Abstract
Auxin is a crucial growth regulator that governs plant development and responses to environmental perturbations. It functions at the heart of many developmental processes, from embryogenesis to organ senescence, and is key to plant interactions with the environment, including responses to biotic and abiotic stimuli. As remarkable as auxin is, it does not act alone, but rather solicits the help of, or is solicited by, other endogenous signals, including the plant hormones abscisic acid, brassinosteroids, cytokinins, ethylene, gibberellic acid, jasmonates, salicylic acid, and strigolactones. The interactions between auxin and other hormones occur at multiple levels: hormones regulate one another's synthesis, transport, and/or response; hormone-specific transcriptional regulators for different pathways physically interact and/or converge on common target genes; etc. However, our understanding of this crosstalk is still fragmentary, with only a few pieces of the gigantic puzzle firmly established. In this review, we provide a glimpse into the complexity of hormone interactions that involve auxin, underscoring how patchy our current understanding is.
The plant hormone auxin is an essential growth regulator central to a wide variety of developmental processes, environmental adaptation, and phenotypic plasticity (for review, see Enders and Strader 2015 and Lavy and Estelle 2016). The name auxin comes from the Greek word “auxein,” meaning “to grow.” The best-studied form of auxin, indole-3-acetic acid (IAA), is synthesized from the amino acid tryptophan (Trp) through a simple, two-step pathway. Trp aminotransferases of the Trp AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1)/TAA1-RELATED (TAR) family convert Trp to indole-3-pyruvic acid (IPyA), which is then metabolized to IAA by flavin-containing monooxygenases, YUCCAs (YUCs). The availability of biologically active IAA is controlled by auxin-catabolizing enzymes of the DIOXYGENASE OF AUXIN OXIDATION (DAO) family that oxidize IAA to 2-oxindole-3-acetic acid, auxin-conjugating enzymes such as the IAA amidosynthetases GRETCHEN HAGEN3 (GH3) and glucosyltransferases that inactivate auxin by linking it to amino acids or sugars, respectively, and by auxin transporters that move auxin in and out of the cell and between cells (for review, see Enders and Strader 2015). Free IAA can enter plant cells passively or be actively imported via AUXIN1 (AUX1)/LIKE AUX1 (LAX) influx carriers. IAA can also exit cells via efflux carriers of the PIN-FORMED (PIN) and P-GLYCOPROTEIN/ATP-BINDING CASETTE transporter families. Auxin perception takes place inside the cell, predominantly in the nucleus, where IAA binds to the TRANSPORT INHIBITOR-RESISTANT1 (TIR1)/AUXIN SIGNALING F-BOX (AFB) family of auxin receptors and promotes their interaction with the auxin coreceptors, Aux/IAAs. In the absence of the hormone, Aux/IAAs associate with AUXIN RESPONSE FACTOR (ARF) transcription factors (TFs) and block their transcriptional activity via the recruitment of TOPLESS (TPL) and chromatin-remodeling machinery. In the presence of IAA, Aux/IAAs undergo ubiquitin-mediated proteasomal degradation triggered by the SKP-CULLIN-F-BOX (SCFTIR) E3-ligase complex, releasing ARFs and enabling ARF-mediated transcriptional regulation of auxin-response genes (for review, see Enders and Strader 2015 and Lavy and Estelle 2016).
While auxin governs multiple aspects of plant development, physiology, and environmental competence, it does not act in isolation. In every process where the contributions of auxin have been explored, it appears to enlist or be enlisted by other endogenous signals and external cues, enabling the plant to tailor its growth and development to the specific conditions it happens to be in. This article aims to present the current state of knowledge in the area of auxin interactions with other plant hormones, specifically abscisic acid (ABA), brassinosteroid (BR), cytokinin (CK), ethylene (ET), gibberellic acid (GA), jasmonate (JA), salicylic acid (SA), and strigolactone (SL). We chose to structure this manuscript by the pairs of plant hormones, with the caveat that in many processes multiple players are involved and we are only beginning to untangle the full complexity of signal crosstalk in plants. We have included graphical representations of key auxin interactions with other hormones during the processes of seed germination, root, shoot, and fruit development in Figures 1–4, respectively. Given the breadth of the auxin interaction network, we were unable to discuss all relevant studies and wish to apologize to those researchers whose work we could not describe in light of space limitations in this article.
Figure 1.
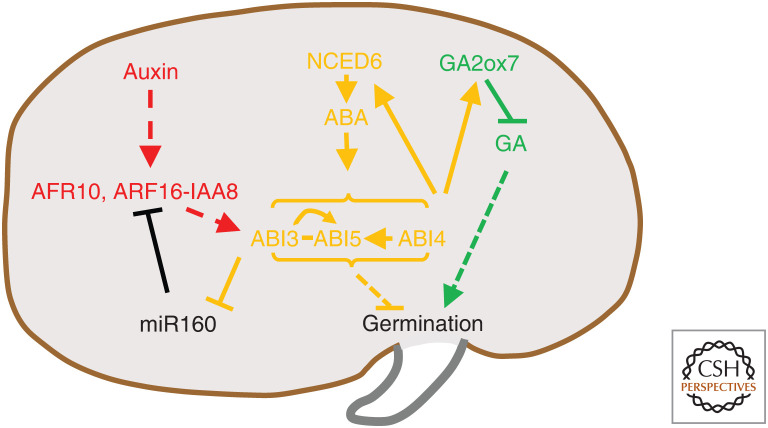
Auxin interactions with abscisic acid (ABA) in the control of seed germination. Seed dormancy is primarily controlled by the balance between ABA and gibberellic acid (GA), which oppose one another in the regulation of seed germination: ABA inhibits, whereas GA promotes seed sprouting. Auxin, acting through IAA8 and ARF10/ARF16, works to enhance seed dormancy by inducing ABI3 expression. ABI3, in turn, down-regulates miR160, an miRNA that inhibits ARF10/16, enabling feedback regulation of ARF activity by ABI3. Transcription factors (TFs) ABI3, ABI4, and ABI5 all contribute to blocking seed germination. ABI4 regulates ABA production and GA metabolism by inducing an ABA biosynthesis gene, NCED6, and a GA catabolism gene, GA2ox7. Additional interactions that take place between ABA and GA, but do not involve auxin, are omitted in this schematic. Arrowheads represent positive regulation; blunt arrows represent negative regulation; solid and interrupted lines represent direct and indirect regulation, respectively; dashes between proteins represent direct interaction. Colors represent auxin (red), ABA (orange), and GA (green) pathway components. Black is used to depict genes/proteins that do not belong to a specific hormone pathway, as well as the developmental processes the network regulates.
Figure 2.
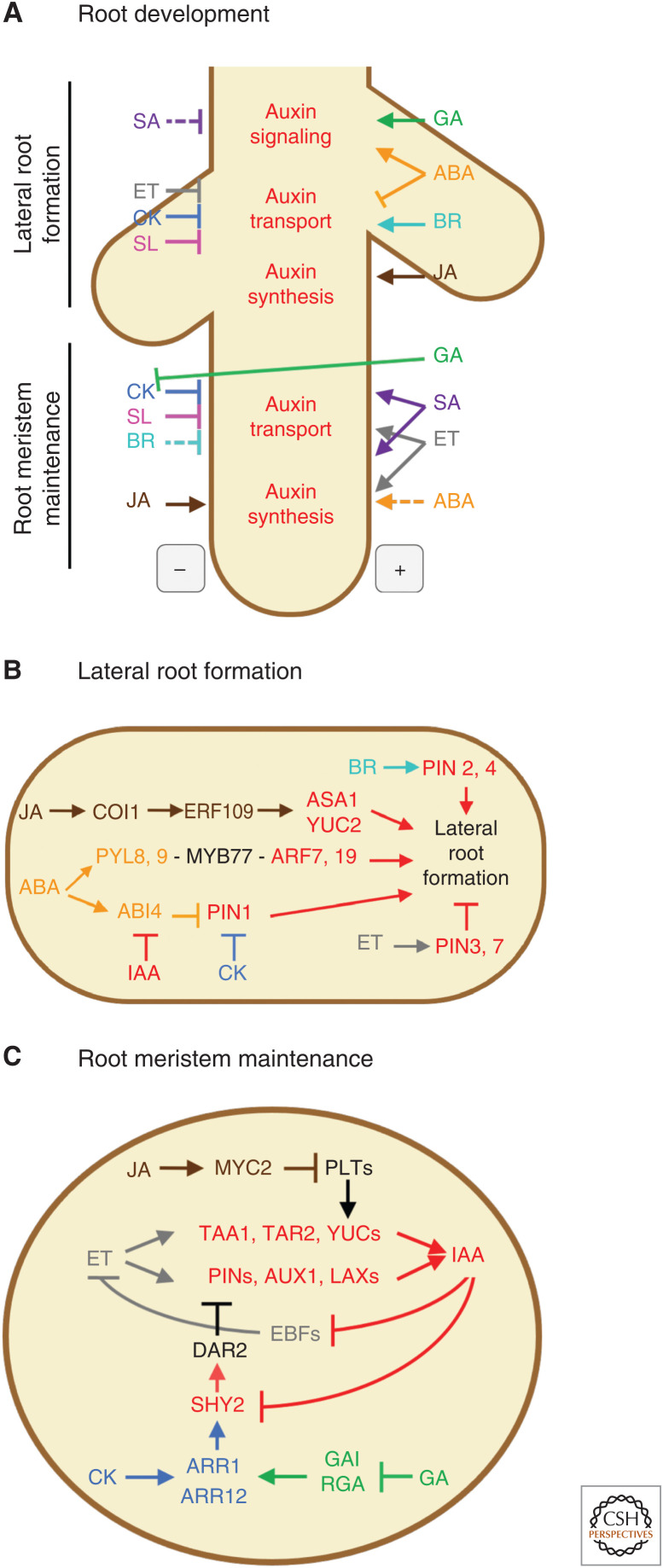
Auxin interactions with other hormones in roots. (A) Hormone interactions regulating auxin biosynthesis, transport, and signaling during lateral root (LR) formation and root meristem maintenance. The minus sign signifies repression of the root processes by the hormones listed on the left side of the root. The plus sign signifies promotion of the root processes by the hormones listed on the right side of the root. (B) Molecular network of the hormone interactions mediating LR formation. Abscisic acid (ABA) induces LR growth through PYL8. The interaction of PYL8, PYL9, with MYB77 promotes the crosstalk with the auxin signaling pathway via ARF7 and ARF19. ABA up-regulates the expression of ABI4, which represses the expression of PIN1 to modify auxin transport. The negative effect of this network on LR formation can be reinforced by the cytokinin (CK)-mediated repression of PIN1 or weakened by auxin-mediated inhibition of ABI4. Ethylene induces the expression of PIN3 and PIN7, which reduce the local accumulation of auxin in the LR initiation sites, and thereby decrease LR formation. Jasmonate (JA) boosts the formation of LRs by inducing the expression of ERF109, which promotes auxin biosynthesis by up-regulating the expression of ASA1 and YUC2. Brassinosteroid (BR) has a positive effect on the formation of LRs by inducing the expression of PIN2 and PIN4. (C) Molecular network of hormone interactions in root meristem maintenance. Ethylene (ET) induces auxin transport and biosynthesis, promoting the accumulation of auxin and enabling root meristem maintenance. Auxin dampens the negative effect of the EBFs on ET, reinforcing meristem function. CK and gibberellic acid (GA) have opposite effects on meristem maintenance by increasing and decreasing the activity of ARR1/ARR12 CK signaling components, respectively, which in turn represses the expression of PINs through SHY2 and DAR2. The JA-mediated repression of PLTs reduces the expression of the YUCs and lowers the production of auxin, thus negatively affecting root meristem maintenance. Arrowheads and blunt arrows represent positive and negative regulation, respectively. Solid and interrupted lines in panel A are used to depict direct and indirect effects, respectively. A dash between two proteins in panel B indicates direct physical interaction. Hormones, protein/genes in these hormonal pathways, and their actions are denoted by the following color pallet, ABA is orange, BR is turquoise, CK is blue, ET is gray, GA is green, IAA is red, JA is brown, salicylic acid (SA) is purple, and strigolactone (SL) is pink. Black is used to depict genes/proteins that do not belong to a specific hormone pathway, as well as the developmental processes the network regulates.
Figure 3.
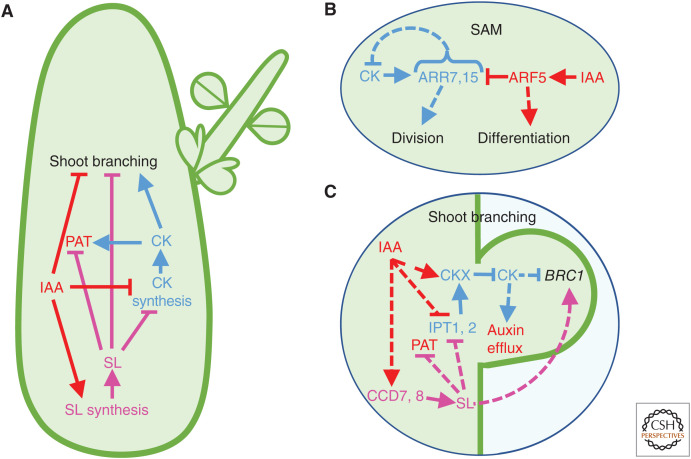
Auxin interactions with other hormones during shoot development. (A) Auxin and cytokinin (CK) oppose one another in the regulation of shoot branching. Strigolactone (SL) acts as a second messenger for auxin signaling: auxin from the shoot promotes SL production in the root and shoot, which then acts to inhibit branching. (B) Auxin acts to promote differentiation in the shoot apical meristem while CK promotes stem cell division. (C) During branching, shoot-derived auxin travels down the main stem where it promotes SL biosynthesis. This SL then travels up the shoot and inhibits CK production and polar auxin transport (PAT) in the shoot, thereby blocking branching. Auxin regulates local metabolism of CK, reducing CK levels, while CK promotes auxin efflux from the developing bud. CK signaling in the bud promotes bud release and branch outgrowth. CK and SL also oppose one another in regulating the expression of BRC1, a negative regulator of bud outgrowth. Red denotes auxin and its actions; blue denotes CK and its actions; magenta denotes SL and its actions. Arrowheads represent positive regulation; blunt arrows represent negative regulation; in panels B and C, dashed lines represent indirect regulation.
Figure 4.
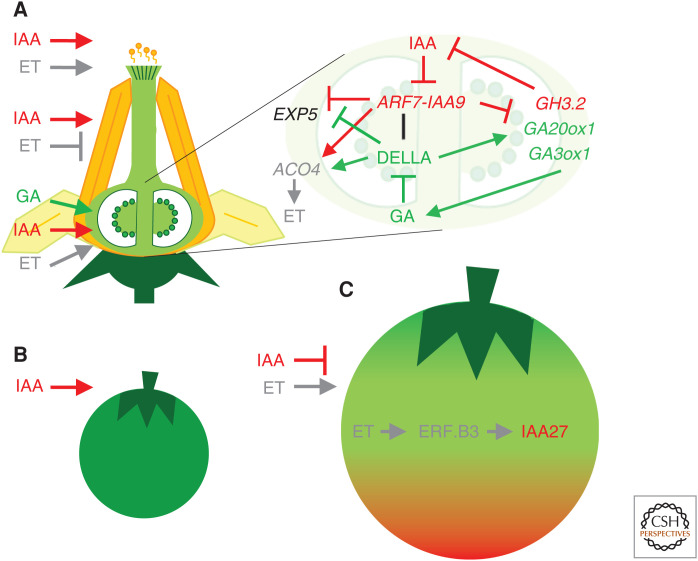
Auxin interactions with other hormones in fruit development. (A) Hormone interactions in flower development and fruit set. Auxin and ethylene (ET) promote pollen germination and pollen tube growth. Auxin induces and ET represses stamen development. ET positively regulates pistil and ovule development. Auxin and gibberellic acid (GA) promote fruit initiation. (Inset) Molecular network of the hormone crosstalk during tomato fruit initiation. Fertilization triggers auxin-mediated GA synthesis. Auxin inhibits the ARF7-IAA9 complex, releasing the repression of key GA biosynthetic genes. GA–auxin interaction promotes fruit growth by inducing EXP5 and reduces the production of ET by repressing ACO4. (B) Auxin regulates early cell division and fruit development phases. (C) Fruit ripening is promoted by ethylene and repressed by auxin. During ripening, a key molecular interaction between ET and IAA is mediated by ERF.B3 and IAA27. Hormones and their actions are denoted by the following colors: ET is gray, GA is green, and IAA is red. Arrowheads and blunt arrows represent positive and negative regulation.
AUXIN–ABA INTERACTIONS
ABA, originally named “abscisin II” for its role in the abscission of cotton fruits (Addicott et al. 1968), is a sequisterpene that belongs to the terpenoid class of metabolites. It is primarily synthesized in the vasculature and guard cells and transported by ABA transporters (for review, see Emenecker and Strader 2020). ABA is perceived by soluble PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABA RECEPTOR proteins, leading to the inhibition of PROTEIN PHOSPHATASE 2C (PP2C). Formation of the ABA-PYR-PP2C complex causes the accumulation of phosphorylated protein kinases in subclass III of the SNF1-RELATED PROTEIN KINASE2 (SnRK2) family, which phosphorylates various target proteins, including ABA-RESPONSIVE ELEMENT-BINDING FACTORs, to achieve the appropriate cellular response (for review, see Emenecker and Strader 2020).
Seed Dormancy and Germination
ABA is the major hormone involved in the establishment and maintenance of seed dormancy (Fig. 1; for review, see Bentsink and Koornneef 2008). Among ABA signaling components in Arabidopsis, TFs ABA-INSENSITIVE3 (ABI3), ABI4, and ABI5 were identified as positive regulators of ABA signaling and negative regulators of seed germination (for review, see Emenecker and Strader 2020).
The B3-domain TF ABI3 is transcriptionally induced by ABA, and disruption of this gene's function reduces seed dormancy and allows germination in the presence of exogenous ABA (Koornneef et al. 1984). Remarkably, ABI3 transcription is also induced by exogenous IAA and positively regulated by ARF10 and ARF16 (Liu et al. 2013b), which are both believed to function as transcriptional repressors (Fig. 1; Wang et al. 2005b). Even though a potential auxin response element (AuxRE) is present in the ABI3 promoter, neither ARF10 nor ARF16 directly bind the ABI3 promoter, suggesting indirect action of these TFs by repressing a repressor of ABI3 (Liu et al. 2013b). In addition, a loss-of-function mutant of IAA8, iaa8-1, shows delayed seed germination, and the IAA8 protein can associate with an AuxRE within the ABI3 promoter, suggesting that IAA8 may bind to the ABI3 promoter via yet unidentified ARFs to regulate seed germination (Hussain et al. 2020). Previous studies revealed the ability of ARFs, including ARF16, to interact with IAA8 (Piya et al. 2014), but it remains to be determined which specific ARF-Aux/IAA combinations bind the ABI3 promoter to regulate its transcription. Interestingly, an older study discovered that ABI3 directly represses the transcription of miR160B, an miRNA that targets ARF10 and ARF16, and thus up-regulates ABI3 through a potentially complex feedback loop (Fig. 1; Tian et al. 2004).
ABI4, an APETALA2 (AP2)-domain TF, controls various developmental processes (for review, see Chandrasekaran et al. 2020). It regulates seed dormancy by directly activating an ABA biosynthetic gene, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE6 (NCED6), and a GA catabolic gene, GA 2-BETA-DIOXYGENASE7 (GA2ox7) (Fig. 1; Shu et al. 2013, 2016). Furthermore, abi4 mutant seeds are insensitive to auxin-mediated inhibition of seed germination and show reduced dormancy (Rohde et al. 2000). The prospective role of ABI4 in mediating auxin effects on germination is further supported by a recent study that showed that YUC4-overexpressing plants display enhanced sensitivity to ABA during seed germination, whereas the same construct in the abi4 background leads to wild-type germination, indicating that ABI4 is required for the ABA hypersensitivity of YUC4-overexpressing lines during germination (Munguia-Rodriguez et al. 2020). Thus, ABI4 is another convergence point between ABA and auxin during inhibition of germination (Fig. 1).
Finally, ABI5, a bZIP TF, functions downstream of ABI3 and is also a substrate of activated SnRK2s (Fig. 1; Yu et al. 2015). It binds ABA-responsive element (ABRE)-containing promoters (Finkelstein and Lynch 2000; Hossain et al. 2010; Zhou et al. 2013). While the Arabidopsis abi5 mutant shows seed germination defects, rice plants with mutations in the ABI5-LIKE1 (OsABL1) gene, which is induced by both ABA and auxin, show normal seed germination but suppressed ABA-triggered root growth inhibition and hypersensitivity to exogenous auxin (Yang et al. 2011). In addition, the OsABL1 protein can directly bind ABREs in vitro and the expression levels of several ABRE-containing genes potentially related to auxin metabolism or signaling are altered in abl1 mutants, indicating that OsABL1 plays a role in the crosstalk between ABA and auxin (Yang et al. 2011).
Root Development
Primary root (PR) growth regulated by auxin depends on Aux/IAA-ARF signaling modules. ARF2 directly binds to AuxREs in the promoter of the zinc finger homeodomain TF HOMEOBOX PROTEIN33 (HB33) and negatively regulates its expression (Wang et al. 2011). Transgenic plants overexpressing HB33 or RNAi lines with reduced HB33 levels are more sensitive and more resistant to ABA respectively in the seed germination and PR growth assays, indicating that HB33 is a positive regulator in the ABA-mediated processes of seed germination and PR growth (Wang et al. 2011). Likewise, in wheat, TaARF4 targets TaHB33 and two TaGH3 genes to concomitantly regulate ABA sensitivity and IAA homeostasis to control root growth (Wang et al. 2019).
Auxin-ABA crosstalk is also involved in lateral root (LR) development (Fig. 2A,B). The ABA receptor mutant pyl8 shows reduced LR growth in the presence of exogenous ABA, indicating that ABA signaling through PYL8 promotes LR growth (Fig. 2B). When pyl8 seedlings are exposed to both ABA and IAA, LR growth is rescued, suggesting that pyl8 seedlings may have an auxin deficiency or reduced auxin response (Zhao et al. 2014). A previous study showed that PYL8 interacts with MYB77 (Arabidopsis Interactome Mapping Consortium 2011) that can promote LR growth by interacting with ARF7 in the SOLITARY ROOT (SLR, IAA14)-ARF7/ARF19 module (Fukaki et al. 2005; Shin et al. 2007). Furthermore, PYL8 is functionally redundant with paralogous PYL9 (Xing et al. 2016) and the two proteins interact with several MYBs, including MYB77, in Arabidopsis to integrate ABA and auxin signals in the regulation of LR growth (Fig. 2B; Zhao et al. 2014).
In addition to controlling LR growth through the PYL–MYB77 interaction, ABA also regulates LRs via the core ABA-SnRK2 signaling pathway (Fig. 2B; for review, see Emenecker and Strader 2020). abi4 mutants possess longer LRs, suggesting that ABI4 inhibits LR growth. ABI4 expression in roots is induced by ABA but repressed by auxin (Shkolnik-Inbar and Bar-Zvi 2010). Furthermore, PIN1 levels are decreased in ABI4-overexpression lines but increased in abi4 mutants. Thus, ABI4 likely mediates ABA-triggered inhibition of LR growth by suppressing PIN1 expression. Similarly, ABI5 also regulates root growth by modulating the accumulation of PIN proteins (Yuan et al. 2014).
Under osmotic/salt stress conditions, another TF, WRKY46, can bind the promoters of the auxin-conjugating enzyme-encoding genes UDP-GLYCOSYLTRANSFERASE 84B2, INDOLE-3-ACETATE BETA-GLUCOSYLTRANSFERASE, and GH3.1, and an ABA signaling gene ABI4. Importantly, loss-of-function wrky46 mutants and overexpression of WRKY46 significantly reduce and increase LR development, respectively, suggesting that WRKY46 modulates LR development through the coregulation of ABA signaling and auxin homeostasis (Ding et al. 2015).
Pathogen Resistance
Black spot disease, a major disease in oilseed Brassica species, is caused by a group of pathogens including Alternaria brassicae, Alternaria brassicicola, and Alternaria raphanin. Numerous ABA and auxin mutants in Arabidopsis show altered susceptibility to A. brassicicola, suggesting that both ABA and auxin are involved in the response against this pathogen (Adie et al. 2007; Qi et al. 2012). A recent study showed that the auxin response factors ARF10, ARF16, and ARF17 are up-regulated in the resistant species Sinapis alba upon challenge with A. brassicicola, but not in the susceptible species Brassica juncea (Mukherjee et al. 2019). Pathogen-induced expression of Arabidopsis ARF10 in B. juncea enhances tolerance to A. brassicicola, and several ABA-responsive genes, including ABI3, ABI4, and ABI5, are up-regulated in the most tolerant transgenic lines. Furthermore, ARF10 interacts with the AuxREs in the ABI5 promoter, suggesting that the binding of ARF10 to ABI5 modulates auxin-ABA crosstalk to regulate resistance to A. brassicicola (Mukherjee et al. 2019).
AUXIN–BR INTERACTIONS
BRs, so named because the first example was identified in rapeseed (Brassica napus), are the only class of steroid hormones found in plants to date. BRs are a group of many compounds that act as extracellular ligands. While there are multiple routes involved in BR production, all of them proceed through triterpenoid pathways from campesterol (for review, see Chung and Choe 2013). BRs are believed to be synthesized in the endoplasmic reticulum and DWARF4 (DWF4) catalyzes a rate-limiting step of BR synthesis (for review, see Planas-Riverola et al. 2019 and Nolan et al. 2020). Unlike other hormones that can be transported throughout the plant, BRs typically act locally. BRs bind to the plasma membrane-localized receptor, BR-INSENSITIVE1 (BRI1), inducing a conformational change that allows interaction with coreceptors, such as BRI1-ASSOCIATED KINASE1 (BAK1). This interaction initiates a signaling cascade that leads to the activation of two TFs, BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1 EMS SUPPRESSOR1 (BES1). In the presence of BR, BZR1 and BES1 are stabilized by a PP2A-dependent dephosphorylation and induce the transcriptional program of BR-dependent genes. In the absence of BR, signaling through the BRI1–BAK1 complex is blocked by autoinhibition and the association of inhibitory proteins, such as BRI1 KINASE INHIBITOR, which disrupts the BRI1–BAK1 interaction. BZR1 and BES1 are inactivated via phosphorylation by BR-INSENSITIVE2 (BIN2), leading to cytoplasmic retention and degradation of these TFs (for review, see Planas-Riverola et al. 2019 and Nolan et al. 2020).
BR-deficient plants have a dwarfed phenotype, consistent with the primary role of BRs in cell elongation. BR signaling has also been implicated in a broad range of plant biological processes, from control of cell division to biotic and abiotic stress responses (for review, see Lv and Li 2020 and Nolan et al. 2020) and, as outlined below, this phytohormone coordinates with auxin to regulate many aspects of plant development.
Root Development
BR signaling is critical for both cell elongation (Mouchel et al. 2004) and cell-cycle progression in the root (Gonzalez-Garcia et al. 2011; Hacham et al. 2011), with both BR-deficient and BR-activated mutants displaying smaller root meristems. In contrast to the synergy reported in shoot elongation, transcriptomic analysis of Arabidopsis root tips revealed that many auxin and BR coresponsive genes are regulated in opposing directions, suggesting these two hormones can act as checks on one another in the root to define different root cell types or functions (Chaiwanon and Wang 2015).
BR has a dose-dependent effect on PR elongation (Clouse et al. 1996; Müssig et al. 2002) and promotes the formation of LRs by regulating polar auxin transport (PAT) (Fig. 2A; Bao et al. 2004). BR treatment enhances shoot and root PAT in rapeseed (Li et al. 2005). Arabidopsis BR biosynthesis and signaling mutants (diminuto1 and bri1) show decreased PAT, while pin1 mutants display decreased sensitivity to BR-induced inhibition of root elongation, and pin2 mutants are deficient in BR-induced LR formation. These results point to the modulation of PAT as a mechanism by which BR regulates root development (Li et al. 2005). Studies in Arabidopsis root meristems implicate transcriptional and posttranscriptional regulation of PIN2 and PIN4 as a potential mechanism for BR-controlled cell growth and proliferation in the root meristem (Hacham et al. 2012).
Another node of auxin–BR interaction during root development is the transcriptional coregulator BREVIS RADIX (BRX), which promotes the expression of a crucial BR biosynthesis gene, CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD). In Arabidopsis, brx mutants have altered root meristems, with increased root branching and shorter roots (Mouchel et al. 2004). These plants also display a BR-deficient phenotype and attenuated auxin transcriptional program (Mouchel et al. 2006). Exogenous BR treatment rescues the BR-deficient phenotype and restores auxin-responsive gene expression. Interestingly, auxin also induces the expression of BRX, thus promoting BR production (Mouchel et al. 2006). More recently, plasma-membrane-associated BRX has been shown to inhibit PIN protein activity in developing Arabidopsis root protophloem sieve elements by negatively regulating PROTEIN KINASE ASSOCIATED WITH BRX (PAX) (which stimulates PIN activity) (Marhava et al. 2018) and by promoting the endocytic removal of PIN1 proteins from the plasma membrane (Marhava et al. 2020). Through its actions on PAX and PIN1, BRX inhibits auxin efflux. In turn, auxin has been shown to promote BRX protein turnover (Scacchi et al. 2009), creating a check on BRX activity if local auxin concentrations become too high.
Shoot Development
BR and auxin act cooperatively to promote leaf lamina bending in rice, and cell elongation in numerous species, including bean, cucumber, maize, pea, rice, squash, and tomato (for review, see Mandava 1988, Clouse and Sasse 1998, and Park 1998). Further studies in Arabidopsis confirmed that these two hormones act synergistically to promote hypocotyl elongation under many conditions (Tanaka, 2003; Nemhauser et al. 2004). Accordingly, Arabidopsis Aux/IAA gain-of-function mutants display reduced BR sensitivity in hypocotyl elongation assays (Nakamura et al. 2006) and BR treatment enhances shoot PAT in rapeseed (Li et al. 2005). Coordinated BR signaling and PAT also control vascular patterning in Arabidopsis shoots, with auxin maxima defining the location and BR signaling determining the number of vascular bundles (Ibanes et al. 2009).
BR signaling is important for proper light responses, and Arabidopsis BR mutants, such as de-etiolated2 and cpd, display a constitutive photomorphogenic phenotype (Li et al. 1996; Szekeres et al. 1996). Both auxin and BR signaling are required for shade avoidance (Keuskamp et al. 2011). In response to low blue light conditions, each hormone regulates a distinct set of genes, the full complement of which is needed for a proper shade avoidance response. While no direct link was established, the NAC TF ATAF2, which regulates auxin biosynthesis (Huh et al. 2012), was also found to regulate BR turnover in Arabidopsis by binding to the promoters and suppressing the expression of two BR catabolic enzyme genes, PHYB-4 ACTIVATION-TAGGED SUPPRESSOR1 and SUPPRESSOR OF PHYB-4 7 (Peng et al. 2015). In this way, ATAF2 promotes hypocotyl elongation by stimulating auxin biosynthesis and inhibiting BR degradation, whereas light and BR suppress ATFAF2 expression, creating a feedback regulatory circuit (Peng et al. 2015).
Unequal auxin distribution leads to the curving of shoots seen in gravitropism, and BR application promotes shoot gravitropism in bean (Meudt 1987; Park 1998). Moreover, BR-mediated enhancement of root gravitropic curving in maize requires PAT (Kim et al. 2000). In Arabidopsis, BR treatment accelerates root and shoot gravitropism and auxin reporter activity following gravistimulation (Li et al. 2005). Following BR treatment, PIN2 expression is enhanced and PIN2 protein localization mimics the protein distribution seen with gravistimulation. These findings suggest that the interaction between BR and auxin in gravitropism converges on PAT.
Another study found that in dark-grown Arabidopsis seedlings, BRs inhibit shoot gravitropism (Vandenbussche et al. 2011) and that the mechanism of this inhibition involves a complex interaction between BR, ET, and auxin signaling (Vandenbussche et al. 2013). In particular, several Aux/IAA proteins and ARFs, ARF7 and ARF19, are implicated in BR-regulated gravitropic responses. However, this inhibition is seen in seedlings grown in low sugar conditions, and the effect is lost when germinating the seedlings on vertical plates or supplementing the growth media with sugar (Vandenbussche et al. 2011). The authors speculate that the impact of BR signaling on gravitropism is due to a weakening of the hypocotyl cell wall. Indeed, further investigation found interactions between glucose, BRs, and potentially PAT during gravitropism and LR development (Singh et al. 2014; Gupta et al. 2015). Clearly, the role of auxin–BR interactions in shoot development is multifaceted, with tissue- and environment-specific effects.
Other Contexts
The first identified BR-responsive gene, BUR1, was found to be also induced by auxin treatment in soybean, albeit at a later time point (18 h vs. 2 h) (Zurek and Clouse 1994). This potential overlap in transcriptional control has been further investigated by many studies (Goda et al. 2002, 2004; Müssig et al. 2002; Yin et al. 2002; Nemhauser et al. 2004). Auxin and BR appear to cooperatively regulate some genes, while they are antagonistic in the control of others; this is fitting, considering the variable roles these two hormones play in different aspects of plant development. While these common transcriptional programs may be due to direct regulation of BR biosynthesis or PAT, one study suggests that these two hormones converge on common promoter motifs found in coregulated genes (Nemhauser et al. 2004) and ChIP data reveal that BZR1/BES1 bind to many auxin-responsive genes (Sun et al. 2010; Yu et al. 2011).
Inhibition of BR biosynthesis impairs the growth-promoting action of auxin (Vert et al. 2008), but together the two hormones enhance and potentiate the expression of common target genes. Closer examination revealed that the BIN2 kinase regulates auxin signaling by phosphorylating ARF2 and suppressing DNA binding by this negative regulator of auxin responses, thereby promoting transcription of its auxin target genes (Vert et al. 2008). BIN2, in turn, is negatively regulated by BR (Peng et al. 2008), supporting the hypothesis that BR blocks the BIN2-mediated activation of the auxin transcriptional program. Additionally, BR signaling is implicated in the control of Aux/IAA gene expression and activity in Arabidopsis (Nakamura et al. 2003, 2006; Cho et al. 2014). During Arabidopsis LR development, the ARF7 and ARF19 proteins become phosphorylated by BIN2, disrupting their interaction with Aux/IAAs and enhancing the auxin response (Cho et al. 2014). A screen for auxin transport mutants in Arabidopsis revealed that BR signaling promotes the nuclear accumulation of auxin by negatively regulating PIN-LIKES mRNA and protein expression (Sun et al. 2020). BR has also been shown to regulate auxin catabolism in barley (Sadura et al. 2019). Mutants in key BR biosynthetic (HvDWARF, HvCPD) and signaling (HvBRI1) genes had wild-type levels of total active auxins but altered levels of methylated and oxidized auxins.
Auxin signaling, in turn, increases BR sensitivity by enhancing expression of the BR receptor BRI1 (Sakamoto et al. 2013). In rice, the OsBRI1 promoter harbors an auxin-responsive element (AuxRE) that is bound by OsARF11 and is required for the up-regulation of OsBRI1 and BR phenotypes, providing a direct connection between auxin signaling and BR perception (Sakamoto et al. 2013). DWF4, which catalyzes a rate-limiting step in BR synthesis, is transcriptionally induced by auxin in Arabidopsis and many auxin-inducible genes are not activated in the presence of a BR inhibitor (Chung et al. 2011), suggesting that the full auxin transcriptional response requires intact BR signaling.
AUXIN–CK INTERACTIONS
Auxin and CK work to balance one another in regulating plant developmental processes. In fact, CK was discovered based on its interaction with auxin to promote the growth and division of cultured plant cells (Miller et al. 1955, 1956). The two hormones are generally antagonistic of one another. For example, while both hormones are required for callus formation in cell culture, an excess of CK promotes shoot development, whereas excess auxin favors root development. These two hormones have a storied scientific past (for review, see Schaller et al. 2015 and Kieber and Schaller 2018). Here, we highlight the many levels of interaction between them. Most of the following studies take place in Arabidopsis, and while it is known that CK and auxin regulate one another in several plant species, the extent to which the observations made in Arabidopsis can be extrapolated to other species remains to be discovered.
CKs are adenine-derived hormones that are synthesized in a series of steps involving ISOPENTENYL TRANSFERASE (IPT), CYTOCHROME P450 FAMILY 735A, and LONELY GUY (LOG) enzymes. CKs are inactivated by conjugation to sugar molecules or by degradation at the hands of enzymes such as the CK OXIDASE/DEHYDROGENASE (CKXs). When CK binds to its receptors, ARABIDOPSIS HISTIDINE KINASEs (AHKs), the receptor autophosphorylates, initiating a phosphorylation cascade. The ARABIDOPSIS HISTIDINE PHOSPHOTRANSMITTER (AHP) proteins transfer phosphates from the AHKs to the type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs), activating them. AHP1,2,3, and 5 are believed to positively regulate CK signaling, while AHP4 may be a negative regulator. AHP6, which lacks the histidine kinase activity of the other AHPs, also inhibits CK signaling through an unclear mechanism. AHP-mediated phosphorylation of the type-B ARRs induces the CK transcriptional response. One family of genes that is rapidly induced by CK are the type-A ARRs, a set of proteins that act in a feedback manner to repress CK signaling (Hutchison et al. 2006; for review, see Kieber and Schaller 2018).
CK induces auxin biosynthesis in several Arabidopsis tissues and this induction requires CK signaling through AHPs and type-A ARRs (Jones et al. 2010). These ARRs have also been shown to be a node of auxin-mediated control of CK signaling (Overvoorde et al. 2005; Müller and Sheen 2008; Lee et al. 2009; Zhao et al. 2010). Using several auxin and CK mutants, mutually inhibitory effects of the two hormones were found in a suite of developmental phenotypes (Kurepa et al. 2019). The authors proposed that the auxin signaling proteins ARF7 and IAA17 promote type-A ARR expression to inhibit CK signaling. However, while this route for auxin control of CK signaling has been previously investigated, the findings are not concordant; thus, the mechanism, and even direction, of type-A ARR regulation by auxin remains in question. It has also been widely demonstrated that CK regulates auxin signaling by altering the expression of PINs (Růžička et al. 2009; for review, see Schaller et al. 2015).
Likewise, auxin influences CK biosynthesis in Arabidopsis and other species via regulation of the IPT genes (Zhang et al. 1995; Miyawaki et al. 2004; Nordstrom et al. 2004; Tanaka et al. 2006; Cheng et al. 2013). As is often the case, the situation is probably far more complex, with auxin shown to also promote CK turnover by enhancing CKX expression (Palni et al. 1988; Werner et al. 2006).
Root Development
CK and auxin generally oppose one another in root development (Fig. 2; Kurepa et al. 2019) and the balance between cell division and differentiation relies on the interaction between auxin and CK (for review, see Jing and Strader 2019). CK promotes a smaller meristem via differentiation of cells in the transition zone, while auxin favors a larger meristem by promoting cell division (Blilou et al. 2005; Dello Ioio et al. 2007; Moubayidin et al. 2010). This effect of CK on meristem size is dependent upon auxin transport, as it is not observed in a pin2/3/7 triple mutant (Dello Ioio et al. 2007). Additionally, Arabidopsis CKX-overexpressing lines have reduced PIN2 and PIN4 expression and abnormal roots with expanded auxin maxima and larger columella cells, suggesting that CK acts to regulate the expression of PIN genes and auxin transport, thereby balancing elongation and cell division (Pernisova et al. 2009).
The Aux/IAA SHORT HYPOCOTYL2 (SHY2, IAA3) has been implicated in the regulation of auxin transport by CK (Fig. 2C). SHY2 transcription is induced by CK, presumably through the type-B ARRs, ARR1 and ARR12 (Dello Ioio et al. 2008; Moubayidin et al. 2010). SHY2 reduces the expression of PIN1, PIN3, and PIN7 transcripts and protein, as assessed by the accumulation of translational fusion reporters (Dello Ioio et al. 2008). The SHY2-dependent decrease in PIN protein levels results in a local redistribution of auxin, cell differentiation, and a smaller meristem (Dello Ioio et al. 2008; Moubayidin et al. 2010). SHY2, in turn, inhibits the auxin-induced expression of an IPT5 promoter reporter fusion construct, completing an auxin-CK regulatory circuit (Dello Ioio et al. 2008).
Auxin, on the other hand, promotes SHY2 degradation (Fig. 2C; Tian et al. 2002), restoring PIN expression, auxin localization, and cell division (Dello Ioio et al. 2008). SHY2 may target PIN genes directly or it could be an indirect interaction through repression of the ubiquitin-binding protein DA1-RELATED PROTEIN2 (DAR2) (Peng et al. 2013). The effects of auxin or CK on root meristem size are lost in dar2 mutants and genetic analyses place DAR2 downstream of CK signaling and SHY2. Further, dar2 mutants have reduced auxin transport toward the root meristem, and reduced CK regulation of PIN3 and PIN7 translational fusions (Peng et al. 2013). Studies of the Arabidopsis root meristem and developing vasculature found that the CK-induced reduction of PIN proteins (via induction of SHY2) involved the transcriptional coregulator BRX (Scacchi et al. 2010). Whereas the mechanism is unclear, it likely involves reciprocal transcriptional repression between SHY2 and BRX and competition for MONOPTEROS (MP, ARF5) binding.
Arabidopsis ARR7 and ARR15, type-A feedback repressors of CK signaling, are up-regulated by auxin in the root stem cell niche (Müller and Sheen 2008). Type-A ARRs, in turn, regulate PIN proteins via an unknown mechanism. The Arabidopsis arr3/4/5/6/7/8/9/15 mutant has a smaller meristem and reduced PIN1, PIN3, and PIN4 protein levels. PIN7 levels in the stele are decreased, but are higher in the root cap, suggesting that CK signaling acts on PIN proteins to alter auxin distribution and regulate meristem size and differentiation (Zhang et al. 2011). Furthermore, Arabidopsis type-B ARR1 and ARR12 inhibit the expression of the auxin influx transporters LAX2 and AUX1, with ARR1 directly binding the promoter of LAX2 (Zhang et al. 2013). ARR1 also promotes auxin biosynthesis, at least in part via the transcriptional activation of ANTHRANILATE SYNTHASE BETA SUBUNIT1 (ASB1) that codes for a rate-limiting enzyme in the biosynthesis of the auxin precursor Trp (Moubayidin et al. 2013). ARR1 suppression by SCARECROW (SCR), a critical regulator of root meristem activity, down-regulates auxin biosynthesis to maintain the root meristem (Moubayidin et al. 2013). Additionally, ARR12 works in concert with RETINOBLASTOMA-RELATED (RBR) protein to activate ARF19 transcription in the root apical meristem, promoting cell differentiation (Rademacher et al. 2011; Perilli et al. 2013).
The role of auxin–CK interactions in LR development has been extensively studied (for review, see Jing and Strader 2019). In Arabidopsis LRs, CK regulates the expression level and pattern of PIN genes (Laplaze et al. 2007), and mutants for several CK signaling components display improper PIN localization (Marhavý et al. 2011; Chang et al. 2013; Moreira et al. 2013). CK was found to direct endocytic recycling of PIN1 toward degradation in LR primordia, thereby reducing the accumulation of PIN1 proteins on the plasma membrane and inhibiting LR initiation (Fig. 2A,B; Marhavý et al. 2011). During later stages of LR formation, CK was found to deplete anticlinal PIN1 protein accumulation, directing auxin flow to promote LR growth (Marhavý et al. 2014). Arabidopsis ahp6 mutant plants display a mild defect in LR emergence and cell division in LR primordia, and have abnormal PIN1 protein localization, all reminiscent of CK treatment effects (Moreira et al. 2013). The ahp6 mutant phenotype is lost in combination with CKX-overexpressing lines, suggesting that AHP6 may function to reduce CK signaling and maintain proper LR cell division. The AHP6 gene is transcriptionally up-regulated by auxin in vascular tissues, pointing to auxin-triggered inhibition of CK signaling (via AHP6) during vascular patterning in Arabidopsis roots (Bishopp et al. 2011). If AHP6 is also auxin regulated in LR primordia, it could serve as a general point of integration for auxin and CK signaling. More recently, CK was shown to exert its effects on LR root formation through TRANSPORTER OF IBA1 (TOB1) that blocks LR formation (Michniewicz et al. 2019). TOB1 is an indole-3-butyruc acid (IBA) transporter transcriptionally induced by CK signaling. IBA is an auxin precursor, suggesting that CK signaling may alter auxin distribution by promoting the relocalization of auxin intermediates.
Auxin plays a well-established role in gravitropism and regulates differential cell expansion to achieve root turning. In Arabidopsis, CK modifies the distribution of auxin by enhancing asymmetric localization of PIN1 proteins to the basal side of cells, redirecting the flow of auxin to promote root gravitropism (Marhavý et al. 2014). Enhancement of an AUX1 fluorescent reporter signal was also reported in CKX2- and CKX3-overexpression lines in Arabidopsis, suggesting that CK also regulates auxin influx (Pernisova et al. 2016).
During LR formation, emerging roots establish an angle of growth with a characteristic displacement from the gravity vector (the so-called gravitropic set point angle). This allows the root system to expand outward from the PR. CK signaling has also been shown to act in the gravitropic response by opposing bending toward the gravity vector (Waidmann et al. 2019). A genome-wide association study (GWAS) screen identified CKX2 variants as a factor in determining LR angle. CK treatment increases the angle of LRs in Arabidopsis and rapeseed, whereas Arabidopsis CK receptor mutants show decreased LR angles. Enhanced CK signaling was observed on the topside of the emerging LR and inhibited root growth, complementing the auxin-mediated growth repression on the underside of the root. Whereas a direct interaction was not defined, treatment with the auxin transport inhibitor naphthylphthalamic acid (NPA) reduced CK reporter activity and blocked the formation of a CK signaling gradient. The balanced repression from auxin and CK promotes radial expansion of the root system by preventing the LR from completing a full 90° turn to align with gravity, demonstrating how these two hormones coordinate to fine-tune LR angles (Waidmann et al. 2019).
Similarly, this auxin-CK competition is believed to be at play in LR hydrotropism. The ER-associated MIZU-KUSSEI1 (MIZ1) protein is essential for a proper hydrotropic response (Kobayashi et al. 2007; Yamazaki et al. 2012). When overexpressed in Arabidopsis, MIZ1 increases CK sensitivity, reduces LR number, and lowers free auxin levels, but exogenous auxin supplementation rescued the LR phenotype (Moriwaki et al. 2011). Interestingly, CK promotes the localization of MIZ1 protein at root primordia, suggesting that MIZ1 serves as a node for auxin-CK crosstalk during LR formation.
Several other genes have been implicated as nodes of auxin-CK communication during root developmental processes. AUXIN UP-REGULATED F-BOX PROTEIN1 is transcriptionally induced by auxin and proposed to mediate CK-regulated cell expansion in the root by regulating PIN expression and, possibly, by targeting ARR1 for degradation (Zheng et al. 2011). The TF TMO5 is transcriptionally up-regulated by auxin (Schlereth et al. 2010) and enhances the expression of the CK-biosynthesis gene, LOG4, helping determine vascular patterning in the Arabidopsis root (De Rybel et al. 2014). Of note, the PLETHORA (PLT) TFs, key regulators of root development that are induced by auxin (Aida et al. 2004; Blilou et al. 2005; Galinha et al. 2007) and regulate both auxin transport and synthesis (Pinon et al. 2013; Santuari et al. 2016), are repressed by CK in Arabidopsis roots (Dello Ioio et al. 2008). Finally, CK has been implicated in promoting auxin responses during nodulation in Lotus japonicas and Medicago truncatula (for review, see Kohlen et al. 2018), with nodule formation defects observed in loss-of-function mutants for CK signaling and auxin biosynthesis genes. On the other hand, CK treatment, CK signaling gain-of-function mutants, and chemical inhibition of PAT lead to the formation of nodule-like structures in the absence of rhizobia in L. japonicas and alfalfa (Medicago sativa) (for review, see Kohlen et al. 2018).
Shoot Development
In contrast to their roles in root meristem development, auxin acts to promote shoot apical meristem differentiation while CK promotes division of undifferentiated cells (for review, see Azizi et al. 2015). There is a general theme of high auxin levels reducing CK biosynthesis or promoting CK turnover, while rising CK levels disrupt auxin distribution by modulating PIN proteins (Fig. 3A). In Arabidopsis hypocotyl explants, CK up-regulates PIN3 and PIN6, but reduces the expression of PIN2, shaping auxin distribution, and thus altering the cell division versus differentiation balance (Pernisova et al. 2009). On the other hand, during shoot induction from callus, auxin production increases, activating ARF3, which in turn suppresses IPT5 (Cheng et al. 2013). In this way, auxin acts to restrict CK activity to the site of future meristems. Accordingly, decapitation of pea plants, thereby removing young expanding leaves, which are a major auxin source in the stem, induces PsIPT1 and PsIPT2, while exogenous auxin inhibits them (Tanaka et al. 2006). In Cremastra appendiculata pseudobulbs, decapitation results in a turnover of auxin and induction of CK biosynthesis (Lv et al. 2018). Treatment with the auxin transport inhibitors NPA and 2,3,5-triiodobenzoic acid up-regulated CaIPT, and reduced the expression of CaCKX, promoting an accumulation of CK and pseudobulb branching.
Auxin and CK are spatially regulated and dependent upon one another to organize the formation of axillary meristems in multiple species (Wang et al. 2014; Dierck et al. 2016; Qiu et al. 2019). To confer apical dominance, auxin blocks axillary growth and CK promotes it. Auxin originating from the shoot apex down-regulates CK biosynthesis in stems, reducing CK levels in axillary buds, and CK regulates auxin efflux from branches (Fig. 3A; for review, see Müller and Leyser 2011). For example, in pea, CK promotes polarization of PIN1 protein in axillary buds (Kalousek et al. 2010) and increases levels of PIN3, 4, and 7 fluorescent fusion proteins in the xylem parenchyma of Arabidopsis main stems (Waldie and Leyser 2018). Correspondingly, CK was found to regulate both auxin transport and bud activation during branching in pea (Kotov and Kotova 2018).
The type-A ARRs ARR7 and ARR15 are induced by CK in the Arabidopsis shoot apical meristem but repressed by ARF5 (Fig. 3B; Zhao et al. 2010). As type-A ARRs inhibit CK signaling, this interaction reveals an important point of coordination for these two hormones in maintenance of the shoot stem cell niche. The roles of CK and auxin in regulating axillary branching are not yet fully elucidated and some studies of type-A and type-B ARRs seem to present paradoxical findings (Müller et al. 2015; Xu et al. 2015; Waldie and Leyser 2018; Zha et al. 2019). How these two hormones coordinate branching is surely complex, involving signals from other hormones and/or nutrients.
CK and auxin signaling also interact during secondary shoot vasculature development in Populus, where CK was found to regulate the auxin gradients that induce cambium formation, but the underlying molecular mechanism was not determined. Overproduction of CK promoted the formation of cambium through increased meristem cell divisions and IAA accumulation (Immanen et al. 2016), while CK treatment of wild-type plants reduced IAA accumulation and favored phloem development following injury (Chen et al. 2019).
The maize aberrant phyllotaxy1 (abph1) mutant displays altered phyllotaxis (Giulini et al. 2004) and reduced auxin levels (Lee et al. 2009). ZmABPH1 encodes a type-A ARR that is up-regulated by auxin and whose action depends on auxin transport (Lee et al. 2009). Both CK and ABPH1 induce ZmPIN1 transcription, and ZmPIN1 localizes to sites of incipient leaf primordium formation, highlighting a link between auxin, CK, and the control of leaf primordia formation (Lee et al. 2009). Leaf shape is also modulated by auxin–CK interaction. Tomato Sliaa9 antisense plants exhibit a range of phenotypes with simpler, rather than compound, leaf morphology and less lobed, even entire, leaf margins (Wang et al. 2014). Further studies revealed that compound leaf development in tomato is regulated by CK signaling, and this effect of CK is dependent upon proper SlPIN localization (Shani et al. 2010).
Regulation of auxin signaling by CK is also critical during flower development. Treatment of Arabidopsis plants with CK or NPA results in similar gynoecium phenotypes with disrupted apicobasal patterns (Zuniga-Mayo et al. 2014). A PIN1 translational GREEN FLUORESCENT PROTEIN (GFP) fusion, which normally displays restricted expression, was detected throughout the developing inflorescence after CK treatment, suggesting that CK mediates its effect on Arabidopsis gynoecium patterning at least in part by modulating PIN1 expression and localization (Zuniga-Mayo et al. 2014). Indeed, Arabidopsis CK receptor mutants show that CK perception is necessary for proper PIN1 expression during ovule development (Bencivenga et al. 2012). This effect on PIN1 is dependent upon the TFs SPOROCYTELESS and BELL1, which are transcriptionally up-regulated and down-regulated by CK, respectively (Bencivenga et al. 2012). More recently, both CK and the TF SPATULA (SPT) were found to induce the expression of the auxin genes TAA1 and PIN3 during Arabidopsis gynoecium development (Reyes-Olalde et al. 2017). The effects of CK were found to be mediated by SPT, which up-regulates the expression of type-B ARR1 and ARR12. These ARRs, in turn, induce the transcription of TAA1 and PIN3, although it is unclear whether they bind alone or cooperatively with SPT.
On the other hand, auxin was found to promote the expression of AHP6, which inhibits CK signaling, to dictate the patterning of developing inflorescences in Arabidopsis (Besnard et al. 2014). And in floral meristems, auxin acts through ARF3 to inhibit CK signaling by repressing IPT, LOG, and AHK4 (Zhang et al. 2018). During sepal formation, DEVELOPMENT RELATED MYB-LIKE1 (DRMY1) helps regulate auxin and CK signaling to define sepal initiation and size (Zhu et al. 2020). Arabidopsis drmy1-2 plants display weaker and more diffuse auxin reporter activity, but stronger and more diffuse CK reporter activity. This lack of proper spatiotemporal signaling results in delayed and smaller sepals (Zhu et al. 2020). These findings paint a complex picture of how auxin and CK regulate one another to create the patterns of hormone signaling required for proper flower development.
Other Contexts
Auxin-CK crosstalk has been implicated in myriad additional processes, including coregulation of common target genes (Hurny et al. 2020) and responses to various biotic (Boivin et al. 2016; Hurny et al. 2020) and abiotic (Wang et al. 2006; Tognetti et al. 2017; Bielach et al. 2017; Yang et al. 2017) factors. Whether these interactions are coincidental or true crosstalk requires further exploration. A greater understanding of the interplay between these two hormones could be informative for adapting crops to unfavorable and changing growth conditions.
AUXIN–ET INTERACTIONS
ET is a gaseous plant hormone first discovered for its effects on leaf abscission and ripening (for review, see Abeles et al. 1973). The crosstalk between ET and auxin is key for proper plant development and manifests itself in a wide range of processes. Plants produce ET from the amino acid L-methionine (Met) that is converted into S-adenosyl-L-methionine (AdoMet) by Ado-Met synthetase (Giovanelli et al. 1985). The first committed and rate-limiting step in the ET biosynthesis pathway is the conversion of AdoMet into 1-aminocyclopropane-1-carboxylate (ACC) by ACC SYNTHASE (ACS). In a second step, ACC OXIDASE (ACO) turns ACC into ET (for review, see Yang and Hoffman 1984). Once synthesized, ET moves from cell to cell by diffusion without requiring specific transporters.
ET is sensed by ER- and Golgi-localized receptors of the ET RECEPTOR/ET RESPONSE SENSOR/ET-INSENSITIVE4 (EIN4) transmembrane protein family (for review, see Binder 2020). In the absence of ET, the receptors continuously activate CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), a serine/threonine protein kinase that phosphorylates and inactivates the transmembrane ER-localized protein EIN2 (for review, see Binder 2020). Phosphorylated EIN2 is targeted for degradation by two F-Box EIN2-TARGETING PROTEINs and the master TFs of the ET response, EIN3/EIN3-LIKE1 (EIL1), are targeted for degradation by the F-Box EIN3-BINDING F-BOX PROTEINs (EBFs) (for review, see Ju and Chang 2015 and Binder 2020). In the presence of ET, the receptors are turned off, inactivating CTR1, reducing the phosphorylation of EIN2, and triggering the cleavage of the carboxy-terminal end of EIN2. The EIN2 C-end blocks translation of EBF1 and EBF2 in the cytoplasm and promotes the activity of EIN3/EIL1 in the nucleus (Merchante et al. 2015; for review, see Binder 2020), eliciting the transcriptional regulation of thousands of downstream genes that mediate ET responses (Binder et al. 2004).
The first clue to the importance of auxin-ET crosstalk came from an observation that a remarkable number of auxin mutants are ET-insensitive, including auxin transport mutants such as aux1 and pin2 (eir1); mutants with impaired auxin biosynthesis, such as weak ET-insensitive2 (wei2), wei7, and wei8; auxin perception mutants, such as tir1; and mutants in components of auxin signal transduction, including axr2 (iaa7) and axr3 (iaa17) (for review, see Merchante and Stepanova 2017). These findings suggest that proper levels of auxin biosynthesis, transport, signaling, and response are required for the normal response of plants to ET.
Root Development
ET promotes local auxin biosynthesis in roots, locally inducing TAA1 and TAR2 and several YUC genes, which contributes to ET-triggered PR shortening (Fig. 2A,C; Růžička et al. 2007; Stepanova et al. 2007; Swarup et al. 2007). The identification of the small molecule L-kynurenine (Kyn) (He et al. 2011) as a potent inhibitor of auxin biosynthesis provides an illustrative example of the intricate crosstalk between auxin and ET. Kyn blocks the conversion of L-Trp into IPyA, catalyzed by TAA1, inhibiting ET-induced auxin production. Kyn suppresses the short-root phenotype of ET-treated wild-type plants and of untreated ctr1 mutants that display constitutive ET responses, supporting the notion that the root growth inhibition triggered by ET is mediated by TAA1-dependent auxin biosynthesis. Elevated levels of auxin, on the other hand, promote the stabilization of EIN3 in the nucleus by suppressing its EBF-dependent degradation. Thus, blocking auxin accumulation with Kyn inhibits EIN3 nuclear accumulation and represses root responses to ET (He et al. 2011).
The epidermis of the root elongation zone is a key site for ET-induced root growth inhibition (Vaseva et al. 2018). ET promotes the transition from the mitotic cycle to endoreduplication, reducing cell division and, therefore, the size of the meristem and root growth (Street et al. 2015). The increased activity of the auxin reporter DR5 in the root elongation zone in ET-treated plants is linked to the ability of ET to arrest cell elongation and, thus, PR growth (Růžička et al. 2007; Stepanova et al. 2007; Swarup et al. 2007). ET inhibits PR growth by transcriptionally up-regulating genes involved in auxin biosynthesis and auxin transport, thereby stimulating auxin translocation from the meristem to the elongation zone and increasing local auxin levels above the physiological threshold required for the cells to become fully sensitized to ET (Růžička et al. 2007; Stepanova et al. 2007; Swarup et al. 2007).
Auxin plays a pivotal role in stimulating LR formation by priming pericycle cells, inducing cell division, and promoting root emergence and elongation (for review, see Lavenus et al. 2013). ET reduces local accumulation of auxin required for LR formation by increasing the expression levels of PIN3 and PIN7 and, thus, shifting auxin away from the initiation sites (Fig. 2A,B). Correspondingly, ET causes a prominent decrease of DR5 reporter levels in the regions of LR emergence, leading to a reduction in LR emergence (Lewis et al. 2011).
Shoot Development
Shortly after germination, seedlings form the apical hook to protect the shoot apical meristem while emerging through the soil. ET up-regulates the expression of auxin biosynthetic genes such as TAA1, TAR2, and several YUC genes in the hypocotyl. ET also modulates auxin transport, inducing AUX1, LAX1, and PIN3 and inhibiting PIN1 and PIN4 expression, to achieve preferential auxin localization on the inner side of the hook. This ET-mediated auxin gradient initiates and maintains the temporary hook structure (Vandenbussche et al. 2010; Zádníková et al. 2010). Consistent with the role of auxin–ET interactions in apical hook formation, the ET-insensitive mutant ein2 displays a defective hook that can be corrected with exogenous auxin application, consistent with the notion that ET induces the boost in auxin production required for proper hook development (Vandenbussche et al. 2010). Moreover, ET stimulates the transcription of an N-acetyltransferase-like gene HOOKLESS1 (HLS1), which down-regulates the expression of ARF2, a repressor of the auxin response (Li et al. 2004). This repression of a repressor results in the general induction of auxin responses. Accordingly, hls1 mutants do not form a hook, highlighting another point of integration of the auxin and ET cues during apical hook development (Lehman et al. 1996).
Fruit Development
Auxin-ET crosstalk is fundamental for the development of the male and female reproductive organs (An et al. 2012). Auxin induces, whereas ET represses, stamen development, and both promote pollen germination and the growth of the pollen tube (Fig. 4A; for review, see An et al. 2020). Genes involved in ET and auxin biosynthesis and signaling are highly expressed during pistil development and specifically in ovules at anthesis, suggesting that these two hormones coregulate the process of fruit set, but act at different time points (for review, see An et al. 2020). In tomato, auxin governs the initial phases of fruit development and ET controls the ripening of the fruit by promoting the degradation of chlorophylls, conversion of xanthophylls to carotenes, initiation of the climacteric ET production, etc. (Fig. 4B,C; Fraser et al. 1994). Fruits treated with auxin exhibit a delay in the climacteric transition to the ET-mediated ripening processes, preservation of high levels of xanthophylls and chlorophyll, and the inhibition of genes involved in carotenoid biosynthesis (Su et al. 2015).
Tomato transgenic lines with reduced expression of SlIAA3 exhibit auxin and ET phenotypes, with delayed ripening and reduced apical dominance, auxin sensitivity, and petiole epinasty, suggesting that SlIAA3 is a link between the auxin and ET response pathways (Chaabouni et al. 2009). Furthermore, both ET and auxin induce the expression of the TF ET RESPONSE FACTOR.B3 (SlERF.B3), a major player in the regulation of ET responses and fruit ripening (Liu et al. 2013a). SlERF.B3 integrates ET and auxin signals by binding to the promoter and inducing the expression of SlIAA27 (Fig. 4C; Liu et al. 2018).
Other Contexts
As mentioned above, in the IPyA pathway of auxin biosynthesis, TAA1 and TARs catalyze the conversion of Trp into IPyA that is subsequently used by the YUCs to produce auxin (Mashiguchi et al. 2011; Stepanova et al. 2011; Won et al. 2011). The VAS1 enzyme directs IPyA away from the YUCs, using it and the ET biosynthetic precursor, Met, to produce Trp and 2-oxo-4-methylthiobutyric acid, reducing IPyA availability and, in turn, auxin production. Correspondingly, vas1 mutants exhibit elevated levels of both IAA and the ET precursor ACC. VAS1 therefore represents a point of interaction at the metabolic level between auxin and ET biosynthesis (Zheng et al. 2013).
AUXIN–GA INTERACTIONS
GAs take their name from the growth-modifying fungus Gibberella fujikuroi that triggers a “foolish seedling disease” in rice. Since GA was first identified in the 1930s, more than 130 GAs have been discovered in plants, fungi, and bacteria. However, only a handful, GA1, GA3, GA4, and GA7, are biologically active, and most nonbioactive GAs are precursors or deactivated catabolites of the bioactive forms. GAs are derived from trans-geranylgeranyl diphosphate (GGPP), a common C20 precursor for diterpenoids. First, GGPP is converted to the tetracyclic hydrocarbon intermediate, ent-kaurene, by the diterprene cyclases ENT-COPALYL DIPHOSPHATE SYNTHASE and ENT-KAURENE SYNTHASE. Then, ent-kaurene is converted to GA12 by a plastid membrane-bound ENT-KAURENE OXIDASE and an endoplasmic reticulum-bound ENT-KAURENOIC ACID OXIDASE. In the third step, the conversion of GA12 to various intermediates and bioactive GAs is mediated in the cytosol by GA20-OXIDASE and GA3-OXIDASE through two parallel pathways. Bioactive GAs can be deactivated by GA2-OXIDASE (GA2ox) (for review, see Binenbaum et al. 2018).
The soluble receptor GIBBERELLIN-INSENSITIVE DWARF1 (GID1) binds to GA in the nucleus triggering a conformational change that promotes association with the transcriptional regulators DELLAs. The formation of the GID1-GA-DELLA complex enhances DELLA binding to GID2/SLY1 F-box proteins, which triggers the degradation of DELLA via the 26S proteasome pathway, activating the GA response. In the absence of GA, DELLA proteins bind to TFs, repressing the GA response (for review, see Hernández-García et al. 2020). As the sites of GA biosynthesis are not always colocalized with the expression domains of GA perception genes, GA movement is thought to be essential. Although GA efflux transporters have not been identified yet, the NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER family of proteins in Arabidopsis have been described as bona fide influx carriers (for review, see Binenbaum et al. 2018). In addition, two members of the SWEET transporter family (SWEET13 and SWEET14) are also capable of transporting GA (for review, see Binenbaum et al. 2018).
While auxin plays essential roles in almost all developmental processes (for review, see Gallei et al. 2020), GAs play important roles in both cell division and cell elongation, such as seed germination, stem/shoot elongation, and floral organ development (Ubeda-Tomás et al. 2012). It is generally considered that auxin acts upstream of GA by activating GA biosynthesis (Hu et al. 2018a), but there are many levels of interaction between these two hormones, as reviewed below.
Root Development
As described above, the auxin-CK regulatory circuit ARR1-SHY2-PIN controls root meristem size by balancing cell differentiation with cell division (Fig. 2C; Moubayidin et al. 2010). ARR1 physically interacts with DELLA proteins GA-INSENSITIVE (GAI) and REPRESSOR OF GA (RGA) and this interaction enhances its transactivation activity (Rosa et al. 2015). Thus, during the meristem growth phase, a high level of GA represses ARR1 expression by promoting degradation of the DELLA proteins, which results in a low level of SHY2, thus promoting cell division (Fig. 2C; Rosa et al. 2015).
Shoot Development
In the absence of light, seedlings undergo skotomorphogenesis/etiolation, resulting in an elongated hypocotyl, presence of an apical hook and small and closed cotyledons (Von Arnim and Deng 1996). Several PHYTOCHROME-INTERACTING FACTORs (PIFs) including PIF1, PIF3, PIF4, and PIF6, play a critical role in etiolation (Huq and Quail 2002; Kim et al. 2003; Huq et al. 2004; Monte et al. 2004). DELLAs physically interact with PIFs, preventing PIF binding to their targets, which results in inhibition of hypocotyl growth (de Lucas et al. 2008; Feng et al. 2008). On the other hand, ARF6 and ARF8 also regulate hypocotyl elongation (Tian et al. 2004; Nagpal et al. 2005). Genome-wide analyses indicate that ARF6 shares a large number of target genes with the BR signaling TF BZR1 and with the light/temperature-regulated TF PIF4 (Oh et al. 2014), two components of the PIF4-BZR1-DELLA module that integrates signals from light, BR, and GA (Bai et al. 2012a,b), suggesting that ARF6 may interact with DELLA. ARF6, ARF7, and ARF8 were confirmed to physically associate with the DELLA protein RGA. Through this interaction, RGA blocks ARF6 binding to the promoters of its target genes, suggesting that GA promotes cell elongation in the Arabidopsis hypocotyl by enhancing auxin/ARF-mediated responses (Oh et al. 2014).
Downstream from DELLA and PIF signaling, the two Arabidopsis paralogous GATA TFs, GATA NITRATE-INDUCIBLE CARBON-METABOLISM INVOLVED (GNC) and GNC-LIKE (GNL), were identified as direct transcriptional targets of PIFs (Richter et al. 2010). Single and double gnc and gnl mutant seeds germinate faster than wild-type, with the double mutant germinating even on plates containing the GA biosynthesis inhibitor paclobutrazol (Richter et al. 2010). Furthermore, gnc gnl partially suppresses the GA biosynthesis mutant ga1, suggesting that GNC and GNL are repressors of GA signaling (Richter et al. 2010). gnc gnl also suppresses arf2 phenotypes, suggesting that, genetically, GNC and GNL act downstream of ARF2 (Richter et al. 2013). Consistent with this idea, ARF2 and ARF7 can directly bind to the promoters of GNC and GNL and inhibit their expression. Thus, GNC and GNL represent another point of convergence for the crosstalk between auxin and GA (Richter et al. 2013).
Another potential node of auxin–GA interaction is miR319, an important player in shoot organ morphogenesis (Curaba et al. 2014) that acts as a positive regulator of auxin signaling by indirectly repressing SHY2 and SMALL AUXIN UP RNA39 in leaf morphogenesis (Tian et al. 2002; Palatnik et al. 2003; Kant et al. 2009). Interestingly, miR319 can affect leaf cell differentiation by targeting LANCEOLATE, which indirectly inhibits GA biosynthesis (Ori et al. 2007; Yanai et al. 2011), thus implicating miR319 in the auxin-GA crosstalk controlling leaf organogenesis (Curaba et al. 2014).
Similar to primary stem growth, cambial growth in secondary stems is also regulated by auxin and GA. Consistent with previous studies in Arabidopsis and in pea (Ross et al. 2000; Frigerio et al. 2006), poplar PttGA20ox1 and PttGA20ox4 transcript levels are decreased in the stem of decapitated trees and induced by IAA, indicating that auxin stimulates the expression of GA biosynthesis genes in cambial growth. Furthermore, GA-only treatment increases cell division in the cambial zone of decapitated poplar trees (i.e., under auxin depletion) (Björklund et al. 2007). GA, in turn, increases local auxin levels by promoting expression of a putative auxin efflux transporter, PttPIN1 (Björklund et al. 2007). A recent study demonstrated that GA can redirect protein trafficking to the plasma membrane, thus coregulating multiple processes, including PIN-dependent auxin fluxes (Salanenka et al. 2018).
GAs not only promote vegetative growth, but also induce developmental phase transitions. In Arabidopsis, the GA pathway plays a major role in flowering time under short-day conditions, promoting flowering through the activation of floral integrator genes such as SUPPRESSION OF OVEREXPRESSION OF CONSTANS1 and LEAFY (LFY) (Blázquez et al. 1998; Bonhomme et al. 2000; Moon et al. 2007). On the other hand, MP (ARF5) plays a critical role in flower primordium initiation (Przemeck et al. 1996) and directly induces LFY (Yamaguchi et al. 2013; Wu et al. 2015). Thus, LFY is yet another point of convergence for the crosstalk between GA and auxin. In addition, MP physically interacts with BRAHMA (BRM) and SPLAYED, two related Arabidopsis SWI/SNF-subgroup ATPases (Wu et al. 2015), and BRM binds the promoters of GA biosynthetic genes such as GA3ox1 as an activator (Archacki et al. 2013). Furthermore, several DELLA proteins physically interact with an SWI/SNF subunit SWI3C (Sarnowska et al. 2013). Thus, crosstalk between GA and auxin during flowering may also be dependent on MP- and DELLA-mediated interactions with chromatin-remodeling complexes.
Fruit Development
Exogenous application of diverse plant growth substances, mainly auxins and GAs, can induce parthenocarpic fruit set and development (Gorguet et al. 2005; Srivastava and Handa 2005). Consistent with these observations, mutations affecting auxin signaling or GA biosynthesis genes (such as AtARF8, SlARF7, SlIAA9, and SlDELLA) can also induce parthenocarpic fruits in Arabidopsis and tomato (Wang et al. 2005a, 2009, 2011; Goetz et al. 2007; de Jong et al. 2009; Mounet et al. 2012). It was proposed that fertilization triggers auxin-mediated GA synthesis (Dorcey et al. 2009). A recent study in tomato showed that SlARF7 and five other activator SlARFs interact with SlDELLA and SlIAA9, and that SlARF7 and SlDELLA directly associate with the promoters of GA biosynthesis (GA20ox1 and GA3ox1) and auxin catabolism (GH3.2) genes (Fig. 4A; Hu et al. 2018a). The direct interaction between the activator SlARF7 and the repressor SlIAA9 may turn the SlARF7/SlIAA9 complex into a transcriptional repressor, whereas SlDELLA blocks SlARF7 binding to the promoters of its target genes, thus antagonizing the repression by SlARF7/SlIAA9 of GA- and auxin-related genes (Hu et al. 2018a). In contrast, the effect of SlARF7/SlIAA9 and SlDELLA on downstream growth-related genes, such as EXPANSIN5 (EXP5) and ACC OXIDASE4 (ACO4), is additive (Fig. 4A). Taken together, these findings reveal that direct crosstalk between SlDELLA-mediated GA and SlARF7/SlIAA9-mediated auxin signaling events coregulates fruit initiation in tomato (Hu et al. 2018a).
Interestingly, this DELLA/ARF-mediated regulation central to fruit initiation in tomato appears to be conserved in grape (Zhang et al. 2019b), also a true botanical fruit derived from ovaries, and in strawberry (Zhou et al. 2020), which makes accessory fruits derived from receptacles. FveARF6 and FveARF8 interact with the DELLA repressor FveRGA1. Like SlARF7 (Hu et al. 2018a; Zhou et al. 2020), FveARF8 interacts with FveRGA1 and FveIAA4 through distinct protein domains, suggesting that FveARF8 may be simultaneously repressed by FveRGA1 and FveIAA4 prior to fertilization (Zhou et al. 2020). Furthermore, FveARF8 can directly bind to the promoter and repress the expression of FveGID1c, suggesting that the auxin-GA crosstalk in strawberry fruits is multifaceted (Zhou et al. 2020).
AUXIN–JA INTERACTIONS
JA is a fatty-acid-derived hormone that takes its name from jasmine oil of Jasminum grandiflorum (Demole et al. 1962). JA regulates not only plant stress responses but also plant growth and development, in part through its tissue-specific interactions with auxin. JA biosynthesis has been extensively reviewed (for review, see Fonseca et al. 2009 and Ruan et al. 2019). Briefly, JAs are lipid-derived molecules produced via two main pathways. The octadecane pathway uses linolenic acid (18:3) as a precursor to produce 12-oxo-phytodienoic acid (OPDA) and the hexadecane pathway uses hexadecatrienoic acid (16:3) yielding dinor-oxo-phytodienoic acid (dn-OPDA) (Chini et al. 2018). These first steps of both pathways take place in the chloroplasts. Further reactions lead to JA production in the peroxisomes and its modification to methyl-JA (MeJA) or conjugation to isoleucine (Ile) to make JA-Ile in the cytoplasm. The bioactive form of JA is JA-Ile, which mediates plant responses to environmental and developmental cues. JA-Ile can promote resistance to a broad range of plant pathogenic bacteria, fungi, and herbivores (Campos et al. 2014; Machado et al. 2016, 2017).
JA biosynthesis and accumulation of JA-Ile are promoted in response to specific endogenous and environmental cues. JA-Ile binds to the CORONATINE-INSENSITIVE1 (COI1) receptor, inducing the degradation of the JASMONATE ZIM-DOMAIN (JAZ) proteins and releasing the MYC2 TFs from the JAZ-MYC2 complex. The MYC2 TFs then induce the expression of JA-responsive genes (Chini et al. 2018).
The JA signaling pathway presents certain similarities to the auxin signaling pathway. COI1 encodes an F-box protein, related to TIR1/AFB family of F-box proteins that bind auxin. Once JA-Ile is bound, COI1 functions in E3-ubiquitin ligase-mediated proteolysis of the targeted JAZ proteins. The JAZ repressors play a negative role in the JA signaling pathway similar to the function of Aux/IAAs in auxin signaling. Likewise, the MYC TFs up-regulate the expression of downstream genes as activator ARFs do in the auxin signaling pathway (Campos et al. 2016). Generally, while auxin is considered to be the growth-promoting hormone, JA is known to repress plant growth.
Root Development
Auxin promotes PR meristem activity and cell division but can also inhibit PR growth by reducing cell elongation in the elongation zone (for review, see Overvoorde et al. 2010 and Vaseva et al. 2018). In contrast, JA treatments inhibit PR meristem activity, decreasing cell number in the root meristem, as well as reduce cell size in both the meristem and the elongation zone, thus leading to the reduction of the overall root growth (Fig. 2A; for review, see Wasternack and Hause 2013). Auxin distribution defines the levels and expression patterns of PLETHORAs (PLTs), AP2-domain TFs that mediate the establishment and maintenance of the root stem cell niche and cell proliferation (Mähönen et al. 2014). ARF2 positively regulates PLT1 and PLT2 expression (Promchuea et al. 2017). In turn, PTLs stimulate the production of auxin by inducing the expression of the auxin biosynthesis genes ANTHRANILATE SYNTHASE ALPHA SUBUNIT1 (ASA1), YUC1, and YUC4 (Pinon et al. 2013). On the other hand, PLTs are also regulated by JA (Chen et al. 2011). Upon JA perception, PLT genes are repressed in the root stem cell niche (Fig. 2C). In JA signaling mutants, such as coi1 or myc2, JA-mediated regulation of PLT is abolished, whereas in a Trp- and auxin-deficient mutant asa1 and in an auxin overproducing mutant yuc1D, PLTs display normal responses to JA (Chen et al. 2011). Furthermore, asa1 and yuc1D mutants are fully sensitive to JA in the PR growth assay. These results argue that the JA-triggered inhibition of the PLTs and the effect of JA on root growth are independent of the effects of PLTs on auxin biosynthesis, and that auxin and JA coregulate root growth via PLTs independently of one another (Chen et al. 2011).
In LR development, there is evidence for direct crosstalk between JA and auxin (for review, see Wasternack and Hause 2013). Exogenous application of JA or an increase in endogenous JA levels caused by the induction of JA biosynthetic genes result in enhanced auxin production and signaling (Fig. 2A; Cai et al. 2015). The coi1-1 JA receptor mutant exhibits an uneven distribution of LRs and is unable to promote the formation of additional LRs in response to JA. In wild-type plants, JA treatment boosts the formation of LRs by inducing the expression of the ERF109 TF in the LR primordium, and in the tip and base of LRs (Cai et al. 2014). ERF109 binds to the promoters of auxin biosynthesis genes such as YUC2 (Cheng et al. 2006) and ASA1 (Sun et al. 2009), increasing auxin levels and promoting the emergence of LRs (Fig. 2B). Consistent with the idea of auxin acting downstream of JA in this process, mutants with compromised auxin signaling, such as solitary root (iaa14) and the double mutant arf7 arf19, fail to increase LR formation in response to JA (Raya-González et al. 2012). These observations support the idea that JA effects in LR development are mediated by auxin.
The formation of adventitious roots (ARs) is another developmental process affected by auxin–JA interactions (Gutierrez et al. 2012). Auxin controls the levels of active JA-Ile by regulating the expression of several GH3 genes, GH3.3, GH3.5, and GH3.6, whose protein products are believed to conjugate JA to amino acids, modulate the levels of both free JA and JA-Ile and, thus, fine-tune AR formation (Gutierrez et al. 2012). Auxin effects are mediated by several ARFs, with ARF6 and ARF8 up-regulating and ARF17 repressing these GH3s (Gutierrez et al. 2012). The formation of JA conjugates by the GH3s reduces the level of free, active JA-Ile and promotes the formation of ARs. Furthermore, JA induces ERF115, a TF that activates CK signaling by up-regulating ARR5 and ARR7 and CK biosynthesis by inducing the expression of IPT3, encoding one of the rate-limiting enzymes in CK production. Thus, JA represses AR formation by modifying CK homeostasis through ERF115 activity (Lakehal et al. 2020).
Upon root damage, the synergistic interaction between JA and auxin signaling pathways favors the activation of the root stem cell division and tissue regeneration (Zhou et al. 2019). In intact roots, ERF109 and ERF115 TFs keep the quiescent center (QC) of the root undifferentiated (Zhou et al. 2019). These TFs work in conjunction with auxin and CYCLIN D6;1 (CYCD6;1) to prevent the QC from dividing, while promoting cell division in other regions of the root meristem. Root wounding triggers the rapid systemic production of JA and the local accumulation of auxin at the sites of injury due to impaired PAT upon tissue damage. JA perception activates MYC2, which directly induces ERF109, which in turn targets and up-regulates CYCD6;1 and promotes auxin biosynthesis by inducing ASA1 (Zhang et al. 2019a) and YUC2 (Cai et al. 2014). JA and auxin cooperatively induce the expression of ERF115, a key TF in tissue regeneration that promotes division of the cells directly surrounding the wounded site to replenish the damaged cells (Ye et al. 2020).
As both IAA and JA-Ile are sensed by SCF E3-ligase complexes, SCFTIR and SCFCOI, respectively, that share multiple components, disruptions of these complexes (e.g., defects in the generic SCF subunits or in upstream players) result in impaired responses to both hormones (Dharmasiri et al. 2007; Moon et al. 2007). For example, single auxin resistant1 (axr1) and double axr1 axr1-like mutants defective in the RUB1-activating enzyme E1 display PR insensitivity to synthetic auxin 2,4-D and to methyl-JA (Dharmasiri et al. 2007). RUB1 modification of the Cullin SCF subunit promotes SCF activity. Thus, the ability to assemble functional SCF complexes is apparently necessary for both JA and auxin signaling.
Shoot Development
Crosstalk between the auxin signaling pathway and JA production regulates the development of floral organs according to external cues and flower phenology (Reeves et al. 2012). The auxin response factors, ARF6 and ARF8, govern the late stages of flower development leading to anthesis. The development of arf6 arf8 mutant flowers is arrested at stage 12, resulting in flowers with short petals and stamen filaments, and immature anthers and gynoecium. The arf6 arf8 flowers never undergo anthesis and are largely male and female sterile. ARF6 and ARF8 regulate flower development, in part through the TEOSINTE BRANCHED/CYCLOIDEA/PCF4-mediated induction of JA synthesis, which in turn up-regulates the expression of the JA-responsive TFs MYB21 and MYB24 that control petal, stamen, and gynoecium development (Reeves et al. 2012). Accordingly, mutants in the JA biosynthesis or signaling pathways often exhibit delayed anther dehiscence, low pollen viability, and compromised filament elongation (for review, see Song et al. 2013). During flower development, the arf6 arf8 mutant produces low levels of JA (Tabata et al. 2010). Consequently, application of exogenous JA can rescue the petal elongation and anther dehiscence defects, but not the stamen filaments or gynoecium developmental deficiencies of the arf6 arf8 double mutant (Nagpal et al. 2005), clearly indicating that JA acts downstream of the auxin signaling and response pathways with respect to petal and stamen development.
Exogenous applications of JA or auxin induce and repress leaf senescence, respectively. JA promotes leaf senescence through the COI1 signaling pathway. The MYC TFs, including MYC2, induce the expression of genes involved in senescence and chlorophyll degradation (Qi et al. 2015; Zhu et al. 2015). The WRKY57 TF plays a major role repressing the expression of senescence-associated genes (Jiang et al. 2014). Upon JA application, WRKY57 transcription is induced, whereas WRKY57 protein is rapidly turned over by the 26S proteasome pathway preventing its accumulation. In contrast, auxin treatment promotes both WRKY57 transcript and protein accumulation. Furthermore, WRKY57 physically interacts with JAZ4 and JAZ8, as well as with IAA29, negative regulators of the JA and auxin signaling pathways, respectively. This competition for WRKY57 between JAZs and IAA29 is thought to also contribute to the antagonistic role of JA and auxin in the regulation of leaf senescence (Jiang et al. 2014).
AUXIN–SA INTERACTIONS
SA is a phenolic compound produced by plants in response to pathogen exposure and, along with JA, plays a central role in plant defenses to biotic stress (for review, see Lefevere et al. 2020). Remarkably, auxin and SA share a common precursor, chorismate, which is produced via the shikimate pathway in the chloroplast (Pérez-Llorca et al. 2019). Chorismate can be converted into Trp to synthesize auxin or into isochorismate to produce SA. The SA biosynthesis pathway is thought to occur through two independent routes, one relying on PHENYLALANINE AMMONIA-LYASE (PAL) and the other on ISOCHORISMATE SYNTHASE (ICS), both residing in the cytosol (Dempsey and Klessig 2017). In the first reaction of the PAL pathway, PAL catalyzes the conversion of phenylalanine into trans-cinnamic acid, which is later metabolized to either ortho-coumaric acid or benzoic acid intermediates to finally produce SA. In the ICS pathway, ICS is responsible for the conversion of chorismate into isochorismate. Then, avrPphB SUSCEPTIBLE3 (PBS3) conjugates isochorismate with glutamate to produce isochorismate-9-glutamate, which can be converted by the acyltransferase ENHANCED PSEUDOMONAS SUSCEPTIBILITY 1 (EPS1) or spontaneously catabolized into SA (for review, see Lefevere et al. 2020).
Members of the NON-EXPRESSOR OF PATHOGENESIS-RELATED (NPR) gene family are postulated to be SA receptors. In the absence of SA, NPR1 is located in the cytoplasm in an oligomeric state. Upon SA binding, NPR1 experiences a conformational change that causes the complexes to dissociate into monomers that migrate to the nucleus where NPR1 interacts with the TGACG-BINDING FACTOR (TGA) family of TFs to induce the expression of PATHOGENESIS-RELATED genes (Wu et al. 2012). NPR3 and NPR4 bind SA and act as adaptor proteins regulating the activity of the CUL3 E3 LIGASE that degrades NPR1 depending on the SA concentration in the cell. In healthy plants, the low levels of SA trigger the degradation of NPR1 by NPR3, NPR4, and the proteasome, thus blocking the induction of defense genes. As SA levels increase upon pathogen attack, SA-bound NPR3 and NPR4 lose their ability to promote the degradation of NPR1, enabling stabilized NPR1 to induce PATHOGENESIS-RELATED genes (Fu et al. 2012).
Besides the NPRs, additional SA-binding proteins may be involved in specific NPR1-independent SA immune responses and in SA-mediated regulation of growth. SA produced by plants is involved not only in plant defenses but also in plant growth and development (Wang et al. 2007). Auxin and SA interact to modulate these processes.
Root Development
Treatments with exogenous SA shape the development of the root, at least in part by affecting auxin production and distribution. PIN efflux transporters are stabilized at the plasma membrane by SA (Du et al. 2013). Low concentrations of SA (below 50 µM) induce the formation of ARs but reduce the size of the PR meristem, whereas high concentrations of SA (above 50 µM) hamper all root developmental processes (Pasternak et al. 2019). SA has an inhibitory effect on root elongation, in part due to the activation of auxin synthesis via TAA1 induction in the epidermis of the elongation zone (Fig. 2A; Pasternak et al. 2019), similar to the expression boost triggered by ET (Stepanova et al. 2008).
The mechanisms governing AR formation are defined by the crosstalk between SA, auxin, ET, and JA (Pasternak et al. 2019). The amidosynthases encoded by the GH3 gene family are able to conjugate not only IAA, but also JA and SA, to amino acids (for review, see Woodward and Bartel 2005 and Zhang et al. 2007). The activity of GH3 enzymes controls the endogenous levels of active hormones, ultimately influencing the number of ARs and other morphometric traits (Staswick et al. 2005; Sorin et al. 2006; Gutierrez et al. 2012). For example, transgenic lines overexpressing GH3.5 exhibit reduced levels of free auxins and SA, but elevated levels of their aspartate conjugated forms, and show dramatic morphological defects including severe dwarfism (Westfall et al. 2016). Furthermore, by regulating the levels of the IAA and SA, GH3s affect plant responses to pathogens (Zhang et al. 2007) and to a wide range of abiotic stresses including drought, freezing, and salinity, consistent with the central role of the GH3 substrates in these processes (Park et al. 2007). In summary, the crosstalk between SA and auxin affects the architecture of the root and its interaction with environmental cues.
Shoot Development
During apical hook formation, ET and GA enhance the uneven distribution of auxin, promoting the formation of an exaggerated apical hook, whereas JA and SA disrupt the differential distribution of auxin and, thus, the formation of the apical hook (Wang and Guo 2019). In the presence of SA, NPR1 monomers migrate to the nucleus and interact with EIN3 and EIL1, blocking the expression of EIN3- and EIL1-target genes, including those involved in auxin biosynthesis and transport and, thus, the formation of the apical hook (Huang et al. 2020). This SA-mediated effect on the formation of the apical hook is NPR1-dependent, with formation of the apical hook being impaired in NPR1 overexpression lines and enhanced in npr1 loss-of-function mutants (Huang et al. 2020).
Pathogen Response
Auxin and SA exhibit antagonistic functions during plant defense (Wang et al. 2007). A number of pathogens are capable of either producing auxin or inducing auxin biosynthesis in the host to modify the plant developmental programs to their own benefit (Chen et al. 2007; for review, see Robert-Seilaniantz et al. 2007). The extra auxin loosens the cell walls and promotes cell elongation, favoring pathogen attack and enabling the development of symptoms.
To counteract pathogens, plants have evolved mechanisms to dampen the effects of the excess auxin produced during an attack (Spaepen et al. 2007). One primary method is to accumulate high levels of SA that inhibit the response to auxin at multiple levels. SA down-regulates the expression of several PIN genes, reducing auxin transport (Armengot et al. 2014). Furthermore, SA dampens auxin signaling by inhibiting the expression of TIR1 and AFBs, stabilizing the Aux/IAA repressors and, thus, blocking the global response to auxin (Wang et al. 2007). In addition, mutants that overaccumulate SA have lower IAA levels relative to wild-type, suggesting that the inhibitory effect of SA on auxin is in part due to a reduction in auxin signaling and response (Wang et al. 2007). SA impairs the production of auxin by inhibiting CATALASE2 (CAT2) activity. CAT2 is down-regulated in response to SA, leading to an increase in hydrogen peroxide that triggers the sulfenylation of TRP SYNTHETASE B SUBUNIT1, reducing the production of Trp and, thus, hampering the biosynthesis of auxin (Yuan et al. 2017). CAT2 coordinates the action of SA not only on auxin but also on JA production, thus affecting plant susceptibility to pathogens (Yuan et al. 2017).
AUXIN–SL INTERACTIONS
SLs are the most recent addition to the phytohormone family. They were discovered based on their involvement in promoting the germination of parasitic plants of the Striga genus (Cook et al. 1966). SLs are produced from carotenoids. While the complete picture of SL synthesis is still unfolding, in general, SL synthesis starts with the conversion of β-carotene to carlactone. This process is catalyzed by three enzymes: DWARF27 (D27), CAROTENOID CLEAVAGE7 (CCD7), and CCD8 (for review, see Omoarelojie et al. 2019). Most SL production takes place in the root, but the shoots also make SL. SL can be transported to the shoot to mediate shoot developmental processes, such as branching (Gomez-Roldan et al. 2008; Umehara et al. 2008), or secreted into the rhizosphere to mediate symbiotic and parasitic relationships (Cook et al. 1966; Akiyama et al. 2005).
The current model for SL signaling is that the SL molecule binds to its receptor, α/β-hydrolase D14, initiating a signaling cascade. In the presence of SL, D14 forms a complex with an F-box protein D3 (in Arabidopsis, MORE AXILLARY BRANCHES, MAX2). This complex directs protein targets, such as rice D53 (in Arabidopsis, three redundant members of the SUPPRESSOR OF MAX2 1-LIKE (SMXL) family) for ubiquitin-mediated degradation. The loss of these target proteins relieves the transcriptional repression of SL-regulated genes, triggering some SL-induced responses (for review, see Omoarelojie et al. 2019 and Bürger and Chory 2020). In the final step, D14 hydrolyzes and inactivates the SL molecule (Seto et al. 2019). There is also evidence for feedback regulation of SL signaling. In pea, the SL pathway mutants ramosus (rms3, rms4, and rms5, defective in the orthologs of rice OsD14, OsD3, and OsCCD7, respectively) show increased expression of PsRMS1 (OsCCD8), suggestive of feedback regulation, but plants harboring the rms2 mutation have reduced expression of PsRMS1 (Foo et al. 2005). PsRMS2 was found to encode an ortholog of the TIR1 auxin receptor that up-regulates SL biosynthesis, providing further support for the connection between auxin response and SL biosynthesis in pea (Ligerot et al. 2017).
Root Development
SL treatment of rapeseed seedlings promotes both shoot and root growth and rapidly reduces endogenous auxin levels, suggesting that auxin–SL interactions are at play both above and below ground (Ma et al. 2020). The Arabidopsis SL mutants max1, max2, and max4 (an ortholog of OsCCD8) have a shorter PR and fewer meristematic cells relative to wild-type, as well as expanded auxin reporter activity (Ruyter-Spira et al. 2011). Application of a synthetic SL to the roots of plants increases the size of the root meristem and transition zone, blocks LR formation and reduces the auxin content of leaves. SL treatment also inhibits the expression of PIN 1, 3, and 7 translational fusions with GFP in the root tip, suggesting that an interaction between SL and auxin transport is critical for balancing root growth (Ruyter-Spira et al. 2011). Similarly, SL promotes PIN2 expression and PIN2 polar localization at the plasma membrane in Arabidopsis root tips, implicating SL in the regulation of PIN-mediated auxin transport in roots (Fig. 2A; Pandya-Kumar et al. 2014).
Arabidopsis max2, max3 (orthologous to OsCCD7), and max4 mutants all display an increased number of LRs, and SL treatment reduces LR number and increases root hair length in max3 and max4 (Kapulnik et al. 2011). SL regulates LR development in rice via inhibition of auxin transport (Fig. 2A; Sun et al. 2019). SL signaling (d3) and biosynthesis (d10) mutants display an increased number of LRs, which is further enhanced by auxin treatment, but attenuated by NPA. The effect of auxin is abrogated by application of SL in d10, but not in d3, providing evidence that SL acts to block auxin-mediated LR growth.
SHY2, involved in auxin and CK signaling, promotes auxin accumulation to inhibit LR development (Goh et al. 2012). The SL signaling mutant max2 displays a similar LR phenotype as a shy2 loss-of-function mutant, and both are insensitive to SL treatment (Koren et al. 2013). Conversely, wild-type plants and a shy2 gain-of-function mutant are both sensitive to exogenous SL. The authors propose that SHY2 acts to integrate signals from multiple hormone pathways, including SL and auxin (Koren et al. 2013).
SL rice mutants have a decreased number of ARs per tiller, but higher levels of endogenous auxin and auxin reporter activity (Sun et al. 2015). Treatment of the d10 mutant with exogenous SL increases the number of ARs, decreases the expression of auxin transport genes, and inhibits auxin reporter activity. Moreover, while NPA treatment does not alter AR number in the d3 or d10 mutants, it does reduce AR number in wild-type plants to a level akin to SL mutants. Together, these data show that, in rice, SL promotes AR formation, which may be mediated by its actions on PAT. However, studies in Arabidopsis and pea suggest that, unlike in rice, SL and auxin might be largely independent of one another in their control of AR development (Rasmussen et al. 2012).
In tomato, SL treatment reduces the abundance and length of root hairs, and blocks auxin-mediated inhibition of root elongation (Koltai et al. 2010). Exogenous auxin is unable to rescue the SL phenotype, but treatment with 2,4-D, a synthetic auxin analog that is a poor substrate for auxin efflux carriers, rescues the root hair and root elongation phenotypes, suggesting that, in tomato, SL modulates root development via effects on auxin transport.
SLs are exuded from plant roots as a signal to stimulate arbuscular mycorrhizal (AM) colonization (Akiyama et al. 2005). In tomato roots, AM fungi induce SlIAA27 expression (Bassa et al. 2013) and silencing of SlIAA27 reduces AM colonization (Guillotin et al. 2017). Interestingly, silencing of SlIAA27 also reduces the expression of SL biosynthesis genes SlNSP1, SlD27, and SlMAX1, and treatment with exogenous SL restores the AM phenotype of SlIAA27-silenced plants (Guillotin et al. 2017). Further evidence in pea supports the idea that auxin and SL interact to promote AM symbiosis. An auxin-deficient mutant, bushy (bsh), exhibits lower PsCCD8 expression, reduced SL in root exudates, and decreased mycorrhizal colonization (Foo 2013). In wild-type plants, blocking endogenous auxin transport to roots by stem girdling also reduces SL levels in root exudates, and exogenous SL treatment restores colonization in the bsh mutant, suggesting that shoot-derived auxin acts through SL to promote AM colonization.
Shoot Development
It has been proposed that SL inhibits branching by acting like a second messenger for auxin: an auxin signal from the young expanding leaves in the primary shoot promotes SL production in the main stem and root, and then SL travels up into axillary meristems to influence branching (Fig. 3A; Brewer et al. 2009). Auxin-mediated apical dominance requires intact SL signaling in pea (Beveridge et al. 2000) and Arabidopsis (Sorefan et al. 2003), and SL treatment prevents decapitation-induced branching in pea (Brewer et al. 2009). Blocking PAT in the bud with NPA takes days to have an effect, whereas SL treatment rapidly inhibits bud outgrowth, suggesting that SL, and not the establishment of auxin export, regulates bud release (Brewer et al. 2009). Auxin has been shown to promote SL production during shoot branching by increasing expression of the genes encoding CCD7 and CCD8 in rice, pea, and Arabidopsis (Fig. 3C; Sorefan et al. 2003; Bainbridge et al. 2005; Foo et al. 2005; Johnson et al. 2006; Zou et al. 2006; Arite et al. 2007; Hayward et al. 2009). In Arabidopsis, auxin-triggered induction of SL biosynthesis is lost in the auxin-insensitive mutant axr1 (Hayward et al. 2009), but synthetic SL can still inhibit axr1 shoot branching (Brewer et al. 2009), supporting the idea that auxin acts through AXR1 to promote SL biosynthesis and, thus, inhibit shoot development.
SLs have been shown to alter PAT by decreasing the expression of PIN genes and/or the membrane abundance and localization of PIN proteins, leading to the inhibition of bud development in Arabidopsis (Fig. 3A,C; Bennett et al. 2006; Crawford et al. 2010; Shinohara et al. 2013). Arabidopsis SL mutants have increased auxin transport, up-regulated PIN protein expression, and enhanced branching (which is lost upon NPA treatment or when an SL mutant is crossed to the pin1 mutant), indicating that SL inhibits branching via regulation of auxin transport (Bennett et al. 2006; Lazar and Goodman 2006). When the pin3/4/7 triple mutant was crossed to highly branched SL mutants (max2 and max4), the quadruple mutants had reduced branching and less auxin transport in the stem than the SL single mutants, bringing their phenotypes closer to wild-type (van Rongen et al. 2019). Likewise, the auxin transport mutant abcb19, when crossed to SL mutants, displayed reduced branching. These findings suggest that several auxin transporters interact with SL to regulate auxin transport and branching. Similar to Arabidopsis, rice SL mutants exhibit increased auxin levels (Arite et al. 2007). However, conflicting experiments in pea found that SL mutants do not have altered auxin transport and SL did not require PAT to inhibit branching (Brewer et al. 2009, 2015). This variation could be due to differences in species, the type of mutants analyzed, or experimental design.
Auxin–SL interactions are at play in several other plant species as well. In tall fescue roots and leaves, expression of the SL signaling component genes FaD3 and FaD14 is up-regulated by exogenous auxin, but down-regulated by NPA, providing evidence for auxin-mediated regulation of SL perception and signal transduction (Hu et al. 2018b, 2019). In peach, both auxin and SL inhibit axillary bud development through actions involving CK (Li et al. 2018). Auxin-treated plants show decreased expression of PpIPTs after decapitation, and SL reduces auxin transport by decreasing PpPIN1 expression (Fig. 3C). In turn, decreased auxin transport inhibits CK biosynthesis and bud outgrowth. Likewise, in apple, SL treatment and NPA treatment have similar phenotypic effects, with both treatments decreasing the expression of MsPIN1, in contrast to CK treatment that enhances MsPIN1 transcript levels (Tan et al. 2019).
Many of the seemingly irreconcilable results may be due to the effects of a third hormone critical to branching and apical dominance: CK. CK and SL are both regulated by auxin, and converge on the TF BRANCHED1 (BRC1) (Dun et al. 2012), known to play a critical role in the SL-mediated control of branching in multiple species (Brewer et al. 2009; Minakuchi et al. 2010; Braun et al. 2012; Dun et al. 2012). CK and SL oppose one another in regulating the expression of PsBRC1 and branching (Fig. 3C; Dun et al. 2012), adding another layer of complexity to the interpretations of the auxin-SL relationship. The interactions between auxin and CK are discussed in more detail in the Auxin–CK section above.
Other Contexts
In Arabidopsis, low inorganic phosphate (Pi) conditions increase root hair density (Ma et al. 2001) and LR density, which coincides with up-regulation of the auxin receptor TIR1 (Mayzlish-Gati et al. 2012). Induction of TIR1 by low Pi is lost in a max2 mutant, but IAA treatment rescues the mutant root hair phenotype. While this effect is specific to a certain developmental time window, it suggests that SL can regulate root hair formation in response to low Pi conditions by up-regulating TIR1 and enhancing auxin signaling. Another study in Arabidopsis found that SL mediates the low Pi response by altering PIN2 polarity (Kumar et al. 2015), suggesting that SL regulates not only auxin perception, but also auxin transport. Similarly, low Pi or low N conditions in rice induce SL biosynthesis (Sun et al. 2014), and mutations in SL signaling and biosynthesis genes (OsD3; OsD10 and OsD27, respectively) lead to reduced root responses to Pi or N deficiency. Low Pi, low N, or SL treatment inhibit IAA transport from shoot to root and reduce the activity of an auxin reporter construct, suggesting that auxin–SL interactions also mediate response to nutrient availability in rice.
In tall fescue under heat stress, auxin inhibits root elongation, but SL promotes it (Hu et al. 2018b). SL and/or NPA treatment result in similar root phenotypes and SL treatment inhibits the expression of auxin transport genes (FaTIR1, FaPIN1, FaPIN2, and FaPIN5) in root tips, especially under heat stress conditions. Nearly identical results were found in tall fescue leaves (Hu et al. 2019), suggesting that SL and auxin coordinate in a general heat stress response.
CONCLUDING REMARKS
The complexity of the hormone crosstalk and the fragmentary view that we have been able to generate up to this point highlight the critical need to balance the efforts of dissecting the function of individual hormones with investigating the molecular mechanisms of hormone interactions. Although studying auxin and other growth regulators individually has proven fruitful, a more holistic outlook on signal interaction is required to better understand the true complexity of the role of these hormones in plant growth and development. What has been holding plant biologists back from generating a more comprehensive view of the hormone interaction network and from uncovering the full complexity of the signal crosstalk? Perhaps, one of the existing limitations is the lack of adequate tools to monitor multiple hormones in parallel with a cellular resolution, which is a prerequisite for identifying and implicating additional players in the process of interest or explaining a pleiotropic phenotype of a mutant. To this end, the development of multihormone reporters or biosensors that enable simultaneous detection of several hormones is clearly a pressing need. The second major roadblock in studying hormone crosstalk is the time and effort required to generate desired multigene mutant combinations to resolve gene functional redundancy and to simultaneously deregulate several interacting pathways. Fortunately, higher-order mutant generation has now been significantly accelerated with the implementation of genome-editing technologies in plants, paving the way to rapidly make mutant combinations that were previously difficult, or even impossible, to generate via traditional crosses (e.g., due to linkage). We hope that the continuous optimization and wide adoption of the latest molecular genetic tools in plant biology, such as base editing, biosensors, and single cell sequencing, will make the study of hormone interactions less daunting, will attract new talent, and shed much-needed light on the web of signal crosstalk in plants.
ACKNOWLEDGMENTS
The work in the Alonso-Stepanova laboratory is supported by the National Science Foundation Grants 1444561 and 1940829 to J.M.A. and A.N.S., and 1750006 to A.N.S.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Abeles FB, Morgan PW, Saltveit ME Jr. 1973. Ethylene in plant biology. Academic, New York. [Google Scholar]
- Addicott FT, Lyon JL, Ohkuma K, Thiessen WE, Carns HR, Smith OE, Cornforth JW, Milborrow BV, Ryback G, Wareing PF. 1968. Abscisic acid: a new name for abscisin II (dormin). Science 159: 1493. 10.1126/science.159.3822.1493 [DOI] [PubMed] [Google Scholar]
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano J, Schmelz EA, Solano R. 2007. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681. 10.1105/tpc.106.048041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H. 2012. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 22: 915–927. 10.1038/cr.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Althiab Almasaud R, Bouzayen M, Zouine M, Chervin C. 2020. Auxin and ethylene regulation of fruit set. Plant Sci 292: 110381. 10.1016/j.plantsci.2019.110381 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607. 10.1126/science.1203877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R, Buszewicz D, Sarnowski TJ, Sarnowska E, Rolicka AT, Tohge T, Fernie AR, Jikumaru Y, Kotlinski M, Iwanicka-Nowicka R, et al. 2013. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS ONE 8: e58588. 10.1371/journal.pone.0058588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029. 10.1111/j.1365-313X.2007.03210.x [DOI] [PubMed] [Google Scholar]
- Armengot L, Marquès-Bueno MM, Soria-Garcia A, Müller M, Munné-Bosch S, Martínez MC. 2014. Functional interplay between protein kinase CK2 and salicylic acid sustains PIN transcriptional expression and root development. Plant J 78: 411–423. 10.1111/tpj.12481 [DOI] [PubMed] [Google Scholar]
- Azizi P, Rafii MY, Maziah M, Abdullah SN, Hanafi MM, Latif MA, Rashid AA, Sahebi M. 2015. Understanding the shoot apical meristem regulation: a study of the phytohormones, auxin and cytokinin, in rice. Mech Dev 135: 1–15. 10.1016/j.mod.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Bai M, Shang J, Oh E, Fan M, Bai Y, Zentella R, Sun T, Wang Z. 2012a. Brassinosteroid, gibberellin, and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817. 10.1038/ncb2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Fan M, Oh E, Wang Z. 2012b. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929. 10.1105/tpc.112.105163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge K, Sorefan K, Ward S, Leyser O. 2005. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J 44: 569–580. 10.1111/j.1365-313X.2005.02548.x [DOI] [PubMed] [Google Scholar]
- Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z. 2004. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134: 1624–1631. 10.1104/pp.103.036897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassa C, Etemadi M, Combier JP, Bouzayen M, Audran-Delalande C. 2013. Sl-IAA27 gene expression is induced during arbuscular mycorrhizal symbiosis in tomato and in Medicago truncatula. Plant Signal Behav 8: e25637. 10.4161/psb.25637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benková E, Colombo L. 2012. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24: 2886–2897. 10.1105/tpc.112.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. 2006. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563. 10.1016/j.cub.2006.01.058 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M. 2008. Seed dormancy and germination. The Arabidopsis Book 6: e0119. 10.1199/tab.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Lainé S, et al. 2014. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505: 417–421. 10.1038/nature12791 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Turnbull CG. 2000. Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulated by genes Rms1 and Rms2. Plant Physiol 123: 689–698. 10.1104/pp.123.2.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielach A, Hrtyan M, Tognetti VB. 2017. Plants under stress: involvement of auxin and cytokinin. Int J Mol Sci 18: 1427. 10.3390/ijms18071427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM. 2020. Ethylene signaling in plants. J Biol Chem 295: 7710–7725. 10.1074/jbc.REV120.010854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Mortimore LA, Stepanova AN, Ecker JR, Bleecker AB. 2004. Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol 136: 2921–2927. 10.1104/pp.104.050393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binenbaum J, Weinstain R, Shani E. 2018. Gibberellin localization and transport in plants. Trends Plant Sci 23: 410–421. 10.1016/j.tplants.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benková E, Mähönen AP, Helariutta Y. 2011. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol 21: 917–926. 10.1016/j.cub.2011.04.017 [DOI] [PubMed] [Google Scholar]
- Björklund S, Antti H, Uddestrand I, Moritz T, Sundberg B. 2007. Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J 52: 499–511. 10.1111/j.1365-313X.2007.03250.x [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Green R, Nilsson O, Sussman MR, Weigel D. 1998. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10: 791–800. 10.1105/tpc.10.5.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Boivin S, Fonouni-Farde C, Frugier F. 2016. How auxin and cytokinin phytohormones modulate root microbe interactions. Front Plant Sci 7: 1240. 10.3389/fpls.2016.01240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme F, Kurz B, Melzer S, Bernier G, Jacqmard A. 2000. Cytokinin and gibberellin activate SaMADS A, a gene apparently involved in regulation of the floral transition in Sinapis alba. Plant J 24: 103–111. 10.1046/j.1365-313x.2000.00859.x [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. 2012. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238. 10.1104/pp.111.182725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. 2009. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493. 10.1104/pp.108.134783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Gui R, Mason MG, Beveridge CA. 2015. Strigolactone inhibition of branching independent of polar auxin transport. Plant Physiol 168: 1820–1829. 10.1104/pp.15.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M, Chory J. 2020. The many models of strigolactone signaling. Trends Plant Sci 25: 395–405. 10.1016/j.tplants.2019.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Xu P, Zhao P, Liu R, Yu L, Xiang C. 2014. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5: 5833. 10.1038/ncomms6833 [DOI] [PubMed] [Google Scholar]
- Cai X, Xu P, Wang Y, Xiang C. 2015. Activated expression of AtEDT1/HDG11 promotes lateral root formation in Arabidopsis mutant edt1 by upregulating jasmonate biosynthesis. J Integr Plant Biol 57: 1017–1030. 10.1111/jipb.12347 [DOI] [PubMed] [Google Scholar]
- Campos ML, Kang J, Howe GA. 2014. Jasmonate-triggered plant immunity. J Chem Ecol 40: 657–675. 10.1007/s10886-014-0468-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. 2016. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7: 12570. 10.1038/ncomms12570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latché A, Pech J, Bouzayen M. 2009. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot 60: 1349–1362. 10.1093/jxb/erp009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY. 2015. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol 25: 1031–1042. 10.1016/j.cub.2015.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran U, Luo X, Zhou W, Shu K. 2020. Multifaceted signaling networks mediated by abscisic acid insensitive 4. Plant Commun 1: 100040. 10.1016/j.xplc.2020.100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. 2013. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J Exp Bot 64: 5021–5032. 10.1093/jxb/ert291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. 2007. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci 104: 20131–20136. 10.1073/pnas.0704901104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. 2011. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. 10.1105/tpc.111.089870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Wang LY, Immanen J, Nieminen K, Spicer R, Helariutta Y, Zhang J, He XQ. 2019. Differential regulation of auxin and cytokinin during the secondary vascular tissue regeneration in Populus trees. New Phytol 224: 188–201. 10.1111/nph.16019 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799. 10.1101/gad.1415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Wang L, Sun W, Zhang Y, Zhou C, Su YH, Li W, Sun TT, Zhao XY, Li XG, et al. 2013. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol 161: 240–251. 10.1104/pp.112.203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Monte I, Zamarreño AM, Hamberg M, Lassueur S, Reymond P, Weiss S, Stintzi A, Schaller A, Porzel A, et al. 2018. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat Chem Biol 14: 171–178. 10.1038/nchembio.2540 [DOI] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. 2014. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol 16: 66–76. 10.1038/ncb2893 [DOI] [PubMed] [Google Scholar]
- Chung Y, Choe S. 2013. The regulation of brassinosteroid biosynthesis in Arabidopsis. Crit Rev Plant Sci 32: 396–410. 10.1080/07352689.2013.797856 [DOI] [Google Scholar]
- Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, Takatsuto S, Tsujimoto M, Kim H, Cho S, et al. 2011. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J 66: 564–578. 10.1111/j.1365-313X.2011.04513.x [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. 1998. BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451. 10.1146/annurev.arplant.49.1.427 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. 1996. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678. 10.1104/pp.111.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. 1966. Germination of Witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190. 10.1126/science.154.3753.1189 [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Muller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913. 10.1242/dev.051987 [DOI] [PubMed] [Google Scholar]
- Curaba J, Singh MB, Bhalla PL. 2014. miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot 65: 1425–1438. 10.1093/jxb/eru002 [DOI] [PubMed] [Google Scholar]
- de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. 2009. The Solanum lycopersicum auxin response factor 7 (Sl ARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170. 10.1111/j.1365-313X.2008.03671.x [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682. 10.1016/j.cub.2007.02.047 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. 10.1126/science.1164147 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière J, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature (London) 451: 480–484. 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- Demole E, Lederer E, Mercier D. 1962. Isolement et détermination de la structure du jasmonate de méthyle, constituant odorant caractéristique de l'essence de jasmin [Isolation and determination of the structure of methyl jasmonate, a characteristic fragrant constituent of jasmine essence]. Helv Chim Acta 45: 675–685. 10.1002/hlca.19620450233 [DOI] [Google Scholar]
- Dempsey DA, Klessig DF. 2017. How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol 15: 23. 10.1186/s12915-017-0364-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Adibi M, Breda AS, Wendrich JR, Smit ME, Novák O, Yamaguchi N, Yoshida S, Van Isterdael G, Palovaara J, et al. 2014. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345: 1255215. 10.1126/science.1255215 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, Estelle M. 2007. AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J 52: 114–123. 10.1111/j.1365-313X.2007.03211.x [DOI] [PubMed] [Google Scholar]
- Dierck R, De Keyser E, De Riek J, Dhooghe E, Van Huylenbroeck J, Prinsen E, Van Der Straeten D. 2016. Change in auxin and cytokinin levels coincides with altered expression of branching genes during axillary bud outgrowth in chrysanthemum. PLoS ONE 11: e0161732. 10.1371/journal.pone.0161732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ. 2015. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J 84: 56–69. 10.1111/tpj.12958 [DOI] [PubMed] [Google Scholar]
- Dorcey E, Urbez C, Blázquez MA, Carbonell J, Perez-Amador M. 2009. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J 58: 318–332. 10.1111/j.1365-313X.2008.03781.x [DOI] [PubMed] [Google Scholar]
- Du Y, Tejos R, Beck M, Himschoot E, Li H, Robatzek S, Vanneste S, Friml J. 2013. Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc Natl Acad Sci 110: 7946–7951. 10.1073/pnas.1220205110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. 2012. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498. 10.1104/pp.111.186783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker RJ, Strader LC. 2020. Auxin-abscisic acid interactions in plant growth and development. Biomolecules 10: 281. 10.3390/biom10020281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders TA, Strader LC. 2015. Auxin activity: past, present, and future. Am J Bot 102: 180–196. 10.3732/ajb.1400285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. 2008. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. 10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R. 2009. The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12: 539–547. 10.1016/j.pbi.2009.07.013 [DOI] [PubMed] [Google Scholar]
- Foo E. 2013. Auxin influences strigolactones in pea mycorrhizal symbiosis. J Plant Physiol 170: 523–528. 10.1016/j.jplph.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA. 2005. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474. 10.1105/tpc.104.026716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Truesdale MR, Bird CR, Schuch W, Bramley PM. 1994. Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105: 405–413. 10.1104/pp.105.1.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio M, Alabadí D, Pérez-Gómez J, García-Cárcel L, Phillips AL, Hedden P, Blázquez MA. 2006. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142: 553–563. 10.1104/pp.106.084871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. 2012. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232. 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. 2005. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44: 382–395. 10.1111/j.1365-313X.2005.02537.x [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. 10.1038/nature06206 [DOI] [PubMed] [Google Scholar]
- Gallei M, Luschnig C, Friml J. 2020. Auxin signalling in growth: Schrödinger's cat out of the bag. Curr Opin Plant Biol 53: 43–49. 10.1016/j.pbi.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Mudd SH, Datko AH. 1985. Quantitative analysis of pathways of methionine metabolism and their regulation in Lemna. Plant Physiol 78: 555–560. 10.1104/pp.78.3.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D. 2004. Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034. 10.1038/nature02778 [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. 2002. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334. 10.1104/pp.011254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. 2004. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573. 10.1104/pp.103.034736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Hooper LC, Johnson SD, Rodrigues JCM, Vivian-Smith A, Koltunow AM. 2007. Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol (Bethesda) 145: 351–366. 10.1104/pp.107.104174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. 2012. Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci 367: 1461–1468. 10.1098/rstb.2011.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. 2008. Strigolactone inhibition of shoot branching. Nature 455: 189–194. 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-Garcia S, Russinova E, Cano-Delgado AI. 2011. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859. 10.1242/dev.057331 [DOI] [PubMed] [Google Scholar]
- Gorguet B, Heusden AW, Lindhout P. 2005. Parthenocarpic fruit development in tomato. Plant Biol (Stuttgart, Germany) 7: 131–139. 10.1055/s-2005-837494 [DOI] [PubMed] [Google Scholar]
- Guillotin B, Etemadi M, Audran C, Bouzayen M, Bécard G, Combier JP. 2017. Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom). New Phytol 213: 1124–1132. 10.1111/nph.14246 [DOI] [PubMed] [Google Scholar]
- Gupta A, Singh M, Laxmi A. 2015. Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol 168: 307–320. 10.1104/pp.114.256313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, Păcurar DI, Novák O, Staswick P, Kowalczyk M, Păcurar M, Demailly H, Geiss G, et al. 2012. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24: 2515–2527. 10.1105/tpc.112.099119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. 2011. Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848. 10.1242/dev.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Sela A, Friedlander L, Savaldi-Goldstein S. 2012. BRI1 activity in the root meristem involves post-transcriptional regulation of PIN auxin efflux carriers. Plant Signal Behav 7: 68–70. 10.4161/psb.7.1.18657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. 2009. Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412. 10.1104/pp.109.137646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. 2011. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960. 10.1105/tpc.111.089029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-García J, Briones-Moreno A, Blázquez MA. 2020. Origin and evolution of gibberellin signaling and metabolism in plants. Semin Cell Dev Biol 109: 46–54. 10.1016/j.semcdb.2020.04.009 [DOI] [PubMed] [Google Scholar]
- Hossain MA, Cho J, Han M, Ahn C, Jeon J, An G, Park PB. 2010. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 167: 1512–1520. 10.1016/j.jplph.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Hu J, Israeli A, Ori N, Sun T. 2018a. The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell 30: 1710–1728. 10.1105/tpc.18.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Zhang S, Huang B. 2018b. Strigolactones and interaction with auxin regulating root elongation in tall fescue under different temperature regimes. Plant Sci 271: 34–39. 10.1016/j.plantsci.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Hu Q, Zhang S, Huang B. 2019. Strigolactones promote leaf elongation in tall fescue through upregulation of cell cycle genes and downregulation of auxin transport genes in tall fescue under different temperature regimes. Int J Mol Sci 20: 1836. 10.3390/ijms20081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Dong Z, Guo P, Zhang X, Qiu Y, Li B, Wang Y, Guo H. 2020. Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 32: 612–629. 10.1105/tpc.19.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SU, Lee SB, Kim HH, Paek KH. 2012. ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol Cells 34: 305–313. 10.1007/s10059-012-0122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH. 2002. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450. 10.1093/emboj/21.10.2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. 2004. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. 10.1126/science.1099728 [DOI] [PubMed] [Google Scholar]
- Hurný A, Cuesta C, Cavallari N, Ötvös K, Duclercq J, Dokládal L, Montesinos JC, Gallemí M, Semerádová H, Rauter T, et al. 2020. SYNERGISTIC ON AUXIN AND CYTOKININ 1 positively regulates growth and attenuates soil pathogen resistance. Nat Commun 11: 2170–2175. 10.1038/s41467-020-15895-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Kim SH, Bahk S, Ali A, Nguyen XC, Yun D, Chung WS. 2020. The auxin signaling repressor IAA8 promotes seed germination through down-regulation of ABI3 transcription in Arabidopsis. Front Plant Sci 11: 111. 10.3389/fpls.2020.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. 2006. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087. 10.1105/tpc.106.045674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanes M, Fabregas N, Chory J, Cano-Delgado AI. 2009. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci 106: 13630–13635. 10.1073/pnas.0906416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immanen J, Nieminen K, Smolander OP, Kojima M, Alonso Serra J, Koskinen P, Zhang J, Elo A, Mähönen AP, Street N, et al. 2016. Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr Biol 26: 1990–1997. 10.1016/j.cub.2016.05.053 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D. 2014. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245. 10.1105/tpc.113.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Strader LC. 2019. Interplay of auxin and cytokinin in lateral root development. Int J Mol Sci 20: 486. 10.3390/ijms20030486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. 2006. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol 142: 1014–1026. 10.1104/pp.106.087676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Gunnerås SA, Petersson SV, Tarkowski P, Graham N, May S, Dolezal K, Sandberg G, Ljung K. 2010. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22: 2956–2969. 10.1105/tpc.110.074856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Chang C. 2015. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol 169: 85–95. 10.1104/pp.15.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek P, Buchtová D, Balla J, Reinöhl V, Procházka S. 2010. Cytokinins and polar transport of auxin in axillary pea buds. Plant J 65: 571–577. [DOI] [PubMed] [Google Scholar]
- Kant S, Bi Y, Zhu T, Rothstein SJ. 2009. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol (Bethesda) 151: 691–701. 10.1104/pp.109.143875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H. 2011. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J Exp Bot 62: 2915–2924. 10.1093/jxb/erq464 [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA, Pierik R. 2011. Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67: 208–217. 10.1111/j.1365-313X.2011.04597.x [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. 2018. Cytokinin signaling in plant development. Development 145: dev149344. 10.1242/dev.149344 [DOI] [PubMed] [Google Scholar]
- Kim SK, Chang SC, Lee EJ, Chung WS, Kim YS, Hwang S, Lee JS. 2000. Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol 123: 997–1004. 10.1104/pp.123.3.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song P, Choi G. 2003. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407. 10.1105/tpc.014498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Takahashi A, Kakimoto Y, Miyazawa Y, Fujii N, Higashitani A, Takahashi H. 2007. A gene essential for hydrotropism in roots. Proc Natl Acad Sci 104: 4724–4729. 10.1073/pnas.0609929104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Ng JLP, Deinum EE, Mathesius U. 2018. Auxin transport, metabolism, and signalling during nodule initiation: indeterminate and determinate nodules. J Exp Bot 69: 229–244. 10.1093/jxb/erx308 [DOI] [PubMed] [Google Scholar]
- Koltai H, Dor E, Hershenhorn J, Joel DM, Weininger S, Lekalla S, Shealtiel H, Bhattacharya C, Eliahu E, Resnick N, et al. 2010. Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J Plant Growth Regul 29: 129–136. 10.1007/s00344-009-9122-7 [DOI] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. 1984. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plantarum 61: 377–383. 10.1111/j.1399-3054.1984.tb06343.x [DOI] [Google Scholar]
- Koren D, Resnick N, Mayzlish Gati E, Belausov E, Weininger S, Kapulnik Y, Koltai H. 2013. Strigolactone signaling in the endodermis is sufficient to restore root responses and involves SHORT HYPOCOTYL 2 (SHY2) activity. New Phytol 198: 866–874. 10.1111/nph.12189 [DOI] [PubMed] [Google Scholar]
- Kotov AA, Kotova LM. 2018. Auxin-cytokinin interactions in the regulation of correlative inhibition in two-branched pea seedlings. J Exp Bot 69: 2967–2978. 10.1093/jxb/ery117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Pandya-Kumar N, Dam A, Haor H, Mayzlish-Gati E, Belausov E, Wininger S, Abu-Abied M, McErlean CS, Bromhead LJ, et al. 2015. Arabidopsis response to low-phosphate conditions includes active changes in actin filaments and PIN2 polarization and is dependent on strigolactone signalling. J Exp Bot 66: 1499–1510. 10.1093/jxb/eru513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Shull TE, Smalle JA. 2019. Antagonistic activity of auxin and cytokinin in shoot and root organs. Plant Direct 3: e00121. 10.1002/pld3.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakehal A, Dob A, Rahneshan Z, Novak O, Escamez S, Alallaq S, Strnad M, Tuominen H, Bellini C. 2020. ETHYLENE RESPONSE FACTOR 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis. New Phytol 228: 1611–1626. 10.1111/nph.16794 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, Maes L, Vanneste S, Swarup R, Weijers D, Calvo V, Parizot B, Herrera-Rodriguez M, et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900. 10.1105/tpc.107.055863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc'h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. 2013. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458. 10.1016/j.tplants.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Lavy M, Estelle M. 2016. Mechanisms of auxin signaling. Development 143: 3226–3229. 10.1242/dev.131870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Goodman HM. 2006. MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci 103: 472–476. 10.1073/pnas.0509463102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Johnston R, Yang Y, Gallavotti A, Kojima M, Travençolo BA, Costa Lda F, Sakakibara H, Jackson D. 2009. Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol 150: 205–216. 10.1104/pp.109.137034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevere H, Bauters L, Gheysen G. 2020. Salicylic acid biosynthesis in plants. Front Plant Sci 10.3389/fpls.2020.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. 1996. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194. 10.1016/S0092-8674(00)81095-8 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK. 2011. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138: 3485–3495. 10.1242/dev.065102 [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. 1996. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401. 10.1126/science.272.5260.398 [DOI] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. 2004. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7: 193–204. 10.1016/j.devcel.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Li L, Xu J, Xu ZH, Xue HW. 2005. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17: 2738–2753. 10.1105/tpc.105.034397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wei Q, Xiao Y, Peng F. 2018. The effect of auxin and strigolactone on ATP/ADP isopentenyltransferase expression and the regulation of apical dominance in peach. Plant Cell Rep 37: 1693–1705. 10.1007/s00299-018-2343-0 [DOI] [PubMed] [Google Scholar]
- Ligerot Y, de Saint Germain A, Waldie T, Troadec C, Citerne S, Kadakia N, Pillot JP, Prigge M, Aubert G, Bendahmane A, et al. 2017. The pea branching RMS2 gene encodes the PsAFB4/5 auxin receptor and is involved in an auxin-strigolactone regulation loop. PLoS Genet 13: e1007089. 10.1371/journal.pgen.1007089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Pirrello J, Kesari R, Mila I, Roustan J, Li Z, Latché A, Pech J, Bouzayen M, Regad F. 2013a. A dominant repressor version of the tomato Sl-ERF.B3 gene confers ethylene hypersensitivity via feedback regulation of ethylene signaling and response components. Plant J 76: 406–419. 10.1111/tpj.12305 [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang H, Luan S, Li J, He Z. 2013b. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci 110: 15485–15490. 10.1073/pnas.1304651110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Chen Y, Chen Y, Shin J, Mila I, Audran C, Zouine M, Pirrello J, Bouzayen M. 2018. The tomato Ethylene Response Factor Sl-ERF.B3 integrates ethylene and auxin signaling via direct regulation of Sl-Aux/IAA27. New Phytol 219: 631–640. 10.1111/nph.15165 [DOI] [PubMed] [Google Scholar]
- Lv M, Li J. 2020. Molecular mechanisms of brassinosteroid-mediated responses to changing environments in Arabidopsis. Int J Mol Sci 21: 2737. 10.3390/ijms21082737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Zhang M, Li X, Ye R, Wang X. 2018. Transcriptome profiles reveal the crucial roles of auxin and cytokinin in the “shoot branching” of Cremastra appendiculata. Int J Mol Sci 19: 3354. 10.3390/ijms19113354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. 2001. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 24: 459–467. 10.1046/j.1365-3040.2001.00695.x [DOI] [Google Scholar]
- Ma N, Wan L, Zhao W, Liu H, Li J, Zhang C. 2020. Exogenous strigolactones promote lateral root growth by reducing the endogenous auxin level in rapeseed. J Integrative Agriculture 19: 465–482. 10.1016/S2095-3119(19)62810-8 [DOI] [Google Scholar]
- Machado RAR, McClure M, Hervé MR, Baldwin IT, Erb M. 2016. Benefits of jasmonate-dependent defenses against vertebrate herbivores in nature. eLife 5: e13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado RAR, Baldwin IT, Erb M. 2017. Herbivory-induced jasmonates constrain plant sugar accumulation and growth by antagonizing gibberellin signaling and not by promoting secondary metabolite production. New Phytol 215: 803–812. 10.1111/nph.14597 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. 10.1038/nature13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava NB. 1988. Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39: 23–52. 10.1146/annurev.pp.39.060188.000323 [DOI] [Google Scholar]
- Marhava P, Bassukas AEL, Zourelidou M, Kolb M, Moret B, Fastner A, Schulze WX, Cattaneo P, Hammes UZ, Schwechheimer C, et al. 2018. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558: 297–300. 10.1038/s41586-018-0186-z [DOI] [PubMed] [Google Scholar]
- Marhava P, Aliaga Fandino AC, Koh SWH, Jelínková A, Kolb M, Janacek DP, Breda AS, Cattaneo P, Hammes UZ, Petrášek J, et al. 2020. Plasma membrane domain patterning and self-reinforcing polarity in Arabidopsis. Dev Cell 52: 223–235.e5. 10.1016/j.devcel.2019.11.015 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Pařezová M, Petrášek J, Friml J, Kleine-Vehn J, et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804. 10.1016/j.devcel.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Marhavý P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benková E. 2014. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr Biol 24: 1031–1037. 10.1016/j.cub.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. 2011. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci 108: 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayzlish-Gati E, De-Cuyper C, Goormachtig S, Beeckman T, Vuylsteke M, Brewer PB, Beveridge CA, Yermiyahu U, Kaplan Y, Enzer Y, et al. 2012. Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol 160: 1329–1341. 10.1104/pp.112.202358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN. 2017. The triple response assay and its use to characterize ethylene mutants in Arabidopsis. Methods Mol Biol 1573: 163–209. 10.1007/978-1-4939-6854-1_13 [DOI] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enríquez P, Binder BM, Heber S, Stepanova AN, Alonso JM. 2015. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697. 10.1016/j.cell.2015.09.036 [DOI] [PubMed] [Google Scholar]
- Meudt WJ. 1987. Investigations on the mechanism of the brassinosteroid response. VI: Effect of brassinolide on gravitropism of bean hypocotyls. Plant Physiol 83: 195–198. 10.1104/pp.83.1.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Ho CH, Enders TA, Floro E, Damodaran S, Gunther LK, Powers SK, Frick EM, Topp CN, Frommer WB, et al. 2019. TRANSPORTER OF IBA1 links auxin and cytokinin to influence root architecture. Dev Cell 50: 599–609.e4. 10.1016/j.devcel.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CO, Skoog F, Von Saltza MH, Strong FM. 1955. Kinetin, a cell division factor from DEOXYRIBONUCLEIC ACID. J Am Chem Soc 77: 1392. 10.1021/ja01610a105 [DOI] [Google Scholar]
- Miller CO, Skoog F, Okumura FS, Von Saltza MH, Strong FM. 1956. Isolation, structure and synthesis of kinetin, a substance promoting cell division. J Am Chem Soc 78: 1375–1380. 10.1021/ja01588a032 [DOI] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, et al. 2010. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51: 1127–1135. 10.1093/pcp/pcq083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. 2004. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138. 10.1046/j.1365-313X.2003.01945.x [DOI] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH. 2004. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci 101: 16091–16098. 10.1073/pnas.0407107101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhao Y, Dai X, Zhang W, Gray WM, Huq E, Estelle M. 2007. A new CULLIN 1 mutant has altered responses to hormones and light in Arabidopsis. Plant Physiol 143: 684–696. 10.1104/pp.106.091439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S, Bishopp A, Carvalho H, Campilho A. 2013. AHP6 inhibits cytokinin signaling to regulate the orientation of pericycle cell division during lateral root initiation. PLoS ONE 8: e56370. 10.1371/journal.pone.0056370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki T, Miyazawa Y, Kobayashi A, Uchida M, Watanabe C, Fujii N, Takahashi H. 2011. Hormonal regulation of lateral root development in Arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiol 157: 1209–1220. 10.1104/pp.111.186270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143. 10.1016/j.cub.2010.05.035 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sozzani R, Pacifici E, Salvi E, Terpstra I, Bao D, van Dijken A, Dello Ioio R, Perilli S, et al. 2013. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell 26: 405–415. 10.1016/j.devcel.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. 2004. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714. 10.1101/gad.1187704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS. 2006. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443: 458–461. 10.1038/nature05130 [DOI] [PubMed] [Google Scholar]
- Mounet F, Moing A, Kowalczyk M, Rohrmann J, Petit J, Garcia V, Maucourt M, Yano K, Deborde C, Aoki K, et al. 2012. Down-regulation of a single auxin efflux transport protein in tomato induces precocious fruit development. J Exp Bot 63: 4901–4917. 10.1093/jxb/ers167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Mazumder M, Jana J, Archana KS, Mondal B, De A, Ghosh S, Saha U, Bose R, Chatterjee S, et al. 2019. Enhancement of ABA sensitivity through conditional expression of the ARF10 gene in Brassica juncea reveals fertile plants with tolerance against Alternaria brassicicola. MPMI 32: 1429–1447. 10.1094/MPMI-05-19-0132-R [DOI] [PubMed] [Google Scholar]
- Müller D, Leyser O. 2011. Auxin, cytokinin and the control of shoot branching. Ann Bot 107: 1203–1212. 10.1093/aob/mcr069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. 10.1038/nature06943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Waldie T, Miyawaki K, To JP, Melnyk CW, Kieber JJ, Kakimoto T, Leyser O. 2015. Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J 82: 874–886. 10.1111/tpj.12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munguía-Rodríguez AG, López-Bucio JS, Ruiz-Herrera LF, Ortiz-Castro R, Guevara-García AA, Marsch-Martínez N, Carreón-Abud Y, López-Bucio J, Martínez-Trujillo M. 2020. YUCCA4 overexpression modulates auxin biosynthesis and transport and influences plant growth and development via crosstalk with abscisic acid in Arabidopsis thaliana. Genet Mol Biol 43: e20190221. 10.1590/1678-4685-gmb-2019-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T. 2002. Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251. 10.1104/pp.011003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. 2005. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118. 10.1242/dev.01955 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S. 2003. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133: 1843–1853. 10.1104/pp.103.030031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Nakajima N, Goda H, Shimada Y, Hayashi K, Nozaki H, Asami T, Yoshida S, Fujioka S. 2006. Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J 45: 193–205. 10.1111/j.1365-313X.2005.02582.x [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. 10.1371/journal.pbio.0020258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan TM, Vukašinović N, Liu D, Russinova E, Yin Y. 2020. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32: 295–318. 10.1105/tpc.19.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G. 2004. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci 101: 8039–8044. 10.1073/pnas.0402504101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J, Ryu H, Hwang I, Wang Z. 2014. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun 5: 4140. 10.1038/ncomms5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoarelojie LO, Kulkarni MG, Finnie JF, Van Staden J. 2019. Strigolactones and their crosstalk with other phytohormones. Ann Bot 124: 749–767. 10.1093/aob/mcz100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. 2007. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791. 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. 2005. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300. 10.1105/tpc.105.036723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. 2010. Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. 10.1101/cshperspect.a001537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Palni LM, Burch L, Horgan R. 1988. The effect of auxin concentration on cytokinin stability and metabolism. Planta 174: 231–234. 10.1007/BF00394775 [DOI] [PubMed] [Google Scholar]
- Pandya-Kumar N, Shema R, Kumar M, Mayzlish-Gati E, Levy D, Zemach H, Belausov E, Wininger S, Abu-Abied M, Kapulnik Y, et al. 2014. Strigolactone analog GR24 triggers changes in PIN2 polarity, vesicle trafficking and actin filament architecture. New Phytol 202: 1184–1196. 10.1111/nph.12744 [DOI] [PubMed] [Google Scholar]
- Park WJ. 1998. Effect of epibrassinolide on hypocotyl growth of the tomato mutant diageotropica. Planta 207: 120–124. 10.1007/s004250050463 [DOI] [PubMed] [Google Scholar]
- Park J, Park J, Kim Y, Staswick PE, Jeon J, Yun J, Kim S, Kim J, Lee Y, Park C. 2007. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282: 10036–10046. 10.1074/jbc.M610524200 [DOI] [PubMed] [Google Scholar]
- Pasternak T, Groot EP, Kazantsev FV, Teale W, Omelyanchuk N, Kovrizhnykh V, Palme K, Mironova VV. 2019. Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol 180: 1725–1739. 10.1104/pp.19.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P, Yan Z, Zhu Y, Li J. 2008. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol Plant 1: 338–346. 10.1093/mp/ssn001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Ma W, Chen L, Yang L, Li S, Zhao H, Zhao Y, Jin W, Li N, Bevan MW, et al. 2013. Control of root meristem size by DA1-RELATED PROTEIN2 in Arabidopsis. Plant Physiol 161: 1542–1556. 10.1104/pp.112.210237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zhao J, Neff MM. 2015. ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development 142: 4129–4138. 10.1242/dev.124347 [DOI] [PubMed] [Google Scholar]
- Pérez-Llorca M, Muñoz P, Müller M, Munné-Bosch S. 2019. Biosynthesis, metabolism and function of auxin, salicylic acid and melatonin in climacteric and non-climacteric fruits. Front Plant Sci 10: 136. 10.3389/fpls.2019.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Perez-Perez J, Di Mambro R, Peris CL, Diaz-Trivino S, Del Bianco M, Pierdonati E, Moubayidin L, Cruz-Ramirez A, Costantino P, et al. 2013. RETINOBLASTOMA-RELATED protein stimulates cell differentiation in the Arabidopsis root meristem by interacting with cytokinin signaling. Plant Cell 25: 4469–4478. 10.1105/tpc.113.116632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisova M, Klima P, Horak J, Valkova M, Malbeck J, Soucek P, Reichman P, Hoyerova K, Dubova J, Friml J, et al. 2009. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci 106: 3609–3614. 10.1073/pnas.0811539106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisova M, Prat T, Grones P, Harustiakova D, Matonohova M, Spichal L, Nodzynski T, Friml J, Hejatko J. 2016. Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol 212: 497–509. 10.1111/nph.14049 [DOI] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez-Perez GF, Scheres B. 2013. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci 110: 1107–1112. 10.1073/pnas.1213497110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piya S, Shrestha SK, Binder B, Stewart CN, Hewezi T. 2014. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5: 744. 10.3389/fpls.2014.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI. 2019. Brassinosteroid signaling in plant development and adaptation to stress. Development 146: dev151894. 10.1242/dev.151894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promchuea S, Zhu Y, Chen Z, Zhang J, Gong Z. 2017. ARF2 coordinates with PLETHORAs and PINs to orchestrate ABA-mediated root meristem activity in Arabidopsis. J Integr Plant Biol 59: 30–43. 10.1111/jipb.12506 [DOI] [PubMed] [Google Scholar]
- Przemeck GH, Mattsson J, Hardtke C, Sung ZR, Berleth T. 1996. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200: 229–237. 10.1007/BF00208313 [DOI] [PubMed] [Google Scholar]
- Qi L, Yan J, Li Y, Jiang H, Sun J, Chen Q, Li H, Chu J, Yan C, Sun X, et al. 2012. Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol 195: 872–882. 10.1111/j.1469-8137.2012.04208.x [DOI] [PubMed] [Google Scholar]
- Qi T, Wang J, Huang H, Liu B, Gao H, Liu Y, Song S, Xie D. 2015. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27: 1634–1649. 10.1105/tpc.15.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Guan SC, Wen C, Li P, Gao Z, Chen X. 2019. Auxin and cytokinin coordinate the dormancy and outgrowth of axillary bud in strawberry runner. BMC Plant Biol 19: 528. 10.1186/s12870-019-2151-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher EH, Möller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. 2011. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 68: 597–606. 10.1111/j.1365-313X.2011.04710.x [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, Brewer PB, Herold S, Agusti J, Geelen D, Greb T, Goormachtig S, Beeckman T, et al. 2012. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol 158: 1976–1987. 10.1104/pp.111.187104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya-González J, Pelagio-Flores R, López-Bucio J. 2012. The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J Plant Physiol 169: 1348–1358. 10.1016/j.jplph.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, Wu M, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al. 2012. A regulatory network for coordinated flower maturation. PLoS Genet 8: e1002506. 10.1371/journal.pgen.1002506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zúñiga-Mayo VM, Serwatowska J, Chavez Montes RA, Lozano-Sotomayor P, Herrera-Ubaldo H, Gonzalez-Aguilera KL, Ballester P, Ripoll JJ, Ezquer I, et al. 2017. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet 13: e1006726. 10.1371/journal.pgen.1006726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. 2010. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104. 10.1101/gad.594910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Zourelidou M, Schwechheimer C. 2013. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc Natl Acad Sci 110: 13192–13197. 10.1073/pnas.1304250110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG. 2007. Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379. 10.1016/j.pbi.2007.06.003 [DOI] [PubMed] [Google Scholar]
- Rohde A, De Rycke R, Beeckman T, Engler G, Van Montagu M, Boerjan W. 2000. ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12: 35–52. 10.1105/tpc.12.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa NM, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Grønlund AL, Wanchoo-Kohli A, Thomas SG, Bennett MJ, et al. 2015. Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet 11: e1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, O'neill DP, Smith JJ, Kerckhoffs LH, Elliott RC. 2000. Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J 21: 547–552. 10.1046/j.1365-313x.2000.00702.x [DOI] [PubMed] [Google Scholar]
- Ruan J, Zhou Y, Zhou M, Yan J, Khurshid M, Weng W, Cheng J, Zhang K. 2019. Jasmonic acid signaling pathway in plants. Int J Mol Sci 20: 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734. 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212. 10.1105/tpc.107.052126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Šimášková M, Duclercq J, Petrášek J, Zažímalová E, Simon S, Friml J, Van Montagu, Marc CE, Benková E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci 106: 4284–4289. 10.1073/pnas.0900060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadura I, Pociecha E, Dziurka M, Oklestkova J, Novak O, Gruszka D, Janeczko A. 2019. Mutations in the HvDWARF, HvCPD and HvBRI1 genes-involved in brassinosteroid biosynthesis/signalling: altered photosynthetic efficiency, hormonal homeostasis and tolerance to high/low temperatures in barley. J Plant Growth Regul 38: 1062–1081. 10.1007/s00344-019-09914-z [DOI] [Google Scholar]
- Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S. 2013. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J 73: 676–688. 10.1111/tpj.12071 [DOI] [PubMed] [Google Scholar]
- Salanenka Y, Verstraeten I, Löfke C, Tabata K, Naramoto S, Glanc M, Friml J. 2018. Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc Natl Acad Sci 115: 3716–3721. 10.1073/pnas.1721760115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L, Gorte M, Prasad K, Bao D, Timmermans-Hereijgers JL, et al. 2016. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28: 2937–2951. 10.1105/tpc.16.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, Fernie AR, Jikumaru Y, Kamiya Y, Franzen R, Schmelzer E, et al. 2013. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol 163: 305–317. 10.1104/pp.113.223933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacchi E, Osmont KS, Beuchat J, Salinas P, Navarrete-Gomez M, Trigueros M, Ferrandiz C, Hardtke CS. 2009. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136: 2059–2067. 10.1242/dev.035444 [DOI] [PubMed] [Google Scholar]
- Scacchi E, Salinas P, Gujas B, Santuari L, Krogan N, Ragni L, Berleth T, Hardtke CS. 2010. Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc Natl Acad Sci 107: 22734–22739. 10.1073/pnas.1014716108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27: 44–63. 10.1105/tpc.114.133595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916. 10.1038/nature08836 [DOI] [PubMed] [Google Scholar]
- Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, et al. 2019. Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10: 191–197. 10.1038/s41467-018-08124-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Ben-Gera H, Shleizer-Burko S, Burko Y, Weiss D, Ori N. 2010. Cytokinin regulates compound leaf development in tomato. Plant Cell 22: 3206–3217. 10.1105/tpc.110.078253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. 2007. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453. 10.1105/tpc.107.050963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. 2013. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. 10.1371/journal.pbio.1001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D. 2010. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573. 10.1105/tpc.110.074641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q. 2013. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9: e1003577. 10.1371/journal.pgen.1003577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu Y, Liu R, Zhang H, Wang P, Li Y, Wang S, Tang S, Liu C, et al. 2016. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J 85: 348–361. 10.1111/tpj.13109 [DOI] [PubMed] [Google Scholar]
- Singh M, Gupta A, Laxmi A. 2014. Glucose control of root growth direction in Arabidopsis thaliana. J Exp Bot 65: 2981–2993. 10.1093/jxb/eru146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Xie D. 2013. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant 6: 1065–1073. 10.1093/mp/sst054 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogne K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474. 10.1101/gad.256603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Negroni L, Balliau T, Corti H, Jacquemot M, Davanture M, Sandberg G, Zivy M, Bellini C. 2006. Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol 140: 349–364. 10.1104/pp.105.067868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. 2007. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31: 425–448. 10.1111/j.1574-6976.2007.00072.x [DOI] [PubMed] [Google Scholar]
- Srivastava A, Handa AK. 2005. Hormonal regulation of tomato fruit development: a molecular perspective. J Plant Growth Regul 24: 67–82. 10.1007/s00344-005-0015-0 [DOI] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627. 10.1105/tpc.104.026690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM. 2007. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185. 10.1105/tpc.107.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie D, Doležal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM. 2011. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973. 10.1105/tpc.111.088047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street IH, Aman S, Zubo Y, Ramzan A, Wang X, Shakeel SN, Kieber JJ, Schaller GE. 2015. Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol 169: 338–350. 10.1104/pp.15.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan J, Bouzayen M, Giuliano G, Chervin C. 2015. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol 15: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. 2009. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511. 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777. 10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G. 2014. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65: 6735–6746. 10.1093/jxb/eru029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tao J, Hou M, Huang S, Chen S, Liang Z, Xie T, Wei Y, Xie X, Yoneyama K, et al. 2015. A strigolactone signal is required for adventitious root formation in rice. Ann Bot 115: 1155–1162. 10.1093/aob/mcv052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Xu F, Guo X, Wu D, Zhang X, Lou M, Luo F, Zhao Q, Xu G, Zhang Y. 2019. A Strigolactone signal inhibits secondary lateral root development in rice. Front Plant Sci 10: 1527. 10.3389/fpls.2019.01527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Feraru E, Feraru MI, Waidmann S, Wang W, Passaia G, Wang ZY, Wabnik K, Kleine-Vehn J. 2020. PIN-LIKES coordinate brassinosteroid signaling with nuclear auxin input in Arabidopsis thaliana. Curr Biol 30: 1579–1588.e6. 10.1016/j.cub.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GTS, Sandberg G, Bhalerao R, Ljung K, Bennett MJ. 2007. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196. 10.1105/tpc.107.052100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. 1996. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182. 10.1016/S0092-8674(00)81094-6 [DOI] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian C, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S. 2010. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51: 164–175. 10.1093/pcp/pcp176 [DOI] [PubMed] [Google Scholar]
- Tan M, Li G, Chen X, Xing L, Ma J, Zhang D, Ge H, Han M, Sha G, An N. 2019. Role of cytokinin, strigolactone, and auxin export on outgrowth of axillary buds in apple. Front Plant Sci 10: 616. 10.3389/fpls.2019.00616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. 2003. Physiological roles of brassinosteroids in early growth of Arabidopsis: brassinosteroids have a synergistic relationship with gibberellin as well as auxin in light-grown hypocotyl elongation. J Plant Growth Regul 22: 259–271. 10.1007/s00344-003-0119-3 [DOI] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. 2006. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036. 10.1111/j.1365-313X.2006.02656.x [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. 2002. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319. 10.1105/tpc.010283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, Yamamoto KT. 2004. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J 40: 333–343. 10.1111/j.1365-313X.2004.02220.x [DOI] [PubMed] [Google Scholar]
- Tognetti VB, Bielach A, Hrtyan M. 2017. Redox regulation at the site of primary growth: auxin, cytokinin and ROS crosstalk. Plant Cell Environ 40: 2586–2605. 10.1111/pce.13021 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Beemster GTS, Bennett MJ. 2012. Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci 17: 326–331. 10.1016/j.tplants.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Petrásek J, Zádníková P, Hoyerová K, Pesek B, Raz V, Swarup R, Bennett M, Zazímalová E, Benková E, et al. 2010. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606. 10.1242/dev.040790 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Suslov D, De Grauwe L, Leroux O, Vissenberg K, Van der Straeten D. 2011. The role of brassinosteroids in shoot gravitropism. Plant Physiol 156: 1331–1336. 10.1104/pp.111.177873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Callebert P, Zadnikova P, Benkova E, Van Der Straeten D. 2013. Brassinosteroid control of shoot gravitropism interacts with ethylene and depends on auxin signaling components. Am J Bot 100: 215–225. 10.3732/ajb.1200264 [DOI] [PubMed] [Google Scholar]
- van Rongen M, Bennett T, Ticchiarelli F, Leyser O. 2019. Connective auxin transport contributes to strigolactone-mediated shoot branching control independent of the transcription factor BRC1. PLoS Genet 15: e1008023. 10.1371/journal.pgen.1008023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva II, Qudeimat E, Potuschak T, Du Y, Genschik P, Vandenbussche F, Van Der Straeten D. 2018. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc Natl Acad Sci 115: E4130–E4139. 10.1073/pnas.1717649115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. 2008. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci 105: 9829–9834. 10.1073/pnas.0803996105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Arnim A, Deng X. 1996. Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol 47: 215–243. 10.1146/annurev.arplant.47.1.215 [DOI] [PubMed] [Google Scholar]
- Waidmann S, Ruiz Rosquete M, Schöller M, Sarkel E, Lindner H, LaRue T, Petřík I, Dünser K, Martopawiro S, Sasidharan R, et al. 2019. Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots. Nat Commun 10: 3540–3544. 10.1038/s41467-019-11483-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldie T, Leyser O. 2018. Cytokinin targets auxin transport to promote shoot branching. Plant Physiol 177: 803–818. 10.1104/pp.17.01691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guo H. 2019. On hormonal regulation of the dynamic apical hook development. New Phytol 222: 1230–1234. 10.1111/nph.15626 [DOI] [PubMed] [Google Scholar]
- Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latché A, Pech JC, Bouzayen M. 2005a. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692. 10.1105/tpc.105.033415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang L, Mao Y, Cai W, Xue H, Chen X. 2005b. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216. 10.1105/tpc.105.033076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Römheld V, Li C, Bangerth F. 2006. Involvement of auxin and CKs in boron deficiency induced changes in apical dominance of pea plants (Pisum sativum L.). J Plant Physiol 163: 591–600. 10.1016/j.jplph.2005.09.014 [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. 2007. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790. 10.1016/j.cub.2007.09.025 [DOI] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech J, Fernie AR, Bouzayen M. 2009. Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452. 10.1105/tpc.108.060830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hua D, He J, Duan Y, Chen Z, Hong X, Gong Z. 2011. Auxin response factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet 7: e1002172. 10.1371/journal.pgen.1002172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y. 2014. The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067. 10.1105/tpc.114.123083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang R, Mao X, Li L, Chang X, Zhang X, Jing R. 2019. TaARF4 genes are linked to root growth and plant height in wheat. Ann Bot 124: 903–915. 10.1093/aob/mcy218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058. 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. 2006. New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8: 371–381. 10.1055/s-2006-923928 [DOI] [PubMed] [Google Scholar]
- Westfall CS, Sherp AM, Zubieta C, Alvarez S, Schraft E, Marcellin R, Ramirez L, Jez JM. 2016. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc Natl Acad Sci 113: 13917–13922. 10.1073/pnas.1612635113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. 2011. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci 108: 18518–18523. 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Ann Bot 95: 707–735. 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C. 2012. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1: 639–647. 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Wu M, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. 2015. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4: e09269. 10.7554/eLife.09269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Zhao Y, Gao J, Xiang C, Zhu JK. 2016. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci Rep 6: 27177. 10.1038/srep27177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zha M, Li Y, Ding Y, Chen L, Ding C, Wang S. 2015. The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Rep 34: 1647–1662. 10.1007/s00299-015-1815-8 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Wu M, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D. 2013. A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell 24: 271–282. 10.1016/j.devcel.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Miyazawa Y, Kobayashi A, Moriwaki T, Fujii N, Takahashi H. 2012. MIZ1, an essential protein for root hydrotropism, is associated with the cytoplasmic face of the endoplasmic reticulum membrane in Arabidopsis root cells. FEBS Lett 586: 398–402. 10.1016/j.febslet.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Yanai O, Shani E, Russ D, Ori N. 2011. Gibberellin partly mediates LANCEOLATE activity in tomato: gibberellin partly mediates LANCEOLATE activity. Plant J 68: 571–582. 10.1111/j.1365-313X.2011.04716.x [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher plants. Ann Rev Plant Physiol 35: 155–189. 10.1146/annurev.pp.35.060184.001103 [DOI] [Google Scholar]
- Yang X, Yang Y, Xue L, Zou M, Liu J, Chen F, Xue H. 2011. Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol 156: 1397–1409. 10.1104/pp.111.173427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZB, Liu G, Liu J, Zhang B, Meng W, Müller B, Hayashi KI, Zhang X, Zhao Z, De Smet I, et al. 2017. Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep 18: 1213–1230. 10.15252/embr.201643806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Shang G, Pan Y, Xu Z, Zhou C, Mao Y, Bao N, Sun L, Xu T, Wang J. 2020. AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 32: 226–241. 10.1105/tpc.19.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. 2011. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646. 10.1111/j.1365-313X.2010.04449.x [DOI] [PubMed] [Google Scholar]
- Yu F, Wu Y, Xie Q. 2015. Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci 20: 569–575. 10.1016/j.tplants.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Yuan TT, Xu HH, Zhang KX, Guo TT, Lu YT. 2014. Glucose inhibits root meristem growth via ABA INSENSITIVE 5, which represses PIN1 accumulation and auxin activity in Arabidopsis. Plant Cell Environ 37: 1338–1350. 10.1111/pce.12233 [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu W, Lu Y. 2017. CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21: 143–155. 10.1016/j.chom.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, et al. 2010. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617. 10.1242/dev.041277 [DOI] [PubMed] [Google Scholar]
- Zha M, Imran M, Wang Y, Xu J, Ding Y, Wang S. 2019. Transcriptome analysis revealed the interaction among strigolactones, auxin, and cytokinin in controlling the shoot branching of rice. Plant Cell Rep 38: 279–293. 10.1007/s00299-018-2361-y [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhang X, Wang J, Letham DS, McKinney SA, Higgins TJV. 1995. The effect of auxin on cytokinin levels and metabolism in transgenic tobacco tissue expressing an ipt gene. Planta 196: 84–94. 10.1007/BF00193221 [DOI] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z. 2007. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145: 450–464. 10.1104/pp.107.106021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, To JP, Cheng CY, Schaller GE, Kieber JJ. 2011. Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J 68: 1–10. 10.1111/j.1365-313X.2011.04668.x [DOI] [PubMed] [Google Scholar]
- Zhang W, Swarup R, Bennett M, Schaller GE, Kieber JJ. 2013. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr Biol 23: 1979–1989. 10.1016/j.cub.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Zhang K, Wang R, Zi H, Li Y, Cao X, Li D, Guo L, Tong J, Pan Y, Jiao Y, et al. 2018. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 30: 324–346. 10.1105/tpc.17.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zhao F, Chen L, Pan Y, Sun L, Bao N, Zhang T, Cui C, Qiu Z, Zhang Y, et al. 2019a. Jasmonate-mediated wound signalling promotes plant regeneration. Nat Plants 5: 491–497. 10.1038/s41477-019-0408-x [DOI] [PubMed] [Google Scholar]
- Zhang W, Abdelrahman M, Jiu S, Guan L, Han J, Zheng T, Jia H, Song C, Fang J, Wang C. 2019b. VvmiR160s/VvARFs interaction and their spatio-temporal expression/cleavage products during GA-induced grape parthenocarpy. BMC Plant Biol 19: 111. 10.1186/s12870-019-1719-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. 2010. Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092. 10.1038/nature09126 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang X, Hou Y, Gao J, Wang P, Duan C, Zhu X, Zhu J. 2014. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal 7: ra53. 10.1126/scisignal.2005051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Miller ND, Lewis DR, Christians MJ, Lee KH, Muday GK, Spalding EP, Vierstra RD. 2011. AUXIN UP-REGULATED F-BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiol 156: 1878–1893. 10.1104/pp.111.179812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Dai X, Zhao Y, Ljung K, Noel JP, Chory J. 2013. Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat Chem Biol 9: 244–246. 10.1038/nchembio.1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Yuan F, Wang M, Guo A, Zhang Y, Xie CG. 2013. Molecular characterization of an ABA insensitive 5 orthologue in Brassica oleracea. Biochem Biophys Res Commun 430: 1140–1146. 10.1016/j.bbrc.2012.12.023 [DOI] [PubMed] [Google Scholar]
- Zhou W, Lozano-Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, Scheres B. 2019. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177: 942–956.e14. 10.1016/j.cell.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Zhou J, Sittmann J, Guo L, Huang X, Pulapaka A, Liu Z. 2020. Gibberellin and auxin signaling genes RGA1 and ARF8 repress accessory fruit initiation in diploid strawberry. Plant Physiol 10.1093/plphys/kiaa087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B. 2015. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J 84: 597–610. 10.1111/tpj.13030 [DOI] [PubMed] [Google Scholar]
- Zhu M, Chen W, Mirabet V, Hong L, Bovio S, Strauss S, Schwarz EM, Tsugawa S, Wang Z, Smith RS, et al. 2020. Robust organ size requires robust timing of initiation orchestrated by focused auxin and cytokinin signalling. Nat Plants 6: 686–698. 10.1038/s41477-020-0666-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. 2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48: 687–698. 10.1111/j.1365-313X.2006.02916.x [DOI] [PubMed] [Google Scholar]
- Zuniga-Mayo VM, Reyes-Olalde JI, Marsch-Martinez N, de Folter S. 2014. Cytokinin treatments affect the apical-basal patterning of the Arabidopsis gynoecium and resemble the effects of polar auxin transport inhibition. Front Plant Sci 5: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD. 1994. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol 104: 161–170. 10.1104/pp.104.1.161 [DOI] [PMC free article] [PubMed] [Google Scholar]