Abstract
The prevalence of GB virus C (GBV-C) in candidate Brazilian blood donors with normal and elevated alanine aminotransferase levels was found to be 5.2% (5 of 95) and 6.5% (5 of 76), respectively. Among Brazilian patients, GBV-C was found in 9.5% (13 of 137) of cases of hepatitis not caused by hepatitis A virus (HAV), HBV, HCV, HDV, or HEV (non-A-E hepatitis) and in 18.2% (8 of 44) of individuals infected with HCV. Molecular characterization of GBV-C by partial sequencing of the NS3 region showed clustering between members of a single family, implying intrafamilial transmission. In conclusion, these results together suggest that contagion mechanisms which facilitate intrafamilial transmission of GBV-C may partially explain the high prevalence of viremic carriers worldwide.
Two different groups have reported the discovery of a novel flavivirus-like agent potentially associated with human hepatitis, designated GB virus C (GBV-C) (18) or hepatitis G virus (HGV) (13). GBV-C transmission by blood transfusion has been clearly demonstrated (1, 13), explaining the high prevalence of this virus among intravenous drug users, polytransfused individuals, hemophiliacs, and hemodialysis patients (13). Vertical transmission from mother to infant has also been reported (6, 21), and epidemiological data further suggest the occurrence of horizontal transmission in confined patients (5). Sexual transmission has been reported through the analysis of patients infected with human immunodeficiency virus (HIV) (8), prostitutes (9), and couples (10). GBV-C has also been documented in cases of hepatitis not caused by hepatitis A virus (HAV), HBV, HCV, HDV, or HEV (non-A-E hepatitis) worldwide (13, 18), including Brazil (17); in HBV- and HCV-infected patients (13); and in cases of aplastic anemia (18). Furthermore, despite its high prevalence in patients with hepatitis, the association between GBV-C and hepatitis, cirrhosis, hepatocellular carcinoma, or end-stage liver disease is uncertain, especially because many infections also occur in healthy individuals (1, 2). The need for additional studies to address this question is clear (14).
Herein, we analyze the prevalence of GBV-C in candidate blood donors and hepatitis patients in Brazil. Through an analysis of viral sequences from infected family members of two patients with index cases, we also provide evidence for intrafamilial transmission of this virus.
Patients.
Samples were obtained from five population groups. Group 1 comprised 137 non-A-E hepatitis patients. These were individuals who had acute or chronic hepatic disorders and for whom viral hepatitis was considered in the differential diagnosis. These patients were always seronegative for hepatitis B surface antigen (HBsAg), antibody to hepatitis B core antigen (anti-HBc), and anti-HCV. Patients with acute cases also tested negative for anti-HAV immunoglobulin M (IgM), anti-HBc IgM, anti-HEV, and HCV RNA. Group 2 consisted of 44 hepatitis C patients, diagnosed by reactive anti-HCV serology and/or HCV RNA by PCR. Group 3 consisted of 95 volunteer blood donors from the Blood Bank of Hemocentro, Campinas, São Paulo State, Brazil, in the order of their acceptance. These blood donors underwent routine screening tests for blood-borne infections (alanine aminotransferase [ALT] levels, HBsAg, and anti-HBc, HCV, HIV, human T-cell leukemia virus types 1 and 2, Treponema pallidum, and Trypanosoma cruzi). Group 4 comprised 76 candidate volunteer blood donors from the same blood bank, in sequential order. These individuals were not accepted, exclusively due to the detection of ALT levels higher than the cutoff value. Familial contacts (two wives and four daughters) of two GBV-C patients made up group 5.
GBV-C RNA detection.
RNA was extracted from 100 μl of serum by using guanidine isothiocyanate. All the extracted material was then used in cDNA synthesis with Moloney murine leukemia virus reverse transcriptase and random hexamers. The cDNA was amplified with primers covering the NS3 region by using a nested PCR protocol. In the first round of amplification, the primers ns3.2-a2 and ns3.2-s1 were used in a “touchdown” PCR (18). One-tenth of the first-round product was then used in the second amplification, with primers gbvc-s1 and gbvc-a1 in another touchdown PCR protocol (12). Positive samples were identified in a 2% agarose gel, and the specificity of each result was confirmed by sequencing, as described below. To avoid false-positive results, strict procedures proposed for nucleic acid amplification diagnostic techniques were followed (11). These procedures include (i) use of completely independent separated areas for RNA extraction, PCR setup, and PCR product analysis; (ii) use of dedicated reagents, equipment, pipettes, plasticware, gloves, and lab coats for each of these areas; and (iii) control of daily flow of personnel only from pre- to postamplification areas. At the preamplification areas, special procedures were adopted: use of aerosol resistance tips, constant changing of gloves, use of paper sheets for opening tubes to avoid aerosols, and working with stock solutions and preparing reaction mixes in separate areas before starting any manipulation with samples on the same day. In every reaction run, one negative control was analyzed for every three samples and a weakly positive control serum, from a patient previously determined to be viremic, was included (17).
Sequencing.
PCR fragments were directly sequenced with fluorescent dye terminators on a Perkin-Elmer Applied Biosystems 377 Prism automated DNA sequencer. Reactions were performed essentially as described by the manufacturer by using commercially available sequencing kits. Sequencing primers were those used in the second PCR amplification. Raw sequence data were assembled into contigs, edited with Sequencher sequence analysis software (Gene Codes, Ann Arbor, Mich.), and exported as ASCII formatted files for subsequent analysis. Double-stranded edited sequences were used for all phylogenetic analyses.
Phylogenetic analysis.
A data set of 246 nucleotides in length (including gaps), with 53 sequences, consisting of 26 GBV-C sequences, representing a wide range of geographical locations, taken from sequence databases and 27 sequences from Brazilian patients, was analyzed. Clinical and epidemiological data for 23 patients are presented in Table 1. Only patients from whom GBV-C sequences were used in the subsequent phylogenetic analysis are shown in Table 1. Phylogenetic trees were reconstructed by using the neighbor-joining method from genetic distances estimated by the HKY85 model of DNA substitution, with values for the transition/transversion ratio (1.98), the shape parameter (α = 0.309) of the gamma distribution which describes rate variation among sites, and the proportion of invariant sites estimated from the empirical data. The robustness of the groupings observed on the phylogenetic tree was assessed by bootstrap resampling (1,000 replications). All analyses were carried out with the PAUP package (beta version 4.0.0d52; kindly provided by David Swofford, Sinauer Associates, Inc.).
TABLE 1.
Geographical origins and epidemiological, clinical, and/or histopathological data for the 23 Brazilian GBV-C-infected patients whose GBV-C sequences were used in the phylogenetic analysis
| Patient | Origina | Clinical and/or histopathological status | Epidemiologyb | Sequence identifier | GenBank accession no. |
|---|---|---|---|---|---|
| 1 | São Paulo, SP | Liver cirrhosis | Frequent traveller to Amazon basin; b. Spain | AMR | AFC124759 |
| 52112 | AFC124765 | ||||
| 67936 | AFC124770 | ||||
| 2 | São Paulo, SP | Periportal fibrosis | Wife of patient 1 | 61871 | AFC124768 |
| 67937 | AFC124771 | ||||
| 3 | São Paulo, SP | Healthy carrier | Daughter of patient 1 | 67945 | AFC124772 |
| 4 | São Paulo, SP | Liver cirrhosis | Amazon basin inhabitant for 7 yr | MPC3 | AFC124758 |
| 5 | Ribeirão Preto, SP | Liver cirrhosis | Blood transfusion | 69251 | AFC124773 |
| 6 | São Paulo, SP | Chronic hepatitis after fulminant hepatitis | Blood transfusion | 76628 | AFC124779 |
| 7 | São Paulo, SP | Steatohepatitis | None | 49592 | AFC124763 |
| 8 | São Paulo, SP | Liver cirrhosis | Blood transfusion | OS | AFC124764 |
| 9 | São Paulo, SP | Acute hepatitis; HCV positive | Acute lymphoid leukemia, blood transfusions | RV | AFC124766 |
| 10 | São Paulo, SP | Chronic hepatitis | None | 71841 | AFC124776 |
| 11 | São Paulo, SP | Healthy carrier | Wife of patient 10; travel to Egypt | 67136 | AFC124769 |
| 12 | São Paulo, SP | Chronic hepatitis | Blood transfusion | 76633 | AFC124780 |
| 13 | Limeira, SP | Chronic active hepatitis | Intravenous drug user | SLS | AFC124784 |
| 14 | São Paulo, SP | Chronic active hepatitis; HCV positive | Cholecystectomy | 77811 | AFC124781 |
| 15 | São Paulo, SP | Liver cirrhosis | None | 70064 | AFC124774 |
| 16 | São Paulo, SP | Liver cirrhosis; HCV positive | Open heart surgery | 49438 | AFC124762 |
| 79956 | AFC124782 | ||||
| 17 | São Paulo, SP | Chronic active hepatitis | None | 48729 | AFC124760 |
| 18 | São Paulo, SP | Liver cirrhosis; HCV positive | Amazon basin inhabitant for 1 yr | 70247 | AFC124775 |
| 19 | São Paulo, SP | Steatohepatitis | Frequent traveller to Amazon basin; b. Italy | 60013 | AFC124767 |
| 20 | Maceió, AL | Chronic hepatitis | Wife with chronic hepatitis | 75503 | AFC124777 |
| 21 | Ribeirão Preto, SP | Chronic hepatitis | Blood transfusion | 86122 | AFC124783 |
| 22 | São Paulo, SP | Liver cirrhosis; HCV positive | Blood transfusion | DVON | AFC124761 |
| 23 | São Paulo, SP | Liver cirrhosis; HCV positive | Blood transfusion | 76282 | AFC124778 |
City and state. SP, São Paulo; AL, Alagoas.
b, born in.
Results and discussion.
We detected GBV-C RNA in 13 of 137 (9.5%) Brazilian non-A-E hepatitis patients, in general agreement with the preliminary data reported by our group (1 of 13; 7.5%) (17). This prevalence is similar to those reported elsewhere—8.3% in Europe (13), 8.7% in the United States (6), and 8% in Japan (19)—although higher levels are observed in China (16.7% [20]) and, most noticeably, in Italy (39% [7]). These cases remain without etiological diagnosis, since no relationship between GBV-C and liver diseases has been proven (1, 2) and no other known causes of hepatitis (such as alcoholism, drug use, autoimmunity, or hereditary diseases) were reported among these patients, with the exception of one with a history of alcoholism.
GBV-C was also detected in 8 of 44 (18.2%) HCV-infected individuals. This is within the range reported in other regions: 8% for Japan (19), 8.2% for China (20), 18.7% for Europe (13), and 20% for the United States (4). The fact that this prevalence is higher than that observed among non-A-E hepatitis patients probably reflects the greater level of exposure to blood products in HCV-infected patients.
In contrast to the situation in patients with hepatitis, our data reveal that Brazil has one of the highest prevalences of GBV-C among volunteer blood donors reported so far: 5 of 95 (5.2%) and 5 of 76 (6.5%), among donors with normal and elevated ALT levels, respectively. A higher prevalence in blood donors was found in Vietnam (7.4%) (3). In China, the GBV-C prevalence was 7.9% in professional blood donors but only 0.6% in healthy volunteers (20). In the United States, the GBV-C prevalence in commercial blood donors was 12.9% (4). Much lower prevalences are found in volunteer blood donor populations from the United States, ranging from 0.8 (4) to 1.7% (13) in donors with normal ALT levels and from 1.5 (13) to 3.9% (4) in donors with elevated ALT levels. The finding of similar prevalence rates among non-A-E hepatitis patients and blood donors with normal and elevated ALT levels reinforces the hypothesis that GBV-C is not pathogenic in most infected individuals (1, 2). It should be noted that the prevalence of this virus in Brazil is probably higher than that shown herein, as other authors have since published reports that by using 5′ noncoding region-derived primers, about one-third more cases could be detected (15, 22). Other studies should be carried out to better analyze GBV-C prevalence in different Brazilian populations. These studies should verify whether the high prevalence in Brazilian candidate volunteer blood donors is also found in the general population, since the former are generally relatives or close friends of hospitalized patients needing blood transfusions. Perhaps this fact introduces an important bias in the selection of Brazilian blood donors and is related to the elevated prevalence found in this study.
GBV-C sequences obtained from the Brazilian patients were checked for any insertion or deletion that would alter the coding capacity of the NS3 gene. We did not find any mutational event that would cause frameshifts or premature termination in the sequences.
It has been previously shown that the utilization of short sequence fragments from the NS3 region did not provide relevant information for classification of this virus (16). Because of a lack of phylogenetic information in the sequence data (reflected in the low bootstrap values), it was not possible to fully resolve the evolutionary relationships of a worldwide sample of GBV-C sequences by using the NS3 amplicon described here. A longer region is clearly required for analyses of this kind.
However, the analysis of our sequence data from the families of two patients with index cases provides important information about the mode of GBV-C transmission (Fig. 1). In one family (F2), no close relationship was found among the sequences obtained from two infected individuals, suggesting that infection in this couple was coincidental (not an unlikely event given the high prevalence of this virus). In another family (F1), the sequences obtained from three infected members (the father, the mother, and the younger of two daughters) form a tight phylogenetic cluster (100% bootstrap support), strongly supporting intrafamilial transmission. No other GBV-C risk factor was reported for any other member of the family. The route of transmission within this family is probably by sexual contact between the parents and by vertical transmission from the mother to her second daughter. Intrafamilial transmission of this kind may therefore represent an important pathway for the propagation of GBV-C and may help to explain the high prevalence among blood donors.
FIG. 1.
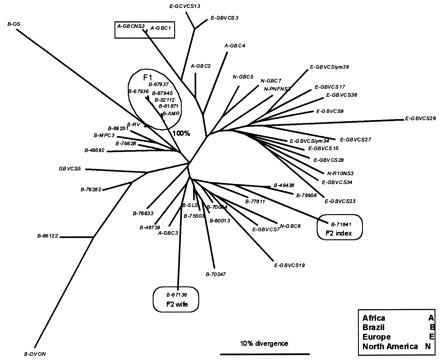
Neighbor-joining phylogenetic tree of GBV-C NS3 sequences. The geographical origin of each sequence is indicated by the first letter in the sequence identifier (see the key). Encircled lineages represent viral samples from members of the two families (F1 and F2). The boxed lineages at the top of the tree are from the same patient. Branch lengths are proportional to the degrees of sequence divergence, estimated by a maximum-likelihood method.
The genetic distances among the sequences obtained from family F1 were 5 to 10 times smaller (data not shown) than the distances among sequences from unrelated patients which appeared adjacent in the tree (e.g., B-MPC3 and B-69251). On the other hand, different sequences from the same patient show a divergence level which is comparable to that observed for F1 (e.g., A-GBCNS3 and A-GBC1 [Fig. 1]). Given the close relatedness among the sequences obtained from viruses circulating in members of the same family (F1), intrafamilial transmission is the most parsimonious explanation, because otherwise one has to postulate several transmission events from an index case not included in this study, which would entail several independent transmission events.
In conclusion, a high prevalence of GBV-C was found among different Brazilian populations, including non-A-E hepatitis and hepatitis C patients, candidate blood donors, and intrafamilial contacts of two patients with index cases. The high prevalence of this virus seems to be sustained by efficient contagion mechanisms that facilitate intrafamilial transmission. Blood transfusion is also another efficient mechanism for sustaining the high GBV-C prevalence, as no screening test specific for this virus is routinely used.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the sequence data reported in this article are AF124758 through AF124784.
Acknowledgments
We thank Edward C. Holmes (Department of Zoology, Oxford University, Oxford, United Kingdom) for suggestions and comments during the preparation of the manuscript.
This work was supported by FAPESP, São Paulo, São Paulo State, Brazil (Proc. 97/04955-0), and by the Laboratório Bioquímico Jardim Paulista.
REFERENCES
- 1.Alter H J, Nakatsuji Y, Melpolder J, Wages J, Wesley R, Shih W-K, Kim J P. The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. N Engl J Med. 1997;336:747–754. doi: 10.1056/NEJM199703133361102. [DOI] [PubMed] [Google Scholar]
- 2.Alter M J, Gallagher M, Morris T M, Moyer L A, Meeks E L, Krawcynski K, Kim J P, Margolis H S for the Sentinel Counties Viral Hepatitis Team. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. N Engl J Med. 1997;336:741–746. doi: 10.1056/NEJM199703133361101. [DOI] [PubMed] [Google Scholar]
- 3.Brown K E, Wong S, Buu M, Binh T V, Be T V, Young N S. High prevalence of GB virus C/hepatitis G virus in healthy persons in Ho Chi Minh City, Vietnam. J Infect Dis. 1997;175:450–453. doi: 10.1093/infdis/175.2.450. [DOI] [PubMed] [Google Scholar]
- 4.Dawson G J, Schlauder G G, Pilot-Matias T J, Thiele D, Leary T P, Murphy P, Rosenblatt J E, Simons J N, Martinson F E A, Gutierrez R A, Lentino J R, Pachucki C, Muerhoff A S, Widell A, Tegtmeier G, Desai S, Mushahwar I K. Prevalence studies of GB virus C infection using reverse transcriptase polymerase chain reaction. J Med Virol. 1996;50:97–103. doi: 10.1002/(SICI)1096-9071(199609)50:1<97::AID-JMV16>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.Egawa K, Yukawa T, Arakawa S, Nakao H, Inoue T, Tanaka T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. Infection with GB virus C in leprous patients in Japan. J Med Virol. 1996;49:110–114. doi: 10.1002/(SICI)1096-9071(199606)49:2<110::AID-JMV7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 6.Feucht H H, Zollner B, Polywka S, Laufs R. Vertical transmission of hepatitis G. Lancet. 1996;347:615–616. [PubMed] [Google Scholar]
- 7.Fiordalisi G, Zanella I, Mantero G. High prevalence of GB virus C infection in a group of Italian patients with hepatitis of unknown etiology. J Infect Dis. 1996;174:181–183. doi: 10.1093/infdis/174.1.181. [DOI] [PubMed] [Google Scholar]
- 8.Fiordalisi G, Bettinardi A, Zanella I, Stellini R, Paraninfo G, Cadeo G, Primi D. Parenteral and sexual transmission of GB virus C and hepatitis C virus among human immunodeficiency virus-positive patients. J Infect Dis. 1997;175:1025–1026. [PubMed] [Google Scholar]
- 9.Kao J H, Chen W, Chen P J, Lai M Y, Lin R Y, Chen D S. GB virus-C/hepatitis G virus infection in prostitutes: possible role of sexual transmission. J Med Virol. 1997;52:381–384. [PubMed] [Google Scholar]
- 10.Kao J H, Liu C J, Chen P J, Chen W, Hsiang S C, Lai M Y, Chen D S. Interspousal transmission of GB virus-C/hepatitis G virus: a comparison with hepatitis C virus. J Med Virol. 1997;53:348–353. [PubMed] [Google Scholar]
- 11.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;239:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 12.Leary T P, Muerhoff A S, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M L, Schlauder G G, Dawson G J, Desai S M, Mushahwar I K. Consensus oligonucleotide primers for the detection of GB virus C in human cryptogenic hepatitis. J Virol Methods. 1996;56:119–121. doi: 10.1016/0166-0934(95)01956-1. [DOI] [PubMed] [Google Scholar]
- 13.Linnen J, Wages J, Zhang-Zeck Z-Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W K, Young L, Piatak M, Jr, Hoover C, Fernandez J, Chen S, Zou J-C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 14.Miyakawa Y, Mayumi M. Hepatitis G virus—a true hepatitis virus or an accidental tourist? N Engl J Med. 1997;336:795–796. doi: 10.1056/NEJM199703133361109. [DOI] [PubMed] [Google Scholar]
- 15.Muerhoff A S, Simons J N, Erker J C, Desai S M, Mushahwar I K. Identification of conserved nucleotide sequences within the GB virus C 5′-untranslated region: design of PCR primers for detection of viral RNA. J Virol Methods. 1996;62:55–62. doi: 10.1016/0166-0934(96)02088-5. [DOI] [PubMed] [Google Scholar]
- 16.Muerhoff A S, Smith D B, Leary T P, Erker J C, Desai S M, Mushahwar I K. Identification of GB virus C variants by phylogenetic analysis of 5′ untranslated and coding region sequences. J Virol. 1997;71:6501–6508. doi: 10.1128/jvi.71.9.6501-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinho J R R, Capacci M L, da Silva L C, Santos C A, Ballarati C A F, Carrilho F J, Pugliesi V, Guz B, Bernardini A P. Hepatitis G virus/GB virus C in Brazil. Preliminary report. Rev Inst Med Trop São Paulo. 1996;38:243–246. doi: 10.1590/s0036-46651996000300016. [DOI] [PubMed] [Google Scholar]
- 18.Simons J N, Leary T P, Dawson G, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 19.Sugai Y, Nakayama H, Fukuda M, Sawada N, Tanaka T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. Infection with GB virus C in patients with chronic liver disease. J Med Virol. 1997;51:175–181. [PubMed] [Google Scholar]
- 20.Wang H-L, Lin D-Y. Prevalence and genotype of hepatitis G virus in Chinese professional blood donors and hepatitis patients. J Infect Dis. 1997;175:1229–1233. doi: 10.1086/593676. [DOI] [PubMed] [Google Scholar]
- 21.Zanetti A R, Tanzi E, Romano L, Principi N, Zuin G, Minola E, Zapparoli B, Palimieri M, Marini A, Ghisotti D, Friedman P, Hunt J, Laffler T. Multicenter trial on mother-to-infant transmission of GBV-C virus. The Lombardy study group on vertical/perinatal hepatitis viruses transmission. J Med Virol. 1998;54:107–112. doi: 10.1002/(sici)1096-9071(199802)54:2<107::aid-jmv7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X H, Shinzawa H, Shao L, Ishibashi M, Saito K, Ohno S, Yamada N, Misawa H, Togashi H, Takahashi T. Detection of hepatitis G virus RNA in patients with hepatitis B, hepatitis C, and non-A-E hepatitis by RT-PCR using multiple primer sets. J Med Virol. 1997;52:385–390. [PubMed] [Google Scholar]


