the malignant clones’ IGH rearrangements are reported in Table 1.
We found that the majority of evaluable cases (7 of 11) from cohort 2 showed a characteristic, previously reported, 9 LP-cell rearrangement containing IGHV3/IGHD3-3/IGHJ6 gene segments except for one case in which the rearrangement contained IGHD2-2 instead (Table 1). Clones with the characteristic IGHV/D/J LP rearrangement showed long CDR3 sequences with a median of 84 nucleotides (termed “longCDR3” rearrangements), while the median length of IGH CDR3 of an average BCR is 51 nucleotides. No “longCDR3” malignant rearrangements were found in patients from cohort 3 (Figure 3, Table 1).
Figure 2.
B lineage repertoire metrics of nodular lymphocyte-predominant Hodgkin lymphoma cohorts 1-3 and control cohorts. (A) Clonality, Shannon diversity and richness for different disease groups. High diversity is represented by high Shannon indices. Bars correspond to mean + standard deviation. Statistical test: unpaired, two-tailed t-test. (B) Mean clonal space distributions plotted according to disease entities. (C) Principal component analysis of IGHD/J usage. Two-dimensional separation is plotted according to similarities between repertoires with small distances between samples reflecting very similar repertoires and long distances between samples reflecting very different repertoire distributions. Statistical test: multivariate analysis of variance (MANOVA) Pillai. DLBCL: diffuse large B-cell lymphoma; THRLBCL: T-cell/histiocyte-rich large B-cell lymphoma; NLPHL: nodular lymphocyte-predominant Hodgkin lymphoma; PTGC: progressive transformation of germinal centers; PCA: principal component analysis.
Figure 3.
B lineage repertoire overlaps of paired tissue samples from the same patient. (A) Cohort 2, cases of nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) with relapse. (B) Cohort 3, cases of NLPHL with transformation. Each dot represents a single B-cell clone and is plotted according to its clone fraction at initial diagnosis (x-axis) and at relapse/transformation (y-axis). Dots below the x-axis (gray area) indicate clones present only at initial diagnosis, dots left of the y-axis (gray area) indicate clones present only at relapse/transformation. Shared clones are plotted in the white area. The malignant lymphocyte-predominant cell clone is highlighted in color. In NLPHL34 and NLPHL41 non-identical clones were assumed although the malignant rearrangement at initial diagnosis was not amplified by FR3 primer sets. NLPHL: nodular lymphocyte-predominant Hodgkin lymphoma; DLBCL: diffuse large B-cell lymphoma.
Figure 4.
Schematic overview of identified malignant rearrangements in cohorts 2 and 3 with nodular lymphocyte-predominant Hodgkin lymphoma. NLPHL: nodular lymphocyte-predominant Hodgkin lymphoma; DLBCL: diffuse large B-cell lymphoma; ID: initial diagnosis; Rel: relapse; Trafo: transformation; LP: lymphocyte predominant.
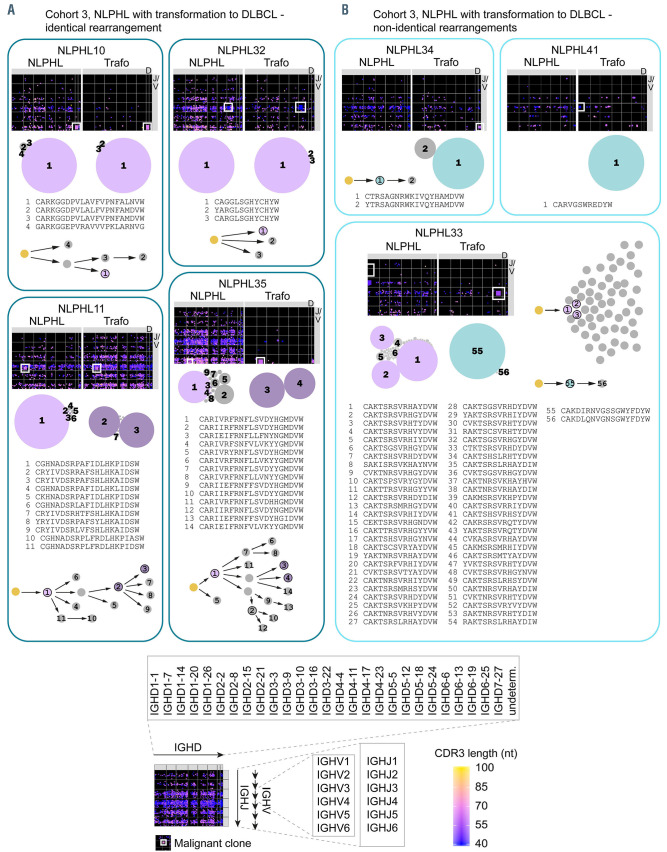
Figure 3 shows overlap plots between repertoires at initial diagnosis and at relapse/transformation, demonstrating the repertoire frequency of the respective malignant clone at the two time points. While all cases from cohort 2 shared the same IGH rearrangement as the malignant LP-cell clone at initial diagnosis and relapse, suggesting the same cell of origin, only four of seven evaluable cases from cohort 3 shared the same IGH rearrangement at initial diagnosis and transformation, suggesting different cells of origin in a substantial percentage of transforming NLPHL cases (Figure 3). Of note, in two of the three cases designated as “non-identical rearrangements”, only one of the malignant IGH rearrangements was detectable with FR3 primers. The absence of this sequence in the respective paired repertoire was interpreted as indirect evidence of different rearrangements even though one of the rearrangements escaped detection by FR3 primers. Compatible with the globally increased B-cell repertoire clonality in NLPHL cases from cohort 3, we found that the frequency of the malignant IGH rearrangement was much higher in NLPHL samples from cohort 3 than in those from cohort 2 (Figure 3). In four out of seven cases from cohort 3, repertoire frequencies of malignant rearrangements were more comparable to typical repertoire frequencies of a malignant NHL clone (>10% of the B-cell repertoire) than to repertoire frequencies of LP-cell rearrangements in non-transforming NLPHL cases (around 1% of the B-cell repertoire). This pointed to further B-cell repertoire similarities between NHL and pre-transformation NLPHL.
Next, we wanted to explore whether relapsing cases with “longCDR3” rearrangements and those carrying average-sized CDR3 as well as transforming cases with identical or non-identical rearrangements at initial diagnosis and transformation had unifying clinical characteristics. While age at diagnosis did not appear to discriminate between these respective subgroups, patients with “longCDR3” rearrangements seemed to relapse earlier than those with average-sized “no longCDR3” (Figure 5A, B). Moreover, identical rearrangements were more likely to be found in patients with earlier transformation (Figure 5B). In addition, IgD positivity was strongly associated with “longCDR3” rearrangements, as previously shown,9 but almost one-third of cases with “longCDR3” rearrangements were IgD-negative demonstrating that these two features were not fully overlapping (Figure 5C). Histomorphological patterns were also distributed differently across the subcohorts, with “longCDR3” cases and cases with transformation from an identical IGH rearrangement showing more atypical patterns (Figure 5C).
Since ongoing somatic hypermutation has been described to occur in NLPHL B-cell receptors38 and antigenic drive by Moraxella antigens has recently been reported in a subset of NLPHL cases,9 we investigated whether NLPHL cases with subsequent transformation showed more intraclonal diversification as indirect evidence for more selective pressure by antigens. Indeed, some level of intraclonal diversification was found in the majority of NLPHL cases with quantitatively more LP subclones in transforming cases of cohort 3 (Figure 5D).
Intraclonal diversification and evolutionary patterns in lymphocyte-predominatn cells from nodular lymphocyte-predominant Hodgkin lymphoma that did or did not subsequently transform
We set out to study intraclonal diversification within the LP-cell clone/NHL malignant clone (LP/NHL clone) clone in more depth in cohorts 2 and 3 in order to identify potentially discriminative evolutionary patterns. More specifically, we intended to show whether relapse/transformation originated from the exact same cell, from a common precursor or from a cell that had undergone clonal evolution since initial diagnosis in our two cohorts.
Table 1.
Characteristics of malignant B-cell clones in paired samples of nodular lymphocyte-predominant Hodgkin lymphoma cohorts 2 and 3.
Figure 5.
Characteristics of the patients with nodular lymphocyte-predominant Hodgkin lymphoma with “longCDR3” versus average CDR3 and identical versus non-identical malignant clones at initial diagnosis and relapse/transformation. (A) Age at initial diagnosis. (B) Years between initial diagnosis and relapse/transformation. (C) Sex, IgD status and histological variant pattern. (D) Number of lymphocyte-predominant/non-Hodgkin lymphoma-related subclones at initial diagnosis in cohorts 2 and 3. Bars correspond to mean + standard deviation. Statistical test: unpaired, two-tailed t-test. ID: initial diagnosis; Rel: relapse/transformation; NLPHL: nodular lymphocyte-predominant Hodgkin lymphoma; DLBCL: diffuse large B-cell lymphoma; LP: lymphocyte-predominant.
To this end, phylogenetic trees were constructed based on somatic hypermutation exclusively for the subclones constituting the malignant LP/NHL clone (Figures 6 and 7). In some patients, the most dominant malignant subclone was completely identical at diagnosis and relapse/transformation (same IGH rearrangement, no difference in somatic hypermutation) – eventually accompanied by a changing environment of clonally related, but less dominant subclones – suggesting that the same cell of origin gave rise to both the initial NLPHL and to the relapse/transformation. This pattern occurred in 11 of 18 cases, but was more frequently observed in relapsing cases (9/11 cases: NLPHL01, NLPHL03, NLPHL04, NLPHL09, NLPHL28, NLPHL13, NLPHL15, NLPHL16, NLPHL20) than in transforming cases (2/7 cases: NLPHL10, NLPHL32). In four of 18 cases, the dominant subclone(s) shared the same IGH rearrangement at diagnosis and relapse/transformation, but differed in somatic hypermutation following a sequential evolutionary trajectory (Figures 6 and 7). In these cases the relapse/transformation clone likely developed from the same cell of origin which, however, had undergone antigenic drive between initial diagnosis and relapse/transformation. This pattern was numerically more frequent in transforming cases (2/7 cases: NLPHL11, NLPHL35) than in relapsing cases (2/11 cases: NLPHL02, NLPHL14). In three out of ten cases, transformations likely arose from a different cell of origin (different IGH rearrangement) (Figure 7). Collectively, these data suggested that transformations more likely originated from cells that were not fully identical or even clonally unrelated to the founder NLPHL clone. Intriguingly, we observed that especially cases that transformed with non-identical or fully unrelated clones showed a highly complex sequential and branching evolution in their initial LP clone suggesting high selective pressure by antigens which potentially extends to other rearrangements in the course of this immune reaction (Figure 7, NLPHL11, NLPHL33, NLPHL35).
Discussion
NLPHL is a rare subtype of Hodgkin lymphoma with a characteristic clinical presentation and disease course, often presenting with localized disease, favorable responses to treatment but a rather high rate of transformation to aggressive NHL. Its pathogenesis and especially the characteristics of malignant LP cells as well as its Bcell environment are of increasing interest to the hematologic community. One of the major limitations to analysis of this entity is the paucity of LP cells in a background of normal B lineage cells that impair high-throughput genetic or immunogenetic studies on these clones. For this reason, analyses of the malignant LP cells’ BCR have been confined to microdissected cases.38-41 An interesting series of NLPHL recently caught attention since it pointed strongly to directional forces through antigenic selection in the pathogenesis of this disease. This interaction study on recombinantly expressed NLPHL BCR derived from microdissected LP cells suggested that bacterial antigens from Moraxella species bind to the lymphoma BCR in a substantial fraction of IgD-positive NLPHL patients.9 This is striking since it suggests that an infectious trigger may drive lymphomagenesis (and potentially also lymphoma evolution) in a subset of patients with NLPHL. The Moraxella-associated cases from this study included only IgD-positive cases with BCR that share common features, such as long CDR3 sequences and specific IGH rearrangements.
Figure 6.
Clonal architecture of B lineage repertoires and lymphocyte-predominant subclones in nodular lymphocyte-predominant Hodgkin lymphoma with relapse (cohort 2). (A) Cases with identical „longCDR3“ lymphocyte-predominant (LP) rearrangement. (B) Cases with identical LP rearrangement and average-sized CDR3. IGHV/D/J maps of the global IGH repertoire are shown for each tissue sample (at initial diagnosis and relapse). Rearrangements of IGHV1-7 main groups and IGHJ1-6 are plotted on the vertical axis. IGHD subgroups are plotted on the horizontal axis. Each dot represents one immunoglobulin rearrangement colored according to its CDR3 length and sized according to clonal fraction. A white square indicates the position of the malignant clone. A schematic overview of intraclonal diversification and evolutionary patterns of LP subclones derived from the same cell of origin are shown below. The sizes of the bubbles reflect relative clone fractions of LP subclones within one repertoire. Arrows indicate somatic hypermutation. Colored clones are characterized further in Table 1. NLPHL: nodular lymphocyte-predominant Hodgkin lymphoma; ID: initial diagnosis; Rel: relapse/transformation; nt: nucleotide.
Figure 7.
Clonal architecture of B lineage repertoires and lymphocyte-predominant-cell clone/ non-Hodgkin lymphoma malignant clone subclones in nodular lymphocyte- predominant Hodgkin lymphoma with transformation to non-Hodgkin lymphoma (cohort 3). (A) Cases with identical rearrangement. (B) Cases with different rearrangements. IGHV/D/J map of the global IGH repertoire are shown. Rearrangement of IGHV1-7 main groups and IGHJ1-6 are plotted on the vertical axis. IGHD subgroups are plotted on the horizontal axis. Each dot represents one immunoglobulin rearrangement colored according to its CDR3 length and sized according to clonal fraction. A white square indicates the position of the malignant clone. A schematic overview of intraclonal diversification and evolutionary patterns of lymphocyte- predominant-cell clone/ non-Hodgkin lymphoma malignant clone (LP/NHL clone) subclones derived from the same cell of origin is shown below. The size of the bubbles reflects relative clone fractions of LP/NHL subclones within one repertoire. Arrows indicate somatic hypermutation. Colored clones are further characterized in Table 1. NLPHL: nodular lymphocyte-predominant Hodgkin lymphoma; Trafo: transformation.
In our study, we confirmed the unique IGHV/D/J rearrangement with a characteristically “longCDR3” configuration in approximately one-third of NLPHL patients, although acknowledging potential selection biases of our cohorts, such as patients with a history of relapse or transformation. The major determinant for antigen recognition is the CDR3 portion of the variable region of the BCR. The very unique characteristics of this region, which are shared between many NLPHL patients, together with the finding of ongoing mutational events in the majority of the studied patients, in our view clearly suggests that antigenic BCR interactions of the LP-cell clone must be pathophysiologically relevant, as reported for other lymphomas.42-46 Our present data do, however, clearly show that almost one-third of IgD-negative NLPHL also share the same characteristic IGHV/D/J rearrangement and might therefore have a similar pathogenesis. Future treatment strategies in early disease stages of cases with this BCR configuration might therefore consist in primary antimicrobial therapy to induce remission (as, for example, in Helicobacter-associated lymphoma) followed by radiation or chemotherapy only if remissions cannot be achieved. This may be especially feasible in NLPHL, since the disease has a rather indolent clinical course providing a “window of opportunity” for strategies to be attempted without compromising the curative potential of more aggressive treatment options.47
We noted a higher occurrence of atypical variant patterns in cases with relapse and transformation, which is in line with previous studies.3,10 However, given the long time until relapse/transformation in a substantial fraction of cases, we cannot exclude the possibility of future recurrences in cohort 1, despite the mean follow-up of 16 years.
From a diagnostic perspective, it was interesting for us to see that the B lineage patterns of NLPHL that can be deduced by IGH NGS can help to discriminate NLPHL cases from other related disorders that – by morphological evaluation – may occasionally pose challenges in differential diagnosis. In our analysis, we were able to distinguish clonal T-cell/histiocyte-rich large B-cell lymphoma repertoires, which resemble those of DLBCL, from the more diverse NLPHL repertoires. Furthermore, DLBCL cases that transformed from NLPHL showed more diverse B-cell repertoires than conventional DLBCL, which might argue that this entity should be kept apart from the general category of DLCBL and would be in line with the frequently observed favorable outcome of these cases after salvage therapy.14,48 As NGS becomes increasingly available, also for routine clinical applications, this technique could be used to support accurate discrimination between lymphoma subgroups in cases that are morphologically challenging. This is especially relevant since these lymphomas often require different treatment modalities.
In addition, we identified a B lineage pattern that was associated with subsequent NLPHL transformation. This pattern was essentially characterized by higher B lineage clonality and lower diversity already at the pre-transformation NLPHL stage of the disease as well as absence of the paradigmatic “longCDR3” IGH rearrangement. When studying the LP-cell clone and its intraclonal variants separately from the bulk B-cell repertoire (which also includes non-malignant bystander cells), we found the product of some sort of intraclonal diversification in the majority of all studied NLPHL cases both at initial diagnosis and at relapse/transformation. However, transforming cases showed a greater magnitude of intraclonal diversification within the LP cell clone as another hallmark of this risk group. Collectively, these data indicate that higher age, IgD-negativity, high B-cell clonality with increased intraclonal diversification and the absence of a “longCDR3” LP rearrangement characterize NLPHL cases with a high risk of transformation to NHL. Clinically, this information could trigger more intense follow-up of patients at increased risk.
Beyond the diagnostic perspective, evolutionary pattern analysis suggested that, compared to transformed cases, relapsing cases more likely originated from a fully identical cell of origin. In transforming cases, intraclonal diversification was much more complex with the founder clones of initial diagnosis and transformation more likely being unrelated or separated by a sequence of mutational events. This, in turn, could be interpreted as higher antigenic selection pressure (potentially via non-Moraxella antigens) acting on the LP-cell clone in the cases that ultimately transformed to NHL.
Of note, our strategy to derive the malignant clone from bulk sequencing of paired samples has the limitation that, unlike microdissection, it is an indirect method that is open to the possibility of misinterpretation. While we can confidently derive the malignant rearrangement from overlapping identical clones with high frequencies, some cases are less absolute. In particular, conclusions on cases NLPHL34 and NLPHL41, for which we did not identify the LP-cell clone but suggested that the transformation arose from a different cell of origin, must be treated with care.
Taken together, our data are strongly indicative of a pathophysiological role of antigens in driving lymphomagenesis and transformation in NLPHL. They provide biological insight into the cell of origin underlying relapse and transformation. Moreover, our data support the diagnostic value of NGS in this type of lymphoma to substantiate the diagnosis in unclear cases and to predict the risk of transformation.
Supplementary Material
Acknowledgments
The authors thank Elena Hartung, Smaro Soworka and Marta Siedlecki for excellent technical assistance.
Funding Statement
Funding: Financial support from the Deutsche Forschungsgemeinschaft (DFG BI 1711/4-1 to MB and DFG HA 6145/3-1 to SH) is acknowledged.
References
- 1.Saarinen S, Pukkala E, Vahteristo P, Mäkinen MJ, Franssila K, Aaltonen LA. High familial risk in nodular lymphocytepredominant Hodgkin lymphoma. J Clin Oncol. 2013;31(7):938-943. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos I, Hansmann ML, Franssila K, et al. European Task Force on Lymphoma project on lymphocyte predominance Hodgkin disease: histologic and immunohistologic analysis of submitted cases reveals 2 types of Hodgkin disease with a nodular growth pattern and abundant lymphocytes. Blood. 2000;96(5):1889-1899. [PubMed] [Google Scholar]
- 3.Hartmann S, Eichenauer DA, Plütschow A, et al. The prognostic impact of variant histology in nodular lymphocyte-predominant Hodgkin lymphoma: a report from the German Hodgkin Study Group (GHSG). Blood. 2013;122(26):4246-4252. [DOI] [PubMed] [Google Scholar]
- 4.WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. (Revised 4th edition). Lyon: International Agency for Research on Cancer; 2017. [Google Scholar]
- 5.Piccaluga PP, Agostinelli C, Gazzola A, et al. Pathobiology of Hodgkin lymphoma. Adv Hematol. 2011;2011:920898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash S, Fountaine T, Raffeld M, Jaffe ES, Pittaluga S.IgD positive L&H cells identify a unique subset of nodular lymphocyte predominant Hodgkin lymphoma. Am J Surg Pathol. 2006;30(5):585-592. [DOI] [PubMed] [Google Scholar]
- 7.Schmid C, Sargent C, Isaacson PG. L and H cells of nodular lymphocyte predominant Hodgkin's disease show immunoglobulin light-chain restriction. Am J Pathol. 1991;139(6):1281-1289. [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann S, Eichenauer DA, Plütschow A, et al. Histopathological features and their prognostic impact in nodular lymphocytepredominant Hodgkin lymphoma--a matched pair analysis from the German Hodgkin Study Group (GHSG). Br J Haematol. 2014;167(2):238-242. [DOI] [PubMed] [Google Scholar]
- 9.Thurner L, Hartmann S, Fadle N, et al. Lymphocyte predominant cells detect Moraxella catarrhalis-derived antigens in nodular lymphocyte-predominant Hodgkin lymphoma. Nat Commun. 2020;11(1):2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Z, Natkunam Y, Bair E, Tibshirani R, Warnke RA. Characterization of variant patterns of nodular lymphocyte predominant Hodgkin lymphoma with immunohistologic and clinical correlation. Am J Surg Pathol. 2003;27(10):1346-1356. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann S, Plütschow A, Mottok A, et al. The time to relapse correlates with the histopathological growth pattern in nodular lymphocyte predominant Hodgkin lymphoma. Am J Hematol. 2019;94(11):1208-1213. [DOI] [PubMed] [Google Scholar]
- 12.Eichenauer DA, Plütschow A, Schröder L, et al. Relapsed and refractory nodular lymphocyte- predominant Hodgkin lymphoma: an analysis from the German Hodgkin Study Group. Blood. 2018;132(14):1519-1525. [DOI] [PubMed] [Google Scholar]
- 13.Al-Mansour M, Connors JM, Gascoyne RD, Skinnider B, Savage KJ. Transformation to aggressive lymphoma in nodular lymphocyte-predominant Hodgkin's lymphoma. J Clin Oncol. 2010;28(5):793-799. [DOI] [PubMed] [Google Scholar]
- 14.Biasoli I, Stamatoullas A, Meignin V, et al. Nodular, lymphocyte-predominant Hodgkin lymphoma: a long-term study and analysis of transformation to diffuse large B-cell lymphoma in a cohort of 164 patients from the Adult Lymphoma Study Group. Cancer. 2010;116(3):631-639. [DOI] [PubMed] [Google Scholar]
- 15.Wickert RS, Weisenburger DD, Tierens A, Greiner TC, Chan WC. Clonal relationship between lymphocytic predominance Hodgkin's disease and concurrent or subsequent large-cell lymphoma of B lineage. Blood. 1995;86(6):2312-2320. [PubMed] [Google Scholar]
- 16.Ohno T, Huang JZ, Wu G, Park KH, Weisenburger DD, Chan WC. The tumor cells in nodular lymphocyte-predominant Hodgkin disease are clonally related to the large cell lymphoma occurring in the same individual. Direct demonstration by single cell analysis. Am J Clin Pathol. 2001;116(4):506-511. [DOI] [PubMed] [Google Scholar]
- 17.Pan LX, Diss TC, Peng HZ, Norton AJ, Isaacson PG. Nodular lymphocyte predominance Hodgkin's disease: a monoclonal or polyclonal B-cell disorder? Blood. 1996;87(6):2428-2434. [PubMed] [Google Scholar]
- 18.Bräuninger A, Yang W, Wacker HH, Rajewsky K, Küppers R, Hansmann ML. Bcell development in progressively transformed germinal centers: similarities and differences compared with classical germinal centers and lymphocyte-predominant Hodgkin disease. Blood. 2001;97(3):714-719. [DOI] [PubMed] [Google Scholar]
- 19.Hansmann ML, Fellbaum C, Hui PK, Moubayed P.Progressive transformation of germinal centers with and without association to Hodgkin's disease. Am J Clin Pathol. 1990;93(2):219-226. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann S, Winkelmann R, Metcalf RA, et al. Immunoarchitectural patterns of progressive transformation of germinal centers with and without nodular lymphocyte-predominant Hodgkin lymphoma. Hum Pathol. 2015;46(11):1655-1661. [DOI] [PubMed] [Google Scholar]
- 21.Kiil K, Bein J, Schuhmacher B, et al. A high number of IgG4-positive plasma cells rules out nodular lymphocyte predominant Hodgkin lymphoma. Virchows Arch. 2018;473(6):759-764. [DOI] [PubMed] [Google Scholar]
- 22.Hartmann S, Eichenauer DA. Nodular lymphocyte predominant Hodgkin lymphoma: pathology, clinical course and relation to Tcell/ histiocyte rich large B-cell lymphoma. Pathology. 2020;52(1):142-153. [DOI] [PubMed] [Google Scholar]
- 23.Schliffke S, Akyüz N, Ford CT, et al. Clinical response to ibrutinib is accompanied by normalization of the T-cell environment in CLL-related autoimmune cytopenia. Leukemia. 2016;30(11):2232-2234. [DOI] [PubMed] [Google Scholar]
- 24.Schliffke S, Sivina M, Kim E, et al. Dynamic changes of the normal B lymphocyte repertoire in CLL in response to ibrutinib or FCR chemo-immunotherapy. Oncoimmunology. 2018;7(4):e1417720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akyüz N, Brandt A, Stein A, et al. T-cell diversification reflects antigen selection in the blood of patients on immune checkpoint inhibition and may be exploited as liquid biopsy biomarker. Int J Cancer. 2017;140(11):2535-2544. [DOI] [PubMed] [Google Scholar]
- 26.Oberle A, Brandt A, Voigtlaender M, et al. Monitoring multiple myeloma by nextgeneration sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica. 2017;102(6):1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mährle T, Akyüz N, Fuchs P, et al. Deep sequencing of bone marrow microenvironments of patients with del(5q) myelodysplastic syndrome reveals imprints of antigenic selection as well as generation of novel T-cell clusters as a response pattern to lenalidomide. Haematologica. 2019;104(7): 1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultheiss C, Paschold L, Simnica D, et al. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53(2): 442-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brüggemann M, Kotrová M, Knecht H, et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia. 2019;33(9):2241-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolotin DA, Poslavsky S, Mitrophanov I, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12(5):380-381. [DOI] [PubMed] [Google Scholar]
- 31.Giudicelli V, Chaume D, Lefranc M-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33(Database issue):D256-D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nazarov VI, Pogorelyy MV, Komech EA, et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics. 2015;16(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dray S, Dufour A-B. The ade4 package: implementing the duality diagram for ecologists. J Stat Software. 2007;22(4):20. [Google Scholar]
- 34.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York; 2016. [Google Scholar]
- 35.Cheng J, Bostock M, Heer J.bubbles: d3 bubble chart htmlwidget. R package version 0.2. [Google Scholar]
- 36.Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. [Google Scholar]
- 37.Evans PAS, Pott C, Groenen PJTA, et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007;21(2): 207-214. [DOI] [PubMed] [Google Scholar]
- 38.Bräuninger A, Küppers R, Strickler JG, Wacker HH, Rajewsky K, Hansmann ML. Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center- derived tumor B cells. Proc Natl Acad Sci U S A. 1997;94(17):9337-9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liso A, Capello D, Marafioti T, et al. Aberrant somatic hypermutation in tumor cells of nodular-lymphocyte-predominant and classic Hodgkin lymphoma. Blood. 2006;108(3):1013-1020. [DOI] [PubMed] [Google Scholar]
- 40.Marafioti T, Hummel M, Anagnostopoulos I, et al. Origin of nodular lymphocyte-predominant Hodgkin's disease from a clonal expansion of highly mutated germinal-center B cells. N Engl J Med. 1997;337(7):453-458. [DOI] [PubMed] [Google Scholar]
- 41.Bräuninger A, Küppers R, Spieker T, et al. Molecular analysis of single B cells from Tcell- rich B-cell lymphoma shows the derivation of the tumor cells from mutating germinal center B cells and exemplifies means by which immunoglobulin genes are modified in germinal center B cells. Blood. 1999;93(8):2679-2687. [PubMed] [Google Scholar]
- 42.Binder M, Léchenne B, Ummanni R, et al. Stereotypical chronic lymphocytic leukemia B-cell receptors recognize survival promoting antigens on stromal cells. PLoS One. 2010;5(12):e15992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binder M, Müller F, Jackst A, et al. B-cell receptor epitope recognition correlates with the clinical course of chronic lymphocytic leukemia. Cancer. 2011;117(9):1891-1900. [DOI] [PubMed] [Google Scholar]
- 44.Binder M, Müller F, Frick M, et al. CLL Bcell receptors can recognize themselves: alternative epitopes and structural clues for autostimulatory mechanisms in CLL. Blood. 2013;121(1):239-241. [DOI] [PubMed] [Google Scholar]
- 45.Thurner L, Hartmann S, Fadle N, et al. LRPAP1 is a frequent proliferation-inducing antigen of BCRs of mantle cell lymphomas and can be used for specific therapeutic targeting. Leukemia. 2019;33(1):148-158. [DOI] [PubMed] [Google Scholar]
- 46.Thurner L, Preuss KD, Bewarder M, et al. Hyper-N-glycosylated SAMD14 and neurabin- I as driver autoantigens of primary central nervous system lymphoma. Blood. 2018;132(26):2744-2753. [DOI] [PubMed] [Google Scholar]
- 47.Borchmann S, Joffe E, Moskowitz CH, et al. Active surveillance for nodular lymphocyte- predominant Hodgkin lymphoma. Blood. 2019;133(20):2121-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kenderian SS, Habermann TM, Macon WR, et al. Large B-cell transformation in nodular lymphocyte-predominant Hodgkin lymphoma: 40-year experience from a single institution. Blood. 2016;127(16):1960-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.