SUMMARY
Spiking immature oligodendrocytes (OLs), referred to as spiking OLs, express voltage-activated Na+ channels (Nav) and K+ (Kv) channels, endowing a subpopulation of OLs with the ability to generate Nav-driven spikes. In this study, we investigate the molecular profile of spiking OLs, using single-cell transcriptomics paired with whole-cell patch-clamp recordings. SCN2A, which encodes the channel Nav1.2, is specifically expressed in spiking OLs in the brainstem and cerebellum, both in mice and in Olive baboons. Spiking OLs express lineage markers of OL progenitor cells (OPCs) and pre-myelinating OLs, indicating they belong to a transitional stage during differentiation. Deletion of SCN2A reduces the Nav current-expressing OL population and eliminates spiking OLs, indicating that SCN2A is essential for spiking in OLs. Deletion of SCN2A does not impact global OL proliferation but disrupts maturation of a subpopulation of OLs, suggesting that Nav1.2 is involved in heterogeneity in OL lineage cells and their development.
Graphical Abstract
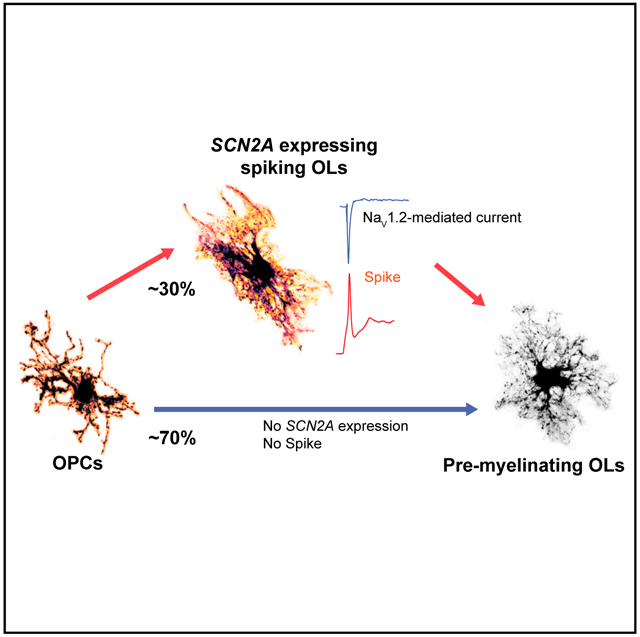
In brief
Using single-cell transcriptomics paired with whole-cell patch-clamp recordings, Gould and Kim demonstrate that Nav1.2 channels, encoded by SCN2A, endow a subpopulation of immature oligodendrocytes (OLs) with the ability to spike, and serve an essential role in OL maturation.
INTRODUCTION
Oligodendrocytes (OLs) all have an ability to produce myelin in the central nervous system (CNS). From development into adulthood, a diverse array of OL lineage cells populate the CNS with distinct phenotypes depending on their differentiation state, genetic profile, morphology, physiologic characteristics, and location (Foerster et al., 2019). Previous studies using single-cell transcriptomic analyses revealed that OL progenitor cells (OPCs) and myelinating OLs have heterogeneous cell populations, highlighting the diversity of immature OL populations (Zhang et al., 2014; Marques et al., 2016; Jäkel et al., 2019; Marisca et al., 2020). Immature OLs show distinct transcriptional profiles for neurotransmitter receptors and various ion channels, which mediate communication with neighboring neurons (Marques et al., 2016; Spitzer et al., 2019). However, the functional relevance of immature OLs with distinct transcriptional profiles remains unclear. In this study, we paired transcriptional analysis and electrophysiological recordings in a single OL to understand the molecular profile of a physiologically distinct subtype of immature OLs in the developing brain.
OL lineage cells undergo dynamic changes in physiological properties as they mature into myelinating OLs (Bakiri et al., 2009; Larson et al., 2016; Berret et al., 2017). A sub-population of immature OLs, including OPCs and pre-myelinating OLs, express voltage-gated sodium (Nav) and potassium (Kv) channels that potentially contribute to action potential generation (Káradóttir et al., 2008; Clarke et al., 2012; Berret et al., 2017). Most studies indicate OPCs (NG2+ or platelet-derived growth factor receptor alpha [PDGFRα+] cells) produce depolarizing transients with smaller amplitude, slower kinetics, and no clear threshold compared with the distinct all-or-none, large-amplitude, rapid spikes of an action potential or spike (De Biase et al., 2010; Clarke et al., 2012). However, several studies have identified a sub-population of OPCs capable of producing large-amplitude spikes with a clear threshold (Chittajallu et al., 2004; Káradóttir et al., 2008; Ge et al., 2009). The idea that OL lineage cells can produce spikes has supported the identification of a sub-population of immature OLs (CNPase+ OLs) that displays an action potential with an all-or-none manner in response to depolarization (Berret et al., 2017).
Spiking OLs are defined by their ability to produce action potentials with a clear threshold, a larger amplitude, and faster repolarization than OPCs (Berret et al., 2017). The amplitude and kinetics of Nav channel currents differed between OPCs and spiking OLs (Berret et al., 2017). Nav channel density is different among OPCs, and the proportion of OPCs with Na+ currents differs across brain regions (Clarke et al., 2012; Berret et al., 2017; Spitzer et al., 2019). Thus, a sub-population of immature OLs with a specific subtype of Nav channel or increased Na+ current density may produce spikes.
Diversity in expression of voltage-gated ion channels could underlie the distinct physiologic properties of spiking OLs. For example, there is a marked increase in expression of slow-inactivating delayed rectifier (KDR) and inward-rectifier (Kir) potassium channels in gray matter OPCs compared with white matter OPCs (Chittajallu et al., 2004). However, currently, there is sparse evidence regarding the diversity of Nav channels in OLs.
Based on transcriptomic studies using single-cell sequencing, four Nav channel subtypes, Nav1.1, Nav1.2, Nav1.3, and Nav1.6, have been identified in OLs, including OPCs (Zhang et al., 2014; Larson et al., 2016; Marques et al., 2016). Immunostaining and pharmacological approaches reveal that spiking in OLs expresses Nav1.2 rather than Nav1.6 in the rat brainstem (Berret et al., 2017). However, whether the expression of SCN2A, the gene encoding the Nav1.2 channel, is specific to spiking OLs or is a key determinant for spiking OLs remains unexplored. Notably, the introduction of SCN2A into cultured OLs confers the ability to spike and enhances myelinating capacity (Jiang et al., 2013). In contrast, the knockdown of Nav1.2 expression using short hairpin RNA (shRNA) in immature OLs reduced myelin basic protein (MBP), indicating the potential role of Nav1.2 in myelination (Berret et al., 2017). Using single-cell transcriptomics paired with whole-cell patch-clamp recordings, we demonstrate that spiking OLs, distinctly expressing SCN2A, are a molecularly distinct population among immature OLs in the developing nervous system. In addition, we found that Nav1.2, encoded by SCN2A, specifically contributes to OL lineage progression and development.
RESULTS
Scn2a expression is restricted to spiking OLs in the mouse brainstem
Using whole-cell patch-clamp recording, we characterized physiological features of OLs in the medial nucleus of the trapezoid body (MNTB) of the mouse brainstem between post-natal days (P) 9 and 12, an onset of rapid myelination (Sinclair et al., 2017). Spiking OLs were identified by the presence of spikes produced by depolarizing current injection (up to 200 pA). Spiking OLs were distinguished from non-spiking OLs by three criteria: (1) >80 mV of spike amplitude (95.9 ± 9.14 mV), (2) overshoot with >0 mV (30.2 ± 11.7 mV of peak potential), and (3) a spike peak much larger than the sustained depolarization following a spike. Physiological properties of spiking OLs from mouse MNTB were comparable with spiking OLs from the rat MNTB (Berret et al., 2017).
We further classified non-spiking OLs into three categories based on physiological and morphological properties: OPC, pre-myelinating OL, and myelinating OL, as described previously (Berret et al., 2017). OPCs display Nav currents and an incomplete spike, which had no clear threshold and failed to reach 0 mV, as previously described (De Biase et al., 2010; Clarke et al., 2012). Pre-myelinating OLs and myelinating OLs did not exhibit distinct Nav currents or spikes (Figure 1A).
Figure 1. Transcription profiling of spiking and non-spiking OLs.

(A) Representative traces of membrane potential changes from spiking (n = 12 cells), OPCs (n = 12 cells), pre-myelinating (n = 7 cells), and myelinating (n = 8 cells) OLs from 10 mice. In current-clamp recordings, spiking OLs show distinct spikes in response to current injections with 150-ms current steps of increasing amplitude (10-pA increments from −70 pA). Scale is consistent across the traces. Inset: a characteristic spike (red) versus OPC transient (gray) as highlighted on the representative trace (box).
(B) Multi-dimensional plot of gene expression. Cells are shown as individual columns. Rows are different genes. Heatmap represents the relative expression level to all genes. Low expression is blue, high expression is red (see scale bar). Gapdh and Actin as reference genes are expressed in all cells at consistent levels (yellow). Bubbles show relative gene expression between different populations (spiking OL = red, non-spiking OLs = blue). Diameter represents the mean of that population relative to the other populations. Notably, Scn2a was not detected in non-spiking OLs.
(C) Scheme of tamoxifen injection (TAM) on mouse pups (at P8) and electrophysiological recording followed by post-immunostaining (EP & Immuno.). Confocal images and associated traces of membrane potential changes from dye-filled TdTomato+ OPCs at day 1 (gray trace) and spiking OLs at day 2 (black trace) after tamoxifen injection. Notably, TdTomato+ spiking OLs do not express PDGFRA. Scale bars, 10 μm.
(D) The percentage of recorded cells that showed characteristic electrophysiological profiles of OPCs (gray) or spiking OLs (black) over the time after tamoxifen administration.
To determine the molecular mechanisms that allow immature OLs to spike, we examined the transcriptional profile of spiking OLs at the single-cell level, using single-cell polymerase chain reaction (qPCR). After evaluating electrophysiological properties using whole-cell recordings, we aspirated cells and collected them with a success rate of 80% (39/49 cells). In total, 47 genes were examined (Figures S1 and S2). Collected OLs were transcriptionally distinct from neurons and showed significantly lower levels of Map2, a neuronal marker (Figures S2A and S2B).
We found that a specific type of Nav channel was associated with a distinct spike from immature OLs. Scn2a, which encodes the alpha subunit of Nav1.2, was expressed in a highly restricted manner with discrete expression in spiking OLs. Overall, 8 of the 12 spiking OLs displayed Scn2a expression (mean RNA level = 226.4 ± 279.8 in spiking OLs). The discrete expression of Scn2a was striking, because other Nav channel genes (Scn1a, Scn3a, and Scn8a) were observed across the immature population without a discrete cell-specific expression (Figure 1; Figures S1A and S2C).
In OPCs, Na+ current density varies among different brain regions, and spiking has been reported in a subpopulation of OPCs (Spitzer et al., 2019). To determine whether spiking OLs represent a specific lineage stage, we analyzed expression of well-known stage markers. OPCs had significantly higher levels of Pdgfra mRNA (spiking OLs: 870.1 ± 1,602; OPCs: 1,184 ± 1,650; pre-myelinating OLs: 32.87 ± 58.45; myelinating OLs: 31.75 ± 78.82; Kruskal-Wallis test, p = 0.011) and Cspg4 (spiking OLs: 1,813 ± 2,830; OPCs: 2017 ± 2581; pre-myelinating OLs: 355.7 ± 545.3; myelinating OLs: 391.4 ± 483.6; Kruskal-Wallis test, p = 0.043) compared with myelinating OLs. Spiking OLs and OPCs expressed Pdgfra and Cspg4 at similar levels (Mann-Whitney, p = 0.33 and p = 0.32, respectively). The cell-cycle gene, Mki67, was highest in the OPC and spiking OL population (spiking OLs: 2,813 ± 5,638; OPCs: 3,686 ± 2,947; pre-myelinating OLs: 622.8 ± 572.8; myelinating OLs: 460.1 ± 688.7; Kruskal-Wallis test, p = 0.0035), indicating that spiking OLs have the potential to proliferate. Notably, Enpp6, an indicator for pre-myelinating OLs, was highly expressed in spiking OLs and non-spiking pre-myelinating OLs compared with OPCs (OPCs: 19.7 ± 48.3; spiking OLs: 1,121 ± 1,640; pre-myelinating: 1,934 ± 2,395; Kruskal-Wallis test, p = 0.02). Spiking OLs expressed RNA markers for OPCs and newly differentiated pre-myelinating OLs, indicating that spiking OLs could be a transient stage between OPCs and pre-myelinating OLs.
Spiking OLs expressed mRNA for myelin genes, including Mag, Mobp, and Plp1. Mag was significantly higher in myelinating OLs than OPCs (OPCs: 294 ± 574; myelinating OLs: 93,781 ± 158,418; Kruskal-Wallis test, p = 0.04). Expression of Mag in spiking OLs was significantly higher than in OPCs (spiking OLs: 6,339 ± 1,806; Kruskal-Wallis test, p = 0.01) and similar to pre-myelinating OLs (1,879 ± 2,144). The presence of mRNA for myelin genes indicates spiking OLs have the potential to myelinate axons. Overall, our results show that Scn2a is a distinct molecular feature of spiking OLs, which have the genetic markers of committed OPCs and newly forming pre-myelinating OLs.
Spiking OLs are in a transient stage between OPCs and pre-myelinating OLs
The presence of Pdgfra and Cspg4 RNA in spiking OLs suggests that the latter could be a subpopulation of OPCs. To test this possibility, we examined whether spiking OLs express PDGFRA protein using whole-cell recoding and post-immunostaining. All dye-filled OPCs (n = 9) with identified physiologic properties were positive for PDGFRA. None of the spiking OLs (n = 10) were positive for PDGFRA in post-immunostaining, indicating that PDGFRA protein is downregulated in spiking OLs (Figure S2E). This result is consistent with the previous finding that spiking OLs do not express NG2 protein, encoded by Cspg4, but express CNP, a marker for pre-myelinating OLs (Berret et al., 2017).
To pinpoint when the spiking phenotype appears during the transition from OPCs, we utilized an inducible Cre line driven by the PDGFRA promoter to label OPCs through expression of TdTomato (Td; MGI: 3809524). At day 1 after tamoxifen injection, all Td+ cells had electrophysiological characteristics of an OPC, displaying an incomplete spike, and were positive on PDGFRA immunostaining (Figures 1C and 1D). No Td+ spiking OLs were identified at 1 day after tamoxifen induction. At day 2, Td+ and PDGFRA− spiking OLs appeared, indicating that a subset of OPCs had transitioned to spiking OLs (30%, 4/12 Td+ cells). The proportion of spiking OLs decreased at day 3 (1/15 Td+ cells; Figure 1D). These studies demonstrate that spiking OLs are a distinct stage from OPCs and suggest that spiking is a functional indicator for a transient stage between OPCs and pre-myelinating OLs.
Scn2a-expressing spiking OLs are present in the mouse cerebellum and the white matter of non-human primates
Scn2a is observed in a subset of OL lineage cells across the CNS (Zhang et al., 2014; Marques et al., 2016). To examine whether spiking OLs exist in other brain regions during early post-natal development, we performed electrophysiological classification and gene profiling in the white matter of the mouse cerebellum at P9–P10. In total, 11 spiking OLs, 7 OPCs, 5 pre-myelinating OLs, and 7 myelinating OLs were collected. Aspiration success rate was 67% (30/41 cells). Spiking OLs displayed spikes with a large amplitude (88.6 ± 8.51 mV) with an overshoot > 0 mV (22.6 ± 8.78 mV of peak potential; Figure 2A). Physiological features of spiking OLs in cerebellar white matter resembled those in the brainstem. Post-recording immunostaining showed that spiking OLs filled with fluorescence dye (Lucifer yellow) were NEUN−/OLIG2+/PDGFRA− OLs in the white matter of the cerebellum (Figure 2B). Spiking OLs had the highest expression of Scn2a compared with non-spiking OLs (spiking OLs: 281.9 ± 78.0 versus OPCs: 0, pre-myelinating OLs: 77.9 ± 107, myelinating OLs: 35.29 ± 94.0; one-way ANOVA, Bennet’s post hoc adjustment, p = 0.01; Figure 2C). The pattern of the other sodium channels was similar to that of the brainstem, with no distinct expression in a single population (Figure S1).
Figure 2. Spiking OLs in the developing cerebellar white matter.

(A) Spiking OLs show a distinct spike in response to current injections of 150-ms current steps of increasing amplitude (10-pA increments from −70 pA).
(B) Post-immunostaining of a spiking OL (Lucifer yellow) in the mouse cerebellum with PDGFRA, OLIG2, and NEUN. (Top) A single focal image with orthogonal views taken at 633 showed an OLIG2+ and PDGFRA− spiking OL. Scale bars, 10 μm. (Bottom) Location of the recorded cell (yellow circle) in white matter with a high density of OLIG2+ OLs (cyan). Scale bars, 100 μm.
(C) Multi-dimensional plot of gene expression. Cells are shown as individual columns. Rows are different genes. Heatmap represents the relative expression level to all genes. Low expression is blue; high expression is yellow. Bubbles show relative gene expression between different populations (spiking OLs = red, nonspiking = blue). Diameter represents the relative mean.
(D) (Top) Representative traces of membrane potential changes of spiking OLs in Olive baboon in response to current injections in current-clamp recording. (Bottom) Multi-dimensional plot of gene expression of spiking OLs and non-spiking OLs from baboon cerebellum.
(E) Immunostaining of baboon cerebellum with Nav1.2 (yellow), NEUN (red), and OLIG2 (cyan). White line delineates white matter (right side) from granule cell layer (left side). Arrows indicate Nav1.2+/NEUN−/OLIG2+ OLs in white matter. Scale bars, 20 μm (top); 10 μm (bottom).
Consistent with rat and mouse brain, spiking OLs are found in the white matter of the non-human primate cerebellum. Slices of cerebellum were made following pre-term delivery of Papio anubis (Olive baboon) at 127–140 days gestation via C-section, a period of active myelination and comparable with mouse cerebellum development (Rees et al., 2009; Takahashi et al., 2014). In whole-cell recordings, ~63% of immature OLs (5/8 recorded cells, 3 baboons) showed distinct spikes in response to depolarizing current injections (Figure 2D). Spiking OLs exhibited spikes with amplitudes of 81.8 ± 13.1 mV from −70 mV and overshoot to 20.2 ± 2.8 mV. Scn2a expression was restricted to spiking OLs (5 spiking OLs, 3 non-spiking OLs; Figure 2D). Other genes encoding Nav channels (Scn8a, Scn3a, and Scn1a) were expressed in spiking OLs and non-spiking OLs (Figure S3). Immunostaining of the baboon cerebellum showed that OLIG2+ OLs express Nav1.2 channels, indicating that the white matter of non-human primate brain exhibits Nav1.2-mediated spiking OLs (Figure 2E).
SCN2A, encoding Nav1.2 channel, is essential for generating spikes in OLs
To determine whether Scn2a expression is required for OLs to produce spikes, we generated a SCN2A conditional knockout (cKO) mouse line using a Cre-Flox system because the conventional deletion of SCN2A is lethal (Planells-Cases et al., 2000). SCN2A-floxed (SCN2Af/f) mice were crossed into an inducible ACTIN-driven Cre line to globally delete SCN2A in the brain. In this global (G)-cKO mouse model, we validated a newly generated Flox line (Figures S4A–S4D) and evaluated the impact of SCN2A loss on OL physiology (Figure 3). In controls, 14 of 41 cells (from 11 control mice) showed spikes, while in G-cKO mice, no recorded OLs (of 31 cells from 7 G-cKO mice) displayed spikes in response to depolarizing current injections (Figure 3A). To examine whether the loss of OL spiking was caused by reduced Nav1.2-mediated currents, we recorded voltage-activated Na+ currents in OLs from control and G-cKO mice (Figure 3B). In controls, 84% of recorded OLs showed Na+ currents (38/45 cells total from 14 mice). In G-cKO mice, 64% of recorded OLs displayed Na+ currents (28/44 cells from 9 G-cKO mice), whereas the rest (36% of OLs) did not display Na+ currents (16/44 cells). Global loss of SCN2A significantly reduced the proportion of OLs exhibiting Na+ currents in the MNTB (chi-square test, p = 0.04; Figure 3B).
Figure 3. SCN2A is required for spiking in OL and OPC differentiation.

(A) Representative traces of membrane potential changes of immature OLs from control (black) and global SCN2A conditional knockout mice (G-cKO, red). Pie chart shows the proportion of spiking OLs (black) to non-spiking OLs (gray) among recorded cells in control (41 cells from 11 mice) and G-cKO mice (31 cells from 7 mice). Numbers indicates spiking OLs/total OLs (top).
(B) In voltage-clamp recordings, Nav currents were recorded (at −50 to −20 mV, Δ = 10 mV) in OLs from control (45 cells from 14 mice) and G-cKO mice (44 cells from 9 mice). Pie chart shows the proportion of cells exhibiting Nav currents (red) and cells without Nav currents (gray) among total recorded cells. Numbers indicate Na+ current-expressing OLs versus total OLs (right).
(C) Summary of INa density at −20 mV in control and G-cKO cells. Line indicates mean ± SD. *p < 0.05.
(D) Immunostaining of mouse brainstem with PDGFRA, OLIG2, and EdU from control (at P9). A single focal image with orthogonal views shows one EdU+ cell that does not express PDGFRA (arrowheads), but the other EdU+ cell (arrows) expresses PDGFRA (cyan). Scale bars, 10 μm.
(E) The proliferation rate in PDGFRA+ OLs and Nav1.2+ OLs is indicated by the percent positive for EdU (blue).
(F) (Top) Immunostaining of mouse brainstem with Nav1.2+, OLIG2, and EdU from control and G-cKO (at P9). (Bottom) The percentage of OLIG2+ OLs labeled with EdU (EdU+OLIG2+/OLIG2+ * 100) did not differ between control (n = 8 mice) and G-cKO mice (n = 8 mice). Data were shown as mean ± SEM.
(G) Immunostaining of mouse brainstem with CC1 and OLIG2 (top), PDGFRA and OLIG2 (middle), and merge with MAP2 (bottom) in control and G-cKO mice at P9. Scale bars, 20 μm.
(H) The ratio of OLIG2+ OLs to MAP2+ neurons decreased in G-cKO mice with a significant reduction in mature CC1+ cells.
(I) The percentage of OLIG2+ OLs expressing PDGFRA increased, whereas the percentage of OLIG2+ OLs expressing CC1+ decreased in G-cKO mice (each 6 mice). Data were shown as mean ± SEM. *p < 0.05.
To verify that the remaining Na+ currents in G-cKO OLs are not mediated by Nav1.2 channel, we examined their sensitivity to μ-conotoxin KIIIA, a blocker of Nav1.2 channels (1 μM) (Zhang et al., 2007, 2010; Berret et al., 2017). In control mice, 5 of 10 recorded OLs showed ~90% reduction of Na+ currents in the presence of conotoxin KIIIA (two-way ANOVA, p < 0.05), while the others (n = 5) were insensitive (Figures S4E–S4G). None of the Na+ currents in G-cKO were sensitive to conotoxin KIIA, indicating SCN2A deletion eliminates Nav1.2-medated Na+ currents in OLs, and that the remaining Na+ currents are mediated by other subtypes of Nav channels in G-cKO mice.
We then examined if the loss of spiking in OLs in G-cKO mice was related to changes in Na+ current density. The density of Na+ currents, calculated by division of the peak currents by membrane capacitance (pA/pF), was significantly smaller in G-cKO mice (−18 ± 9.2 pA/pF at −20 mV, 27 cells/9 mice) than controls (−24 ± 13 pA/pF at −20 mV, 36 cells/14 mice, repeated-measures ANOVA, p < 0.001; Figure 3C). There was no difference in membrane capacitance between groups (15.2 ± 0.94 pF in control versus 16.6 ± 1.57 pF in G-cKO). Deletion of SCN2A significantly reduced the density of Na+ currents in immature OLs without changing cell size.
Loss of SCN2A impairs OL differentiation
Next, we investigated the impact of SCN2A loss on both proliferation and differentiation of immature OLs. To determine whether the proliferating cells (EdU+) include both PDGFRA+ OPCs and PDGFRA− OLs, we counted EdU+ cells that were PDGFRA+ (Figure 3D). In control mice, most EdU+ cells were PDGFRA+ OPCs, with ~26% of PDGFRA+ OPCs incorporating EdU (31/117 cells, 3 mice; Figure 3E). Only ~8% of PDGFRA− OLs proliferated within the same period (9/117 cells, 3 mice). A few Nav1.2+ OLs (~14%) proliferated within 2 h (6/43 cells, 6 mice; Figures 3E and 3F), indicating that most Nav1.2+ OLs do not proliferate in control mice. OLIG2+ OLs that incorporated EdU were similar in controls and G-cKO mice, suggesting that the absence of SCN2A does not have a broad impact on OL proliferation (G-cKO: 6.7% ± 0.72%; control: 8.8% ± 1.0%, 8 mice/genotype; t test, p = 0.12; Figure 3F). The results demonstrate that Nav1.2+ OLs contribute to a small percentage of proliferating OLs in the MNTB, and that SCN2A deletion has no impact on global OL proliferation.
To examine the impact of SCN2A deletion on OL differentiation, we evaluated the numbers of OPCs (PDGFRA+ cells) and mature OLs (CC1+ cells) in the auditory brainstem from P9 control and G-cKO mice (Figure 3G). SCN2A deletion in the mouse brainstem led to a reduction of CC1+ differentiated OLs, as shown by the proportion of CC1+ OLs to MAP2+ neurons (G-cKO: 0.58 ± 0.04, n = 6; control: 1.0 ± 0.06, n = 6; t test, p < 0.001; Figure 3H) and the percentage of OLIG2+ OLs that express CC1 (G-cKO: 59 ± 1.6; control: 74 ± 2.2, t test, p < 0.001; Figure 3I). The percentage of OLIG2+ OLs that expressed the OPC marker PDGFRA increased in G-cKO mice (G-cKO: 21 ± 2.1 versus control: 9.1 ± 0.76, t test, p < 0.001; Figure 3I). Taken together, this result indicates that SCN2A deletion impairs differentiation of a subpopulation of OPCs.
The cell-autonomous role of SCN2A in development of OL lineage cells
Because SCN2A is expressed in both neurons and OLs in the CNS, global deletion of SCN2A may influence OL lineage cell maturation via alterations in neuronal activity. To examine whether SCN2A serves a cell-autonomous role in OL differentiation, we crossed the SCN2A flox line to an inducible CRE line driven by the PDGFRA promoter (OL-specific cKO [OL-cKO]). In OL-cKO mice, MNTB principal neurons showed no changes in action potential generation (Figure S5). The OL-specific deletion of SCN2A led to a loss of spiking OLs (Figure 4A). In control mice, 10 cells among a total of 37 cell recordings showed spikes, and 26 of 35 cells displayed Na+ currents (9 mice). However, in OL-cKO mice, only 1 of 44 cells displayed spikes (9 mice). Na+ currents were observed in 20 cells of 42 (47%) recorded cells in OL-cKO mice (9 mice), indicating a significant reduction in recorded OLs with Na+ currents (chi-square, p < 0.01; Figure 4B). The density of Na+ currents in the cKO (−4.19 ± 3.7 pA/pF, n = 11 cells from 5 mice) was significantly reduced compared with control mice (−10.1 ± 6.7 pA/pF, n = 11 cells from 5 mice).
Figure 4. Role of oligodendroglia SCN2A in OL lineage cell development.

(A) Representative traces of membrane potential changes of immature OLs from control and OL-specific cKO (OL-cKO) mice. Most OLs in OL-cKO mice did not show spikes.
(B) Pie chart shows the proportion of spiking OLs (black) to non-spiking OLs (gray) and the proportion of OLs with Na+ currents (red) among recorded cells in control (37 cells from 9 mice) and OL-cKO mice (42 cells from 9 mice). Numbers indicates spiking OLs/total OLs (top) or Na+ current-expressing OLs/total OLs (bottom).
(C) Immunostaining of mouse brainstem with PDGFRA, CC1, and OLIG2 in control (top) and OL-cKO mice (bottom) at P9. Scale bars, 10 μm.
(D) The percentage of OLIG2+ OLs expressing PDGFRA (PDGFRA+ and OLIG2+/OLIG2+ * 100) or CC1+ (CC1+ and OLIG2+/OLIG2+ * 100) in control and OL-cKO mice (at P9, n = 4 mice for each genotype). Data were shown as mean ± SEM. *p < 0.05.
(E) Immunostaining of mouse brainstem with OLIG2, EdU, and PDGFRA in control (left) and OL-cKO mice (right) at P9. Scale bars, 25 μm. Inset: EdU+ PDGFRA+ OPCs. Scale bars, 10 μm.
(F) The percentage of total OLIG2+ cells labeled with EdU+ in control and PDGFRA-OL-cKO mice.
(G) The percentage of total PDGFRA+ OPCs labeled with EdU+. *p < 0.05.
(H) The percent of total EdU+ cells that were PDGFRA+ versus PDGFRA− in control and OL-cKO mice.
Similar to G-cKO mice, PDGFRA+ OPC density was increased, while CC1+ OLs were reduced in OL-cKO mice (PDGFRA: OL-cKO, 39% ± 3.6%; control, 23% ± 3.2%; 5 mice/genotype; t test, p = 0.01; CC1: cKO, 44% ± 0.9%; control, 63.5% ± 7.5%; 4 mice/genotype; t test, p = 0.04; Figures 4C and 4D). This increase in OPC density was not due an increase in proliferating OPCs. OL-cKO mice did not exhibit a change in global OL proliferation. 47/551 OLIG2+ cells (8.5%) were EdU+ in control mice (n = 6 mice), and 29/498 OLIG2+ cells (6.0%) were EdU+ in OL-cKO mice (n = 6; chi-square test, p = 0.14; Figures 4E and 4F). Furthermore, fewer PDGFRA+ OPCs incorporated EdU in OL-cKO compared with control mice (OL-cKO: 29/180 OLIG2+ OLs, 16%; control: 31/86 OLIG2+ OLs, 36%; 4 mice/genotype; chi-square test, p = 0.007; Figure 4G). There was no difference in the proliferation of PDGFRA- EDU+ OLs (OL-cKO: 7/76 cells, 9.2%; control: 9/95 cells 9.4%; 4 mice/genotype, chi-square, p = 1; Figure 4H). Thus, increased PDGFRA+ OPCs of OL-cKO mice is not caused by an increased proliferation rate, but rather SCN2A deletion impairs differentiation of a subpopulation of OPCs, which retain a progenitor state. Taken together, our results indicate that SCN2A serves a cell-autonomous role in OL differentiation by contributing to OL excitability.
DISCUSSION
SCN2A expression is restricted to a subpopulation of OLs with the distinct ability to spike
Single-cell transcriptomics reveal that SCN2A is expressed in a cell-specific manner, with the highest expression in spiking OLs in the developing auditory brainstem (Figure 1). Scn2a-expressing spiking OLs were observed across brain regions, including brainstem and cerebellum, and across species, including mice, rats, and the non-human primate, Olive baboons. The results are consistent with previous studies using single-cell transcriptomics demonstrating that Scn2a is identified in OLs across brain regions and in both mice and humans (Zhang et al., 2014; Marques et al., 2016; Jäkel et al., 2019). The identification of SCN2A as a specific transcriptional signature in spiking OLs suggests that spiking OLs are a genetically distinct subpopulation.
We demonstrated that spiking OLs expressed Pdgfra and Cspg4 RNA at the single-cell level but did not express PDGFRA protein, consistent with an absence of NG2 protein in spiking OLs in the rat brainstem at post-natal days 7–14 (Berret et al., 2017). Our single-cell transcriptomic analyses indicate that spiking OLs have higher levels of Enpp6, a marker for pre-myelinating OLs, and myelin genes compared with OPCs. We interpret the overlap of OPCs and pre-myelinating OLs RNA in spiking OLs to mean that a spiking state indicates a transition between them. This idea is supported by transcriptional profiling of single OLs showing that the highest level of expression of Scn2a is observed in newly formed OLs (Marques et al., 2016).
When OPCs proliferate, PDGFRA is rapidly downregulated in daughter cells, leading to asymmetric cell fates within a single day after proliferation (Boda et al., 2015). In addition, genetically deleting PDGFRA leads to loss of protein within 2 days, suggesting rapid turnover of PDGFRA protein (Ðặng et al., 2019). To resolve whether spiking OLs represent a transitional state that arises from OPCs, we assessed genetically labeled OPCs as they matured over time using PDGFRA-driven Td-tomato tracing (Figures 1C and 1D). We found that spiking OLs arise from OPCs with rapidly downregulated protein levels of OPC markers within 2 days of expressing PDGFRA. Our electrophysiology data at 2–3 days past an OPC state suggest that electrophysiological properties change during the critical time window of OPC transition to pre-OLs. Five days after an OPC state is established, most OLs have reduced Na+ currents with no spiking OLs, consistent with global reduction in Na+ channels as OLs mature within myelinating cells (De Biase et al., 2010).
Some non-spiking OLs express Nav channels, including OPCs (Figure 1), but it is unclear why not all OLs with Nav channels produce spikes. Thus, we examined whether a distinct Nav channel subtype specifically contributes to the ability to produce spikes. In cKO mice, deletion of SCN2A eliminated spiking OLs and significantly reduced the portion of OLs exhibiting Na+ currents, indicating SCN2A is the primary determinant of Na+ currents in spiking OLs. Some OLs from cKO mice exhibit reduced Na+ currents, mediated by other types of Na+ channels, and could not generate spikes. It is possible that non-spiking immature OLs, including OPCs, exhibit Na+ currents via Nav1.1, Nav1.3, and Nav1.6, but those currents may not be large enough to generate a spike in OLs (Spitzer et al., 2019). SCN2A, encoding Nav1.2 channels, is an additional source of Na+ current for membrane discharge to generate spikes. However, without specifically isolating spiking OLs in cKO mice, the result may underestimate the contribution of SCN2A to Na+ currents. In a future study, it would be worthwhile to identify the specific electrophysiologic profile of SCN2A-expressing OLs utilizing a SCN2A-driven reporter in both cKO and control mice.
The functional significance of SCN2A-expressing immature OLs during development
Nav channel expression in immature OLs coincides with the onset of active myelination and is reduced as myelination tapers off, suggesting that Nav channels are important for development of myelination (Berret et al., 2017; Spitzer et al., 2019). Proliferating OPCs have a relatively large Na+ current compared with non-cycling OPCs, and Na+ currents may regulate cell-cycle dynamics (Spitzer et al., 2019). However, the physiological relevance of the Nav1.2-mediated spiking in immature OLs is unclear. We found that only a small fraction of Nav1.2+ OLs or PDGFRA− OLs was actively proliferating, representing few proliferating OLs. Instead, most proliferating cells in the brainstem were PDGFRA+ OPCs. Both the global and OL-specific loss of SCN2A did not impair proliferation or production of OPCs, but critically impacted differentiation. OPCs expressed higher levels of Nav1.3 (SCN3A) and Nav1.6 (SCN8A) channels, which may contribute to OL proliferation, while Nav1.2 (SCN2A) in spiking OLs likely plays a distinct role in differentiation (Marques et al., 2018). This suggests different subtypes of Nav channels serve distinct roles during OL development.
Nav1.2 channels in spiking OLs appear to influence OL maturation, because the cell-specific loss of SCN2A reduces the number of CC1+ OLs. Introducing Scn2a into cultured OLs increases their myelinating capacity, while deletion of Scn2a in vivo with shRNA in rats alters global MBP levels in the brainstem (Jiang et al., 2013; Berret et al., 2017). The differentiation of only a subpopulation of OLs depends on SCN2A, because CC1+ OLs are still observed in SCN2A cKO mice, indicating functional heterogeneity among OLs. However, it remains unanswered if OL differentiation deficits in the cKO are specific to the spiking OL population, or whether the absence of a spiking OL population influences widespread OL differentiation deficits.
In the zebrafish spinal cord, two subpopulations of OPCs showed different myelinating potential. OPCs with high process motility and low calcium signals differentiated more easily than other OPCs (Marisca et al., 2020). Spiking OLs express mRNA for myelin genes, including Mag, Mobp, and Plp1 (Figure 1), and SCN2A mediates OL process extension and myelination in OLs in rat brainstem (Berret et al., 2017). Thus, spiking OLs could represent a population with high process motility driven by Nav1.2-dependent signaling that readily differentiates into myelinating OLs. Our results demonstrate that Nav1.2 channels encoded by SCN2A serve an essential role in maturation of a subpopulation of newly formed OLs by increasing Na+ current density and promoting the ability to spike.
Limitations of study
Identifying a molecular signature for spiking OLs would require a more comprehensive understanding of the specific transcriptional profile of spiking OLs. As of now, there are technical limitations in isolating spiking OLs and tracking their development over time without a specific reporter line for spiking OLs. In future studies, pairing patch-clamp electrophysiology with RNA sequencing could provide a more comprehensive transcriptional profile of spiking OLs (Lipovsek et al., 2021).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jun Hee Kim (kimjh@uthscsa.edu).
Materials availability
Materials generated in this study will be available upon request to the lead contact.
Data and code availability
The datasets in this study are available from the lead contact.
Single-cell qPCR data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table.
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-OLIG2 | Milipore | Cat#AB9610; RRID:AB_2299035 |
| Mouse IgG1 anti-NEUN | Milipore | Cat#MAB377; RRID:AB_2298772 |
| Mouse IgG2b anti-CC1 | Abcam | Cat# ab16794; RRID:AB_443473 |
| Mouse IgG2a anti-Nav1.2 cline K69/3 | Neuromab | ; RRID:AB_2184030 |
| Rat anti-PDFRA | Abcam | Cat#Ab90967; RRID:AB_2049372 |
| Biological samples | ||
| Universal mouse RNA | BioChain | Cat#R4334566-1 |
| Chemicals, peptides, and recombinant proteins | ||
| μ-conotoxin KIIIA | Alomone Labs | Cat#C-280 |
| 32% paraformaldehyde | Electron Microscopy Sciences | Cat#15714S |
| Critical commercial assays | ||
| Cells DirectTM one-step RT-PCR kit | Invitrogen | Cat#11753-100 |
| 1X SsoFast Eva-Green supermix with low ROX | Bio-Rad | Cat#PN172-5211 |
| 1X DNA binding dye sample loading reagent | Fluidigm | Cat#PN100-3738 |
| Deposited data | ||
| Single cell qPCR data | This study | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181630 Accession# GSE181634 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6, ACTIN-CreERT | (Hayashi and McMahon, 2002) | Manzoor Bhat, PhD. |
| Mouse: C57BL/6, SCN2A FLOX | Transviragen, Inc. Chapel Hill, NC, USA | N/A |
| Mouse: B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J | The Jackson Laboratory, Bar Harbor, ME, USA | 018280 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14 (CAG-tdTomato)Hze/J | The Jackson Laboratory, Bar Harbor, ME, USA | 007914 |
| Non-human primate: Baboon (Papio Anubis) | The Texas Biomedical Research Institute in San Antonio | N/A |
| Oligonucleotides | ||
| List of oligonucleotides in Table S1 | N/A | N/A |
| Software and algorithms | ||
| Prism | GraphPad | N/A |
| Igor Pro 8 | Wavemerics | N/A |
| Patch master | HEKA, Elektronik, Lambrecht/Pfalz | N/A |
| RStudio R 3.6.0 | R | N/A |
| Other | ||
| BioMark HD MX/HX system | Fluidigm | Cat#BMKHDPKG- MH |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal procedures and maintenance were performed in accordance with the guidelines approved by the University of Texas Health Science Center, San Antonio (UTHSCSA) Institutional Animal Care and Use Committee protocols.
Mouse
BL/6 mice aged 9–12 days were used for single-cell studies. To label OPCs, B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice (stock# 007914, The Jackson Laboratory, Bar Harbor, ME, USA) were crossed with B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J (PDGFRA-CreERT, stock# 018280, The Jackson Laboratory). At P8, a single i.p. injection of (Z)-4-hydroxytamoxifen (5 mg/kg; HB2508, Hello Bio Inc., Princeton, NJ, USA) was used to induce TdTomato expression. SCN2A+/fl mice were created by inserting LoxP sites around exons 3–6 of the SCN2A gene (Transviragen, Inc. Chapel Hill, NC, USA). Guide RNAs were developed to target LoxP insertion sites in intron 2 (gRNA 5sg81B, protospacer sequence 5′-TATTCTAGGGGTCACCC-3′) and intron 6 (gRNA 3sg66T, protospacer sequence 5′-GGATTGTTAGGACAATG). The guide RNAs were cloned separately into a plasmid vector designed to guide RNA expression from the U6-promoter and co-expression of Cas9 and EGFP. Separate donor vectors were produced for insertion of a FRT-Hygro-FRT-LoxP cassette at the 5sg81B cut site and a F3-Neo-F3-LoxP cassette a the 3sg66T cut site. The Cas9-guide RNA expression plasmids and circular LoxP donor vectors were co-transfected into C57BL/6N mouse embryonic stem cells and clones were selected for hygromycin and G418 resistance.
Clones were screened by PCR and Southern blot analysis to identify those with correct insertion of the LoxP cassettes in the cis position. Two correctly-targeted ES cell clones were microinjected in C57BL/6-Albino blastocysts to generate chimeras. Chimeras were mated to Rosa26-Flpe mice on a C57BL/6N-Albino genetic background for simultaneous germ-line transmission of the targeted allele and removal of the hygromycin and neomycin selection cassettes. Chimeras from both ES cell clones transmitted the floxed allele through the germline with selectable markers removed. Mice were screened with a deletion screen using PCR that generates a 402 bp product (Forward: CCAGAACTTAGTAAGCCATTTGACCTAG, Reverse: ACTTCATACATGGACTAAGAATAGCATAC). Heterozygotes and homozygotes were determined by PCR with two additional primers (F2: ACTTCATACATGGACTAAGAATAGCATAC R2: TGCTTCTGTCCCTTCATCTCCAC). Wild-type mice exhibit a product at 399 bp, while LoxP sites exhibit a product at 498 bp. SCN2Afl/fl mice were crossed with ACTIN-CreERT (provided by Dr. Bhat, UTHSCSA) and PDGFRA-Cre-ERT (Hayashi and McMahon, 2002). SCN2A deletion was induced by administering 40 mg/kg tamoxifen i.p. at postnatal (P) days 4, 5 and 6.
Baboon
A total of 3 brains from pre-term baboons of either sex were used. Preterm baboon were delivered at 127–140 days gestation via C-section and immediately euthanized at the Texas Biomedical Research Institute in San Antonio.
METHOD DETAILS
Slice preparation
All procedures followed approved UTHSCSA animal care protocols. Transverse brainstem slices were prepared from P9–12 mice. Coronal sections of the cerebellum were prepared from P9–12 mice and a baboon aged 127–140 days. Following dissection, the tissue was quickly immersed in ice-cold artificial cerebrospinal fluid (aCSF) with low calcium containing (in mM): 125 NaCl, 2.5 KCl, 3 MgCl2, 0.1 CaCl2, 25 glucose, 25 NaHCO3, 1.25 NaH2PO4, 0.4 ascorbic acid, 3 myoinositol, and 2 Na-pyruvate, pH 7.3–7.4 when bubbled with carbogen (95% O2, 5% CO2; osmolarity of 310–320 mOsm). Then, 200-μm-thick sections were sliced using a vibratome (VT1200S, Leica, Germany). Slices were incubated in a chamber that contained normal aCSF bubbled with carbogen at 35°C for 30 min and then were kept at room temperature. The normal aCSF was the same as the low-calcium aCSF, except that 3 mM MgCl2 and 0.1 mM CaCl2 were replaced with 1 mM MgCl2 and 2 mM CaCl2.
Electrophysiology
Slices were perfused at 2 ml/min and visualized using an infrared differential interference contrast microscope (AxoExaminer, Zeiss, Oberkochen, Germany) with a 60x water-immersion objective and a CMOS camera (ORCA-Flash2.8, Hamamatsu, Japan). Whole-cell patch-clamp recordings were performed in the normal aCSF at room temperature (22–24°C) using an EPC-10 amplifier controlled by Patch master software (HEKA, Elektronik, Lambrecht/Pfalz, Germany). For OL spike recordings, the pipette solution contained (in mM): 130 K-gluconate, 20 KCl, 5 Na2-phosphocreatine, 10 HEPES, 4 Mg-ATP, 0.2 EGTA, and 0.3 GTP, pH 7.3 (adjusted with KOH). For Na+ current recordings in OLs, the pipette solution contained (in mM):130 Cs-methanesulphonate, 10 CsCl, 5 mM Na2-phosphocreatine, 10 HEPES, 4 Mg-ATP, 5 EGTA, 10 TEA-HCl, and 0.3 GTP, pH 7.3 (adjusted with CsOH). The external solution contained 2 mM 4-aminopyridine, 10 mM TEA-Cl, and 0.2 mM CdCl2 to block K+ and Ca2+ channels. μ-conotoxin KIIIA (Alomone Labs, C-280) was applied at a concentration of 1 μM. Lucifer yellow (2 mM, L453, Fisher) was added to the internal solution for fluorescence dye filling to visualize cell shapes. Recordings were not corrected for the predicted liquid junction potential of 4 mV (CsCl based internal solution) or 11 mV (K-gluconate-based internal solution). Patch electrodes had resistances of 5–6 MΩ. For voltage clamp experiments, series resistance was < 20 MΩ and was compensated 80%. Signals were filtered at 2.9 kHz and acquired at a sampling rate of 10–50 μs.
Single-cell qPCR
Primers were selected from previous literature and the Harvard Primer Database (Spandidos et al., 2008). Primers were validated using qPCR on RNA isolated from homogenates from either mice or baboon brains. All primers selected passed validation, which included logarithmic amplification plots and the production of a single product, consistent with the validation protocol from the Harvard Primer Database. All primers showed logarithmic amplification in single-cell qPCR.
Isolated single cells were collected by applying light suction until the cell cytoplasm had been aspirated and the cell rose with the pipette. Elevating the pipette above the slice, increased suction was used to collect the entire cell body into the pipette. Patch pipettes with nonspecific cell debris stuck from surrounding cells were discarded. The contents of the pipette were collected using positive pressure into an RNase-free PCR tube containing 4 μL 2X buffer. These cells were immediately transferred and stored at −80°C until further processing for single-cell qPCR. Single-cell gene expression analysis was performed by the Bioanalytic and Single Cell Core (BASiC) at UT Health at San Antonio.
Single-cell qRT-PCR was carried out using Cells DirectTM one-step RT-PCR kit (cat #11753–100, Invitrogen) and a microfluidics device, BioMark HD MX/HX system (cat #BMKHDPKG- MH, Fluidigm, Inc., South San Francisco, CA). Single cells were collected in 4 μL 2X buffer and stored at −80°C. For qPCR, the cells were thawed, mixed well, and spun down before lysed at 75°C for 10 min. Genomic DNA contamination was reduced by treating cells with DNase I. PCR primers of selected genes were divided into two panels to fit BioMark 12 × 12 chips. Reverse transcription, preamplification, and PCR amplification were carried out according to the protocol of single-cell gene expression.
Target genes were amplified using a BioMark HB MX/HX system with 1X SsoFast Eva-Green supermix with low ROX (cat #PN172–5211, Bio-Rad, Hercules, CA) and 1X DNA binding dye sample loading reagent (cat #PN 100–3738, Fluidigm). In each mouse chip assay, universal mouse RNA (200 pg) from mouse normal tissues (cat #R4334566–1, BioChain, Newark, CA) was utilized as a positive control and human embryonic kidney cells and no template controls were utilized as negative controls. In the baboon chip assay, baboon RNA isolated from whole tissue served as a positive control and mouse RNA plus no template controls were negative controls. Quantitative PCR products were detected using a Fluidigm BioMark HD system according to the protocol for gene expression with the FlexSix IFC using Delta Gene assays (Fluidigm PN 100–7717 B1). The GE Flex Six Fast PCR+Melt v1 program was used to collect the cycle threshold (CT) values. CT values were converted to mRNA levels using 2^(25-CT) (Ståhlberg et al., 2013).
Immunohistochemistry
Tissue slices (200 μM) were post-fixed with 4% (wt/vol) paraformaldehyde (15714S, Electron Microscopy Sciences) in phosphate buffer solution (PBS) for 15 min and washed three times for 10 minutes in PBS (pH 7.4). Free-floating sections were blocked in 3% goat serum and 0.3% Triton X-100 in PBS for 1 h. For immunohistochemistry, slices were incubated with the primary antibody (please see the following table) overnight at 4°C. Following washing 3 times for 10 minutes in PBS, slices were incubated in secondary antibody diluted in blocking solution for 1–2 hours at room temperature. Slices were washed once for 10 min with PBS and then once for 10 min in water. Slices were mounted with media (H-1000, Vectashield). Stained slices were imaged with laser lines at 488 nm, 568 nm, and 647 nm using a 40x or 60x oil-immersion objective on a confocal laser-scanning microscope (LSM-510, Zeiss).
Antibodies
The following commercially available antibodies were used (source, catalog#, dilution): rabbit polyclonal anti-OLIG2 (Milipore, AB9610, 1:1000), mouse IgG1 monoclonal anti-NEUN (Millipore, MAB377, 1:1000), mouse IgG2b monoclonal anti-CC1 (Abcam, ab16794, 1:1000), mouse IgG2a monoclonal anti-Nav1.2 (Neuromab, 75–024, 1:400), rat polyclonal anti-PDGFRA (Abcam, Ab90967, 1:400).
EDU labeling
Mice (aged P9) were injected with EdU (25 mg/kg). For labeling of cells generated within 1–2 cell cycles, mice were sacrificed 2 hours after a single i.p. injection of EdU, and proliferating cells (EdU+) were evaluated. An EDU click-it kit was used to visualize EdU labeling and was used according to the manufacturer’s directions (C10340, ThermoFisher).
Quantitative PCR
RNA extraction.
RNA isolation was performed using a GeneJET RNA Purification kit with no modifications to procedure (K0732, ThermoFisher). This kit includes DNase treatment. RNA was quantified using a nanodrop (ND-1000, ThermoFisher). Purity was assessed by A260/A280 ratios with values ranging from 1.83 to 1.98. 100 ng of RNA was utilized for each reverse transcription reaction. Reverse transcription was performed using Superscript III First-strand synthesis with 1 μL Oligo (dT) primers, 1 μL 10 mM dNTP mix (180180–051, Invitrogen). cDNA was stored at −20°C for up to 48 hr. qPCR was performed using the PowerUp SYBR green master mix (A25742, ThermoFisher). Primers were used at 0.25 μM. Reactions were manually loaded into optical plates with covers (plates:4309849, covers:4360954, Applied Biosystems). The plate was spun for 30 s. qPCR was performed using 7900HT Fast Real-Time PCR system (Applied Biosystems), data was analyzed using SDS v2.4 (Applied Biosystems) and the determined CT was used for analysis. No outliers were removed. No template controls and no reverse transcription controls did not produce measurable CT values. GAPDH was chosen as a reference. Technical replicates were done in triplicate. Delta CT (meanGene – meanGAPDH) was used to determine Delta Delta CT (Delta CTPos-Delta CTNeg) or (Delta CTcko-Delta CTCTL); relative gene expression was calculated as 2^(-deltadeltaCT) and normalized to 100%.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using Graphpad Prism Version 8 and RStudio R 3.6.0. For experiments where data were quantified as proportions, a chi-square test was used. For all other analyses, we tested whether the values were normally distributed using D’Agostino–Pearson omnibus and Shapiro–Wilk normality tests. When data were normally distributed, we used parametric tests; otherwise, we used non-parametric tests. α values were set to 0.05, and all comparisons were two-tailed. To compare two groups, unpaired t tests or Mann-Whitney U tests were carried out. For three or more groups, a one-way or two-way ANOVA with post hoc Tukey’s multiple comparison test was used. Significance was set at p < 0.05. Data collected as raw values are shown as mean ± SEM or mean ± SD with n values representing the number of animals per experimental group or the number of cells per group where indicated. Details of statistical methods are reported in the text.
Supplementary Material
Highlights.
SCN2A, encoding Nav1.2 channel, is essential for spiking in OLs
SCN2A-expressing spiking OLs are present in the brainstem and cerebellum
Spiking OLs are in a transitional stage between OPCs and pre-myelinating OLs
Deletion of SCN2A impacts maturation of a subpopulation of immature OLs
ACKNOWLEDGMENTS
We thank Dr. Manzoor Bhat for providing the Cre mouse line. This work was supported by a grant from the National Institute of Neurological Disorders and Stroke (NINDS; R21 NS116573), SFARI Pilot grant (#609186) to J.H.K., and The National Heart, Lung, and Blood Institute (NHLBI, T32 HL007446-39) to E.G.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109653.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bakiri Y, Attwell D, and Káradóttir R (2009). Electrical signalling properties of oligodendrocyte precursor cells. Neuron Glia Biol. 5, 3–11. [DOI] [PubMed] [Google Scholar]
- Berret E, Barron T, Xu J, Debner E, Kim EJ, and Kim JH (2017). Oligodendroglial excitability mediated by glutamatergic inputs and Nav1.2 activation. Nat. Commun 8, 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boda E, Di Maria S, Rosa P, Taylor V, Abbracchio MP, and Buffo A (2015). Early phenotypic asymmetry of sister oligodendrocyte progenitor cells after mitosis and its modulation by aging and extrinsic factors. Glia 63, 271–286. [DOI] [PubMed] [Google Scholar]
- Chittajallu R, Aguirre A, and Gallo V (2004). NG2-positive cells in the mouse white and grey matter display distinct physiological properties. J. Physiol 561, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Young KM, Hamilton NB, Li H, Richardson WD, and Attwell D (2012). Properties and fate of oligodendrocyte progenitor cells in the corpus callosum, motor cortex, and piriform cortex of the mouse. J. Neurosci 32, 8173–8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ðặng TC, Ishii Y, Nguyen V, Yamamoto S, Hamashima T, Okuno N, Nguyen QL, Sang Y, Ohkawa N, Saitoh Y, et al. (2019). Powerful Homeo-static Control of Oligodendroglial Lineage by PDGFRα in Adult Brain. Cell Rep. 27, 1073–1089.e5. [DOI] [PubMed] [Google Scholar]
- De Biase LM, Nishiyama A, and Bergles DE (2010). Excitability and synaptic communication within the oligodendrocyte lineage. J. Neurosci 30, 3600–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster S, Hill MFE, and Franklin RJM (2019). Diversity in the oligodendrocyte lineage: Plasticity or heterogeneity? Glia 67, 1797–1805. [DOI] [PubMed] [Google Scholar]
- Ge W-P, Zhou W, Luo Q, Jan LY, and Jan YN (2009). Dividing glial cells maintain differentiated properties including complex morphology and functional synapses. Proc. Natl. Acad. Sci. USA 106, 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, and McMahon AP (2002). Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol 244, 305–318. [DOI] [PubMed] [Google Scholar]
- Jäkel S, Agirre E, Mendanha Falcão A, van Bruggen D, Lee KW, Knuesel I, Malhotra D, Ffrench-Constant C, Williams A, and Castelo-Branco G (2019). Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Chen C, Liu XB, Selvaraj V, Liu W, Feldman DH, Liu Y, Pleasure DE, Li RA, and Deng W (2013). Generation and characterization of spiking and nonspiking oligodendroglial progenitor cells from embryonic stem cells. Stem Cells 31, 2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Káradóttir R, Hamilton NB, Bakiri Y, and Attwell D (2008). Spiking and nonspiking classes of oligodendrocyte precursor glia in CNS white matter. Nat. Neurosci 11, 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson VA, Zhang Y, and Bergles DE (2016). Electrophysiological properties of NG2+ cells: Matching physiological studies with gene expression profiles. Brain Res. 1638, 138–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek M, Bardy C, Cadwell CR, Hadley K, Kobak D, and Tripathy SJ (2021). Patch-seq: Past, Present, and Future. J. Neurosci 41, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisca R, Hoche T, Agirre E, Hoodless LJ, Barkey W, Auer F, Castelo-Branco G, and Czopka T (2020). Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat. Neurosci 23, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, van Bruggen D, Vanichkina DP, Floriddia EM, Munguba H, Väremo L, Giacomello S, Falcão AM, Meijer M, Björklund ÅK, et al. (2018). Transcriptional Convergence of Oligodendrocyte Lineage Progenitors during Development. Dev. Cell 46, 504–517.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planells-Cases R, Caprini M, Zhang J, Rockenstein EM, Rivera RR, Murre C, Masliah E, and Montal M (2000). Neuronal death and perinatal lethality in voltage-gated sodium channel alpha(II)-deficient mice. Biophys. J 78, 2878–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees SM, Loeliger MM, Munro KM, Shields A, Dalitz PA, Dieni S, Thomson MA, Coalson J, and Inder T (2009). Cerebellar development in a baboon model of preterm delivery: impact of specific ventilatory regimes. J. Neuropathol. Exp. Neurol 68, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair JL, Fischl MJ, Alexandrova O, Heb M, Grothe B, Leibold C, and Kopp-Scheinpflug C (2017). Sound-Evoked Activity Influences Myelination of Brainstem Axons in the Trapezoid Body. J. Neurosci 37, 8239–8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, and Seed B (2008). A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics 9, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer SO, Sitnikov S, Kamen Y, Evans KA, Kronenberg-Versteeg D, Dietmann S, de Faria O Jr., Agathou S, and Káradóttir RT (2019). Oligodendrocyte Progenitor Cells Become Regionally Diverse and Heterogeneous with Age. Neuron 101, 459–471.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhlberg A, Rusnakova V, Forootan A, Anderova M, and Kubista M (2013). RT-qPCR work-flow for single-cell data analysis. Methods 59, 80–88. [DOI] [PubMed] [Google Scholar]
- Takahashi E, Hayashi E, Schmahmann JD, and Grant PE (2014). Development of cerebellar connectivity in human fetal brains revealed by high angular resolution diffusion tractography. Neuroimage 96, 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M-M, Green BR, Catlin P, Fiedler B, Azam L, Chadwick A, Terlau H, McArthur JR, French RJ, Gulyas J, et al. (2007). Structure/function characterization of micro-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J. Biol. Chem 282, 30699–30706. [DOI] [PubMed] [Google Scholar]
- Zhang M-M, Han TS, Olivera BM, Bulaj G, and Yoshikami D (2010). μ-conotoxin KIIIA derivatives with divergent affinities versus efficacies in blocking voltage-gated sodium channels. Biochemistry 49, 4804–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets in this study are available from the lead contact.
Single-cell qPCR data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table.
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-OLIG2 | Milipore | Cat#AB9610; RRID:AB_2299035 |
| Mouse IgG1 anti-NEUN | Milipore | Cat#MAB377; RRID:AB_2298772 |
| Mouse IgG2b anti-CC1 | Abcam | Cat# ab16794; RRID:AB_443473 |
| Mouse IgG2a anti-Nav1.2 cline K69/3 | Neuromab | ; RRID:AB_2184030 |
| Rat anti-PDFRA | Abcam | Cat#Ab90967; RRID:AB_2049372 |
| Biological samples | ||
| Universal mouse RNA | BioChain | Cat#R4334566-1 |
| Chemicals, peptides, and recombinant proteins | ||
| μ-conotoxin KIIIA | Alomone Labs | Cat#C-280 |
| 32% paraformaldehyde | Electron Microscopy Sciences | Cat#15714S |
| Critical commercial assays | ||
| Cells DirectTM one-step RT-PCR kit | Invitrogen | Cat#11753-100 |
| 1X SsoFast Eva-Green supermix with low ROX | Bio-Rad | Cat#PN172-5211 |
| 1X DNA binding dye sample loading reagent | Fluidigm | Cat#PN100-3738 |
| Deposited data | ||
| Single cell qPCR data | This study | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181630 Accession# GSE181634 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6, ACTIN-CreERT | (Hayashi and McMahon, 2002) | Manzoor Bhat, PhD. |
| Mouse: C57BL/6, SCN2A FLOX | Transviragen, Inc. Chapel Hill, NC, USA | N/A |
| Mouse: B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J | The Jackson Laboratory, Bar Harbor, ME, USA | 018280 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14 (CAG-tdTomato)Hze/J | The Jackson Laboratory, Bar Harbor, ME, USA | 007914 |
| Non-human primate: Baboon (Papio Anubis) | The Texas Biomedical Research Institute in San Antonio | N/A |
| Oligonucleotides | ||
| List of oligonucleotides in Table S1 | N/A | N/A |
| Software and algorithms | ||
| Prism | GraphPad | N/A |
| Igor Pro 8 | Wavemerics | N/A |
| Patch master | HEKA, Elektronik, Lambrecht/Pfalz | N/A |
| RStudio R 3.6.0 | R | N/A |
| Other | ||
| BioMark HD MX/HX system | Fluidigm | Cat#BMKHDPKG- MH |


