Abstract
Background
COVID-19 has threatened the health of tens of millions of people all over the world. Massive research efforts have been made in response to the COVID-19 pandemic. Utilization of clinical data can accelerate these research efforts to combat the pandemic since important characteristics of the patients are often found by examining the clinical data. Publicly accessible clinical data on COVID-19, however, remain limited despite the immediate need.
Objective
To provide shareable clinical data to catalyze COVID-19 research, we present Columbia Open Health Data for COVID-19 Research (COHD-COVID), a publicly accessible database providing clinical concept prevalence, clinical concept co-occurrence, and clinical symptom prevalence for hospitalized patients with COVID-19. COHD-COVID also provides data on hospitalized patients with influenza and general hospitalized patients as comparator cohorts.
Methods
The data used in COHD-COVID were obtained from NewYork-Presbyterian/Columbia University Irving Medical Center’s electronic health records database. Condition, drug, and procedure concepts were obtained from the visits of identified patients from the cohorts. Rare concepts were excluded, and the true concept counts were perturbed using Poisson randomization to protect patient privacy. Concept prevalence, concept prevalence ratio, concept co-occurrence, and symptom prevalence were calculated using the obtained concepts.
Results
Concept prevalence and concept prevalence ratio analyses showed the clinical characteristics of the COVID-19 cohorts, confirming the well-known characteristics of COVID-19 (eg, acute lower respiratory tract infection and cough). The concepts related to the well-known characteristics of COVID-19 recorded high prevalence and high prevalence ratio in the COVID-19 cohort compared to the hospitalized influenza cohort and general hospitalized cohort. Concept co-occurrence analyses showed potential associations between specific concepts. In case of acute lower respiratory tract infection in the COVID-19 cohort, a high co-occurrence ratio was obtained with COVID-19–related concepts and commonly used drugs (eg, disease due to coronavirus and acetaminophen). Symptom prevalence analysis indicated symptom-level characteristics of the cohorts and confirmed that well-known symptoms of COVID-19 (eg, fever, cough, and dyspnea) showed higher prevalence than the hospitalized influenza cohort and the general hospitalized cohort.
Conclusions
We present COHD-COVID, a publicly accessible database providing useful clinical data for hospitalized patients with COVID-19, hospitalized patients with influenza, and general hospitalized patients. We expect COHD-COVID to provide researchers and clinicians quantitative measures of COVID-19–related clinical features to better understand and combat the pandemic.
Keywords: COVID-19, open data, electronic health record, data science, research, data, access, database, symptom, cohort, prevalence
Introduction
COVID-19 has threatened the health of tens of millions of people all over the world. The global pandemic caused by COVID-19 has sparked massive research efforts in the fight against the novel disease, including characterizing the disease and clinical progression, identifying risk factors for hospitalization, and finding drugs that can be repurposed to lessen disease severity [1-3]. Utilization of clinical data from different institutions, hospitals, and nations can accelerate these research efforts since important characteristics of the patients are often found by examining the shared clinical data. Although many studies sharing epidemiological data [4,5], public health [6], and social measures [7] for COVID-19 research have been conducted, publicly accessible clinical data on COVID-19 remain limited despite the immediate need [8], mainly owing to the potential risk to patient privacy that can still exist even after deidentification of the data [9].
Recognizing the need for publicly accessible electronic health record (EHR)–derived data in a broad range of clinical and translational research, we previously developed Columbia Open Health Data (COHD). COHD provides open and easy access to prevalence and co-occurrence statistics on conditions, drugs, procedures, and demographics derived from structured EHR data from NewYork-Presbyterian/Columbia University Irving Medical Center (NYP/CUIMC) [10], which serves the large and diverse population of NYC and its surrounding areas. Since its deployment, COHD has accelerated biomedical research by providing 2 informative resources, prevalence and co-occurrence statistics, and their derived association metrics [10-12].
NYC was one of the first epicenters of COVID-19 in the United States with the first confirmed case on February 29, 2020 [13]. As one of the largest academic medical centers in NYC, NYP/CUIMC has admitted more than 4000 patients as of September 1, 2020. We aim to provide shareable clinical data to catalyze future COVID-19 research by presenting Columbia Open Health Data for COVID-19 Research (COHD-COVID), a publicly accessible database providing clinical concept prevalence, the clinical concept prevalence ratio between cohorts, clinical concept co-occurrence, and clinical symptom prevalence for a cohort of hospitalized patients with COVID-19 and comparator cohorts (a cohort of hospitalized patients with influenza and a general hospitalized patient cohort) derived from NYP/CUIMC’s electronic health records. In addition to providing publicly accessible data files via the Figshare data repository, we also developed the COHD-COVID web application programming interface (API) for easy access and better usability.
Methods
Methods Overview
We used the term “concept” to refer to clinical entities and events such as conditions (ie, diagnosis), drugs, and procedures. The concepts and their names are defined by the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM). When concepts are referenced in this paper, the name of the concept is styled in italics (eg, Disorder of respiratory system) to distinguish the formalized concepts from regular text. We also styled entities in the OMOP CDM (eg, person_id column in condition_occurrence table) in italics.
Figure 1 depicts the overall workflow to create COHD-COVID. Columbia’s clinical data warehouse was converted to the OMOP CDM. We first filtered the EHR data in accordance with the cohort definitions, and then EHR for each patient’s inpatient visits were identified. Condition, drug, and procedure concepts were obtained from the identified visits. Concept prevalence, concept prevalence ratio, concept co-occurrence, and symptom prevalence analyses followed using the obtained concepts. To protect patient privacy, we excluded rare concepts observed in 10 or fewer visits and perturbed the true counts using Poisson randomization. Perturbation of the true counts and exclusion of rare concepts reduce the uniqueness of individual in the data, which can minimize the risk of reidentification [9]. Perturbed counts generated by the Poisson randomization process do not show a significant difference from the true counts [10]. The resulting data were stored in a MySQL database and made publicly available via the COHD-COVID web API [14]. All analyses were conducted using Python 3.5.2. This study received institutional review board approval with a waiver for informed consent.
Figure 1.
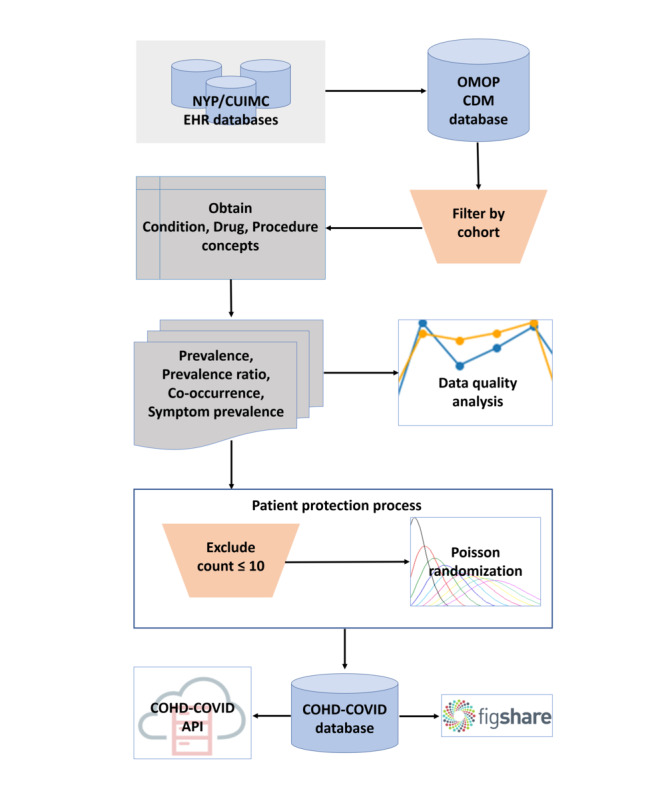
Overall workflow of Columbia Open Health Data for COVID-19 Research (COHD-COVID). API: application programming interface; NYP/CUIMC: NewYork-Presbyterian/Columbia University Irving Medical Center; OMOP CDM: Observational Medical Outcomes Partnership Common Data Model.
Data Source
We utilized EHR data from the NYP/CUIMC’s clinical data warehouse, where inpatient and outpatient data dating back to 1985 are stored. EHR data were collected during routine clinical care. Patients were notified of potential uses of their data for research at the point of care and data collection. NYP/CUIMC has converted its clinical data warehouse to OMOP CDM on a regular basis. Since NYP/CUIMC covers NYC and the surrounding area, which has a diverse population of 8.2 million people and was an early COVID-19 epicenter, the EHR data from NYP/CUIMC can provide a diverse and large sample of patients with COVID-19.
Three different patient cohorts were used in this study. The COVID-19 cohort was defined as hospitalized patients aged ≥18 years with a COVID-19–related condition diagnosis or a confirmed positive COVID-19 test result during their hospitalization period or within the prior 21 days. Patients identified with the COVID-19 cohort definition from March 1, 2020, to September 1, 2020, were included in the COVID-19 cohort. The influenza cohort was similarly defined as patients aged ≥18 years who had at least 1 occurrence of influenza conditions or precoordinated positive measurements or positive influenza test results during their hospitalization period or within the prior 21 days. The general cohort was defined as all hospitalized patients aged ≥18 years. Patient visits from calendar years 2014 to 2019 were included for the influenza and general cohorts. All cohorts were divided into subcohorts stratified by sex (male vs female) and age (adults aged 18-64 years vs seniors aged >65 years) for further investigation. The COVID-19 and influenza cohort definitions were adapted from the cohort definitions created by the Observational Health Data Science and Informatics’ (OHDSI’s) international network study for COVID-19 [3]. Summary statistics of the cohorts are provided in Table 1.
Table 1.
Basic statistics of 3 cohorts in the Columbia Open Health Data for COVID-19 Research database. The statistics summarized here are based on the data from NewYork-Presbyterian/Columbia University Irving Medical Center as of September 1, 2020.
|
|
COVID-19 cohort | Influenza cohort | General cohort |
| Time range | March 1, 2020, to August 31, 2020 | January 1, 2014, to December 12, 2019 | January 1, 2014, to December 12, 2019 |
| Patients, n | 4127 | 3261 | 175,930 |
| Total inpatient visits, n | 4846 | 3860 | 314,680 |
| Male patients; inpatient visits, n (%) | 2103 (51.0); 2518 (52.0) | 1454 (44.6); 1732 (44.9) | 67,662 (38.5); 128,908 (41.0) |
| Female patients; inpatient visits, n (%) | 2024 (49.0); 2328 (48.0) | 1807 (55.4); 2128 (55.1) | 108,268 (61.5); 185,772 (59.0) |
| Adult patients (aged 18-64 years); inpatient visits, n (%) | 2147 (52.0); 2511 (51.8) | 1315 (40.3); 1553 (40.2) | 104,020 (59.1); 173,843 (55.2) |
| Senior patients (>65 years); inpatient visits, n (%) | 1980 (48.0); 2335 (48.2) | 1946 (59.7); 2307 (59.8) | 71,910 (40.9); 140,837 (44.8) |
Patients belonging to the cohorts based on the 3 cohort definitions above were identified using the unique person_id from the person table in the OMOP CDM. Condition, drug, and procedure concepts observed in these patients during inpatient visits were extracted from the condition_occurrence, drug_exposure, and procedure_occurrence tables in the OMOP CDM, respectively. Inpatient visits of patients and the concepts in these visits were identified using person_id along with visit_occurrence_id from the visit_occurrence table. We used visit-based counts instead of patient-based counts in the following analyses for robust comparison between cohorts that had different observation windows. For example, if we use patient-based counts, the patients included in cohorts with longer observation windows are likely to be observed with more concepts than the patients in cohorts with shorter observation windows, which could inject bias into the metrics used in the analyses. Thus, we used visit-based counts to mitigate the effect of different observation window lengths to reduce the bias.
Concept Prevalence and Concept Prevalence Ratio Analysis
We calculated the concept prevalence in each cohort as detected from the EHR. The concept prevalence is defined as in equation (1).
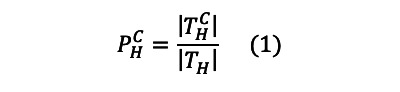 |
where 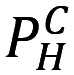 is the prevalence of concept C in cohort H,
is the prevalence of concept C in cohort H, 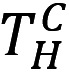 is the set of unique inpatient visits of patients in cohort H observed with concept C, and TH is the set of unique inpatient visits of patients in cohort H. We also calculated hierarchical concept prevalence by defining
is the set of unique inpatient visits of patients in cohort H observed with concept C, and TH is the set of unique inpatient visits of patients in cohort H. We also calculated hierarchical concept prevalence by defining 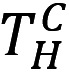 as the set of unique visits of patients observed with concept C or any of concept C’s descendant concepts as defined in the concept_ancestor table in the OMOP CDM. For example, the hierarchical count for Ibuprofen (OMOP concept ID 1177480) not only includes entries where the specific concept Ibuprofen was used, but also includes entries using other descendant concepts, such as Ibuprofen 600 MG Oral Tablet (OMOP concept ID 19019073). Taking hierarchical relationships into account mitigates some of the issues with coding variations across time and practices as different concepts with minor semantic differences can be aggregated into higher-level concepts.
as the set of unique visits of patients observed with concept C or any of concept C’s descendant concepts as defined in the concept_ancestor table in the OMOP CDM. For example, the hierarchical count for Ibuprofen (OMOP concept ID 1177480) not only includes entries where the specific concept Ibuprofen was used, but also includes entries using other descendant concepts, such as Ibuprofen 600 MG Oral Tablet (OMOP concept ID 19019073). Taking hierarchical relationships into account mitigates some of the issues with coding variations across time and practices as different concepts with minor semantic differences can be aggregated into higher-level concepts.
The concept prevalence ratio indicates how frequently concept C occurs in cohort A relative to cohort B. The natural logarithm of the concept prevalence ratio is defined as in equation (2).
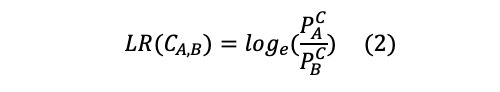 |
where LR(CA,B) is the log ratio of the prevalence of concept C for cohort A to cohort B, 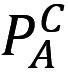 is prevalence of concept C in cohort A, and
is prevalence of concept C in cohort A, and 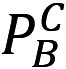 is prevalence of concept C in cohort B. Hierarchical concept prevalence ratio can be calculated by using hierarchical concept prevalence
is prevalence of concept C in cohort B. Hierarchical concept prevalence ratio can be calculated by using hierarchical concept prevalence 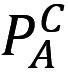 and
and 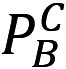 .
.
Concept Co-occurrence Analysis
Concept co-occurrence represents how frequently a specific concept pair appears in a cohort. We defined concept co-occurrence prevalence as in equation (3).
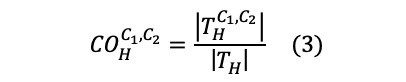 |
where 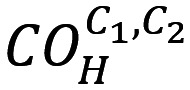 is the co-occurrence prevalence of concepts C1 and C2 in cohort H,
is the co-occurrence prevalence of concepts C1 and C2 in cohort H, 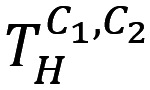 is the set of unique visits of patients observed with concept C1 and C2 in cohort H, and TH is the set of unique visits of patients in the cohort H. We also calculated hierarchical concept co-occurrence using the hierarchy of concepts as described above.
is the set of unique visits of patients observed with concept C1 and C2 in cohort H, and TH is the set of unique visits of patients in the cohort H. We also calculated hierarchical concept co-occurrence using the hierarchy of concepts as described above.
COVID-19 Symptom Prevalence Analysis
Since clinical symptoms often include multiple granular clinical concepts, a set of related concepts for a symptom is required to calculate the prevalence of the symptom. For example, dyspnea, which is one of the major symptoms of COVID-19, can be detected as standard concept Dyspnea or Acute respiratory distress in different patients. The 2 concepts do not have any hierarchical relationship and thus will not be aggregated by the hierarchical prevalence analyses. Thus, a concept set containing both Dyspnea and Acute respiratory distress is needed for accurate calculation of the prevalence of dyspnea. We defined symptom prevalence as in equation (4).
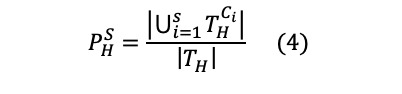 |
where 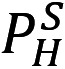 is prevalence of symptom S in cohort H,
is prevalence of symptom S in cohort H, 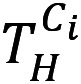 is the set of unique visits of patients observed with concept Ci, TH is the set of unique visits of patients observed in the cohort H, s is the number of the unique concepts in the concept set for symptom S, and ∪ is the union operator. Hierarchy between concepts is not considered in the symptom prevalence analysis since a concept set of a symptom already reflects hierarchy for that symptom.
is the set of unique visits of patients observed with concept Ci, TH is the set of unique visits of patients observed in the cohort H, s is the number of the unique concepts in the concept set for symptom S, and ∪ is the union operator. Hierarchy between concepts is not considered in the symptom prevalence analysis since a concept set of a symptom already reflects hierarchy for that symptom.
We used concept sets for 11 major symptoms of COVID-19 (Cough, Chills, Abdominal pain, Diarrhea, Dyspnea, Fatigue, Fever, Myalgia, Nausea and vomiting, Tachypnea, and Throat pain), which have been defined by OHDSI to calculate symptom prevalence. The concepts included in each symptom are available in Figshare [15].
Data Quality Analysis
Assessing the quality of EHR data is critical for its effects on secondary analysis for research in the health care and medical domains [16]. We calculated the sum of nonrandomized counts of all concepts on a monthly basis for the COVID-19 cohort and on a yearly basis for the general and influenza cohorts. For each of the condition, drug, and procedure domains, we examined the total counts of concepts with the number of visits to detect any issues regarding data quality and temporal plausibility of the EHR data we used in the study [17]. We also calculated the annual mean (SD) values of concept prevalence and concept co-occurrence for the general and influenza cohorts to assess the temporal variance of each concept and concept co-occurrence pair.
Results
Results Overview
In this section, we show a sample of the results of analyses using the data from COHD-COVID. Since COHD-COVID contains massive amounts of data covering several thousand concepts, it is worth noting that only a small sample of the results is shown in this section. Users can obtain the results of interest in addition to the results shown in this section by using the COHD-COVID API [14] or by downloading the flat data files from Figshare [15]. The results of all analyses, concept definitions, and concepts included in each symptom are available as tab-delimited flat files in Figshare, except the concept prevalence ratio analyses, since they can be directly computed from concept prevalence data. COHD-COVID API [14] provides all results.
Concept Prevalence Analysis
Figure 2 shows the prevalence of 10 condition (Figure 2A) and drug concepts (Figure 2B) in the COVID-19 cohort, influenza cohort, and general cohort. We chose the 10 most prevalent condition and drug concepts in the COVID-19 cohort for this use case. For drug concepts, we chose the most prevalent drug ingredient concepts using hierarchical analysis to count multiple drugs that have different brand names, dosages, and formulations but are based on the same ingredients together. The condition concepts were chosen without hierarchical analysis to identify the top 10 specific conditions. Fever showed the highest prevalence (0.2619) among all condition concepts for the COVID-19 cohort, followed by Cough (0.2491) and Dyspnea (0.2594). Acetaminophen was the most prevalent (0.7912) drug ingredient used for patients with COVID-19, followed by Enoxaparin (0.5803) and Glucose (0.4424). Figures 3 and 4 show the prevalence of 10 condition and drug concepts, respectively, in the COVID-19 cohort stratified by age (Figures 3A and 4A) and gender (Figure 4A and 4B). The 10 condition and drug concepts were the top 10 most prevalent concepts in the full COVID-19 cohort without stratification.
Figure 2.
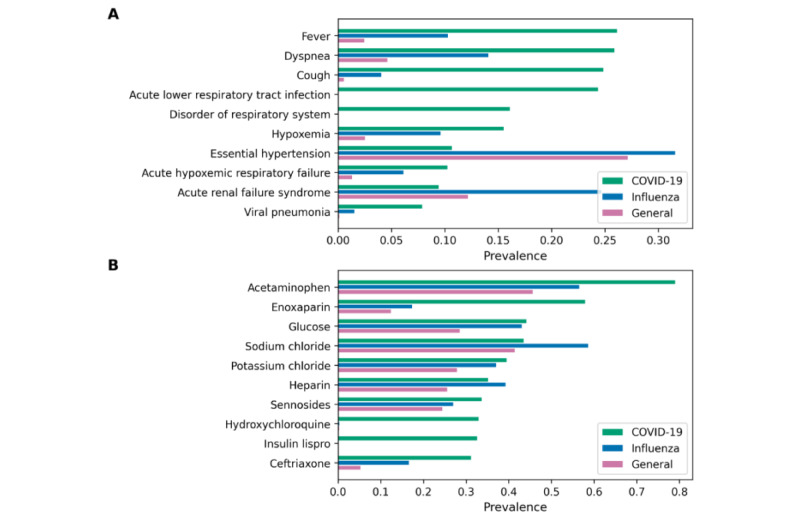
(A) Condition and (B) drug concept prevalence in the COVID-19 cohort, influenza cohort, and general cohort.
Figure 3.
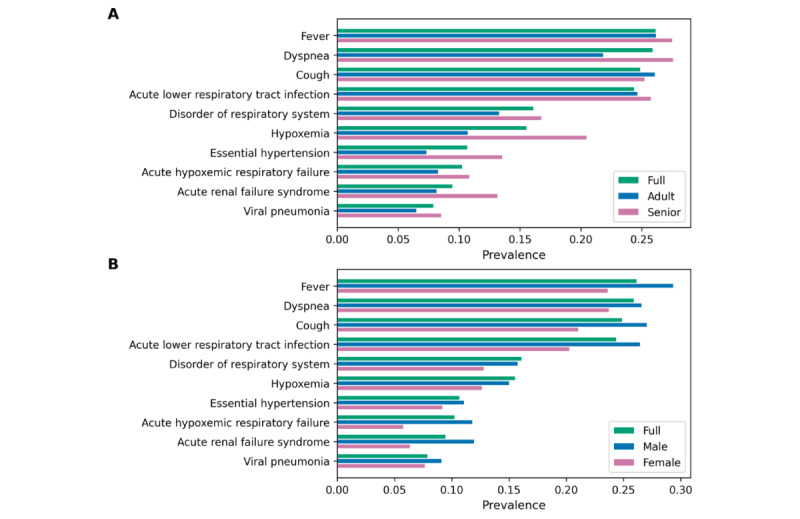
Condition concept prevalence in (A) age and (B) sex sub-cohorts of the COVID-19 cohort. The full COVID-19 cohort indicates original COVID-19 cohort without stratification.
Figure 4.
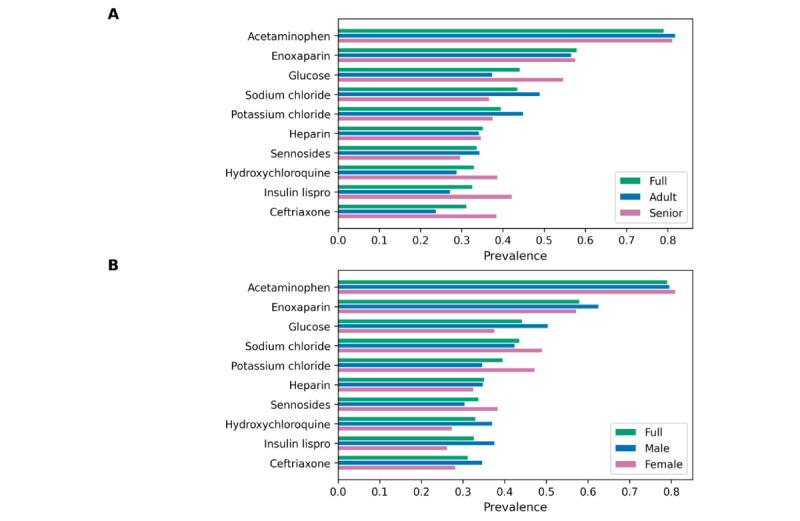
Drug concept prevalence in (A) age and (B) sex subcohorts of the COVID-19 cohort. The full COVID-19 cohort indicates original COVID-19 cohort without stratification.
Concept Prevalence Ratio Analysis
Table 2 shows the top 10 condition concepts that showed the highest concept prevalence ratio for the COVID-19 cohort relative to the comparator cohorts (general cohort and influenza cohort). Disease due to Coronavirus and Acute lower respiratory tract infection showed the highest concept prevalence ratio for the COVID-19 cohort relative to the influenza cohort and the general cohort, respectively.
Table 2.
Top 10 condition concepts that show the highest concept prevalence ratio for the COVID-19 cohort relative to the general and influenza cohorts.
| Influenza cohort (prevalence ratio) | General cohort (prevalence ratio) |
| Disease due to Coronavirus (3.28) | Acute lower respiratory tract infection (7.30) |
| Disorientated (2.44) | Disorder of respiratory system (5.56) |
| Blood chemistry abnormal (2.25) | General finding of observation of patient (5.17) |
| Acute respiratory distress syndrome (2.16) | Outcome of delivery – finding (5.13) |
| Cerebral infarction (1.84) | Chest pain on breathing (5.09) |
| Cough (1.81) | Disease due to Coronavirus (4.95) |
| Viral pneumonia (1.61) | Patient status finding (4.60) |
| Acute respiratory distress (1.52) | Unplanned pregnancy (4.23) |
| Heart disease (1.32) | Acute respiratory distress syndrome (4.17) |
| Delivery normal (1.28) | Deliveries by cesarean (3.98) |
Concept Co-occurrence Analysis
Table 3 shows the top 10 most frequently co-occurring concepts with Acute lower respiratory tract infection in the COVID-19 cohort. Acute lower respiratory tract infection was chosen among the 10 most prevalent condition concepts in the COVID-19 cohort. We used nonhierarchical concept co-occurrence to obtain the result.
Table 3.
The 10 concepts that most frequently co-occurred with Acute lower respiratory tract infection in the full COVID-19 cohort.
| Concept name | Co-occurrence prevalence |
| Disease due to Coronavirus | 0.2329 |
| Radiologic examination, chest; single view | 0.2214 |
| Infectious agent detection by nucleic acid (DNA or RNA); severe acute respiratory syndrome coronavirus 2 | 0.2205 |
| Acetaminophen 325 MG Oral Tablet | 0.2037 |
| 0.4 ML Enoxaparin sodium 100 MG/ML Prefilled Syringe | 0.1504 |
| Hydroxychloroquine Sulfate 200 MG Oral Tablet | 0.1246 |
| 50 ML Glucose 500 MG/ML Prefilled Syringe | 0.1085 |
| Ceftriaxone 1000 MG Injection | 0.1073 |
| Blood typing, serologic; ABO | 0.1044 |
| Glucose 0.4 MG/MG Oral Gel | 0.0970 |
COVID-19 Symptom Prevalence Analysis
Figure 5 shows the prevalence of the 11 major COVID-19 symptoms for all 3 cohorts. In the COVID-19 cohort, Dyspnea showed the highest prevalence among the 11 symptoms followed by Fever and Cough.
Figure 5.
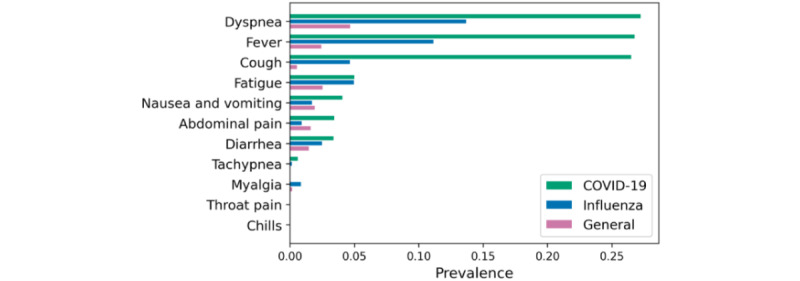
Symptom prevalence of 11 major symptoms in patients with COVID-19 for all 3 cohorts.
Data Quality Analysis
Figure 6 shows the total counts across all concepts in the condition domain (Figure 6A), drug domain (Figure 6B), procedure domain (Figure 6C), and the total visits per month between March 2020 and August 2020 (Figure 6D) for the COVID-19 cohort. The total counts of conditions, drugs, procedures, and total visits all show steep increases in March and April 2020, when the number of COVID-19 cases surged in NYC (Figure 6E) [18]. The total counts of all domains and the total visits decreased starting May 2020 as the number of patients with COVID-19 in NYC decreased.
Figure 6.
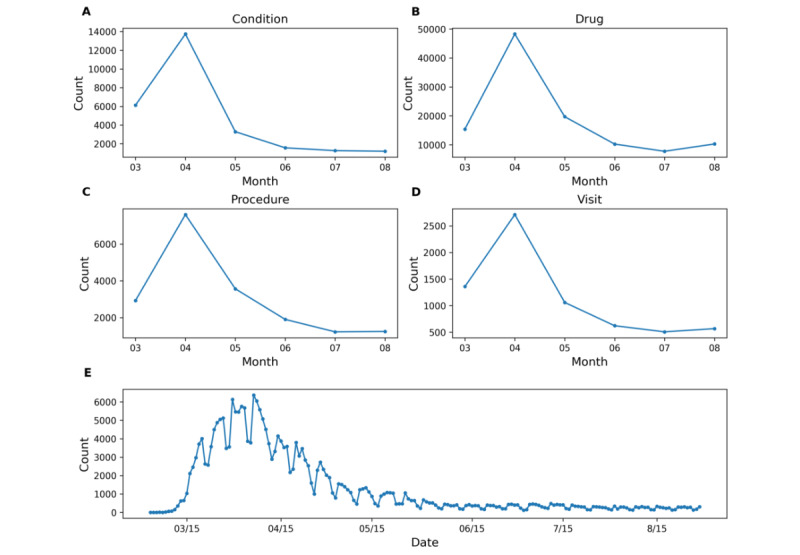
Total counts across all concepts in the (A) condition domain, (B) drug domain, (C) procedure domain, and (D) the total visits per month between March 2020 and August 2020 for the COVID-19 cohort. (E) Total COVID-19–positive cases per day in New York City from March 2020 to August 2020.
Figure 7 shows the total counts per year across all concepts in the condition domain (Figure 7A), drug domain (Figure 7B), procedure domain (Figure 7C), and the total visits per year between 2014 and 2019 (Figure 7D) for the general cohort and influenza cohort. The total counts of conditions, drugs, procedures, and total visits per year for the 2 cohorts show consistent trends.
Figure 7.
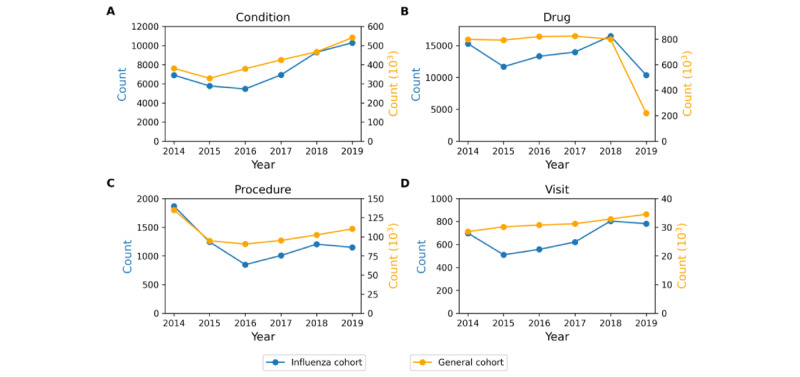
Total counts across all concepts in the (A) condition domain, (B) drug domain, (C) procedure domain, and (D) the total visits per year between 2014 and 2019 for the general and influenza cohort.
The annual mean (SD) value of concept prevalence and concept co-occurrence for the general and influenza cohorts are available in Figshare [15] to assess the temporal variance of each concept and concept co-occurrence pair. The mean and standard deviation of annual concept prevalence and co-occurrence should only be compared to each other to assess the stability of the concept over the given time period of the data set.
A change in the EHR system can affect the quality and characteristics of EHR data collected for secondary research. NYP/CUIMC changed its EHR system as of February 1, 2020, from Allscripts to Epic, which might affect the COVID-19 cohort data and its characteristics as opposed to the influenza and general cohorts, which were collected prior to the EHR change. To detect the impact of the change, we performed a t test for all concepts reported in the COVID-19 cohort between the counts from a pre-Epic period (January 1, 2020, to January 31, 2020) and from a post-Epic period (February 1, 2020, to February 29, 2020). The post-Epic period was chosen to minimize the inclusion of dates when COVID-19 would have impacted clinical practices in NYC. The counts of concepts were recorded on a daily basis. We considered that there would be no difference between the 2 periods if the counts for the concept from the 2 periods are the same (ie, all 0 counts for the 2 periods). Of all 1066 unique concepts reported in the COVID-19 cohort, 119 (11.2%) concepts showed a significant difference (P≤.01) between the pre- and post-Epic periods. The P values of the t tests for all individual concepts are available in Figshare [15] to allow users to factor in these data quality considerations for each concept.
Discussion
Principal Findings
In this study, we present a publicly accessible database providing clinical concept prevalence, clinical concept co-occurrence, and clinical symptom prevalence for hospitalized patients with COVID-19, hospitalized patients with influenza, and general hospitalized patients. We showed the utility of the data for future data-driven studies by exploring the results of the concept prevalence analysis, concept prevalence ratio analysis, concept co-occurrence analysis, and symptom prevalence analysis. Results from the analyses aligned with published findings and can be used to find novel hypotheses as described in the following discussion. The results of all analyses were provided as flat files in Figshare [15] and also easily accessible through web API [14].
Concept prevalence analysis on the 3 cohorts can be used to determine the clinical characteristics of the COVID-19 cohort. A comparison of the prevalence of these concepts between the COVID-19 cohort and the influenza or general cohorts provides contextual evidence regarding whether the concept is associated with COVID-19 or if it is common among hospitalized patients. Figure 2A shows that Fever, Cough, Dyspnea, and Acute lower respiratory syndrome were more highly prevalent in the COVID-19 cohort than in the 2 comparator cohorts. The concepts highly prevalent in the COVID-19 cohort were well-known symptoms of COVID-19, concurrent with existing studies. As shown in Figure 2B, Hydroxychloriquine, which was one of the drugs widely administered to patients with COVID-19 [19] during the early months of the pandemic, showed high prevalence in the COVID-19 cohort.
Concept prevalence analysis on the COVID-19 cohort and its subcohorts can be used to determine the clinical characteristics of the subcohorts and determine if these characters differ between the sexes or between adult and older patients. Figure 3A shows that the senior cohort had higher prevalences in all 10 condition concepts than the full and adult cohorts and also shows that Essential hypertension and Hypoxemia were particularly more prevalent in senior patients with COVID-19, which indicates that senior patients are more likely to have these apparent symptoms of COVID-19 than adult patients. Figure 4A shows that drug ingredients related to type 2 diabetes (eg, Glucose and Insulin Lispro) showed higher prevalence in senior patients than in adult patients, which indicates that type 2 diabetes is a more common comorbidity among senior patients with COVID-19.
Concept prevalence ratio analysis can be used to unveil how often specific concepts appeared in the COVID-19 cohort compared to the comparator cohorts. Table 2 shows that common and general symptoms of COVID-19 (eg, Chest pain on breathing, Cough, and Disease due to coronavirus) showed a high prevalence ratio in the COVID-19 cohort relative to the general cohort. In contrast, more severe symptoms of COVID-19 (eg, Disoriented, Acute respiratory distress syndrome, Viral pneumonia, and Blood chemistry abnormal) showed a higher prevalence ratio in the COVID-19 cohort than in the influenza cohort. Since both cohorts include hospitalized patients, these results may indicate that the aforementioned conditions are more strongly associated with the SARS-CoV-2 infection than with influenza. We also observed high prevalence ratios in concepts related to delivery in the COVID-19 cohort relative to the general cohort. Regrading Outcome of delivery – finding; for example, we observed a visit prevalence rate of 1.18% in the COVID-19 cohort, but only 0.0070% in the general cohort, yielding a prevalence ratio of 5.13. During normal times, deliveries only account for a small fraction of all inpatient visits. However, during the pandemic, there was a dramatic decrease in elective surgeries and hospitalizations besides those related to COVID-19 since hospital capacity was diverted their focus toward patients with COVID-19. In contrast, patients going into labor cannot be rescheduled and were regularly tested for SARS-CoV-2, which increased their representation within the COVID-19 cohort.
Potential associations between specific concepts of interest can be found through co-occurrence analysis. For instance, Table 3 confirms that the concepts associated with SARS-CoV-2 tests (ie, Disease due to Coronavirus, Radiologic examination, chest; single view, and Infectious agent detection by nucleic acid [DNA or RNA]; severe acute respiratory syndrome coronavirus 2) showed high co-occurrence prevalence with Acute lower respiratory tract infection, which shows natural strong associations between testing concepts for COVID-19 and one of the most prevalent concepts in the COVID-19 cohort.
Symptom prevalence analysis can be used to examine symptom-level characteristics of the cohorts. The COVID-19 cohort showed higher prevalence in dyspnea, fever, and cough symptoms than the other 2 comparator cohorts, which is concurrent with the known characteristics and symptoms of COVID-19 [20]. In contrast, a few of the known COVID-19 symptoms (eg, myalgia, throat pain, and chills) did now show high prevalence in the COVID-19 cohort, which might indicate that those symptoms are not clinically distinctively prevalent in patients with COVID-19.
Most of the results from the analyses align with those of existing studies, thus empirically validating the utility of COHD-COVID. COHD-COVID also can be used to find novel hypotheses related COVID-19. COHD-COVID can be used as cross-institutional data to validate or support other COVID-19 studies. For instance, the high prevalence ratio of Cerebral infarction in the COVID-19 cohort compared to that in the influenza cohort (Table 2) corroborates with reports from a few studies that SARS-CoV-2 might be more likely to cause thrombotic vascular events, including stroke, than other coronavirus and seasonal infectious diseases [21,22].
While we admit that the aggregated concept-level analyses may not be suitable to answer some clinical research questions, COHD-COVID will be useful for hypothesis generation and for validating emerging newly published findings on COVID-19 using real-world data. COHD, the precedent study of COHD-COVID, has been integrated into the National Center for Advancing Translational Sciences Biomedical Data Translator program, where the EHR-based data associations from COHD are linked to other sources of knowledge via knowledge graphs, allowing automated algorithms to perform reasoning on these knowledge graphs to answer biomedical questions and suggest novel hypotheses [23,24].
Limitations
There are several limitations to this study, which should be noted. One of the limitations is that analyses performed in this study can be affected by the factors included in the data acquisition process (eg, change in the EHR system, human biases, and errors during entry). For example, coding trends and patterns (ie, the trend and pattern of frequently used concepts) can be changed through a shift in system. Another limitation is that multiple visits from the same patient can be used to calculate the metrics in the analyses since we used visit-based counts instead of patient-based counts for the analyses. This could affect the results of the specific concepts appearing in patients who are hospitalized more frequently. We also admit that some of results from the analyses cannot be validated by existing or up-to-date findings, considering rapidly growing and changing knowledge related to COVID-19. Thus, it is desirable to conduct a literature search before utilizing the data and results. We will update the data and results on a regular basis to further alleviate this limitation.
The EHR data used in this study were obtained from a single site: NYP/CUIMC. Even though NYP/CUIMC is a large academic medical center whose services cover the city and its surrounding areas, we admit that performing the analyses on the basis of the EHR data across multiple institutions and nations will be beneficial since multiple sites can diversify the population, improve accuracy, increase power and sensitivity to rare conditions, validate results by comparing across sites, and reduce variance that might exist in specific locations. Since the OMOP CDM provides the fundamentals to perform the same analyses on clinical data across different sites, we hope to collaborate with future studies sharing clinical characteristics of patients with COVID-19 and to generate a larger, richer, and more robust database that can be leveraged in translational research on COVID-19.
Conclusions
In this study, we present COHD-COVID, a publicly accessible database providing useful clinical data for hospitalized patients with COVID-19, hospitalized patients with influenza, and general hospitalized patients. The analyses using the data from COHD-COVID confirmed the well-known clinical characteristics of patients with COVID-19 and can also be used to find novel hypotheses related to COVID-19. We expect COHD-COVID will provide researchers and clinicians quantitative measures of COVID-19–related clinical features to better understand and combat the pandemic.
Acknowledgments
This study was supported by National Library of Medicine grant R01LM012895-03S1 and National Center for Advancing Translational Science grant 1OT2TR003434-01.
Abbreviations
- API
application programming interface
- COHD
Columbia Open Health Data
- COHD-COVID
Columbia Open Health Data for COVID-19 Research
- EHR
Electronic health records
- NYP/CUIMC
NewYork-Presbyterian/Columbia University Irving Medical Center
- OHDSI
Observational Health Data Science and Informatics
- OMOP CDM
Observational Medical Outcomes Partnership Common Data Model
Footnotes
Conflicts of Interest: None declared.
References
- 1.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 Mar 28;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. http://europepmc.org/abstract/MED/32171076 .S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah B, Modi P, Sagar SR. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020 Jul 01;252:117652. doi: 10.1016/j.lfs.2020.117652. http://europepmc.org/abstract/MED/32278693 .S0024-3205(20)30400-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burn E, You SC, Sena AG, Kostka K, Abedtash H, Abrahão MTF, Alberga A, Alghoul H, Alser O, Alshammari TM, Aragon M, Areia C, Banda JM, Cho J, Culhane AC, Davydov A, DeFalco FJ, Duarte-Salles T, DuVall S, Falconer T, Fernandez-Bertolin S, Gao W, Golozar A, Hardin J, Hripcsak G, Huser V, Jeon H, Jing Y, Jung CY, Kaas-Hansen BS, Kaduk D, Kent S, Kim Y, Kolovos S, Lane JCE, Lee H, Lynch KE, Makadia R, Matheny ME, Mehta PP, Morales DR, Natarajan K, Nyberg F, Ostropolets A, Park RW, Park J, Posada JD, Prats-Uribe A, Rao G, Reich C, Rho Y, Rijnbeek P, Schilling LM, Schuemie M, Shah NH, Shoaibi A, Song S, Spotnitz M, Suchard MA, Swerdel JN, Vizcaya D, Volpe S, Wen H, Williams AE, Yimer BB, Zhang L, Zhuk O, Prieto-Alhambra D, Ryan P. Deep phenotyping of 34,128 adult patients hospitalised with COVID-19 in an international network study. Nat Commun. 2020 Oct 06;11(1):5009. doi: 10.1038/s41467-020-18849-z. doi: 10.1038/s41467-020-18849-z.10.1038/s41467-020-18849-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B, Gutierrez B, Mekaru S, Sewalk K, Goodwin L, Loskill A, Cohn EL, Hswen Y, Hill SC, Cobo MM, Zarebski AE, Li S, Wu C, Hulland E, Morgan JD, Wang L, O'Brien K, Scarpino SV, Brownstein JS, Pybus OG, Pigott DM, Kraemer MUG. Epidemiological data from the COVID-19 outbreak, real-time case information. Sci Data. 2020 Mar 24;7(1):106. doi: 10.1038/s41597-020-0448-0. doi: 10.1038/s41597-020-0448-0.10.1038/s41597-020-0448-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasell J, Mathieu E, Beltekian D, Macdonald B, Giattino C, Ortiz-Ospina E, Roser M, Ritchie H. A cross-country database of COVID-19 testing. Sci Data. 2020 Oct 08;7(1):345. doi: 10.1038/s41597-020-00688-8. doi: 10.1038/s41597-020-00688-8.10.1038/s41597-020-00688-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Q, Jones FK, Leavitt SV, Ung L, Labrique AB, Peters DH, Lee EC, Azman AS, HIT-COVID Collaboration HIT-COVID, a global database tracking public health interventions to COVID-19. Sci Data. 2020 Aug 27;7(1):286. doi: 10.1038/s41597-020-00610-2. doi: 10.1038/s41597-020-00610-2.10.1038/s41597-020-00610-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desvars-Larrive A, Dervic E, Haug N, Niederkrotenthaler T, Chen J, Di Natale A, Lasser J, Gliga DS, Roux A, Sorger J, Chakraborty A, Ten A, Dervic A, Pacheco A, Jurczak A, Cserjan D, Lederhilger D, Bulska D, Berishaj D, Tames EF, Álvarez FS, Takriti H, Korbel J, Reddish J, Grzymała-Moszczyńska J, Stangl J, Hadziavdic L, Stoeger L, Gooriah L, Geyrhofer L, Ferreira MR, Bartoszek M, Vierlinger R, Holder S, Haberfellner S, Ahne V, Reisch V, Servedio VDP, Chen X, Pocasangre-Orellana XM, Garncarek Z, Garcia D, Thurner S. A structured open dataset of government interventions in response to COVID-19. Sci Data. 2020 Aug 27;7(1):285. doi: 10.1038/s41597-020-00609-9. doi: 10.1038/s41597-020-00609-9.10.1038/s41597-020-00609-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Argenziano MG, Bruce SL, Slater CL, Tiao JR, Baldwin MR, Barr RG, Chang BP, Chau KH, Choi JJ, Gavin N, Goyal P, Mills AM, Patel AA, Romney MS, Safford MM, Schluger NW, Sengupta S, Sobieszczyk ME, Zucker JE, Asadourian PA, Bell FM, Boyd R, Cohen MF, Colquhoun MI, Colville LA, de Jonge JH, Dershowitz LB, Dey SA, Eiseman KA, Girvin ZP, Goni DT, Harb AA, Herzik N, Householder S, Karaaslan LE, Lee H, Lieberman E, Ling A, Lu R, Shou AY, Sisti AC, Snow ZE, Sperring CP, Xiong Y, Zhou HW, Natarajan K, Hripcsak G, Chen R. Characterization and clinical course of 1000 patients with coronavirus disease 2019 in New York: retrospective case series. BMJ. 2020 May 29;369:m1996. doi: 10.1136/bmj.m1996. http://www.bmj.com/lookup/pmidlookup?view=long&pmid=32471884 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benitez K, Malin B. Evaluating re-identification risks with respect to the HIPAA privacy rule. J Am Med Inform Assoc. 2010;17(2):169–177. doi: 10.1136/jamia.2009.000026. http://europepmc.org/abstract/MED/20190059 .17/2/169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ta CN, Dumontier M, Hripcsak G, Tatonetti NP, Weng C. Columbia Open Health Data, clinical concept prevalence and co-occurrence from electronic health records. Sci Data. 2018 Nov 27;5:180273. doi: 10.1038/sdata.2018.273. doi: 10.1038/sdata.2018.273.sdata2018273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahalt SC, Chute CG, Fecho K, Glusman G, Hadlock J, Taylor CO, Pfaff ER, Robinson PN, Solbrig H, Ta C, Tatonetti N, Weng C, Biomedical Data Translator Consortium Clinical Data: Sources and Types, Regulatory Constraints, Applications. Clin Transl Sci. 2019 Jul;12(4):329–333. doi: 10.1111/cts.12638. http://europepmc.org/abstract/MED/31074176 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fecho K, Ahalt SC, Arunachalam S, Champion J, Chute CG, Davis S, Gersing K, Glusman G, Hadlock J, Lee J, Pfaff E, Robinson M, Sid E, Ta C, Xu H, Zhu R, Zhu Q, Peden DB, Biomedical Data Translator Consortium Sex, obesity, diabetes, and exposure to particulate matter among patients with severe asthma: Scientific insights from a comparative analysis of open clinical data sources during a five-day hackathon. J Biomed Inform. 2019 Dec;100:103325. doi: 10.1016/j.jbi.2019.103325. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(19)30244-8 .S1532-0464(19)30244-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyal NP. First Case of COVID-19 in NYC, First Death Reported in Washington State. Infectious Disease Advisor. 2020. Mar 03, [2020-11-01]. https://www.infectiousdiseaseadvisor.com/home/topics/respiratory/first-case-of-covid-19-in-nyc-first-death-reported-in-washington-state/
- 14.Columbia Open Health Data - COVID-19. [2021-09-20]. https://covid.cohd.io/ [DOI] [PMC free article] [PubMed]
- 15.Lee J, Kim J, Liu C, Hripcsak G, Ta C, Weng C. COHD-COVID: Columbia Open Health Data for COVID-19 Research. Figshare. [2021-09-20]. [DOI] [PMC free article] [PubMed]
- 16.Ta CN, Weng C. Detecting Systemic Data Quality Issues in Electronic Health Records. Stud Health Technol Inform. 2019 Aug 21;264:383–387. doi: 10.3233/SHTI190248. http://europepmc.org/abstract/MED/31437950 .SHTI190248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn MG, Callahan TJ, Barnard J, Bauck AE, Brown J, Davidson BN, Estiri H, Goerg C, Holve E, Johnson SG, Liaw S, Hamilton-Lopez M, Meeker D, Ong TC, Ryan P, Shang N, Weiskopf NG, Weng C, Zozus MN, Schilling L. A Harmonized Data Quality Assessment Terminology and Framework for the Secondary Use of Electronic Health Record Data. EGEMS (Wash DC) 2016;4(1):1244. doi: 10.13063/2327-9214.1244. http://europepmc.org/abstract/MED/27713905 .egems1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.COVID-19 Daily Counts of Cases, Hospitalizations, and Deaths. NYC Open Data. [2021-09-20]. https://data.cityofnewyork.us/resource/rc75-m7u3 .
- 19.Geleris J, Sun Y, Platt J, Zucker J, Baldwin M, Hripcsak G, Labella A, Manson DK, Kubin C, Barr RG, Sobieszczyk ME, Schluger NW. Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N Engl J Med. 2020 Jun 18;382(25):2411–2418. doi: 10.1056/NEJMoa2012410. http://europepmc.org/abstract/MED/32379955 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen K, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang J, Liang Z, Peng Y, Wei L, Liu Y, Hu Y, Peng P, Wang J, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N, China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020 Apr 30;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. http://europepmc.org/abstract/MED/32109013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fifi JT, Mocco J. COVID-19 related stroke in young individuals. Lancet Neurol. 2020 Sep;19(9):713–715. doi: 10.1016/S1474-4422(20)30272-6. http://europepmc.org/abstract/MED/32822622 .S1474-4422(20)30272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oxley TJ, Mocco J, Majidi S, Kellner CP, Shoirah H, Singh IP, De Leacy RA, Shigematsu T, Ladner TR, Yaeger KA, Skliut M, Weinberger J, Dangayach NS, Bederson JB, Tuhrim S, Fifi JT. Large-Vessel Stroke as a Presenting Feature of Covid-19 in the Young. N Engl J Med. 2020 May 14;382(20):e60. doi: 10.1056/NEJMc2009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biomedical Data Translator Consortium Toward A Universal Biomedical Data Translator. Clin Transl Sci. 2019 Mar;12(2):86–90. doi: 10.1111/cts.12591. http://europepmc.org/abstract/MED/30412337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biomedical Data Translator Consortium The Biomedical Data Translator Program: Conception, Culture, and Community. Clin Transl Sci. 2019 Mar;12(2):91–94. doi: 10.1111/cts.12592. http://europepmc.org/abstract/MED/30412340 . [DOI] [PMC free article] [PubMed] [Google Scholar]


