Abstract
Immunological memory and exhaustion are fundamental features of adaptive immunity. Recent advances reveal increasing heterogeneity and diversity among CD8 T-cell subsets, resulting in new subsets to annotate and understand. Here, we review our current knowledge of differentiation and maintenance of memory and exhausted CD8 T cells, including phenotypic classification, developmental paths, transcriptional and epigenetic features, and cell intrinsic and extrinsic factors. Additionally, we use this outline to discuss the nomenclature of effector, memory, and exhausted CD8 T cells. Finally, we discuss how new findings about these cell types may impact the therapeutic efficacy and development of immunotherapies targeting effector, memory, and/or exhausted CD8 T cells in chronic infections and cancer.
EFFECTOR AND MEMORY CD8 T-CELL DIFFERENTIATION
Following acutely resolved infection or vaccination, naive CD8 T cells are activated, undergo robust clonal expansion, and give rise to a diverse pool of effector CD8 T (Teff) cells. If activated by signals 1 (antigen), 2 (costimulation), and 3 (inflammation), most CD8 T cells differentiate into short-lived effector cells (SLECs). Although SLECs are heterogeneous, they often express KLRG1 and/or CX3CR1, produce cytokines and cytotoxic molecules, express activation markers, and acquire inflammatory homing properties. The majority of Teff cells, mostly SLECs, die in the weeks following the peak of expansion, usually (though not always) corresponding to clearance of antigen. However, a small subset of this activated, clonally expanded pool, known as memory precursors (Tmp cells), has the potential to differentiate into long-lived memory CD8 T (Tmem) cells. Some SLECs also persist, but these do not further differentiate into Tmem cells. These Tmem cells persist long term and acquire the ability to undergo antigen-independent self-renew via interleukin (IL)-7 and IL-15. This Tmem cell differentiation is accompanied by dynamic transcriptional, epigenetic, and metabolic reprogramming (Cui and Kaech 2010; Chang et al. 2014; Zehn and Wherry 2015; Jameson and Masopust 2018). Although this general outline describes the broad features of events following acute infection, there is considerable cellular and population heterogeneity at each stage of differentiation. Moreover, if antigen is not cleared, the development of exhausted CD8 T (Tex) cells occurs instead of Tmem cells, a topic discussed further below. Although parallels exist in concept for CD4 T-cell memory, there is clearly more complexity for CD4 T cells including the differentiation of Th1, Th2, Th17, and T follicular helper subsets. We will focus the following discussion on CD8 T cells. As a note, historically, the term effector and memory have been used in different contexts and meanings, such as in the context of time, function, and lineage, which are summarized in Table 1.
Table 1.
Terminology of “effector” and “memory”
Teff AND Tmem CELL SUBSETS
Immunological memory is defined as the ability of the immune system to remember specific antigenic encounters and then mount enhanced responses upon reexposure. As such, Tmem cells are a key part of cellular immunological memory. Tmem cell subsets were originally defined based on homing and migration markers together with effector functions (Hamann et al. 1997; Sallusto et al. 1999). Cells expressing the lymph node (LN) homing receptors CCR7 and CD62L and possessing a high proliferative capacity but low cytotoxicity were termed central memory (Tcm) cells. Those lacking LN homing capacity and possessing inflammatory migration potential as well as effector function were termed effector memory (Tem) cells. In the subsequent two decades, concepts of Tem and Tcm cells have evolved to include additional subsets, such as effector memory rheumatoid arthritis (RA) (so called because they express CD45RA; Temra), transitional memory, memory stem (Tscm) cells, and tissue-resident memory (Trm) cells (Chang et al. 2014; Jameson and Masopust 2018). Our understanding of CD8 T-cell heterogeneity has advanced along with technology; increasingly large flow cytometry panels have generated new T-cell subsets. An early CyTOF study resolved ∼200 different subpopulations of T cells based on high dimensional combinations of protein expression (Newell et al. 2012). It remains unclear whether these subpopulations represent distinct T-cell identities or simply different phenotypic states of a smaller number of T-cell fates. Nevertheless, it is clear that there are at least some major CD8 T-cell subtypes that can be defined by the combination of phenotype, behavior, and function. Some of these are outlined here.
Central Memory
Tcm cells are quiescent memory T cells that express high levels of molecules, such as CD62L and CCR7, that allow efficient homing to LNs, and low levels of cytotoxic proteins like granzymes and perforin. Functionally, Tcm cells possess low ex vivo cytotoxic activity and a high proliferative capacity (Wherry et al. 2003). These cells can produce interleukin (IL)-2 and, in some settings, interferon γ (IFN-γ) upon stimulation. Tcm cells also acquire the ability to self-renew in response to IL-7 and IL-15. This homeostatic proliferation is unique to memory T cells and, in the CD8 T-cell compartment, resides mainly in the Tcm subset. Tcm cells can rapidly proliferate and expand after reencountering antigen and can generate effector progeny upon antigen restimulation (Wherry et al. 2003). In humans, Tcm cells are often phenotypically characterized as CD45RA−CD45RO+CCR7+CD27+CD28+ (Hamann et al. 1997; Mahnke et al. 2013), whereas in mice these cells are usually defined simply by high expression of CD62L and/or CCR7 with markers of antigen experience such as high CD44.
Effector Memory
Tem cells do not express LN homing receptors CD62L and CCR7, and thus cannot home to LNs. Rather, Tem cells often express molecules that allow homing to nonlymphoid tissues and/or inflammatory sites. The concept of “effector memory” has evolved considerably since the original definition, and today there is clearly both phenotypic and conceptual heterogeneity. In early studies, Tem cells were simply defined as a pool of memory cells that retained the ability to rapidly produce effector functions and were thus capable of providing a first line of defense against reinfection. For example, Tem cells were defined to produce mainly IFN-γ and tumor necrosis factor (TNF), but not IL-2, and were capable of rapid cytotoxicity ex vivo (Hamann et al. 1997; Sallusto et al. 1999). These cells were found in blood and possessed inflammatory homing molecules, and the model was proposed that Tem cells therefore surveyed peripheral sites for new reinfection. In the two decades since they were first described, it has become clear that the Tem cell compartment is more complex, with additional heterogeneity or subsets existing and the concept of tissue-resident memory (Trm) cells (Schenkel and Masopust 2014) that provide protection in peripheral tissues has emerged (see below). In the absence of antigen stimulation, a subset of Tem cells can convert to Tcm cells and, in so doing, acquire properties of self-renewal and long-term homeostatic maintenance (Wherry et al. 2003). This intersubset conversion suggests flexibility in Tcm and Tem cell lineages. However, not all Tem cells can convert to Tcm cells, suggesting the existence of other paths that give rise to Tcm cells. In humans, Tem cells are phenotypically characterized as CD45RA−CD45RO+CCR7−CD27− (Hamann et al. 1997; Mahnke et al. 2013). In mice, Tem cells are often denoted by markers of antigen experience (i.e., CD44Hi) and the absence of CD62L and/or CCR7 expression.
Effector Memory RA (Temra) Cells
Temra cells are a subset of human CD8 T cells that reexpress CD45RA in the absence of CD27 (i.e., CD27−CD45RA+; Hamann et al. 1997; Mahnke et al. 2013). Temra cells also do not express CCR7, CD62L, or CD28 (CCR7− CD62L−CD28−) (Lugli et al. 2010). However, these cells express high levels of cytotoxic molecules and produce proinflammatory cytokines. They often accumulate with repetitive stimulation, as occurs during cytomegalovirus (CMV) infection in humans (Weekes et al. 1999; Patin et al. 2018). Temra cells are terminally differentiated or senescent and display the shortest telomeres among human T cells (Romero et al. 2007). These cells express markers of terminal differentiation/senescence such as KLRG-1 and/or CD57 (Brenchley et al. 2003; Henson and Akbar 2009). Terminally differentiated Teff cells in mice, marked by high KLRG1, are perhaps the closest counterpart to the Temra cells found in humans. However, KLRG1Hi Teff cells arise from a single infectious or stimulation event, not repetitive stimulation that gives rise to Temra cells in humans.
Memory Stem (Tscm) Cells
Tscm cells are a small subset of CD8 T cells, defined in humans as CD45RA+CCR7+CD27+ CD28+CD62L+IL7R+, but distinguished from naive T cells by expression of CD95, CD122, CXCR3, and/or LFA-1 (Gattinoni et al. 2011). Tscm cells are reportedly long-lived, have a stem cell–like ability to self-renew, and the multipotent capacity to differentiate into memory and effector CD8 T-cell subsets (Gattinoni et al. 2017). However, Tscm cells are rare and phenotypically appear less differentiated than Tcm cells. Although the precise ontogeny of Tscm cells remains to be defined, some evidence indicates that Tscm cells may arise prior to full differentiation into Teff or Tem cells, whereas many Tcm cells can arise from Tem cells.
Tissue-Resident Memory (Trm) Cells
Originally considered part of the Tem cell pool, Trm cells were first identified in the intestinal mucosa (Masopust et al. 2001), but now have been found in nearly all tissues in humans and mice (Schenkel and Masopust 2014; Thome et al. 2014). Trm cells primarily exist as residents of tissues without recirculating back to blood and function as a first line of defense against reinfection (Masopust et al. 2006; Schenkel and Masopust 2014; Mueller and Mackay 2016; Szabo et al. 2019). In many tissues, Trm cells express CD103 and CD69. They can be operationally defined as cells that are protected from labeling with intravenously injected antibody and do not equilibrate between hosts in parabiosis experiments.
Exhausted T (Tex) Cells
Tex cells were originally defined in chronic viral infections as antigen-specific CD8 T cells with decreased effector function (Gallimore et al. 1998; Zajac et al. 1998). Tex cells coexpress high levels of multiple inhibitory receptors (IRs), including PD-1, and have a unique transcriptional and epigenetic program (Barber et al. 2006; Wherry et al. 2007; Blackburn et al. 2009; Pauken et al. 2016; Sen et al. 2016). Tex cells are found in chronic infections and cancer in both humans and mice. In these settings of chronic disease, Tex cells likely provide some level of containment or delay in disease progression but are incapable of full control of infection or tumors (Wherry and Kurachi 2015; McLane et al. 2019). Based on canonical markers in humans, Tex cells often fall into the Tem or Tcm cell fraction of CD45RA+CD27+ cells but are distinguished by high expression of PD-1 and other IRs. In tissues, Tex cells may share features and/or have phenotypic overlap with Trm cells. Additional heterogeneity in the Tex cell compartment will be discussed in more detail below.
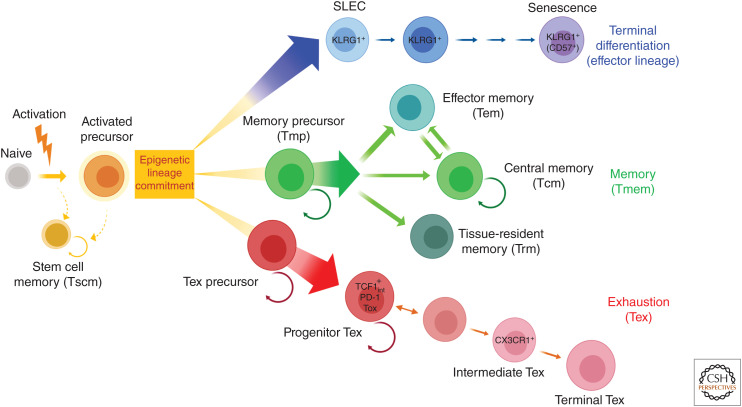
The overview of CD8 T-cell subsets and terminology is summarized in Figure 1 and Table 2. (Epigenetic lineage commitment will be further discussed in more detail below.)
Figure 1.
Schematic of CD8 T-cell differentiation and subsets. (Blue cells) terminally differentiated Teff cell lineage, (green cells) Tmem cell lineage, and (red cells) Tex cell lineage. The color scheme is used throughout this review.
Table 2.
Characteristics of T-cell subsets
HETEROGENEITY IN THE Teff CELL POOLS
Developmental heterogeneity arises following the initial activation, differentiation, and clonal expansion of Teff cells (Cui and Kaech 2010; Chang et al. 2014; Herndler-Brandstetter et al. 2018). Heterogeneity may arise as early as the first cell division, with asymmetric partitioning of key signaling molecules reinforcing divergent developmental circuits (Chang et al. 2007; Reiner and Adams 2014). As the Teff cell pool continues to develop, two major populations, committing to distinct differentiation lineages, emerge. These two populations are defined as KLRG1HiIL-7RLo SLECs and KLRG1LoIL-7RHi memory precursor (Tmp) cells (Kaech et al. 2003; Joshi et al. 2007).
KLRG1HiIL-7RLo SLECs comprise the majority of the Teff cell population. They have potent effector functions and produce cytokines, kill their targets, and disseminate widely. However, these SLECs become terminally differentiated and cannot form long-term Tcm cells. Many SLECs die during the contraction phase, but a subset may persist into the memory phase in the form of “terminal Tem” cells (Joshi et al. 2007). These terminal Tem cells, which are essentially resting SLECs, can persist for ∼2–3 months or more in the absence of antigen in some settings. These cells remain CD62LLo in the post-effector phase and, therefore, could fall into the category of Tem cells but will not undergo conversion to CD62LHi Tcm cells. Rather, these SLEC-derived terminal Tem cells represent a pool of residual cells that may be retained for systemic surveillance and rapid protection from reinfection. Such a scenario, however, suggests at least two distinct types of Tem cells. One type is Tem cells that can convert to Tcm cells and is derived from Tmp cells (see below), and another is derived from surviving KLRG1Hi SLECs that eventually die off over time.
In contrast, the IL-7RHi Tmp cell subset preferentially forms Tmem cells, including CD62LLo Tem cells that are then capable of differentiating into CD62LHi Tcm cells. It is the latter population that responds to both IL-7 and IL-15 and undergoes antigen-independent self-renewal. In addition to giving rise to the systemic and circulating pools of Tcm and some Tem cells, Tmp cells can also give rise to Trm cells (Schenkel and Masopust 2014; Mueller and Mackay 2016). However, the potential to generate Trm cells peaks before the end of clonal expansion, typically around day 5 postinfection in mice (Masopust et al. 2001). Once in tissues, Tmp cells acquire a new residency program, up-regulate CD103 and/or CD69, and durably occupy these tissues (Kaech et al. 2003; Joshi et al. 2007; Sarkar et al. 2008; Obar and Lefrançois 2010; Angelosanto et al. 2012; Mackay et al. 2013).
Although this Tmp/SLEC model has helped clarify Teff and Tmem cell differentiation, additional complexities in Teff and Tem (and likely Tcm) cell populations exist. For example, studies using reporter mice suggest that KLRG1+ Teff cells (i.e., SLECs) might possess developmental plasticity because a subset appeared to down-regulate KLRG1 and give rise to Tcm, Tem, and Trm cells (Herndler-Brandstetter et al. 2018). In other studies, CX3CR1 expression can further subdivide the Teff and early Tem cell populations into CX3CR1Lo, CX3CR1Int, and CX3CR1Hi populations, each with different phenotypic and functional properties (Böttcher et al. 2015; Gerlach et al. 2016). Whereas the CX3CR1Lo and CX3CR1Hi populations were Teff-like (e.g., SLEC), the CX3CR1Int population formed a transitional or Tmp-like pool that seeded the long-term Tmem cell pool. The CX3CR1Hi subset is mainly a recirculating, vascular-patrolling population (Gerlach et al. 2016). These mouse studies indicate considerable complexity in Teff→ Tem→Tmem subset biology. Although future studies are required to fully delineate the complexity and key lineage relationships underlying this heterogeneity, it is likely that the general concept of “effector memory” has value as an umbrella term under which there are likely multiple additional subsets.
REGULATION OF EFFECTOR VERSUS MEMORY CD8 T-CELL DEVELOPMENT
Defining the signals and mechanisms that regulate Teff and Tmem cell differentiation remains a major goal in the field of immunology. A considerable body of work has demonstrated that qualitative and quantitative differences in signals (antigen, cytokine, costimulation, CD4 T-cell help, metabolism, etc.) play an important role in memory versus effector CD8 T-cell differentiation (Chang et al. 2014).
Tmem cell differentiation is influenced by signals received during priming, including antigen levels, clonal competition, and/or the duration of stimulation (Sarkar et al. 2007). In particular, antigen recognition and TCR stimulation (e.g., signal 1) have been shown to differentially regulate Tmem cell development. Weaker TCR signaling upon initial antigen-recognition promotes a faster transition from Tem to a Tcm cell, and strong TCR signaling promotes terminal differentiation (Wherry et al. 2003; Teixeiro et al. 2009; Zehn et al. 2009; Smith-Garvin et al. 2010).
Costimulatory molecules (signal 2), such as CD28 and other members of the tumor necrosis factor receptor (TNFR) family, have also been shown to both play important roles in the primary response, and also regulate Tmem generation, function, and survival (Boise et al. 1995; Hendriks et al. 2000; Bertram et al. 2004; Borowski et al. 2007; Fuse et al. 2008; Garidou et al. 2009; Dong et al. 2012; Esensten et al. 2016; Schildberg et al. 2016).
CD4 T-cell help plays a key role in regulating Teff and Tmem cell differentiation, and early studies showed that development of Tmem and Tcm cells was compromised in the absence of CD4 T-cell help (Janssen et al. 2003; Shedlock and Shen 2003; Sun and Bevan 2003). These effects are associated with an increase in SLEC generation, dependent on the TF T-bet (Intlekofer et al. 2007). The mechanisms by which CD4 T cells influence Tmem cell differentiation remain unclear and may differ depending on the setting. Mechanisms may include licensing of dendritic cells (DCs) (Castellino et al. 2006) via CD40L (Bennett et al. 1998; Ridge et al. 1998; Schoenberger et al. 1998), production of IL-2 (Kalia et al. 2010; Makedonas et al. 2010; Pipkin et al. 2010), production of IL-21 (Cui et al. 2011), and supporting the production of effective antibodies (Bachmann et al. 2004).
In addition, inflammation (signal 3) modulates the balance between Teff/SLEC and Tmp/Tmem cell lineage commitment (Joshi et al. 2007; Harty and Badovinac 2008). For example, IL-12 (Joshi et al. 2007; Xiao et al. 2009), IFN-γ (Badovinac et al. 2000), and IL-27 (Yoshida and Hunter 2015) can enhance effector-like differentiation—particularly during SLEC generation—whereas IL-10 (Foulds et al. 2006; Laidlaw et al. 2015), IL-21 (Cui et al. 2011), and transforming growth factor β (TGF-β) (Zhang and Bevan 2013; Guan et al. 2018) can promote Tmem cell differentiation.
TFs and transcriptional circuits underlie these differentiation events, guiding Teff and Tmem cells. A series of TFs have been implicated in the formation of Tmem, Tmp, and/or Tcm cells, including Id3 (Yang et al. 2011), TCF-1 (Zhou et al. 2010; Utzschneider et al. 2016; Kratchmarov et al. 2018), Bcl6 (Ichii et al. 2002), STAT3 (Cui et al. 2011), Foxo1 (Utzschneider et al. 2018), Eomes (Banerjee et al. 2010), and Zeb1 (Guan et al. 2018; Scott and Omilusik 2019). On the other hand, the formation of Teff, SLECs, and/or Tem cells is promoted by T-bet (Intlekofer et al. 2007; Joshi et al. 2007), Id2 (Cannarile et al. 2006; Yang et al. 2011; Masson et al. 2013), Blimp-1 (Kallies et al. 2009; Rutishauser et al. 2009), STAT4 (Mollo et al. 2014), and Zeb2 (Guan et al. 2018; Scott and Omilusik 2019). Many of these TFs function in opposing pairs, such as T-bet and Eomes, Id2 and Id3, and Blimp1 and Bcl6. In these cases, one TF drives Teff-like differentiation and the other promotes Tmem-like differentiation. Many additional TFs are necessary for the initiation stages of T-cell activation, including BATF and IRF4 (Kurachi et al. 2014), c-Myb (Chen et al. 2017), Runx3 (Wang et al. 2018), as well as other signal-dependent TFs directly downstream of TCR and/or costimulation including NFATs, Fos, Jun, and other AP-1 family members, NF-κB, and others (Daniels and Teixeiro 2015; Chen et al. 2018).
Most TFs function by binding to DNA in regions of epigenetically remodeled and/or open chromatin. Thus, the mechanisms of epigenetic regulation of Teff and Tmem cell differentiation are of interest (Araki et al. 2009; Zediak et al. 2011; Akondy et al. 2017; Abdelsamed et al. 2018; Carty et al. 2018; Tough et al. 2020). Histone modifications such as acetylation and methylation and associated alterations in chromatin accessibility have been implicated in differentiation of Tmem and Teff cells (Northrop et al. 2006; Araki et al. 2008; Zediak et al. 2011; Shin et al. 2013). Moreover, enzymes catalyzing epigenetic changes such as DNMT3a (Ladle et al. 2016) and TET2 (Carty et al. 2018), both of which are involved in regulating DNA methylation, are suggested to regulate early Teff cell differentiation and Teff versus Tmem cell fate decisions.
In addition to TFs and epigenetic changes, many other factors can influence Teff and Tmem cell differentiation, including metabolism (Pollizzi and Powell 2014; Buck et al. 2017), the microbiome (Bachem et al. 2019), aging (Eberlein et al. 2016), social stress (Weber et al. 2017), and neuroimmune interactions (Slota et al. 2015). Future studies are needed to determine whether some of these effects are linked through the TF pathways outlined above or whether their effects are mediated through distinct, potentially novel mechanisms.
Although much of the information discussed above has been derived from mouse models, it is becoming increasingly possible to interrogate these questions in humans. Studies using yellow fever vaccine (YFV) have led to seminal observations about human CD8 T-cell memory. The use of YFV in healthy subjects, combined with in vivo deuterium labeling, allowed detailed, longitudinal analysis of YFV-specific CD8 Teff and Tmem cells in humans in the absence of antigen reexposure (Akondy et al. 2017). These analyses showed that Tmem cells originated from extensively dividing CD8 T cells generated during the first 2 weeks after vaccination (i.e., derived from Teff cells). These Tmem cells were maintained and largely quiescent but divided once every year (doubling time of over 450 days). Epigenetic analysis revealed that the long-lived, YFV-specific Tmem cells retained epigenetic signatures of their Teff cell history. Moreover, these Teff cell epigenetic imprints were detected in Tmem cells decades after the initial vaccination (Akondy et al. 2017). Thus, these studies indicate one development pathway in which long-term Tmem cells develop from Teff cells can occur in humans.
TISSUE-RESIDENT MEMORY CELLS
Tissue-resident memory T (Trm) cells are a subset of Tmem cells that reside mainly in nonlymphoid tissues (although they can also be found in lymphoid tissues) and do not recirculate through blood and lymph. Trm cells function as a first line of defense against reinfection at barrier sites and can provide effective protective immunity (Masopust et al. 2006; Schenkel and Masopust 2014; Mueller and Mackay 2016; Szabo et al. 2019). Trm cells are often defined by expression of CD69 and CD103, each of which can contribute to tissue residency, but the expression of these molecules varies across tissues (Masopust et al. 2006; Schenkel and Masopust 2014; Mueller and Mackay 2016; Szabo et al. 2019). Compared to circulatory Tmem cells, Trm cells display distinct transcriptional (Wakim et al. 2012; Mackay et al. 2013, 2016; Milner and Goldrath 2018) and epigenetic programs (Milner et al. 2017), consistent with Trm cells representing a distinct subset of Tmem cells (Wakim et al. 2012; Mackay et al. 2013, 2016; Milner et al. 2017). Trm cells appear to arise from early Teff cells—likely Tmp cells that enter tissues and acquire additional environmental and transcriptional reprogramming—at least in some cases driven by TGF-β signaling (Zhang and Bevan 2013). Hobit, Blimp-1 (Mackay et al. 2016), and Runx3 (Milner et al. 2017) are key TFs expressed in Trm cells. Loss of TF KLF2 and subsequent down-regulation of the exit-controlling receptor S1P1 (Skon et al. 2013) is crucial for Trm cell identity. Thus, Trm cells are a distinct functional subset of Tmem cells endowed with tissue-specific residency, a unique role in barrier and tissue immunity, and a transcriptional program linked to their function.
MEMORY STEM (Tscm) CELLS AND RELATIONSHIP TO OTHER Tmem SUBSETS
T memory stem (Tscm) cells are a rare subset of Tmem cells, endowed with the long-lived, stem cell-like ability to self-renew and the multipotent capacity to reconstitute a wide spectrum of Tmem and Teff cell subsets (Gattinoni et al. 2017). Tscm cells have a phenotype that overlaps with naive and Tcm cells in mice (CD44Lo CD62LHi and high Sca-1, CD122, and Bcl-2) and humans (CD45RA+CD27+ and CCR7+, CD95+, CXCR3+/−, IL-2Rβ+, CD58+ and CD11a+) (Zhang et al. 2005; Gattinoni et al. 2011). These Tscm cells also express the TF TCF1 (Gautam et al. 2019). However, whereas studies have shown that at least some Tcm cells derive from Tem cells (Wherry et al. 2003) and cells that have gone through an effector phase (Akondy et al. 2017), studies on Tscm cells suggest that these cells may arise prior to full Teff cell differentiation (Gattinoni et al. 2009; Lugli et al. 2013). Future studies should help clarify the precise developmental origins of Tscm versus Tcm cells, test the comparative developmental potential to give rise to other subsets, and/or serially self-renew as well as define the epigenetic control mechanisms that might distinguish these cell types.
EXHAUSTION
During chronic infections or cancer, when antigen stimulation persists, activated T cells can leave the Teff and Tmem cell differentiation path and instead develop into exhausted CD8 T (Tex) cells (Wherry and Kurachi 2015). Tex cells are characterized by reduced (though not completely absent) effector function, high coexpression of IRs such as PD-1, TIM3, LAG3, CTLA4, and TIGIT, altered metabolism, and impaired proliferation when stimulated (Wherry and Kurachi 2015; McLane et al. 2019). These IRs restrain Tex cells and blockade of the PD-1 pathway reinvigorates Tex cells, thus enhancing disease control (Barber et al. 2006). Despite poor proliferative potential, Tex cells are maintained by constant in vivo cell division by a hierarchy of Tex cell subsets with progenitor or “stem-like” properties (Wherry et al. 2007; Blackburn et al. 2008; Paley et al. 2012). In addition, Tex cells have a distinct transcriptional and epigenetic landscape compared to Teff and Tmem cell subsets. Thus, Tex cells are a CD8 T-cell lineage distinct from Teff or SLEC and Tmem cell subsets (Wherry et al. 2007; Doering et al. 2012; Pauken et al. 2016; Scott-Browne et al. 2016; Sen et al. 2016; Philip et al. 2017; Chen et al. 2019). Several studies subsequently identified the high mobility group TF Tox as the key factor that initiates the epigenetic program and fate commitment of Tex cells by both promoting accessibility to Tex cell–specific open chromatin regions and suppressing the epigenetic program of the terminally differentiated Teff cell fate (Alfei et al. 2019; Khan et al. 2019; Scott et al. 2019; Seo et al. 2019; Yao et al. 2019). These data indicate that Tex cells are distinct from other mature CD8 T-cell types, an observation with implications for predicting how immunotherapies might target Teff versus Tex cells.
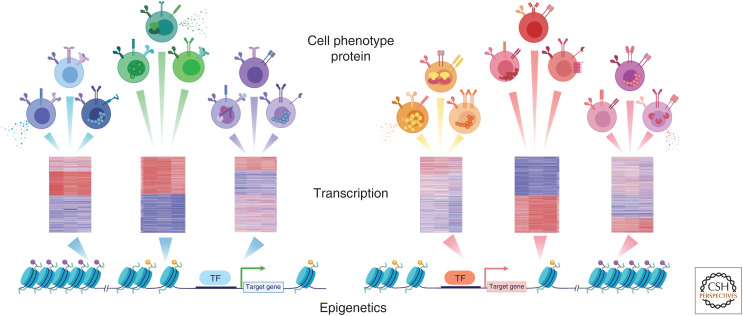
Tex cells are the major responding cell types following PD-1 pathway blockade in cancer and chronic infections (Im et al. 2016; Pauken et al. 2016; Utzschneider et al. 2016; Wu et al. 2016; Huang et al. 2017). Much of the clinical benefit of PD-1/PD-L1 blockade, and perhaps other checkpoint blockades, is likely mediated by reinvigoration of these cells (Topalian et al. 2015; Huang et al. 2017, 2019; LaFleur et al. 2018). Technological advances continually facilitate a deeper understanding of this cell type. Profiling open chromatin regions by ATAC-seq demonstrated that Tex cells differ from Teff and Tmem cells by approximately 6000 open chromatin regions (Pauken et al. 2016; Scott-Browne et al. 2016; Sen et al. 2016; Philip et al. 2017). The magnitude of this difference is similar to the ∼5000–7000 open chromatin regions that differ between developing myeloid cells and B cells (Lara-Astiaso et al. 2014; Monticelli and Natoli 2017). It was remarkable that this global open chromatin landscape did not change appreciably upon PD-1 pathway blockade, despite robust transcriptional reengagement of Teff cell–like genes (Pauken et al. 2016). These observations have two implications. First, the reinvigorated Tex cells did not convert to Teff cells, but instead retained their Tex cell identity while temporarily reacquiring Teff cell functions. Second, these observations highlight the notion that cells can exist in multiple transcriptional and/or phenotypic states despite harboring a single epigenetic state (Fig. 2). This concept is important for defining criteria that determine cell identities or subsets.
Figure 2.
From open chromatin landscape to diverse cell phenotype. Observations highlight the notion that cells can exist in multiple transcriptional and/or phenotypic states despite harboring a single epigenetic state. (TF) Transcription factor.
SUBSETS OF Tex CELLS
Tex cells in cancer and chronic infection are heterogeneous with multiple subsets (Wherry and Kurachi 2015; McLane et al. 2019). Tex cell subsets were first described based on different expression levels of PD-1 and CD44 and these subsets were linked to differential responsiveness to PD-1 pathway blockade (Blackburn et al. 2008). Subsequent studies demonstrated that Tex cell progenitor and terminal subsets were related in a proliferative hierarchy with the Tex cell progenitor population undergoing constant proliferation in response to persisting antigen to give rise to a more numerically abundant terminal Tex cell population (Paley et al. 2012). This progenitor or “stem-like” Tex cell subset is controlled by TCF1 (Im et al. 2016; Utzschneider et al. 2016; Wu et al. 2016) (see below). Notably, all Tex cell subsets express the Tex TF Tox (Alfei et al. 2019; Khan et al. 2019; Scott et al. 2019; Seo et al. 2019; Yao et al. 2019), but other TFs have sequentially important roles throughout the Tex proliferative hierarchy (Wherry and Kurachi 2015; McLane et al. 2019). Although Tex cell subset biology is consistent across studies, different nomenclature has been used. An attempt is made here to connect some of this nomenclature to the common underlying Tex cell subset biology.
Progenitor Tex Cells
Tex cell subsets were first defined by PD-1 and CD44 expression, with PD-1Int CD44Hi Tex cells readily distinguished from PD-1Hi CD44Int Tex cells in chronic lymphocytic choriomeningitis virus (LCMV) infection (Blackburn et al. 2008). When purified and tested separately, PD-1Int CD44Hi cells retained proliferative potential and were the only subset of Tex cells capable of responding to PD-1 pathway blockade (Blackburn et al. 2008). This PD-1Int pool was then shown to be maintained over time, retain some self-renewal capacity, and directly give rise to the more terminal PD-1Hi population through cell division in vivo (Paley et al. 2012). Thus, the PD-1Int subset of Tex cells possessed “progenitor” activity within the Tex cell population, whereas the PD-1Hi subset was terminal (Paley et al. 2012). These studies also defined a role for the TFs T-bet and Eomes in this process. Loss of T-bet and increase in Eomes was associated with a transition from the PD-1Int progenitor pool to the more terminal PD-1Hi Tex cell subset (Paley et al. 2012; Odorizzi et al. 2015). Network analysis also suggested a potential role for Wnt signaling and/or TCF1 associated with the PD-1Int Tex cell subset (Doering et al. 2012). Indeed, a series of subsequent studies refined the definition of Tex cell progenitors using CXCR5 and/or TCF1 (and Ly108 as a surrogate) to identify smaller populations that possessed progenitor activity and the ability to respond to PD-1 blockade (Im et al. 2016; Utzschneider et al. 2016; Wu et al. 2016). It was clear from these studies that TCF1 is a critical TF for this Tex cell progenitor subset. Some of these studies used the term “stem-like” Tex cells for the population that expressed TCF1 and possessed the ability to respond to PD-1 blockade. Likely, this stem-like Tex cell subset is a subpopulation of the original Tex cell progenitor subset. Additional heterogeneity in this TCF1+ progenitor population has also been identified; one subpopulation of the TCF1+ Tex cell progenitor (“Texprog1”) is quiescent and confined to lymphoid tissue, whereas a second subpopulation of TCF1+ Tex progenitors (“Texprog2”) displays more blood accessibility, has robustly up-regulated cell cycle genes, and is actively dividing (Beltra et al. 2020). At the Texprog2 stage, TCF1 expression begins to decrease and T-bet expression begins to increase (Beltra et al. 2020). These Tex cell progenitors have been identified in blood (Huang et al. 2017, 2019; Bengsch et al. 2018) and tumors (Sade-Feldman et al. 2018; Li et al. 2019; Miller et al. 2019) and have direct implications for antitumor immunity in patients because of their responsiveness to PD-1 pathway blockade. However, in human tumors, the identification of tumor-specific Tex cell progenitors remains elusive.
Intermediate Tex Cells
This Tex cell subset expresses T-bet, but not TCF1, and has evidence of recent proliferation (Hudson et al. 2019; Zander et al. 2019; Beltra et al. 2020). These cells also express CX3CR1 and possess robust circulatory or homing capacity. There is evidence that this T-bet+ Tex cell intermediate subset gives rise to the more terminal Tex cell population (Hudson et al. 2019; Beltra et al. 2020), although some recent data suggests that perhaps in some settings this Tex cell intermediate subset might not generate terminal Tex cells (Zander et al. 2019). Discrepancies in the relationships between Tex cell subsets could relate to antigen load, inflammation, and/or CD4 T-cell help and should be clarified by future studies including lineage tracing.
Terminal Tex Cells
Early studies of Tex cell subsets defined terminal Tex cells as PD-1HiCD44Int cells that lacked proliferative potential and were unresponsiveness to antigen stimulation or PD-1 pathway blockade (Blackburn et al. 2008). These cells also had a relatively short half-life in vivo and were characterized by high Eomes (Paley et al. 2012). Terminal Tex cells also expressed higher amounts of many IRs and were less polyfunctional by cytokine production (Blackburn et al. 2009; Wherry and Kurachi 2015; McLane et al. 2019). This terminal subset has also been identified as TCF1−, CXCR5−, and/or TIM3+ (He et al. 2016; Im et al. 2016), although some of these phenotyping approaches capture only partially overlapping populations (Beltra et al. 2020). However, this Tex cell terminal subset retains cytotoxic killing activity (Blackburn et al. 2009; Wherry and Kurachi 2015; McLane et al. 2019) and, because it is numerically abundant in most tissues, may play a role in at least temporary or partial disease control. Moreover, terminal Tex cells are also present in and accumulate to different degrees in nonlymphoid tissues, in some cases with phenotypic and operational (i.e., lack of blood accessibility) similarity to Trm cells (Schenkel and Masopust 2014; He et al. 2016; Im et al. 2016; Mueller and Mackay 2016; Jansen et al. 2019; Szabo et al. 2019).
Tex Cell Precursor
A major question has been: from what precursor population do Tex cells develop? One possibility was that Tex cells arise from overstimulated Teff cells (SLECs). However, lineage-tracing experiments demonstrated that Tex cells did not arise from SLECs, but rather were only generated from the pool of CD127+ Tmp cells present in the effector phase (Angelosanto et al. 2012). In recent studies, scRNA-seq of cells isolated during the early phase of chronic LCMV (clone 13) infection identified a branched trajectory at an early stage CD8 T-cell differentiation where Tex versus Teff cell development diverged (Chen et al. 2019). Together with lineage tracing experiments, a TCF-1+Ly108+PD-1+ CD8 T-cell population present in the effector phase was identified as the Tex cell precursor that seeds development of mature Tex cells (Chen et al. 2019). These studies also distinguished this Tex cell precursor from the Tmp cell population (Chen et al. 2019). These early Tex cell precursors are not yet developmentally fixed, however, and can give rise to Tmem cells if removed from chronic antigen stimulation (Angelosanto et al. 2012), likely because the full epigenetic program of mature Tex cells is not established for 2–3 weeks following infection (Pauken et al. 2016; Sen et al. 2016; Philip et al. 2017; Khan et al. 2019). Tex cell precursors require Tox and TCF1 (Chen et al. 2019), with Tox directly controlling TCF1 expression. Tox is dispensable for TCF1 expression in the Tmem cell lineage (Khan et al. 2019; Scott et al. 2019). Tox also directly represses the SLEC Teff cells to help enable Tex cell precursor development and formation of the Tex cell lineage (Alfei et al. 2019; Khan et al. 2019; Scott et al. 2019; Seo et al. 2019; Yao et al. 2019). Moreover, PD-1 is required to preserve this initial Tex cell precursor population and allows formation of the initial pool of Tex cell progenitors (Chen et al. 2019), perhaps explaining why the Tex cell population is not sustained long term in the genetic absence of PD-1 (Odorizzi et al. 2015).
Thus, Tex cell subsets represent a multistage developmental hierarchy with Tex cell progenitor subsets (and likely biologically distinct Texprog1 and Texprog2 populations) giving rise to a Tex cell intermediate and then finally a terminal Tex cell subset. Additional heterogeneity may exist. Future profiling of Tex cell subset heterogeneity in humans, linked to tumor specificity, will help improve immunotherapies targeting Tex cells such as immune checkpoint blockade.
FROM EPIGENETIC SIGNATURES TO DIVERSE T-CELL PHENOTYPE
Given the critical roles of epigenetics and chromatin remodeling in lineage commitment at the early stage of T-cell differentiation, we propose the following model for Tmem and Tex cell differentiation (Fig. 1; Table 2). After naive T cells are activated and primed by antigen stimulation, they develop into a common precursor population with the potential to commit to multiple CD8 T-cell lineages. Transcriptional and initial epigenetic rewiring then establishes gene expression programs biased toward the development of each lineage, generating the lineage-committed progenitors (Tmem, Teff/SLEC, and Tex cells). These different initial lineage-biased precursors initially retain fate flexibility, but then become progressively more committed as the epigenetic regulation becomes more fixed over time. These epigenetic changes induce transcriptional circuits and establish the full lineage program. However, the open chromatin landscape of these major lineages determines the potential for gene expression, and each major lineage may access different components of this epigenetic and gene expression potential depending on environmental cues. Thus, CD8 T cells of a single lineage fate could display distinct protein or mRNA phenotypes depending on recent signals received and yet have the same underlying open chromatin landscape (Fig. 2). Such is the case, for example, for Tex cells following PD-1 blockade. Upon PD-1 blockade, Tex cells change transcriptionally and functionally but retain largely the same open chromatin profile (Pauken et al. 2016).
THERAPEUTIC IMPLICATIONS
The biology of subsets of Teff, Tmem, and Tex cells has direct clinical relevance for protection from infectious disease, vaccination, autoimmunity, and cancer immunotherapy. Immune memory, a hallmark of effective vaccination, is maintained in the T-cell compartment by subsets of Tmem cells that can give rise to protective activities, including direct effector function or generation of downstream progeny including Teff cells. Tex cells may be critical to establishing a host–pathogen stalemate and providing limited pathogen (or tumor) control while avoiding immunopathology. Moreover, understanding the diverse biological features of these cell populations should help improve efficacy of vaccines and cellular and biological therapies for infectious disease and cancer.
SUMMARY AND OPEN QUESTIONS
Despite tremendous advances in our understanding of the diversity of Teff, Tmem, and Tex cells in the past decade, a number of questions and challenges for the field remain. For example, how many subsets of Teff, Tmem, and Tex cells exist? What defines a discreet subset? How can we as a field harmonize the information about Teff, Tmem, and Tex cell biology even if nuances and distinctions (sometimes important ones) exist between studies?
CD8 T-cell differentiation may operate like fractal geometry, filling the existing biological “space” yet having distinct features, the deeper one examines the cells. Perhaps a key future goal will be to attempt to delineate a set of biologically relevant, measurable, and stable features of CD8 T-cell subsets and link these to nomenclature, knowing that under any subset designation there will be additional phenotypic, functional, transcriptional, and epigenetic diversity. This is not simply about naming cell types. These immune cell types are central to a wide variety of human diseases and our nomenclature implies relationships that may be important for vaccines and therapeutic approaches. In addition, because CD8 T cells can behave extraordinarily differently, determining which subset is desirable (or undesirable) to target could be the difference between clinical success and failure. Thus, establishing a comprehensive understanding of CD8 T-cell heterogeneity and development will significantly impact a wide variety of human disease and treatment, including vaccine development for infectious diseases, immunosuppressants for autoimmunity, and checkpoint blockade for cancer.
COMPETING INTEREST STATEMENT
E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Roche, Pieris, Elstar, and Surface Oncology. E.J.W. has a patent licensing agreement on the PD-1 pathway with Roche/Genentech. E.J.W. is a founder of Surface Oncology and Arsenal Biosciences. Y.M. has no COI to disclose.
ACKNOWLEDGMENTS
We thank Wherry laboratory members (especially A.R. Greenplate, Z. Chen, J.-C. Beltra, D. Mathew, and M. Abdel-Hakeem) for their comments and discussions. This work is supported by grants from the National Institutes of Health (NIH) to E.J.W. (AI105343, AI082630, AI112521, AI115712, AI117718, AI108545, and AI117950) and Stand Up 2 Cancer (SU2C). E.J.W. is a member of the Parker Institute for Cancer Immunotherapy, which supports the UPenn Cancer Immunotherapy Program. Y.M. was a Stand Up 2 Cancer, Society of Immunotherapy of Cancer (SU2C-SITC) Convergence Scholar, and a recipient of the KANAE Foundation for the Promotion of Medical Science Grant Award.
Footnotes
Editors: David Masopust and Rafi Ahmed
Additional Perspectives on T-Cell Memory available at www.cshperspectives.org
REFERENCES
- Abdelsamed HA, Zebley CC, Youngblood B. 2018. Epigenetic maintenance of acquired gene expression programs during memory CD8 T cell homeostasis. Front Immunol 9: 6. 10.3389/fimmu.2018.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. 2017. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552: 362–367. 10.1038/nature24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, et al. 2019. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571: 265–269. 10.1038/s41586-019-1326-9 [DOI] [PubMed] [Google Scholar]
- Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. 2012. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol 86: 8161–8170. 10.1128/JVI.00889-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Fann M, Wersto R, Weng NP. 2008. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B). J Immunol 180: 8102–8108. 10.4049/jimmunol.180.12.8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Wang Z, Zang C, Wood WH, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, et al. 2009. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30: 912–925. 10.1016/j.immuni.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dähling S, Kastenmüller W, et al. 2019. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51: 285–297.e5. 10.1016/j.immuni.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Bachmann MF, Hunziker L, Zinkernagel RM, Storni T, Kopf M. 2004. Maintenance of memory CTL responses by T helper cells and CD40-CD40 ligand: antibodies provide the key. Eur J Immunol 34: 317–326. 10.1002/eji.200324717 [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Tvinnereim AR, Harty JT. 2000. Regulation of antigen-specific CD8+ T cell homeostasis by perforin and interferon-γ. Science 290: 1354–1357. 10.1126/science.290.5495.1354 [DOI] [PubMed] [Google Scholar]
- Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. 2010. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 185: 4988–4992. 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Beltra J-C, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, Casella V, Ngiow SF, Khan O, Huang YJ, et al. 2020. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52: 825–841. 10.1016/j.immuni.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengsch B, Ohtani T, Khan O, Setty M, Manne S, O'Brien S, Gherardini PF, Herati RS, Huang AC, Chang KM, et al. 2018. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity 48: 1029–1045.e5. 10.1016/j.immuni.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SRM, Carbone FR, Karamalis F, Flavell RA, Miller JFAP, Heath WR. 1998. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature 393: 478–480. 10.1038/30996 [DOI] [PubMed] [Google Scholar]
- Bertram EM, Dawicki W, Sedgmen B, Bramson JL, Lynch DH, Watts TH. 2004. A switch in costimulation from CD28 to 4-1BB during primary versus secondary CD8 T cell response to influenza in vivo. J Immunol 172: 981–988. 10.4049/jimmunol.172.2.981 [DOI] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Freeman GJ, Wherry EJ. 2008. Selective expansion of a subset of exhausted CD8 T cells by αPD-L1 blockade. Proc Natl Acad Sci 105: 15016–15021. 10.1073/pnas.0801497105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DAA, Wherry EJ. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10: 29–37. 10.1038/ni.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boise LH, Minn AJ, Noel PJ, June CH, Accavitti MA, Lindsten T, Thompson CB. 1995. CD28 costimulation can promote T cell survival by enhancing the expression of Bcl-XL. Immunity 3: 87–98. 10.1016/1074-7613(95)90161-2 [DOI] [PubMed] [Google Scholar]
- Borowski AB, Boesteanu AC, Mueller YM, Carafides C, Topham DJ, Altman JD, Jennings SR, Katsikis PD. 2007. Memory CD8+ T cells require CD28 costimulation. J Immunol 179: 6494–6503. 10.4049/jimmunol.179.10.6494 [DOI] [PubMed] [Google Scholar]
- Böttcher JP, Beyer M, Meissner F, Abdullah Z, Sander J, Höchst B, Eickhoff S, Rieckmann JC, Russo C, Bauer T, et al. 2015. Functional classification of memory CD8+ T cells by CX3CR1 expression. Nat Commun 6: 8306. 10.1038/ncomms9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Karandikar NJ, Betts MR, Ambrozak DR, Hill BJ, Crotty LE, Casazza JP, Kuruppu J, Migueles SA, Connors M, et al. 2003. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood 101: 2711–2720. 10.1182/blood-2002-07-2103 [DOI] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, Pearce EL. 2017. Metabolic instruction of immunity. Cell 169: 570–586. 10.1016/j.cell.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. 2006. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol 7: 1317–1325. 10.1038/ni1403 [DOI] [PubMed] [Google Scholar]
- Carty SA, Gohil M, Banks LB, Cotton RM, Johnson ME, Stelekati E, Wells AD, Wherry EJ, Koretzky GA, Jordan MS. 2018. The loss of TET2 promotes CD8+ T cell memory differentiation. J Immunol 200: 82–91. 10.4049/jimmunol.1700559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. 2006. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell–dendritic cell interaction. Nature 440: 890–895. 10.1038/nature04651 [DOI] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315: 1687–1691. 10.1126/science.1139393 [DOI] [PubMed] [Google Scholar]
- Chang JT, Wherry EJ, Goldrath AW. 2014. Molecular regulation of effector and memory T cell differentiation. Nat Immunol 15: 1104– 1115. 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Stelekati E, Kurachi M, Yu S, Cai Z, Manne S, Khan O, Yang X, Wherry EJ. 2017. miR-150 regulates memory CD8 T cell differentiation via c-Myb. Cell Rep 20: 2584–2597. 10.1016/j.celrep.2017.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zander R, Khatun A, Schauder DM, Cui W. 2018. Transcriptional and epigenetic regulation of effector and memory CD8 T cell differentiation. Front Immunol 9: 2826. 10.3389/fimmu.2018.02826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, et al. 2019. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51: 840–855.e5. 10.1016/j.immuni.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Kaech SM. 2010. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev 236: 151–166. 10.1111/j.1600-065X.2010.00926.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Liu Y, Weinstein Jason S, Craft J, Kaech Susan M. 2011. An interleukin-21- interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35: 792–805. 10.1016/j.immuni.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E. 2015. TCR signaling in T cell memory. Front Immunol 6: 617. 10.3389/fimmu.2015.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ. 2012. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37: 1130–1144. 10.1016/j.immuni.2012.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Franklin NA, Roberts DJ, Yagita H, Glennie MJ, Bullock TNJ. 2012. CD27 Stimulation promotes the frequency of IL-7 receptor-expressing memory precursors and prevents IL-12–mediated loss of CD8+ T cell memory in the absence of CD4+ T cell help. J Immunol 188: 3829–3838. 10.4049/jimmunol.1103329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlein J, Davenport B, Nguyen T, Victorino F, Haist K, Jhun K, Karimpour-Fard A, Hunter L, Kedl R, Clambey ET, et al. 2016. Aging promotes acquisition of naive-like CD8+ memory T cell traits and enhanced functionalities. J Clin Invest 126: 3942–3960. 10.1172/JCI88546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esensten Jonathan H, Helou Ynes A, Chopra G, Weiss A, Bluestone Jeffrey A. 2016. CD28 costimulation: from mechanism to therapy. Immunity 44: 973–988. 10.1016/j.immuni.2016.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds KE, Rotte MJ, Seder RA. 2006. IL-10 is required for optimal CD8 T cell memory following Listeria monocytogenes infection. J Immunol 177: 2565–2574. 10.4049/jimmunol.177.4.2565 [DOI] [PubMed] [Google Scholar]
- Fuse S, Zhang W, Usherwood EJ. 2008. Control of memory CD8+ T cell differentiation by CD80/CD86–CD28 costimulation and restoration by IL-2 during the recall response. J Immunol 180: 1148–1157. 10.4049/jimmunol.180.2.1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore A, Glithero A, Godkin A, Tissot AC, Plückthun A, Elliott T, Hengartner H, Zinkernagel R. 1998. Induction and exhaustion of lymphocytic choriomeningitis virus–specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I–peptide complexes. J Exp Med 187: 1383–1393. 10.1084/jem.187.9.1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garidou L, Heydari S, Truong P, Brooks DG, McGavern DB. 2009. Therapeutic memory T cells require costimulation for effective clearance of a persistent viral infection. J Virol 83: 8905–8915. 10.1128/JVI.00027-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, et al. 2009. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 15: 808–813. 10.1038/nm.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. 2011. A human memory T cell subset with stem cell–like properties. Nat Med 17: 1290– 1297. 10.1038/nm.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L, Speiser DE, Lichterfeld M, Bonini C. 2017. T memory stem cells in health and disease. Nat Med 23: 18– 27. 10.1038/nm.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam S, Fioravanti J, Zhu W, Le Gall JB, Brohawn P, Lacey NE, Hu J, Hocker JD, Hawk NV, Kapoor V, et al. 2019. The transcription factor c-Myb regulates CD8+ T cell stemness and antitumor immunity. Nat Immunol 20: 337–349. 10.1038/s41590-018-0311-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, von Andrian UH. 2016. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45: 1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef P, Buchholz VR, Stemberger C, Flossdorf M, Henkel L, Schiemann M, Drexler I, Höfer T, Riddell SR, Busch DH. 2014. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8+ central memory T cells. Immunity 41: 116–126. 10.1016/j.immuni.2014.05.018 [DOI] [PubMed] [Google Scholar]
- Gray SM, Amezquita RA, Guan T, Kleinstein SH, Kaech SM. 2017. Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity 46: 596–608. 10.1016/j.immuni.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Dominguez CX, Amezquita RA, Laidlaw BJ, Cheng J, Henao-Mejia J, Williams A, Flavell RA, Lu J, Kaech SM. 2018. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J Exp Med 215: 1153–1168. 10.1084/jem.20171352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann D, Baars PA, Rep MHG, Hooibrink B, Kerkhof-Garde SR, Klein MR, van Lier RAW. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med 186: 1407–1418. 10.1084/jem.186.9.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, Badovinac VP. 2008. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol 8: 107–119. 10.1038/nri2251 [DOI] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. 2016. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature 537: 412– 416. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
- Hendriks J, Gravestein LA, Tesselaar K, van Lier RAW, Schumacher TNM, Borst J. 2000. CD27 is required for generation and long-term maintenance of T cell immunity. Nat Immunol 1: 433–440. 10.1038/80877 [DOI] [PubMed] [Google Scholar]
- Henson SM, Akbar AN. 2009. KLRG1—more than a marker for T cell senescence. Age (Dordr) 31: 285–291. 10.1007/s11357-009-9100-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndler-Brandstetter D, Ishigame H, Shinnakasu R, Plajer V, Stecher C, Zhao J, Lietzenmayer M, Kroehling L, Takumi A, Kometani K, et al. 2018. KLRG1+ effector CD8+ T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity 48: 716–729.e8. 10.1016/j.immuni.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al. 2017. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545: 60–65. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Orlowski RJ, Xu X, Mick R, George SM, Yan PK, Manne S, Kraya AA, Wubbenhorst B, Dorfman L, et al. 2019. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med 25: 454–461. 10.1038/s41591-019-0357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin JX, Konieczny BT, Im SJ, Freeman GJ, et al. 2019. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51: 1043–1058.e4. 10.1016/j.immuni.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T. 2002. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol 3: 558–563. 10.1038/ni802 [DOI] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537: 417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, Shen H, Wherry EJ, Reiner SL. 2007. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med 204: 2015–2021. 10.1084/jem.20070841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin JX, Leonard WJ, Greenleaf WJ, Ahmed R, Goronzy JJ. 2019. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci 116: 14113–14118. 10.1073/pnas.1903520116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. 2018. Understanding subset diversity in T cell memory. Immunity 48: 214–226. 10.1016/j.immuni.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, Cardenas M, Wilkinson S, Lake R, Sowalsky AG, et al. 2019. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576: 465–470. 10.1038/s41586-019-1836-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421: 852–856. 10.1038/nature01441 [DOI] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. 2007. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27: 281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4: 1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. 2010. Prolonged interleukin-2Rα Expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32: 91–103. 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Kallies A, Xin A, Belz GT, Nutt SL. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity 31: 283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang AC, Alexander KA, Wu JE, et al. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571: 211–218. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MV, Ouyang W, Liao W, Zhang MQ, Li MO. 2013. The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 39: 286–297. 10.1016/j.immuni.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratchmarov R, Magun AM, Reiner SL. 2018. TCF1 expression marks self-renewing human CD8+ T cells. Blood Adv 2: 1685–1690. 10.1182/bloodadvances.2018016279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, et al. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol 15: 373– 383. 10.1038/ni.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladle BH, Li KP, Phillips MJ, Pucsek AB, Haile A, Powell JD, Jaffee EM, Hildeman DA, Gamper CJ. 2016. De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc Natl Acad Sci 113: 10631–10636. 10.1073/pnas.1524490113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur MW, Muroyama Y, Drake CG, Sharpe AH. 2018. Inhibitors of the PD-1 pathway in tumor therapy. J Immunol 200: 375–383. 10.4049/jimmunol.1701044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Cui W, Amezquita RA, Gray SM, Guan T, Lu Y, Kobayashi Y, Flavell RA, Kleinstein SH, Craft J, et al. 2015. Production of IL-10 by CD4+ regulatory T cells during the resolution of infection promotes the maturation of memory CD8+ T cells. Nat Immunol 16: 871–879. 10.1038/ni.3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. 2014. Chromatin state dynamics during blood formation. Science 345: 943–949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen JBAG, Blank CU, et al. 2019. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 176: 775–789.e18. 10.1016/j.cell.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli E, Goldman CK, Perera LP, Smedley J, Pung R, Yovandich JL, Creekmore SP, Waldmann TA, Roederer M. 2010. Transient and persistent effects of IL-15 on lymphocyte homeostasis in nonhuman primates. Blood 116: 3238–3248. 10.1182/blood-2010-03-275438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli E, Dominguez MH, Gattinoni L, Chattopadhyay PK, Bolton DL, Song K, Klatt NR, Brenchley JM, Vaccari M, Gostick E, et al. 2013. Superior T memory stem cell persistence supports long-lived T cell memory. J Clin Invest 123: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon M-L, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. 2013. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat Immunol 14: 1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Mackay LK, Minnich M, Kragten NAM, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352: 459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. 2013. The who's who of T-cell differentiation: human memory T-cell subsets. Eur J Immunol 43: 2797–2809. 10.1002/eji.201343751 [DOI] [PubMed] [Google Scholar]
- Makedonas G, Hutnick N, Haney D, Amick AC, Gardner J, Cosma G, Hersperger AR, Dolfi D, Wherry EJ, Ferrari G, et al. 2010. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8+ T cells. PLoS Pathog 6: e1000798. 10.1371/journal.ppat.1000798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Marzo AL, Lefrançois L. 2001. Preferential localization of effector memory cells in nonlymphoid tissue. Science 291: 2413–2417. 10.1126/science.1058867 [DOI] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. 2006. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol 176: 2079–2083. 10.4049/jimmunol.176.4.2079 [DOI] [PubMed] [Google Scholar]
- Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, Speed TP, Busslinger M, Nutt SL, Belz GT. 2013. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol 190: 4585–4594. 10.4049/jimmunol.1300099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane LM, Abdel-Hakeem MS, Wherry EJ. 2019. CD8 T cell exhaustion during chronic viral infection and cancer. Ann Rev Immunol 37: 457–495. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20: 326–336. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JJ, Goldrath AW. 2018. Transcriptional programming of tissue-resident memory CD8+ T cells. Curr Opin Immunol 51: 162–169. 10.1016/j.coi.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 552: 253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollo SB, Ingram JT, Kress RL, Zajac AJ, Harrington LE. 2014. Virus-specific CD4 and CD8 T cell responses in the absence of Th1-associated transcription factors. J Leukoc Biol 95: 705–713. 10.1189/jlb.0813429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli S, Natoli G. 2017. Transcriptional determination and functional specificity of myeloid cells: making sense of diversity. Nat Rev Immunol 17: 595–607. 10.1038/nri.2017.51 [DOI] [PubMed] [Google Scholar]
- Mueller SN, Mackay LK. 2016. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol 16: 79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. 2012. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36: 142–152. 10.1016/j.immuni.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop JK, Thomas RM, Wells AD, Shen H. 2006. Epigenetic remodeling of the IL-2 and IFN-γ loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol 177: 1062–1069. 10.4049/jimmunol.177.2.1062 [DOI] [PubMed] [Google Scholar]
- Obar JJ, Lefrançois L. 2010. Early signals during CD8+ T cell priming regulate the generation of central memory cells. J Immunol 185: 263–272. 10.4049/jimmunol.1000492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ. 2015. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J Exp Med 212: 1125–1137. 10.1084/jem.20142237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, et al. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338: 1220–1225. 10.1126/science.1229620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin E, Hasan M, Bergstedt J, Rouilly V, Libri V, Urrutia A, Alanio C, Scepanovic P, Hammer C, Jönsson F, et al. 2018. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat Immunol 19: 302–314. 10.1038/s41590-018-0049-7 [DOI] [PubMed] [Google Scholar]
- Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, et al. 2016. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354: 1160–1165. 10.1126/science.aaf2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, et al. 2017. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545: 452– 456. 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32: 79–90. 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Powell JD. 2014. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol 14: 435–446. 10.1038/nri3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner SL, Adams WC. 2014. Lymphocyte fate specification as a deterministic but highly plastic process. Nat Rev Immunol 14: 699–704. 10.1038/nri3734 [DOI] [PubMed] [Google Scholar]
- Ridge JP, Di Rosa F, Matzinger P. 1998. A conditioned dendritic cell can be a temporal bridge between a CD4+ T-helper and a T-killer cell. Nature 393: 474–478. 10.1038/30989 [DOI] [PubMed] [Google Scholar]
- Romero P, Zippelius A, Kurth I, Pittet MJ, Touvrey C, Iancu EM, Corthesy P, Devevre E, Speiser DE, Rufer N. 2007. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol 178: 4112–4119. 10.4049/jimmunol.178.7.4112 [DOI] [PubMed] [Google Scholar]
- Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. 2009. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31: 296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. 2018. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175: 998–1013.e20. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401: 708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Teichgräber V, Kalia V, Polley A, Masopust D, Harrington LE, Ahmed R, Wherry EJ. 2007. Strength of stimulus and clonal competition impact the rate of memory CD8 T cell differentiation. J Immunol 179: 6704–6714. 10.4049/jimmunol.179.10.6704 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med 205: 625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, Masopust D. 2014. Tissue-resident memory T cells. Immunity 41: 886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildberg FA, Klein SR, Freeman GJ, Sharpe AH. 2016. Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 44: 955–972. 10.1016/j.immuni.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberger SP, Toes REM, van der Voort EIH, Offringa R, Melief CJM. 1998. T-cell help for cytotoxic T lymphocytes is mediated by CD40–CD40L interactions. Nature 393: 480–483. 10.1038/31002 [DOI] [PubMed] [Google Scholar]
- Scott CL, Omilusik KD. 2019. ZEBs: novel players in immune cell development and function. Trends Immunol 40: 431–446. 10.1016/j.it.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, et al. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571: 270–274. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM. 2016. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45: 1327–1340. 10.1016/j.immuni.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, et al. 2016. The epigenetic landscape of T cell exhaustion. Science 354: 1165–1169. 10.1126/science.aae0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Chen J, González-Avalos E, Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO, Zhang W, Onodera A, et al. 2019. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc Natl Acad Sci 116: 12410–12415. 10.1073/pnas.1905675116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock DJ, Shen H. 2003. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300: 337–339. 10.1126/science.1082305 [DOI] [PubMed] [Google Scholar]
- Shin HM, Kapoor VN, Guan T, Kaech SM, Welsh RM, Berg LJ. 2013. Epigenetic modifications induced by Blimp-1 regulate CD8+ T cell memory progression during acute virus infection. Immunity 39: 661–675. 10.1016/j.immuni.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. 2013. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol 14: 1285–1293. 10.1038/ni.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slota C, Shi A, Chen G, Bevans M, Weng NP. 2015. Norepinephrine preferentially modulates memory CD8 T cell function inducing inflammatory cytokine production and reducing proliferation in response to activation. Brain Behav Immunity 46: 168–179. 10.1016/j.bbi.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin JE, Burns JC, Gohil M, Zou T, Kim JS, Maltzman JS, Wherry EJ, Koretzky GA, Jordan MS. 2010. T-cell receptor signals direct the composition and function of the memory CD8+ T-cell pool. Blood 116: 5548–5559. 10.1182/blood-2010-06-292748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Bevan MJ. 2003. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300: 339–342. 10.1126/science.1083317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PA, Miron M, Farber DL. 2019. Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol 4: eaas9673. 10.1126/sciimmunol.aas9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, Palmer E. 2009. Different T cell receptor signals determine CD8+ memory versus effector development. Science 323: 502–505. 10.1126/science.1163612 [DOI] [PubMed] [Google Scholar]
- Thome JJC, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, Farber DL. 2014. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159: 814–828. 10.1016/j.cell.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, Pardoll DM. 2015. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27: 450–461. 10.1016/j.ccell.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tough DF, Rioja I, Modis LK, Prinjha RK. 2020. Epigenetic regulation of T cell memory: recalling therapeutic implications. Trends Immunol 41: 29–45. 10.1016/j.it.2019.11.008 [DOI] [PubMed] [Google Scholar]
- Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. 2016. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45: 415–427. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Utzschneider DT, Delpoux A, Wieland D, Huang X, Lai CY, Hofmann M, Thimme R, Hedrick SM. 2018. Active maintenance of T cell memory in acute and chronic viral infection depends on continuous expression of FOXO1. Cell Rep 22: 3454–3467. 10.1016/j.celrep.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, Bevan MJ. 2012. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J Immunol 189: 3462–3471. 10.4049/jimmunol.1201305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, Yu B, Crotty S, Goldrath AW, Pipkin ME. 2018. The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48: 659–674.e6. 10.1016/j.immuni.2018.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MD, Godbout JP, Sheridan JF. 2017. Repeated social defeat, neuroinflammation, and behavior: monocytes carry the signal. Neuropsychopharmacology 42: 46–61. 10.1038/npp.2016.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes MP, Wills MR, Mynard K, Hicks R, Sissons JGP, Carmichael AJ. 1999. Large clonal expansions of human virus-specific memory cytotoxic T lymphocytes within the CD57+CD28−CD8+ T-cell population. Immunology 98: 443–449. 10.1046/j.1365-2567.1999.00901.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Kurachi M. 2015. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15: 486–499. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 4: 225–234. 10.1038/ni889 [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27: 670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1: eaai8593. 10.1126/sciimmunol.aai8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF. 2009. Programming for CD8 T cell memory development requires IL-12 or type I IFN. J Immunol 182: 2786–2794. 10.4049/jimmunol.0803484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D'Cruz LM, et al. 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol 12: 1221–1229. 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, Heuston EF, Kirby M, Anderson S, Cheng J, et al. 2019. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat Immunol 20: 890–901. 10.1038/s41590-019-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hunter CA. 2015. The immunobiology of interleukin-27. Annu Rev Immunol 33: 417–443. 10.1146/annurev-immunol-032414-112134 [DOI] [PubMed] [Google Scholar]
- Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, Pereira RM, Crotty S, Chang JT, Pipkin ME, et al. 2017. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat Immunol 18: 573–582. 10.1038/ni.3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJD, Suresh M, Altman JD, Ahmed R. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188: 2205–2213. 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, Cui W. 2019. CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity 51: 1028–1042.e4. 10.1016/j.immuni.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zediak VP, Johnnidis JB, Wherry EJ, Berger SL. 2011. Cutting edge: persistently open chromatin at effector gene loci in resting memory CD8+ T cells independent of transcriptional status. J Immunol 186: 2705–2709. 10.4049/jimmunol.1003741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D, Wherry EJ. 2015. Immune memory and exhaustion: clinically relevant lessons from the LCMV model. In Crossroads between innate and adaptive immunity V (ed. Schoenberger SP, Katsikis PD, Pulendran B), pp. 137–152. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Zehn D, Lee SY, Bevan MJ. 2009. Complete but curtailed T-cell response to very low-affinity antigen. Nature 458: 211–214. 10.1038/nature07657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. 2013. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity 39: 687–696. 10.1016/j.immuni.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Joe G, Hexner E, Zhu J, Emerson SG. 2005. Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat Med 11: 1299–1305. 10.1038/nm1326 [DOI] [PubMed] [Google Scholar]
- Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. 2010. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33: 229–240. 10.1016/j.immuni.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]