Abstract
The hydrolytic processes occurring at the surface of silicon nitride (Si3N4) bioceramic have been indicated as a powerful pathway to instantaneous inactivation of SARS-CoV-2 virus. However, the virus inactivation mechanisms promoted by Si3N4 remain yet to be elucidated. In this study, we provide evidence of the instantaneous damage incurred on the SARS-CoV-2 virus upon contact with Si3N4. We also emphasize the safety characteristics of Si3N4 for mammalian cells. Contact between the virions and micrometric Si3N4 particles immediately targeted a variety of viral molecules by inducing post-translational oxidative modifications of S-containing amino acids, nitration of the tyrosine residue in the spike receptor binding domain, and oxidation of RNA purines to form formamidopyrimidine. This structural damage in turn led to a reshuffling of the protein secondary structure. These clear fingerprints of viral structure modifications were linked to inhibition of viral functionality and infectivity. This study validates the notion that Si3N4 bioceramic is a safe and effective antiviral compound; and a primary antiviral candidate to replace the toxic and allergenic compounds presently used in contact with the human body and in long-term environmental sanitation.
Keywords: Silicon nitride, Virus, Surface chemistry, Ammonia, SARS-CoV-2, Hydrolysis
Graphical abstract
1. Introduction
In a previous study on the effect of silicon nitride bioceramic powder on SARS-CoV-2, we found that simple contact of the powder with the virus in a dilute aqueous suspension resulted in instantaneous and complete inactivation of the virus [1]. The degree of instantaneous inactivation recorded was similarly observed for other single-stranded RNA (ssRNA) viruses [2]. Subsequently, it was hypothesized that the agent responsible for this effect is ammonia. Conversion into virucidal ammonia from ammonium increasingly occurs with increasing the pH of the aqueous environment [3]. Since the pH at the biological interface between solid Si3N4 and the virion surface likely exceed the pK value [4], a steep concentration gradient is established at the solid/virion interface with a far greater percentage of indigenous ammonia in the virucidal state at the local contact. We used this argument to explain why ssRNA virus inactivation is greatly accelerated upon contact with Si3N4 particles in suspension.
Studies on the inactivation rates of enterovirus [5] and other enteric viruses [6] with ammonia have been reported, and locate the viral sensitivity to this agent as a general property of viruses belonging to the enterovirus group with few exceptions (e.g., the reovirus, an enteric virus insensitive to ammonia) [[5], [6], [7]]. It should be immediately apparent from the structural features that the interaction of viruses exposed to Si3N4 with hydrolytically eluted ammonia results in structural and chemical damage.
Si3N4 is a biocompatible ceramic with the unique property of concurrently supporting cell-cycle progression in eukaryotic cells and counteracting pathogens [4,[8], [9], [10], [11], [12], [13], [14]]. Such dual (and uncommon) behavior is the result of its peculiar surface chemistry, which features a cell-proliferation supportive cation (silicon) and the formation of ammonia/ammonium (and nitrogen radicals) as byproducts of the hydrolytic interaction between surface secondary amines and aqueous environment at the biological interface [4]. Such reactions in turn induce a robust pH buffering effect. Accordingly, optimal biological properties in Si3N4 strongly depend on its surface stoichiometry, which could be modulated to enhance either cell proliferation or pathogen lysis. The different capacity of eukaryotic cells and pathogens of metabolizing un-ionized species of ammonia and, thus, of resisting under NH3 stress is key in the observed “antibiotic-like” behavior of Si3N4. In the specific context of the present study, we build upon the widely accepted notion that ssRNA virions have in general no capacity to resist inactivation by ammonia attack to their RNA and viral proteins [2,15].
With the scenarios by which SARS-CoV-2 could actually have arisen and its immunoevasive mechanisms yet under discussion [16], we looked for a safe antiviral material with wide effectiveness and, concurrently, for an analytical tool capable of screening the virions’ structure at the molecular scale. Accordingly, the main emphasis of this study was placed on elucidating the mechanisms of virus inactivation by means of Raman vibrational analyses of SARS-CoV-2 virions before and after exposure to Si3N4 micrometric particles (5 vol%) suspended in aqueous solution. We then compared the results with the susceptibility of other viruses to the same Si3N4 powder. The present spectroscopic characterizations, which are in line with our previous in situ Raman studies in microbiology [17], focused on the antiviral mechanisms, and clarified the pathway by which eluted ammonia and related reactive nitrogen species inactivate the virus, in comparison with other ssRNA viruses. Understanding the virucidal effects of Si3N4 at the molecular scale is key in controlling and optimizing its surface stoichiometry for the development of advanced environmental sanitation approaches capable to counteract the COVID-19 pandemic without affecting human health even in the long term.
2. Experimental procedures
SARS-CoV-2 viral stock of the original Japanese isolate (JPN/TY/WK-521) was obtained from the Japanese National Institute of Infectious Diseases. The viral stock was propagated using VeroE6/TMPRSS2 cells at 37 °C for 2 days and harvested by centrifugation at 4800 rpm for 20 min. Samples for Raman spectroscopy were supernatant virions with a concentration of 3.0–4.5 × 109 TCID50 laid on a glass sheet with an area of ∼38.5 mm2. Five volume percent (5 vol%) of Si3N4 powder was dispersed in 1 mL of Phosphate Buffer Solution (PBS(−)), followed by the addition of the viral suspension (2 × 105 median tissue culture infectious dose (TCID50) in 20 μL). The Si3N4 powder (SINTX Technologies, Inc., Salt Lake City, UT USA) had an average particle size of 0.8 ± 1.0 μm. Mixing was gently performed at room temperature for 1 min by slow manual rotation. After exposure, the powders were pelleted by centrifugation (2400 rpm for 2 min) followed by filtration through a 0.22 μm filter (Hawach Sterile PES Syringe Filter, HAWACH SCIENTIFIC CO., LTD., Xi'an, China). Supernatants were collected, aliquoted, and subjected to TCID50 assays, real-time RT-PCR, and fluorescence assays. The full details of sample preparation and analyses were described in a previous study [1]. SARS-CoV-2 virus experiments were conducted in a biosafety level 3 (BSL-3) biocontainment facility using BSL-3 work practices.
A Raman spectrometer (LabRAM HR800, Horiba/Jobin-Yvon, Kyoto, Japan) operating with a 50x optical lens was used for in situ spectral analysis of the virus sample. The preparation procedure of the isolate JPN/TY/WK-521 sample is described in the Supplementary Information. The spectroscope was set in confocal mode and used a holographic notch filter, which concurrently provided high-efficiency and high-resolution spectral acquisitions. The incoming light was from a 532 nm solid-state laser source operating at 10 mW. A spectral resolution of ∼1 cm−1 was achieved upon analyzing the Raman scattered light by a double monochromator connected with an air-cooled charge-coupled device (CCD) detector (Andor DV420-OE322; 1024 × 256 pixels). The acquisition time of one spectrum was ∼15 s. To obtain an average spectrum, thirty spectra were collected at different locations over areas of ∼2 mm2 and averaged.
The average Raman spectrum was treated with a baseline subtraction and automatically deconvoluted into series of Voigtian sub-bands using commercial software (LabSpec 4.02, Horiba/Jobin-Yvon, Kyoto, Japan). Baseline subtraction applied the criterion of polynomial fitting. For spectral analysis, we applied a machine-learning algorithm specially crafted for the Raman spectrum. This consisted of an automatic solver exploiting a linear polynomial expression of Voigtian functions. An algorithm searching for the minimum value of the difference between the experimental and the fitted spectrum was then applied through a computer program. The program chose a series of Voigtian sub-bands from the deconvoluted spectra of pre-selected compounds from a database of key biomolecules in aqueous solution and in a solid state according to the chemical and structural peculiarities of the virions. According to the spectra of pre-selected compounds, the algorithm pinpointed the closest matches to the experimental spectra according to established criteria of relative intensity, spectral position and bandwidth. These conditions provided the required mathematical constraints to univocally deconvolute the experimental spectrum. Additional details about the spectral deconvolution procedure and the machine-learning algorithm were given in a previous report [12].
3. Experimental results
3.1. Evidences of the antiviral behavior of Si3N4
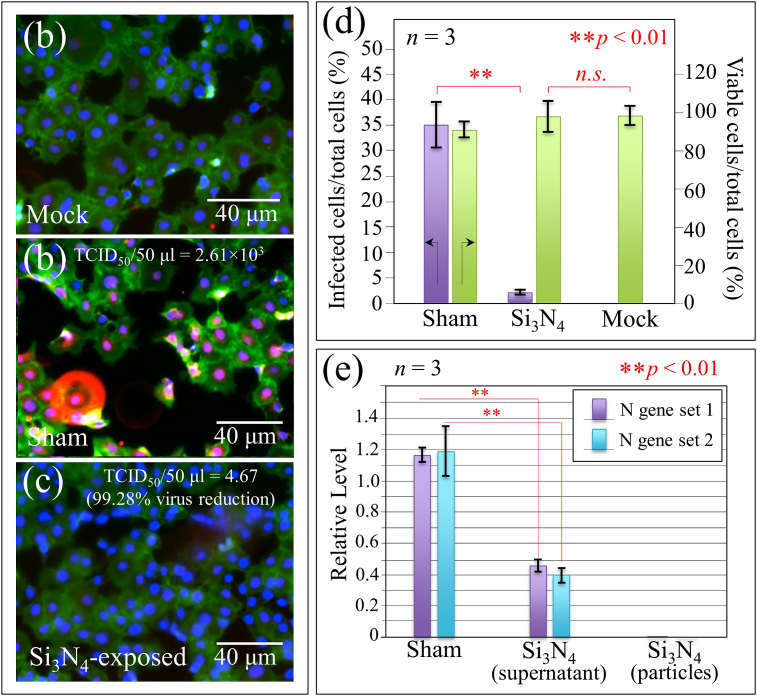
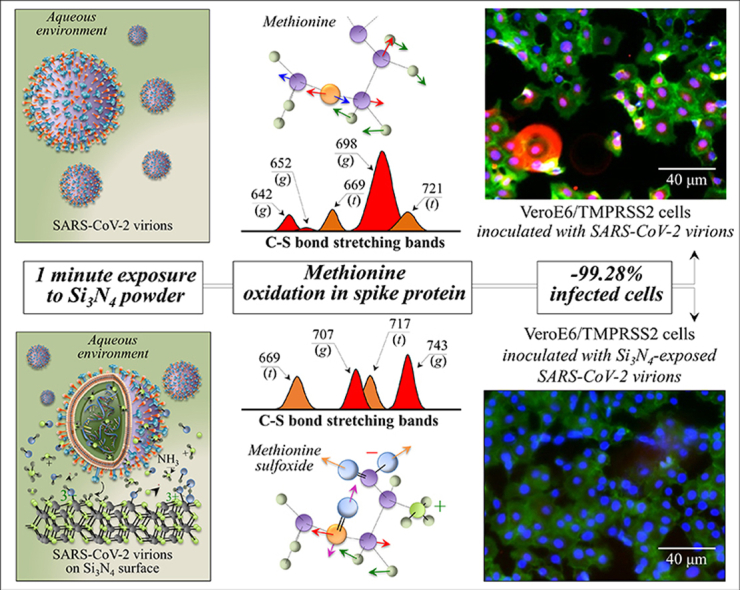
Fig. 1 summarizes the salient immunochemistry evidences so far collected on the antiviral behavior of Si3N4 powder vs. SARS-CoV-2 [1]. Details of the procedures adopted are given in the Supplementary Information. Immunofluorescence images of VeroE6/TMPRSS2 cells are shown for (a) non-inoculated cells (mock), (b) cells inoculated with virions unexposed to the Si3N4 powder (sham), and (c) cells inoculated with supernatant virions exposed for 1 min to 5 vol% Si3N4 powder, respectively. In the images, the envelope antibody of the anti-SARS coronavirus is stained red, the viable cell F-actin (Phalloidin-stained) green, and the cell nuclei (DAPI-stained) blue. The red fluorescent signal is thus a marker for the synthesis of viral protein. As expected, the as-cultured cells unexposed to virions (mock sample) showed no red staining (cf. Fig. 1(a)), while the images of the sham cell sample revealed extensive cell infection by the virus (cf. Fig. 1(b)). Remarkably, the cells inoculated with supernatant virions treated with Si3N4 were viable, showed no infected cells, and even appeared to further proliferate (cf. Fig. 1(c)). The plot in Fig. 1(d), which quantifies a series of fluorescence images, and shows ∼35% fraction of cells in the sham sample (negative control) and only 2% of cells inoculated with Si3N4–exposed supernatant virions. Moreover, the fraction of viable cells was up to 10% higher than that in the mock sample although lacked statistical significance. Data by fluorescence imaging were confirmed by testing with TCID50 assay (cf. data in inset to Fig. 1(b) and (c)). Compared with the negative control (sham sample), the Si3N4 powder produced >99% effective inactivation of SARS-CoV-2 virions.
Fig. 1.
Fluorescence micrographs of (a) non-inoculated cells (mock), (b) cells inoculated with virions unexposed to Si3N4 powder (sham), and (c) cells inoculated with supernatant virions exposed for 1 min to a suspension of ∼5 vol% Si3N4 powder (results of TCID50 assay in inset); (d) quantification of fluorescence microscopy data given as % of infected cells on total cells (i.e., % of red-stained cells with respect to the total number of blue-stained nuclei), and % of viable cells on total cells (i.e., % of green-stained cells with respect to the total number of blue-stained nuclei). Labels in inset specify statistics by the unpaired two-tailed Student's test with n = 3. In (e), results of RT-PCR tests to evaluate viral RNA.
The fragmentation of viral RNA upon 1-min contact with the Si3N4 powder was evaluated by means of RT-PCR experiments on the virions N-gene sequence (Fig. 1(e)). Both supernatant virions and virions on pellets were tested. Unlike the case of powder-unexposed control supernatant (sham sample), the viral RNA underwent severe damage after Si3N4 contact, and viral RNA could not be detected on pelleted powder.
For further confirmation, the above characterizations were repeated for virions exposed for 10 min to Si3N4 particles (n = 3). The results, shown inFig. S-1 of the Supplementary Information, validated the trend observed at 1 min exposure as shown in Fig. 1. The combination of fluorescence spectroscopy and RT-PCR results unequivocally proves the occurrence of SARS-CoV-2 inactivation by Si3N4 bioceramic powder. Based on this evidence and our previous work on the antipathogenic properties of the Si3N4 [1,2,4,12,13,18], we hypothesized that ammonia likely plays a fundamental role in inactivating SARS-CoV-2 and we attempted to prove this hypothesis using Raman spectroscopy.
3.2. Raman spectroscopic results
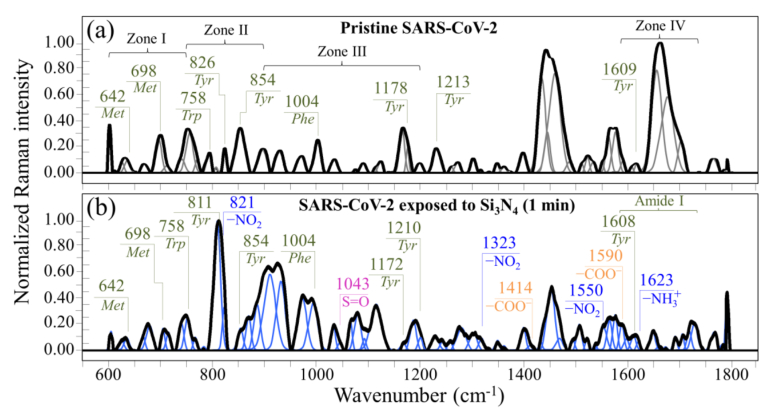
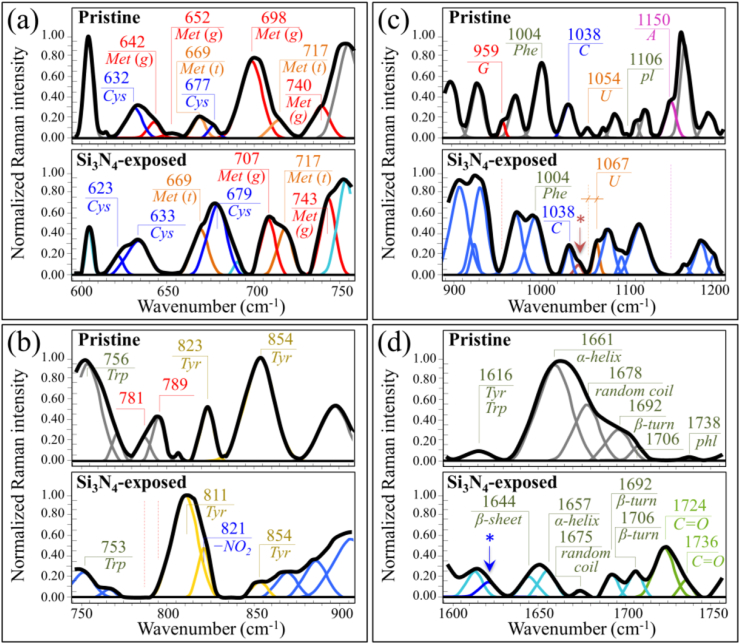
Fig. 2 shows normalized and deconvoluted Raman spectra as collected in the frequency interval 600–1800 cm−1 on SARS-CoV-2 Japanese isolate JPN/TY/WK-521 virions (a) before and (b) after 1 min exposure to Si3N4 powder in aqueous solution. The spectra collected before and after Si3N4 exposure appeared very different to each other, thus proving that the Raman spectrum captured the fundamental differences caused to the virus structure upon contact with Si3N4 particles at the molecular level. Precise analyses of spectral differences are discussed with respect to four selected spectral zones labeled as Zones I ∼ IV and located at 600–750 cm−1, 750–900 cm−1, 900–1200 cm−1, and 1600–1750 cm−1. Raman spectra in the selected spectral zones were collected with high spectral resolution, normalized, and deconvoluted into sub-band components according to the procedure discussed in Section 2. Frequencies at maximum and proposed vibrational origins are given in the Supplementary Information (Fig. S-2 and Tables S-I(a)∼(d)).
Fig. 2.
Raman spectra in the frequency interval 600–1800 cm−1 of (a) the original SARS-CoV-2 Japanese isolate JPN/TY/WK-521, and (b) of the same isolate after exposure for 1 min to 5 vol% Si3N4 particles in aqueous suspension. Spectra are normalized to their maximum signal and deconvoluted into Voigtian band components. Four Zones are emphasized in (a) and labels show frequencies at maximum of selected bands (Met, Tyr, Trp, and Phe are abbreviations for methionine, tyrosine, tryptophan and phenylalanine, respectively). In (b), specific vibrations related to structural modification upon Si3N4 powder treatment are emphasized and discussed in the text.
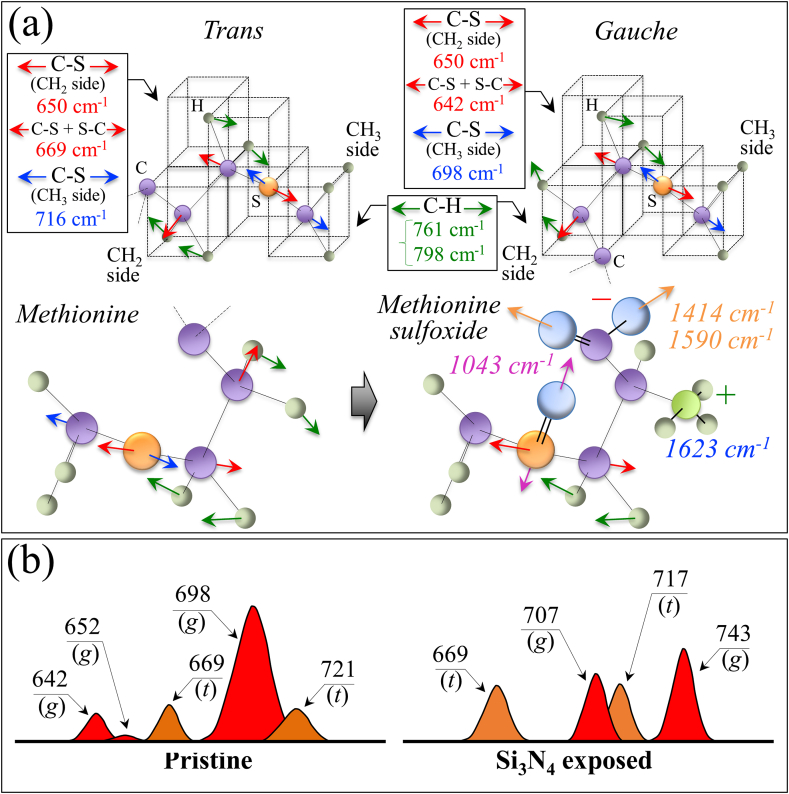
Fig. 3(a)–(d) show the spectra collected in Zone I, II, III, and IV. The upper and lower spectra in each section are from virions before and after exposure (cf. labels). Zone I is dominated by vibrational signals related to the C–S bond [[19], [20], [21]], and thus belongs to methionine and cysteine rotamers, namely, the only amino acids incorporated into viral proteins that contain sulfur. Zone II was selected because it contains a doublet from tyrosine (often referred to as “Fermi doublet”) at 854 and 826 cm−1. The doublet intensity ratio, referred to as I854/I826, is diagnostic of an H-bonding environment around the tyrosine units; the lower the ratio the more hydrophobic the environment in which the tyrosine residue is embedded [22,23]. Zone III contains a prominent Raman signal from phenylalanine at 1004 cm−1 (symmetric ring breathing) and incorporates ring-related signals assignable to individual RNA purine and pyrimidines. Cytosine (C) and uracil (U) pyrimidine bands from C–N–C in-plane deformation of heterocyclic aromatic ring care entered at 1038 cm−1 and 1054 cm−1, respectively [[24], [25], [26]]; guanine purine shows the same vibrational mode at 959 cm−1, while the C–N stretching mode at 1150 cm−1 is cumulative of the imidazole and pyridine rings of the adenine (A) [27,28]. Finally, Zone IV designates the Amide I frequency region, which is representative of the secondary structure of viral proteins. The deconvoluted Amide I band components are assigned to β-sheet (βs; at 1638–1640), α-helix (αh; 1657–1661), random coil (rc; 1675–1679), and β-turn rotamers (βt-I and βt-II; at 1692–1698 and 1713–1716 cm−1, respectively).
Fig. 3.
Enlarged Raman spectral zones of the JPN/TY/WK-521 isolate before and after exposure to 5 vol% Si3N4 micrometric powder in aqueous suspension: Zone I (600–750 cm−1), Zone II (750–900 cm−1), Zone III (900–1200 cm−1), and Zone IV (1600–1750 cm−1); spectra are deconvoluted into a sequence of Voigtian sub-bands (frequencies for selected bands shown in inset). The abbreviations Met and Cys refer to methionine and cysteine, respectively (with (t) and (g) locate trans and gauche rotamers, respectively); the abbreviations G, C, U, pl, and A refer to guanine, cytosine, uracil, phosphodiester linkages, and adenosine, respectively.
Structural changes in methionine residues are among the most important post-translational modifications of the proteins engendered by the presence of non-radical and free radical species [29,30]. Raman spectroscopy is uniquely posed to capture such structural modifications because of its high sensitivity to C–S bond vibrations, which strongly display in Zone I. Fig. 4(a) shows a draft of the vibrational modes in different methionine rotamers. The C–S stretching modes on the CH3 side of the molecule give rise to two Raman bands at ∼716 and ∼698 cm−1, which are attributed to the trans and gauche rotamers, respectively [31]. On the other hand, the C–S signal on the CH2 side found at ∼650 cm−1 belongs to both rotamers. However, the C–S + S–C in-phase stretching mode appears at distinct frequencies in different rotamers (669 and 642 cm−1 for trans and gauche configurations, respectively). In the Raman spectrum of the pristine SARS-CoV-2 virus, the most intense methionine band at ∼698 cm−1 suggests that methionine residues are mainly in the gauche rotameric configuration with respect to the –C(α)–S bond (cf. labels in Fig. 4(a)). Exposure to the Si3N4-surface environment stressed the structure and induced a change in the population ratio of the two rotameric forms, as shown by the significant decrease in the intensity ratio, I700/I721 (from 3.5 to ∼1; cf. Fig. 3(a) and the schematic draft in Fig. 4(b)). The complete disappearance of the gauche C–S + S–C in-phase stretching band at 642 cm−1 confirms this trend. However, additional fingerprint features could be found in the methionine spectrum, which were induced by environmental interactions with Si3N4. They corresponded to a clear shift toward higher frequencies of the C–S stretching band from 698 to 707 cm−1 and a more than two fold increase in the relative intensity of the C–S + S–C in-phase stretching signal at ∼740 cm−1. Concurrently, two new bands appeared at 1043 and 1623 cm−1 (cf. arrowed asterisks in Fig. 3(c) and (d), respectively). These two signals can be assigned to S O stretching and NH3+ in-plane bending, respectively; both vibrations appeared as shoulder bands, but were clearly missing in the spectrum of unexposed virions. According to a vibrational spectroscopy study by Torreggiani et al. [32], the appearance of the above S O and NH3+ signals coupled with a reduction in the C–S band at 698 cm−1, which was found replaced by a more intense signal at 707 cm−1, reveals the formation of methionine sulfoxide (cf. Fig. 4(b)). This molecule, depicted in Fig. 4(a), is a post-translational product of methionine, which forms upon oxidation and occurs as a consequence of environmental stress exposure operated by non-radical and free radical species. In support of this interpretation, one could note a significant enhancement of two bands from –COO– terminal bonds, whose symmetric and asymmetric stretching vibrations display at 1414 and 1590 cm−1, respectively (cf. Fig. 2(b)). This latter band also overlaps in-plane bending vibrations of the NH3+ terminal group in methionine sulfoxide [32]. Moreover, both marked shift and signal intensification of C–S stretching at 698 cm−1, and of C–S + S–C in-phase stretching at ∼740 cm−1 (cf. Fig. 4(b)) could conceivably be the consequences of variations in bond stiffness/flexibility in the oxidized molecule. The next section will discuss which non-radical or radical reaction(s) have caused methionine oxidation at the surface of Si3N4 bioceramics.
Fig. 4.
(a) Structures and C–S stretching vibrational modes/frequencies of trans and gauche methionine rotamers (cf. labels) and structural and vibrational variations after sulfoxidation of the methionine structure; (b) Voigtian components representing signals from different rotameric configurations of the methionine structure before and after exposure to Si3N4 powder (labels in inset give band frequencies and types of rotamer).
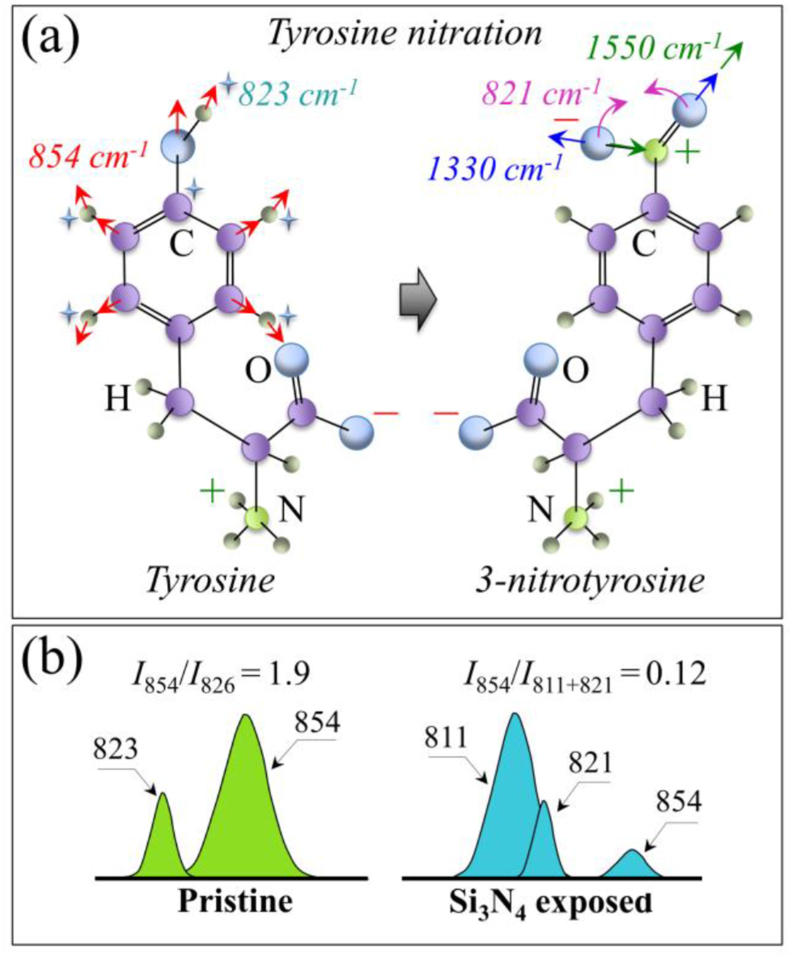
Fig. 5 represents the main expected molecular structure modifications and the spectroscopic characteristics revealed by the Raman analysis of Zone II. Fig. 5(a) depicts the vibrational origins of the tyrosine doublet as related to independent modes of the phenol ring: in-plane ring breathing and out-of-plane C–H bending at 854 cm−1 and 826 cm−1, respectively [23]. The intensity ratio of these two signals, I854/I826, has been indicated as a sensor of hydrophobic/hydrophilic balance in environmental interactions involving the tyrosine phenol ring [23]. Ammonia is known to induce protein tyrosine nitration in biological systems [33], and to be the cause of several pathologies [34]. The process of tyrosine nitration (depicted in Fig. 5(a)), which leads to 3-nitrotyrosine formation, has been studied using Raman spectroscopy [35]. The intensity ratio of the phenol ring tyrosine doublet, I854/I826, is a useful nitration indicator. Additional markers of nitration are signals from the nitro group, –NO2, (cf. Fig. 5(a)): stretching (i.e., symmetric and asymmetric at ∼1330 cm−1 and ∼1550 cm−1, respectively) and in-plane bending vibrations (i.e., at ∼821 cm−1) [36,37]. Upon nitration, the relative intensity of the tyrosine doublet undergoes a trend inversion (cf. Fig. 5(b)) due to the contribution of –NO2 bending at ∼821 cm−1 [35]. The spectroscopic signals of Fig. 3(b) in the frequency interval of the tyrosine doublet before and after exposure to Si3N4 powder are schematically reproduced in Fig. 5(b). We indeed observed a trend inversion for the tyrosine doublet upon exposure to Si3N4, with the intensity ratio, I854/I826, changing from 1.9 to 0.12. This bold variation is partly due to overlap with the –NO2 signal (the low-frequency band of the doublet appears split into two sub-bands), but predominantly arises from a perturbation of benzene ring symmetry (causing intensity increase and shift of the low-frequency band toward lower wavenumbers). Confirmation of the tyrosine nitration process was also obtained by the enhancement of the relative intensity of –NO2 symmetric and asymmetric stretching bands at 1330 and 1550 cm−1, respectively (cf. labels in Fig. 2(b)). Nitration of tyrosine residues is also expected to induce severe alterations of protein secondary structure [38].
Fig. 5.
(a) Structure of tyrosine and 3-nitrotyrosine (cf. labels): shown together with in-plane ring breathing and out-of-plane C–H bending vibrational modes/frequencies of the phenol ring in the former and symmetric stretching and in-plane bending modes/frequencies in the nitro group, –NO2, of the latter; (b) Voigtian components representing signals of the tyrosine doublet components as detected before and after exposure of the virions to Si3N4 powder (Raman ratios I854/I826 and I854/I811+821 given in inset).
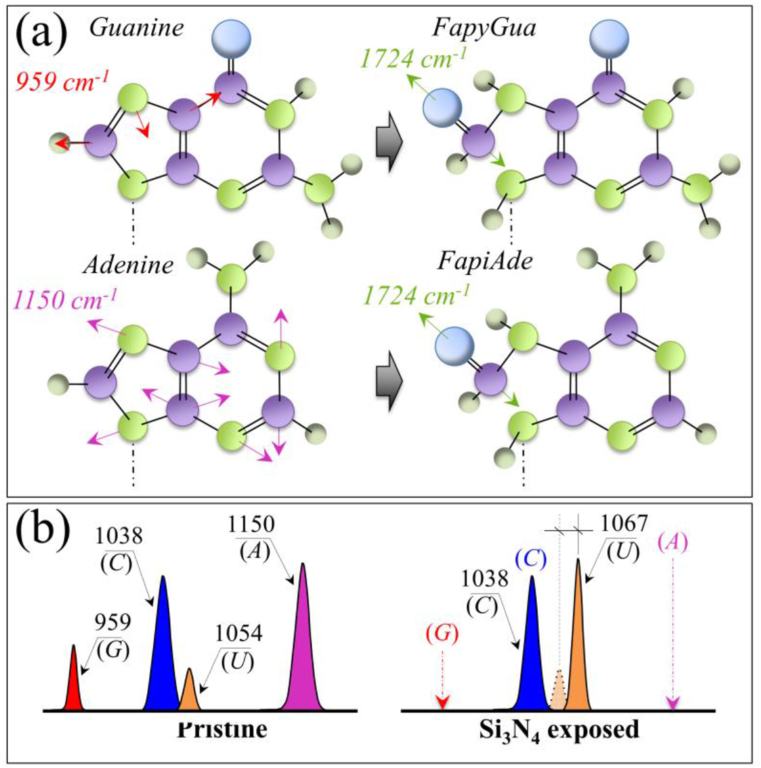
An additional bold feature in Zone II is the complete disappearance of two bands at ∼781 and 789 cm−1 in the spectrum of virions exposed to Si3N4 (cf. Fig. 3(b)). These two bands are cumulatively contributed to by the ring vibrations of the RNA bases and C–O–P–O–C phosphodiester bonds [39]; and their disappearance is an unambiguous proof that the viral RNA has been damaged upon contact with Si3N4. This finding is in line with the severe fragmentation of viral RNA found upon screening the virions N-gene sequence by means of RT-PCR after 1-min contact with the Si3N4 powder (cf. Fig. 1(e)). Additional details of RNA damage upon short-term exposure to Si3N4 powder could be found in Zone III (Fig. 6(a)). The Raman signals selected as markers of RNA purines in this zone (C–N–C in-plane ring deformation in guanine and C–N ring stretching mode in adenine; at 959 cm−1 and 1150 cm−1, respectively) completely disappeared in the spectrum of Si3N4-exposed virions (cf. Fig. 3(c)). The selected spectroscopic marker of uracil base in Zone II was found ∼4-fold stronger in relative intensity and shifted by ∼13 cm−1 toward higher wavenumbers. An exception in the above prospect of RNA damage was represented by the cytosine signal (C–N–C in-plane deformation of heterocyclic aromatic ring) centered at ∼1038 cm−1, which remained unaltered (cf. Fig. 3(c)). These trends are schematically drawn in Fig. 6(b). It is known that ssRNA viruses are the viruses most prone to ammonia attack, because their genome type is more susceptible to degradation, while other more stable genome types present a slower inactivation, mainly driven by degradation of viral proteins [6]. However, a complete disappearance of ring vibrations in both guanine and adenine purines, as observed in this study, can only be explained by invoking breakage of the imidazole ring to form formamidopyrimidines (Fapy) [40]. Such substantial structural changes are not obviously assignable to any direct interaction with ammonia molecules; instead, they can only be caused by off-stoichiometric reactions with oxygen/nitrogen radicals. Note that also the Raman results of post-translational modifications in the methionine structure (in spectral Zone I) pointed to the action of free radicals (cf. above). Work by Douki and Cadet [41] showed that peroxynitrite anion (O NOO−) can exhibit pronounced oxidizing properties toward purine moieties, which leads to the production of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dGuo) at a very low yield. In a successive work, Douki et al. [42] linked the presence of 8-oxo-dGuo to the formation of 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua), and also extended the findings to 4,6-diamino-5-formamidopyrimidine (FapyAde) as an oxidation product of the adenine base. The hypothesized structural variations, as induced upon oxidation by O NOO−, are schematically shown in Fig. 6(a). In support of this hypothesis, one could invoke a significant enhancement in C O stretching vibration in Zone IV (cf. Fig. 3(d)) as a consequence of the formation of Fapy molecules (cf. newly formed C O bonds in open Fapy rings as depicted in Fig. 6(a)).
Fig. 6.
(a) Schematic draft of guanine and adenine RNA purines with their respective ring vibrational fingerprints/frequencies (C–N–C in-plane ring deformation at 959 cm−1 and C–N ring stretching mode at 1150 cm−1 for guanine and adenine, respectively), and their transformation into formamidopyrimidines upon opening of the imidazole ring and oxidation with a new C O stretching mode appearing at 1724 cm−1. In (b), a comparison is shown of Voigtian components representing fingerprint signals (and respective frequencies) of RNA purine and pyrimidines before and after exposure of the virions to Si3N4 powder.
Besides the evidence of ring disruption in purines, the present Raman study also reveals the behavior of pyrimidine bases in viral RNA upon exposure to Si3N4 powder. The evidence of no variations in the cytosine signal indeed supports the hypothesis of a radical species effect on RNA degradation. Degradation of the cytosine structure to form 6-hydroxy-5,6-dihydrocytosine (cytosine photohydrate) only arises from the photohydration reaction of cytosine [43]. This mechanism requires a high-energy radiative, oxygen-independent process [44], which is not involved with the hydrolytic reactions in object here. Nonoyama et al. [45] have reported that both cytosine and uracil are almost inert to the peroxynitrite anion. Their study thus supports our finding of cytosine inertness in the hydrolytic environment of Si3N4. However, an explanation for the observed vibrational behavior of uracil pyrimidine requires further elaboration. According to ultraviolet and ultrasonic absorption results [46,47], the uracil structure primarily exists in the lactam (amide) form in neutral aqueous solution. However, in an alkaline environment (pK∼9.2), ionization takes place and forms hydroxyl groups to replace the double-bonded oxygens forming the lactim (imidic acid) structure [48]. The hydrolytic reactions taking place at the solid surface of Si3N4 in an aqueous environment induce a robust pH buffering at the interface with virions with local pH values comparable with the uracil ionization pK value. Accordingly, it is conceivable to hypothesize that the uracil structure underwent an amide-imidic acid tautomeric shift. This structural change could explain both the increased intensity and the shift toward the higher frequency of the uracil ring vibration in Zone III (cf. Fig. 6(b)).
The Amide I Zone IV at 1600–1750 cm−1 (Fig. 3(d)) gives information about the secondary structure of proteins. A comparison between pristine and Si3N4-exposed virions reveals marked variations, with a relative increase in β-sheet fraction mainly at the expenses of random coil, and a reshuffling in the balance of β-turn rotamers. The usefulness of Raman spectroscopy in assessing post-translational modifications in proteins is widely exploited in therapeutic production, a field in which subtle changes are associated with key structural rearrangements hardly detected by conventional methods of protein analysis [49]. Post-translational modifications of Coronavirus proteins, including spike, envelope, membrane, and nucleocapsid proteins have recently been reviewed by Fung and Liu [50], with emphasis on their impact on viral replication and pathogenesis. Although the present Raman spectroscopic assessments do not provide enough information to link changes in protein secondary structure to specific proteins responsible for the observed loss of viral infectivity, the rotameric variations observed in S-containing amino acids are the key to interpreting protein structural changes and amply justify viral inactivation; since disulfide bond formation is fundamental for a correct folding, trafficking, and trimerization of the spike protein. The observed rotameric modifications in Zone I can be connected to the unfolding of the α-helical structure after coming in contact with Si3N4, because methionine has an especially high helix-forming propensity [51]. On the other hand, a recent computational simulation study of the impact of methionine oxidation on the dynamics and energetics of proteins’ α-fold conformation has shown that the sulfoxidation of helical methionines is a strong trigger for the destabilization of the native α-fold configuration and leads to protein conformational switch in the β-sheet structure [52]. This change gives a more flexible secondary structure, but it also favors alternative states which maybe essential in the observed viral inactivation pathway.
4. Discussion
4.1. Post-translational modifications uncovered by Raman spectroscopy
Taken together, these results provide a set of unequivocal spectroscopic markers of the main structural changes occurring to SARS-CoV-2 virions as a consequence of their exposure to Si3N4-eluted ammonia and related radicals. Despite the impressive experimental methods presently available in virology, there are currently no techniques that can provide molecular-scale insights into rotameric modifications at specific residues. However, Raman spectroscopy, with its unique capacity to unfold differences in molecular symmetry, has revealed rotameric transactions and the formation of sulfoxidated helical methionines destabilizing the structure of the spike receptor binding domain (RBD) recombinant proteins. Methionine residues are highly susceptible to oxidation and form methionine sulfoxide [53]. Oxidation modifications of methionine residues within proteins is mediated by a variety of oxygen and nitrogen radicals, including peroxynitrite, O NOO− [54]. Oxidation of free and peptide-bound methionine to sulfoxide results in significant conformational and functional changes in proteins. Dado and Weissbach [55] have shown that methionine oxidation serves as a conformational switch toward increasing the fraction of β-sheet structure, which is indeed what we observed in this study. The rationale for the formation of peroxynitrite at the virion/Si3N4 interface is given in the next section in discussing the formation of Fapy structures.
An additional finding in the RBD was the nitration of tyrosine residues, which are dominant component of the SARS-CoV-2 spike structure [56]. An important feature of the interface between the spike RBD protein and angiotensin-converting enzyme (ACE 2) receptor (i.e., the entry point for the virus to hook into human cells) is its dense network of hydrophilic interactions. With thirteen hydrogen bonds and two salt bridges at the SARS-CoV-2 RBD/ACE2 interface [56], the involvement of multiple tyrosine residues in forming hydrogen-bonding interactions with the polar hydroxyl groups is key in determining the level of infectivity. The present Raman assessments revealed the formation of nitrotyrosine and, consequently, a dramatic impairment of hydrophobic/hydrophilic balance in phenol ring environmental interactions.
Methionine sulfoxidation and tyrosine nitration represent oxidative post-translational modifications of the viral spike protein that block the SARS-CoV-2 virions from entering cells. Post-translational modifications of the spike protein create this block as the virions insert the wrong key inside the cell lock. These findings exemplify well the antiviral pathway and justify the loss of infectivity of the viral strain upon contact with Si3N4. As an additional finding of this study, we suggest that Raman assessments of rotameric methionine fractions, and the degree of nitration in tyrosine residues could represent promising spectroscopic protocols for assessing the effectiveness of antiviral therapies.
4.2. Formamidopyrimidine formation and the origin of free radicals
As mentioned in Section 3.2, breakage of the imidazole ring in RNA purines to form Fapy structures necessarily requires the presence of oxygen/nitrogen radicals, and the peroxynitrite anion is believed to be the main agent of purine oxidation because of its strong oxidation capacity. The formation of O NOO− at the Si3N4/pathogen biological interface has been proved in a previous investigation using stimulated emission depletion microscopy of fungal pathogens and explained according to a cascade of off-stoichiometric reactions starting from the formation of ammonia [12]. Briefly, the homolytic cleavage of Si–N bonds occurring upon hydrolysis at the Si3N4 surface concurrently liberates nitrogen anions and unpaired electrons [57]; free electrons split the surrounding water molecules to form oxygen radicals, which in turn catalyze NH3 oxidation into hydroxylamine and initiate a cascade of off-stoichiometric reactions leading to the formation of reactive nitrogen species (i.e., including NO, NO2, and O NOO−) [58,59]. Note that the present study supports the above interpretation by documenting the presence of nitrotyrosine residues as a proof for the existence of peroxynitrite at the virions’ biological interface. This follows the same argument used by Beckman et al. [60] proving the formation of peroxynitrite in arterial plaques, since tyrosine is nitrated by peroxynitrite, but not directly by nitric oxide [61]. A successive study showed that O NOO− does not directly react with tyrosine [62], but promotes tyrosine nitration through reactions with the peroxynitrite-derived hydroxyl radicals and nitrogen dioxide.
For a similarly definitive degree of viral inactivation, a comparison of the effects of Si3N4 hydrolytic reactions on SARS-CoV-2 and Influenza A virions [2] revealed similarities, but also interesting differences in the mechanisms of viral inactivation. Radical driven post-translational alterations of methionine C–S bonds upon contact with Si3N4 were a common feature in both viral strains. However, rather than methionine sulfoxidation as in the case of SARS-CoV-2, C–S bond alterations in Influenza A virions pointed to the formation of thioether groups in methionine residues. Deprived of the C–S bond on the CH3 side, methionine transformed into homocysteine. Regarding the effect of reactive nitrogen species on RNA bases, ring opening was also observed for Influenza A, but only in the guanine base (to form FapyGua), not in the adenine base. On the other hand, the amide-imidic acid tautomeric shift of uracil pyrimidine similarly appeared in the spectra of both SARS-CoV-2 and Influenza A virions. The observed differences are in line with the notion that different viruses can exhibit different inactivation pathways and kinetics when treated with the same biocide [63]. Structural and genomic differences have a significant impact on type and extent of structural damage that a virus can sustain before losing its infectivity. The resistance to biocidal inactivation of a specific virus remains related to its ability to cope with and adapt to the applied stressor. However, taken together, the present results suggest that the Si3N4 inactivation pathways have a broad effectiveness, and preserve chemical similarities among quite dissimilar ssRNA viral strains.
4.3. Comparison of Si3N4 inactivation kinetics vs. other viral strains
Aqueous ammonia, NH3(aq), is the neutral dissolved form of the total ammonia (NH4+/NH3) released by the Si3N4 surface upon hydrolysis. NH3(aq) possesses biocidal activity against most pathogenic microorganisms through a number of different mechanisms [[64], [65], [66]]. It was also recognized as the main agent responsible for virus die-off in sludge [67]. In a previous comparative study of Influenza A-H1N1, A-H2N3, and B/98 strains [17], the structural characteristics of methionine components (including the matrix protein M1, which plays a key role in nuclear import and viral replication) were found to greatly differ for different viral strains. Similar to the present case of SARS-CoV-2 virions, Si3N4 was found to be effective as a solid-state virus inactivator by various concurrent mechanisms including thioether cleavage of methionine residues, RNA phosphodiester bond cleavage, and nucleotide damage [2]. However, clear differences in inactivation kinetics could be detected among different ssRNA viruses. Although the effect of differences in the genomic structure between different viruses might have a role (i.e., longer genome lengths involve a higher probability of RNA cleavage), variations in inactivation kinetics are believed to mainly depend on the different levels of electrical attraction of virions toward the Si3N4 particles; namely, the probability that a virion enters in contact with a Si3N4 particle for a given volume fraction of particles in aqueous solution. This hypothesis is supported by studies of the isoelectric point (IEP) of different viruses [68,69]. For example, the reported IEP of Feline Calicivirus (FCV) is ∼3.9,[70] which is very close to that of Si3N4 (∼4.4) [71]. An electrical charge very close to that of the Si3N4 surface over a wide range of pH makes the FCV experience a low probability of encountering a Si3N4 particle in suspension due to weak electrical attraction. This ultimately results in a slower inactivation kinetics of FCV in an homeostatic aqueous dispersion of Si3N4 particles [2]. On the other hand, Influenza A (H1N1) and Enterovirus (EV-71) virions possess IEPs significantly greater than that of Si3N4 (i.e., 7.47 and 10.9, respectively) [72,73]. Accordingly, they undergo a faster inactivation kinetics [2], because of the high electrical attraction enhancing the probability of virions entering in contact with a Si3N4 particle in aqueous suspension. The IEP values of SARS-CoV-2 virions have been measured as ∼10.1 in nucleoproteins, ∼9.5 in membrane proteins, and ∼6.2 in the spike glycoproteins [74]. In all cases, IEP values are far from that of Si3N4. Accordingly, the attraction toward Si3N4 particles in aqueous environments at homeostatic pH could be strong, which fully justifies the quite fast inactivation kinetics reported in Fig. 1. Once in contact with the surface of Si3N4 particles, the virions become almost instantaneously “poisoned” by the eluted NH3(aq) molecules. Such a two-steps antiviral mechanism has been branded as the “catch-and-kill” effect [1].
4.4. The importance of a safe solid-state “non-nano” antiviral agent
As the COVID-19 pandemic continues to pose significant risks worldwide, antiviral materials with “smart” surface chemistry, capable of multiple and broad mechanisms of viral inactivation, are being actively searched for. Environmental disinfection, sterilization of surfaces, and the sanitization of protective equipment have traditionally been pursued by means of chemical disinfectants, such as chlorines, quaternary amines, alcohols, and peroxides [75]. However, chemical disinfectants generally exhibit low durability and require high concentrations in order to reach a full viral inactivation. Additionally, while being often an inevitable choice, they represent a severe risk factor for public health and environmental pollution [76]. In an attempt to foster long-term persistence and to reach high effectiveness at low doses, nano-sized solid-state particles, both metallic and non-metallic, have been proposed, which include Ag, Cu, TiO2, and graphene [77,78]. Nanoparticles possess an inherently broad antiviral effectiveness, which originates from the generation of reactive oxygen species at their surface. However, the majority (if not the totality) of nanomaterials also present adverse effects on both human health and the environment [79]. In vitro, in vivo, and genomic studies have clearly shown how nanoparticles, due to their small size, can easily enter and remain trapped in the human body upon inhalation or even through the skin; their toxicity arises from strong chemical interactions with proteins and enzymatic molecules that alter gene expression and biological functions at the cellular and subcellular levels [80].
Based on previous and present data of viral inactivation by micrometer-sized Si3N4 powder, we challenge here the commonplace idea that a solid-state antiviral agent should be “nano” to be broad, effective, and efficient. Moreover, we propose the use of a “smart” antiviral chemistry employing a volume fraction of powder as low as 5 vol% and based on nitrogen rather than oxygen radicals. This new approach becomes highly efficient in terms of virus inactivation kinetics upon exploiting the above-mentioned “catch-and-kill” effect. Note that nanotechnology-based approaches are generally proclaimed as the most promising antipathogenic formulations since benefiting from high surface-to-volume ratio and enhanced surface reactivity [81,82]. Accordingly, even the most recent reviews in the field of functional materials continue to list silver, TiO2, and copper-derived nano-sized chemicals, which are toxic to human cells, as the only possible alternatives to counteract the COVID-19 pandemic [83]. These are indeed options that feature an old chemistry, while “smarter” inorganic materials have already been reported in the literature, which exploit differently efficient mechanisms [84]. The present Si3N4 approach belongs to the category that does not need to exploit a nano-scale dimension to be highly effective. The physiological gap in nitrogen radical metabolization existing between eukaryotic cells and pathogens (especially viruses) indeed renders Si3N4 supportive of the former and fatal to the latter (cf. Fig. 1) [4]. As a solid-state compound, Si3N4 matches the Ehrlich's concept of “magic bullet”, namely, a chemical compound with selective toxicity: toxic for the infecting microbe but safe for the human host [85].
Si3N4 has been used for more than 15 years in the human body in contact with bone and soft tissue without any adverse effect being so far reported [86]. Moreover, several in vitro studies have shown the compatibility of Si3N4 with different cells lines, including mature osteoblasts [87], mesenchymal cells [4,88,89], and primary odontoblasts [90]. As a fine micrometric powder, Si3N4 can remain suspended in the air for some period, but its effect on human health is comparable to inert dust [91]. Si3N4 presents enormous potential as a disinfectant against SARS-CoV-2 and other ssRNA virions, due to the intrinsic anti-viral property exerted by reactive nitrogen species in aqueous (or moist) environments. Lacking the adverse effects of nanomaterials on human health and the environment, micron-sized Si3N4 bioceramic powder is a primary candidate to replace toxic and allergenic compounds in long-term environmental sanitation.
In a recent review [92], an highlight was given of the contribution of materials science to the development of personal protective equipment to counteract the COVID-19 pandemic. Methods were also reviewed for the design of simple, accurate, and low-cost virus-detection devices. This study matches both these two fundamental items by proposing a potent and safe antiviral bioceramic and by showing the power of Raman spectroscopy in detecting the related mechanisms of viral inactivation.
4.5. Comparison with other nitrides and possible future developments
The mechanism of hydrolysis and nitrogen elution at the biological interface between virions and Si3N4 particles is, in principle, also exploitable by other inorganic nitride compounds, although with different kinetics. For example, aluminum nitride (AlN), which shares with Si3N4 the chemical similarity of N atoms with strong electronegativity, is also known to undergo hydrolysis and to liberate nitrogen species in aqueous environment [93]. AlN has shown similar antiviral effect against SARS-CoV-2 [1]. However, the concurrent formation of aluminols at the solid surface and the possible liberation of aluminium salts to cellular environment makes this compound less safe than Si3N4 with respect to eukaryotic cells. From this point of view, Si3N4 has the important advantage of having a cation (Si) completely compatible with (and even supportive of) eukaryotic cells [4]. Another important factor in view of future applications is represented by the kinetics of nitrogen elution. The evolution of NH3 upon immersion of AlN powder in aqueous environment is quite abrupt and aggressive, with a pH buffering up to >10 [93]. However, the almost immediate formation of a thin shell of a stable aluminum oxide or hydroxide, which acts as a hydrophobic coating, locks the particle surface by preventing further reactions of AlN with water [94,95]. Conversely, Si3N4 experiences a slower kinetics as compared to AlN. Its pH buffering effect is milder (maximum pH∼8.5), but yet suffices to fight viruses and other pathogens, while being retained over several runs of environmental exposure [1,2,4,13]. This behavior is consequence of the enhanced solubility of surface silanols in the alkaline environment developed when nitrogen leaves the solid surface [96]. In summary, the uniqueness of Si3N4 as an antiviral compound is the result of two concurrent and synergic factors: the safety of its Si cation toward eukaryotic cells and the slow and durable kinetics of its hydrolytic reactions. Possible applications of the antiviral surface chemistry of Si3N4 include its introduction as micrometric powder in facemasks, surgical drapes and fabrics, as dispersoids in protective polymeric coatings for doorknobs or digital screens, and as loading particles in disinfectant sprays with long-term efficacy. In this perspective, Si3N4 surface chemistry could be listed among “resilient and agile” materials engineering solutions designed to address the challenge of the COVID-19 pandemic [97].
Looking for other possible nitride candidates, graphitic carbon nitride (g-C3N4) might deserve special attention for antiviral applications because of its peculiar catalytic behavior [98]. Already used for degrading pollution, as an antimicrobial agent, and in hydrolysis to generate hydrogen and oxygen, g-C3N4 appears to possess a potent surface chemistry capable to instantaneously inactivate viruses upon irradiation exposure both in presence and in absence of aqueous environment. The antiviral application of g-C3N4 could also represent a brand new development with respect to the already reported theranostics approaches using inorganic nitride compounds [[99], [100], [101], [102]].
5. Conclusion
Following the discovery of the strong antiviral effect of Si3N4 micrometer powder [1,2], this study delved into the molecular chemistry mechanisms of this unambiguous phenomenon. High spectral resolution Raman spectroscopy enabled us to form a picture of the viral inactivation pathways. We discovered two distinct types of structural degenerative effects on SARS-CoV-2 virions: (i) oxidative post-translational modifications of the viral spike protein blocking the SARS-CoV-2 virions from entering cells; and, (ii) breakage of the imidazole ring to form formamidopyrimidine structures, causing a severe damage to the ssRNA genomic structure. Both types of degradation originated from the formation of reactive nitrogen species at the virion/Si3N4 interface, and held similarities with the structural damages previously observed in other ssRNA viral strains.
From a more general viewpoint, the originality of this work resided in debunking the widely spread idea that an inorganic powder to be effective against pathogens must exploit nanometer dimensions. We instead put forward here the new concept of Si3N4 as a long-term “solid-state antibiotic” matching the Ehrlich's selective toxicity concept of “magic bullet”: toxic for the infecting virus but safe for human host cells. An additionally original achievement here is the demonstration that high-resolution Raman spectroscopy aided by a machine-learning algorithm specially crafted for the Raman spectrum is capable to provide invaluable insight into molecular-scale mechanisms behind viral inactivation.
In summary, the use of micron-sized Si3N4 particles could easily and safely be implemented in disinfectant sprays and coatings. Direct embedment in personal protective equipment fabrics, including facemasks, surgical drapes, and other garments, could help limit viral transmission in hospitals and nursing homes. As neither anion- nor cation-side surface chemistry of Si3N4 will affect human health and the environment, even in the long term, this unique bioceramic could become an invaluable tool in fighting SARS- CoV-2 and future pandemics.
CRediT authorship contribution statement
G. Pezzotti: Project administration, Writing – original draft, Conceptualization, Visualization. F. Boschetto: Formal analysis, Investigation. E. Ohgitani: Resources, Formal analysis, Investigation. Y. Fujita: Investigation. M. Shin-Ya: Resources. T. Adachi: Data curation. T. Yamamoto: Resources. N. Kanamura: Resources. E. Marin: Resources. W. Zhu: Data curation. I. Nishimura: Supervision, Data curation. O. Mazda: Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
SARS-CoV-2 strain (JPN/TY/WK-521) was kindly provided by the National Institute of Infectious Diseases. Raman experiments were conducted at the Kyoto Integrated Science & Technology Bio-Analysis Center. This work was supported in part by grants fro the Tokuyama Science Foundation and AMED under Grant Number 20he1122006j0001.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2021.100144.
Contributor Information
G. Pezzotti, Email: pezzotti@kit.ac.jp.
O. Mazda, Email: mazda@koto.kpu-m.ac.jp.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Pezzotti G., Ohgitani E., Sin-Ya M., Adachi T., Marin E., Boschetto F., Zhu W., Mazda O. Instantaneous “catch-and-kill” inactivation of SARS-CoV-2 by nitride ceramics. Clin. Transl. Med. 2020;10 doi: 10.1002/ctm2.212. e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pezzotti G., Boschetto F., Ohgitani E., Fujita Y., Zhu W., Marin E., McEntire B.J., Bal B.S., Mazda O. Silicon nitride: a potent solid-state bioceramic inactivator of ssRNA viruses. Sci. Rep. 2021;11:2977. doi: 10.1038/s41598-021-82608-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward R.L. Mechanism of poliovirus inactivation by ammonia. J. Virol. 1978;26:299–305. doi: 10.1128/jvi.26.2.299-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pezzotti G. Silicon nitride: a bioceramic with a gift. ACS Appl. Mater. Interfaces. 2019;11:26619–26636. doi: 10.1021/acsami.9b07997. [DOI] [PubMed] [Google Scholar]
- 5.Ward R.L., Ashley C.S. Identification of the virucidal agent in wastewater sludge. Appl. Environ. Microbiol. 1977;33:860–864. doi: 10.1128/aem.33.4.860-864.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decrey L., Kazama S., Kohn T. Ammonia as an in situ sanitizer: influence of virus genome type on inactivation. Appl. Environ. Microbiol. 2016;82:4909–4920. doi: 10.1128/AEM.01106-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz B.D., Margolin A.B. Inactivation of hepatitis A HM-175/18f, reovirus T1 Lang and MS2 during alkaline stabilization of human bio- solids. J. Appl. Microbiol. 2007;103:2225–2233. doi: 10.1111/j.1365-2672.2007.03463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pezzotti G. A spontaneous solid-state NO donor to fight antibiotic resistant bacteria. Mater. Today Chem. 2018;9:80–90. [Google Scholar]
- 9.Neumann A., Reske T., Held M., Jahnke K., Ragoss C., Maier H.R. Comparative investigation of the biocompatibility of various silicon nitride ceramic qualities in vitro. J. Mater. Sci. Mater. Med. 2004;15:1135–1140. doi: 10.1023/B:JMSM.0000046396.14073.92. [DOI] [PubMed] [Google Scholar]
- 10.Santos C., Ribeiro S., Daguano J.K.M.F., Rogero S.O., Strecker K., Silva C.R.M. Development and cytotoxicity evaluation of SiAlONs ceramics. Mater. Sci. Eng. 2007;C27:148–153. [Google Scholar]
- 11.Howlett C.R., McCartney E., Ching W. The effect of silicon nitride ceramic on rabbit skeletal cells and tissue. Clin. Orthop. Relat. Res. 1989;244:293–304. [PubMed] [Google Scholar]
- 12.Pezzotti G., Asai T., Adachi T., Ohgitani E., Yamamoto T., Kanamura N., Boschetto F., Zhu W., Zanocco M., Marin E., Bal B.S., McEntire B.J., Makimura K., Mazda O., Nishimura I. Antifungal activity of polymethyl methacrylate/Si3N4 composites against Candida albicans. Acta Biomater. 2021;126:259–276. doi: 10.1016/j.actbio.2021.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Pezzotti G., Fujita Y., Boschetto F., Zhu W., Marin E., Vandelle E., McEntire B.J., Bal S.B., Giarola M., Makimura K., Polverari A. Activity and mechanism of action of the bioceramic silicon nitride as an environmentally friendly alternative for the control of the grapevine downy mildew pathogen Plasmopara viticola. Front. Microbiol. 2020;11:610211. doi: 10.3389/fmicb.2020.610211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pezzotti G., Bock R.M., McEntire B.J., Adachi T., Marin E., Boschetto F., Zhu W., Mazda O., Bal S.B. In vitro antibacterial activity of oxide and non-oxide bioceramics for arthroplastic devices: I. In situ time-lapse Raman spectroscopy. Analyst. 2018;15:3708–3721. doi: 10.1039/c8an00233a. [DOI] [PubMed] [Google Scholar]
- 15.Decrey L., Kazama S., Kohn T. Ammonia as an in situ sanitizer: influence of virus genome type on inactivation. Appl. Environ. Microbiol. 2016;82:4909–4929. doi: 10.1128/AEM.01106-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pezzotti G. Raman spectroscopy in cell biology and microbiology. J. Raman Spectrosc. 2021 doi: 10.1002/jrs.6204. [DOI] [Google Scholar]
- 18.Pezzotti G. Bioceramics are not bioinert. Mater. Today. 2017;20:395–398. [Google Scholar]
- 19.Podstawka E., Ozaki Y., Proniewicz L.M. Part II: surface-enhanced Raman spectroscopy investigation of methionine containing heterodipeptides adsorbed on colloidal silver. Appl. Spectrosc. 2004;58:581–590. doi: 10.1366/000370204774103417. [DOI] [PubMed] [Google Scholar]
- 20.Cao X., Fischer G. Conformational and infrared spectral studies of L-methionine and its N-deuterated isotopomer as isolated zwitterions. J. Phys. Chem. 2002;106:41–50. [Google Scholar]
- 21.Hernandez B., Pfluger F., Adenier A., Kruglik S.K., Ghomi M. Side chain flexibility and protonation states of sulfur atom containing amino acids. Phys. Chem. Chem. Phys. 2011;13:17284–17294. doi: 10.1039/c1cp21054h. [DOI] [PubMed] [Google Scholar]
- 22.Hildebrandt P.G., Copeland R.A., Spiro T.G., Otlewski J., Laskowski M., Jr., Prendergast F.G. Tyrosine hydrogen-bonding and environmental effects in proteins probed by ultraviolet resonance Raman spectroscopy. Biochem. 1988;27:5426–5433. doi: 10.1021/bi00415a007. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez B., Coic Y.-M., Pluger F., Kruglik S.G., Ghomi M. All characteristic Raman markers of tyrosine and tyrosinate originate from phenol ring fundamental vibrations. J. Raman Spectrosc. 2016;47:210–220. [Google Scholar]
- 24.Sanchez Cortes S., Garcia-Ramos J.V. SERS of cytosine and its methylated derivatives on metal colloids. J. Raman Spectrosc. 1992;23:61–66. [Google Scholar]
- 25.Mathlouthi M., Seuvre A.M., Koenig J.L. F.T.-I.R. and laser-Raman spectra of cytosine and cytidine. Carbohydr. Res. 1986;146:1–13. doi: 10.1016/0008-6215(86)85019-4. [DOI] [PubMed] [Google Scholar]
- 26.Madzharova F., Heiner Z., Guehlke M., Kneipp J. Surface-enhanced hyper-Raman spectra of adenine, guanine, cytosine, thymine, and uracil. J. Phys. Chem. C. 2016;120:15415–15423. doi: 10.1021/acs.jpcc.6b02753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes R.P., Marques M.P.M., Valero R., Tomkinson J., Batista de Carvalho L.A.E. Guanine: a combined study using vibrational spectroscopy and theoretical methods. Spectrosc. Int. J. 2012;27:273–292. [Google Scholar]
- 28.Lopes R.P., Valero R., Tomkinson J., Marques M.P.M., Batista de Carvalho L.A.E. Applying vibrational spectroscopy to the study of nucleobases – adenine as a case study. New J. Chem. 2013;37:2691–2699. [Google Scholar]
- 29.Davies M.J. The oxidative environment and protein damage. Biochim. Biophys. Acta - Proteins and Proteomics. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Schoneich C. Methionine oxidation by reactive oxygen species: reaction mechanisms and relevance to Alzheimer's disease. Biochim. Biophys. Acta - Protein Proteomics. 2005;1703:111–119. doi: 10.1016/j.bbapap.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Lord R.C., Yu N.-t. Laser-excited Raman spectroscopy of biomolecules. I. Native lysozyme and its constituent amino acids. J. Mol. Biol. 1970;50:509–524. doi: 10.1016/0022-2836(70)90208-1. [DOI] [PubMed] [Google Scholar]
- 32.Torreggiani A., Barata-Vallejo S., Chatgilialoglu C. Combined Raman and IR spectroscopic study on the radical-based modifications of methionine. Anal. Bioanal. Chem. 2011;401:1231–1239. doi: 10.1007/s00216-011-5203-0. [DOI] [PubMed] [Google Scholar]
- 33.Schliess F., Gorg B., Fisher R., Desjardins D., Bidmon H.J., Herrmann A., Butterworth R.F., Zilles K., Haussinger D. Ammonia induces MK-801-sensitive nitration and phosphorylation of protein tyrosine residues in rat astrocytes. FASEB J. 2002;16:739–741. doi: 10.1096/fj.01-0862fje. [DOI] [PubMed] [Google Scholar]
- 34.Auron A., Brophy P.D. Hyperammonemia in review: pathophysiology, diagnosis, and treatment. Pediatr. Nephrol. 2012;27:207–222. doi: 10.1007/s00467-011-1838-5. [DOI] [PubMed] [Google Scholar]
- 35.Niederhafner P., Safarik M., Neburkova J., Keiderling T.A., Bour P., Sebestik J. Monitoring peptide tyrosine nitration by spectroscopic methods. Amino Acids. 2021;53:517–532. doi: 10.1007/s00726-020-02911-7. [DOI] [PubMed] [Google Scholar]
- 36.Tuma R. Raman spectroscopy of proteins: from peptides to large assemblies. J. Raman Spectrosc. 2005;36:307–319. [Google Scholar]
- 37.Rygula A., Majzner K., Marzec K.M., Kaczor A., Pilarczyk M., Baranska M. Raman spectroscopy of proteins: a review. J. Raman Spectrosc. 2013;44:1061–1076. [Google Scholar]
- 38.Radi R. Protein tyrosine nitration: biochemical mechanisms and structural basis of its functional effects. Acc. Chem. Res. 2013;46:550–559. doi: 10.1021/ar300234c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notingher I., Green C., Dyer C. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J. Royal Soc. Interface. 2004;1:79–90. doi: 10.1098/rsif.2004.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides. Nucl. Acid Res. 2003;31:517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douki T., Cadet J. Peroxynitrite mediated oxidation of purine bases in nucleosides and DNA. Free Rad. Res. Commun. 1996;24:369–380. doi: 10.3109/10715769609088035. [DOI] [PubMed] [Google Scholar]
- 42.Boorstein R.J., Hilbert T.P., Cadet J., Cunningham R.P., Teebor G.W. UV-induced pyrimidine hydrates in DNA are repaired by bacterial and mammalian DNA glycosylase activities. Biochemistry. 1989;28:6164–6170. doi: 10.1021/bi00441a007. [DOI] [PubMed] [Google Scholar]
- 43.Cadet J., Vigny P. In: Morrison H., editor. vol. 1. Wiley and Sons; New York: 1990. The photochemistry of nucleic acids; pp. 1–272. (Bioorganic Photochemistry: Photochemistry and the Nucleic Acids). [Google Scholar]
- 44.Cadet J., Douki T., Ravanat J.-L. Oxidatively generated damage to cellular DNA by UVB and UVA radiation. Photochem. Photobiol. 2015;91:140–155. doi: 10.1111/php.12368. [DOI] [PubMed] [Google Scholar]
- 45.Nonoyama N., Oshima H., Shoda C., Suzuki H. The reaction of peroxynitrite with organic molecules bearing a biologically important functionality. The multiplicity of reaction modes as exemplified by hydroxylation, nitration, nitrosation, dealkylation, oxygenation, and oxidative dimerization and cleavage. Bull. Chem. Soc. Jpn. 2001;74:2385–2395. [Google Scholar]
- 46.Shugar D., Fox J.J. Spectrophotometric studies of nucleic acid derivatives and related compounds as a function of pH. I. Pyrimidines. Biochim. Biophys. Acta. 1952;9:199–218. doi: 10.1016/0006-3002(52)90147-9. [DOI] [PubMed] [Google Scholar]
- 47.Lang J., Strum J., Zana R. Ultrasonic absorption in aqueous solutions of nucleotides and nucleosides. I. Effect of pH and concentration. J. Phys. Chem. 1973;77:2329–2334. doi: 10.1021/j100638a600. [DOI] [PubMed] [Google Scholar]
- 48.Brown D.J., Evans R.F., Cowden W.B., Fenn M.D. John Wiley & Sons; New York, USA: 1994. The Pyrimidine; pp. 96–106. [Google Scholar]
- 49.McAvan B.S., Bowsher L.A., Powell T., O'Hara J.F., Spitali M., Goodacre R., Doig A.J. Raman spectroscopy to monitor post-translational modifications and degradation in monoclonal antibody therapeutics. Anal. Chem. 2020;92:10381–10389. doi: 10.1021/acs.analchem.0c00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colombo G., Meli M., Morra G., Gabizon R., Gasset M. Methionine sulfoxides on prion protein helix-3 switch on the α-fold destabilization required for conversion. PloS ONE. 2009;4 doi: 10.1371/journal.pone.0004296. e4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogt W. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic. Biol. Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 54.Perrin D., Koppenol W.H. The quantitative oxidation of methionine to methionine sulfoxide by peroxynitrite. Arch. Biochem. Biophys. 2000;377:266–272. doi: 10.1006/abbi.2000.1787. [DOI] [PubMed] [Google Scholar]
- 55.Dado G.P., Weissbach H. Redox control of secondary structure in a designed peptide. J. Am. Chem. Soc. 1993;115:12609–12610. [Google Scholar]
- 56.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 57.Arp D.J., Stein L.Y. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Critical Rev. Biochem. Mol. Biol. 2003;38:471–495. doi: 10.1080/10409230390267446. [DOI] [PubMed] [Google Scholar]
- 58.Squadrito G.L., Pryor W.A. The formation of peroxynitrite in vivo from nitric oxide and superoxide. Chemico-Biol. Interactions. 1995;96:203–206. doi: 10.1016/0009-2797(94)03591-u. [DOI] [PubMed] [Google Scholar]
- 59.Fang F.C. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beckman J.S., Ye Y.Z., Anderson P., Chen J., Accavitti M.A., Tarpey M.M., White C.R. Extensive nitration of protein tyrosines in human atherosclerosis detected by immunohistochemistry. Biol. Chem. Hoppe-Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 61.Van Der Vliet A., O'Neill C.A., Halliwell B., Cross C.E., Kaur H. Aromatic hydroxylation and nitration of phenylalanine and tyrosine by peroxynitrite. Evidence for hydroxyl radical production from peroxynitrite. FEBS Lett. 1994;339:89–92. doi: 10.1016/0014-5793(94)80391-9. [DOI] [PubMed] [Google Scholar]
- 62.Alvarez B., Ferrer-Sueta G., Freeman B.A., Radi R. Kinetics of peroxynitrite reaction with amino acids and human serum albumin. J. Biol. Chem. 1999;274:842–848. doi: 10.1074/jbc.274.2.842. [DOI] [PubMed] [Google Scholar]
- 63.Cromeans T.L., Kahler A.M., Hill V.R. Inactivation of adenoviruses, enteroviruses, and murine norovirus in water by free chlorine and monochloramine. Appl. Environ. Microbiol. 2010;76:1028–1033. doi: 10.1128/AEM.01342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emmoth E., Ottoson J., Albihn A., Belak S., Vinneras B. Ammonia disinfection of hatchery waste for elimination of single-stranded RNA viruses. Appl. Environ. Microbiol. 2011;77:3960–3966. doi: 10.1128/AEM.02990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jenkins M.B., Bowman D.D., Ghiorse Wc W.C. Inactivation of Cryptosporidium parvum oocysts by ammonia. Appl. Environ. Microbiol. 1998;64:784–788. doi: 10.1128/aem.64.2.784-788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fidjeland J., Nordin A., Pecson B.M., Nelson K.L., Vinnerås B. Modeling the inactivation of Ascaris eggs as a function of ammonia con- centration and temperature. Water Res. 2015;83:153–160. doi: 10.1016/j.watres.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 67.Ward R.L., Ashley C.S. Identification of virucidal agent in waste- water sludge. Appl. Environ. Microbiol. 1977;33:860–864. doi: 10.1128/aem.33.4.860-864.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2010;109:388–397. doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- 69.Joonaki E., Hassanpouryouzband A., Heldt C.L., Areo O. Surface chemistry can unlock drivers of surface stability of SARS-CoV-2 in a variety of environmental conditions. Chem. 2020;6:2135–2146. doi: 10.1016/j.chempr.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samandoulgou I., Fliss I., Jean J. Zeta potential and aggregation of virus-like particle of human Norovirus and feline calicivirus under different physicochemical conditions. Food Environ. Virol. 2015;7:249–260. doi: 10.1007/s12560-015-9198-0. [DOI] [PubMed] [Google Scholar]
- 71.Bock R.M., McEntire B.J., Bal B.S., Rahaman M.N., Boffelli M., Pezzotti G. Surface modulation of silicon nitride ceramics for orthopaedic applications. Acta Biomater. 2015;26:318–330. doi: 10.1016/j.actbio.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 72.Influenza research database (file EF541421) https://www.fludb.org/brc/fluSegmentDetails.spg?ncbiGenomicAccession=EF541421&decorator=influenza.
- 73.Schein C.H., Ye M., Paul A.V., Oberste M.S., Chapman N., van der Heden van Noort G.J., Filippov D.V., Choi K.H. Sequence specificity for uridylylation of the viral peptide linked to the genome (VPg) of enteroviruses. Virol. 2015;484:80–85. doi: 10.1016/j.virol.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scheller C., Krebs F., Minkner R., Astner I., Gil-Moles M., Watzig H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation, and attenuation, vaccine formulation and quality control. Electrophoresis. 2020;41:1137–1151. doi: 10.1002/elps.202000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Talebian S., Wallace G.G., Schroeder A., Stellacci F., Conde J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020;15:618–624. doi: 10.1038/s41565-020-0751-0. [DOI] [PubMed] [Google Scholar]
- 76.Wang J., Shen J., Ye D., Yan X., Zhang Y., Yang W., Li X., Wang J., Zhang L., Pan L. Disinfection technology of hospital wastes and wastewater: suggestions for disinfection strategy during coronavirus Disease 2019 (COVID-19) pandemic in China. Environ. Pollut. 2020;262:114665. doi: 10.1016/j.envpol.2020.114665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sportelli M.C., Izzi M., Kukushkina E.A., Hossain S.I., Picca R.A., Ditaranto N., Cioffi N. Can nanotechnology and materials science help the fight against SARS-CoV-2? Nanomaterials. 2020;10:802. doi: 10.3390/nano10040802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palmieri V., Pap M. Can graphene take part in the fight against COVID-19? Nano Today. 2020;33:100883. doi: 10.1016/j.nantod.2020.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pogribna M., Hammons G. Epigenetic effects of nanomaterials and nanoparticles. J. Nanobiotechnol. 2021;19:2. doi: 10.1186/s12951-020-00740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jamuna B.A., Ravishankar R.V. In: Nanomaterials for Environmental Protection. Kharisov B.I., Kharissova O.V., Rasika Dias H.V., editors. John Wiley & Sons; Hoboken, NJ, USA: 2014. Environmental risk, human health, and toxic effects of nanoparticles; pp. 523–535. [Google Scholar]
- 81.Rai M., Deshmukh S.D., Ingle A.P., Gupta I.R., Galdiero M., Galdiero S. Metal nanoparticles: the protective nanoshield against virus infection. Critical Rev. Microbiol. 2016;42:46–56. doi: 10.3109/1040841X.2013.879849. [DOI] [PubMed] [Google Scholar]
- 82.Sportelli M.C., Izzi M., Kukushkina E.A., Hossain S.I., Picca R.A., Ditaranto N., Cioffi N. Can nanotechnology and materials science help the fight against SARS-CoV-2? Nanomater. 2020;10:802. doi: 10.3390/nano10040802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balasubramaniam B., Prateek, Ranjan S., Saraf M., Kar P., Singh S.P., Thakur V.K., Singh A., Gupta R.K. Antibacterial and antiviral functional materials: chemistry and biological activity toward tackling COVID-19-like pandemics. ACS Pharmacol. Transl. Sci. 2021;4:8–54. doi: 10.1021/acsptsci.0c00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji X., Ge L., Liu C., Tang Z., Xiao Y., Chen W., Lei Z., Gao W., Blake S., De D., Shi B., Zeng X., Kong N., Zhang X., Tao W. Capturing functional two-dimensional nanosheets from sandwich-structure vermiculite for cancer theranostics. Nat. Commun. 2021;12:1124. doi: 10.1038/s41467-021-21436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan S.Y., Grimes S., Ehrlich Paul. (1854-1915): man with the magic bullet. Singapore Med. J. 2010;51:842–843. [PubMed] [Google Scholar]
- 86.Ball H.T., McEntire B.J., Bal B.S. Accelerated cervical fusion of silicon nitride versus PEEK spacers: a comparative clinical study. J. Spine. 2017;6:396. [Google Scholar]
- 87.Pezzotti G., Marin E., Adachi T., Rondinella A., Boschetto F., Zhu W., Sugano N., Bock R.M., McEntire B., Bal S.B. Bioactive silicon nitride: a new therapeutic material for osteoarthropathy. Sci. Rep. 2017;7:44848. doi: 10.1038/srep44848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pezzotti G., Adachi T., Boschetto F., Zhu W., Zanocco M., Marin E., Bal B.S., McEntire B.J. Off-stoichiometric reactions at the cell-substrate biomolecular interface of biomaterials: in situ and ex situ monitoring of cell proliferation, differentiation, and bone tissue formation. Int. J. Mol. Sci. 2019;20:4080. doi: 10.3390/ijms20174080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pezzotti G., Bock R.M., Adachi T., Rondinella A., Boschetto F., Zhu W., Marin E., McEntire B., Bal B.S., Mazda O. Silicon nitride surface chemistry: a potent regulator of mesenchymal progenitor cell activity in bone formation. Appl. Mater. Today. 2017;9:82–95. [Google Scholar]
- 90.Y. Gong, Y. Honda, T. Adachi, E. Marin, K. Yoshikawa, G. Pezzotti, K. Yamamoto, Optimization of Silicon Nitride Surface Chemistry for Facilitating Osteogenic Differentiation of Rat Dental Pulp Cells, (manuscript in preparation). [DOI] [PMC free article] [PubMed]
- 91.Lal S., Caseley E.A., Hall R.M., Tipper J.L. Biological impact of silicon nitride for orthopaedic applications: role of particle size, surface composition and donor variation. Sci. Rep. 2018;8:9109. doi: 10.1038/s41598-018-27494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang Z., Kong N., Zhang X., Liu Y., Hu P., Mou S., Liljestroem P., Shi J., Tan W., Kim J.S., Cao Y., Langer R., Leong K.W., Farokhzad O.C., Tao W. A materials-science perspective on tackling COVID-19. Nat. Rev. Mater. 2020;5:847–860. doi: 10.1038/s41578-020-00247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fukumoto S., Hookabe T., Tsubakino H. Hydrolysis behavior of aluminum nitride in various solutions. J. Mater. Sci. 2000;35:2743–2748. [Google Scholar]
- 94.Bowen P., Highfield J.G., Mocellin A., Ring T.A. Degradation of aluminum nitride powder in an aqueous environment. J. Am. Ceram. Soc. 1990;73:724–728. [Google Scholar]
- 95.Highfield J.G., Bowen P. Diffuse-reflectance Fourier-transform infrared spectroscopic studies of the stability of aluminum nitride powder in an aqueous environment. Anal. Chem. 1989;61:2399–2402. [Google Scholar]
- 96.Alexander G.B., Heston W.M., Iler R.K. The solubility of amorphous silica in water. J. Phys. Chem. 1954;58:453–455. [Google Scholar]
- 97.Goel S., Hawi S., Goel G., Thakur V.K., Agrawal A., Hoskins C., Pearce O., Hussain T., Upadhyaya H.M., Cross G., Barber A.H. Resilient and agile engineering solutions to address societal challenges such as coronavirus pandemic. Mater. Today Chem. 2020;17:100300. doi: 10.1016/j.mtchem.2020.100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sano T., Tsutsui S., Koike K., Hirakawa T., Teramoto Y., Negishi N., Takeuchi K. Activation of graphitic carbon nitride (g-C3N4) by alkaline hydrothermal treatment for photocatalytic NO oxidation in gas phase. J. Mater. Chem. A. 2013;1:6489–6496. [Google Scholar]
- 99.Leng K., Mai W., Zhang X., Liu R., Lin X., Huang J., Lou H., Xie Y., Fu R., Wu D. Construction of functional nanonetwork-structured carbon nitride with Au nanoparticle yolks for highly efficient photocatalytic applications. Chem. Commun. 2018;52:7159–7162. doi: 10.1039/c8cc03095b. [DOI] [PubMed] [Google Scholar]
- 100.Gao D., Guo X., Zhang X., Chen S., Wang Y., Chen T., Huang G., Gao Y., Tian Z., Yang Z. Multifunctional phototheranostic nanomedicine for cancer imaging and treatment. Mater. Today Bio. 2020;5:100035. doi: 10.1016/j.mtbio.2019.100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Y., Wei X., Zhang N., Zheng J., Chen X., Wen Q., Luo X., Lee C.-Y., Liu X., Zhang X., Chen J., Tao C., Zhang W., Fan X. A non-printed integrated-circuit textile for wireless theranostics. Nat. Commun. 2021;12:4876. doi: 10.1038/s41467-021-25075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tang Z., Zhang X., Shu Y., Guo M., Zhang H., Tao W. Insights from nanotechnology in COVID-19 treatment. Nano Today. 2021;36:101019. doi: 10.1016/j.nantod.2020.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.