Summary
Genomic discovery and characterization of risk loci for type 2 diabetes (T2D) have been conducted primarily in individuals of European ancestry. We conducted a multiethnic genome-wide association study of T2D among 53,102 cases and 193,679 control subjects from African, Hispanic, Asian, Native Hawaiian, and European population groups in the Population Architecture Genomics and Epidemiology (PAGE) and Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortia. In individuals of African ancestry, we discovered a risk variant in the TGFB1 gene (rs11466334, risk allele frequency (RAF) = 6.8%, odds ratio [OR] = 1.27, p = 2.06 × 10−8), which replicated in independent studies of African ancestry (p = 6.26 × 10−23). We identified a multiethnic risk variant in the BACE2 gene (rs13052926, RAF = 14.1%, OR = 1.08, p = 5.75 × 10−9), which also replicated in independent studies (p = 3.45 × 10−4). We also observed a significant difference in the performance of a multiethnic genetic risk score (GRS) across population groups (pheterogeneity = 3.85 × 10−20). Comparing individuals in the top GRS risk category (40%–60%), the OR was highest in Asians (OR = 3.08) and European (OR = 2.94) ancestry populations, followed by Hispanic (OR = 2.39), Native Hawaiian (OR = 2.02), and African ancestry (OR = 1.57) populations. These findings underscore the importance of genetic discovery and risk characterization in diverse populations and the urgent need to further increase representation of non-European ancestry individuals in genetics research to improve genetic-based risk prediction across populations.
Keywords: Type 2 Diabetes, Genetic Risk Score
We report a multiethnic genome-wide association study of type 2 diabetes and evaluate the performance of a genetic risk score (GRS) in five ethnic groups. Drawing upon large-scale resources, our study explores how a multiethnic derived GRS predicts T2D risk in European American, African American, Hispanic, Native Hawaiian, and Asian subgroups.
Introduction
The global burden of type 2 diabetes (T2D; MIM: 125853) has risen substantially in the last 20 years, affecting more than 415 million individuals worldwide.1 The rapid growth of this epidemic is a reflection of socioeconomic trends (including changes in environmental factors, such as high calorie intake and low levels of physical activity) and a rising incidence of obesity, with individuals from Asian, African, and Hispanic populations having higher risk of developing T2D than individuals from European populations.2 The pathogenesis of T2D is characterized by loss of β-cell function and rising insulin resistance, with disease severity also differing by race and ethnicity.3
Individual risk of T2D and inter-individual variability of glycemic traits are, at least in part, influenced by genetic factors.4 To date, T2D genome-wide association studies (GWASs) have discovered ∼600 independent loci comprised primarily of common variants with modest effects, with genome-wide chip heritability explaining 19% of T2D risk.5 While the largest T2D GWAS consortia have been largely based on European ancestry populations,6 ancestry-specific and multiethnic GWASs have emerged.5,7 Further, the recent development of genetic imputation reference panels and genotyping arrays that capture common variation in ancestrally diverse groups allows for better characterization of the genetic architecture of T2D across race and ethnicity.8
The Population Architecture Genomics and Epidemiology (PAGE) study was assembled to study the genetic architecture of complex traits across racial and ethnic populations in the US.9 In the current study, we performed a multiethnic GWAS meta-analysis of T2D, combining population-specific GWAS results for T2D in PAGE with European ancestry GWAS results from the Diabetes Genetics Replication and Meta-analysis (DIAGRAM) consortium to discover risk loci for T2D. Findings were investigated in independent ancestry-specific T2D studies and consortia. We also constructed genetic risk scores (GRSs) using T2D risk variants previously identified in a large multiethnic population5 to examine whether such scores effectively stratify T2D risk across racial and ethnic groups.
Material and methods
Studies/consortia
The multiethnic GWAS meta-analysis included studies with genotype data from the PAGE study (Table S1) and publicly available summary statistics for European ancestry populations from the DIAGRAM consortium.10 GRS analyses included non-European ancestry populations in PAGE with Illumina MEGA array data (described below) and European ancestry populations from the UK Biobank.11 A flow chart of research studies contributing to GWAS and GRS analyses is shown in Figure S1.
The PAGE study consists of large, ongoing population-based studies including the Multiethnic Cohort (MEC) study,12 the Women’s Health Initiative (WHI),13 Atherosclerosis Risk in Communities (ARIC),14 Coronary Artery Risk Development in Young Adults (CARDIA),15 the Hispanic Community Health Study/Study of Latinos (HCHS/SOL),16 and the Mount Sinai School of Medicine’s (MSSM) Electronic Health Record (EHR)-linked biobank (BioMe).17 The PAGE study has been described elsewhere,18 with additional detail found in the Supplemental methods.
T2D cases were defined as individuals with (1) a T2D diagnosis by a physician/medical professional and use of medication for treatment of diabetes, and/or (2) a fasting (≥8 h) blood glucose measurement ≥126 mg/dL indicated in examination records. In BioMe, T2D diagnosis and treatment was assessed using electronic medical record diagnosis codes, abnormal laboratory values, or diagnosis by a physician. For MEC, T2D cases were based on self-report of a T2D diagnosis and medication use, diabetes registries of health plans, the Chronic Condition Data Warehouse of Medicare, and California hospital discharge records. Control subjects were participants who did not self-report a previous diagnosis of T2D or the use of diabetes medications and who did not have any evidence of diabetes in linked health plan data. For studies with fasting blood glucose measurements (all studies except MEC), participants with levels >126 mg/dL were excluded as control individuals. Individuals who were pregnant at blood draw, had type 1 diabetes, or had an age of diabetes diagnosis below 20 years were excluded from case and control groups. Additional details about T2D case and control selection criteria for each PAGE study and replication studies are described in the Supplemental methods and elsewhere.19
The 18 studies comprising stage 1 of the DIAGRAM consortium have been previously described.10 Briefly, T2D case/control status varied by contributing study based on a combination of diagnostic fasting glucose or hemoglobin A1c (HbA1c) level cutoffs, hospital discharge diagnosis, use of oral diabetes medication, International Classification of Disease, 9th Revision (ICD-9) T2D codes ranging from 249–250.99, or self-report.
In the UK Biobank, which was used exclusively for GRS analyses, T2D status was defined as previously described20 and included having an ICD-10 code of E11.X or a self-reported diagnosis in an interview with a trained nurse.
Genotyping
In PAGE, 48,781 individuals were genotyped on the Illumina MEGA array, which includes ∼2 million variants. The MEGA array was designed in PAGE to improve discovery in diverse ancestral populations and account for genetic heterogeneity across ethnic groups.9 The backbone includes highly informative variants for GWAS analyses in European and East Asian descent populations for compatibility with other genotyping arrays. The MEGA backbone was enhanced with content (83%) to improve coverage of common variation in African ancestry and Hispanic/Latino populations. An additional 53,600 PAGE participants were genotyped in previous studies using a variety of other genotyping arrays, as described in Table S1. Genotyping details are described in the Supplemental methods.
European ancestry T2D studies in DIAGRAM were genotyped using several commercial genome-wide arrays, as described in detail along with quality control procedures elsewhere.10
The UK Biobank included genotypes for 488,377 participants; 438,427 participants were genotyped using the Applied Biosystems UK Biobank Axiom Array, and 49,950 participants from the UK Biobank Lung Exome Variant Evaluation (UK BiLEVE) study were genotyped using the Applied Biosystems UK BiLEVE Axiom Array by Affymetrix. These arrays share 95% of marker content.11
Imputation
In PAGE, allele dosages for autosomal variants were imputed using the 1000 Genomes phase 3 reference panel (October 2014 release).21 Genotype dosage imputation was performed using IMPUTE2.22 Variants were filtered out if they were monomorphic or had an imputation info score < 0.4. For the PAGE MEGA array imputed data, additional variants were filtered if the effective n (effn, calculated as 2 × minor allele frequency (MAF) × (1 − MAF) × n × info score) was <30.
In DIAGRAM, imputation was performed using IMPUTE2 and minimac using the 1000 Genomes phase 1 reference panel (March 2012 release). Variants were excluded if they had an info score < 0.40 or were missing in 50% (n = 79,093) or more of the total DIAGRAM sample. Of the 11.7 million genotyped or imputed variants with summary statistics available in DIAGRAM, 10.8 million were also observed in the PAGE European ancestry studies and evaluated in the meta-analysis.
For UK Biobank, the Haplotype Reference Consortium reference panel was merged together with the UK10K and 1000 Genomes phase 3 reference panels, and imputation was carried out using IMPUTE2.
Statistical analysis
To account for population stratification in PAGE, principal components (PCs) were calculated separately for each genotyping array/population group using Eigenstrat23 with common and uncorrelated variants.
The multiethnic GWAS included 246,781 participants, 87,573 of whom were from PAGE (African American: 8,591 cases and 16,887 control subjects; Asian: 3,124 cases and 4,313 control subjects; Native Hawaiian: 1,642 cases and 2,152 control subjects; Hispanic: 9,913 cases and 22,958 control subjects; European American: 3,156 cases and 14,837 control subjects; Table S1) and 159,208 of whom were of European ancestry from DIAGRAM (26,676 cases and 132,532 control subjects).10
In PAGE, ∼28 million genotyped and imputed variants were analyzed for association with T2D risk. Analyses were performed separately by race/ethnicity (based on self-report) and PAGE sub-study. A total of 30 groups (combinations of race and sub-study) were included in the meta-analysis described below (Table S1).
One of the sub-studies in PAGE (the SOL study) included pairs or large clusters of closely related individuals by design.16 We used a modified version of the generalized estimating equations (GEE) method implemented in SUGEN24 to correct for these known relationships in SOL. For other studies, we used SUGEN to perform traditional logistic regression analysis, without GEE correction.
In PAGE, logistic regression analyses were performed assuming an additive genetic model for each variant while adjusting for age, sex, body mass index (BMI, kg/m2), and the first 10 PCs. Analyses were performed separately for each population. For DIAGRAM, stage 1 T2D GWAS meta-analysis summary statistics were used, which were adjusted for age, sex, BMI, and six PCs.10
Meta-analysis
Population-specific and multiethnic meta-analyses were conducted across PAGE studies and DIAGRAM (Table S1) for the 28 million SNPs. Summary statistics from each contributing GWAS were combined across studies to form a single combined log odds ratio (OR) estimate, standard error, and Wald test for each variant, using a fixed effects model weighted by the inverse variances of the log OR estimates, as implemented by METAL.25 Associations from population-specific and multiethnic meta-analyses were variants that reached genome-wide significance (p < 5 × 10−8) and were not reported in previous GWASs or within 500 kb of established T2D risk variants.
Replication
Variants associated with T2D risk in ancestry-specific or multiethnic analyses at p < 5 × 10−8 were examined in independent studies utilizing logistic regression adjusting for the same covariates as in the discovery stage (except 23andMe, which did not adjust for BMI). Replication was performed in 23andMe, where T2D case status was based on self-report (European American: 109,274 cases and 154,3466 control subjects; Hispanic/Latino: 13,985 cases and 237,247 control subjects; African American: 7,521 cases and 82,988 control subjects; East Asian: 2,237 cases and 72,819 control subjects; South Asian: 1,171 cases and 18,874 control subjects).26 Additional replication studies included Hispanics/Latinos from the DIAMANTE Hispanic/Latino Consortium (4,806 cases and 6,515 control subjects)27 and the Slim Initiative for Genomic Medicine in the Americas (SIGMA) T2D Consortium (1,817 cases and 2,284 control subjects),28 Asians from the Asian Genetic Epidemiology Network (AGEN; 77,418 cases and 356,122 control subjects),29 and African Americans from the MEta-analysis of type 2 DIabetes in African Americans (MEDIA) Consortium (10,160 cases and 13,231 control subjects).30 We also accessed publicly available summary results from a published T2D study in Africa (2,633 cases and 1,714 control subjects).31 Replication studies/consortia were meta-analyzed using METAL25 and described in detail in the Supplemental methods.
Annotation
We annotated risk variants with whole genome sequencing annotator (WGSA), RegulomeDB, and HaploReg 4.1.32, 33, 34 Based on the evidence that the A risk allele of rs11466334 affected binding profiles of CCCTC-binding factor (CTCF), we also examined CTCF chromatin immunoprecipitation sequencing (ChIP-seq) data from the Encyclopedia of DNA Elements (ENCODE) project across 25 tissue types and chromatin looping in pancreas and lymphoblastoid tissues from the WashU Epigenome Browser.
GRS
The GRS construction was based on 582 T2D risk variants and their corresponding effect estimates as previously reported in the largest multiethnic T2D GWAS reported to date.5 Of the 582 variants, 579 were present in both PAGE and the UK Biobank and had imputation info scores > 0.45. The median info score in both the UK Biobank and PAGE (MEGA array) was 0.99.
GRSs were calculated for 467,951 participants, which included 44,222 from PAGE (African American: 5,972 cases and 9,637 control subjects; Asian: 2,004 cases and 2,572 control subjects; Native Hawaiian: 1,534 cases and 2,017 control subjects; Hispanic: 4,137 cases and 16,349 control subjects) and 423,729 individuals of European ancestry from the UK Biobank (19,786 cases and 403,943 control subjects).
The GRS was computed as a sum of the number of risk alleles carried by the individual, weighted by variant-specific effect estimates. The GRS was then categorized into the following percentiles: 0%–10%, 10%–20%, 20%–30%, 30%–40%, 40%–60%, 60%–70%, 70%–80%, 80%–90%, and 90%–100%. Additional analyses were performed splitting the bottom and top deciles into two categories to obtain the GRS risk for the bottom 1% and top 1%: 0%–1%, 1%–10%, 90%–99%, and 99%–100%. GRS thresholds were determined using the observed distribution among control individuals for the corresponding population. Genetic risk of T2D was estimated within each population group by comparing participants in each of the defined GRS percentiles to those in the 40%–60% category using logistic regression models adjusted for age, sex, BMI, study (for non-Europeans), and ten PCs. We also evaluated effect modification of the GRS on T2D risk by body size. This was evaluated by stratifying participants into low- and high-BMI groups based on the median BMI per population. Logistic regression was then performed with T2D as the outcome and GRS categories as the independent predictors, adjusting for age, sex, BMI, study (for non-Europeans), and ten PCs. In order to evaluate whether the effect of the GRS significantly differed between the high and low BMI models, similar logistic regression models were evaluated in all participants with an added interaction term for GRS category × BMI (low/high). We report the p values for each resulting interaction (between each GRS category and the dichotomized BMI variable) as well as the p value for a likelihood ratio test, comparing a model with and without the added interaction term.
To further assess the discriminative ability of the GRS, the area under the curve (AUC) was calculated for each population using the “pROC” R package.35 Heterogeneity of the effect of the GRS across population groups was assessed by calculating Cochran’s Q in the R package “meta.”36
For each population, we also estimated the proportion of familial relative risk of T2D explained by the 582 known variants as previously described.37 This calculation used population-specific risk allele frequencies (using the risk allele identified in Vujkovic et al.5) and the per-allele odds ratios estimated in our GWAS and assumed a 2.77 relative risk to first-degree relatives of T2D individuals.38
Results
Multiethnic GWAS
Genome-wide association analyses of T2D were carried out in 244,936 individuals from PAGE (Table S1) and DIAGRAM (52,204 cases and 192,732 control subjects), with ∼10.8 million overlapping variants tested. Genomic inflation ranged from λ = 1.03 in the African ancestry GWAS to λ = 1.07 in the Hispanic GWAS, with the overall multiethnic GWAS having λ = 1.07 (Figure S2). Genome-wide significant associations were observed for 39 of the 582 known risk loci in the multiethnic meta-analysis, 20 of which were also significant in population-specific analyses: 18 in the European ancestry GWAS, 8 in Hispanics, 3 in African Americans, and 2 in Asians (Table S2). The most statistically significant associations among known risk loci were rs35011184 near TCF7L2 (MIM: 602228) (OR = 1.31, 95% confidence interval [CI] = 1.27–1.34, p = 3.32 × 10−102) in the multiethnic meta-analysis, which was also the most significant variant in European (OR = 1.34, 95% CI = 1.29–1.38, p = 1.55 × 10−75) and African populations (OR = 1.29, 95% CI = 1.20–1.39, p = 2.42 × 10−11), followed by rs2237897 near KCNQ1 (MIM: 607542) (OR = 1.29, 95% CI = 1.25–1.33, p = 2.08 × 10−54), which was the most significant variant in the Hispanic (OR = 1.36, 95% CI = 1.29–1.43, p = 4.70 × 10−31) and Asian populations (OR = 1.34, 95% CI = 1.24–1.45, p = 7.54 × 10−13) (Table S2). We detected genome-wide significant associations in four regions located >500 kb of previously known T2D loci, two of which were only found in African and Hispanic populations and one of which was only found in Asian and Native Hawaiian populations (Tables S3 and S4). Among the 582 known variants reported in Vujkovic et al.,5 in our results, 338 variants (58.1%) had a multiethnic with a p value < 0.05, and 548 (94.2%) had consistent directions of effect (results for the 582 variants are shown in Table S2).
To confirm associations with these four putative risk loci, we examined associations in independent population-specific consortia (replication results shown in Table S4). Of the four signals, associations with rs11466334 in TGFB1 (meta-analyzed across three replication studies in individuals of African ancestry, p = 6.26 × 10−23) and rs13052926 in BACE2 (meta-analyzed across multiple replication studies p = 3.45 × 10−4) replicated in one or more studies at p < 0.05 (Table 1; see Materials and methods).
Table 1.
T2D risk variants from discovery PAGE + DIAGRAM meta-analysis and replication studies
| Chromosome: position (rs #) | Nearest gene | R/O allelea | Stage | Study | Ancestry group | Cases/control subjects | Risk allele frequencyb | OR (95% CI) | p value |
|---|---|---|---|---|---|---|---|---|---|
| 19: 41847737 (rs11466334)c | TGFB1 | A/G | discovery | PAGE | African | 8,591/16,887 | 0.068 | 1.27 (1.18–1.35) | 2.06E−08 |
| replication | MEDIA | African | 10,160/13,231 | 0.075 | 1.32 (1.19–1.46) | 2.83E−07 | |||
| replication | 23andMe | African | 7,521/82,988 | 0.062 | 1.36 (1.26–1.48) | 4.02E−14 | |||
| replication | Africa T2D | African | 2,663/1,714 | 0.079–0.116dd | 1.41 (1.23–1.63) | 1.20E−06 | |||
| replication | meta-analysis | African | 20,344/97,933 | 1.35 (1.26–1.39) | 6.26E−23 | ||||
| 21:42585088 (rs13052926) | BACE2 | A/G | discovery | PAGE+DIAGRAM | multiethnic | 53,102/193,679 | 0.726–0.957d | 1.08 (1.05–1.10) | 5.75E−09 |
| discovery | PAGE | European | 29,832/147,369 | 0.831 | 1.09 (1.05–1.13) | 1.77E−06 | |||
| discovery | PAGE | Hispanic | 9,913/22,958 | 0.830 | 1.06 (1.01–1.12) | 0.026 | |||
| discovery | PAGE | Asian | 3,124/4,313 | 0.966 | 1.31 (1.07–1.59) | 0.007 | |||
| discovery | PAGE | African | 8,591/16,887 | 0.726 | 1.05 (1.01–1.10) | 0.028 | |||
| discovery | PAGE | Native Hawaiian | 1,642/2,152 | 0.912 | 1.02 (0.84–1.23) | 0.869 | |||
| replication | AGEN | Asian | 77,418/356,122 | 0.960 | 1.04 (0.99–1.08) | 0.080 | |||
| replication | MEDIA | African | 10,160/13,231 | 0.726 | 1.01 (0.97–1.08) | 0.484 | |||
| replication | DIAMANTE/LATINO | Hispanic | 4,806/6,515 | 0.869 | 1.01 (0.90–1.13) | 0.877 | |||
| replication | SIGMA | Hispanic | 1,817/2,284 | 0.884 | 0.96 (0.83–1.11) | 0.542 | |||
| replication | 23andMe | European | 109,274/1,543,466 | 0.838 | 1.02 (1.00–1.03) | 0.013 | |||
| replication | 23andMe | African | 7,521/82,988 | 0.739 | 1.00 (0.96–1.05) | 0.880 | |||
| replication | 23andMe | East Asian | 2,237/72,819 | 0.957 | 1.04 (0.89–1.23) | 0.587 | |||
| replication | 23andMe | South Asian | 1,171/18,874 | 0.848 | 1.36 (1.18–1.56) | 1.79E−05 | |||
| replication | 23andMe | Latino | 13,985/237,247 | 0.841 | 1.03 (0.99–1.08) | 0.104 | |||
| replication | meta-analysis | 228,389/2,333,546 | 1.02 (1.01–1.03) | 3.45E−04 |
R/O allele denotes risk/other allele.
Calculated from T2D MEGA control subjects or WHI T2D control subjects for EUR.
RAF ≤ 1% in all non-African populations.
RAF range across sub-studies.
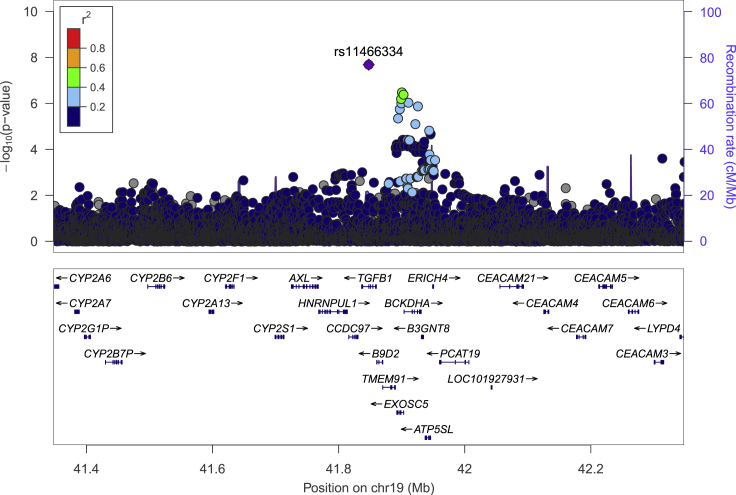
Variant rs11466334, located in intron 3 of the transforming growth factor beta-1 (TGFB1, MIM: 190180) gene, is common in the African ancestry (6.8%) and Hispanic (1.3%) populations and is rare in the other groups (<1%). The association with rs11466334 was statistically significant in the African ancestry (OR = 1.27, 95% CI = 1.17–1.38, p = 2.06 × 10−8; Table 1; Figure 1) and also in African/Hispanic multiethnic analysis (OR = 1.26, 95% CI = 1.16–1.35, p = 3.61 × 10−9). It was replicated in the MEDIA African ancestry T2D consortium (RAF = 7.5%, OR = 1.32, 95% CI = 1.19–1.46, p = 2.83 × 10−7); in a study conducted in South Africa, Nigeria, Ghana, and Kenya (RAF = 7.9%–11.6%, OR = 1.41, 95% CI = 1.23–1.63, p = 1.20 × 10−6);31 and in individuals of African ancestry from 23andMe (RAF = 6.2%, OR = 1.36, 95% CI = 1.26–1.48, p = 4.02 × 10−14).
Figure 1.
Locus zoom plot of rs11466334 region
Locus-zoom plot of PAGE African ancestry T2D associations with surrounding variants (color coded by r2 bin) and replicated index variant, rs11466334 (denoted by purple diamond), using African linkage disequilibrium (LD). The x axis represents the chromosomal position, the left y axis represents −log10 (p value) of the association between the genetic variant and T2D, and the right y axis represents recombination rate.
The risk allele (A) of rs1146634 is predicted to alter binding affinity of a number of transcription factors (e.g., CTCF, Maf, Rad21, SMC3, TAL1, and YY1) by inducing a motif change, as evidenced by differential position weight matrix scores > 10 from HaploReg 4.1 (Table S5). The variant is also located in a predicted CTCF peak, based on ChIP-seq data from multiple cell lines (pancreas, transverse colon, esophagus squamous epithelium, adrenal gland) within ENCODE, which suggests it may be biologically functional (Figure S3). Variant rs11466334 was found to be associated with the expression of a nearby gene, ATP5SL (MIM: 617262), within whole blood (p = 2.9 × 10−7) in GTEx (version 8). HiC chromatin interaction maps indicate that this variant possibly regulates genes HNRNPUL1 (MIM: 605800), CCDC97 (HGNC: 28289), TGFB1 (MIM: 190180), B9D2 (MIM: 611951), EXOSC5 (MIM: 606492), BCKDHA (MIM: 608348), B3GNT8 (MIM: 615357), and ATP5SL (MIM: 617262) (Figure S4).
Variant rs13052926 is located in intron 3 of the Beta-secretase 2 (BACE2, MIM: 065668) gene and was common in all ancestry groups (RAF = 72.5%–95.7%). A genome-wide significant association for this variant was observed in the multiethnic meta-analysis (OR = 1.08, 95% CI = 1.05–1.10, p = 5.75 × 10−9; Table 1; Figure 2), with the strongest associations observed in the European population (OR = 1.09, 95% CI = 1.05–1.13, p = 1.77 × 10−6), followed by the Asian (OR = 1.31, 95% CI = 1.07–1.59, p = 0.0074), Hispanic (OR = 1.06, 95% CI = 1.01–1.12, p = 0.026), and African populations (OR = 1.05, 95% CI = 1.01–1.10, p = 0.28), with no evidence of association observed in Native Hawaiians (Table S4).
Figure 2.
Locus zoom plot of rs13052926
Locus-zoom plot of PAGE + DIAGRAM multiethnic T2D associations with surrounding variants (color coded by r2 bin) and the replicated index variant, rs13052926 (denoted by purple diamond), using European LD. The x axis represents the chromosomal position, the left y axis represents −log10 (p value) of the association between the genetic variant and T2D, and the right y axis represents recombination rate.
GRS
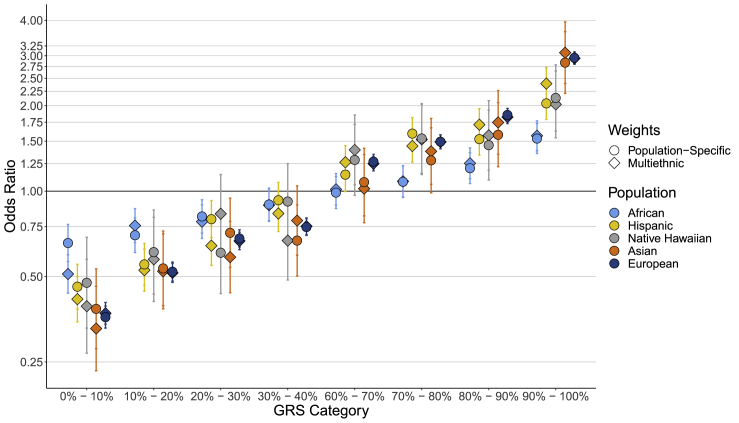
Results for the multiethnic- and population-specific-weighted GRSs constructed using 579 previously identified variants are shown in Figure 3 and Table S6. Comparing individuals in the top 10% of the multiethnic-weighted GRSs to those at average genetic risk in the 40%–60% GRS category, we observed significant differences in effect estimates by population (heterogeneity p = 3.85 × 10−20), with T2D risk being greatest for individuals from Asians (OR = 3.08, 95% CI = 2.40–3.95), followed by European (OR = 2.94, 95% CI = 2.80–3.08), Hispanic (OR = 2.39, 95% CI = 2.10–2.73), Native Hawaiian (OR = 2.02, 95% CI = 1.54–2.65), and African populations (OR = 1.57, 95% CI = 1.39–1.77). A similar pattern was observed for the top 1% of the multiethnic-weighted GRS, with greater T2D risk observed for individuals in Asians (OR = 5.68, 95% CI = 3.11–10.35), followed by European (OR = 5.02, 95% CI = 4.58–5.50), Native Hawaiian (OR = 3.92, 95% CI = 2.18–7.05), Hispanic (OR = 3.48, 95% CI = 2.61–4.64), and African populations (OR = 2.46, 95% CI = 1.86–3.25) (heterogeneity p = 2.06 × 10−5).
Figure 3.
GRS decile category by odds ratio per population
Associations between GRS deciles (relative to the 40%–60% GRS category) and T2D by population. GRSs are weighted by multiethnic (denoted by diamonds) and population-specific (denoted by circles) effect estimates and corresponding error bars. The GRS is based on 582 variants and corresponding weights reported in Vujkovic et al.5
AUCs for the multiethnic-weighted GRS ranged from 0.568 in African ancestry populations to 0.659 in European ancestry populations (without adjustment for covariates; see Material and Methods for analytic details). When including covariates in the model with the multiethnic-weighted GRS, AUCs ranged from 0.670 in African ancestry populations to 0.841 in Asian ancestry populations (Table S7). Interestingly, the multiethnic-weighted GRSs typically had stronger effect estimates and AUCs when compared to the population-specific-weighted GRSs across populations (Tables S6 and S7; Figure 3).
We found significant suggestive differences in the multiethnic-weighted GRS effects on T2D risk between BMI strata (stratified at population-specific median cutoffs), particularly comparing BMI strata within the top 10% GRS decile (Table S8). In particular, the top GRS decile had a greater effect on T2D risk for those with lower BMI (<median) compared to those with higher BMI in all populations except Asians and Native Hawaiians, with a significant difference observed in European populations (OR = 3.42, 95% CI = 3.06–3.82 in those with lower BMI versus OR = 2.78, 95% CI = 2.63–2.93 in those with higher BMI; Ptest for interaction = 0.003).
In aggregate, the 582 variants used in the GRS were estimated to explain 15.3% (European), 11.8% (African), 13.3% (Asian), 15.3% (Hispanics), and 14.3% (Native Hawaiian) of the familial relative risk of T2D in these populations (Table S9).
Discussion
Our multiethnic GWAS meta-analysis of T2D in men and women from European, Asian, African, Hispanic, and Native Hawaiian populations led to the identification of two T2D risk loci that replicated in independent samples. One of the risk variants, rs11466334 within intron 3 of the TGFB1 gene, was only found in African ancestry populations, highlighting the importance of conducting multiethnic studies in order to identify genetic loci that are not otherwise discoverable in individuals of European ancestry. A robust effect size of the rs11466334 variant within TGFB1 was similar (OR ranged from 1.27 to 1.41) to the effect associated with variant rs7903146 in the TCF7L2 gene (OR = 1.37).6
Notably, the African-ancestry-specific T2D risk variant rs11466334 strongly replicated in three additional African ancestry studies. Within the African ancestry discovery and replication studies, the risk allele frequency ranged from 6% in African Americans to 12% in African continental subgroups31 and was either monomorphic or had a very low frequency (<1%) in non-African populations. The variant is predicted to disrupt the CTCF binding motif, changing the position weight matrix logarithm of the odds score from 11.5 to −0.02. CTCF is enriched at chromatin interaction anchors, potentially regulating the communication between an enhancer region and a promoter, and has a role in shaping three-dimensional chromatin organization.39 Chromosomal interactions via looping may affect gene expression in the region.40 Comparing the CTCF binding profiles of 25 tissue types near rs11466334, we found the CTCF signal present in stomach, transverse colon, spleen, thyroid gland, and pancreas tissues, suggesting that this CTCF peak has a regulatory role in tissues involved in T2D (Figure S3). Further investigation of rs11466334 in GTEx showed that this variant significantly predicted tissue expression of the nearby gene ATP synthase subunit s-like protein (ATP5SL) but not TGFB1. Furthermore, in a study mapping quantitative trait loci associated with T2D in adipose and muscle tissue of African American individuals from the MEDIA consortia, a cis-eSNP (rs7259208) within the ATP5SL gene was significantly associated with ATP5SL expression (p = 1.20 × 10−5).41 The protein encoded from the ATP5SL gene is required for assembly of the mitochondrial complex I; however, the role of ATP5SL with T2D is unknown. These two variants in TGFB1 and ATP5SL are moderately correlated (linkage disequilibrium calculations based on the Yoruba in Ibadan, Nigeria [YRI] population between rs11466334 and rs7259208, r2 = 0.30 and D′ = 0.57). Together, these data suggest that allele change of rs11466334 may disrupt CTCF binding to alter expression of nearby target genes; however, additional studies will be needed to understand the biological association with T2D risk.
The TGFB1 mechanistic link to diabetes may involve the disruption of the adaptive β cell expansion from TGF-β signaling, as evidenced in both cell line and mouse experimental studies.42 The TGFB1 gene encodes a secreted ligand that binds to TGF-β receptors leading to activation of the SMAD pathway and thereby mediating TGF-β signaling. Inhibition of TGF-β signaling promotes human pancreatic β cell replication, which is an adaptive response to changing insulin demands. From in vivo mouse and human islet cell experiments, Dhawan et al.42 emphasized the importance of epigenetic pathways involved in the inhibition of TGF-β signaling and regulation of the CDKN2A/B locus (Ink4a/Arf).
Variant rs13052926, identified in the multiethnic analysis, is located within the BACE2 gene, which encodes beta-secretase 2, an enzyme that cleaves amyloid precursor protein into amyloid beta peptide. BACE2 is expressed in the brain and pancreas in human cell lines43 and may play a role in amyloidogenic diseases, such as Alzheimer disease and T2D via islet amyloid polypeptide deposits inducing glucose tolerance defects.44 Based on knockout mouse models and beta cell experiments,43,44 BACE2 inhibitors are deemed a promising drug target for T2D by promoting beta cell survival.45 While variants within BACE2 have not previously been identified in T2D GWASs, a variant in the gene (rs6517656) has been associated with fasting C-peptide levels among pregnant women (∼28 weeks gestation) in multiple ancestral groups.46 Although rs6517656 has not been GWAS-associated with T2D, the variant is in close proximity to (1,400 base pairs) and is in moderate to strong linkage disequilibrium with the T2D-associated variant rs13052926 (r2 = 0.41 and D′ = 0.997 in all populations, and r2 = 0.97 and D′ = 1.0 for European ancestry).
We also evaluated a multiethnic GRS of T2D and found the performance to be greater in European and Asian ancestry populations, likely the result of the larger sample sizes of these two populations in the Vujkovic et al.5 data used to develop the GRS. Interestingly, the multiethnic GRS typically performed better than the population-specific GRS, a finding that has been reported in previous studies and partially attributed to multiethnic weights being more likely to converge on the true causal effect of a variant, in addition to being estimated from larger sample sizes.47,48 The OR for T2D comparing the highest GRS decile to the average 40%–60% GRS category based on multiethnic weights ranged from 1.57 (95% CI = 1.39–1.77) for African ancestry individuals to 2.94 (95% CI = 2.80–3.08) for European and 3.08 (95% CI = 2.40–3.95) for Asian populations. The discriminative ability of the multiethnic GRS estimated by AUCs led to similar conclusions, with the highest AUCs seen in European (AUC = 0.659) and lowest in African population groups (AUC = 0.568). We also found that the proportion of familial relative risk of T2D explained by the aggregate 582 variants used to construct the GRS was lowest in African ancestry populations (11.8%) and highest in European and Hispanic populations (15.3%).
We compared the effect of the GRS on T2D stratified by BMI and found that the GRS had a larger effect in those with lower BMI, suggesting that the role of genetics in T2D risk may be stronger in individuals at low non-genetic risk. Given the suggestive findings of BMI modifying the effect of the GRS on T2D risk, it will be important for future studies to investigate whether other risk factors modify the effect of the GRS on T2D risk, which may account for some of the population differences observed in the discriminative ability of the GRS.
Possible limitations of our study include potential misclassification of type 1 diabetes and T2D, as the exclusion of type 1 diabetes was not routinely conducted among all contributing cohorts. It is possible that a difference in covariate adjustment in the 23andMe statistical replication model, which did not include BMI, may have skewed some replication results.
Our multiethnic investigation identified T2D genetic risk variants and evaluated the most up-to-date GRS in multiple populations. Future GRS studies will benefit from larger population-specific studies to provide robust weights for non-European ancestry populations49 and contribute to fine-mapping efforts, as the current set of GWAS risk variants may not be the best surrogates of the underlying biologically functional allele in all populations. A T2D GRS that is predictive of risk across populations could have future utility in motivating individuals to modify established risk factors and behaviors (i.e., quitting smoking, dietary changes, and BMI reduction) and/or seek regular screening.
Acknowledgments
This research was supported by National Institutes of Health (NIH) National Human Genome Research Institute (NHGRI) grant R56HG010297 and was conducted using the UK Biobank Resource under application number 42195. Study-specific acknowledgments can be found in the Supplemental acknowledgments.
Declaration of interests
W.W. and P.F. are employees of 23andMe Inc. and have stock, stock options, or both in 23andMe. For L.S.P.: Diasyst, Inc. co-founder (equity and stock) Board of Directors or officer Diasyst, Inc. President, secretary, chief medical and scientific officer royalties from Emory. President, secretary, chief medical and scientific officer industry funds to Emory and/or the Atlanta Research and Education Foundation, Inc. (related to the Atlanta VA Medical Center) for research support. Eli Lilly, Novartis, Merck, Amylin, Novo Nordisk, Diasome, Roche, AbbVie, Sanofi-Aventis, Vascular Pharma, Janssen, GlaxoSmithKline, Pfizer, Kowa Investigator - diabetes studies (research support only), Boehringer-Ingelheim, Merck, Novartis, Takeda, Janssen, Profil Research Institute Speaker, and/or Scientific Advisory Board, and/or Consultant. All other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100029.
Contributor Information
Linda M. Polfus, Email: polfus@usc.edu.
Christopher A. Haiman, Email: christopher.haiman@med.usc.edu.
Data and code availability
Example code for the GRS analysis performed in this study is available on GitHub. Code for analyses using other indicated software is readily available from the websites of the corresponding software. Summary-level (PAGE) and individual-level (ARIC [Atherosclerosis Risk in Communities], HCHS/SOL [Hispanic Community Health Study/Study of Latinos], MESA, and PAGE) data are available at DbGaP: phs000090.v1.p1 (ARIC), phs000810.v1.p1 (HCHS/SOL), phs000293.v1 (MESA), phs000.56.v1.p1 (PAGE), and phs000200.v1 (Women’s Health Initiative).
Web resources
DIAGRAM 1000G GWAS meta-analysis with adjustments for BMI Stage 1 Summary statistics, https://diagram-consortium.org/downloads.html
ENCODE, https://www.encodeproject.org/
GTEx Portal, http://gtexportal.org/home/index.html
OMIM, https://www.omim.org
WashU Epigenome Browser, https://epigenomegateway.wustl.edu/
Supplemental information
References
- 1.IDF Diabetes Atlas Group Update of mortality attributable to diabetes for the IDF Diabetes Atlas: Estimates for the year 2013. Diabetes Res. Clin. Pract. 2015;109:461–465. doi: 10.1016/j.diabres.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 2.Xu G., Liu B., Sun Y., Du Y., Snetselaar L.G., Hu F.B., Bao W. Prevalence of diagnosed type 1 and type 2 diabetes among US adults in 2016 and 2017: population based study. BMJ. 2018;362:k1497. doi: 10.1136/bmj.k1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karter A.J., Schillinger D., Adams A.S., Moffet H.H., Liu J., Adler N.E., Kanaya A.M. Elevated rates of diabetes in Pacific Islanders and Asian subgroups: The Diabetes Study of Northern California (DISTANCE) Diabetes Care. 2013;36:574–579. doi: 10.2337/dc12-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willemsen G., Ward K.J., Bell C.G., Christensen K., Bowden J., Dalgård C., Harris J.R., Kaprio J., Lyle R., Magnusson P.K., et al. The Concordance and Heritability of Type 2 Diabetes in 34,166 Twin Pairs From International Twin Registers: The Discordant Twin (DISCOTWIN) Consortium. Twin Res. Hum. Genet. 2015;18:762–771. doi: 10.1017/thg.2015.83. [DOI] [PubMed] [Google Scholar]
- 5.Vujkovic M., Keaton J.M., Lynch J.A., Miller D.R., Zhou J., Tcheandjieu C., Huffman J.E., Assimes T.L., Lorenz K., Zhu X., et al. HPAP Consortium. Regeneron Genetics Center. VA Million Veteran Program Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020;52:680–691. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spracklen C.N., Horikoshi M., Kim Y.J., Lin K., Bragg F., Moon S., Suzuki K., Tam C.H.T., Tabara Y., Kwak S.H., et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature. 2020;582:240–245. doi: 10.1038/s41586-020-2263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook J.P., Morris A.P. Multi-ethnic genome-wide association study identifies novel locus for type 2 diabetes susceptibility. Eur. J. Hum. Genet. 2016;24:1175–1180. doi: 10.1038/ejhg.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bien S.A., Wojcik G.L., Zubair N., Gignoux C.R., Martin A.R., Kocarnik J.M., Martin L.W., Buyske S., Haessler J., Walker R.W., et al. PAGE Study Strategies for Enriching Variant Coverage in Candidate Disease Loci on a Multiethnic Genotyping Array. PLoS ONE. 2016;11:e0167758. doi: 10.1371/journal.pone.0167758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott R.A., Scott L.J., Mägi R., Marullo L., Gaulton K.J., Kaakinen M., Pervjakova N., Pers T.H., Johnson A.D., Eicher J.D., et al. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolonel L.N., Henderson B.E., Hankin J.H., Nomura A.M., Wilkens L.R., Pike M.C., Stram D.O., Monroe K.R., Earle M.E., Nagamine F.S. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Women’s Health Initiative Study Group Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 14.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 15.Hughes G.H., Cutter G., Donahue R., Friedman G.D., Hulley S., Hunkeler E., Jacobs D.R., Jr., Liu K., Orden S., Pirie P., et al. Recruitment in the Coronary Artery Disease Risk Development in Young Adults (Cardia) Study. Control. Clin. Trials. 1987;8(4, Suppl):68S–73S. doi: 10.1016/0197-2456(87)90008-0. [DOI] [PubMed] [Google Scholar]
- 16.Sorlie P.D., Avilés-Santa L.M., Wassertheil-Smoller S., Kaplan R.C., Daviglus M.L., Giachello A.L., Schneiderman N., Raij L., Talavera G., Allison M., et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol. 2010;20:629–641. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman O., Kuivaniemi H., Tromp G., Faucett W.A., Li R., Manolio T.A., Sanderson S.C., Kannry J., Zinberg R., Basford M.A., et al. eMERGE Network The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matise T.C., Ambite J.L., Buyske S., Carlson C.S., Cole S.A., Crawford D.C., Haiman C.A., Heiss G., Kooperberg C., Marchand L.L., et al. PAGE Study The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am. J. Epidemiol. 2011;174:849–859. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haiman C.A., Fesinmeyer M.D., Spencer K.L., Buzková P., Voruganti V.S., Wan P., Haessler J., Franceschini N., Monroe K.R., Howard B.V., et al. Consistent directions of effect for established type 2 diabetes risk variants across populations: the population architecture using Genomics and Epidemiology (PAGE) Consortium. Diabetes. 2012;61:1642–1647. doi: 10.2337/db11-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson L.E. Partitioning large-sample microarray-based gene expression profiles using principal components analysis. Comput. Methods Programs Biomed. 2003;70:107–119. doi: 10.1016/s0169-2607(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 24.Lin D.Y., Tao R., Kalsbeek W.D., Zeng D., Gonzalez F., 2nd, Fernández-Rhodes L., Graff M., Koch G.G., North K.E., Heiss G. Genetic association analysis under complex survey sampling: the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2014;95:675–688. doi: 10.1016/j.ajhg.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Multhaup M.L., Kita R., Krock B., Eriksson N., Fontanillas P., Aslibekyan S., Del Gobbo L., Shelton J.F., Tennen R.I., Lehman A., et al. 2019. Estimating the likelihood of developing type 2 diabetes with polygenic models.https://permalinks.23andme.com/pdf/23_19-Type2Diabetes_March2019.pdf [Google Scholar]
- 27.Parra E.J., Below J.E., Krithika S., Valladares A., Barta J.L., Cox N.J., Hanis C.L., Wacher N., Garcia-Mena J., Hu P., et al. Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican-American sample from Starr County, Texas. Diabetologia. 2011;54:2038–2046. doi: 10.1007/s00125-011-2172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams A.L., Jacobs S.B., Moreno-Macías H., Huerta-Chagoya A., Churchhouse C., Márquez-Luna C., García-Ortíz H., Gómez-Vázquez M.J., Burtt N.P., Aguilar-Salinas C.A., et al. SIGMA Type 2 Diabetes Consortium Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho Y.S., Chen C.H., Hu C., Long J., Ong R.T., Sim X., Takeuchi F., Wu Y., Go M.J., Yamauchi T., et al. DIAGRAM Consortium. MuTHER Consortium Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat. Genet. 2011;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng M.C. Genetics of Type 2 Diabetes in African Americans. Curr. Diab. Rep. 2015;15:74. doi: 10.1007/s11892-015-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J., Sun M., Adeyemo A., Pirie F., Carstensen T., Pomilla C., Doumatey A.P., Chen G., Young E.H., Sandhu M., et al. Genome-wide association study of type 2 diabetes in Africa. Diabetologia. 2019;62:1204–1211. doi: 10.1007/s00125-019-4880-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., White S., Peng B., Johnson A.D., Brody J.A., Li A.H., Huang Z., Carroll A., Wei P., Gibbs R., et al. WGSA: an annotation pipeline for human genome sequencing studies. J. Med. Genet. 2016;53:111–112. doi: 10.1136/jmedgenet-2015-103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balduzzi S., Rücker G., Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid. Based Ment. Health. 2019;22:153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Profile Study. Australian Prostate Cancer BioResource (APCB) IMPACT Study. Canary PASS Investigators. Breast and Prostate Cancer Cohort Consortium (BPC3) PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium. Cancer of the Prostate in Sweden (CAPS) Prostate Cancer Genome-wide Association Study of Uncommon Susceptibility Loci (PEGASUS) Genetic Associations and Mechanisms in Oncology (GAME-ON)/Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) Consortium Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemminki K., Li X., Sundquist K., Sundquist J. Familial risks for type 2 diabetes in Sweden. Diabetes Care. 2010;33:293–297. doi: 10.2337/dc09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K., et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S., Yu N.K., Kaang B.K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015;47:e166. doi: 10.1038/emm.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sajuthi S.P., Sharma N.K., Chou J.W., Palmer N.D., McWilliams D.R., Beal J., Comeau M.E., Ma L., Calles-Escandon J., Demons J., et al. Mapping adipose and muscle tissue expression quantitative trait loci in African Americans to identify genes for type 2 diabetes and obesity. Hum. Genet. 2016;135:869–880. doi: 10.1007/s00439-016-1680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhawan S., Dirice E., Kulkarni R.N., Bhushan A. Inhibition of TGF-β Signaling Promotes Human Pancreatic β-Cell Replication. Diabetes. 2016;65:1208–1218. doi: 10.2337/db15-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rulifson I.C., Cao P., Miao L., Kopecky D., Huang L., White R.D., Samayoa K., Gardner J., Wu X., Chen K., et al. Identification of Human Islet Amyloid Polypeptide as a BACE2 Substrate. PLoS ONE. 2016;11:e0147254. doi: 10.1371/journal.pone.0147254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alcarraz-Vizán G., Castaño C., Visa M., Montane J., Servitja J.M., Novials A. BACE2 suppression promotes β-cell survival and function in a model of type 2 diabetes induced by human islet amyloid polypeptide overexpression. Cell. Mol. Life Sci. 2017;74:2827–2838. doi: 10.1007/s00018-017-2505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh A.K., Brindisi M., Yen Y.C., Lendy E.K., Kovela S., Cárdenas E.L., Reddy B.S., Rao K.V., Downs D., Huang X., et al. Highly Selective and Potent Human β-Secretase 2 (BACE2) Inhibitors against Type 2 Diabetes: Design, Synthesis, X-ray Structure and Structure-Activity Relationship Studies. ChemMedChem. 2019;14:545–560. doi: 10.1002/cmdc.201800725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayes M.G., Urbanek M., Hivert M.F., Armstrong L.L., Morrison J., Guo C., Lowe L.P., Scheftner D.A., Pluzhnikov A., Levine D.M., et al. HAPO Study Cooperative Research Group Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes. 2013;62:3282–3291. doi: 10.2337/db12-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Márquez-Luna C., Loh P.R., Price A.L., South Asian Type 2 Diabetes (SAT2D) Consortium. SIGMA Type 2 Diabetes Consortium Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol. 2017;41:811–823. doi: 10.1002/gepi.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grinde K.E., Qi Q., Thornton T.A., Liu S., Shadyab A.H., Chan K.H.K., Reiner A.P., Sofer T. Generalizing polygenic risk scores from Europeans to Hispanics/Latinos. Genet. Epidemiol. 2019;43:50–62. doi: 10.1002/gepi.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coram M.A., Fang H., Candille S.I., Assimes T.L., Tang H. Leveraging Multi-ethnic Evidence for Risk Assessment of Quantitative Traits in Minority Populations. Am. J. Hum. Genet. 2017;101:638. doi: 10.1016/j.ajhg.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Example code for the GRS analysis performed in this study is available on GitHub. Code for analyses using other indicated software is readily available from the websites of the corresponding software. Summary-level (PAGE) and individual-level (ARIC [Atherosclerosis Risk in Communities], HCHS/SOL [Hispanic Community Health Study/Study of Latinos], MESA, and PAGE) data are available at DbGaP: phs000090.v1.p1 (ARIC), phs000810.v1.p1 (HCHS/SOL), phs000293.v1 (MESA), phs000.56.v1.p1 (PAGE), and phs000200.v1 (Women’s Health Initiative).