SUMMARY
Sexual development in filamentous fungi is a complex process that relies on the precise control of and interaction between a variety of genetic networks and pathways. The mating-type (MAT) genes are the master regulators of this process and typically act as transcription factors, which control the expression of genes involved at all stages of the sexual cycle. In many fungi, the sexual cycle typically begins when the mating pheromones of one mating type are recognized by a compatible partner, followed by physical interaction and fertilization. Subsequently, highly specialized sexual structures are formed, within which the sexual spores develop after rounds of meiosis and mitosis. These spores are then released and germinate, forming new individuals that initiate new cycles of growth. This review provides an overview of the known genetic networks and pathways that are involved in each major stage of the sexual cycle in filamentous ascomycete fungi.
KEYWORDS: sexual reproduction, Pezizomycotina, MAT genes, pheromones, fertility, pheromones
INTRODUCTION
Sexual reproduction is an essential component of the life cycle in most eukaryotes. This is particularly true for metazoans, where sexual reproduction is usually the only process that allows for the production of offspring (1). Fungi are no exception and have maintained complex and active sexual cycles, in addition to being capable of asexual reproduction (2). Combined with the energy intensive nature of sexual development, this suggests that regular cycles of sexual development are important to the long-term survival of fungal species, resulting in benefits beyond simply the production of genetically unique offspring (3).
Sexual reproduction has been described in many species across the phylum Ascomycota (4, 5). Although the members of this group show immense diversity in morphology and ecology, their sexual cycles remain highly conserved. In most species, sexual reproduction is centrally controlled by two sets of major regulators (6): the mating-type (MAT) genes and the pheromone response pathway (7). While the sexual cycles of only a subset of species have been thoroughly investigated, their conserved nature, even between distantly related taxa, suggests that these processes are likely to be similar in most fungal lineages. Despite these similarities, there are some differences with regard to the functions of the genes involved and the role of external processes in the sexual cycles of fungi.
Ascomycete fungi reside in three subphyla: Saccharomycotina, Taphrinomycotina, and Pezizomycotina (8). The first two groups accommodate yeasts and dimorphic species, which have been extensively studied, particularly with regard to their sexual cycles (9, 10). In contrast, the Pezizomycotina is comprised entirely of filamentous fungi. Representing the largest subphylum and encompassing a wide diversity of important fungi, the Pezizomycotina accommodates both detrimental and beneficial fungi. Examples include the human-pathogenic opportunist Aspergillus fumigatus (11) and the dermatophyte Trichophyton rubrum (12), as well as plant pathogens such as the notorious chestnut blight fungus, Cryphonectria parasitica (13), and the causal agent of pine pitch canker, Fusarium circinatum (14). A number of ecologically important species have also been described, including those that are important for wood and litter decay, as well as those that form associations with other organisms, including lichens (15, 16).
Importantly, the subphylum Pezizomycotina also accommodates a number of important model species, including Neurospora crassa. Research on this species has significantly contributed to our understanding of many fundamental genetic processes and molecular mechanisms (as reviewed in reference 17). The “one gene, one enzyme” hypothesis that gave birth to molecular biology was developed using nutritional mutants of N. crassa in 1941. Since then, this species been used as a model to study the molecular components of circadian rhythms, the Repeat-Induced Point (RIP) mutation genome defense mechanism, RNA interference (RNAi) and epigenetics. Together with other model Pezizomycotina organisms, such as Aspergillus nidulans, fungi have provided insights into a variety of important eukaryotic processes, including photosensing (18), secondary metabolism (19, 20), cellular synthesis pathways (21), asexual reproduction (22, 23), and sexual development (24–27). In this review, we consider the genes and genetic networks that govern sexual reproduction in a wide variety of species from the Pezizomycotina, considering research done in both model and non-model fungi.
MATING IN FILAMENTOUS FUNGI
Mating in ascomycete fungi is relatively well conserved, particularly with regard to the genetic mechanisms that govern sexual development (28). In both filamentous fungi and the yeasts, sexual reproduction is regulated by the mating-type (MAT) genes, mating pheromones, and various signal transduction pathways. In fact, much of our understanding of sexual reproduction in filamentous fungi is based on similar pathways and processes that were first identified and studied in the yeasts (28). For example, the MAT genes and the pheromone genes were first described in Saccharomyces cerevisiae (29–31). Later, similar genes were identified in the genomes of many filamentous fungi (32–37) and, in some cases, these genes can fulfill their roles when heterologously expressed in S. cerevisiae (38, 39). This alone illustrates the phenomenal level of conservation in the regulatory pathways of sexual development across diverse ascomycete fungi.
Despite the regulatory and genetic conservation of sexual reproduction in the yeasts and the filamentous fungi, there are notable differences, particularly with regard to the sexual tissues that are formed during the sexual cycle. In S. cerevisiae, compatible mating partners typically fuse and undergo karyogamy to form a diploid cell. Under conducive environmental conditions, this is followed by meiosis and the production of sexual progeny cells within a simple ascus as reviewed by Haber (40). In contrast, filamentous fungi form highly complex and multicellular sexual structures, including the ascomata (Fig. 1). Fungi residing in different orders produce ascomata of different shapes and sizes, but all of these structures contain asci in which the sexual ascospores are produced (reviewed in reference 41).
FIG 1.
Generalized sexual cycle of heterothallic Pezizomycotina fungi. The cycle begins (step 1) with an interaction between compatible isolates, with male structures from one of the partners secreting pheromones (step 2), which are recognized by the female structures from a suitable mating partner (step 3). This recognition results in growth of the trichogyne toward the male cells (step 4), resulting in physical contact between the two partners; fertilization (step 5); and the subsequent production of a protoascoma (step 6). Within the ascus, multiple rounds of meiosis and mitosis produce the mature ascospores (steps 7 to 12), which are subsequently released (step 13) from the mature ascoma (step 14). This cycle would be identical in homothallic species, except that an opposite mating partner would not be required to initiate the process. The different ascomatal types are depicted below the sexual cycle, illustrating the morphological diversity that can be observed in the Pezizomyctonia.
While ascomatal structures are morphologically diverse across the Pezizomycotina, the intermediate steps that result in the production of these tissues are relatively well conserved (Fig. 1). In many fungi, particularly self-sterile fungi, sexual reproduction begins with the fertilization of a female structure (ascogonium) by male structures, in the form of conidia or spermatia (42). Fertilization initiates the production of highly specialized cells and tissues, such as the protoascomata that mature into ascomata which bear asci. Also, within the asci, ascospores are produced via numerous rounds of meiosis and mitosis. The culmination of sexual development occurs when the ascospores are released into the environment to germinate.
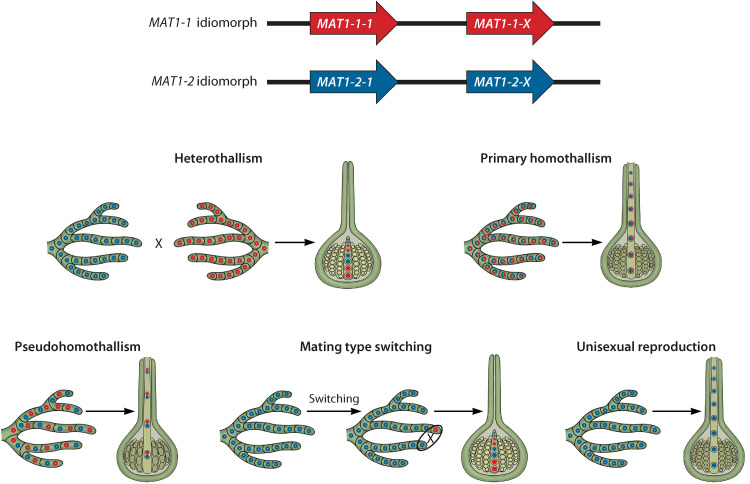
The initiation of sexual reproduction, i.e., the step(s) before fertilization, differs from species to species. The requirements for initiation can be used to describe the four major sexual strategies found in the Pezizomycotina, i.e., heterothallism, primary homothallism, secondary homothallism, and unisexuality (Fig. 2). Individuals that exhibit heterothallic behavior are self-sterile, requiring a partner to engage in sexual reproduction (Fig. 1, step 1). Each of the mating partners carries a unique version of the MAT locus, and these must be different for sexual reproduction to occur. Each partner thus has a unique sexual identity, referred to as mating type, and this is conferred by the type and distribution of the MAT genes (Fig. 2).
FIG 2.
Mating-type idiomorphs, MAT genes, and their distribution in fungi utilizing the various sexual strategies exhibited by Pezizomycotina fungi. Open circles represent nuclei in each cell, while closed circles represent the ascospores produced. Red indicates the presence of genes from the MAT1-1 idiomorph, and blue indicates the presence of genes from the MAT1-2 idiomorph.
It is worth noting that while the sexual and mating identities in heterothallic fungi can be described using mating type, these fungi also independently produce cells and structures that can be classified as male or female. Thus, in addition to being of the opposite mating type, compatible partners must also produce complementary male and female structures (43, 44). These two identities are not linked, and many heterothallic fungi are hermaphroditic, producing both male cells and female structures. Mating type is specifically determined by the genes present at the MAT locus, as discussed in detail below. In contrast, male and female structures are produced independently of the MAT information present. Importantly, a successful sexual interaction only requires the recognition of one set of male/female cells. Thus, male cells from an individual of the MAT1-1 type must interact with the female structures from an individual of the MAT1-2 type or vice versa (43, 44). Mating type and male/female identity primarily interact upon the expression of the mating pheromones, a process that is discussed in detail later in this review.
It is somewhat more difficult to distinguish between the different forms of homothallism at the physiological level because in all cases, a single isolate is capable of independent sexual development and is considered self-fertile (Fig. 2). These can, however, be differentiated genetically by studying the distribution of the MAT genes (Fig. 2) (45). In primary homothallism, all of the necessary MAT genes are present and expressed by a single individual and within a single nucleus (46). Secondary homothallism describes two different strategies: pseudohomothallism and mating type switching. In the former, the entire MAT complement is expressed by a single individual, but the MAT information is distributed between two independent nuclei that produce a heterokaryotic mycelium (47). Species capable of mating type switching harbor two versions of the mating information. One of these versions has a copy of all the MAT genes but can be altered to produce the second variant, which has a different gene complement (48, 49). Each of these two variants has a different mating specificity, and it is the expression of the MAT genes that drives compatibility between the two, thereby resulting in self-fertility in a single isolate (50). Unisexuality describes a system where a self-fertile individual retains and expresses only mating information from a single mating type (51).
The complex physiological processes underpinning sexual reproduction are stringently controlled by a huge number of genes. The primary regulators are encoded by the MAT genes, which regulate the expression of hundreds of sex-related genes (38, 52, 53), such as those involved in mating partner recognition. In addition, many non-MAT-regulated genes, including those involved in male and female fertility, ascomatal maturation and ascosporegenesis, are essential for the completion of the sexual cycle. The following sections provide details of the roles that each of these genes play.
MAT GENES
MAT1 Locus
The mating-type locus contains the MAT genes and has a primary role in the sexual process. This locus was formally described as MAT1 based on the assumption that all ascomycetes would have a single copy of this region (54). Two versions of the MAT1 locus are present in the diallelic mating system of heterothallic fungi, and this is recognized by naming these MAT1-1 and MAT1-2, with the use of capital letters acknowledging the lack of dominance between the two “alleles” (Fig. 2) (54). The nonallelic nature of these versions was formally acknowledged in 1990, when the term idiomorph was suggested by John Wyatt and introduced by Metzenberg and Glass (55) to describe these two versions of the MAT1 locus. Although the two different idiomorphs can easily be recognized in most heterothallic species, the term does not extend to their homothallic counterparts. Most homothallic species have a single MAT1 locus that is defined based on the presence of homologous mating-type genes (56), although in some cases more than one, often unlinked, MAT locus can be present within a single genome (32, 57, 58).
Mating-Type Genes
The precise definition of what constitutes a MAT gene is centered around their position in one of the two mating-type idiomorphs (56). This definition is broadly accepted, but an alternative description holds that a true mating-type gene is involved in establishing the mating-type identity of the nuclei after fertilization and that its protein product would localize only to the nucleus (59). Although such a definition may be considered the golden standard, it is impractical because many mating-type genes have not been the subject of functional studies. There is also convincing experimental and computational evidence that at least one of the known mating-type proteins, MAT1-1-2, localizes only to the cytoplasm and not the nucleus (60, 61). Furthermore, some homologous MAT genes have different functions in different species, precluding the application of function as an indication of mating identity. In the absence of a more suitable option, a system that defines mating-type genes as those present at the mating-type locus remains the most intuitive.
The locus-based definition for mating-type genes has been subject to valid criticism. Named MAT genes are widely considered to be the master regulators of sexual reproduction, responsible for conferring sexual identity and orchestrating processes linked to the sexual cycle. These roles can be validated only through functional studies (56, 62), which are lacking for most MAT genes. This implies that newly described MAT genes are mating-type genes in name only. This disparity is now being addressed, with new functional studies of diverse MAT genes being published regularly (63–65). Another criticism of a locus-based MAT gene definition is that the dynamic nature of this region can result in the inclusion of genes with no known role in mating into the MAT1 locus (66, 67). This contradicts the position-based definition of a mating-type gene, a shortcoming highlighted in a recent revision of the MAT gene nomenclature system (56). Assigning a novel, MAT-specific name to a gene within the MAT1 locus, only when no known homolog exists, can resolve this issue (56). This approach has been successfully applied to many novel MAT genes (68–71). In cases where homologs of known genes have been observed, these are significantly diverged to the point of representing MAT-specific versions of these genes (67, 72). Future studies must consider whether these genes have gained novel, MAT-specific functions.
Primary Mating-Type Genes
Two MAT genes are almost universally present in all studied Pezizomycotina classes (Table 1). MAT1-1-1 was named as the first gene (MAT1-1-1) from the MAT1-1 (MAT1-1-1) idiomorph (73, 74), and its presence defines that idiomorph. Similarly, the MAT1-2-1 gene typifies the MAT1-2 idiomorph as the first described gene from this idiomorph (73, 75). The protein products of both these genes have conserved domains involved in DNA binding (76). In addition, a conserved intron is present in the MAT1-2-1 HMG-box domain positioned across a serine codon, a characteristic that is unique to mating-type genes that have this domain (56, 77, 78). The core roles of MAT1-1-1 and MAT1-2-1 in the sexual process, as well as their wide distribution across the Pezizomycotina, has led to these genes being known as the core, or primary, mating-type genes (56, 62).
TABLE 1.
MAT genes described from the Pezizomycotina
| Gene | Representative taxa | Representative protein NCBI accession no. | Conserved protein domain | Reference(s) |
|---|---|---|---|---|
| Primary MAT genes | ||||
| MAT1-1-1 | Neurospora crassa | ESA43845 | PF04769–MATα1 HMG-box | 74, 202 |
| MAT1-2-1 | Neurospora crassa | AAA33598 | PF00505–HMG box | 55, 75 |
| Secondary MAT genes | ||||
| MAT1-1-2 | Neurospora crassa | EAA35087 | PF17043–MAT1-1-2 domain of unknown function | 203 |
| MAT1-1-3 | Neurospora crassa | EAA35088 | PF00505–HMG box | 204 |
| MAT1-1-4 | Pyrenopeziza brassicae | CAA06845 | None | 204 |
| MAT1-1-5 | Botrytis cinerea | AHX22633 | None | 205 |
| MAT1-1-6 | Pseudogymnoascus destructans | AIG95729 | None | 206 |
| MAT1-1-7 | Coccidioides immitis | ABS19617 | None | 67 |
| MAT1-1-8 | Diplodia sapinea | AHA91691 | None | 207 |
| MAT1-1-9a | ||||
| MAT1-1-10 | Morchella importuna | MG681026 | None | 68 |
| MAT1-1-11 | Morchella importuna | MG681027 | None | 68 |
| MAT1-1-12 | Teratosphaeria zuluensis | QEJ80698 | None | 69 |
| MAT1-2-2 | Neurospora crassa | -b | None | 85, 208 |
| MAT1-2-3 | Coccidioides immitis | KMP00264 | PF11051–mannosyl trans3 | 67 |
| MAT1-2-4 | Aspergillus fumigatus | XP_754990 | None | 63 |
| MAT1-2-5 | Diplodia sapinea | AHA91682 | None | 207 |
| MAT1-2-6 | Magnaporthe grisea | BAE66607 | PF00505–HMG box | 209 |
| MAT1-2-7 | Huntiella omanensis | AOY41711 | None | 71 |
| MAT1-2-8 | Villosiclava virens | AKE48512 | None | 210 |
| MAT1-2-9 | Fusarium fujikuroi | AEP03799 | None | 211 |
| MAT1-2-10 | Botrytis cinerea | CDF43998 | None | 205 |
| MAT1-2-11 | Pseudogymnoascus destructans | AIG95713 | None | 206 |
| MAT1-2-12 | Teratosphaeria zuluensis | QEJ80704 | None | 69 |
| MAT1-2-13 | Calonectria pauciramosa | QKY89075 | None | 70 |
| MAT1-2-14 | Letharia columbiana | QEL51134 | None | 81 |
| MAT1-2-15 | Colletotrichum fructicola | XM_032032861 | None | 80 |
Secondary Mating-Type Genes
Apart from MAT1-1-1 and MAT1-2-1, 11 additional MAT1-1-specific and 14 additional MAT1-2-specific genes have been described in the Pezizomycotina (56, 68, 70, 79, 80) and are all considered secondary MAT genes (Table 1). Although a formal definition for secondary mating-type genes does not exist, these genes are present within the confines of the mating-type locus, have no known homologs outside the mating-type genes, and tend to show a lineage-specific distribution (56). MAT1-1-2 and MAT1-1-3 have been identified from species in the Sordariomycetes and Leotiomycetes (56), while homologs of MAT1-1-7 have been reported among the Lecanoromycetes and Eurotiomycetes (81), making these three genes the most widely spread secondary MAT genes. In a small number of Sordariomycetes, the MAT1-1-2 gene is unusually present in the MAT1-2 idiomorph (66, 82, 83), although this does not impact the sexual cycle in these fungi. Most other MAT genes have been identified in only a single order, family, or genus (56).
Since the last overview and revision of all known mating-type genes (56), four secondary MAT1-1 genes have either had names assigned or amended. A homolog of the known MAT1-1-7 gene was recently identified in several families of the Lecanoromycetes (81). Through positional conservation and phylogenetic analysis, it was shown that this gene is also homologous to the MAT1-1-9 gene described from members of the Eurotiomycetes (81), leaving the MAT1-1-9 gene name vacant (Table 1). MAT1-1-10 and MAT1-1-11 have been described from the MAT loci of Morchella species (Pezizomycetes) (68). Another putative MAT1-1-10 gene was described in the Dothideomycete genus Teratosphaeria (69, 84) and appears also to be present in the MAT1 locus of other members of the Mycosphaerellaceae (69). There is no apparent homology between the two MAT1-1-10 genes, and they follow a lineage specific distribution.
Four novel genes have been identified from the MAT1-2 mating-type idiomorph. Two of these have been named MAT1-2-12—one in Teratosphaeria species (69, 84) and the other in Calonectria species (70). There is no homology between the two MAT1-2-12 genes or their encoded proteins (B. Wingfield, unpublished data). According to the guidelines of Wilken et al. (56), the Teratosphaeria MAT1-1-10 (69, 84) should be renamed MAT1-1-12, while the Calonectria MAT1-2-12 (70) should be known as MAT1-2-13. A novel MAT gene was also described in species of the Lecanoromycetes and has been named MAT1-2-14 (81), while another gene identified in Colletotrichum species was added as MAT1-2-15 (80). None of these genes have any conserved domains that might suggest their possible functions.
The complexity in naming novel mating-type genes, together with the need to constantly reevaluate already established MAT gene names, highlights the need for a dynamic, curated database of mating-type gene names and homologs. A web-based MAT gene server to assist in mating-type gene identification is currently being developed (B. D. Wingfield, unpublished data) and will go some way to address these challenges.
Functions of Mating-Type Genes
Functional analyses remain the only true test for the role of a gene in the biology of a fungus, but such studies are lacking for most of the MAT genes. It is not surprising that the majority of available functional studies have targeted the core MAT1-1-1 and MAT1-2-1 genes, largely due to their wide distribution and central role in the sexual process (25, 52, 53, 60, 85–87). More recently, some studies have emerged that target the secondary MAT genes, illustrating the complex interactions that underlie sexual reproduction (63, 64, 85, 86, 88).
The history of functional studies on MAT genes has closely followed technological advances. Early studies relied on the transformation of cloned DNA into a complementary strain (33, 89). Later, mutations were induced in target genes by employing techniques such as the RIP mechanism that silences duplicate sequence products (25). The commercialization of molecular techniques relying on PCR amplification has strongly advanced new technologies used in functional studies. These include transcription profiling during sexual development (53, 87), comparisons of whole expression profiles between wild-type and mutant strains (53), antisense RNA approaches to disrupt gene function (87), and microarray experiments (52). Genome-based technologies have further expanded the available toolbox, with techniques such as CHIP-Seq analysis (90) and transcriptomics (91) becoming more prominent. The advent of novel, easy-to-use gene editing technologies, including the CRISPR-Cas9 system, promises further progress and has already led to the functional analysis of some mating-type genes (60, 64).
Functional studies on the MAT1-1-1 and MAT1-2-1 genes and their core domains are too numerous to cover in detail here. They have, however, provided an impression of conserved functions intermingled with great adaptability. Generally, both primary mating-type genes rarely impact vegetative growth directly (25, 52, 87, 88, 92), although they are necessary for successful completion of the sexual cycle in both homothallic and heterothallic fungi (25, 38, 52, 60, 75, 87, 92, 93). MAT genes control the sexual process by conferring sexual identity to individual isolates (34, 75), mediating the formation of male and female reproductive tissues (88, 93, 94), regulating the mating pheromone and receptor system (35, 52, 53, 88), and controlling ascomatal and ascospore development (34, 38, 88, 95). Although these functions appear to be well conserved, even among distantly related species, it is worth noting that these effects may not be universal. For example, in the smut fungus Villosiclava virens that causes false smut in rice, deletion of the MAT1-1-1 gene has a significant impact on the vegetative growth rate and conidial structure (60). Whether this species is an exception or provides evidence of as-yet-undiscovered complexity in the functions of the mating genes remains to be discovered.
Among the secondary MAT genes, MAT1-1-2 and MAT1-1-3 have been the subject of most of the functional studies. Such studies have shown that these genes are generally not essential for sexual reproduction but rather function to fine-tune the sexual process. Early work on the model species N. crassa showed that both MAT1-1-2 and MAT1-1-3 influence the expression of sex-related genes, with mutants showing reduced fertility (25, 43). In contrast, in Fusarium graminearum both genes are important for male and female fertility, as well as ascospore formation (96). Once again, there are exceptions to this generalization, such as the Sordaria macrospora MAT1-1-2 gene, which is essential for completion of the sexual cycle (85). In some cases, this gene is also involved in functions attributed to the core MAT genes, including nuclear recognition, control of the pheromone response pathway, and ascomatal development (43, 59, 85). Deletion of the S. macrospora MAT1-1-3 gene shows no observable phenotype (85), although deletion of the homologous gene in some homothallic Fusarium species abolishes self-fertility (97) and leads to significantly reduced fertility in Podospora anserina (43).
An interesting finding that has emerged from studies on MAT1-1-1, MAT1-1-2, MAT1-1-3, and MAT1-2-1 genes is that many nonmating functions are also under the control of these genes. These include general metabolic processes and energy production (52, 53), stress responses (60), the production of secondary metabolites (90), virulence (60), vegetative growth (64), and other diverse biological processes. This should not be surprising since all of these genes encode proteins that are characterized by conserved DNA-binding domains that are associated with transcription factors (56, 98). Therefore, it was plausible that these proteins would be involved in regulating a complex network of gene expression to accomplish the sexual process (99). In addition, sexual reproduction is an involved process that relies on specialized tissue differentiation in response to environmental and genetic cues (62). Unfortunately, many functional studies only evaluate the effect of MAT gene disruption on the sexual process and do not screen for pleiotropic effects. As new functional studies on the MAT genes are conducted, the effect of the MAT genes on nonmating functions will likely become more clear.
Apart from MAT1-1-2 and MAT1-1-3, only five additional secondary MAT genes have been functionally characterized. Deletion of the MAT1-1-5 gene in Sclerotinia sclerotiorum reduces the production of spermatia and completely blocks ascomatal development, halting the sexual process (88). The MAT1-2-10 gene in the same species is involved in regulation of the cytoskeleton, with deletion mutants showing delayed ascomatal development and, when mature, produce aberrant numbers of ascospores (88). In the closely related species Botrytis cinerea, the roles of MAT1-1-5 and MAT1-2-10 are different, with both proteins jointly regulating the transition to karyogamy (92). The MAT1-2-9 gene in F. graminearum appears not to be essential for sexual reproduction, but its transcription patterns do mirror those of the primary MAT genes (97). The MAT1-2-4 gene in A. fumigatus is another essential regulator of sexual reproduction, but deletion of this gene did not allow for the elucidation of a specific function in sexual reproduction (63). A gene knockout study of the Huntiella omanensis MAT1-2-7 gene showed that this gene is essential for maturation of the ascomata, a function shared with other secondary mating-type genes (64).
The mating-type genes are themselves subject to genetic regulation, mostly as a response to environmental cues. Studies on the model species P. anserina have revealed a network of HMG-box motif genes that regulate sexual development, some through direct transcriptional control of the mating-type genes and their downstream targets (99). Two genes, ste11 and hmg8 were identified as directly controlling MAT gene expression. A third gene, pro1, connects environmental cues, such as nutrient starvation, directly to MAT gene expression by activating hmg8 (100). In the heterothallic species Trichoderma reesei, the effect of light on the sexual process is mediated by the env1 and vel1 genes through direct transcriptional regulation of MAT1-2-1 (101, 102). Many other environmental response genes have been linked either directly or indirectly to MAT gene expression, and these have been extensively reviewed by Wilson et al. (62). Although these studies have provided some clarity, a thorough understanding of the genes that control MAT gene expression is still needed to fully appreciate the complexities of sexual reproduction in the Pezizomycotina.
Despite great progress having been made in recent times to identify and characterize mating-type genes, substantial gaps remain in our knowledge of this subject. For example, MAT genes have not been formally described in many classes of fungi, and even in the classes with known mating-type genes, some orders lack representation (Fig. 3). It has been shown that variations in mating strategy can be present among even members of the same genus (58, 66), and this is often linked to interesting variations in the mating-type genes or MAT1 locus structure (71, 82, 83). Examples exist of gene conversions (e.g., MAT1-1-1 to MAT1-2-7 [103]) and locus expansions (66, 67) that have helped to shape the MAT1 region, but much remains unknown regarding the evolution and maintenance of this important region. Future studies on MAT gene function, as well as the description of the MAT1 loci in underrepresented groups (Fig. 3), will contribute to addressing current knowledge gaps.
FIG 3.
Cladogram of the Pezizomycotina highlighting the lack of studies on sexual reproduction in this group. Classes or orders shown in red have no described MAT genes and/or no published functional studies relating to the sexual process. Orders shown in white have at least one species with described MAT genes and/or at least one functional study related to sexual reproduction. Details can be found in Table S1 in the supplemental material. The cladogram was constructed in phyloT using NCBI taxonomic identification numbers and was edited in iTol v6 (https://itol.embl.de/).
FEMALE AND MALE FERTILITY
Regardless of their mating type, most heterothallic fungi are capable of producing both male and female structures. Conidia or other spermatizing cells act as the male cells, which act by fertilizing the female structures known as ascogonia (Fig. 1) (104). Given that these structures are produced independently of one another, there are genes whose functions are exclusively linked to either male or female fertility. As a direct consequence, certain isolates can be female fertile and male sterile or vice versa (105). Isolates that are considered male sterile are typically unable to produce spermatizing agents or are incapable of fertilizing the ascogonia of the female partner. In contrast, isolates considered female sterile are either unable to form ascogonia or protoascomata or produce protoascomata that are unable to mature.
Some of the most important genes that influence male and female fertility are the mating pheromones and their cognate receptors, respectively (37, 106, 107). In general, the pheromones are expressed by the male cells and are recognized by their receptors, which are present on the cells of the female ascogonia (107). Understandably, deletion of the pheromone factors or receptors results in either a decrease or complete loss of fertility and the entire sexual pathway. Given the complexity and importance of this pathway in sexual development, a later section of this review is devoted entirely to the pheromone response pathway.
Very few genes have been associated with male fertility in the filamentous fungi. Instead, many of the genes that have been shown to contribute to the early steps of the sexual cycle predominantly regulate female fertility (108–111). Thus, apart from certain MAT genes (96) and the pheromone genes (109, 112), only the mcm1 gene in Magnaporthe oryzae has thus far been described as being essential to male fertility (113). Encoding a MADS box transcription factor, this gene is also required for the production of microconidia, which likely act as the spermatizing agents in this fungus. It is thus not surprising that Δmcm1 mutants are male sterile (113).
In P. anserina, four genes have been shown to contribute significantly to female fertility; nox1 is important for the production of maternal tissues (114), mpk1 is important for the development of the underlying mycelium that surrounds and nourishes the growing ascomata (115), and the two IDC genes, idc2 and idc3, are involved in the communication between these two tissue types (116). It is thought that nox1, which encodes an NADPH oxidase, is responsible for providing the developing protoascomata with fresh nutrients via reactive oxygen species (ROS)-mediated signaling, and its deletion results in very small protoascomata and delayed development (114). Similarly, deletion of either of the IDC genes results in isolates that can differentiate into both male and female structures but exhibit delayed protoascomatal maturation after fertilization. In these isolates, some ascomata will develop, but only a few ascospores are released (116).
In N. crassa, three genes coding for proteins involved in the calcium ion signaling pathway are important for female fertility (117): a phospholipase (plc1), a secretory phospholipase (splA2), and a calcium-hydrogen ion exchanger (cpe1). Two double mutants, Δplc1/ΔsplA2 and Δplc1/Δcpe1, are entirely female sterile and produce no ascomata. It is thought that these phenotypes are the result of a significant decrease in the expression of the two mating pheromones, as well as a female/male fertility gene (fmf-1) in the double mutants (117). A fourth gene, fig1, is involved in a calcium ion uptake system and is essential for female fertility in MAT1-2 isolates of N. crassa (118), illustrating not only that calcium uptake and signaling are important for fertility, but that the role of a protein can change based on the mating type of the isolate.
RECOGNITION OF A SUITABLE MATING PARTNER
The sexual cycle, particularly in heterothallic species, begins with the identification and recognition of an appropriate mating partner (28). This process is facilitated in many species by the pheromone response pathway (7). Partners of opposite mating type express one of the two mating pheromones, which are secreted into the environment by male cells, and are subsequently recognized by a suitable opposite mating type partner via pheromone receptors (Fig. 1). Upon recognition of these diffusible peptides, specialized hyphae known as trichogynes grow from the female partner toward the male cell. Once the male and female cells interact physically, fertilization takes place, and the sexual cycle begins. At a molecular level, the pheromone factor/receptor recognition activates a heterotrimeric G-protein, thereby initiating the signal transduction pathway, which culminates in the transcription of genes related to mating (Fig. 4).
FIG 4.
Pheromone response pathway in Neurospora crassa. The MAT1-1-1 and MAT1-2-1 proteins control the expression of the α- and a-factor pheromones, respectively. Recognition of the pheromones by their respective cognate receptors results in activation of the coupled G-protein and dissociation of its subunits. These subunits subsequently activate one of the three MAPK cascades, MAK2, that ultimately results in protoascomatal development. Two other MAPK cascades, MAK1 and OS-2, are also important for protoascomatal development but respond to different environmental signals.
Mating Pheromones
The two most commonly utilized fungal mating pheromones are the α- and a-factors (7, 119). The genes encoding these small proteins are transcriptionally regulated by the MAT1-1-1 and MAT1-2-1 proteins, respectively. Consequently, the pheromone genes are expressed in a mating type-dependent manner in most heterothallic species, allowing for recognition between individuals of opposite mating type (35, 91, 120, 121).
Genes encoding the two pheromones have been described in species with different sexual strategies. These genes have been found in diverse heterothallic species such as N. crassa (35), H. omanensis (91), C. parasitica (120), and Magnaporthe grisea (121). Similarly, the homothallic species F. graminearum (122), A. nidulans (36), and S. macrospora (123, 124) all harbor genes encoding mating pheromones. Even species exhibiting the more unique strategies, such as unisexuality (Huntiella moniliformis [69] and Neurospora africana [125]) and pseudohomothallism (P. anserina [37]) express mating pheromones.
Interestingly, there are groups of fungi where only a single mating pheromone has been identified. This is despite a wealth of genomic and transcriptomic data and a significant effort to identify these genes. In A. nidulans (36) and other Eurotiomycete fungi (125), for example, the gene encoding a protein with similarity to the a-factor pheromone has remained elusive, while the α-factor pheromone has been identified. Similarly, species in the Pezizomycetes, including Tuber melanosporum (126), also harbor only the α-factor encoding gene. Whether a second factor exists, with or without similarity to the a-factor, remains to be discovered. These genes, being highly variable and typically very short, are notoriously difficult to identify. Their identification consequently requires gene sequences from very close relatives to be used as BLAST queries against the genomes of newly investigated species. Alternatively, when such sequences are not available, it is possible to make use of the microsynteny that exists between more distantly related species (125). In this way, pheromone-flanking genes in species where the pheromones are known can be used to identify the locus harboring the pheromone, after which the pheromone genes can be manually annotated. In addition, the conserved domains of the pheromones, as detailed below, can also be used to search through and filter protein-coding genes in these genomes. This makes it possible to identify proteins that share structural similarities to the mating pheromones.
Given their important role in partner recognition, it is not surprising that the pheromone genes are essential for sexual reproduction in many model heterothallic species (Table 2). In N. crassa (106) and C. parasitica (112, 127), both pheromone types are indispensable for male fertility, with deletion mutants unable to complete the sexual cycle. In N. crassa, the importance of each pheromone in male fertility is mating type dependent (106). Thus, a-factor deletion inhibits sexual development in a MAT1-2 strain, while α-factor deletion precludes sexual development in a MAT1-1 strain. In addition, the a-factor pheromone also plays an important role in female fertility in N. crassa (109), a role that is not restricted to either mating type. Similarly, the α-factor is important for male fertility (112) in C. parasitica, while the a-factor pheromone is essential for female fertility (120).
TABLE 2.
Comparison between the functions of proteins involved in the pheromone response pathway and MAPK cascades in heterothallic and homothallic species
| Protein |
Neurospora crassa (heterothallic) |
Sordaria macrospora (homothallic) |
||||
|---|---|---|---|---|---|---|
| Gene | Mutant phenotypesa | Gene | Mutant phenotypesb | |||
| α-Factor pheromone | ccg4 | Male sterility* in MAT1-1 partner (106) | ppg1 | No sexual defects (128) | Δppg1/Δppg2: reduced ascomatal production (128) | Δppg1/Δpre1: greatly reduced ascomatal production (128) |
| a-Factor pheromone | mfa-1 | Male sterility in MAT1-2 partner (106); aberrant female development (108) | ppg2 | No sexual defects (128) | Δppg2/Δpre2: greatly reduced ascomatal production (128) | |
| α-Factor receptor | pre2 | No protoascomatal maturation in MAT1-2 partner (24) | pre2 | No sexual defects (128) | Δpre1/Δpre2: no protoascomatal maturation (128) | Δppg2/Δpre2: greatly reduced ascomatal production (128) |
| a-Factor receptor | pre1 | No protoascomatal maturation in MAT1-1 partner (107) | pre1 | No sexual defects (128) | Δppg1/Δpre1: greatly reduced ascomatal production (128) | |
| Gα subunit | gna1 | Unusually shaped protoascomata that do not grow towards spermatia (150) | gsa1 | Delayed and reduced ascomatal production (152) | Δgsa1/Δgsa2 and Δgsa1/Δgsa3: greatly reduced protoascomatal production and no protoascomatal maturation (152) | |
| gna2 | No sexual defects (151) | gsa2 | No sexual defects (152) | |||
| gsa3 | Greatly reduced ascospore germination (152) | |||||
| Gβ subunit | gnb1 | No protoascomatal maturation (111, 153) | ||||
| Gγ subunit | gng1 | No protoascomatal maturation† (153) | ||||
| MAPKKK | nrc1 | No protoascomatal production‡ (212) | mik2 | No protoascomatal maturation (160) | ||
| MAPKK | mek2 | No protoascomatal maturation (160) | ||||
| MAPK | mak2 | No protoascomatal production (155) | mak2 | No protoascomatal maturation (160) | ||
| Transcriptional regulator | pp-1 | No protoascomatal production (155) | ste12 | Reduced asci production, fragile walls of asci and ascospores (193) | ||
| MAPKKK | mik1 | No protoascomatal production (110) | mik1 | No protoascomatal maturation (159) | ||
| MAPKK | mek1 | No protoascomatal production (110) | mek1 | No protoascomatal maturation (159) | ||
| MAPK | mak1 | No protoascomatal production (110) | mak1 | No protoascomatal maturation (159) | ||
| Rrg-1 | No protoascomatal production (156) | |||||
| MAPKKK | os-4 | No protoascomatal production (157) | ||||
| MAPKK | os-5 | No protoascomatal production (157) | ||||
| MAPK | os-2 | No protoascomatal production (156) | ||||
| Adenylyl cyclase | cr-1 | Delayed ascomatal development (213) | sac1 | Reduced ascomatal size and ascospore germination (152) | ||
*, Male sterility indicates that an isolate is unable to act as the male partners; †, no protoascomatal maturation indicates that protoascomata are produced but do not mature into ascomata; ‡, no protoascomatal production indicates that protoascomata are not formed at all.
Wild-type (proto)ascomatal production > reduced (proto)ascomatal production > greatly reduced (proto)ascomatal production.
The mating pheromones play an important role in the sexual development of the homothallic species, F. graminearum and S. macrospora (Table 2) (122, 128). While not essential for this pathway, gene deletion mutants in both species display decreased fertility, producing significantly fewer mature ascomata than wild-type isolates. This phenotype is observed in F. graminearum when the α-factor gene or both the α- and a-factor genes are deleted (122). In contrast, only the deletion of both pheromone genes induces this phenotype in S. macrospora (128). In both species, deletion of only the a-factor pheromone results in fully self-fertile isolates (122, 128), suggesting that this gene is not important in homothallic mating.
The strict association of the α-factor with the MAT1-1-1 protein and the a-factor with the MAT1-2-1 protein has some interesting implications and may play an important role in establishing the sexual strategies observed in various fungal species. This association is typically strongest in heterothallic species, where the pheromones are used to communicate the mating type of an individual to potential mating partners, thereby facilitating partner recognition (35, 91, 120, 121). This association is less stringent in homothallic species, with both the primary and secondary MAT proteins regulating pheromone expression. In S. macrospora, MAT1-1-1 positively regulates the expression of both pheromones, MAT1-1-2 negatively regulates a-factor expression while MAT1-2-1 positively regulates a-factor expression (52, 85). Similarly, in the homothallic F. graminearum, the MAT1-1-1, MAT1-1-2, MAT1-1-3, and MAT1-2-1 proteins all negatively regulate the expression of both pheromone factors to various degrees (96). The lack of a requirement to attract and recognize partners of opposite mating type has evidently changed the MAT-dependent regulation of the pheromones, even while these mating factors remain important for sexual development in these species.
Pheromone expression and regulation is particularly noteworthy in species with the more unique mating strategies. Both pheromones are expressed in the two unisexual species, H. moniliformis and N. africana, despite the absence of MAT1-1-associated genes in H. moniliformis and MAT1-2-associated genes in N. africana (91, 125). Similarly, species of the genus Colletotrichum are known to exhibit various mating strategies, including mating type switching (129) and unbalanced heterothallism, a type of self-sterility in which suitable mating partners have complementary mutations in genes involved in sexual reproduction (130). This is despite the fact that these species harbor only genes associated with the MAT1-2 idiomorph (80). These species have also been shown to have undergone concerted gene loss events associated with both pheromone factors, with some species harboring only one of the two pheromones and some species harboring neither (80). This suggests that the pheromones in these species may be regulated in a MAT-independent manner that may, at least partly, explain their divergence from the typical mating strategies.
Fungal mating pheromones are produced as larger preproteins that are subsequently processed into smaller, mature pheromone factors that are functionally active (Fig. 5) (7). Much of our understanding regarding the processing of mating pheromones in fungi arises from research conducted on S. cerevisiae (7). However, many of the processing proteins found in this model yeast have homologs in filamentous fungi and are thought to play similar roles in pheromone processing.
FIG 5.
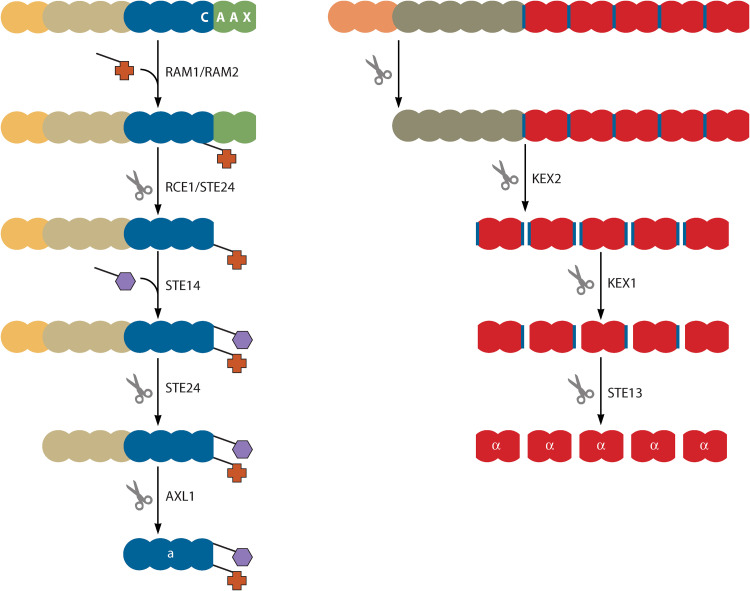
Structure and processing of a- and α-factor mating pheromones. Both fungal mating pheromones are initially expressed as large preproteins which are processed into small, mature pheromone factors. The first step in the processing of the a-factor includes the addition of a farnesyl (indicated by the “+” symbol) by RAM1/RAM2. This is followed by cleavage of the terminal AAX amino acids by RCE1/STE24 and the addition of a carboxymethyl group (indicated by a hexagon symbol) by STE14. Two final cleavage events, facilitated by STE24 and AXL1, release the mature pheromone. The first step in α-factor processing is the removal of the signal peptide (indicated in dark orange). KEX2-, KEX1-, and STE13-mediated cleavage releases numerous copies of the mature pheromone.
Processing of the a-factor.
The defining feature of the a-factor preprotein is a terminal CaaX domain, a region comprised of a cysteine residue followed by two aliphatic residues and an amino acid of any type (Fig. 5, left) (131). This highly conserved domain is posttranslationally modified by a variety of other proteins before the mature pheromone factor is secreted (reviewed in reference 7). The first posttranslational modification occurs when RAM1 and RAM2 proteins farnesylate the cysteine residue of the CaaX domain. The three C-terminal residues are then removed by RCE1 or STE24, where after STE14 adds a carboxymethyl group to the now-terminal C residue. The final step is the proteolysis of the N-terminal region by STE24 and AXL1. This produces a mature factor which is subsequently secreted by the STE6 transport protein.
The a-factor pheromone processing pathway has not been elucidated in any filamentous fungi, but many of these fungi contain genes encoding homologs of the S. cerevisiae processing proteins. Almost all of these genes have been identified in the genomes of species as diverse as A. fumigatus (132), A. cristatus (133), A. niger (134), M. oryzae (135), N. crassa (109), Penicillium marneffei (141), P. anserina (53), and Verticillium dahliae (136). An interesting exception is axl1, which has only been described in N. crassa (109). The fact that most of these processing proteins, as well as the a-factor pheromone, have all been maintained in the Pezizomycotina strongly implies that there may be some conservation in the entire processing pathway.
Unfortunately, very little research has been conducted on the function of the pheromone processing genes in the Pezizomycotina. Consequently, it remains unclear whether they play the same roles in pheromone processing as they do in S. cerevisiae. Experimental evidence does suggest that the two RAM proteins play a far more general role in the biology of these fungi than simply being involved in pheromone processing. The deletion of ram1 and ram2 is lethal in P. anserina (53) and M. oryzae (135), respectively, whereas ram1 disruption in M. oryzae affects processes as diverse as growth, stress resistance, and virulence (135). This is perhaps not surprising because the RAM proteins are posttranslational modifiers that add farnesyl groups to proteins in order to ensure their appropriate cellular localization (137). This form of modification affects a large number of proteins, particularly those involved in signal transduction, and consequently affects a variety of biological processes not limited to mating (135).
Manipulation of the A. cristatus MAT1-1-1 and MAT1-2-1 genes provides further experimental evidence to support the importance of the a-factor pheromone processing proteins in the sexual development of filamentous fungi. The rce1 and ste14 genes, both involved in some of the final processing steps, are significantly downregulated in MAT1-1-1 and MAT1-2-1 mutants, suggesting that these genes are (in)directly under the control of these two master regulators (133). Similarly, under conditions of high osmolarity, where sexual reproduction is precluded, rce1 is downregulated in A. cristatus (138). While such results do not confirm a function for these proteins in pheromone processing, they certainly imply a role for these genes during sexual development.
Processing of the α-factor.
The α-factor preprotein harbors a hydrophobic N-terminal secretion signal, a proregion and repeating units of the mature pheromone sequence (Fig. 5, right) (reviewed in reference 7). The first step in processing is the removal of the secretion signal by a signal peptidase in the endoplasmic reticulum. This is followed by proteolytic cleavage and removal of the proregion by the KEX2 protein. Once the N terminus has been removed, the preprotein is left containing repeats of a mature α-factor. Release of the mature pheromone peptides relies on the presence of two dipeptide motifs that flank these repeats. The first, either the dipeptide KR or RR, is a KEX2-like peptidase recognition site. KEX2 activity releases the numerous immature pheromone peptides, which are then subjected to terminal amino acid removal by KEX1. The final step in the processing of this pheromone is due to STE13-like aminopeptidase activity, which recognizes the second dipeptide motif, either XA or XP. Finally, STE13 activity releases the mature pheromone, which is subsequently packaged into vesicles and secreted via the classical secretion pathway.
Genes encoding the KEX1, KEX2, and STE13 processing proteins have been found in the genomes of various filamentous fungi, including S. macrospora (39, 139), various Trichoderma species (140), A. fumigatus (132), A. nidulans (32), N. crassa (35), P. marneffei (141), H. omanensis (91), and H. moniliformis (91). Similarly, the processing sites for these proteins are present in the α-factor pheromone preproteins of these species, suggesting conservation in the processing of these proteins across yeast and filamentous fungi. This has been experimentally proven in S. macrospora, where the α-factor preprotein in this species can be properly processed, secreted, and is functional in S. cerevisiae MATα cells (39).
Gene deletion and silencing studies in N. crassa and C. parasitica have further supported the role of these processing proteins in the sexual development of filamentous fungi. Fertility in N. crassa is influenced by KEX1, where deletion of the kex1 homolog, scp, results in greatly reduced male fertility (35, 142). Unsurprisingly, this effect is only apparent in the α-factor-expressing MAT1-1 cells, while MAT1-2 cells are unaffected by the gene’s deletion. Similarly, kex2-silencing in C. parasitica also results in reduced male fertility and an inability to produce ascospore-bearing ascomata (143). Lastly, a genome-wide gene expression study in N. crassa also showed that the expression patterns of ste13 closely mimic those of the α-factor pheromone, both increasing during ascomatal development (144). This hints at a role for this protein in the sexual cycle.
Cognate Receptors
Each of the mating pheromones is recognized by a specific cognate receptor. Both receptors are seven-transmembrane domain proteins, which are comprised of an extracellular recognition domain, seven membrane spanning domains and an intracellular domain that is linked to a G-protein (Fig. 4) (reviewed in references 145 and 146). The a-factor pheromone is recognized by the PRE1 protein, which has similarity to the STE3 protein from S. cerevisiae (107). Similarly, the PRE2 protein, which is similar to the S. cerevisiae STE2 protein, is the α-factor’s cognate receptor (38).
As with the pheromone factors themselves, the pheromone receptor genes have been identified and functionally characterized in many filamentous ascomycete fungi (24, 53, 107, 122, 128). While these genes are found in both heterothallic and homothallic fungi, their relative importance in sexual development differs from species to species (Table 2). In some species, both of the receptor genes are essential for sexual development, while in others these genes are dispensable, although their deletion may result in decreased fertility (53, 128, 147, 148).
An interesting characteristic of the two pheromone receptor proteins is that they often have mating type-dependent roles in heterothallic species. In these species, the PRE1 protein plays an important role in female fertility in MAT1-1 isolates, while female fertility in the MAT1-2 isolates depends on the PRE2 protein. This is understandable given that MAT1-1 isolates express the α-factor and need to recognize the a-factor via the expression of pre1, while the converse is true of the MAT1-2 isolates. This mating type-dependent effect has been observed in P. anserina (53), N. crassa (24, 107), and T. reesei (147), where Δpre1 MAT1-1 and Δpre2 MAT1-2 isolates are female sterile-producing protoascomata that are incapable of being fertilized and thus do not mature into ascospore-bearing ascomata. Importantly, Δpre1 MAT1-2 and Δpre2 MAT1-1 isolates interact normally with wild-type partners and produce fully mature ascomata and viable ascospores.
The pheromone receptors are important, but typically not essential, for the sexual cycle in a number of homothallic species (Table 2). In both S. macrospora (128) and A. nidulans (148), the deletion of both pre1 and pre2 results in the complete loss of the sexual cycle by precluding the production of ascomata. However, deletion of a single receptor produces no detectable effect in S. macrospora (128) but does result in reduced fertility in A. nidulans, with mutants producing smaller ascomata and fewer ascospores (148). In F. graminearum, pre1 is dispensable for sexual development, with Δpre1 isolates sporulating normally (122). In contrast, single Δpre2 mutants and double Δpre1/Δpre2 mutants exhibit subnormal fertility levels (122).
Signal Transduction Pathways
As G-protein coupled receptors (GPCRs), the pheromone receptors are linked to a G-protein via their terminal intracellular domains (Fig. 4) (149). Prior to pheromone recognition, the G-protein exists as a trimer of three protein subunits: Gα, Gβ, and Gγ. Once the pheromone has physically bound to the receptor, the G-protein is activated, resulting in dissociation of the subunits into a Gα monomer and a Gβ-Gγ dimer, each of which is capable of initiating a downstream signal transduction cascade (149).
Given their close association with the pheromone receptors, it is not surprising that all three subunits play an important role in female fertility (Table 2). Two genes, gna1 (150) and gna2 (151), encode Gα-subunits that have been functionally characterized with respect to their roles in mating in the heterothallic N. crassa. Deletion of gna1 results in mutant isolates that produce unusually shaped protoascomata (150) and are incapable of growth toward the spermatia (84). There is consequently little or no maturation of these structures into mature ascomata. Interestingly, gna2 deletion mutants do not produce particularly notable sexual defects, but Δgna1/Δgna2 mutants are more severely affected than single gna1 deletion mutants (151). This fact illustrates that while the two α-subunits may have overlapping functions, the GNA1 protein can compensate for GNA2 but not vice versa. Deletion of the Gα-subunit gene in S. macrospora, results in a similar phenotype; though, while protoascomatal production is delayed and reduced in Δgsa1 mutants, mature ascomata are produced after sufficient incubation (152). This could reflect a change in the importance of the pheromone response pathway in the homothallic S. macrospora compared to the heterothallic N. crassa (Table 2).
The N. crassa β- and γ-subunits, encoded by gnb1 and gng1, respectively, are essential for the maturation of normal ascomata. Single and double mutants of the gnb1 and gng1 genes are all female sterile, producing immature protoascomata that do not develop further, even when adequately fertilized (111, 153). On the rare occasion where mature ascomata are produced, they remain small and are unable to eject the ascospores (111). It is perhaps not surprising that deletion of the Gα-, Gβ-, and Gγ-subunits produce similar phenotypes and, in fact, the expressions of the three subunit genes are controlled by one another. For example, gnb1 deletion results in the complete downregulation of gna1 and gng1 expression (111, 153), whereas gng1 deletion results in significant downregulation of gnb1 (153).
After dissociation, the Gα and Gβ-Gγ subunits often act by activating mitogen-activated protein (MAP) kinase signal transduction pathways that are capable of eliciting a variety of effects, including the initiation of sexual reproduction via the activation or repression of relevant transcription factors (Fig. 4) (reviewed in reference 154). In general, the first component of this kind of transduction pathway is a MAP kinase kinase kinase (MAPKKK), followed by a MAPKK, a terminal MAPK and, finally, the targeting protein, which is often a transcription factor. In N. crassa, three of these pathways are involved in sexual development and are named after their terminal kinases, i.e., MAK1 (110), MAK2 (155), and OS2 (156, 157) (Fig. 4). Intriguingly, while all three pathways have been implicated in protoascomatal development, only the MAK2 pathway is directly linked to the mating pheromone receptors (Fig. 4) (158). This suggests that the cross talk between these three pathways is essential for sexual development in N. crassa. Thus, the deletion of even a single gene in one of these pathways results in female sterility (Table 2) (110). The MAK1 (159) and MAK2 (160) pathways are also important for sexual reproduction in S. macrospora, where single gene deletions also preclude sexual development (Table 2).
There are some important differences between heterothallic and homothallic fungi relating to the pheromone response pathway and various MAPK cascades. To illustrate these differences, the mutant phenotypes of genes involved in the heterothallic N. crassa and homothallic S. macrospora are shown in Table 2. What is evident is that the proteins that play a role during the initiation of the pheromone response pathway, i.e., the pheromone factors, their receptors, and the coupled G-proteins, are less important for sexual development in S. macrospora than they are in N. crassa. In fact, single gene deletions in S. macrospora either result in no sexual defects or simply delay sexual development. In contrast, similar deletions in N. crassa drastically affect male and/or female fertility. When the MAPK cascades are considered, however, the effects of single gene deletions in the heterothallic and homothallic species are comparable. This suggests that while the process of initiating sexual recognition is different in heterothallic and homothallic fungi, the downstream processes are the same.
PROTOASCOMATAL DEVELOPMENT AND ASCOMATAL MATURATION
After partner recognition occurs, male cells are capable of fertilizing the female ascogonia, which will begin to develop into a protoascoma [reviewed in reference (41)]. It is within this immature sexual structure that the various stages of nucleus migration, karyogamy, mitosis and meiosis will commence. During this time, the protoascoma develops into a mature ascoma containing asci and the sexual ascospores. Three protein complexes and/or pathways (the STRIPAK complex, the PACC pathway and the COP9 signalosome) have been specifically linked to ascomatal development in the filamentous fungi (Fig. 6).
FIG 6.
Important protein complexes and pathways involved in protoascomatal maturation and ascomatal development in various filamentous fungi. (A) The STRIPAK is important for sexual development in the homothallic Sordaria macrospora. (Based on data from reference 161.) (B) The PACC pathway plays an essential role in protoascomatal development in the heterothallic Neurospora crassa. (Adapted from reference 175.) (C) The COP9 signalosome is important for sexual development and other developmental processes in the homothallic Aspergillus nidulans. (Based on data from references 166 and 186.) Protein names and/or abbreviations: GPI1 (glycosylphosphatidylinositol-anchored protein); MOB3 (monopolar spindle one-binder protein); PP2Ac and PP2Aa (protein phosphatase subunits); KIN3 and KIN24 (kinases 3 and 24); CLA4 (from the p21-activated kinase family); PALA, PALB, PALC, PALF, and PALH (various proteins from the PAL/RIM pathway); PACC (terminal transcription factor of the PACC pathway); and CSN1-8 (COP9 signalosome proteins 1 to 8).
STRIPAK Complex
One of the most important protein complexes that controls the development of the ascoma is the striatin-interacting phosphatases and kinases (STRIPAK) complex (Fig. 6) (161, 162). In fungi, this complex is made up of a protein phosphatase (PP2AA), a striatin (PRO11), a striatin-interacting protein (PRO22), a phocein (MOB3), kinases (KIN3, KIN24), a membrane-anchoring protein (PRO45), and many other proteins (162). This complex is highly conserved across the eukaryotes and is thought to be involved in various signal transduction pathways important for the differentiation of multicellular structures (161).
The first fungal STRIPAK subunits were identified in N. crassa (163), and some research has been conducted to elucidate the roles that these proteins play in this species (163–165). However, this complex, its functions, and the protein-protein interactions are much better understood in S. macrospora (reviewed in reference 162). The S. macrospora STRIPAK complex appears to be involved in diverse biological processes due to its primary function being the (de)phosphorylation of a variety of target proteins (162). Many of these targets are known to play important roles in sexual development (166). For example, the α-factor pheromone processing proteins KEX1 and KEX2 are both (de)phosphorylation targets of the STRIPAK complex in S. macrospora.
The importance of the STRIPAK complex and its subunits in sexual development has been experimentally shown in S. macrospora. Deletion of the genes encoding STRIPAK proteins commonly results in similar phenotypes, particularly during the development of the immature ascomata. In Δpro11 (167), Δpro22 (161), ΔPP2Ac1 (168), Δkin24 (169), and Δpro45 (170) mutants, protoascomata are produced normally but do not become pigmented over time and do not develop into mature sexual structures. In strains where mob3 (171) or pro11 (161) are deleted, an even more extreme phenotype is apparent, with the complete absence of any sexual tissues. Furthermore, the deletion of PP2Ac2, the catalytic subunit of the phosphatase, is lethal (168).
The precise manner in which the STRIPAK complex regulates ascomatal development is not fully understood. However, CLA4, a STRIPAK target might provide some insight into this pathway (166). This protein is a member of the p21-activated kinase family and is important for sexual reproduction in F. graminearum, N. crassa, and M. grisea (172–174). In S. macrospora specifically, Δcla4 strains produce deformed ascomata and significantly fewer asci and ascospores than wild-type strains (166). The role that CLA4 plays in sexual development in S. macrospora is heavily dependent on a phosphorylation site within its catalytic subunit, a site that is directly regulated by STRIPAK. In fact, when this site was mutated so that it could not be dephosphorylated by STRIPAK, the production of asci and ascospores was greatly reduced, producing a phenotype comparable to the Δcla4 mutant (166). Further investigation into the other STRIPAK target proteins is likely to identify additional developmental pathways regulated by this complex.
PACC Pathway
A second pathway that appears to be important in the development of the protoascomata in filamentous fungi, particularly in N. crassa, is the PACC pathway. This name reflects the final step of the pathway that involves the activation of the PACC transcription factor (175). This pathway has been implicated in pH response, the regulation of xylanase, cellulolytic and endoglucanase activities, and metabolism of glycogen and trehalose in N. crassa (176).
The PACC pathway has been particularly well-characterized in S. cerevisiae and A. nidulans, where it is activated when there are significant pH changes in the environment (Fig. 5) (177–179). The PALH protein acts as the pH sensor and is physically bound to the PALF protein, which is phosphorylated upon sensor stimulation. This results in PALH being released and joining a complex of proteins, including PALA, PALB, PALC, and PACC. This in turn stimulates the activation of PACC via PALB-mediated cleavage. The activated PACC transcription factor is then translocated into the nucleus, where it brings about transcriptional change (175).
The genes that encode the PACC pathway proteins in A. nidulans do not appear to play an important role in sexual development. In contrast, genes encoding the six proteins of this pathway have been identified in the N. crassa genome, and all are essential for the production of protoascomata (175). Deletion of any of the six genes that encode PACC pathway proteins result in mutants which, when grown on synthetic nitrogen-limited, crossing media, are unable to produce ascogonia or protoascomata. This phenotype was rescued when mutants were grown on medium containing the carbohydrates typically required for sexual development in N. crassa (175). Given that the pH-dependent PACC pathway is known to regulate the accumulation and metabolism of glycogen and trehalose in N. crassa (180), it is possible that the production of the highly complex protoascomata, which is an energetically intensive process, relies on the metabolism of these energy-rich carbohydrates, hence the requirement of a functional PACC pathway.
The PACC pathway is also essential in N. crassa for steps downstream of protoascomatal production (175). In single gene mutants where protoascomatal formation had taken place, fertilization resulted in melanization and growth of the protoascomata into mature ascomata. However, the maturing ascomata did not reach full size and did not harbor mature asci, and no ascospores were produced, a phenotype that could not be rescued under different environmental conditions. This might be explained by the fact that the S. cerevisiae Rim1p, a pacC homolog, is important for meiosis (181, 182). Given that most of this pathway is conserved in N. crassa and S. cerevisiae (175), it is likely that the PACC pathway is also important for meiosis in N. crassa. Overall, this indicates that the PACC pathway is important in both the initial and the later stages of sexual development.
COP9 Signalosome
The COP9 signalosome (CSN) is another evolutionarily conserved complex that regulates sexual development in fungi. The complex regulates this pathway via its roles in response to oxidative stress, the regulation of redox status and cell wall rearrangement (183). The complex is made up of a variety of proteins, such as those involved in CSN assembly (e.g., PCI-domain proteins) (184, 185) and those with enzymatic activities (e.g., metalloproteases) (184, 186). Together, these proteins act in posttranslational processes, such as protein ubiquitylation and subsequent degradation (186). Via these important enzymatic activities, it is thought that the CSN complex plays an important role as a regulator of multicellularity (184, 187). While the specific manner in which this complex regulates cellular differentiation remains to be understood, it appears that ubiquitin-dependent protein degradation is important for the development of complex, multicellular structures with specific spatial requirements (184).
While the CSN can be made up of as few as six or seven proteins, as is the case in Schizosaccharomyces pombe and N. crassa (186), the most common conformation of this eukaryotic complex is exemplified by A. nidulans (Fig. 6) (184). The A. nidulans CSN is comprised of eight proteins, which are encoded by genes cnsA to csnH. Four of the eight genes have been functionally characterized with respect to their role in sexual development, including csnA (184), csnB (184), csnD (185), and csnE (185). Given the importance of this complex in the development of multicellular structures, it is perhaps not surprising that all four of these genes are important for the production of protoascomata. Furthermore, their deletion results in mutant isolates capable of forming only the initial structures (primordia) associated with sexual development, but these initials do not mature into protoascomata or any other mature sexual structures. Remarkably, even a single gene deletion results in this extreme phenotype, strongly suggesting that the functioning of this complex relies on all of the eight proteins (184, 185).
ASCOSPORE PRODUCTION AND GERMINATION
The final stages of sexual reproduction include the production of the ascospores, their release into the environment, and subsequent germination.
Ascosporegenesis
The production of ascospores relies on multiple rounds of cell division. Meiotic divisions within the ascus result in four haploid nuclei that are typically arranged in a linear conformation, each of which undergo a round of mitosis to produce the final eight ascospores (115). Further ascospore development takes place to ensure the eight spores are arranged, partitioned, and delineated correctly in the ascus.
There are various genes in A. nidulans and N. crassa that, when deleted, produce empty asci. This phenotype indicates that the appropriate sexual tissues are present but that meiosis cannot be initiated within the mature asci. Four of these genes are tubB (188) and grrA (189) in A. nidulans and sad1 (190) and sad2 (191) in N. crassa. The two A. nidulans genes have very different functions; the tubB gene is involved in microtubule assembly (188), while grrA plays an important role in protein degradation (189). In contrast, both of the N. crassa genes are important for meiotic silencing (190, 191) and, importantly, SAD2 ensures the proper localization and thus functioning of SAD1 (191). Despite difference in their precise functions, each of these genes enables the commencement of the meiotic cycle within the sexual structures.
The ski8 gene in S. macrospora is essential for many different stages of ascosporegenesis, with functions as early as ascomatal production and ascus emergence and as late as meiosis (192). Importantly, it is thought that the SKI8 protein is particularly important for the production of double stand breaks in the DNA during meiosis, since Δski8 mutants do not produce chiasmata, and they undergo meiosis aberrantly with delayed karyogamy. These mutants typically contain many asci in which unfused nuclei are common and mature ascospores have not been produced (192).
The S. macrospora STE12, a transcription factor that interacts directly with the MAT1-1-1 protein, is important for ascus and ascospore formation (193). When the ste12 gene is deleted, mutants produce ascomata that appear normal but with very few asci, and those that are produced have very thin and fragile cell walls that burst easily. Similarly, the number of reproductive structures, including hooked cells and meiosis-specific cell stages, are significantly reduced in mutants compared to wild-type isolates (193).
Several genes that encode proteins involved in autophagy in S. macrospora are important for production of asci and ascospores (194–196). Two such examples are nbr1 and lon2, which encode an autophagy cargo receptor and a Lon protease, respectively (194, 195). Given that autophagy results in the reconstitution of cellular components and may thereby act as a mechanism of nutrient provision, particularly during periods of cellular development, it is not surprising that this pathway directly affects the production of highly specialized sexual tissues. The deletion of nbr1 resulted in the production of immature, malformed asci which have significantly fewer ascospores than wild-type asci (195). Similarly, Δlon2 mutants produced fewer ascomata, most of which contained asci housing only immature ascospores (194). Both mutants were more sensitive to stressors as well, with the Δnbr1 mutants not producing fruiting bodies under temperature and nutritional stress and Δlon2 mutants not producing fruiting bodies during starvation (194, 195). These results strongly suggest that the autophagous pathway is important in the environmental stress response, while maintaining the nutritional status necessary for sexual development.
In N. crassa, the asd1 gene partially regulates ascosporegenesis once meiosis and mitosis have already taken place (197). This gene encodes a rhamnogalacturonase, which is thought to play a role in the regulation of sexual development, via the production of various polysaccharides. In Δasd1 mutants, the asci form normally but despite going through meiosis as usual, the spores do not delineate, and the haploid nuclei remain unpackaged (197). The sec22 gene plays a similar role in S. macrospora where it is involved in ascospore maturation (198). In wild type isolates, the eight spores present in the asci develop at a uniform rate before being ejected together. In Δsec22 mutants, however, these spores mature at different rates and some are ejected well before maturation (198).
Germination
Ascospore germination has been studied in detail in S. macrospora, and a number of genes that directly influence germination rate have been described. In addition to the role played by the autophagous pathway in the production of asci as described above, autophagy is also important for ascospore germination in S. macrospora (196). Two genes, atg4 and atg8, encode autophagy-related proteins, and both directly affect the germination rate of ascospores in this species (196). Deletion of either atg4 or atg8 results in a germination rate reduction of about 50%. Since germination involves the growth of a mycelium from a spore, it is reasonable to speculate that autophagy may play an important role in providing nutrients to the germ tube. Given this role, it is not surprising that these two genes are also involved in the development of the maturing ascomata as well (196).
A third gene involved in ascospore germination in S. macrospora is sec22 (198). Interestingly, this gene, which encodes a membrane-associated protein involved in membrane fusion, may also be involved in autophagy (199), further supporting the role of this process in germination. In Δsec22 mutants, a mixture of immature and mature spores is released from the asci (198). The immature spores never germinate, and the mature spores germinate at a lower rate than the wild type (198).
As with atg4 and atg8, sec22 appears to have multiple roles in sexual development, ranging from spore production, pigmentation, and maturation to ascospore germination postrelease (196, 198). Interestingly, deletion of sec22 resulted in the deregulation of genes involved in numerous developmental pathways, including those putatively involved in vesicle transport (198). Given the large number of diverse molecular processes that rely on this cellular transport pathway, it is not surprising that the sec22 gene is involved at numerous stages in the sexual cycle.
Another two genes in S. macrospora that influence germination are important for G-protein-coupled signal transduction (152). These genes, gsa3 and sac1, encode the Gα-subunit and the adenylyl cyclase protein, respectively. Upon activation of the GPCR, the Gα-subunit activates the adenylyl cyclase, thereby initiating an appropriate transduction cascade, which, in this case, would result in the formation of the germ tube. Deletion of the gsa3 gene results in normal ascomata with ascospore-bearing asci, but only approximately 5% of these ascospores germinate (152). Similarly, Δsac1 mutants also exhibit a significant decrease in germination but also produce much smaller ascomata during sexual development (152).
LOOKING TO THE FUTURE
Sexual reproduction in fungi has been studied for decades and the progress that has been made is nothing short of remarkable. From the original descriptions of different fungal fruiting bodies and mating behaviors to the identification of the diffusible mating pheromones and the elucidation of the MAT genes, our understanding of this pathway has grown considerably, stretching from physiological observations to the characterization of the underlying genetic mechanisms that govern the process.
Studies on the pathways that govern sexual reproduction relied heavily on the functional characterization of single genes and their direct effect on the sexual capabilities of model fungi. This approach has proven very useful to shape our understanding of this process but is plagued by some significant drawbacks. The first of these is that such a complex pathway is necessarily governed by extensive, multifaceted gene and protein networks that need to be characterized with respect to, and within the context of, one another. In addition, a significant level of redundancy often exists when it comes to the function of these genes, with two or three proteins having overlapping functions and thus compensating for one another. This essentially masks the effects and importance of certain genes when they are knocked out. Lastly, it is often not possible to fully characterize the function of a gene if it is essential for numerous steps in a particular pathway. For example, if a gene is essential for both fertilization and the production of asci, its role in ascus production will be masked because the block in fertilization will preclude any further sexual development. Thus, single gene knockouts do not always accurately allow for the characterization of the functions of these genes and cannot be used to describe all forms of interconnected complexity. The advent of more powerful tools such as CRISPR-Cas9 and NGS promises to change this. Apart from the ability to produce single gene knockouts, CRISPR can efficiently produce multigene knockouts in a single organism, allowing complex pathways to be better dissected. With transformation protocols becoming more prevalent across the Pezizomycotina, functional studies in underrepresented taxa will become an exciting source of new information in future.
Currently, extensive data are available linking the functions of individual genes and genetic pathways to various steps of the sexual cycle in model species such as N. crassa, A. nidulans, S. macrospora, and the other species discussed above (Fig. 3). However, a significant step forward in our understanding of sexual development in the Pezizomycotina has been the broadening of the field to include non-model species. Over the past few years, the sexual cycles of non-model species like V. virens (60, 65) and H. omanensis (64) have also been subjected to genetic inquiry. This has enabled the comparison of gene function of homologs from different species, which in turn enables further dissection of the evolutionary conservation of these processes. This is particularly important if we seek to continue using fungi as multicellular representatives of the Eukarya and extrapolating the conclusions drawn from fungi to other eukaryotes.
A number of gaps exist in the literature (Fig. 3), paving the way for future investigations and a better understanding of sexual development in the Pezizomycotina. Some key genes and pathways remain to be elucidated, including the identification of the a-factor-like pheromone in the Eurotiomycetes and related fungi, as well as the targets of the evolutionarily conserved protein complexes, STRIPAK and CSN. In addition, much work remains to done on how various environmental cues regulate sexual development. For example, the effect of light on the sexual cycles of N. crassa and A. nidulans is very well understood both in terms of the environmental stimuli and the genetic and physiological responses. However, the importance of other environmental factors such as pH, temperature, and nutrient availability are less well understood in filamentous species, particularly with regard to the genes and proteins involved in regulating these pathways.
The role of posttranscriptional and posttranslational modification clearly requires further investigation. It is known, for example, that RNA editing is important for ascosporegenesis and ascus maturation in both F. graminearum (200) and N. crassa (201). Similarly, posttranslational modifications are known to be essential for the functioning of both pheromone factors. However, the extent to which these types of modifications are necessary and essential for the sexual cycle is not yet known. Whether such of modifications occur in all of the Pezizomycotina also remains a topic open for debate.
CONCLUDING REMARKS
Fungi provide fascinating and effective models for studies focused on elucidating a process as complicated as sexual reproduction. Fungi exhibit numerous distinct and, in some cases, unusual sexual strategies and there are a growing number of model species available for study and comparison. The fungi also include an unusually wide diversity of morphologies, sexual structures, and preferred environments, providing a wide array of opportunities to study sexual development in depth.
New and emerging technologies are driving opportunities to better understand sexual reproduction in the fungi. For example, genome editing, whole-genome sequencing, and comparative transcriptomics are becoming increasingly accessible and available for such investigations. These are likely to reveal a substantially improved understanding of sexual development in a diverse array of model and non-model fungal species. The results will make it possible to extrapolate emerging knowledge beyond the fungal kingdom to the other eukaryotes.
ACKNOWLEDGMENTS
This project was supported by the University of Pretoria, the Department of Science and Innovation (DSI)/National Research Foundation (NRF) Centre of Excellence in Plant Health Biotechnology (CPHB). The project was further supported by the DSI/NRF SARChI chair in Fungal Genomics (grant 98353) held by B.D.W. The grant holders acknowledge that the opinions, findings and conclusions, or recommendations expressed are theirs and that the funding bodies accept no liability in this regard.
We declare there are no conflicts of interest.
We thank Glenda Brits of the Department of Education Innovation at the University of Pretoria and Patrick Lane of ScEYEnce Studios for producing the figures used in this review.
Biographies

Andi M. Wilson is a postdoctoral researcher at the Forestry and Agricultural Biotechnology Institute at the University of Pretoria, South Africa. She also completed her Ph.D. at this institute under the primary supervision of Prof. B. D. Wingfield. Andi’s primary research focus is on elucidating the underlying genetic mechanisms that control sexual development and mating in ascomycete fungi. Using a combination of comparative genomics and transcriptomics as well as classical genetics and CRISPR-Cas9 gene editing, she has unraveled some of the pathways that allow for unisexual development in filamentous ascomycete fungi.

P. Markus Wilken completed his Ph.D. (Genetics) at the University of Pretoria. He studied the unidirectional mating type switching system as the reproductive mode for many Ceratocystidaceae species. Thereafter, he completed two postdoctoral fellowships under the SARChI Chair in Fungal Genomics run by Prof. B. D. Wingfield, where he used genomics to characterize the mating system of other Ceratocystidaceae species. Currently, Markus is a lecturer in the Department of Biochemistry, Genetics, and Microbiology at the University of Pretoria, where he runs a research program linked to studying sexual reproduction in plant-pathogenic fungi.

Michael J. Wingfield, the founding director of the Forestry and Agricultural Biotechnology Institute, serves as an advisor to the University of Pretoria Executive. His research focuses on the health of forest trees, having published widely on this topic together with more than a hundred past Ph.D. students. He served a five-year term as President of the International Union of Forestry Research Organizations (IUFRO) and has been widely recognized for his research, including receiving the Kwame Nkrumah Science Award from the African Union and being awarded honorary doctorates from the University of British Columbia (Canada) and North Carolina State University (USA).

Brenda D. Wingfield has made the study of the global movement and evolution of fungal pathogens, particularly those on trees, her research focus for the past 30 years. Professor Wingfield holds the DSI-NRF SARChI chair in Fungal Genomics. She pioneered fungal genomics at the University of Pretoria and was responsible for the first fungal genome to be sequenced in Africa. Her success is reflected in the internationally recognized work of many of her past Ph.D. students. She is currently the Secretary General of the International Society of Plant Pathology and was the vice president of the Academy of Science of South Africa.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Speijer D, Lukeš J, Eliáš M. 2015. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc Natl Acad Sci USA 112:8827–8834. 10.1073/pnas.1501725112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nieuwenhuis BPS, James TY. 2016. The frequency of sex in fungi. Philos Trans R Soc Lond B Biol Sci 371:20150540. 10.1098/rstb.2015.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieuwenhuis BPS, Aanen DK. 2012. Sexual selection in fungi. J Evol Biol 25:2397–2411. 10.1111/jeb.12017. [DOI] [PubMed] [Google Scholar]
- 4.Bennett RJ, Turgeon BG. 2016. Fungal sex: the Ascomycota. Microbiol Spectr 4:1–28. 10.1128/microbiolspec.FUNK-0005-2016. [DOI] [PubMed] [Google Scholar]
- 5.Wallen RM, Perlin MH. 2018. An overview of the function and maintenance of sexual reproduction in dikaryotic fungi. Front Microbiol 9:503. 10.3389/fmicb.2018.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler G. 2007. The evolution of MAT: the Ascomycetes, p 3–18. In Heitman J, Kronstad JW, Taylor J, Casselton LA (ed), Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC. [Google Scholar]
- 7.Jones SK, Bennett RJ. 2011. Fungal mating pheromones: choreographing the dating game. Fungal Genet Biol 48:668–676. 10.1016/j.fgb.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lumbsch HT, Huhndorf SM. 2010. Myconet, volume 14. Part One. Outline of Ascomycota—2009. Part Two. Notes on Ascomycete systematics. Nos. 4751–5113. Myconet Fieldiana Life Earth Sci 1:1–64. 10.3158/1557.1. [DOI] [Google Scholar]
- 9.Usher J. 2019. The mechanisms of mating in pathogenic fungi: a plastic trait. Genes 10:831–818. 10.3390/genes10100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun S, Lin X, Coelho MA, Heitman J. 2020. Evolution of yeast mating systems: all roads lead to selfing. Curr Biol 29:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latgé JP, Chamilos G. 2019. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev 33:75. 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nenoff P, Krüger C, Ginter-Hanselmayer G, Tietz HJ. 2014. Mycology: an update. Part 1. Dermatomycoses: causative agents, epidemiology, and pathogenesis. JDDG 12:188–210. [DOI] [PubMed] [Google Scholar]
- 13.Rigling D, Prospero S. 2018. Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol Plant Pathol 19:7–20. 10.1111/mpp.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wingfield MJ, Hammerbacher A, Ganley RJ, Steenkamp ET, Gordon TR, Wingfield BD, Coutinho TA. 2008. Pitch canker caused by Fusarium circinatum: a growing threat to pine plantations and forests worldwide. Austral Plant Pathol 37:319–334. 10.1071/AP08036. [DOI] [Google Scholar]
- 15.Gueidan C, Hill D, Miadlikowska J, Lutzoni FL. 2015. Pezizomycotina: Lecanoromycetes, p 89–120. In McLaughlin D, Spatafora J (ed), Systematics and evolution, 2nd ed. Springer, Berlin, Germany. [Google Scholar]
- 16.Spatafora JW, Sung G-H, Johnson D, Hesse C, O’Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J, Reeb V, Gueidan C, Fraker E, Lumbsch T, Lücking R, Schmitt I, Hosaka K, Aptroot A, Roux C, Miller AN, Geiser DM, Hafellner J, Hestmark G, Arnold AE, Büdel B, Rauhut A, Hewitt D, Untereiner WA, Cole MS, Scheidegger C, Schultz M, Sipman H, Schoch CL. 2006. A five-gene phylogeny of Pezizomycotina. Mycologia 98:1018–1028. 10.1080/15572536.2006.11832630. [DOI] [PubMed] [Google Scholar]
- 17.Roche CM, Loros JJ, McCluskey K, Glass N. 2014. Neurospora crassa: looking back and looking forward at a model microbe. Am J Bot 101:2022–2035. 10.3732/ajb.1400377. [DOI] [PubMed] [Google Scholar]
- 18.Bayram Ö, Braus GH, Fischer R, Rodriguez-Romero J. 2010. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet Biol 47:900–908. 10.1016/j.fgb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Bayram Ö, Braus GH. 2012. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36:1–24. 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 20.Bayram ÖS, Dettmann A, Karahoda B, Moloney NM, Ormsby T, McGowan J, Cea-Sánchez S, Miralles-Durán A, Brancini GTP, Luque EM, Fitzpatrick DA, Cánovas D, Corrochano LM, Doyle S, Selker EU, Seiler S, Bayram Ö. 2019. Control of development, secondary metabolism and light-dependent carotenoid biosynthesis by the velvet complex of Neurospora crassa. Genetics 212:691–710. 10.1534/genetics.119.302277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel PK, Free SJ. 2019. The genetics and biochemistry of cell wall structure and synthesis in Neurospora crassa, a model filamentous fungus. Front Microbiol 10:2294. 10.3389/fmicb.2019.02294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkhayyat F, Chang Kim S, Yu JH. 2015. Genetic control of asexual development in Aspergillus fumigatus. Adv Appl Microbiol 90:93–107. 10.1016/bs.aambs.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Ruger-Herreros C, Corrochano LM. 2020. Conidiation in Neurospora crassa: vegetative reproduction by a model fungus. Int Microbiol 23:97–105. 10.1007/s10123-019-00085-1. [DOI] [PubMed] [Google Scholar]
- 24.Kim H, Wright SJ, Park G, Ouyang S, Krystofova S, Borkovich KA. 2012. Roles for receptors, pheromones, G-proteins, and mating-type genes during sexual reproduction in Neurospora crassa. Genetics 190:1389–1404. 10.1534/genetics.111.136358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira AVB, An Z, Metzenberg RL, Glass NL. 1998. Characterization of matA-2, matA-3, and ΔmatA mating-type mutants of Neurospora crassa. Genetics 148:1069–1079. 10.1093/genetics/148.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dyer PS, O’Gorman CM. 2012. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev 36:165–192. 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- 27.Lim JY, Kang EH, Park YH, Kook JH, Park HM. 2020. Survival factor SvfA plays multiple roles in differentiation and is essential for completion of sexual development in Aspergillus nidulans. Sci Rep 10:5586. 10.1038/s41598-020-62455-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debuchy R, Berteaux-Lecellier V, Silar P. 2010. Mating systems and sexual morphogenesis in Ascomycetes, p 501–535. In Borkovich KA, Ebbole DJ (ed), Cellular and molecular biology of filamentous fungi. ASM Press, Washington, DC. [Google Scholar]
- 29.Astell CR, Ahlstrom-Jonasson L, Smith M, Tatchell K, Nasmyth KA, Hall BD. 1981. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell 27:15–23. 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
- 30.Brake A, Brenner C, Najarian R, Laybourn P, Merryweather J. 1985. Structure of genes encoding precursors of the yeast peptide mating pheromone a-factor, p 103–108. In Gething M (ed), Protein transport and secretion. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 31.Kurjan J, Herskowitz I. 1982. Structure of a yeast pheromone gene (MFα): a putative α-factor precursor contains four tandem copies of mature α-factor. Cell 30:933–943. 10.1016/0092-8674(82)90298-7. [DOI] [PubMed] [Google Scholar]
- 32.Galagan JE, Calvo SE, Cuomo C, Ma L-J, Wortman JR, Batzoglou S, Lee S-I, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 33.Glass NL, Vollmer SJ, Staben C, Grotelueschen J, Metzenberg RL, Yanofsky C. 1988. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science 241:570–573. 10.1126/science.2840740. [DOI] [PubMed] [Google Scholar]
- 34.Debuchy R, Coppin E. 1992. The mating types of Podospora anserina: functional analysis and sequence of the fertilization domains. Mol Gen Genet 233:113–121. 10.1007/BF00587568. [DOI] [PubMed] [Google Scholar]
- 35.Bobrowicz P, Pawlak R, Correa A, Bell-Pedersen D, Ebbole DJ. 2002. The Neurospora crassa pheromone precursor genes are regulated by the mating-type locus and the circadian clock. Mol Microbiol 45:795–804. 10.1046/j.1365-2958.2002.03052.x. [DOI] [PubMed] [Google Scholar]
- 36.Dyer PS, Paoletti M, Archer DB. 2003. Genomics reveals sexual secrets of Aspergillus. Microbiology (Reading) 149:2301–2303. 10.1099/mic.0.C0119-0. [DOI] [PubMed] [Google Scholar]
- 37.Coppin E, de Renty C, Debuchy R. 2005. The function of the coding sequences for the putative pheromone precursors in Podospora anserina is restricted to fertilization. Eukaryot Cell 4:407–420. 10.1128/EC.4.2.407-420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Böhm J, Hoff B, O’Gorman CM, Wolfers S, Klix V, Binger D, Zadra I, Kürnsteiner H, Pöggeler S, Dyer PS, Kück U. 2013. Sexual reproduction and mating type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci USA 110:1476–1481. 10.1073/pnas.1217943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayrhofer S, Pöggeler S. 2005. Functional characterization of an α-factor-like Sordaria macrospora peptide pheromone and analysis of its interaction with its cognate receptor in Saccharomyces cerevisiae. Eukaryot Cell 4:661–672. 10.1128/EC.4.4.661-672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haber JE. 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191:33–64. 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pöggeler S, Nowrousian M, Kück U. 2018. Fruiting-body development in Ascomycetes, p 1–56. In Anke T, Schüffler A (ed), Physiology and genetics, 2nd ed. Springer International Publishing, Basel, Switzerland. [Google Scholar]
- 42.Coppin E, Debuchy R, Arnaise S, Picard M. 1997. Mating types and sexual development in filamentous Ascomycetes. Microbiol Mol Biol Rev 61:411–428. 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnaise S, Zickler D, Le Bilcot S, Poisier C, Debuchy R. 2001. Mutations in mating-type genes of the heterothallic fungus Podospora anserina lead to self-fertility. Genetics 159:545–556. 10.1093/genetics/159.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiu PKT, Glass NL. 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous Ascomycetes. Curr Opin Microbiol 3:183–188. 10.1016/S1369-5274(00)00073-4. [DOI] [PubMed] [Google Scholar]
- 45.Wilson AM, Markus Wilken P, Van Der Nest MA, Steenkamp ET, Wingfield MJ, Wingfield BD. 2015. Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus 6:207–214. 10.5598/imafungus.2015.06.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin X, Heitman J. 2007. Mechanisms of homothallism in fungi and transitions between heterothallism and homothallism, p 35–57. In Heitman J, Kronstad JW, Taylor JW, Casselton LA (ed), Sex in fungi. ASM Press, Washington, DC. [Google Scholar]
- 47.Merino ST, Nelson MA, Jacobson DJ, Natvig DO. 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143:789–799. 10.1093/genetics/143.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chitrampalam P, Inderbitzin P, Maruthachalam K, Wu BM, Subbarao KV. 2013. The Sclerotinia sclerotiorum mating-type locus (MAT) contains a 3.6-kb region that is inverted in every meiotic generation. PLoS One 8:e56895. 10.1371/journal.pone.0056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilken PM, Steenkamp ET, Wingfield MJ, De Beer ZW, Wingfield BD. 2014. DNA loss at the Ceratocystis fimbriata mating locus results in self-sterility. PLoS One 9:e92180. 10.1371/journal.pone.0092180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins D. 1987. Mating type switching in filamentous Ascomycetes. Genetics 115:215–216. 10.1093/genetics/115.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roach KC, Feretzaki M, Sun S, Heitman J. 2014. Unisexual reproduction, p 255–305. In Friedmann T, Dunlap JC, Goodwin SF (ed), Advances in genetics, 85th ed. Academic Press, Waltham, MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pöggeler S, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Kück U. 2006. Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol Genet Genomics 275:492–503. 10.1007/s00438-006-0107-y. [DOI] [PubMed] [Google Scholar]
- 53.Bidard F, Benkhali J, Coppin E, Imbeaud S, Grognet P, Delacroix H, Debuchy R. 2011. Genome-wide gene expression profiling of fertilization competent mycelium in opposite mating types in the heterothallic fungus Podospora anserina. PLoS One 6:e21476. 10.1371/journal.pone.0021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoder OC, Valent B, Chumley FG. 1986. Genetic nomenclature and practice for plant pathogenic fungi. Phytopathology 76:383–385. 10.1094/Phyto-76-383. [DOI] [Google Scholar]
- 55.Metzenberg RL, Glass NL. 1990. Mating type and mating strategies in Neurospora. Bioessays 12:53–59. 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 56.Wilken PM, Steenkamp ET, Wingfield MJ, de Beer ZW, Wingfield BD. 2017. Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered. Fungal Biol Rev 31:199–211. 10.1016/j.fbr.2017.05.003. [DOI] [Google Scholar]
- 57.Petters-Vandresen DAL, Rossi BJ, Groenewald J, Crous P, Machado MA, Stukenbrock E, Glienke C. 2020. Mating-type locus rearrangements and shifts in thallism states in Citrus-associated Phyllosticta species. Fungal Genet Biol 144:103444. 10.1016/j.fgb.2020.103444. [DOI] [PubMed] [Google Scholar]
- 58.Nagel JH, Wingfield MJ, Slippers B. 2018. Evolution of the mating types and mating strategies in prominent genera in the Botryosphaeriaceae. Fungal Genet Biol 114:24–33. 10.1016/j.fgb.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Arnaise S, Debuchy R, Picard M. 1997. What is a bona fide mating-type gene? Internuclear complementation of mat mutants in Podospora anserina. Mol Gen Genet 256:169–178. 10.1007/pl00008611. [DOI] [PubMed] [Google Scholar]
- 60.Yong M, Yu J, Pan X, Yu M, Cao H, Song T, Qi Z, Du Y, Zhang R, Yin X, Liu W, Liu Y. 2020. Two mating-type genes MAT1-1-1 and MAT1-1–2, with significant functions in conidiation, stress response, sexual development, and pathogenicity of rice false smut fungus Villosiclava virens. Curr Genet 66:989–1002. 10.1007/s00294-020-01085-9. [DOI] [PubMed] [Google Scholar]
- 61.Debuchy R, Turgeon B. 2006. Mating-type structure, evolution, and function in Euascomycetes, p 293–323. In Kües U, Fischer R (ed), The Mycota: growth, differentiation and sexuality. Springer, Berlin, Germany. [Google Scholar]
- 62.Wilson A, Wilken P, van der Nest M, Wingfield M, Wingfield B. 2019. It’s all in the genes: the regulatory pathways of sexual reproduction in filamentous ascomycetes. Genes 10:330. 10.3390/genes10050330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu Y, Amich J, Will C, Eagle CE, Dyer PS, Krappmann S. 2017. The novel Aspergillus fumigatus MAT1-2-4 mating-type gene is required for mating and cleistothecia formation. Fungal Genet Biol 108:1–12. 10.1016/j.fgb.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Wilson AM, Wilken PM, van der Nest MA, Wingfield MJ, Wingfield BD. 2020. The novel Huntiella omanensis mating gene, MAT1-2-7, is essential for ascomatal maturation. Fungal Genet Biol 137:103335. 10.1016/j.fgb.2020.103335. [DOI] [PubMed] [Google Scholar]
- 65.Yong M, Yu J, Pan X, Yu M, Cao H, Qi Z, Du Y, Zhang R, Song T, Yin X, Chen Z, Liu W, Liu Y. 2020. MAT1-1-3, a mating-type gene in the Villosiclava virens, is required for fruiting bodies and sclerotia formation, asexual development and pathogenicity. Front Microbiol 11:1337. 10.3389/fmicb.2020.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanzi AM, Steenkamp ET, Van der Merwe NA, Wingfield BD. 2019. The mating system of the Eucalyptus canker pathogen Chrysoporthe austroafricana and closely related species. Fungal Genet Biol 123:41–52. 10.1016/j.fgb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 67.Mandel MA, Barker BM, Kroken S, Rounsley SD, Orbach MJ. 2007. Genomic and population analyses of the mating-type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot Cell 6:1189–1199. 10.1128/EC.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chai H, Chen W, Zhang X, Su K, Zhao Y. 2019. Structural variation and phylogenetic analysis of the mating-type locus in the genus Morchella. Mycologia 111:551–562. 10.1080/00275514.2019.1628553. [DOI] [PubMed] [Google Scholar]
- 69.Aylward J, Havenga M, Dreyer LL, Roets F, Wingfield BD, Wingfield MJ. 2020. Genomic characterization of mating-type loci and mating type distribution in two apparently asexual plantation tree pathogens. Plant Pathol 69:28–37. 10.1111/ppa.13094. [DOI] [Google Scholar]
- 70.Li JQ, Wingfield BD, Wingfield MJ, Barnes I, Fourie A, Crous PW, Chen SF. 2020. Mating genes in Calonectria and evidence for a heterothallic ancestral state. Persoonia 45:163–176. 10.3767/persoonia.2020.45.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson AM, Godlonton T, van der Nest MA, Wilken PM, Wingfield MJ, Wingfield BD. 2015. Unisexual reproduction in Huntiella moniliformis. Fungal Genet Biol 80:1–9. 10.1016/j.fgb.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 72.Kanematsu S, Adachi Y, Ito T. 2007. Mating-type loci of heterothallic Diaporthe spp.: homologous genes are present in opposite mating types. Curr Genet 52:11–22. 10.1007/s00294-007-0132-3. [DOI] [PubMed] [Google Scholar]
- 73.Turgeon BG, Yoder OC. 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet Biol 31:1–5. 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- 74.Glass NL, Grotelueschen J, Metzenberg RL. 1990. Neurospora crassa A mating-type region. Proc Natl Acad Sci USA 87:4912–4916. 10.1073/pnas.87.13.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Staben C, Yanofsky C. 1990. Neurospora crassa a mating-type region. Proc Natl Acad Sci USA 87:4917–4921. 10.1073/pnas.87.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Štros M, Launholt D, Grasser KD. 2007. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci 64:2590–2606. 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arie T, Christiansen SK, Yoder OC, Turgeon BG. 1997. Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box. Fungal Genet Biol 21:118–130. 10.1006/fgbi.1997.0961. [DOI] [PubMed] [Google Scholar]
- 78.Arie T, Kaneko I, Yoshida T, Noguchi M, Nomura Y, Yamaguchi I. 2000. Mating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Mol Plant Microbe Interact 13:1330–1339. 10.1094/MPMI.2000.13.12.1330. [DOI] [PubMed] [Google Scholar]
- 79.Aylward J. 2017. Comparative genomics of Knoxdaviesia species in the Core Cape Subregion. Stellenbosch University, Stellenbosch, South Africa. [Google Scholar]
- 80.Wilson AM, Lelwala RV, Taylor PWJ, Wingfield MJ, Wingfield BD. 2021. Unique patterns of mating pheromone presence and absence could result in the ambiguous sexual behaviours of Colletotrichum species. G3 Genes Genomes Genet 10.1093/g3journal/jkab187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ament-Velásquez SL, Tuovinen V, Bergström L, Spribille T, Vanderpool D, Nascimbene J, Yamamoto Y, Thor G, Johannesson H. 2021. The plot thickens: haploid and triploid-like thalli, hybridization, and biased mating type ratios in Letharia. Front Fungal Biol 2. 10.3389/ffunb.2021.656386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krämer D, Lane FA, Steenkamp ET, Wingfield BD, Wilken PM. 2021. Unidirectional mating-type switching confers self-fertility to Thielaviopsis cerberus, the only homothallic species in the genus. Fungal Biol 125:427–434. 10.1016/j.funbio.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 83.Wilken PM, Steenkamp ET, van der Nest MA, Wingfield MJ, de Beer ZW, Wingfield BD. 2018. Unexpected placement of the MAT1–1-2 gene in the MAT1-2 idiomorph of Thielaviopsis. Fungal Genet Biol 113:32–41. 10.1016/j.fgb.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 84.Havenga M, Wingfield BD, Wingfield MJ, Roets F, Dreyer LL, Tatham CT, Duong TA, Wilken PM, Chen SF, Aylward J. 2020. Mating strategy and mating type distribution in six global populations of the Eucalyptus foliar pathogen Teratosphaeria destructans. Fungal Genet Biol 137:103350. 10.1016/j.fgb.2020.103350. [DOI] [PubMed] [Google Scholar]
- 85.Klix V, Nowrousian M, Ringelberg C, Loros JJ, Dunlap JC, Pöggeler S. 2010. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot Cell 9:894–905. 10.1128/EC.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Debuchy R, Arnaise S, Lecellier G. 1993. The mat allele of Podospora anserina contains three regulatory genes required for the development of fertilized female organs. Mol Gen Genet 241:667–673. 10.1007/BF00279909. [DOI] [PubMed] [Google Scholar]
- 87.Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, Dyer PS. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol 17:1384–1389. 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 88.Doughan B, Rollins JA. 2016. Characterization of MAT gene functions in the life cycle of Sclerotinia sclerotiorum reveals a lineage-specific MAT gene functioning in apothecium morphogenesis. Fungal Biol 120:1105–1117. 10.1016/j.funbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 89.Pöggeler S, Risch S, Kück U, Osiewacz HD. 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147:567–580. 10.1093/genetics/147.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Becker K, Beer C, Freitag M, Kück U. 2015. Genome-wide identification of target genes of a mating-type α-domain transcription factor reveals functions beyond sexual development. Mol Microbiol 96:1002–1022. 10.1111/mmi.12987. [DOI] [PubMed] [Google Scholar]
- 91.Wilson AM, van der Nest MA, Wilken PM, Wingfield MJ, Wingfield BD. 2018. Pheromone expression reveals putative mechanism of unisexuality in a saprobic ascomycete fungus. PLoS One 13:e0192517. 10.1371/journal.pone.0192517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rodenburg SYA, Terhem RB, Veloso J, Stassen JHM, van Kan JAL. 2018. Functional analysis of mating-type genes and transcriptome analysis during fruiting body development of Botrytis cinerea. mBio 9:e01939-17. 10.1128/mBio.01939-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee J, Lee T, Lee YW, Yun SH, Turgeon BG. 2003. Shifting fungal reproductive mode by manipulation of mating-type genes: obligatory heterothallism of Gibberella zeae. Mol Microbiol 50:145–152. 10.1046/j.1365-2958.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 94.Böhm J, Dahlmann TA, Gümüşer H, Kück U. 2015. A MAT1-2 wild-type strain from Penicillium chrysogenum: functional mating-type locus characterization, genome sequencing and mating with an industrial penicillin-producing strain. Mol Microbiol 95:859–874. 10.1111/mmi.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim W, Cavinder B, Proctor RH, O’Donnell K, Townsend JP, Trail F. 2019. Comparative genomics and transcriptomics during sexual development gives insight into the life history of the cosmopolitan fungus Fusarium neocosmosporiellum. Front Microbiol 10:1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zheng Q, Hou R, Juanyu Zhang Ma J, Wu Z, Wang G, Wang C, Xu JR. 2013. The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PLoS One 8:e66980. 10.1371/journal.pone.0066980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim HK, Cho EJ, Lee S, Lee YS, Yun SH. 2012. Functional analyses of individual mating-type transcripts at MAT loci in Fusarium graminearum and Fusarium asiaticum. FEMS Microbiol Lett 337:89–96. 10.1111/1574-6968.12012. [DOI] [PubMed] [Google Scholar]
- 98.Dyer PS, Inderbitzin P, Debuchy R. 2016. Mating-type structure, function, regulation, and evolution in the Pezizomycotina, p 351–385. In Wendland J (ed), Growth, differentiation and sexuality, 3rd ed. Springer International Publishing, Geneva, Switzerland. [Google Scholar]
- 99.Ait Benkhali J, Coppin E, Brun S, Peraza-Reyes L, Martin T, Dixelius C, Lazar N, van Tilbeurgh H, Debuchy R. 2013. A network of HMG-box transcription factors regulates sexual cycle in the fungus Podospora anserina. PLoS Genet 9:e1003642. 10.1371/journal.pgen.1003642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gautier V, Tong L, Nguyen T-S, Debuchy R, Silar P. 2018. PaPro1 and IDC4, two genes controlling stationary phase, sexual development and cell degeneration in Podospora anserina. J Fungi 4:85. 10.3390/jof4030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bazafkan H, Dattenböck C, Böhmdorfer S, Tisch D, Stappler E, Schmoll M. 2015. Mating type-dependent partner sensing as mediated by VEL1 in Trichoderma reesei. Mol Microbiol 96:1103–1118. 10.1111/mmi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seibel C, Tisch D, Kubicek CP, Schmoll M. 2012. ENVOY is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei). Eukaryot Cell 11:885–895. 10.1128/EC.05321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aylward J, Steenkamp ET, Dreyer LL, Roets F, Wingfield MJ, Wingfield BD. 2016. Genetic basis for high population diversity in Protea-associated Knoxdaviesia. Fungal Genet Biol 96:47–57. 10.1016/j.fgb.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 104.Nauta MJ, Hoekstra RF. 1992. Evolution of reproductive systems in filamentous ascomycetes. I. Evolution of mating types. Heredity 68:405–410. 10.1038/hdy.1992.60. [DOI] [PubMed] [Google Scholar]
- 105.Leslie JF, Klein KK. 1996. Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics 144:557–567. 10.1093/genetics/144.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim H, Borkovich KA. 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot Cell 5:544–554. 10.1128/EC.5.3.544-554.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim H, Borkovich KA. 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol Microbiol 52:1781–1798. 10.1111/j.1365-2958.2004.04096.x. [DOI] [PubMed] [Google Scholar]
- 108.Cano-Domínguez N, Álvarez-Delfín K, Hansberg W, Aguirre J. 2008. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell 7:1352–1361. 10.1128/EC.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim H, Metzenberg RL, Nelson MA. 2002. Multiple functions of mfa-1, a putative pheromone precursor gene of Neurospora crassa. Eukaryot Cell 1:987–999. 10.1128/EC.1.6.987-999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park G, Pan S, Borkovich KA. 2008. Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa. Eukaryot Cell 7:2113–2122. 10.1128/EC.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang Q, Poole SI, Borkovich KA. 2002. A G-protein β subunit required for sexual and vegetative development and maintenance of normal Gα protein levels in Neurospora crassa. Eukaryot Cell 1:378–390. 10.1128/EC.1.3.378-390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Turina M, Prodi A, Van Alfen NK. 2003. Role of the Mf1–1 pheromone precursor gene of the filamentous ascomycete Cryphonectria parasitica. Fungal Genet Biol 40:242–251. 10.1016/S1087-1845(03)00084-7. [DOI] [PubMed] [Google Scholar]
- 113.Zhou X, Liu W, Wang C, Xu Q, Wang Y, Ding S, Xu JR. 2011. A MADS-box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae. Mol Microbiol 80:33–53. 10.1111/j.1365-2958.2011.07556.x. [DOI] [PubMed] [Google Scholar]
- 114.Malagnac F, Lalucque H, Lepère G, Silar P. 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet Biol 41:982–997. 10.1016/j.fgb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 115.Silar P. 2014. Simple genetic tools to study fruiting body development in fungi. TOMYCJ 8:148–155. 10.2174/1874437001408010148. [DOI] [Google Scholar]
- 116.Lalucque H, Malagnac F, Green K, Gautier V, Grognet P, Chan Ho Tong L, Scott B, Silar P. 2017. IDC2 and IDC3, two genes involved in cell non-autonomous signaling of fruiting body development in the model fungus Podospora anserina. Dev Biol 421:126–138. 10.1016/j.ydbio.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 117.Barman A, Tamuli R. 2017. The pleiotropic vegetative and sexual development phenotypes of Neurospora crassa arise from double mutants of the calcium signaling genes plc-1, splA2, and cpe-1. Curr Genet 63:861–875. 10.1007/s00294-017-0682-y. [DOI] [PubMed] [Google Scholar]
- 118.Cavinder B, Trail F. 2012. Role of Fig1, a component of the low-affinity calcium uptake system, in growth and sexual development of filamentous fungi. Eukaryot Cell 11:978–988. 10.1128/EC.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bolker M, Kahmann R. 1993. Sexual pheromones and mating responses in fungi. Plant Cell 5:1461–1469. 10.1105/tpc.5.10.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang L, Baasiri RA, Van Alfen NK. 1998. Viral repression of fungal pheromone precursor gene expression. Mol Cell Biol 18:953–959. 10.1128/MCB.18.2.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shen WC, Bobrowicz P, Ebbole DJ. 1999. Isolation of pheromone precursor genes of Magnaporthe grisea. Fungal Genet Biol 27:253–263. 10.1006/fgbi.1999.1151. [DOI] [PubMed] [Google Scholar]
- 122.Lee J, Leslie JF, Bowden RL. 2008. Expression and function of sex pheromones and receptors in the homothallic ascomycete Gibberella zeae. Eukaryot Cell 7:1211–1221. 10.1128/EC.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pöggeler S. 2000. Two pheromone precursor genes are transcriptionally expressed in the homothallic ascomycete Sordaria macrospora. Curr Genet 37:403–411. 10.1007/s002940000120. [DOI] [PubMed] [Google Scholar]
- 124.Pöggeler S, Kück U. 2001. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280:9–17. 10.1016/s0378-1119(01)00786-7. [DOI] [PubMed] [Google Scholar]
- 125.Wilson AM, Gabriel R, Singer SW, Schuerg T, Wilken PM, van der Nest MA, Wingfield MJ, Wingfield BD. 2021. Doing it alone: unisexual reproduction in filamentous ascomycete fungi. Fungal Biol Rev 35:1–13. 10.1016/j.fbr.2020.12.003. [DOI] [Google Scholar]
- 126.Martin F, Kohler A, Murat C, Balestrini R, Coutinho PM, Jaillon O, Montanini B, Morin E, Noel B, Percudani R, Porcel B, Rubini A, Amicucci A, Amselem J, Anthouard V, Arcioni S, Artiguenave F, Aury JM, Ballario P, Bolchi A, Brenna A, Brun A, Buée M, Cantarel B, Chevalier G, et al. 2010. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464:1033–1038. 10.1038/nature08867. [DOI] [PubMed] [Google Scholar]
- 127.Zhang L, Churchill AC, Kazmierczak P, Kim DH, Van Alfen NK. 1993. Hypovirulence-associated traits induced by a mycovirus of Cryphonectria parasitica are mimicked by targeted inactivation of a host gene. Mol Cell Biol 13:7782–7792. 10.1128/mcb.13.12.7782-7792.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mayrhofer S, Weber JM, Pöggeler S. 2006. Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics 172:1521–1533. 10.1534/genetics.105.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liang X, Yao L, Kong Y, Li B, Hao X, Lin Y, Cao M, Dong Q, Zhang R, Rollins JA, Sun G. 2021. Molecular dissection of perithecial mating line development in Colletotrichum fructicola, a species with a nontypical mating system featured by plus-to-minus switch and plus-minus mediated sexual enhancement. Appl Environ Microbiol 87:e00474-21. 10.1128/AEM.00474-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wheeler H. 1954. Genetics and evolution of heterothallism in Glomerella. Phytopathology 44:342–345. [Google Scholar]
- 131.Anderegg RJ, Betzll R, Carr SA, Crabb JW, Duntzell W, Chern WJB. 1988. Structure of Saccharomyces cerevisiae mating hormone a-factor. J Biol Chem 263:18236–18240. 10.1016/S0021-9258(19)81351-0. [DOI] [PubMed] [Google Scholar]
- 132.Pöggeler S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr Genet 42:153–160. 10.1007/s00294-002-0338-3. [DOI] [PubMed] [Google Scholar]
- 133.Ge YY, Yu FM, Yang ZJ, Tan YM, Xiang T, Chen X, Yu YX, Chen L, Liu ZY. 2019. Genetic basis and function of mating-type genes in Aspergillus cristatus. Mycosphere 10:622–633. 10.5943/mycosphere/10/1/12. [DOI] [Google Scholar]
- 134.Pál K, Van Diepeningen AD, Varga J, Debets AJM, Hoekstra RF. 2008. Sexual genes in the asexual filamentous fungus Aspergillus niger and related aspergilli. Aspergillus Genomic Era 2008:107–131. [Google Scholar]
- 135.Aboelfotoh Hendy A, Xing J, Chen X, Chen XL. 2019. The farnesyltransferase β-subunit RAM1 regulates localization of RAS proteins and appressorium-mediated infection in Magnaporthe oryzae. Mol Plant Pathol 20:1264–1278. 10.1111/mpp.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Short DPG, Gurung S, Hu X, Inderbitzin P, Subbarao KV. 2014. Maintenance of sex-related genes and the co-occurrence of both mating types in Verticillium dahliae. PLoS One 9:e112145. 10.1371/journal.pone.0112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maurer-Stroh S, Washietl S, Eisenhaber F. 2003. Protein prenyltransferases. Genome Biol 4:212. 10.1186/gb-2003-4-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ge Y, Yu F, Tan Y, Zhang X, Liu Z. 2017. Comparative transcriptome sequence analysis of sporulation-related genes of Aspergillus cristatus in response to low and high osmolarity. Curr Microbiol 74:806–814. 10.1007/s00284-017-1250-x. [DOI] [PubMed] [Google Scholar]
- 139.Nowrousian M, Stajich JE, Chu M, Engh I, Espagne E, Halliday K, Kamerewerd J, Kempken F, Knab B, Kuo HC, Osiewacz HD, Pöggeler S, Read ND, Seiler S, Smith KM, Zickler D, Kück U, Freitag M. 2010. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet 6:e1000891. 10.1371/journal.pgen.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hernández-Chávez MJ, González-Hernández RJ, Trujillo-Esquivel JE, Hernández-Cervantes A, Mora-Montes HM. 2014. The secretory pathway in the filamentous fungus Trichoderma, p 115–121. In Gupta V, Schmoll M, Herrera-Estrella A, Upadhyay R, Druzhinina I, Tuohy M (ed), Biotechnology and biology of Trichoderma. Elsevier, New York, NY. [Google Scholar]
- 141.Woo PCY, Chong KTK, Tse H, Cai JJ, Lau CCY, Zhou AC, Lau SKP, Yuen KY. 2006. Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett 580:3409–3416. 10.1016/j.febslet.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 142.Kato E. 1998. Characterization of a serine carboxypeptidase in Neurospora crassa, homologous to the KEX1 gene of S. cerevisiae. University of Hawaii, Honolulu, HI. [Google Scholar]
- 143.Jacob-Wilk D, Turina M, Kazmierczak P, Van Alfen NK. 2009. Silencing of Kex2 significantly diminishes the virulence of Cryphonectria parasitica. Mol Plant Microbe Interact 22:211–221. 10.1094/MPMI-22-2-0211. [DOI] [PubMed] [Google Scholar]
- 144.Wang Z, Lopez-Giraldez F, Lehr N, Farré M, Common R, Trail F, Townsend JP. 2014. Global gene expression and focused knockout analysis reveals genes associated with fungal fruiting body development in Neurospora crassa. Eukaryot Cell 13:154–169. 10.1128/EC.00248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xue C, Hsueh YP, Heitman J. 2008. Magnificent seven: roles of G-protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev 32:1010–1032. 10.1111/j.1574-6976.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fischer MS, Glass NL. 2019. Communicate and fuse: how filamentous fungi establish and maintain an interconnected mycelial network. Front Microbiol 10:619. 10.3389/fmicb.2019.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seibel C, Tisch D, Kubicek CP, Schmoll M. 2012. The role of pheromone receptors for communication and mating in Hypocrea jecorina (Trichoderma reesei). Fungal Genet Biol 49:814–824. 10.1016/j.fgb.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seo JA, Han KH, Yu JH. 2004. The gprA and gprB genes encode putative G-protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol Microbiol 53:1611–1623. 10.1111/j.1365-2958.2004.04232.x. [DOI] [PubMed] [Google Scholar]
- 149.Brown NA, Schrevens S, Van Dijck P, Goldman GH. 2018. Fungal G-protein-coupled receptors: mediators of pathogenesis and targets for disease control. Nat Microbiol 3:402–414. 10.1038/s41564-018-0127-5. [DOI] [PubMed] [Google Scholar]
- 150.Ivey FD, Hodge PN, Turner GE, Borkovich KA. 1996. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol Biol Cell 7:1283–1297. 10.1091/mbc.7.8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Baasiri RA, Lu X, Rowley PS, Turner GE, Borkovich KA. 1997. Overlapping functions for two G protein α subunits in Neurospora crassa. Genetics 147:137–145. 10.1093/genetics/147.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kamerewerd J, Jansson M, Nowrousian M, Pöggeler S, Kück U. 2008. Three α-subunits of heterotrimeric G-proteins and an adenylyl cyclase have distinct roles in fruiting body development in the homothallic fungus Sordaria macrospora. Genetics 180:191–206. 10.1534/genetics.108.091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Krystofova S, Borkovich KA. 2005. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot Cell 4:365–378. 10.1128/EC.4.2.365-378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xu JR. 2000. MAP kinases in fungal pathogens. Fungal Genet Biol 31:137–152. 10.1006/fgbi.2000.1237. [DOI] [PubMed] [Google Scholar]
- 155.Li D, Bobrowicz P, Wilkinson HH, Ebbole DJ. 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170:1091–1104. 10.1534/genetics.104.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jones C, Greer-Phillips S, Borkovich K. 2007. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol Biol Cell 18:2123–2136. 10.1091/mbc.e06-03-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fujimura M, Ochiai N, Oshima M, Motoyama T, Ichiishi A, Usami R, Horikoshi K, Yamaguchi I. 2003. Putative homologs of SSK22 MAPKK kinase and PBS2 MAPK kinase of Saccharomyces cerevisiae encoded by os-4 and os-5 genes for osmotic sensitivity and fungicide resistance in Neurospora crassa. Biosci Biotechnol Biochem 67:186–191. 10.1271/bbb.67.186. [DOI] [PubMed] [Google Scholar]
- 158.Frawley D, Bayram Ö. 2020. The pheromone response module, a mitogen-activated protein kinase pathway implicated in the regulation of fungal development, secondary metabolism and pathogenicity. Fungal Genet Biol 144:103469. 10.1016/j.fgb.2020.103469. [DOI] [PubMed] [Google Scholar]
- 159.Teichert I, Steffens EK, Schnass N, Fränzel B, Krisp C, Wolters DA, Kück U. 2014. PRO40 is a scaffold protein of the cell wall integrity pathway, linking the MAP kinase module to the upstream activator protein kinase C. PLoS Genet 10:e1004582. 10.1371/journal.pgen.1004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schmidt S, Märker R, Ramšak B, Beier-Rosberger AM, Teichert I, Kück U. 2020. Crosstalk between pheromone signaling and NADPH oxidase complexes coordinates fungal developmental processes. Front Microbiol 11:1722. 10.3389/fmicb.2020.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bloemendal S, Bernhards Y, Bartho K, Dettmann A, Voigt O, Teichert I, Seiler S, Wolters DA, Pöggeler S, Kück U. 2012. A homologue of the human STRIPAK complex controls sexual development in fungi. Mol Microbiol 84:310–323. 10.1111/j.1365-2958.2012.08024.x. [DOI] [PubMed] [Google Scholar]
- 162.Kück U, Beier AM, Teichert I. 2016. The composition and function of the striatin-interacting phosphatases and kinases (STRIPAK) complex in fungi. Fungal Genet Biol 90:31–38. 10.1016/j.fgb.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 163.Xiang Q, Rasmussen C, Louise Glass N. 2002. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160:169–180. 10.1093/genetics/160.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Simonin AR, Rasmussen CG, Yang M, Glass NL. 2010. Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet Biol 47:855–868. 10.1016/j.fgb.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 165.Fu C, Iyer P, Herkal A, Abdullah J, Stout A, Free SJ. 2011. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot Cell 10:1100–1109. 10.1128/EC.05003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Märker R, Blank-Landeshammer B, Beier-Rosberger A, Sickmann A, Kück U. 2020. Phosphoproteomic analysis of STRIPAK mutants identifies a conserved serine phosphorylation site in PAK kinase CLA4 to be important in fungal sexual development and polarized growth. Mol Microbiol 113:1053–1069. 10.1111/mmi.14475. [DOI] [PubMed] [Google Scholar]
- 167.Pöggeler S, Kück U. 2004. A WD40 repeat protein regulates fungal cell differentiation and can be replaced functionally by the mammalian homologue striatin. Eukaryot Cell 3:232–240. 10.1128/EC.3.1.232-240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Beier A, Teichert I, Krisp C, Wolters DA, Kück U. 2016. Catalytic subunit 1 of protein phosphatase 2A is a subunit of the STRIPAK complex and governs fungal sexual development. mBio 7:e00870-16. 10.1128/mBio.00870-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Frey S, Reschka EJ, Pöggeler S. 2015. Germinal center kinases SmKIN3 and SmKIN24 are associated with the Sordaria macrospora striatin-interacting phosphatase and kinase (STRIPAK) complex. PLoS One 10:e0139163–27. 10.1371/journal.pone.0139163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nordzieke S, Zobel T, Fränzel B, Wolters DA, Kück U, Teichert I. 2015. A fungal sarcolemmal membrane-associated protein (SLMAP) homolog plays a fundamental role in development and localizes to the nuclear envelope, endoplasmic reticulum, and mitochondria. Eukaryot Cell 14:345–358. 10.1128/EC.00241-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bernhards Y, Pöggeler S. 2011. The phocein homologue SmMOB3 is essential for vegetative cell fusion and sexual development in the filamentous ascomycete Sordaria macrospora. Curr Genet 57:133–149. 10.1007/s00294-010-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li L, Xue C, Bruno K, Nishimura M, Xu JR. 2004. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol Plant Microbe Interact 17:547–556. 10.1094/MPMI.2004.17.5.547. [DOI] [PubMed] [Google Scholar]
- 173.Park G, Servin JA, Turner GE, Altamirano L, Colot HV, Collopy P, Litvinkova L, Li L, Jones CA, Diala FG, Dunlap JC, Borkovich KA. 2011. Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot Cell 10:1553–1564. 10.1128/EC.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang C, Zhang S, Hou R, Zhao Z, Zheng Q, Xu Q, Zheng D, Wang G, Liu H, Gao X, Ma JW, Kistler HC, Kang Z, Xu JR. 2011. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog 7:e1002460. 10.1371/journal.ppat.1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chinnici JL, Fu C, Caccamise LM, Arnold JW, Free SJ. 2014. Neurospora crassa female development requires the PACC and other signal transduction pathways, transcription factors, chromatin remodeling, cell-to-cell fusion, and autophagy. PLoS One 9:e110603. 10.1371/journal.pone.0110603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Virgilio S, Bertolini MC. 2018. Functional diversity in the pH signaling pathway: an overview of the pathway regulation in Neurospora crassa. Curr Genet 64:529–534. 10.1007/s00294-017-0772-x. [DOI] [PubMed] [Google Scholar]
- 177.Lamb TM, Xu W, Diamond A, Mitchell AP. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276:1850–1856. 10.1074/jbc.M008381200. [DOI] [PubMed] [Google Scholar]
- 178.Arst HN, Peñalva MA. 2003. pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet 19:224–231. 10.1016/S0168-9525(03)00052-0. [DOI] [PubMed] [Google Scholar]
- 179.Peñalva MA, Tilburn J, Bignell E, Arst HN. 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol 16:291–300. 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 180.Virgilio S, Cupertino FB, Ambrosio DL, Bertolini MC. 2017. Regulation of the reserve carbohydrate metabolism by alkaline pH and calcium in Neurospora crassa reveals a possible cross-regulation of both signaling pathways. BMC Genomics 18:457. 10.1186/s12864-017-3832-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Su SSY, Mitchell AP. 1993. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics 133:67–77. 10.1093/genetics/133.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li W, Mitchell AP. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63–73. 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nahlik K, Dumkow M, Bayram Ö, Helmstaedt K, Busch S, Valerius O, Gerke J, Hoppert M, Schwier E, Opitz L, Westermann M, Grond S, Feussner K, Goebel C, Kaever A, Meinicke P, Feussner I, Braus GH. 2010. The COP9 signalosome mediates transcriptional and metabolic response to hormones, oxidative stress protection and cell wall rearrangement during fungal development. Mol Microbiol 78:964–979. 10.1111/j.1365-2958.2010.07384.x. [DOI] [PubMed] [Google Scholar]
- 184.Busch S, Schwier EU, Nahlik K, Bayram O, Helmstaedt K, Draht OW, Krappmann S, Valerius O, Lipscomb WN, Braus GH. 2007. An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proc Natl Acad Sci USA 104:8089–8094. 10.1073/pnas.0702108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Busch S, Eckert SE, Krappmann S, Braus GH. 2003. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol Microbiol 49:717–730. 10.1046/j.1365-2958.2003.03612.x. [DOI] [PubMed] [Google Scholar]
- 186.Braus GH, Irniger S, Bayram Ö. 2010. Fungal development and the COP9 signalosome. Curr Opin Microbiol 13:672–676. 10.1016/j.mib.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 187.Christmann M, Schmaler T, Gordon C, Huang X, Bayram Ö, Schinke J, Stumpf S, Dubiel W, Braus GH. 2013. Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet 9:e1003275. 10.1371/journal.pgen.1003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kirk KE, Morris NR. 1991. The tubB α-tubulin gene is essential for sexual development in Aspergillus nidulans. Genes Dev 5:2014–2023. 10.1101/gad.5.11.2014. [DOI] [PubMed] [Google Scholar]
- 189.Krappmann S, Jung N, Medic B, Busch S, Prade RA, Braus GH. 2006. The Aspergillus nidulans F-box protein GrrA links SCF activity to meiosis. Mol Microbiol 61:76–88. 10.1111/j.1365-2958.2006.05215.x. [DOI] [PubMed] [Google Scholar]
- 190.Shiu PK, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell 107:905–916. 10.1016/S0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 191.Shiu PKT, Zickler D, Raju NB, Ruprich-Robert G, Metzenberg RL. 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc Natl Acad Sci USA 103:2243–2248. 10.1073/pnas.0508896103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci USA 100:12865–12870. 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nolting N, Pöggeler S. 2006. A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol Microbiol 62:853–868. 10.1111/j.1365-2958.2006.05415.x. [DOI] [PubMed] [Google Scholar]
- 194.Werner A, Otte K, Stahlhut G, Hanke LM, Pöggeler S. 2021. The glyoxysomal protease LON2 is involved in fruiting-body development, ascosporogenesis and stress resistance in Sordaria macrospora. J Fungi 7:82. 10.3390/jof7020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Werner A, Herzog B, Voigt O, Valerius O, Braus GH, Pöggeler S. 2019. NBR1 is involved in selective pexophagy in filamentous ascomycetes and can be functionally replaced by a tagged version of its human homolog. Autophagy 15:78–97. 10.1080/15548627.2018.1507440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Voigt O, Pöggeler S. 2013. Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora. Autophagy 9:33–49. 10.4161/auto.22398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nelson MA, Merino ST, Metzenberg RL. 1997. A putative rhamnogalacturonase required for sexual development of Neurospora crassa. Genetics 146:531–540. 10.1093/genetics/146.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Traeger S, Nowrousian M. 2015. Functional analysis of developmentally regulated genes chs7 and sec22 in the ascomycete Sordaria macrospora. G3 (Bethesda) 5:1233–1245. 10.1534/g3.115.017681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ, Klionsky DJ. 2011. SNARE proteins are required for macroautophagy. Cell 146:290–302. 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cao S, He Y, Hao C, Xu Y, Zhang H, Wang C, Liu H, Xu JR. 2017. RNA editing of the AMD1 gene is important for ascus maturation and ascospore discharge in Fusarium graminearum. Sci Rep 7–:4617. 10.1038/s41598-017-04960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Liu H, Li Y, Chen D, Qi Z, Wang Q, Wang J, Jiang C, Xu JR. 2017. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc Natl Acad Sci USA 114:E7756–E7765. 10.1073/pnas.1702591114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Glass NL, Metzenberg RL, Raju NB. 1990. Homothallic Sordariaceae from nature: the absence of strains containing only the a mating type sequence. Exp Mycol 14:274–289. 10.1016/0147-5975(90)90025-O. [DOI] [Google Scholar]
- 203.Ferreira AVB, Saupe S, Glass NL. 1996. Transcriptional analysis of the mt A idiomorph of Neurospora crassa identifies two genes in addition to mt A-1. Mol Gen Genet 250:767–774. 10.1007/BF02172989. [DOI] [PubMed] [Google Scholar]
- 204.Singh G, Ashby AM. 1998. Cloning of the mating-type loci from Pyrenopeziza brassicae reveals the presence of a novel mating type gene within a discomycete MAT1–2 locus encoding a putative metallothionein-like protein. Mol Microbiol 30:799–806. 10.1046/j.1365-2958.1998.01112.x. [DOI] [PubMed] [Google Scholar]
- 205.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier J-M, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7:e1002230. 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Palmer JM, Kubatova A, Novakova A, Minnis AM, Kolarik M, Lindner DL. 2014. Molecular characterization of a heterothallic mating system in Pseudogymnoascus destructans, the fungus causing white-nose syndrome of bats. G3 (Bethesda) 4:1755–1763. 10.1534/g3.114.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bihon W, Wingfield MJ, Slippers B, Duong TA, Wingfield BD. 2014. MAT gene idiomorphs suggest a heterothallic sexual cycle in a predominantly asexual and important pine pathogen. Fungal Genet Biol 62:55–61. 10.1016/j.fgb.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 208.Pöggeler S, Kück U. 2000. Comparative analysis of the mating-type loci from Neurospora crassa and Sordaria macrospora: identification of novel transcribed ORFs. Mol Gen Genet 263:292–301. 10.1007/s004380051171. [DOI] [PubMed] [Google Scholar]
- 209.Kanamori M, Kato H, Yasuda N, Koizumi S, Peever TL, Kamakura T, Teraoka T, Arie T. 2007. Novel mating type-dependent transcripts at the mating-type locus in Magnaporthe oryzae. Gene 403:6–17. 10.1016/j.gene.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 210.Yu J, Sun W, Yu M, Yin X, Meng X, Zhao J, Huang L, Huang L, Liu Y. 2015. Characterization of mating-type loci in rice false smut fungus Villosiclava virens. FEMS Microbiol Lett 362:fnv014. 10.1093/femsle/fnv014. [DOI] [PubMed] [Google Scholar]
- 211.Martin SH, Wingfield BD, Wingfield MJ, Steenkamp ET. 2011. Structure and evolution of the Fusarium mating type locus: new insights from the Gibberella fujikuroi complex. Fungal Genet Biol 48:731–740. 10.1016/j.fgb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 212.Kothe GO, Free SJ. 1998. The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 149:117–130. 10.1093/genetics/149.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ivey FD, Kays AM, Borkovich KA. 2002. Shared and independent roles for a Gαi protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa. Eukaryot Cell 1:634–642. 10.1128/EC.1.4.634-642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download MMBR.00020-21-s0001.xlsx, XLSX file, 0.02 MB (17.1KB, xlsx)