Abstract
SARS-CoV-2 virions are surrounded by a lipid bilayer that contains membrane proteins such as spike, responsible for target-cell binding and virus fusion. We found that during SARS-CoV-2 infection, spike becomes lipid modified, through the sequential action of the S-acyltransferases ZDHHC20 and 9. Particularly striking is the rapid acylation of spike on 10 cytosolic cysteines within the ER and Golgi. Using a combination of computational, lipidomics, and biochemical approaches, we show that this massive lipidation controls spike biogenesis and degradation, and drives the formation of localized ordered cholesterol and sphingolipid-rich lipid nanodomains in the early Golgi, where viral budding occurs. Finally, S-acylation of spike allows the formation of viruses with enhanced fusion capacity. Our study points toward S-acylating enzymes and lipid biosynthesis enzymes as novel therapeutic anti-viral targets.
Keywords: SARS, coronavirus, spike, ZDHHC, S-palmitoylation, lipid sorting, virus, lipid microdomains, viral envelope
Graphical abstract

Mesquita et al. report that SARS-CoV-2 spike protein undergoes massive lipid modification during viral biogenesis. This controls the membrane, in particular cholesterol, composition of the envelope and enhances infectivity of the viruses.
Introduction
β-coronaviruses (CoVs) are positive-stranded RNA enveloped viruses that infect a wide range of hosts (V’kovski et al., 2021). Human-tropic species can cause from mild respiratory symptoms to life-threatening forms of atypical pneumonia termed severe acute respiratory syndrome (SARS) (V’kovski et al., 2021; Wang et al., 2020), such as COVID-19 caused by SARS-CoV-2 (SARS coronavirus 2) (Bojkova et al., 2020; Gordon et al., 2020).
The CoV membrane envelope has 3 primary structural proteins: spike (S), membrane (M), and envelope (E) (Boson et al., 2021; Siu et al., 2008; V’kovski et al., 2021) as well as accessory proteins such as Orf3a (Ito et al., 2005; Tan et al., 2004). Viral assembly only requires E and M (Boson et al., 2021; Siu et al., 2008; Vennema et al., 1996). Yet, S is essential for infectivity as it mediates attachment to host cell receptors, angiotensin-converting enzyme 2 (ACE2) for SARS-CoV-2 (Hoffmann et al., 2020b; Letko et al., 2020), and fusion between viral and target cell membranes.
S, E, and M are synthesized in the ER, where folding and quaternary assembly occurs. They may also undergo post-translation modifications such as N-glycosylation on their luminal domains (Watanabe et al., 2020) and S-acylation (commonly referred to as S-palmitoylation) on their cytosolic parts, which occurs for spike and E from different CoVs (Boscarino et al., 2008; Lopez et al., 2008; McBride and Machamer, 2010; Petit et al., 2007; Thorp et al., 2006; Tseng et al., 2014). S-acylation, which consists in the covalent attachment of medium-chain fatty acids (frequently palmitate, C16) to the sulfur atom of cytosolic cysteines, is very frequent in mammalian cells and a hallmark of viral envelope proteins (Gadalla and Veit, 2020; Schmidt, 1982). S-acylation of spike was proposed to mediate its association to lipid microdomains, promote syncytia formation, and increase infectivity of mouse hepatitis virus (MHV) and pseudotyped particles (McBride and Machamer, 2010; Nguyen et al., 2020; Sanders et al., 2020; Thorp et al., 2006).
Here, we present a comprehensive study of spike S-acylation, the identification of the host enzymes involved, and its importance for viral biogenesis and infection. We identify ZDHHC20 as the dominant acyltransferase responsible for lipidation of spike, which plays several roles. In the infected cell, it promotes spike biogenesis by protecting it from premature ER degradation, increases its half-life, and controls the lipid organization of its immediate membrane environment. Once the virus has formed, spike S-acylation controls fusion with the target cell. Lipidomic analyses of SARS-CoV-2 viruses reveal a unique lipid composition, enriched in specific early Golgi sphingolipids. Finally, we show that S-acylation and sphingolipid metabolism promote SARS-CoV-2 infection, pointing toward S-acylation and lipid biogenesis related enzymes as novel potential anti-viral targets.
Results and discussion
Acylation of SARS-CoV-2 viral proteins by ZDHHC20
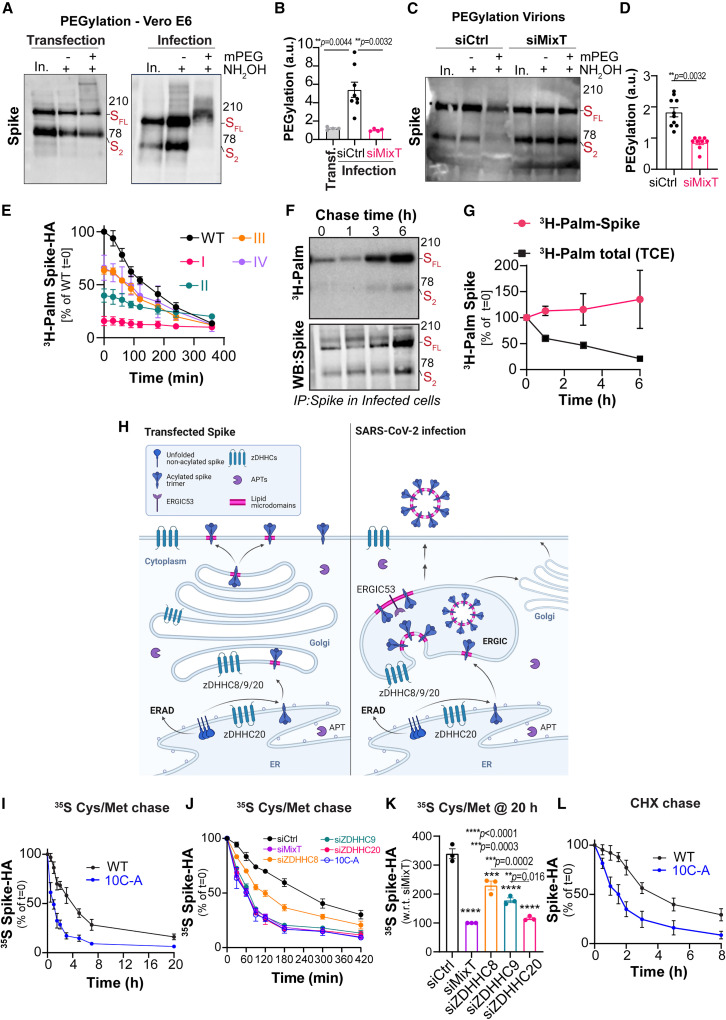
Consistent with findings on SARS-CoV-1 (McBride and Machamer, 2010), spike from SARS-CoV-2 undergoes S-acylation when ectopically expressed in Hela or monkey kidney Vero E6 cells (Figures 1A and 1B) and during infection of Vero E6 cells with SARS-CoV-2 (Figures 1C and 1D), when a large proportion of infected cells displayed high levels of spike (Figure S1A). Infections were performed with the SARS-CoV-2 D614G variant, which appeared early during the pandemic and rapidly spread worldwide (Korber et al., 2020). To monitor protein acylation, we used two established assays. The acyl-resin assisted capture assay (Acylrac) allows the capture of S-acylated proteins using thiol-reactive sepharose beads. Cysteine thiol groups become exposed by a cleavage step with hydroxylamine, which breaks the thioester bond between target proteins and the fatty-acid moiety (Figures 1A and 1C). The second assay consists in incubating cells with radioactive 3H-palmitate and monitoring its incorporation into spike using immunoprecipitation and autoradiographic analysis (Figures 1B and 1D). Several spike bands can be observed because, to become fusion competent, spike undergoes two sequential cleavage steps, mediated by host proteases, at the S1-S2 and S2′ sites. The S2 fragment contains the membrane fusion subunit and the transmembrane anchor (Hoffmann et al., 2020a, 2020b). Western blot analysis of total extracts from infected or spike-transfected cells also showed a small C-terminal fragment (Figures 1A and 1C). This band, which contains the transmembrane domain and the cytosolic tail, has been observed previously and has unknown function (Westerbeck and Machamer, 2019). Acylation of all spike fragments could be readily detected (Figures 1A–1D). Given its small size and high number of cysteines, the small C-terminal fragment showed an accelerated migration when deacylated (Figures 1A and 1C).
Figure 1.
Identification of the spike-modifying ZDHHC acyltransferases
See also Figure S1.
(A) Acylrac capture assay in Vero E6 expressing Spike WT (24 h). Western blot (WB) of total cell extracts (in), S-acylated (plus hydroxylamine +NH2OH) and control fractions (–NH2OH) show full length (FL), S2, and C-terminal spike (C-term).
(B) WB and autoradiography (3H-Palm) of spike-immunoprecipitated (IP) fractions IP from 3H-palmitic acid-labeled Hela or Vero E6 expressing WT or D614G (D/G) spike.
(C and D) Vero E6 infected with SARS-CoV-2, (MOI ≈ 0.1, 24 h) processed for Acylrac (C) or metabolic labeling (for 24 h of infection) (D) as in (B).
(E) Spike-incorporated radioactivity, as in (B), in spike-expressing (48–72 h) siRNA-targeted as indicated HeLa, normalized to control cells (ctrl).
(F) Same as in (E) in HAP-1 knockout for the indicated ZDHHCs.
(G) Same as (E) in Vero E6 siRNA targeted as indicated and infected as in (D).
(H) Same as (F) or (E) in Vero E6 knockout for the indicated ZDHHCs.
Ctrl2 and WT2 refer to independent controls. Results are mean ± SEM, n ≥ 3; p values obtained by one-way ANOVA.
Although S-acylation of enveloped virus proteins has been known for decades, the mechanisms and mediators remain ill-defined. S-acylation is catalyzed by a family of 23 protein acyltransferases defined by the presence of a conserved zinc-finger domain with a DHHC motif (ZDHHCs) (Zaballa and Goot, 2018). To identify the enzyme modifying spike, we individually depleted all ZDHHC enzymes in Hela cells (ZDHHC1 to 24, ZDHHC10 does not exist). Downregulation of three family members, ZDHHC8, 9, and 20, led to a >50% drop in 3H-palmitate incorporation into ectopically expressed spike (Figures 1E and S1B). A subset screen, in near-haploid HAP-1 cells knocked out (KO) for individual ZDHHCs, confirmed these findings and indicated a principal role for ZDHHC20 (Figures 1F and S1C). These screens did not pick up ZDHHC5, which during the course of this study was proposed as a spike acyltransferase based on overexpression experiments (Wu et al., 2021). Spike S-acylation upon depletion of the ZDHHC8, 9, or 20 could be restored by expression of the correspondent siRNA-resistant myc-tagged human enzyme, but not a random acyltransferase (Figure S1D).
We next tested the involvement of the identified ZDHHC enzymes during infection in Vero E6 cells. ZDHHC enzymes are largely ubiquitous, even if their expression levels vary significantly between cell types (Figure S1E). All cell lines we tested, including the human lung cell line Calu-3, expressed ZDHHC20, 9, and 8 (Figure S1E). We first jointly targeted ZDHHC8, 9, and 20 using a monkey-specific siRNA mix (MixT). Acylated spike could no longer be retrieved using the Acylrac capture assay (Figure S1F). We then silenced the enzymes individually and included additional ZDHHCs as controls (Figures 1G and S1G). Unfortunately, efficient depletion (below 50%) of monkey ZDHHC8 also led to a decrease in monkey ZDHHC20 mRNA, an effect we did not observed in human cell lines (Figure S1H). We therefore omitted siZDHHC8 from further analysis in Vero cells but maintained the use of MixT. Knocking down ZDHHC9 and 20 led to a strong decrease in spike S-acylation during infection (Figure 1G). We also observe a mild, although significant, effect for siZDHHC5 (Figures 1G and S1G). This effect is likely indirect since it was not observed in spike-transfected cells and was concomitant to a strong decrease in viral infection and/or spike expression (Figure S1G). Finally, we generated Vero E6 KO cells for each of the 3 enzymes using CRISPR-Cas9. Knocking out one transferase did not affect the levels of others (Figure S1I). S-acylation was monitored during transfection and infection of these cells, leading to the conclusion that ZDHHC20 has the major role of modifying spike during infection, and ZDHHC9 also contributes (Figures 1H, S1J, and S1K).
Rapid and extensive acylation during spike biogenesis
Spike proteins contain a conserved cysteine-rich stretch of amino acids in their cytosolic tail (Figure 2 A) (Gelhaus et al., 2014; Thorp et al., 2006). Spike from SARS-CoV-2 has 10 cysteines within its first 20 cytosolic amino acids (Figure 2A) and is one of the most cysteine-rich proteins encoded by animal viruses (Sanders et al., 2020). We generated four HA-tagged spike mutants changing groups of 2–3 adjacent cysteines to alanine (Figure 2A). In transfected cells, mutation of the two most membrane-proximal cysteines (group I) led to an 80% drop in 3H-palmitate incorporation (Figures 2B and 2C), as previously observed for SARS-CoV-1 spike (Petit et al., 2007). Mutation of the next three cysteines (group II) also had an effect, albeit lower, with a ≈40% drop in 3H-palmitate incorporation (Figures 2B and 2C). Mutating cysteines from both groups I and II had an impact almost equivalent to that of changing all 10 to alanine (Figures 2B and 2C). Yet, cysteines from groups III and IV must also undergo S-acylation, since mutating both groups alone or in conjunction with group II still significantly reduced 3H-palmitate incorporation (Figures 2B and 2C). This analysis suggests that all 10 cysteines are targets of S-acylation, which is primarily controlled by those located adjacent to the membrane. Possibly, this initial S-acylation brings the more distal cysteines closer to the membrane, more accessible to the membrane-localized ZDHHC active site.
Figure 2.
ZDHHC20 initiates acylation on the two juxtamembranous cysteines
See also Figure S2.
(A) Alignment of the C-terminal regions of spike (top-human, bottom-animal species). Transmembrane domains-blue and cysteines-yellow split into 4 groups: I, II, III, IV.
(B and C) Western blot (WB) or autoradiography (3H-Palm) of spike-HA-immunoprecipitation (IP) fractions from 3H-palmitic-acid-labeled Hela expressing spike-HA variants (WT, or Cys-to-Ala mutants individual or combined as depicted in (A); in 10C-A all cysteines were mutated).
(C) Spike-HA-incorporated radioactivity normalized to WT samples (WT2). Results are mean ± SEM, n ≥ 4.
(D) Same as in (C) in cells labeled as indicated. Values for WT at 15 min were set to 100%. Results are mean ± SD, n = 3.
(E and F) Same as in (D) in Vero E6 infected with SARS-CoV-2 (MOI ≈ 0.5–1) for 4 h. Values were normalized to the increasing levels of IP-spike and set to 100% for t = 30 min. Results are mean ± SD, n = 3. Parallel experiments as in (D) using untagged WT spike transfected in HeLa are also displayed.
(G) Same as in (B) in Hela pre-treated (1 h) with cycloheximide (CHX), brefeldin A (BrfA), or Mock-medium before metabolic labeling as in (C). Values for Mock-treated cells were set to 100% for each variant. Results are mean ± SEM, n = 4.
(H) Same as in (D) in HeLa expressing untagged spike and siRNA-targeted as indicated. Results are mean ± SD, n = 3.
(I) Same as (B) in Vero ZDHHC-KO cells processed as in (G) expressing spike-HA variant III-IV (with Cys-groups I and II). Values for Mock-treated cells were set to 100% for each KO-cell line. Results are mean ± SEM, n = 3. p values were obtained by (C) one-way or (G and I) two-way ANOVA.
We next measured 3H-palmitate incorporation kinetics in HeLa cells expressing various HA-tagged spike mutants. Incorporation of 3H-palmitate into WT spike was rapid, reaching a plateau within about 3 h, in great contrast with group-I and group-II mutated variants, where it was very slow and limited (Figure 2D). Mutation of groups III and IV cysteines had no major effect on S-palmitoylation kinetics but resulted in lower final levels of incorporation, consistent with the removal of 3 or 2 acceptor sites, respectively (Figure 2D).
We also compared 3H-palmitate incorporation into spike between transfection in HeLa cells and single-round infection in Vero E6 cells (Figures 2E, 2F, and S2A). We used a higher MOI for infection (≈0.5 to 1) and added radioactive 3H-palmitate 4 h post-viral inoculation, when spike started being detected by flow cytometry and significant viral RNA replication was observed by qPCR (Figures S2B–S2D). Remarkably, the kinetics of spike 3H-palmitate incorporation were essentially identical for infected and transfected cells (Figure 2F). Note that the presence of a C-terminal HA tag had no consequences on these measurements (Figures 2D and 2F).
Next, we investigated where spike acylation occurs during viral biogenesis. After synthesis, folding, and assembly, spike trimers are transported to the ER-Golgi intermediate compartment (ERGIC) where viral assembly and budding take place (de Haan and Rottier, 2005; Klein et al., 2020; Stertz et al., 2007). To determine if spike acylation occurs during protein biogenesis, and/or after transport from the ER to the early, pre-Golgi, we monitored 3H-palmitate incorporation, into various HA-tagged spike versions, under the effects of the protein synthesis inhibitor cycloheximide (CHX), and the Golgi-disrupting drug brefeldin A (BrfA). Palmitate incorporation into WT spike was decreased by ≈50% when cells were treated with either CHX or BrfA (Figure 2G), suggesting that S-palmitoylation is initiated shortly after protein synthesis and continues once the protein has exited the ER. This is consistent with previous reports on SARS-CoV-1 and MHV spike proteins indicating that their palmitoylated versions were present in both pre- and post-Golgi compartments (McBride and Machamer, 2010; van Berlo et al., 1987). We then compared the effect of the drugs on the S-palmitoylation of two additional mutants, bearing respectively only the most membrane-proximal 2 (groups II, III, and IV mutated) or 5 cysteines (groups III and IV mutated) (Figure 2G). With the former mutant, spike sensitivity to BrfA was lost, suggesting that acylation of the 2 juxta-membrane cysteines occurs in the ER, whereas the latter mutant, with the 5 first cysteines, had a profile similar to that of WT indicating that more distal cysteines can be modified in downstream compartments.
We also monitored the kinetics of 3H-palmitate incorporation when silencing the spike-modifying ZDHHC enzymes. ZDHHC20 has a broad distribution throughout the entire biosynthetic pathway (Figure S2E), ZDHHC9 appears to be more restricted to the ER, and ZDHHC8 is more concentrated in the perinuclear-Golgi region (Figure S2E). Silencing any of the three enzymes in HeLa cells had a drastic effect on 3H-palmitate incorporation, but silencing ZDHHC20 caused the earliest and most dramatic effect (Figure 2H), phenocopying the group-I cysteine mutant (Figure 2D). Finally, we monitored 3H-palmitate incorporation of the mutant harboring only the 5 most membrane-proximal cysteines (III-IV mutant) in our Vero E6 KO cells. As in HeLa cells (Figure 2G), 3H-palmitate incorporation was diminished by ≈ 40% upon BrfA treatment of control cells (Figure 2I). When ZDHHC20 was knocked out, BrfA further suppressed palmitoylation, indicating that exit from the ER was required for optimal acylation. In contrast, as soon as ZDHHC20 was present, but either ZDHHC 8 or 9 were absent, the BrfA sensitivity was lost (Figure 2I).
These results indicate that following synthesis, spike can undergo acylation on its two membrane-adjacent cysteines in the ER by ZDHHC20, a step required for the modification of the more distal cysteines by ZDHHC20 and 9 in the ER and Golgi. While ZDHHC8 has the ability to modify spike, this does not occur during infection of Vero E6 cells (Figure 1H).
The above experiments demonstrate that S-acylation occurs, but they do not indicate what percentage of spike molecules undergo modification. A variant of Acylrac has been developed in which after removal of the acyl chains using hydroxylamine, the newly freed up cysteines were labeled with maleimide-PEG (5 kDa - PEGylation), leading to molecular weight shifts (Howie et al., 2014; Percher et al., 2017). For small proteins, each PEGylation reaction leads to an approximate 5-kDa shift in the SDS-PAGE migration pattern. When proteins are large and have a large number of acylation sites, the molecular weight shifts are less predictable. When spike was transfected into Vero E6 cells, we observed a weak high molecular-weight smear, but the full-length or S2 spike bands remained essentially unaltered (Figure 3 A, left panel). In contrast, PEGylation of proteins from infected cells led to the disappearance of the S2 band (Figure 3A, right panel). It is unclear where exactly the S2 band shifted to. It could be the band migrating slightly higher than the full-length form. Extensive PEGylation may also hinder the full-length spike from entering into the SDS gels. Our interpretation of the disappearance of the S2 band is that all spike molecules become highly acylated following synthesis, during infection, before they are cleaved into S2. A kinetic analysis of PEGylation indicated that spike S-acylation occurred as early as 2 h after viral inoculation (Figure S3A), consistent with the rapid dynamics observed previously (Figures 2E and 2F). PEGylation on the virion containing supernatants of infected Vero E6 cells showed similar shifts (Figures 3C and 3D), which were abolished when cells were silenced with MixT or ZDHHC20 siRNA (Figures 3B–3D and S3B). Correspondingly, capturing of acylated spike from supernatants of infected cells was also abrogated when cells were silenced with MixT (Figure S3C). Thus, the entire spike population produced by infected cells undergoes massive and rapid acylation, apparently on all 10 cysteines.
Figure 3.
S-acylation of spike during infection is extensive and stable
See also Figure S3.
(A) Pegylation in Vero E6 expressing untagged WT spike (24 h, left) or infected with SARS-CoV-2, MOI = 0.1, 24 h (right). Western blot (WB) of input cell extracts (In), Mass-tagged proteins (+mPEG), and control (−mPEG) after hydroxylamine treatment (NH2OH).
(B) Quantification of spike-PEG band shifts shown in (A) and siRNA-targeted as indicated. Results are mean ± SEM, n ≥ 4.
(C and D) Pegylation of pre-cleared, filtered supernatants from infected samples as in (B). Results are mean ± SEM, n = 10.
(E) Spike-HA-incorporated radioactivity from 3H-palmitic acid-labeled Hela cells, 24 h after transfection with the indicated HA-tagged spike mutant variants, chased in complete medium. Values for WT at T = 0 min set to 100%. Results are mean ± SD, n = 3.
(F) Same as (E) in Vero E6 infected with SARS-CoV-2, (MOI ≈ 0.5–1) labeled from 1 to 14 h post-viral inoculation (p.i.). Cells were chased for up to 6 h (until 20 h p.i.). Values, normalized to IP-spike levels, were set to 100% for T = 0 min. Values of radioactivity incorporated by total cell extracts (TCE) are also displayed (see Figure S3F). Results are mean ± SD, n = 3.
(H) Model of the cellular trajectories of spike during transfection versus SARS-CoV-2 infection (BioRender.com).
(I and J) Hela expressing spike-HA (WT or 10C-A mutant) metabolically labeled with 35S-Met/Cys for 4 h and chased as indicated. In (J) cells were siRNA-treated as indicated. Quantification of IP-spike-HA-incorporated radioactivity was set to 100% for T = 0 min for each sample. Results are mean ± SD, (I) n = 4 and (J) n = 3.
(K) Same as in (J), following 20 h of chase normalized (with respect to [w.r.t.]) to 100% for siMixT displayed as mean ± SD, n = 3.
(L) HeLa expressing spike as in (I) were treated with cycloheximide (CHX) and extracts analyzed at indicated time points by WB. Spike full length (FL) expressed relative to T = 0 set as 100%. Results are mean ± SD, n = 4. All p values were obtained by one-way ANOVA or (D) by unpaired Student’s t test.
Spike acylation is stable during infection
S-acylation can be reversed by the action of deacylating enzymes, the acyl protein thioesterases (APTs) (Zaballa and Goot, 2018). Using a metabolic 3H-palmitate pulse-and-chase approach in transfected HeLa cells, we found that WT HA-tagged spike lost its palmitate with a half-life of about 3 h, as did group III and IV mutants (Figures 3E and S3D). This process could be blocked using the APT inhibitor palmostatin B (Figures S3D and S3E).
We performed a similar 3H-palmitate decay analysis during SARS-CoV-2 infection. 3H-Palmitate was added from 1 to 14 h post-inoculation. At 14 h, infected Vero E6 cells were further incubated in label-free medium. While depalmitoylation of cellular proteins (detected by autoradiography of total cell extracts) was readily observed (Figures S3F and S3G), there was no detectable loss of palmitate from spike (Figures 3F and 3G), indicating that spike deacylation barely occurs during infection. Possibly, spike is rapidly segregated from acyl thioesterases during formation of virions in the early Golgi, while the spike cysteines remain cytosolically exposed to deacylation in transfected cells (Figure 3H). Alternatively, the virus could affect the activity of APTs.
S-acylation promotes the biogenesis of spike and reduces its turnover rate
We next tested whether S-acylation modified the turnover of spike. We used two methods: 35S Cys/Met metabolic pulse-chase experiments, which monitor the evolution of a protein starting from its synthesis, and cycloheximide chase, which monitors the degradation of fully folded mature proteins. The 35S Cys/Met pulse period was chosen based on the plateau of palmitate incorporation, i.e., 4 h (Figure 2D). The decay in the 35S signal observed for WT spike indicated an apparent half-life of ≈4 h, while that observed for the 10C-A acylation-deficient mutant was ≈1 h (Figures 3I and S3G). The accelerated early decay for the 10C-A mutant indicates that this variant is rapidly targeted to degradation in the ER, whereas acylation protects the WT protein. A previous study did not reveal an effect of S-acylation on the degradation of spike from SARS-CoV-1 (McBride and Machamer, 2010). In these experiments, the 35S Cys/Met labeling time was short, 20 min, but we know now this is insufficient to significantly populate the acylated spike species (Figure 2D) (Abrami et al., 2019). Acceleration of spike degradation following its synthesis was also observed when silencing ZDHHC9 and/or 20 (Figure 3J). Comparison of the total levels 35S Cys/Met-labeled spike at 20 h of chase showed that acylation, primarily by ZDHHC20, leads to a 3-fold increase in total amount of spike (Figure 3K). In addition, through the cyloheximide chase approach, we found that S-acylation also slows down the turnover of mature, fully folded spike, increasing the half-life from about 2 h to more than 4 h (Figures 3L and S3H).
Therefore, by increasing the flux of newly synthesized spike through the ER quality control gateway and by slowing down the turnover of the fully folded protein, S-acylation dually increases the levels of spike.
Spike S-acylation modifies the organization of the lipid bilayer
Following synthesis, spike assembles into trimers (Delmas and Laude, 1990), which for SARS-CoV-2 can bear 30 fatty acids. Based on cryo-electron tomography studies (Bar-On et al., 2020; Ke et al., 2020; Klein et al., 2020), 25 to 130 spike trimers are present per virion. With an 80–90-nm virus diameter and an average headgroup surface of 70 Å2 per phospholipid, a “back-of-the-envelop” calculation indicates that the acyl chains attached to spike could represent 2%–12% of the lipid molecules of the inner leaflet of the viral membrane.
Given this very significant contribution of palmitate, we analyzed the possible impact of spike acylation on the surrounding lipid organization, using coarse-grained (CG) molecular dynamics (MDs) simulations. We compared the behavior of palmitoylated and non-palmitoylated spike in a model membrane. We chose a membrane composition known to capture lipid nanodomain formation (Lorent et al., 2017; Risselada and Marrink, 2008): 50% dipalmitoylphosphatidylcholine (DPPC), 30% dilinoleylphosphatidylcholine (DLiPC), and 20% cholesterol (CHOL), and parameterized it using the MARTINI CG force field. We first reproduced the results by Lorent et al. (2017) of microdomain formation (a liquid-disordered DLiPC-rich phase and a liquid-ordered DPPC+CHOL-rich phase) within ≈1 μs when starting from randomly distributed lipids. We subsequently prepared two systems, again with lipids distributed randomly, and introduced the transmembrane domain (TMD) of spike followed by the cytosolic tail residues (residues 1,201 to the end, Uniprot P0DTC2), either fully S-palmitoylated or not at all. We ran each system ten times for ≈3.7–3.8 μs in CG MD simulations, starting from different random seeds (Figure 4 A). Analysis of the MD simulations led to two predictions. While the cytoplasmic tails of the protomers were initially introduced in an extended form, acylation led to the collapse of the cytosolic tails, bringing the palmitate moieties in close proximity of the TMD and, leading to a ≈9-fold local increase in the local acyl chain concentration (Figures 4A and 4B). Such compaction is not an artifact of the CG description of the protein, as it was also observed in atomistic MD simulations using the CHARMM36m force field (Figure 4B). Spike preferentially associated with the disordered phase when unmodified, but rather partitioned into the ordered phase when S-palmitoylated. This preference was dictated by the cytosolic half of the TMD (Figures 4A, 4B, and S4A). In contrast, the luminal half of the TMD interacted preferentially with the disordered phase, irrespective of spike acylation (Figures 4B and S4A). These simulations raised the interesting possibility, not tested here, that acylated spike could drive trans-bilayer lipid asymmetry.
Figure 4.
S-Acylation controls the lipid environment of spike
See also Figure S4.
(A) Coarse-grained (CG) model of spike’s transmembrane (TM) helix + C-terminal region (residues 1,201-end in UniProt P0DTC2) in trimeric form, inserted into a DPPC-(yellow)-DLiPC-(red)-cholesterol-(green) membrane model. Left, starting conditions: random lipid positions and stretched cytoplasmic tail; center, representative end state of same view after 3.75 μs of CG MD simulation of the S-palmitoylated protein depicts phase separation and spike partitioning. Right, side view of the same model after 2 μs of atomistic MD simulation in the S-palmitoylated form in a POPC membrane.
(B) Quantification of contacts at equilibrium between the full TM helix, the luminal half, or the cytosolic half, and each lipid phase. Results are means ± SD (n = 10).
(C) Vero E6 expressing spike-HA WT surface labeled with biotin and processed for fractionation. Western blot (WB) of biotinylated proteins (Surface-Strp. Pull-down) precipitated from each fraction were compared to unbound fractions (intracellular-unbound).
(D) Quantification of spike-HA (all forms) in each fraction relative to the total signal in all 6 fractions. Results are mean ± SD, n = 3.
(E and F) High-throughput automated microscopy of Vero E6 expressing spike-HA (WT, or 10C-A mutant) for 24 h immunolabeled for HA and stained with Filipin. (E) Total levels of filipin staining within HA-positive cells (or per cell—mock sample) quantified per number of nuclei. The data were averaged for 49 frames over 8 wells per condition. Each dot represents one independent well, and results are mean ± SEM. Equivalent results were obtained for three independent assays.
(G) Vero E6 infected with SARS-CoV-2 24 h, MOI ≈ 0.1 were incubated with beta-methylcyclodextrin (βMCD) or not (Ctrl) for 30 min. Cell extracts were fractionated as in (C). DRM marker Caveolin1 (Cav1)-positive control, transferrin receptor (TfR)-negative control.
(H and I) Quantification of spike forms (H) and caveolin 1 (I) signals as in (C). Results are mean ± SD, n = 3.
(J) Vero E6 transfected with indicated siRNA (siCtrl or siMixT pool), infected and fractionated as in (G).
(K–M) Quantification of (K) and (L). Spike (all forms) and (M) ERGIC53 in each fraction as in (C). Results are mean ± SD, n = 3. All p values were obtained by two-way (except for E, one-way) ANOVA.
S-acylation of spike drives its association with ordered lipid nanodomains
Given the predicted compaction of the saturated acyl chains around the spike TMD, the local lipid organization is expected to be ordered. Consistently, spike from SARS-CoV-2 associates with detergent-resistant membranes (DRMs), in a process that depends on its cytosolic cysteine residues (Figures S4B and S4C) as observed for SARS-CoV-1 (McBride and Machamer, 2010). DRMs, of which caveolin-1 is a marker, have been considered a useful biochemical readout for the association of proteins with cholesterol-rich lipid domains, even though not equivalent to the ordered lipid domains in cells (Levental et al., 2020). Such domains are predominantly found in the plasma membrane, the endosomal system and the late Golgi (van Meer et al., 2008). The ER and the early Golgi are presumed to be devoid of such domains since their cholesterol and sphingolipid contents are very low (Levental et al., 2020). Therefore, the ERGIC, the intermediate compartment between the ER and the Golgi, into which CoVs bud (Klein et al., 2020), is not expected to contain cholesterol-rich lipid domains.
To probe the lipid environment of spike, we first tested whether transfected intracellular spike could be found in DRMs. For this, we biotinylated the surface proteins of transfected cells, prepared DRMs, and performed streptavidin pull-downs on each fraction to separate plasma membrane (bound) from intracellular (unbound) proteins. Intracellular WT spike was exclusively found in the DRM fractions, while cell surCace WT spike was distributed throughout the gradient (Figures 4C and 4D). The intracellular 10C-A mutant, in contrast, was detergent soluble (Figures S4B–S4E) and less expressed at the cell surface (Figures S4F and S4G).
We next analyzed the distribution of cholesterol in spike-expressing cells using the cholesterol-binding fungal metabolite, filipin. Filipin staining was more than 2-fold higher in WT than in acylation-deficient 10C-A Vero E6 cells expressing spike (Figures 4E and 4F). Staining was atypically concentrated in the perinuclear area (Figure 4F) and expression of WT, but not 10C-A spike, promoted cell-to-cell fusion leading to the formation of large multi-nucleated syncytia, as reported (Hoffmann et al., 2020a; McBride and Machamer, 2010) (Figures 4F and S4H).
We next performed the DRM analysis on SARS-CoV-2-infected cells, and spike was exclusively found in DRMs (Figures 4G and 4H). Spike-DRM association appeared mostly intracellular as indicated by its resistance to cholesterol extraction from the plasma membrane using addition of β-methylcyclodextrin (ßMCD) (Figures 4G and 4H). Partitioning of the viral M protein also remain unaltered, while ßMCD treatment did solubilize caveolin-1, which is predominantly found at the plasma membrane (Figures 4G and 4I). The DRM association of spike was lost when cells were treated with the MixT siRNA pool or ZDHHC20 siRNA (Figures 4J–4L and S4I). In contrast, silencing of ZDHHC9 had no effect (Figures 4L and S4I). Thus ZDHHC20-mediated acylation of spike in SARS-CoV-2-infected cells leads to it association with DRMs, presumably derived from the ERGIC and newly formed intracellular virions.
As mentioned, the ERGIC is not expected to have cholesterol- and sphingolipid-rich, ordered domains. We therefore hypothesized that acylation of spike might drive the formation of such domains in the ERGIC. To test this, we monitored the behavior of ERGIC53, a transmembrane protein that accumulates in the ERGIC. In control cells, ERGIC53 was detergent soluble (Figure S4J) as expected. In contrast, ERGIC53 was mostly detected in DRMs when cells were infected with SARS-CoV-2 (Figures 4J and 4M). This was due to protein acylation since ERGIC53 was detergent soluble in infected cells treated with the siRNA MixT pool, or siZDHHC20 but not siZDHHC9 (Figures 4J, 4M, and S4I). Thus, during infection, S-acylated spike drives the formation of ordered lipid domains in the ERGIC.
S-acylation of spike controls the local lipid environment in viral-like particles
We next investigated whether S-acylation of spike influences its lipid environment in the context of two minimal viral particle assembly systems: an HIV-derived lentivector-based system to generate spike harboring pseudo particles (PPs) (Fenwick et al., 2021) (Figure S5A) and a SARS-CoV-based system to generate viral-like particles (VLPs) (Kumar et al., 2021; Qing et al., 2020, 2021). In both systems, the particles produced include a built-in luminescence-based reporter system, which allows for evaluation of particle production and subsequent infectivity. VLPs and PPs were produced in similar quantities irrespective of spike acylation (Figures 5A and S5B). Incorporation (S to N or M ratios) of spike and cleavage status in CoV VLPs was also equivalent for WT and palmitoylation-deficient (10C-A) (Figures 5B–5D). The presence of the cytosolic cysteine residues did however influence incorporation of spike and cleavage status in PPs (Figures S5C–S5E). Of note, the interaction between spike and M within cells expressing the CoV VLP system was not dependent on spike acylation (Figure S5F) as proposed for SARS-CoV-1 spike (McBride and Machamer, 2010). Acylation of spike was confirmed in both PPs and VLPs (Figures 5E and S5G), and spike associated with DRMs in both types of particles, in a cysteine-dependent manner (Figures 5F, 5G, S5H, and S5I). Presence of S-acylated spike also promoted the localization of both M and E to DRMs (Figures 5F and 5H). Correspondently, in PPs, the lentiviral protein P17 was found in DRMs when these where pseudo-typed with WT spike (Figures S5H and S5J). Finally, we observed that ERGIC53 also became DRM-associated in cells producing VLPs (Figures 5I and 5J), further indicating that spike can alter the organization of its local lipid environment.
Figure 5.
Spike S-acylation controls lipid organization of VLPs
See also Figure S5.
(A) Untyped (Bald) or spike-HA (WT or 10C-A) pseudotyped VLPs purified from supernatants of HEK293T (see STAR Methods). Normalized VLP released (mean ± SEM) was determined by quantification of Nluc-bioluminescence from 5 independent preparations.
(B–D) Western blot (WB) of cell extracts and VLPs: non-typed (bald) or VLPs pseudotyped with spike-HA (WT or 10C-A mutant). GAPDH used as cellular-control.
(C) Spike-HA levels in VLPs normalized against N or M and set to 1 for WT.
(D) Spike cleavage in VLPs quantified as the ratio between S2 and S2 + S full length. Results are mean ± SEM, n = 3.
(E) Acylrac capture assay in purified VLPs and correspondent cell extracts isolated as in (B). Total cell extracts (input, In), S-palmitoylated proteins (+NH2OH), and control fractions (−NH2OH) analyzed by WB against HA (spike).
(F) WB of fractions from concentrated VLPs processed for fractionation and DRM isolation.
(G and H) Quantification of (G). Spike-HA (all forms) or (H) M in each fraction divided by the total signal in the 6 fractions. Results are mean ± SD, n = 3.
(I and J) Same as in (F) from lysates of VLP-producing HEK293T. Fractions were analyzed by WB and ERGIC53 levels in each fraction quantified as in (G) and (H). Results are mean ± SD, n = 3. p values were obtained by two-way ANOVA.
Unusual lipid composition of SARS-CoV-2 virions
We next extended our biochemical analyses to SARS-CoV-2 virions. Virions were purified from infected Vero E6 cells (48 h post-inoculation) silenced for acyltransferase expression using MixT. Similar levels of spike and M in virions and cells were observed irrespective of acylation, with a small, yet, significant decrease of the spike to M ratio in siMixT-derived virions (Figures 6A and 6B). The relative proportion of the S2 fragment compared with the total amount of spike was also largely unaltered by siMixT (Figure 6C). Well-defined viruses were observed by cryoelectron microscopy under both conditions, with spike complexes in the characteristic prefusion conformation (Figure 6D). The virions obtained from siMixT-treated cells, however, appeared less homogeneous in size and slightly but significantly larger on average (Figures 6D and 6E). We modeled the tails of the distribution of virion diameters using a generalized Pareto distribution, which enables the analysis of the exceedances of a particular dataset. The shape factor (ξ) for virions derived from control cells was negative, of −0.20 ±0.19, indicating a low propensity to generate extreme diameters (Figures 6E and S6A). In contrast, for virions derived from siMixT-treated cells, ξ was 0.64 ± 0.34, clearly showing an increased frequency of extreme diameters (Figures 6E and S6B). Thus, the ZDHHC acyltransferases 8, 9, and 20 contribute to the tight control of viral particle dimensions.
Figure 6.
Impact of S-acylation on SARS-CoV-2 virion biogenesis
See also Figure S6. SARS-CoV-2 virion suspensions purified and concentrated from infected (48 h, MOI ≈ 0.1) Vero E6 siRNA-targeted as indicated (siMixT, pool-ZDHHC8/9/20).
(A) Samples analyzed by western blot (WB) for spike, M, and GAPDH.
(B and C) Quantification of the spike/M ratio, set up to 1 for controls, and the ratio between S2 and S2 + S full length in samples obtained in (A). Results are mean ± SEM, n = 3.
(D) PFA-fixed virion suspensions, concentrated as in (A) and analyzed by cryoelectronic microscopy. Scale bars, 100 nm.
(E) Frequency distribution of virion diameters (Bin width 5 nm), quantified over 300 (siCtrl) and 336 virions (siMixT) harvested from 2 independent preparations. Results are mean ± SD. A generalized Pareto distribution analysis of the tail frequency shows an increase of extreme diameters (shape factor ξ) for siMixT samples.
(F) Concentrated virions processed for fractionation and DRM isolation and analyzed by WB.
(G) Quantification of spike (all forms) in each fraction obtained in (F) divided by the total spike signal in all 6 fractions. Results are mean ± SD from n = 3.
(H) Volcano plots of lipid molecular species levels in producer Vero E6 vs SARS-CoV-2 virions. The log2 fold change (FC) is plotted against the −log10 p value. Dots represent color coded specific lipid species: HexCer, Hexasil ceramide; SM, sphingomyelin; Cer, ceramides; PA, phosphatic acid; TAG, triglycerides, PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PG, phosphatidylglycerol; PI, phosphatidylinositol; and other SL, sphingolipids.
(I and J) Volcano plots as in (H) from cells transfected with siRNA oligos as in (A) siCtrl (I) or siMixT (J) with virion-specific sphingolipid enrichments (red).
(K) Virion/cell lipid ratios of samples produced in (I) and (J).
(L) Fluorometric cholesterol and cholesterol-ester analysis of lysates from virions produced in (I) and (J). Equal volumes analyzed with comparable levels of M protein as in (A). Results are mean ± SEM, n = 3. All p values were obtained by (B), (C), (E), and (L) Student’s t test and (G) two-way ANOVA.
We next analyzed the DRM association of spike within SARS-CoV-2 envelopes. Virions were submitted to detergent solubilization in the cold. Note that for both VLPs and virions, the detergent to membrane ratio is higher than when solubilizing cells, due to the low amount of material, which generates a high risk of over-solubilization. Nevertheless, spike was found in DRMs, as observed for VLPs and PPs, in a process that was indeed dependent on S-acylation, since sensitive to MixT (Figures 6F and 6G).
We next determined the lipid composition of SARS-CoV-2 virions and compared it with that of parental cells. Virions were deprived of non-membrane lipids (triglycerides >80% less represented), poor in glycerolipids (25% less represented), such as phosphatidylserine, and enriched in sphingolipids (40% more represented) (Figures 6H and S6C–S6G). They were also enriched in phosphatidic acid, which displays a small headgroup and a bulky acyl tail that contributes to negative membrane curvature and could influence intraluminal vesicle budding (Egea-Jimenez and Zimmermann, 2018; Zhukovsky et al., 2019). Most striking, hexosylceramide (HexCer) species (most likely glucosylceramide-GlcCer) were found to be on average 330% more represented in virions than in parental cells (Figures 6H and S6C). GlcCer, the precursor of glycosphingolipids, is produced on the cytosolic side of early Golgi cisternae where GlcCer synthase resides (Halter et al., 2007). Once produced, GlcCer is translocated to the lumenal leaflet of either Golgi cisternae or, following non-vesicular transport, of the TGN (D’Angelo et al., 2007, 2013). In the luminal leaflet of the Golgi/TGN membrane, GlcCer is readily converted to lactosylceramide and complex glycosphingolipids. Thus, GlcCer primarily populates the early segment of the biosynthetic pathway. It is widely accepted that virions retain/select a lipid composition for their envelope characteristic of their budding site, and this has been used as a mean to determine the lipid composition of the plasma membrane of polarized cells, for example (Heaton and Randall, 2011; Ketter and Randall, 2019). Our analysis therefore provides insight into the lipid composition of the SARS-CoV-2-budding site and shows biochemically that these viruses bud in the early secretory pathway, consistent with previous morphological studies (Klein et al., 2020; Stertz et al., 2007). The overall lipid composition of the virions was not significantly modified by MixT (Figures 6I, 6K, and S6C–S6G; Table S3). Silencing of the acyltransferases did however lead to a major drop, 30%, of the relative cholesterol amounts in virions and VLPs, (Figures 6L and S6H), fully consistent with the observation that spike S-acylation promotes the formation of ordered lipid nanodomains in the ERGIC (Figure 4). These observations suggest that acyl moieties recruit cholesterol in close proximity to spike, triggering the formation of nanodomains, in particular in the inner leaflet of the bilayer, by attracting lipids such as sphingolipids from the neighboring membrane. Indeed, the overall sphingolipid composition was not altered.
Thus, S-acylation of spike does not drastically affect the overall lipid composition of SARS-CoV-2 viruses but instead modifies the lipid environment in its vicinity, promoting the formation of cholesterol-rich lipid domains. This local effect is consistent the sparse distribution of spike trimers in the viral membrane (Ke et al., 2020; Klein et al., 2020; Yao et al., 2020).
S-acylation is essential for SARS-CoV-2 fusion with the host cell
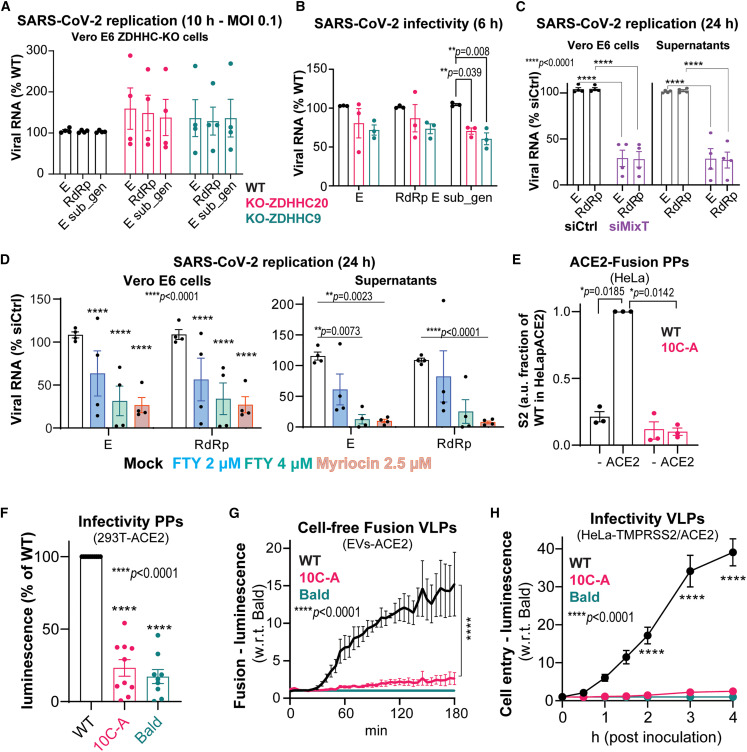
Having determined the importance of spike S-acylation for the lipid organization of SARS-CoV-2 virions, our aim was to investigate its role in infection. In view of generating virions from ZDHHC20 and ZDHHC9 KO cells, we first analyzed whether these cells were still competent for SARS-CoV-2 infection, i.e., could the virus enter and deliver its genome for replication. For this, we choose a 10-h infection at a MOI of ∼0.1. We monitored the viral RNA for E and ribosomal dependent RNA polymerase (RdRP), as well as sub-genomic E RNA (E-sub_gen). We found no inhibition of viral replication in any of the ZDHHC-KOs (Figure 7 A). The slight increase observed at 10 h in ZDHHC20-KO cells was lost at later times of infection (Figure S7A). Thus, ZDHHC9 or 20 activity is not required in the target cells for viral entry, fusion, and early genome replication.
Figure 7.
S-acylation enhanced infectivity of spike-typed VLP and SARS-CoV-2
See also Figure S7.
(A) Vero E6, WT, or knockout as indicated were infected with SARS-CoV-2, MOI ≈ 0.1 for 10 h. Total viral RNA (E, RdRp) and sub-genomic viral RNA (E, sub-gen) quantified by qPCR. Results are mean ± SEM, and each dot represent the average RNA levels of an independent experiment.
(B) Virion supernatants harvested from cells infected as in (A) adjusted to comparable viral RNA content used to re-infect naive Vero E6 using ≈10 viral E copies per cell. Infectivity assessed by quantifying viral RNA as in (A) 6 h post-inoculation (p.i.). Results are mean ± SEM, n = 3.
(C) Quantification of viral RNA from Vero E6, and correspondent supernatants, siRNA-targeted as indicated (siMixT: pool siZDHHC8/9/20) and infected with SARS-CoV-2 MOI ≈ 0.1 for 24 h.
(D) Quantification of viral RNA as in (C) in Vero E6 treated with 2.5 μM of myriocin for ≈5 days before and during infection or treated with the indicated concentrations of FTY720 from 1 hpi. Control cells were treated with equivalent volume of DMSO. For (C) and (D), results are mean ± SEM, n = 4.
(E) Spike-HA-pseudotyped (WT or 10C-A mutant) PPs suspensions harvested from HEK293T, with adjusted spike and p17 content were used for synchronized fusion assays in HeLa or HeLa-ACE2. Surface-attached particles were removed by acid wash and internalized spike S2 signal quantified by western blot (see Figure S7C) and expressed as fraction of WT-PPs signal in HeLa-ACE-2. Results are mean ± SEM, n = 3.
(F) PPs suspensions processed as in (E) and adjusted to p24 content were used to transduce 293T-ACE2. Transduction efficiencies quantified and normalized to WT are mean ± SEM, n = 7.
(G and H) VLPs suspensions with adjusted HiBiT-N input multiplicities used for (G) Cell-free VLP-extracellular vesicles fusion assays or (H) cell entry assays into ACE2-LgBiT/hTMPRSS2 target cells. Time course data relative to the fusion or cell entry levels of control untyped (bald) VLPs are mean ± SEM, n = 3. p values were obtained by two-way ANOVA.
We next analyzed the infectivity of viruses produced by the ZDHHC-KO cells. Cell supernatants were adjusted to comparable amounts of viral RNA (≈ 10 E copies per target cell). The infection levels were analyzed 6 h after viral inoculation, focusing on the sub-genomic E RNA, a bona fide marker of intracellular viral RNA replication. The infectivity of virions produced from either ZDHHC20 and ZDHHC9 KO cells was 30% to 40% lower than that of control virions, based on sub-genomic E RNA, respectively, with a similar trend for RdRp and E RNA (Figure 7B). Thus, virions produced from cells lacking either ZDHHC20 or 9 are not as infectious as virions produced from control cells. To affect both enzymes together, we performed a longer infection, 24 h, i.e., involving consecutive rounds of entry, replication, viral biogenesis, and release (>2 to 3), at an MOI of 0.1, on cells treated with MixT. Both the RNAs of E and RdRp dropped by ≈ 75% (Figure 7C). The viral RNAs also dramatically dropped in the supernatants of these cells, which contain released virions (Figure 7C). These observations confirm the importance of ZDHHC20 and 9 for SARS-CoV-2 infectivity.
Next, we tested whether pharmacological inhibition of sphingolipid biogenesis, which prevents formation of cholesterol and sphingolipid-rich domains, could also affect SARS-CoV-2 infection. We used the fungal metabolite myriocin, which targets serine palmitoyltransferase-1 at early steps of sphingolipid biosynthesis and fully depletes sphingolipids in cells in 5 days. We also tested Fingolimod (FTY720), a drug that derives from myriocin and is approved for treatment of multiple sclerosis. FTY720 modulates the intracellular levels of sphingolipids, whithin shorter treatment times, by targeting ceramide synthases, which are downstream serine palmitoyltransferase-1 (Berdyshev et al., 2009; Lahiri et al., 2009). Vero E6 cells were either treated with myriocin for 5 days and subsequently infected with SARS-CoV-2 for 24 h or first infected with SARS-CoV-2 and exposed to FTY720 1 h after inoculation for the remaining 24 h. Both treatments, myriocin and FTY720, phenocopied the MixT treatment, leading to a drastic reduction of viral RNA both in the cells and in supernatants after 24 h of infection (Figure 7D). Thus, disruption of sphingolipid cascades recapitulates the inhibition of S-acylation during infection.
Finally, in order to specifically assess the role of spike acylation, we made use of the two viral particle systems, PPs and SARS-CoV VLPs. To analyze binding to the ACE-2 receptor, we compared the competitive binding between spike-pseudotyped PPs (WT or 10C-A) and purified spike receptor-binding domain (RBD), using a modified human ACE2 ELISA kit. Both WT and 10-CA pseudotyped PPs displayed equivalent levels of competition with purified RBD, whereas bald PPs did not display significant binding (Figure S7B). Next, we performed synchronized fusion assays by incubating ACE-2-expressing HeLa cells with PP supernatants at 4°C and allowing subsequent fusion at 37°C for 30 min. Prior to cell harvesting, surface-attached particles were removed using a stringent acid-wash and fused and/or internalized particles were monitored by western blot. While it was possible to detect ACE2-dependent fusion of WT spike-S2, no spike 10C-A was detected (Figures 7E and S7C), indicating that 10C-A-pseudotyped PPs were unable to fuse with target cells. We also monitor the infectivity of these PPs by transducing them into ACE2-expressing HEK293. The infectivity of WT spike-pseudotyped PPs was more than four times higher than that of the same amount of 10C-A containing PPs, or bald non-typed particles (Figure 7F). This effect was far more pronounced when compared with the difference in infectivity between spike-pseudotyped PPs produced from control or ZDHHC20-silenced cells (Figure S7D). However, silencing ZDHHC20 did not affect the infectivity of VSVG-pseudotyped PPs (Figure S7E).
We next analyzed the importance of spike acylation for the infectivity of SARS-CoV VLPs. We used a recently described split reporter system engineered into the N genes of the CoV VLPs (Kumar et al., 2021; Qing et al., 2021). Fusion dynamics were monitored by measuring bioluminescence as a function of time, using either TMPRSS2-expressing target cells or a cell-free system based on ACE-2-containing extracellular vesicles, both harboring the complementary reporter fragment tagged to ACE2. For these assays, in order to avoid interference of the HA-epitope tags on viral fusion, VLPs were produced with untagged spike. Spike incorporation (S/N ratio) was unaffected by mutation of the acylation sites (Figure S7F). In both the cell-free fusion assay and the HeLa-ACE2 entry system, mutation of the spike acylation sites drastically reduced VLP fusion, by 20- to 40-fold (Figures 7G and 7H) confirming that spike acylation is essential for efficient viral entry into, and fusion with, target cells.
Conclusion
Here, based on a combination of functional genomics, biochemical, kinetic, and computational studies, we show that during SARS-CoV-2 biogenesis in infected cells, spike protein is rapidly and efficiently S-acylated following its synthesis in the ER, through the sequential action of the ZDHHC20 and ZDHHC9 enzymes. Acylation prevents premature degradation promoting the biogenesis of spike, which is subsequently transported to the ERGIC where it arrives with up to 30 acyl chains decorating each trimer. The presence of these saturated lipid attracts cholesterol (Lenventhal et al., 2020), driving the formation of specific domains around spike. Using VLPs pseudotyped with WT or acylation-deficient spike, we could show that acylation is required for efficient viral fusion. Our findings are consistent and complementary to those by Brangwynne and co-workers who recently identified that cholesterol present in the membrane of PPs is critical for spike-mediated fusion (Sanders et al., 2020). Our study shows that S-acylation constitutes a promising drug target for coronavirus infection. ZDHHC20 is also involved in the acylation of the hemagglutinin of influenza virus (Gadalla et al., 2020), and the recent elucidation of its structure indicates that this enzyme should be specifically druggable (Rana et al., 2018). This study sets the ground for a re-emergent interest in the study of S-acylation and also lipid biosynthetic pathways as important regulatory mechanisms of infection by coronaviruses and enveloped viruses in general.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ACE2 | ABCAM | RRID: AB_301861 Cat#ab15348 |
| CALNEXIN | MILLIPORE | RRID: AB_2069152 Cat#MAB3126 |
| CAVEOLIN-1 | SANTA CRUZ | RRID: AB_2072042 Cat#sc-894 |
| E | gift from Machamer lab | RRID: N/A |
| ERGIC53 | ProSci | RRID: AB_796913 Cat#PSC-PM-7213 |
| FILIPIN | SIGMA | Cat#F4767 |
| FLAG | SIGMA | RRID: AB_259529 Cat#F3165 |
| GAPDH | ThermoFisher | RRID: AB_2533438 Cat#398600 |
| HA-HRP | ROCHE | RRID: AB_390917 Cat#12013819001 |
| HA | ROCHE | RRID: AB_390918 Cat#11867423001 |
| HA gel beads | ROCHE | RRID: AB_390914 Cat#1815016001 |
| LRP6 | Cell signaling | RRID: AB_10831525 Cat#2560 |
| M | gift from Machamer lab | N/A |
| MYC | SIGMA | RRID: AB_439694 Cat#M4439 |
| protein G al beads | GEHealthcare | Cat#17-0618-01 |
| P17 | home made, gift fom Trono lab | N/A |
| SPIKE | LIFESPAN | RRID: AB_840148 Cat#LS-C19510 |
| streptavidin beads | SIGMA | Cat#S1638 |
| Transferrin Receptor | ThermoFisher | RRID: AB_2533029 Cat#13-6800 |
| alexa-647-d.anti-mouse | ThermoFisher | RRID: AB_2536183 Cat#A31573 |
| alexa-488 anti-rat | Jackson Immunoresearch | RRID: 712.545.153 |
| anti-mouse HRP | GEHealthcare | RRID: AB_772210 Cat#NA931V |
| anti-rabbit-HRP | GEHealthcare | RRID: AB_772206 Cat#NA934V |
| anti-ZDHHC8 | Millipore | RRID : AB_11211894 Cat#AB15242 |
| anti-ZDHHC9 | SIGMA | RRID : AB_10747302 Cat#SAB4502104 |
| anti-ZDHHC20 | SIGMA | RRID : AB_10744838 Cat#SAB4501054 |
| Bacterial and virus strains | ||
| hCoV-19/Switzerland/GE9586/2020 | Switzerland-Geneve | EPI_ISL_414022|2020-02-27 |
| Chemicals, peptides, and recombinant proteins | ||
| BREFELDIN A | BioLegend | Cat#BLG-420601 |
| CYCLOHEXIMIDE | SIGMA | Cat#C7698 |
| FTY720 | SIGMA | Cat#SML0700 |
| Hydroxylamine | SIGMA | Cat#55460 |
| beta-methyl-cyclodextrin | SIGMA | Cat#C4555 |
| MYRIOCIN | CAYMAN Chemical | Cat#TRCM884400 |
| Palmostatin B | CALBIOCHEM | Cat#178501 |
| Critical commercial assays | ||
| Amplex red cholesterol assay | ThermoFisher | Cat#A12216 |
| Amplify NAMP100 | GEHealthcare | Cat#GENAMP100 |
| AVICEL GP3515 | DUPONT | Cat#41094401 |
| Bright-GLo Luciferase assay | PROMEGA | Cat#E2610 |
| Crystal violet | SIGMA | Cat#V5265 |
| EPISERF Medium-1 | ThermoFisher | Cat#10732022 |
| DMEM | ThermoFisher | Cat#31966021 |
| DMEM w/o cystein, methionin | Bioconcept | Cat#1-26S59 |
| ECL reagent spray-HRP | Witec | Cat#K-12049-D50 |
| EZ-Link Sulfo-NHS-SS-biotin | PIERCE | Cat#PIER21326 |
| E.Z.N.A viral RNA kit | OMEGA Bio-teck | Cat#R6874-02 |
| GMEM for IM | ThermoFisher | Cat#11710035 |
| HBS-EP buffer | CYTIVIA | Cat#BR100188 |
| iSCRPT | BIO-Rad | Cat#1708891 |
| Lacey carbon film EM grid | Agar Scientific UK | Cat#POL24924-25 |
| MALEIMIDE | Fluorochem | Cat#11793 |
| MEM | SIGMA | Cat#M4655 |
| MIRUS TRANSIT-X2RTM | MIRUS Bio Corporation | Cat#MIR6000 |
| MMTS | SIGMA | Cat#64306 |
| OPTIMEM-reduced serum medium | ThermoFisher | Cat#31985062 |
| OPTIPREP | AXISSHIELD | Cat#1114542 |
| Paraformaldehyde | Electron Microscopy Science | Cat#15714 |
| PBS | BioConcept | Cat#3-05F29-I |
| PBS-EDTA | BioConcept | Cat#5-32F00-I |
| ProLond mounting medium | ThermoFisher | Cat#P36934 RRID: SCR_015961 |
| protease inhibitor Complete | ROCHE | Cat#11836153001 |
| P24 ELISA | Zeptometrix | Cat#801200 |
| QUICKchange II XL | AGILENT | Cat#200522 |
| SARS--Cov2 surrogate virus neutralization kit | GENSCRIPT | Cat#L00847 |
| SYBR green Master Mix | Roche | Cat#4913914001 |
| TCEP | SIGMA | Cat#C4706 |
| thiopropyl sepharose beads | CreativeBiomart | Cat#Thio-001A |
| TX-100 | PIERCE | Cat#PIER28314 |
| Water Ambion | ThermoFisher | Cat#AM9906 |
| Zebra spin desalting columns | PIERCE | Cat#PIER89882 |
| 3H-palmitic acid | American Radio Labelled Chemicals | Cat#ART0129-25 |
| 35S-methionin-cysteine | Hartmann Analytic GmbH | Cat#IS103-185 |
| 5kDa PEG | SIGMA | Cat#63187 |
| Nano-Glo-Hibit | Promega | Cat#N2420 |
| Passive Lysis buffer | Promega | Cat#E1941 |
| Experimental models: Cell lines | ||
| HEK-293T | ATCC | CVCL_1926 |
| HELA | ATCC | CVCL_0030 |
| HAP1 | Horizon discovery | CVCL_XU08 |
| VERO E6 | ATCC | CVCL_0574 |
| 293T-ACE2 | Generated in this study | NA |
| Oligonucleotides | ||
| Supplemental information | This paper | N/A |
Resource availability
Lead contact
All resource and materials will be made available upon reasonable request to Francisco Mesquita (Francisco.mesquita@epfl.ch).
Materials availability
All data that support the findings of this study are available within the article and supplemental information.
Experimental model and subject detail
Cell culture
HAP1 cells were obtained from Horizon Discovery, Vero E6 cells were obtained from the American Type Culture Collection (ATCC) and maintained in DMEM medium supplemented with 10% FCS, penicillin and streptomycin; HELA cells were maintained in MEM medium supplemented with 10% FCS and complemented with 2 mM glutamine and 1% of non-essential amino-acids. For lentiviral vector production, 293T cells were cultured in DMEM supplemented with 10% FBS for maintenance and transfection, and in EpiSerf for production. All cells were kept at 37°C in a humidified atmosphere of 5% CO2 and were tested negative for mycoplasma. The 293T-ACE2 stable cell line was generated by transduction with an ACE2-ires-puro encoding lentivector (produced as described below and at a MOI of 0.5) followed by puromycin selection and maintained in DMEM 10% FCS with 1μg/ml puromycin. VERO E6 ZDHHC knockout cells were constructed using CRIPR/Cas9 system with plasmids pX459 encoding SpCas9-HF1 and puromycin resistance (Addgene); gene deletions were obtained with the monkey guide RNAs described in Table S1. Transfected cells were singly isolated in 96-well plates, selected with puromycin (1μg/ml) and knockout clones detected by PCR and confirmed by Western blots.
Viral Stock production and titration with plaque-based assays
All viral stocks were produced and isolated from supernatants of Vero E6 cells, cultured in T75 culture flasks to a confluency of 80-90%, and infected with an original passage 2 (P2) SARS-CoV-2 virus, for 48 or 72 h, at MOI≈0.05, in 10 ml DMEM supplemented with 2.5% FCS. Original stocks were obtained from the following strain: hCoV-19/Switzerland/GE9586/2020|EPI_ISL_414022|2020-02-27. Passage 3 supernatants were harvested, clear of cell debris by centrifugation (500 g 10 min) and filtration (0.45 μm), aliquoted and stored at -80 C. Viral titers were quantified by determining the number of individual plaque forming units after 48 h of infection in confluent Vero E6 cells. In brief, viral stocks were serially diluted (10-fold) in serum-free medium and (400 μl) inoculated in triplicate 48 wells, confluent Vero E6 cells (2.5 x 105) cells per well. After 1 h, inoculums were discarded and cells overlaid with a mixture of 0.4% of Avicel-3515 (Dupont) (from 2% stock) in DMEM supplemented with 5% FCS and pen/strep for additional 48 h. Overlays were discarded and cells fixed in 4% PFA for 30 min at RT. Fixed cells were washed in PBS and stained with 0.1% crystal violet solution (in 20% ethanol/water) for 15 min. Staining solution was discard and wells washed twice in water. Plates were allowed to dry and analyzed for quantification of the cytopathic effect as number of individual Plaque forming units (PFU) per ml (Avg PFU∗1/Volume∗1/dilution factor). Such un-concentrated viral stocks yielded between 0.5 to 3x106 PFU/ml.
Oligonucleotides, plasmids and sequences
HDM-IDTSpike-fixK (BEI catalog number NR-52514): plasmid expressing under a CMV promoter the Spike from SARS-CoV-2 strain Wuhan-Hu-1 (Genbank NC_045512) codon-optimized using IDT and the Kozak sequence in the plasmid fixed compared to an earlier version of this plasmid was obtained from Bloom Lab - Fred Hutchinson Cancer Research Center. Spike with mutation D614G was generated from HDM-IDTSpike-fixK using mutated oligonucleotides annealing. Plasmids expressing epitope-tagged SARS-CoV-2 Spike proteins were cloned in pcDNA6.2 with HA-tag in C-terminal using the Gateway system, and the donor plasmid pDONR223_SARS-CoV-2_Spike (Fritz Roth, Addgene plasmid # 149329; http://n2t.net/addgene:149329; RRID: Addgene_149329). Cysteine to Alanine substitutions were also generated with Quickchange. Plasmids expressing human ZDHHC proteins were cloned in pcDNA3.1 with MYC-tag (provided by the Fukata lab). The ACE2 sequence (from pHAGE2-EF1aInt-ACE2-WT a kind gift from J.D. Bloom) was swapped with the LacZ cassette of the pRRL-PGK-lacZ-ires-puro lentivector to generate the pRRL-ACE2-ires-puro vector. All oligonucleotides, namely QPCR primers and siRNA oligos used in this study are described in detail in Tables S1 and S2.
Spike sequences used for alignments in Figure 2A were retrieved from accession numbers: SARS2 (A0A7D6HN98), SARS (P59594), MERS (K9N5Q8), OC43 (P36334), 5HKU1 (A0A451ERM8), NL63 (H9EJ93), 229E (A0A1YOEV30), BCov (P25191), MHV-JHM (P11225), MHV2 (Q77NQ7), IBV (P11223), TGEV (P07946), FIPV (P10033), RATG13 (A0A6B9WHD3).
Method details
Transfection for plasmids and siRNA
Unless otherwise indicated, plasmids were transfected into 293T, Hela or Vero cells for 24 or 48 hours (3 μg/ 9.6cm2 plate) using Transit-X2RTM (Mirus) transfection reagent. For control transfections, we used an empty pcDNA6.2 plasmid. siRNA for Hela cells or for Vero cells were purchased from Qiagen (see Tables S1 and S2). For gene silencing, cells were transfected for 72 hours with (15 pmol/ 9.6cm2 plate) using Transit-X2RTM (Mirus) transfection reagent. Control siRNA and species-specific siRNA oligos targeting human or monkey ZDHHCs are described in Table S1.
SARS CoV-2 infections
All infections for experimental analysis were done using passage 3 SARS-CoV-2 stocks. Vero E6 cells seeded to a confluency of 90 to 100%, were, washed twice in warm serum-free medium and inoculated with the indicated MOI of SARS-CoV-2, diluted in serum free medium (5 ml for T75; 2 ml for T25; 1 ml for 6-well plates). 1 hour after inoculation cells were washed with complete medium and infection allowed to proceed for the indicated time points in DMEM supplemented with 2.5% FCS, penicillin and streptomycin (unless otherwise indicated: 10 ml for T75; 4 ml for T25; 2 ml for 6-well plates). Cells cultured in T75/T25 flasks for biochemical analysis, or 6-well plates for qPCR and Flow cytometry experiments, were infected with a MOI of ≈0.1, except for time-course experiments measuring: radioactive 3H-Palmitic acid -incorporation; -decay; and PEGylation dynamics, in which cells were infected with a MOI of ≈0.5 to 1. For infections involving 3H-Palmitic acid, medium without serum IM (see below) was used for all washes, viral inoculation and throughout infection.
Immunoprecipitation
For immunoprecipitations, cells were lysed 30 min at 4°C in IP buffer (0.5%NP40, 500 mM Tris-HCl pH 7.4, 20 mM EDTA, 10 mM NaF, 2 mM benzamidine, and a cocktail of protease inhibitors), centrifuged 3 min at 2000 g and supernatants were precleared with protein G-agarose conjugated beads and supernatants were incubated 16 h at 4°C with specified antibodies and beads. For Spike-M immunoprecipitations cells were lysed in 30 min at 4°C in PBS-1% triton-X, 0.5% SDS, and a cocktail of protease inhibitors, and processed as described.
Radiolabeling 3H-palmitic acid incorporation
To follow S-acylation, transfected or infected cells were incubated 1 hour in medium without serum IM (Glasgow minimal essential medium buffered with 10 mM Hepes, pH 7.4), followed by 2 hours or indicated hours at 37°C in IM with 200 μCI /ml 3H palmitic acid, washed with IM and incubated different times at 37°C with complete medium prior immunoprecipitation overnight. Beads were incubated 5 min at 90°C in reducing sample buffer prior to SDS-PAGE. Immunoprecipitates were split into two, run on 4-20% gels and analyzed either by autoradiography (3H-palmitate) after fixation (25% isopropanol, 65% H2O, 10% acetic acid), gels were incubated 30 min in enhancer Amplify NAMP100, and dried; or Western blotting. Autoradiograms and western blotting were quantified using the Typhoon Imager (Image QuantTool, GE healthcare).
Radiolabeling 35S-cys/met incorporation
For metabolic labeling, transfected cells were washed with methionine /cysteine free medium, incubated 4hours pulse at 37°C with 50 μCi/ml 35S-methionine/cysteine, washed and further incubated for different times at 37°C in complete medium with a 10-fold excess of non-radioactive methionine and cysteine. Proteins were immunoprecipitated and analyzed by SDS-PAGE. Autoradiography and western blotting were quantified using the Typhoon Imager (Image QuantTool, GE healthcare).
Drug treatments
Protein synthesis was blocked by 1 hour or different indicated time treatment with 10 μg/ml cycloheximide at 37°C in serum free medium. To block protein transport from ER to Golgi, we used Brefeldin A at 5 μg/ml for 1 hour at 37°C in serum free medium.
To sequester cholesterol, beta-metyl-cyclodextrin was used at 10 mM for 30 min in serum free medium. As inhibitor of depalmitoylation, Palmostatin B was used at 10 μM for 4 hours in serum free medium. To block serine palmitoyltransferase, denovo sphingolipid biosynthesis, cells were cultured either in Myriocin at 2.5 μM, 5 days prior infection and during the course of infection, or with a synthetic myriocin analogue, Fingolimod-FTY20 at 2 μM or 4 μM during infection.
Acyl-RAC capture assay
Protein S-palmitoylation was assessed by the Acyl-RAC assay as previously described (Werno and Chamberlain, 2015) with some modifications. Cells or supernatants were lysed in 400 μl buffer (0.5% Triton-X100, 25 mM HEPES, 25 mM NaCl, 1 mM EDTA, pH 7.4, and protease inhibitor cocktail). Cell lysis were incubated 30 min at RT with 10mM TCEP. Then, 200 μl of blocking buffer (100 mM HEPES, 1 mM EDTA, 87.5 mM SDS, and 1.5% [v/v] methyl methanethiosulfonate (MMTS)) was added to the lysates and incubated for 4 h at 40°C to block free the SH groups with MMTS. Proteins were acetone precipitated and resuspended in buffer (100 mM HEPES, 1 mM EDTA, 35 mM SDS). For treatment with hydroxylamine (NH2OH) and capture by Thiopropyl Sepharose beads, 2 M of hydroxylamine was added together with the beads (previously activated for 15 min with water) to a final concentration of 0.5 M of hydroxylamine and 10% (w/v) beads. As a negative control, 2 M Tris was used instead of hydroxylamine. These samples were then incubated overnight at room temperature on a rotating wheel. After washes, the proteins were eluted from the beads by incubation in 40 μl SDS sample buffer with ß-mercaptoethanol for 5 min at 95°C. Finally, samples were separated by SDS-PAGE and analyzed by Western blot. A fraction of the cell lysate (total cell extract) was saved as the input.
Acyl-Peg-exchange
This PEG switch assay is used to PEGylate previously palmitoylated cysteines following removal of hydroxylamine as previously described (Plain et al., 2020). To block free cysteine, cells or supernatant or purified virions were lysed and incubated in 400 μl buffer (2.5% SDS, 100 mM HEPES, 1 mM EDTA, 100mM Maleimide pH 7.5, and protease inhibitor cocktail) for 4 h at 40°C. To remove excess unreacted maleimide, proteins were acetone precipitated and re-suspended in buffer (100 mM HEPES, 1 mM EDTA, 1% SDS, pH 7.5). Previously palmitoylated cysteines were revealed by treatment with 250 mM hydroxylamine (NH2OH) for 1 hour at 37°C. Cell lysates were desalted using Zebra spin columns and incubated 1 hour at 37°C with 2mM 5kDa PEG: methoxypolyethylene glycol maleimide. Reaction was stopped by incubation in SDS sample buffer with ß-mercaptoethanol for 5 min at 95°C. Samples were separated by SDS-PAGE and analyzed by Western blot. A fraction of the cell lysate was saved before the addition of 5 kDa PEG as the input.
Isolation of detergent-resistant membranes (DRMs)
Approximately 1 × 107 cells were re-suspended in 0.5 ml cold TNE buffer (25 mMTris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100) with a tablet of protease inhibitors (Roche). Membranes were solubilized in a rotating wheel at 4 °C for 30 min. DRMs were isolated using an Optiprep™ gradient: the cell lysate was adjusted to 40% Optiprep™, loaded at the bottom of a TLS.55 Beckman tube, overlaid with 600 μl of 30% Optiprep™ and 600 μl of TNE, and centrifuged for 2 hours at 55,000 rpm at 4 °C for cells or 4 hours at 55,000 rpm at 4 °C for VLPs and PPs. Six fractions of 400 μl were collected from top to bottom. DRMs were found in fractions 1 and 2. Equal volumes from each fraction were analyzed by SDS-PAGE and western blot analysis using anti-Spike, anti-M, HRP-conjugated anti-HA, ERGIC53, caveolin1 and transferrin receptor antibodies.
Surface biotinylation
Surface biotinylation was performed on transfected cells. Cells were allowed to cool down shaking at 4°C for 15 min to arrest endocytosis. Cells were then washed three times with cold PBS and treated with EZ-Link Sulfo-NHS-SS-Biotin No weight for 30 min shaking at 4°C. Cells were then washed 3 times for 5 min with 100mM NH4Cl and lysed in 1% Tx-100 to do DRMs or in IP Buffer for 1h at 4°C. Lysate were then centrifuged for 5 min at 5000rpm and the supernatant incubated with streptavidin agarose beads overnight on a wheel at 4°C. Beads were washed with IP buffer 5 times and the proteins were eluted from the beads by incubation in SDS sample buffer with ß-mercaptoethanol for 5 min at 95°C. Samples were separated by SDS-PAGE and analyzed by immunoblotting. Western blots were developed using the ECL protocol and imaged on a Fusion Solo from Vilber Lourmat. Densitometric analysis was performed using the software Bio-1D from the manufacturer.
Lentiviral pseudotyped particle (PPs) production
Plasmids expressing SARS-CoV-2 Spike WT-HA or SARS-CoV-2 Spike 10C-A-HA proteins, pHAGE2-CMV-Luc-ZSgreen, Hgpm2, REV1b, Tat1b (a kind gift from J.D. Bloom) were co-transfected into 293T cells for 24 hours with the following ratio 3/9/2/2/2 (18 μg/ 56.7cm2 plate) using Fugene transfection reagent. The following day, cells were transferred in EpiSerf medium, and cell supernatants were collected after 8 hours and 16 hours. Harvested supernatants were pooled, clarified by low-speed centrifugation, filtered to remove cell debris (5 ml were kept at this stage for infectivity assays) and concentrated by centrifugation at 47000g through a 20%(w/v) sucrose cushion for 90min. Concentrated PPs were immediately resuspended in 300 μl of cold TNE buffer (25mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100) for biochemical experiments.
Binding Assay ELISA
Binding between Spike-pseudotyped PPs and ACE-2 was determining using an ELISA-based SARS-CoV-2 Surrogate Virus Neutralization Test Kit with a modified protocol. In brief, Non-concentrated PPs suspensions adjusted to comparable Spike and p17 content (monitor by western blot) were used to pre-incubate human-ACE2-coated ELISA plates for 30 min at 37°C. After 30 min, 100 μl of diluted (1/2000) Horseradish peroxidase (HRP)-conjugated Spike Receptor binding domain RBD was added to each well for further incubation, 15 min at 37°C. Plates were then washed 4x in the Kits washing solution and developed with 100 μl of substrate solution for 10 min in the dark, followed by a quenching step with 50 μl of stop solution for each well. Absorbance was monitored at 450 nm immediately and inversely correlated with the binding of Spike PPs to ACE-2. Eppiserf medium was used as negative control, and results expressed as % binding = (1 - (Sample OD/ Neg.Control OD)) normalized to 1 for the mean of WT-Spike-pseudotyped PPs.
Synchronized fusion assay
ACE-expressing HeLa or standard HeLa cells seeded in six well plates were washed 2 times in cold PBS and incubated on ice in cold Eppiserf medium for 2 to 5 min. Cells were then incubated with equivalent volumes of non-concentrated PPs suspensions adjusted to comparable Spike content (monitored by western blot) for 30 min on ice to allow binding. Cells were subsequently washed 3 times in cold PBS and incubated in warm Eppiserf medium at 37°C for 30 min to allow fusion. Next, cells were washed 2 times in cold PBS and incubated in Acid-wash buffer (145mM NaCl, 20mM MES-Tris pH 4.5) for 10 min with gentle rocking, to remove unfused surface attached particles. Cells were then lysed 30 min at 4°C in IP buffer, centrifuged 3 min at 2000 g and supernatants resuspended in SDS sample buffer with ß-mercaptoethanol for 5 min at 95°C. Samples were separated by SDS-PAGE and analyzed by Western blot.
PPs infectivity
Non-concentrated PPs suspensions were adjusted for p24 content (as monitored by ELISA) and used to transduce 293T-ACE2 cells (technical replicates n=12). Transduced cells were analyzed 3 days later with the Bright-Glo Luciferase assay system.
VLPs production
HiBiT-N-tagged virus-like particles (VLPs) were produced as described previously (Ref). Briefly, equimolar amounts of full-length CoV S, E (envelope), M (membrane), and HiBiT-N-encoding plasmids (total, 10mg) were transfected into 107 HEK293T cells. To produce Bald (“no-S”) VLPs, the S expression plasmids were replaced with empty vector plasmids. At 6 h post-transfection, cells were replenished with fresh DMEM-10% FBS. HiBiT-N VLP suspensions were collected in FBS-free DMEM or Eppiserf from 24 to 48 h post-transfection. Suspensions containing HiBiT-N VLPs were clarified by centrifugation (300g, 4°C, 10 min; 3,000g, 4°C, 10 min). To obtain purified viral particles, clarified VLP suspensions were concentrated 100-fold by overlaid onto 20%, wt/wt, sucrose cushions and particles purified via slow-speed pelleting (SW28, 6500 rpm, 4°C, 24 h or SW32 8000 rpm, 4°C, 24 h). The resulting pellet was resuspended in FBS-free DMEM or HBSS to 1/100 of the original medium volumes. VLPs were stored at -80°C or analyzed promptly.
VLP cell entry assay
HeLa target cells were LipoD transfected with pcDNA3.1-hACE2-LgBiT and pCAGGS-TMPRSS2FLAG. At 2 days posttransfection, cells were incubated with a live-cell Nluc substrate (Nano-Glo Endurazine; Promega), and 2 h later, HiBiT-N VLPs were inoculated at equivalent HiBiT input multiplicities. HiBiT-N VLPs lacking S proteins (no S) served as negative controls. At hourly intervals following VLP inoculation, Nluc levels were quantified using a Veritas microplate luminometer. For data presentation, the Nluc recordings in cultures inoculated with bald spikeless (no-S) VLPs were normalized to values of 1.0, and the fold increases over this control condition were calculated and plotted as “relative entry.”
Cell-free fusion assay
Cell-free fusion assays required ACE2-LgBiT extracellular vesicles (EVs). To obtain these EVs, HEK293T target cells were transfected with pcDNA3.1-hACE2-LgBiT. At 6 h posttransfection, transfection media were removed, rinsed, and replaced with FBS-free DMEM. Media were collected at 48 h posttransfection, clarified (300xg, 4°C, 10 min; 3,000xg, 4°C, 10 min), and concentrated 100-fold by ultrafiltration (Amicon; 100 kDa). EVs were then purified using SEC (qEV original; Izon, Inc.) using phosphate- buffered saline (PBS; pH 7.4) as the eluant. Peak EV fractions were identified by the addition of HiBiT-containing detergent and subsequent Nluc measurement by luminometry. EVs were stored at 4°C. Cell-free fusion assays were performed by mixing HiBiT-N VLPs, each introduced at equivalent HiBiT concentrations, with hACE2-LgBiT EVs, the NanoLuc substrate, and trypsin (Sigma; 10 ng/ml or as indicated) in 384-well multiwell plates. After 5 min at 4°C, sample plates were loaded into a GloMax luminometer maintained at 37°C. VLP-EV cell-free fusions were quantified as Nluc accumulations over time. For data presentation, the Nluc recordings from samples containing control Bald (no-S) VLPs were normalized to values of 1.0, and the fold increases over levels for this control condition were calculated and plotted as fold fusion.
Analysis of intracellular viral RNA-Q-PCR
For time-course optimization (Figure S2), 10 h and 24 h replication experiments (Figure 7) Infected Vero-E6 cells and correspondent supernatants (150 μl), cultured in 6-well plates (2 ml culture medium per well - 3 independent wells per condition) were harvested at indicated time points and lysed in 500 μl of mastermix QVL lysis buffer from E.Z.N.A. Viral RNA kit used for Viral RNA extraction according to manufacturer’s instructions. RNA concentration was measured and 500 ng or 1000 ng of total RNA was used for cDNA synthesis using iScript. A 1:5 dilution of cDNA was used to perform quantitative real-time PCR (QPCR) using Applied Biosystems SYBR Green Master Mix on 7900 HT Fast QPCR System (Applied Biosystems) with SDS 2.4 Software. Primers used are described in Table S2. All data (always in triplicate) were normalized to Ct values from three housekeeping (HK) genes ALAS-1, Guss and TBP, except for supernatants from replication experiments at 24 h, which Ct data were normalized using Ct values from correspondent cellular HK genes. Results were expressed as 2ˆ(-ΔΔCt)∗100%.
Titration of viral E copies
For titration of viral RNA, equivalent volumes of RNA extracted from infected culture supernatants or RNA from serial dilutions of in vitro transcribed RNA for Wuhan coronavirus (2019-nCoV) targeting region of the E gene standards (E-standards - European Virus Archive platform - Ref. 026N-03866) were used for cDNA synthesis. Samples were then used to perform QPCR analysis (using E specific primers) as described. A standard curve was generated by plotting the Ct values against the number of E copies per ml indicated by the serial dilution of the E-standards (stock at 1e5 copies / μl) and used to extrapolate the number of E copies per ml in samples.
SARS-CoV-2 single round infections and infectivity analysis
Supernatants harvested from 10 h infections were processed for QPCR analysis (150 μl), aliquoted and stored at -80°C until titration of viral E copies. After titration and adjustment to viral E RNA copies non-concentrated SARS-CoV-2 supernatants were used to infect confluent Vero E6 cell monolayers cultured in 6 well plates using an approximate ratio of 5 to 10 E copies per cultured host cell. At least 3 wells were infected per supernatant per condition. Infection was done as described until 6 h post inoculation when cells were washed lysed and processed for QPCR analysis as described
Analytical flow cytometry of SARS-CoV-2 infected cells
Vero E6 cells were infected in 6-well plates [as mentioned previously]. Cells washed and scrapped in 1 ml PBS and recovered by mild centrifugation 400g 3 min. Supernatants were discard and cells were fixed in 4% PFA for at least 30 min, washed twice in PBS, and permeabilized for 5 min with 0.1% Triton in PBS. The following blocking, antibody, and washing steps were all done with FACS buffer (2% FBS in PBS/EDTA) primarily at RT. Blocking was done for 15 min, primary antibody incubation with mouse anti-Spike antibody (LC-C19510) was performed at 1:200 for 30 min at RT or at 1:500 ON at 4 °C, and cells were washed twice. Alexa-647-conjugated donkey anti-mouse was used as a secondary antibody at 1:600 for 30 min at RT, after which cells were washed three times and kept cold before flow cytometry acquisition. Analytical flow cytometry was performed using an LSRII or LSR Fortessa (BD; Becton Dickinson) instrument and results were analyzed using the FlowCore package in R.
Immunohistochemistry
Vero E6 cells seeded in glass coverslips in 24-well plates and transfected with the indicated myc-ZDHHC or HA-Spike constructs (for 24 h) were fixed in 4% paraformaldehyde (15 min), quenched with 50 mM NH4Cl (30 min) and permeabilized with 0.1% Triton X-100 (5 min). Antibodies were diluted in PBS containing 1% BSA. Coverslips blocked in PBS 1% BSA, for 30 min, washed and incubated with primary antibodies for up to 2 hours RT or overnight at 4°C, washed three times in PBS, and incubated 45 min with secondary antibodies. Coverslips were mounted onto microscope slides with ProLong™ Gold Antifade Mountant. Images were collected using a confocal laser-scanning microscope (Zeiss LSM 700) and processed using Fiji™ software.
SARS CoV-2 isolation for biochemical, LPDX, Cryo-EM, infectivity assays
Vero E6 cells cultured in T75 flasks, transfected with the indicated siRNAs were infected as described. 1 h after viral inoculation cells were washed twice and incubated in EpiSerf medium (serum free) for the remaining of the infection. Culture supernatants (10 ml per flask) were harvested after 48 h, clarified by low-speed centrifugation (500 g 10 min) and filtration (0.45 μm); and stored at -80 C (for infectivity assays), or further processed for biochemical, LPDX or CryO EM experiments.
Electronic microscopy
At least 20 ml (10 ml per T75 flask per condition per preparation) of pre-cleared culture supernatants from 48 h infections were inactivated with paraformaldehyde (PFA; final concentration 4%) for > 2 h and concentrated by centrifugation at 47000g through a 20%(w/v) sucrose cushion for 90min 16°C. Concentrated virions were immediately re-suspended in cold 50 μl of EM-gradel Hepes buffered saline for subsequent EM processing. A Lacey carbon film EM grid was held in tweezers and 4-5 μL of sample solution was applied on the grid. The tweezers are mounted in an automatic plunge freezing apparatus (Vitrobot, ThermoFisher) to control humidity and temperature. After blotting, the grid was immersed in a small metal container with liquid ethane that is cooled from outside by liquid nitrogen. The speed of cooling is such that ice crystals do not have time to form. Observation was made at -170°C in a Tecnai F 20 microscope (ThermoFisher) operating at 200 kV equipped with a cryo-specimen holder Gatan 626 (AMETEK). Digital images were recorded with a Falcon III (ThermoFisher) camera 4098 X 4098 pixels. Magnification of 50000X with a pixel size of 0.2nm, using a defocus range from -2μm to -3μm.
Concentration of SARS-CoV-2 virions for LPDX and biochemical assays
Approximately 10 ml (per T75 flask) of pre-cleared culture supernatants from 48 h infections were aliquoted into 1.5 ml microtubes and concentrated by centrifugation at 47000g through a 20%(w/v) sucrose cushion (200 μl) for 90min. Medium and sucrose supernatants were discard, pellets allowed to dry and re-suspended and pooled in: 400 μl of cold 1%-Triton X TNE buffer for DRM analysis; or 500 μl 1:1 mixture of methanol:chloroform for LPDX analysis.
Lipid extraction
Total lipid extracts were prepared using a standard methyl-tert-butyl ether (MTBE) protocol followed by a methylamine treatment for total lipid analysis by mass spectrometry. Briefly, cell pellets or viral fractions were resuspended in 100 μL H2O. 360 μL methanol and 1.2 mL of MTBE were added and samples were placed for 10 min on a vortex at 4°C followed by incubation for 1 h at room temperature on a shaker. Phase separation was induced by addition of 200 μL of H2O. After 10 min at room temperature, samples were centrifuged at 1000 g for 10 min. The upper (organic) phase was transferred into a glass tube and the lower phase was re-extracted with 400 μL artificial upper phase [MTBE/methanol/H2O (10:3:1.5, v/v/v)]. The combined organic phases were dried in a vacuum concentrator. Lipids where then resuspended in 500 μL of ChCl3.
LC-MS untargeted lipidomics
Lipid extracts (2 μL injection volume in ChCl3:MeOH 2:1) were separated over an 8 min gradient at a flow rate of 200 μL/min on a HILIC Kinetex Column (2.6lm, 2.1 × 50 mm2) on a Shimadzu Prominence UFPLC xr system (Tokyo, Japan). Mobile phase A was acetonitrile:methanol 10:1 (v/v) containing 10 mM ammonium formate and 0.5% formic acid while mobile phase B was deionized water containing 10 mM ammonium formate and 0.5% formic acid. The elution of the gradient began with 5% B at a 200 μL/min flow and increased linearly to 50% B over 7 min, then the elution continued at 50% B for 1.5 min and finally, the column was re-equilibrated for 2.5 min. MS data were acquired in full-scan mode at high resolution on a hybrid Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany). The system was operated at 240,000 resolution (m/z 400) with an AGC set at 1.0E6 and one microscan set at 10-ms maximum injection time. The heated electrospray source HESI II was operated in positive mode at a temperature of 90 C and a source voltage at 4.0KV. Sheath gas and auxiliary gas were set at 20 and 5 arbitrary units, respectively, while the transfer capillary temperature was set to 275 °C. Mass spectrometry data were acquired with LTQ Tuneplus2.7SP2 and treated with Xcalibur 4.0QF2 (Thermo Fisher Scientific). Lipid identification was carried out with Lipid Data Analyzer II (LDA v. 2.6.3, IGB-TUG Graz University) (Hartler et al., 2011). The LDA algorithm identifies peaks by their respective retention time, m/z and intensity. Care was taken to calibrate the instrument regularly to ensure a mass accuracy consistently lower than 3 ppm thereby leaving only few theoretical possibilities for elemental assignment. Lipid levels were calculated by normalization over internal standards. Enrichments in viral particles over cells were calculated according to the formula: (viral_Lipid x / viral_PtdCho 34:1)/ (cellular_Lipid x / cellular_PtdCho 34:1).
Cholesterol analysis
To detect free cholesterol and cholesteryl esters in samples we used a fluorometric method based on an enzyme-coupled reaction, Cholesteryl esters are hydrolyzed by cholesterol esterase into cholesterol, which is then oxidized by cholesterol oxidase to yield H2O2: Amplex Red Cholesterol Assay kit (Thermofisher).
Automated microscopy analysis of Filipin staining
Vero cells were plated at 10000 cells per well and transfected, with plasmids expressing HA-tagged Spike (WT or 10C-A), for 24 h using TransIT-X2® Transfection Reagent (Mirus) in Ibidi 96-well μplates. Cells were fixed with 3% PFA and permeabilized with 0.05% saponin. Anti-HA antibody (Roche Diagnostics) was used at 1:100 together with RNase (Qiagen) for primary staining. After washes with automated plate washer (BioTek EL406), cells were incubated for half an hour with Alexa488 anti-Rat and filipin diluted 1:50 (Sigma), nuclei were stained by PI. Imaging was acquired on a Molecular Devices™ IXM-C with a 40x plan Apo objective, 49 images were acquired in each well. We had 8 replicas for each condition and for each of 3 independent experiments.
To analyze and quantify free cholesterol (filipin) staining, we used the MetaXpress Custom Module editor software from Molecular Devices, as in previous publications (Larios et al., 2020; Moreau et al., 2019). We first segmented each individual nucleus using the PI channel, we then used the HA channel to mask the expressing cells. The final masks were applied to all original fluorescent images and measurements per cell (nucleus) and average per well were extracted. The same analysis pipeline was applied to all images.
Molecular modeling and simulations
The model of trimeric Spike’s TM helix + C-terminal region (residues 1201-end in Uniprot P0DTC2) was built by extending laterally the C-terminus of model 1_1_2 built from PDB 6VSB by (Woo et al., 2020). Membrane insertion was carried out with CHARMM-GUI’s web tool in atomistic or coarse-grained (CG) lipid bilayers as required (Hsu et al., 2017; Jo et al., 2008; Qi et al., 2015). For CG molecular dynamics (MD) simulations, we parametrized, also using CHARMM-GUI, the protein with the MARTINI 2.2p CG force field (Marrink et al., 2007), the DPPC, DLiPC and cholesterol with MARTINI CG force field for lipids (Marrink et al., 2004) (notice that DLiPC corresponds to MARTINI’s DIPC lipid), and S-palmitoylated cysteines with the parameters provided by (Atsmon-Raz and Tieleman, 2017). Membranes for CG MD simulations contained 50% DPPC, 30% DLiPC and 20% cholesterol, which we verified forms rafts within 1 μs of CG MD simulation without proteins inserted, as shown by (Lorent et al., 2017). For atomistic MD simulations, we parameterized the protein, POPC membrane and palmitoylated cysteine with CHARMM36m as implemented in CHARMM-GUI. All MD simulations were run in Gromacs (Abraham et al., 2015) using standard settings from CHARMM-GUI for CG and atomistic membrane simulations, at 303 K. In atomistic MD we used a 12 Å cutoff for nonbonded interactions, PME, NVT equilibration followed by NPT production with semi-isotropic pressure coupling to 1 atm and 2 fs timestep; in CG MD we used standard MARTINI equilibration for membranes followed by NPT with semi-isotropic pressure coupling to 1 atm and 20 fs timestep. Simulations were visualized in VMD (Humphrey et al., 1996) and quantitative analysis were carried out through custom VMD Tcl and bash scripts. Membrane perturbation maps were built with MEMBPLUGIN for VMD (Guixà-González et al., 2014).
Quantification and statistical analysis
All data were repeated independently at least three times. Unless otherwise stated each data-point indicates one independent experiment with consistent results. Statistical analysis was carried using Prism software. Data representations and statistical details can be found in figure legends. For ANOVA analysis p values were obtained by post hoc tests used to compare every mean or pair of means (Tukey’s & Sidak’s) or to compare every mean to a control sample (Dunnet’s). For the Pareto Distribution analysis all details can be found in correspondent figure legends.
Acknowledgments
We thank Machamer laboratory for M and E antibodies (Cohen et al., 2011; McBride et al., 2007); D. Demurtas and G. Knott from the BioEM EPFL Core Facility; S. Vossio and D. Moreau from ACCESS Geneva (the Swiss National Centre of Competence in Research [NCCR] Chemical Biology); V. Cagno, C. Tapparel, and I. Eckerle from University of Geneva for viral technical support; V. Chavez from University of Lausanne for the statistical analysis; all the members of the VDG-lab for discussions; P. Nattawadee for the model design (created with BioRender.com); and C. Ernst for primer sharing. This work was supported by the Swiss National Science Foundation (SNSF) Corona Call, a foundation advised by CARIGEST S.A., the EPFL Corona Research task force and the NCCR Chemical Biology to F.G.v.d.G., by SNSF (/205321_192371) from the SNSF and PRACE COVID19 fast track project #17 (pr97) from the Swiss Supercomputing Center (SDSC) to M.D.P. MD simulations were carried out on the Piz Daint computer at the SDSC.
Author contributions
Conceptualization, F.S.M., L.A., D.T., G.D., and F.G.v.d.G.; investigation, F.S.M., L.A., O.S., P.T., B.K., C.R., J.P.M., E.Q., L.A.A., and G.D.; funding acquisition, F.S.M., M.D.P., and F.G.v.d.G.; writing – original draft, F.S.M., L.A., G.D., L.A.A., and F.G.v.d.G.; writing – review & editing, F.S.M., L.A., O.S., P.T., J.P.M., L.A.A., M.D.P., T.G., D.T., G.D., and F.G.v.d.G.; resources, B.K. and O.S.
Declaration of interests
The authors declare no competing interests.
Published: October 1, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2021.09.016.
Supplemental information
Analysis of all individual lipid molecular species detected in SARS-CoV-2 virions and respective target cells, used for the graphs depicted in Figures 6 and S6. Lipid nomenclature is presented as: lipid class abbreviation (e.g., HexCer), and variable components (fatty acids, number of C atoms:number of double bonds–e.g., HexCer_34:1).
Data availability
Other specific enquiries and data sets are available from lead contact upon reasonable request. All data that support the findings of this study are available within the article and supplemental information. Table S3 is also publicly available at: https://data.mendeley.com/datasets/47gsfkkjjz/1.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Abrami L., Denhardt-Eriksson R.A., Hatzimanikatis V., van der Goot F.G. Dynamic radiolabeling of S-Palmitoylated proteins. Methods Mol. Biol. 2019;2009:111–127. doi: 10.1007/978-1-4939-9532-5_9. [DOI] [PubMed] [Google Scholar]
- Atsmon-Raz Y., Tieleman D.P. Parameterization of palmitoylated cysteine, farnesylated cysteine, geranylgeranylated cysteine, and myristoylated glycine for the martini force field. J. Phys. Chem. B. 2017;121:11132–11143. doi: 10.1021/acs.jpcb.7b10175. [DOI] [PubMed] [Google Scholar]
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9:e57309. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdyshev E.V., Gorshkova I., Skobeleva A., Bittman R., Lu X., Dudek S.M., Mirzapoiazova T., Garcia J.G.N., Natarajan V. FTY720 inhibits ceramide synthases and up-regulates dihydrosphingosine 1-phosphate formation in human lung endothelial cells. J. Biol. Chem. 2009;284:5467–5477. doi: 10.1074/jbc.M805186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008;82:2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boson B., Legros V., Zhou B., Siret E., Mathieu C., Cosset F.L., Lavillette D., Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021;296:100111. doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.R., Lin L.D., Machamer C.E. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J. Virol. 2011;85:5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo G., Uemura T., Chuang C.C., Polishchuk E., Santoro M., Ohvo-Rekilä H., Sato T., Di Tullio G., Varriale A., D’Auria S., et al. Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature. 2013;501:116–120. doi: 10.1038/nature12423. [DOI] [PubMed] [Google Scholar]
- D’Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C.C., van der Spoel A.C., et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- de Haan C.A.M., Rottier P.J.M. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Jimenez A.L., Zimmermann P. Phospholipase D and phosphatidic acid in the biogenesis and cargo loading of extracellular vesicles. J. Lipid Res. 2018;59:1554–1560. doi: 10.1194/jlr.R083964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick C., Turelli P., Pellaton C., Farina A., Campos J., Raclot C., Pojer F., Cagno V., Nusslé S.G., D’Acremont V., et al. A high-throughput cell- and virus-free assay shows reduced neutralization of SARS-CoV-2 variants by COVID-19 convalescent plasma. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abi8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla M.R., Abrami L., van der Goot F.G., Veit M. Hemagglutinin of Influenza A, but not of Influenza B and C viruses is acylated by ZDHHC2, 8, 15 and 20. Biochem. J. 2020;477:285–303. doi: 10.1042/BCJ20190752. [DOI] [PubMed] [Google Scholar]
- Gadalla M.R., Veit M. Toward the identification of ZDHHC enzymes required for palmitoylation of viral protein as potential drug targets. Expert Opin. Drug Discov. 2020;15:159–177. doi: 10.1080/17460441.2020.1696306. [DOI] [PubMed] [Google Scholar]
- Gelhaus S., Thaa B., Eschke K., Veit M., Schwegmann-Weßels C. Palmitoylation of the Alphacoronavirus TGEV spike protein S is essential for incorporation into virus-like particles but dispensable for S–M interaction. Virology. 2014;464–465:397–405. doi: 10.1016/j.virol.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guixà-González R., Rodriguez-Espigares I., Ramírez-Anguita J.M., Carrió-Gaspar P., Martinez-Seara H., Giorgino T., Selent J. MEMBPLUGIN: studying membrane complexity in VMD. Bioinformatics. 2014;30:1478–1480. doi: 10.1093/bioinformatics/btu037. [DOI] [PubMed] [Google Scholar]
- Halter D., Neumann S., van Dijk S.M., Wolthoorn J., de Mazière A.M., Vieira O.V., Mattjus P., Klumperman J., van Meer G., Sprong H. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J. Cell Biol. 2007;179:101–115. doi: 10.1083/jcb.200704091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartler J., Trötzmüller M., Chitraju C., Spener F., Köfeler H.C., Thallinger G.G. Lipid Data Analyzer: unattended identification and quantitation of lipids in LC-MS data. Bioinformatics. 2011;27:572–577. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie J., Reilly L., Fraser N.J., Walker J.M.V., Wypijewski K.J., Ashford M.L.J., Calaghan S.C., McClafferty H., Tian L., Shipston M.J., et al. Substrate recognition by the cell surface palmitoyl transferase DHHC5. Proc. Natl. Acad. Sci. USA. 2014;111:17534–17539. doi: 10.1073/pnas.1413627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.C., Bruininks B.M.H., Jefferies D., Cesar Telles de Souza P.C.T. de, Lee J., Patel D.S., Marrink S.J., Qi Y., Khalid S., Im W. CHARMM-GUI Martini Maker for modeling and simulation of complex bacterial membranes with lipopolysaccharides. J. Comput. Chem. 2017;38:2354–2363. doi: 10.1002/jcc.24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27. doi: 10.1016/0263-7855(96)00018-5. 33–8. [DOI] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketter E., Randall G. Virus impact on lipids and membranes. Annu. Rev. Virol. 2019;6:319–340. doi: 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B., Hawkins G.M., Kicmal T., Qing E., Timm E., Gallagher T. Assembly and entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV2): evaluation using virus-like particles. Cells. 2021;10:853. doi: 10.3390/cells10040853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S., Park H., Laviad E.L., Lu X., Bittman R., Futerman A.H. Ceramide synthesis is modulated by the sphingosine analog FTY720 via a mixture of uncompetitive and noncompetitive inhibition in an acyl-CoA chain length-dependent manner. J. Biol. Chem. 2009;284:16090–16098. doi: 10.1074/jbc.M807438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larios J., Mercier V., Roux A., Gruenberg J. ALIX- and ESCRT-III–dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I., Levental K.R., Heberle F.A. Lipid rafts: controversies resolved, mysteries remain. Trends Cell Biol. 2020;30:341–353. doi: 10.1016/j.tcb.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L.A., Riffle A.J., Pike S.L., Gardner D., Hogue B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008;82:3000–3010. doi: 10.1128/JVI.01914-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent J.H., Diaz-Rohrer B., Lin X., Spring K., Gorfe A.A., Levental K.R., Levental I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017;8:1219. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink S.J., de Vries A.H., Mark A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108:750–760. doi: 10.1021/jp036508g. [DOI] [Google Scholar]
- Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- McBride C.E., Li J., Machamer C.E. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 2007;81:2418–2428. doi: 10.1128/JVI.02146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C.E., Machamer C.E. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell–cell fusion but not interaction with M protein. Virology. 2010;405:139–148. doi: 10.1016/j.virol.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau D., Vacca F., Vossio S., Scott C., Colaco A., Paz Montoya J., Ferguson C., Damme M., Moniatte M., Parton R.G., et al. Drug-induced increase in lysobisphosphatidic acid reduces the cholesterol overload in Niemann–Pick type C cells and mice. EMBO Rep. 2019;20:e47055. doi: 10.15252/embr.201847055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.T., Zhang S., Wang Q., Anang S., Wang J., Ding H., Kappes J.C., Sodroski J. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. bioRxiv. 2020 doi: 10.1101/2020.10.22.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A., Thinon E., Hang H. Mass-tag labeling using acyl-PEG Exchange for the determination of endogenous protein S-fatty acylation. Curr. Protoc. Protein Sci. 2017;89:14.17.1–14.17.11. doi: 10.1002/cpps.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C.M., Chouljenko V.N., Iyer A., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Palmitoylation of the cysteine-rich endodomain of the SARS–coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360:264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plain F., Howie J., Kennedy J., Brown E., Shattock M.J., Fraser N.J., Fuller W. Control of protein palmitoylation by regulating substrate recruitment to a zDHHC-protein acyltransferase. Commun Biol. 2020;3:411. doi: 10.1038/s42003-020-01145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Ingólfsson H.I., Cheng X., Lee J., Marrink S.J., Im W. CHARMM-GUI martini Maker for coarse-grained simulations with the martini force field. J. Chem. Theory Comput. 2015;11:4486–4494. doi: 10.1021/acs.jctc.5b00513. [DOI] [PubMed] [Google Scholar]
- Qing E., Hantak M., Perlman S., Gallagher T. Distinct roles for sialoside and protein receptors in coronavirus infection. mBio. 2020;11 doi: 10.1128/mBio.02764-19. e02764-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E., Kicmal T., Binod K., Grant M.,H., Emily T., Stanley P., Gallagher T. Dynamics of SARS-CoV-2 spike proteins in cell entry: control elements in the amino-terminal domains. mBio. 2021;12:e0159021. doi: 10.1128/mBio.01590-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M.S., Kumar P., Lee C.J., Verardi R., Rajashankar K.R., Banerjee A. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science. 2018;359:eaao6326. doi: 10.1126/science.aao6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risselada H.J., Marrink S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D.W., Jumper C.C., Ackerman P.J., Bracha D., Donlic A., Kim H., Kenney D., Castello-Serrano I., Suzuki S., Tamura T., et al. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. bioRxiv. 2020 doi: 10.1101/2020.12.14.422737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.F.G. Acylation of virol. spike glycoproteins: a feature of enveloped RNA viruses. Virology. 1982;116:327–338. doi: 10.1016/0042-6822(82)90424-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S.M., et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H.P., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp E.B., Boscarino J.A., Logan H.L., Goletz J.T., Gallagher T.M. Palmitoylations on murine coronavirus spike proteins are essential for Virion assembly and infectivity. J. Virol. 2006;80:1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.T., Wang S.M., Huang K.J., Wang C.T. SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production. J. Biomed. Sci. 2014;21:34. doi: 10.1186/1423-0127-21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo M.F., van den Brink W.J., Horzinek M.C., van der Zeijst B.A.M. Fatty acid acylation of viral proteins in murine hepatitis virus-infected cells. Brief report. Arch. Virol. 1987;95:123–128. doi: 10.1007/BF01311339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werno M.W., Chamberlain L.H. S-acylation of the insulin-responsive aminopeptidase (IRAP): quantitative analysis and Identification of Modified cysteines. Sci. Rep. 2015;5:12413. doi: 10.1038/srep12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J.W., Machamer C.E. The infectious bronchitis coronavirus envelope protein alters Golgi pH to protect the spike protein and promote the release of infectious virus. J. Virol. 2019;93 doi: 10.1128/JVI.00015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H., Park S.J., Choi Y.K., Park T., Tanveer M., Cao Y., Kern N.R., Lee J., Yeom M.S., Croll T.I., et al. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Zhang Z., Wang X., Zhang J., Ren C., Li Y., Gao L., Liang X., Wang P., Ma C. Palmitoylation of SARS-CoV-2 S protein is essential for viral infectivity. Signal Transduct. Target. Ther. 2021;6:231. doi: 10.1038/s41392-021-00651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaballa M.E., Goot F.G. van der. The molecular era of protein S-acylation: spotlight on structure, mechanisms, and dynamics. Crit. Rev. Biochem. Mol. Biol. 2018;53:420–451. doi: 10.1080/10409238.2018.1488804. [DOI] [PubMed] [Google Scholar]
- Zhukovsky M.A., Filograna A., Luini A., Corda D., Valente C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019;593:2428–2451. doi: 10.1002/1873-3468.13563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of all individual lipid molecular species detected in SARS-CoV-2 virions and respective target cells, used for the graphs depicted in Figures 6 and S6. Lipid nomenclature is presented as: lipid class abbreviation (e.g., HexCer), and variable components (fatty acids, number of C atoms:number of double bonds–e.g., HexCer_34:1).
Data Availability Statement
Other specific enquiries and data sets are available from lead contact upon reasonable request. All data that support the findings of this study are available within the article and supplemental information. Table S3 is also publicly available at: https://data.mendeley.com/datasets/47gsfkkjjz/1.