Abstract
Programmed cell death ligand 1 (PD‐L1) is a major immunosuppressive checkpoint protein expressed by tumor cells to subvert anticancer immunity. Recent studies have shown that ionizing radiation (IR) upregulates the expression of PD‐L1 in tumor cells. However, whether an IR‐induced DNA damage response (DDR) directly regulates PD‐L1 expression and the functional significance of its upregulation are not fully understood. Here, we show that IR‐induced upregulation of PD‐L1 expression proceeds through both transcriptional and post‐translational mechanisms. Upregulated PD‐L1 was predominantly present on the cell membrane, resulting in T‐cell apoptosis in a co‐culture system. Using mass spectrometry, we identified PD‐L1 interacting proteins and found that BCLAF1 (Bcl2 associated transcription factor 1) is an important regulator of PD‐L1 in response to IR. BCLAF1 depletion decreased PD‐L1 expression by promoting the ubiquitination of PD‐L1. In addition, we show that CMTM6 is upregulated in response to IR and participates in BCLAF1‐dependent PD‐L1 upregulation. Finally, we demonstrated that the ATM/BCLAF1/PD‐L1 axis regulated PD‐L1 stabilization in response to IR. Together, our findings reveal a novel regulatory mechanism of PD‐L1 expression in the DDR.
Keywords: BCLAF1, PD‐L1, post‐translational modification, radiation, ubiquitination
IR‐induced PD‐L1 upregulation expression proceeded through post‐translational mechanisms. We identified BCLAF1 as a new PD‐L1 stabilization regulator in response to IR.

1. Introduction
The PD‐L1/PD‐1 (Programmed cell death ligand 1/Programmed cell death 1) interaction is an immunosuppressive co‐signaling pathway acquired by tumor cells to escape from the host immunity in the tumor microenvironment. Recent studies have shown that PD‐L1/PD‐1 signal‐blockade antibodies can reactivate immune surveillance and exhibit a dramatic antitumor efficacy in patients with certain types of cancer. Radiation therapy is pivotal in cancer treatment. Mounting evidence has revealed that radiotherapy can elicit an anticancer immune response by upregulating MHC tumor‐associated antigen complexes, enhancing tumor antigen cross‐presentation, and increasing T‐cell infiltration into the tumor.1, 2, 3, 4 The synergistic combination of radiotherapy and immunotherapy may provide marked therapeutic efficacies in tumors. In clinical trials comparing the combination of radiotherapy and anti‐PD‐1/PD‐L1 antibody with single traditional therapy,5, 6 patients who received anti‐PD‐1/PD‐L1 antibodies had significantly longer median progression‐free survival and overall survival rate. These observations indicate that radiotherapy serves as an immune priming event. However, the mechanism involved is not fully understood. One mechanism for the synergistic activity of irradiation and PD‐L1/PD‐1 blockade antibody is that ionizing radiation (IR) upregulates PD‐L1 expression in tumor cells. Previous studies have revealed that IFN‐γ produced by T cells is responsible for mediating PD‐L1 upregulation in tumor cells after radiotherapy in vivo.7, 8 Deletion of T cells or inhibition of IFN‐γ abrogates radiation efficacy. IFN‐γ could be an exogenous factor of radiation‐induced PD‐L1 upregulation in tumor cells. However, whether IR directly increases PD‐L1 expression in tumor cells needs to be explored. Sato et al.9 showed that the DNA double‐stranded break repair pathway regulates PD‐L1 expression in cancer cells at the transcriptional level.
Programmed cell death ligand 1 post‐translational modification is involved in many critical biological processes. However, it is not known whether the DNA damage response (DDR) can induce post‐translational modification and, if so, how it functions. In our initial observation, we observed an IR‐induced PD‐L1 post‐translational modification in a Bcl‐2‐associated transcription factor 1 (BCLAF1)‐dependent manner. BCLAF1 was identified as a binding protein for the adenoviral Bcl2 homolog E1B 19K and expressed in the nucleus and cytoplasm.10 Although initial studies showed that BCLAF1 overexpression triggers apoptosis and repressed transcription,10 many studies have suggested that BCLAF1 plays a role in a wide range of processes including lung development,11 T‐cell activation,12 mRNA metabolism,12 posttranscriptional processes,13 tumorigenic roles in colon cancer14 and DNA repair.15 In breast cancer, depletion of BCLAF1 results in sensitivity to DNA damage and defective DNA repair induced by radiation.16 In hepatocellular carcinoma, BCLAF1 promotes tumor progression by manipulating c‐MYC mRNA stability.17
In this report, we demonstrate that IR upregulates PD‐L1 expression at both transcriptional and post‐translational levels. Mass spectrometry findings revealed that BCLAF1 is a PD‐L1‐interacting protein and a regulator of PD‐L1 de‐ubiquitination after DDR. CMTM6, which is a critical PD‐L1 regulator, is involved in the BCLAF1 regulatory pathway. These findings establish BCLAF1 as a new avenue to regulate tumor immune resistance and may contribute to improving the efficacy of therapies, particularly when radiotherapy is considered.
2. Materials and methods
2.1. Cell lines, reagents, and transfection
MDA‐MB‐231, PC‐3, HT1080, HEK293T, and Jurkat cell lines used in this study were obtained from the American Type Culture Collection. They were cultured in DMEM or RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. The transient transfection reagent was Lipofectamine 3000 (Life Technologies). siRNA duplexes targeting PD‐L1 were synthesized and purified by OriGene (OriGene Technologies). siRNA duplexes targeting ATM, BCLAF1, and CMTM6 were synthesized and purified by GenePharma. The following siRNA sequences were used:
PD‐L1#1, 5′‐rGrCrArArUrArUrGrArCrArArArUrUrGrArArUrGrCrArArATT‐3′;
#2, 5′‐rGrCrCrGrArCrUrArCrArArGrCrGrArArUrUrArCrUrGrUGA‐3′;
#3, 5′‐rGrGrArCrCrUrArUrArUrGrUrGrGrUrArGrArGrUrArUrGGT‐3′;
ATM#1, 5′‐AAUGUCUUUGAGUAGUAUGUUTT‐3′;
#2, 5′‐AAGCACCAGUCCAGUAUU GGCTT‐3′;
BCLAF1#1, 5′‐GAUGAAGAGUCUAGAGUAUTT‐3′;
#2, 5′‐GGTTCACTTCGTATCAGAA‐3′;
CMTM6#1, 5′‐CCUUUCUUCUGAGUCUC CUUA‐3′;
#2, 5′‐CUUUCUUCUGAGUCUCCUUAU‐3′.
PD‐L1, PD‐L1‐GFP BCLAF1, CMTM6, and ubiquitin‐HA overexpression plasmids were constructed in our laboratory and confirmed by enzyme digestion and DNA sequencing. Cycloheximide (CHX) was purchased from Selleckchem. Anti‐PD‐1 blocking antibody (10 µg/ml) was purchased from Invitrogen (eBioscience, J116). Recombinant human IFN‐γ (10 ng/ml, Novus. #NBP2‐34992) was added into the medium for additional culturing for 48 h.
2.2. Irradiation
IR was delivered using an X‐RAD 320 X‐ray irradiator (Precision X‐ray Inc.) at a dose rate of 3.1 Gy/min.
2.3. Western blotting, cell fraction isolation, and immunoprecipitation
Cells were harvested and lysed in lysis buffer (Thermo Fisher Scientific, #78503) after washing with phosphate‐buffered saline (PBS). Pierce BCA Protein Assay (Thermo Fisher Scientific, #PI‐23227) was used to measure the protein concentration. Immunoblotting was performed with primary antibodies against PD‐L1 (Cell Signaling Technology; #13684), ATM (Cell Signaling Technology; #2873), BCLAF1 (Abcam, #ab181240), CMTM6 (Abcam, #ab198284), Phospho‐SHP‐2 (Cell Signaling Technology; #3751), caspase‐3 (Cell Signaling Technology; #9662), Ub (Santa Cruz, #sc‐166553), GFP tag antibody (Proteintech, 50430‐2‐AP), HA Tag antibody (Proteintech, 66006‐2‐Ig), Na+/Ka+ ATPase (Abcam, #ab76020), Lamin A/C (Cell Signaling Technology, #4777) and GAPDH (Cell Signaling Technology; #5174). The cell membrane protein was isolated using the Mem‐PER Plus Membrane Protein Extraction Kit (Thermo Fisher Scientific, #89842), and the assay was performed in accordance with the manufacturer’s protocol.
For immunoprecipitation, the cells were lysed in buffer (20 mM Tris‐HCl pH 8.0, 250 mM NaCl, 0.2% NP40, 10% glycerol, 5 mM EDTA, protease, and phosphatase inhibitor cocktail) on ice. After sonication in water bath (55 Hz for 3 min), the lysates were centrifuged at 16 000 ×g for 30 min to remove debris. The cleared lysates were subjected to immunoprecipitation with PD‐L1 antibody (Cell Signaling Technology; #13684) or IgG (Cell Signaling Technology; #2729) and incubated on a head‐over‐head rotator overnight at 4°C. Protein A/G plus‐agarose beads (Santa Cruz, #sc‐2003) were added and incubated further for 2 h at 4°C. After 4 washes in lysis buffer, the samples were eluted in 2× Laemmli sample buffer (BIO‐RAD, #161‐0737) for 10 min at 95°C.
2.4. Immunofluorescence
The MDA‐MB‐231 cells were cultured on coverslips overnight and treated with or without 5 Gy radiation. After the indicated time points, the cells were washed with cold PBS 3 times and fixed with methanol for 5 min, permeabilized and blocked with the Perm/Block solution (0.1% Triton X‐100, 10% normal goat serum in PBS) for 30 min at room temperature. The samples were stained with PD‐L1 antibody (Abcam, #213524) overnight at 4°C. The next day, the cells were washed with PBS 3 times at room temperature and stained with secondary antibody (Invitrogen, #A‐11012) and DAPI at room temperature for 30 min in the dark. A confocal microscope (Carl Zeiss) was used for image analysis.
2.5. Mass spectrometry analysis
Duplicate PD‐L1 and IR‐treated samples (and IgG controls) protein eluates were sent to the University of Alabama at Birmingham mass spectrometry core for analysis. The samples were loaded onto a 10% Bis‐Tris gel. The gel was stained overnight with Colloidal Coomassie (Invitrogen). Each lane was cut into 6‐molecular weight fractions, and each fraction was digested with trypsin (Promega) in accordance with the manufacturer’s instructions. Peptide digests were analyzed using an nLC LTQ Velos Pro Orbitrap mass spectrometer (Thermo Fisher Scientific). The data were analyzed by Professor Mobley James at the University of Alabama, Birmingham.
2.6. Quantitative reverse transcription polymerase chain reaction assays
The cells were lysed, and total RNA was isolated using the RNeasy Mini Kit (Qiagen, #74104) in accordance with the manufacturer’s instructions. cDNA was synthesized through reverse transcription from purified RNA using High‐Capacity cDNA Reverse Transcription Kits (Applied Biosystems). qRT‐PCR was performed in the CFX96 Real‐Time System (Bio‐Rad) using the primers below. All data analyses were performed using the comparative Ct method. Results were normalized to the internal control β‐actin mRNA:
PD‐L1‐Forward: 5′‐GGTGCCGACTACAAGCGAAT‐3′,
PD‐L1‐Reverse: 5′‐AGCCCTCAGCCTGACATGTC‐3′;
BCLAF1‐Forward: 5′‐TCACCTGAGCAGGTAAAGTCTGA‐3′,
PD‐L1‐Reverse: 5′‐AGTCAAGGAA GCAGGTCTGTTAG‐3′;
β‐actin‐Forward: 5‐CATGTACGTTGCTATCCAGGC‐3′,
β‐actin‐Reverse: 5′‐CTCCTTAATGTCACGCACGAT‐3′.
2.7. Flow cytometry
To detect apoptosis, Jurkat cells in the culture medium were collected, centrifuged, and washed with cold PBS. The Tali Apoptosis Kit‐Annexin V Alexa Fluor 488 and propidium iodide (Thermo Fisher Scientific, #A10788) were used to stain the phosphatidyl serine (PS) and nucleic acids of cells separately. Stained cells were subjected to flow cytometric analysis using an Intellicyt iQue+ screener (Intellicyt). The data acquired by the Intellicyt were analyzed using Forecyt analysis software.
2.8. Jurkat and PC‐3 co‐culture systems
PC‐3 tumor cells were plated into 6‐well plates and treated with 0.1 Gy radiation. siRNA‐PD‐L1 was transfected into PC‐3 cells for 48 h before radiation. Subsequently, Jurkat cells were added to PC‐3 cells at a ratio of 1:2 Jurkat:PC‐3. Jurkat cells in the medium were collected after 24 h of co‐culture and were harvested by centrifugation. The cell pellet was lysed to detect phospho‐SHP‐2 (pSHP‐2) by western blot or resuspended in binding buffer for detecting apoptosis by flow cytometry.
2.9. Tissue samples and immunohistochemistry
Archival formalin‐fixed, paraffin‐embedded esophageal squamous cell carcinoma (ESCC) samples were collected from patients diagnosed with primary ESCC after radical esophagectomy at the Cancer Institute and Hospital of Tianjin Medical University from January 2005 to December 2009. Patients who had a history of malignant disease or had received preoperative treatment (chemotherapy and/or radiotherapy) were excluded from this study. The retrieval of tissue and clinical data was approved by the Ethical Institutional Review Board of Tianjin Medical University Cancer Institute and Hospital.
In total, 132 ESCC samples were constructed into a tissue microarray on 1 slide. The tissue microarray was cut into 5‐μm thicknesses and, heated at 70°C for 1 h, deparaffinized in xylene, and rehydrated in a graded series of ethanol. Antigen retrieval was performed using a pressure cooker to heat tissue sections in ethylenediamine tetra‐acetic acid buffer (pH 8.0) for 10 min, and then cooled to room temperature. After incubation with peroxidase blocking reagent (3% H2O2 solution) for 30 min, the tissues were washed with PBS solution 3 times and incubated with primary PD‐L1 antibody (Spring Bioscience, #SP142) or primary BCLAF1 antibody (Abcam, #ab181240) in a humidified chamber overnight at 4°C. After washing the tissue slices with PBS 3 times, they were incubated with HRP‐polymer anti‐mouse/rabbit secondary antibody (Fuzhou Maixin Biotech) for 30 min. Antibody detection was visualized using a DAB detection kit (ZSGB‐BIO, #ZLI‐9018). The slides were counterstained with hematoxylin.
Scoring was evaluated by 2 blinded pathologists. PD‐L1 immunostaining was evaluated as previously described.18 PD‐L1 positivity was defined as >10% PD‐L1 positive cells. PD‐L1 expression was scored in at least 5 microscopic fields. The scoring system of BCLAF1 was assessed as previously described.17, 19, 20 The intensity of immunostaining was scored as negative, weak, moderate, and strong. We defined moderate and strong intensity as positive staining.
2.10. Analysis of BCLAF1 RNA levels in breast cancer, and correlation between BCLAF1 and PD‐L1 RNA levels in The Cancer Genome Atlas (TCGA) samples
We used GEPIA (http://gepia.cancer‐pku.cn/), which is an interactive web application for gene expression analysis based on 9736 tumors and 8587 normal samples from TCGA and the GTEx databases.21 Correlation coefficients and associated P‐values between BCLAF1 and PD‐L1 were computed on this website using Pearson’s method. We used UALCAN (http://ualcan.path.uab.edu/) and Kaplan‐Meier Plotter (https://kmplot.com/analysis/) to analyze the impact of BCLAF1 mRNA expression on survival rate in 1081 breast cancer patients and 211 breast cancer patients with chemotherapy using the Kaplan‐Meier method.
2.11. Statistical analysis
All independent Student t tests were two‐tailed for comparing continuous variables. The correlation between BCLAF1 and PD‐L1 mRNA was analyzed using Pearson’s method. The Spearman method was performed to investigate the association between BCLAF1 and PD‐L1 protein expression. A Kaplan‐Meier estimation was used to compare the differences in overall survival. Values of P < .05 were considered to indicate statistical significance.
3. Results
3.1. Upregulated PD‐L1 after IR induces T‐cell apoptosis
Previous studies have shown that IR upregulates PD‐L1 expression in an IFN‐γ‐dependent manner. However, whether IR directly induces PD‐L1 expression is not known. To test this possibility, we isolated membrane and cytoplasmic or nuclear fractions of MDA‐MB‐231 cells after 5‐Gy IR treatment. We found that IR upregulated PD‐L1 expression in MDA‐MB‐231 cells in the membrane fraction (Figures 1A and S1a). This phenomenon was confirmed in the PC‐3 prostate cancer cell line (Figures 1B and S1b). To examine whether IR‐induced PD‐L1 affected T‐cell function, we pretreated PC‐3 cells with a low dose of IR (0.1 Gy) and then co‐cultured them with Jurkat cells for 24 h. The Jurkat cell line is an immortalized T lymphocyte cell line and has most often been used as a prototypical T‐cell line to study T‐cell signaling. The Jurkat cell line has been used to model and characterize signaling events in T‐cell activation. We detected phosphorylated Src homology region 2 domain‐containing phosphatase‐2 (phospho‐SHP‐2) expression in Jurkat cells. The recruitment of SHP‐2 to the ITSM tyrosine of PD‐1 is critical for inhibiting PD‐1 function,22 in which phosphorylation of SHP‐2 induces its phosphatase activity and regulates the negative signal of PD‐1.23 As shown in Figure 1B, after co‐culturing Jurkat cells with PC‐3 cells containing upregulated PD‐L1, the expression of pSHP‐2 and cleaved caspase‐3 in Jurkat cells increased, indicating that upregulated PD‐L1 activated the PD‐1 downstream pathway. Deletion of PD‐L1 expression in PC‐3 cells with siRNA‐PD‐L1 or blockade of the PD‐L1/PD‐1 interaction with PD‐1 antibodies abrogated the upregulation of pSHP‐2 and cleaved caspase‐3 in Jurkat cells after co‐culture (Figure 1C). These observations were further confirmed by flow cytometry analysis. Co‐culture with PD‐L1‐depleted PC‐3 cells exposed to IR, Jurkat cells did not show cell death (Figure 1D). Taken together, these results indicated that IR‐upregulated PD‐L1 activates T‐cell downstream molecular signals to induce apoptosis.
FIGURE 1.
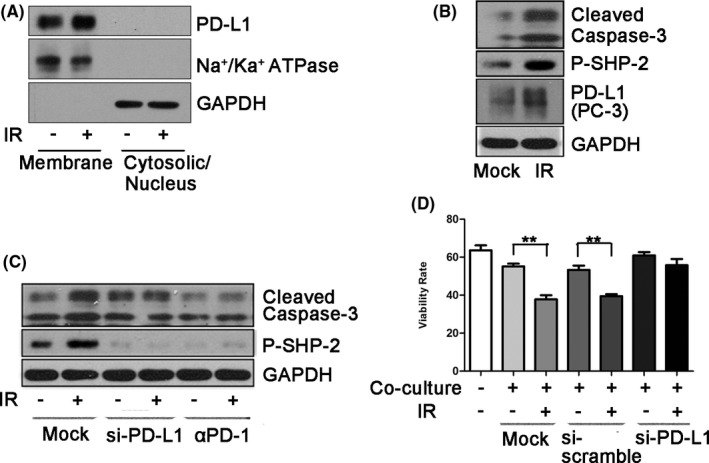
Ionizing radiation induces cancer immunosuppression through upregulation of PD‐L1. A, Cell‐surface expression of PD‐L1 was examined using cell fractionation in MDA‐MB‐231 cells at 2 h after exposure to 5 Gy radiation. Na+/Ka+ ATPase was used as a plasma membrane marker. B, After exposure to 0.1 Gy, PC‐3 cells were co‐cultured with Jurkat cells for 24 h. Cleaved caspase‐3 and pSHP‐2 levels were examined by western blotting (WB) after harvesting Jurkat cells. PD‐L1 was examined after harvesting PC‐3 cells. C, Jurkat cells were co‐cultured with PC‐3 cells treated with 0.1 Gy for 24 h. Cleaved caspase‐3 and pSHP‐2 levels in Jurkat cells were examined by WB after knocking down PD‐L1 expression of PC‐3 cells using siRNA‐PD‐L1 or using an anti‐PD‐1 blocking antibody. The siRNA‐PD‐L1 was transfected into PC‐3 cells for 48 h before co‐culture. D, Flow cytometry was performed to detect annexin V‐FITC/7‐AAD staining to determine the viability of Jurkat cells after co‐culture with PC‐3 cells treated with 0.1 Gy. siRNA‐PD‐L1 or anti‐PD‐1 blocking antibodies were also used in this co‐culture system. Data are presented as the mean ± standard deviation (SD) of triplicate independent experiments. Statistical significance was determined using Student two‐tailed t test. *P < .05, **P < .01
3.2. IR upregulates PD‐L1 expression at both the transcriptional and post‐translational levels
To further study PD‐L1 regulation in the IR response, we examined changes in PD‐L1 protein and mRNA expression in MDA‐MB‐231, HT1080, and PC‐3 cells. As shown in Figure 2A,B, PD‐L1 protein expression was upregulated by 5Gy IR after 2 h, but PD‐L1 mRNA expression did not show any significant change; PD‐L1 expression induced by IR was dose independent (Figure S2a). A time course experiment was performed in MDA‐MB‐231 cells, and both immunoblot and immunofluorescence analyses showed that PD‐L1 protein expression was upregulated from 2 h after IR (Figures 2C and S2b). However, PD‐L1 mRNA expression in MDA‐MB‐231 cells began to increase only at a later time point (8 h) but not at 2 h (Figure 2d). These data suggested that IR upregulated PD‐L1 protein expression by not only inducing PD‐L1 mRNA but also stabilizing the PD‐L1 protein. IR‐induced PD‐L1 stabilization was also observed in an experiment using cycloheximide. At 2‐4 h after IR, PD‐L1 protein expression was still upregulated even when cycloheximide blocked the PD‐L1 mRNA translational process (Figures 2E and S2c).
FIGURE 2.
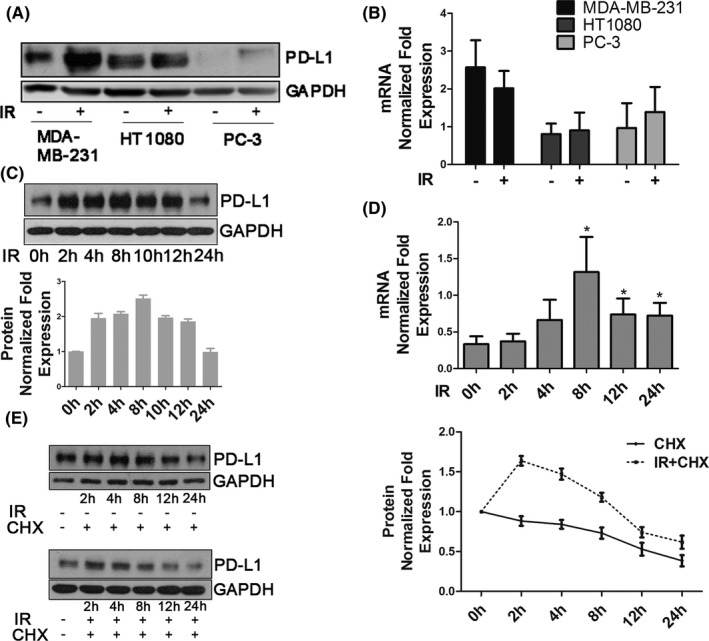
Ionizing radiation increases PD‐L1 expression of cancer cells at the transcriptional and post‐translational levels. A, PD‐L1 protein expression and (B) PD‐L1 mRNA expression were examined in MDA‐MB‐231, HT1080, and PC‐3 cells 2 h after 5 Gy IR. C, PD‐L1 protein expression was examined in MDA‐MB‐231 cells at the indicated time points after 5 Gy IR. Quantification of bands using ImageJ software for PD‐L1 expression is shown in the bar chart below. Error bars are expressed as mean ± SD of 3 independent experiments. D, PD‐L1 mRNA expression was examined in MDA‐MB‐231 cells at the indicated time points after 5 Gy IR. E, Protein stability of PD‐L1 in MDA‐MB‐231 cells. Cells were treated with 20 µM cycloheximide (CHX) at indicated intervals with or without IR (5 Gy) and analyzed by western blot (left). Quantification of bands for PD‐L1 expression is shown as a line graph on the right. Error bars are expressed as mean ± SD of 3 independent experiments
3.3. BCLAF1 mediates IR‐induced PD‐L1 stabilization
To identify the regulatory element controlling IR‐induced PD‐L1 stabilization, we performed liquid chromatography‐tandem mass spectrometry analysis to analyze changes in PD‐L1‐binding proteins after IR. Protein samples were obtained using immunoprecipitation with an anti‐PD‐L1 antibody. Among the 102 binding proteins identified, we selected proteins whose fold change of the quantitative values was more than 1.5, which were located on the membrane or in the cytoplasm of cells. We then ranked these proteins based on the protein mean quantitative value of the untreated group. Among the top 10 proteins related to PD‐L1 regulation shown in Table 1, the first protein was BCLAF1. The interaction between BCLAF1 and PD‐L1 was confirmed by co‐immunoprecipitation (Figure 3A).
TABLE 1.
Results of PD‐L1 binding proteins related to IR by mass spectrometry
| Identified proteins | Accession number | Quantitative values of MOCK group | Unique peptide count of MOCK group | Fold change |
|---|---|---|---|---|
| Bcl‐2‐associated transcription factor 1 | Q9NYF8 | 20.77 | 14 | 1.9 |
| Programmed cell death 1 ligand 1 | Q9NZQ7 | 12.11 | 6 | 1.7 |
| Polyadenylate‐binding protein 4 | Q13310 (+1) | 8.57 | 3 | ‐ |
| Protein FAM208B | Q5VWN6 | 7.70 | 7 | 6.0 |
| Fragile X mental retardation syndrome‐related protein 1 | P51114 | 6.07 | 5 | 2.0 |
| Sperm‐specific antigen 2 | P28290 | 5.83 | 4 | 1.7 |
| Filamin‐B | O75369 | 5.2 | 4 | 2.9 |
| DNA topoisomerase 2‐alpha | P11388 | 4.87 | 4 | 2.0 |
| N‐terminal kinase‐like protein | Q96KG9 | 4.37 | 2 | 5.7 |
| Eukaryotic initiation factor 4A‐III | P38919 | 3.87 | 3 | 1.9 |
FIGURE 3.
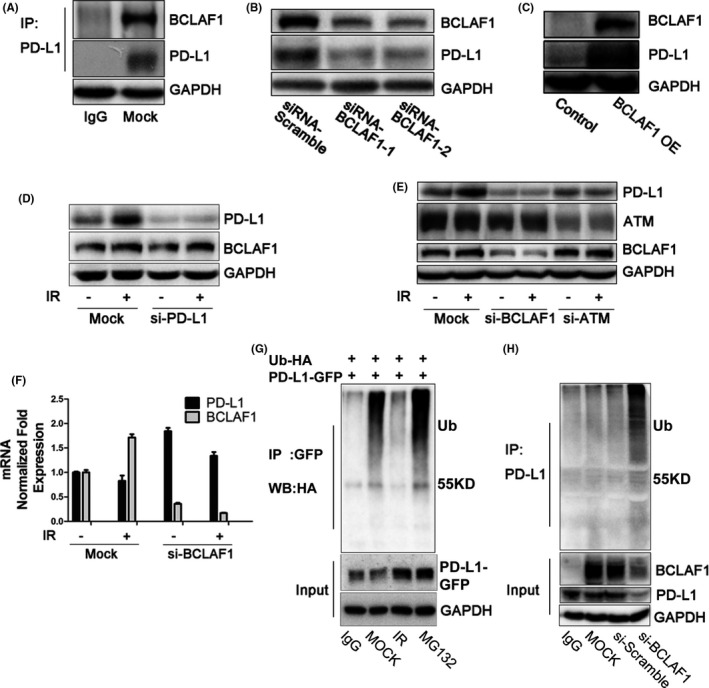
BCLAF1 mediates PD‐L1 stabilization through deubiquitination in response to IR. A, Interaction between endogenous PD‐L1 and BCLAF1 in MDA‐MB‐231 cells. B, PD‐L1 expression after BCLAF1 depletion using 2 different siRNA sequences in MDA‐MB‐231 cells. PD‐L1 was examined 48 h after transfection with siRNA‐BCLAF1. C, HEK293T cells transiently transfected with the PD‐L1 expression plasmid in the presence or absence of the BCLAF1 expression plasmid. PD‐L1 and BCLAF1 levels were analyzed by western blot. D, Western blot analysis of BCLAF1 and PD‐L1 expression in mock and PD‐L1‐knockdown MDA‐MB‐231 cells with or without IR (5 Gy, 2 h). E, Western blot analysis of ATM, BCALF1, and PD‐L1 expression in mock and ATM‐knockdown, BCLAF1‐knockdown MDA‐MB‐231 cells with or without exposure to IR (5 Gy, 2 h). F, Quantitative PCR analysis of BCLAF1 and PD‐L1 mRNA level in mock and BCLAF1‐knockdown MDA‐MB‐231 cells with or without exposure to IR (5 Gy, 2 h). G, After transient co‐transfection of the PD‐L1‐GFP expression plasmid and ubiquitin‐HA plasmid into HEK293T cells, cells were treated with IR (5 Gy for 2 h) or MG132 (10 µM) for 6 h. Ubiquitinated PD‐L1 was immunoprecipitated (IP) with the anti‐GFP antibody and subjected to western blot analysis with an anti‐HA antibody. H, MDA‐MB‐231 cells were transiently transfected with siRNA‐BCLAF1 or scramble siRNA for 48 h. Ubiquitinated PD‐L1 was IPed with an anti‐PD‐L1 antibody and subjected to western blot with an anti‐ubiquitin antibody
Downregulation of BCLAF1 expression with 2 different siRNAs reduced PD‐L1 expression in MDA‐MB‐231 cells (Figure 3B). Overexpression of BCLAF1 increased PD‐L1 expression in HEK293T cells, which expressed exogenous PD‐L1 (Figure 3C). However, downregulation of PD‐L1 with a siRNA did not influence BCALF1 expression (Figures 3D and S2d). As shown in Figure 3E, PD‐L1 upregulation induced by IR was abolished by BCLAF1 knockdown. Similar results were observed in PC‐3 cells (Figure S1b,c). These data suggested that PD‐L1 expression induced by IR is dependent on BCLAF1. Because ATM is the most important transducer of DNA damage signaling and BCLAF1 is a substrate of ATM,24 we detected PD‐L1 and BCLAF1 expression after IR in the presence or absence of a specific ATM siRNA. The results showed that BCLAF1 and PD‐L1 protein expression induced by IR was depleted when ATM was knocked down (Figure 3E). It should be noted that BCLAF1 did not affect PD‐L1 mRNA expression because BCLAF1 knockdown using siRNA did not downregulate PD‐L1 mRNA expression in response to IR (Figure 3F).
Next, we investigated whether BCLAF1‐dependent PD‐L1 upregulation in response to IR is through the ubiquitination pathway. We performed exogenous and endogenous immunoprecipitation experiments. We observed that ubiquitination was increased after MG132 treatment and decreased after IR in HEK293T cells overexpressing PD‐L1‐GFP and ubiquitin‐HA (Figure 3G). Similarly, ubiquitination of endogenous PD‐L1 was also decreased after IR in MDA‐MB‐231 cells (Figure S3a). After BCLAF1 knockdown, we found that PD‐L1 ubiquitination increased (Figure 3H). These results indicated that BCLAF1 mediates deubiquitination of PD‐L1 in response to IR.
3.4. BCLAF1 is required for CMTM6‐mediated PD‐L1 protein stabilization
CKLF‐like MARVEL transmembrane domain‐containing protein 6 (CMTM6) is a critical PD‐L1 regulator that deubiquitinates PD‐L1.25, 26 Whether CMTM6 plays a role in IR‐induced PD‐L1 stabilization is unknown. To test this, we examined CMTM6 protein expression levels by immunoblotting in MDA‐MB‐231, PC‐3, HeLa, and BT‐483 cell lines. We found that CMTM6 expression in these 4 cell lines was upregulated after IR (Figures 4A and S3b,c). We also found that this increased expression was dependent on ATM (Figure S3d). CMTM6 knockdown abolished IR‐induced PD‐L1 upregulation but did not affect BCLAF1 expression (Figure 4B). Similar results were obtained in PC‐3 cells (Figure S1d). However, we found that knocking down BCLAF1 downregulated CMTM6 (Figures 4C and S3e). Furthermore, PD‐L1 expression levels did not decrease further when co‐transfecting BCLAF1 siRNA and CMTM6 siRNA (Figure 4C), suggesting that BCLAF‐1 and CMTM6 might share the same pathway to regulate PD‐L1. The changes in IR‐induced PD‐L1 expression after knocking down BCLAF1/CMTM6 were similar to those of the above results (Figure S3e). However, restoring CMTM6 expression in MDA‐MB‐231 cells after knockdown of BCLAF1 failed to restore the increase in PD‐L1 expression compared with the CMTM6 overexpression group (Figure 4D). These results indicated that BCLAF1 is required for CMTM6‐mediated PD‐L1 protein stabilization.
FIGURE 4.
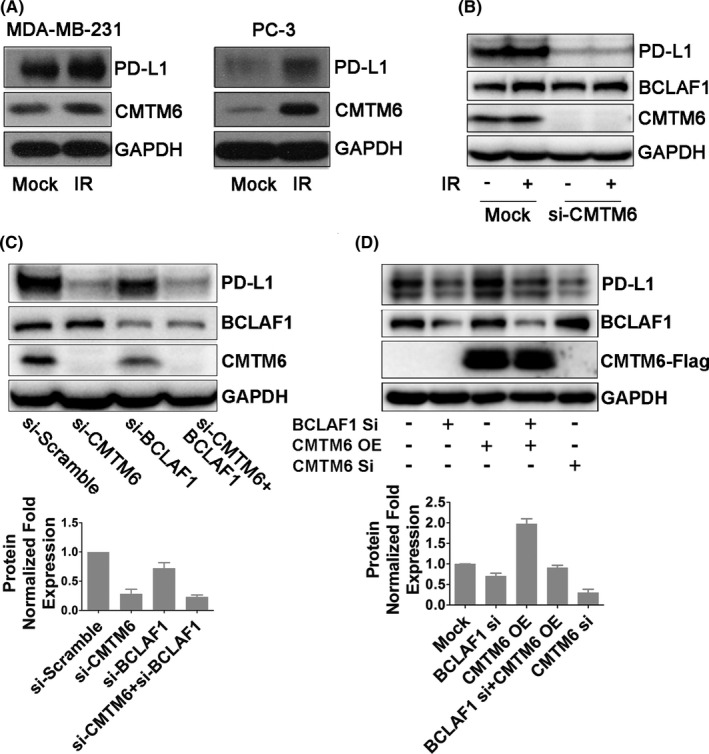
BCLAF1 is required for CMTM6‐mediated PD‐L1 protein stabilization. A, CMTM6 and PD‐L1 expression in MDA‐MB‐231 and PC‐3 cells 2 h after exposure to 5 Gy of IR. B, Western blot analysis of BCLAF1, CMTM6, and PD‐L1 expression in control or CMTM6‐knockdown MDA‐MB‐231 cells in the absence of presence of IR (5 Gy, 2 h). C, Western blot analysis of BCLAF1, CMTM6, and PD‐L1 expression in MDA‐MB‐231 cells transiently transfected with siRNA‐CMTM6, siRNA‐BCLAF1, or both. Quantification of bands for PD‐L1 expression is shown as a bar chart below. Error bars are expressed as mean ± SD of 3 independent experiments. D, Western blot analysis of PD‐L1 after overexpression of CMTM6 in BCLAF1‐knowdown MDA‐MB‐231 cells. Quantification of bands for PD‐L1 expression is shown as a bar chart below. Error bars are expressed as mean ± SD of 3 independent experiments
3.5. Correlation of BCLAF1 and PD‐L1 expression in human tumor tissues
To further validate the correlation between BCLAF1 and PD‐L1, we analyzed BCLAF1 and PD‐L1 RNA expression in tumor samples from TCGA. The RNA expression levels of BCLAF1 and PD‐L1 were positively correlated in 15 types of cancer (Figures 5A and S4a). The Pearson correlation coefficient in 5 types of cancer was more than 0.2 (Figure 5A). To further examine the association of BCLAF1 and PD‐L1 at the protein level, immunohistochemical staining was performed for 132 ESCC specimens. BCLAF1 was detected in 79 of the 86 specimens with high PD‐L1 expression, indicating that there was a positive correlation between BCLAF1 and PD‐L1 protein expression (r = 0.495, P = .000) (Figure 5B). These results suggested a relationship between BCLAF1 and PD‐L1. As a new regulator of PD‐L1, we also investigated the survival predictive value of BCLAF1 in TCGA. High expression of BCLAF1 mRNA was associated with a poor overall survival rate in 1081 breast cancer patients (P = .00014) (Figure 5C). In 211 breast cancer patients with chemotherapy, higher expression of BCLAF1 mRNA was also correlated with worse survival rate (P = .042) (Figure S4b).
FIGURE 5.
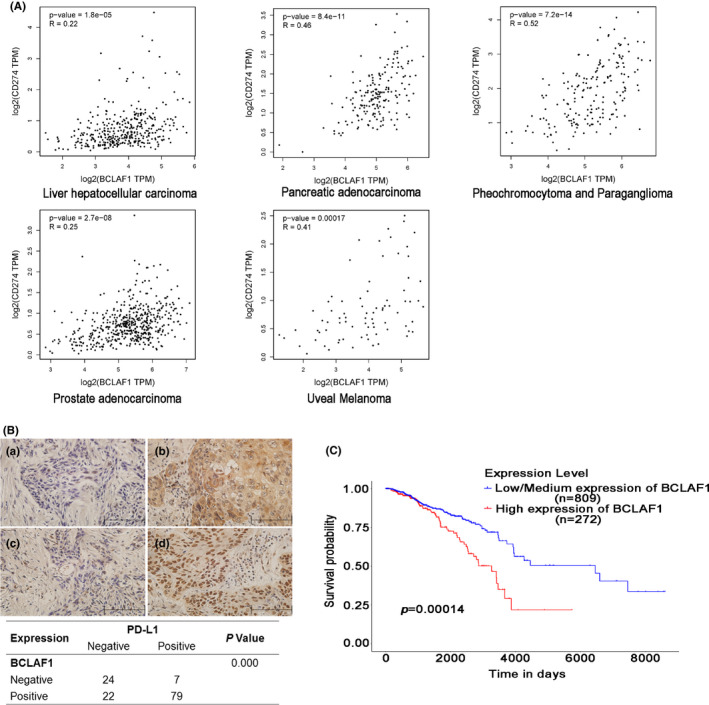
BCLAF1 is positively correlated with PD‐L1 in human cancer tissues and a potential survival predictor for breast cancer. A, From TCGA database, BCLAF1 was correlated with PD‐L1 at the mRNA level in 15 types of cancers. Pearson’s correlation coefficients (r) are shown along with associated P‐values. Correlation coefficients above 0.2 between BCLAF1 and PD‐L1 of cancer are shown. B, In 132 esophageal squamous cancer tissues, the expression of BCLAF1 and PD‐L1 was examined by immunohistochemistry, and the relationship between BCLAF1 and PD‐L1 is shown in the table. C, Kaplan‐Meier analysis indicates a correlation between BCLAF1 overexpression and survival in breast cancer from TCGA database
4. Discussion
Our findings demonstrate that IR‐induced PD‐L1 expression happens at both the transcriptional and post‐translational levels. At the transcriptional level, our data are consistent with a previous report9 showing that the DNA double‐stranded break repair pathway regulates PD‐L1 mRNA expression. At the post‐translational level, we identify BCLAF1 as the key regulator for PD‐L1 stabilization in response to IR. These findings provide a novel mechanism for understanding the regulatory system of PD‐L1, especially in tumor cells treated with DNA damaging agents, including radiotherapy.
Our data demonstrated that BCLAF1 not only participates in DDR‐induced PD‐L1 expression, but is also involved in PD‐L1 upregulation stimulated by IFN‐γ (Figure S2e). These 2 mechanisms might have a compounding effect on tissue levels. How BCLAF1 is directly involved in the processes remains to be discovered. As a transcriptional repressor, BCLAF1 might be involved in transcriptional regulation of CMTM6 and other proteins directly involved in PDL‐1 stabilization. BCLAF1 was originally identified as a protein that interacts with the E1B 19K protein, which is functionally similar to anti‐apoptotic members of the Bcl2 family. Because of its apoptotic activity, BCLAF1 has been proposed to be a tumor suppressor.10 Subsequent studies have shown that BCLAF1 plays a role in a wide range of processes that are not normally associated with the actions of Bcl2 family members.12 Lee et al 15 revealed that BCLAF1 is involved in the γH2AX‐mediated regulation of apoptosis and DNA repair. BCLAF1 contributed to mRNA splicing and export of some genes involved in DNA damage signaling and maintained the genomic stability.16, 27 More recently, BCLAF1 has been reported to have oncogenic features in some tumors. For example, BCLAF1 promotes cancer progression by activating autophagy in bladder cancer.28 BCLAF1 stimulated cell proliferation and colony formation in vitro and promoted the tumorigenic capacity of colon cancer cells in mice.14 In hepatocellular carcinoma, BCLAF1 promoted tumor progression by manipulating c‐MYC mRNA stability17 and regulating HIF‐1α.29 In our study, high expression of BCLAF1 was associated with a poor overall survival rate in breast cancer. Therefore it is possible that BCLAF1 positively regulates the immune checkpoint PD‐L1, which helps the tumor cells escape immune surveillance. We believe that BCLAF1 may exert an oncogenic function in breast cancer by regulating PD‐L1.
Our immune‐histochemical study in ESCC shows a positive correlation between BCLAF1 and PD‐L1 protein expression, indicating a clinical relevance of our findings. In addition, we analyzed 211 breast cancer patients with chemotherapy alone from TCGA database and found that high expression of BCLAF1 was associated with a poor overall survival rate in the subgroup, supporting the notion that BCALF‐1 is related to DNA damage‐based cancer therapy and a potential biomarker.
CMTM6 has been identified as a master regulator of PD‐L1 by reducing PD‐L1 ubiquitination and increasing its protein half‐life.26 PD‐L1 can be induced by IR, however little information is known as to whether IR affects CMTM6 expression. In our study, we found that CMTM6 expression was upregulated in response to IR. Furthermore, knocking down of CMTM6 abolished IR‐induced PD‐L1 expression, indicating that CMTM6 is required for the process. Because BCLAF1‐mediated PD‐L1 stabilization through deubiquitination, we investigated whether CMTM6 played a role in this process. BCLAF1 was found to be an upstream regulator of CMTM6, and BCLAF1 is required for CMTM6‐mediated PD‐L1 stabilization.
In summary, we demonstrated a mechanism of immunosuppression in cancer cells through BCLAF1‐mediated stabilization of PD‐L1. This regulatory event is critical for DDR‐induced PD‐L1 expression. Importantly, inhibition of PD‐L1 stabilization through BCLAF1 not only inhibited PD‐L1 expression induced by DDR but also blocked PD‐L1 mRNA expression stimulated by exogenous IFN‐γ. Therefore, targeting cancer cell PD‐L1 stabilization through BCLAF1 inhibition may represent a potential strategy to restore immune surveillance.
Disclosure
The authors declare that they have no conflict of interest.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Acknowledgments
This work was supported by the fund from the National Natural Science Foundation of China (81672743 and 81974464), the US NIH grant R01CA133093, the Beijing Tianjin Hebei basic research cooperation project (19JCZDJC64500(Z)), the Scientific Research Starting Foundation for Doctoral Scientists of Tianjin Medical University Cancer Institute and Hospital (B1812) and the Fundamental Research Funds for Universities in Tianjin (2020KJ134).
We thank James Mobley of the University of Alabama at Birmingham for his help in mass spectrometry data analysis. We also thank Rebecca J Boohaker of Southern Research Institute for her technical assistance.
Ma Z, Wang H, Meng F, et al. Role of BCLAF‐1 in PD‐L1 stabilization in response to ionizing irradiation. Cancer Sci. 2021;112:4064–4074. 10.1111/cas.15056
Zhentao Yu and Bo Xu jointly supervised this work.
References
- 1.Sharabi AB, Nirschl CJ, Kochel CM, et al. Stereotactic radiation therapy augments antigen‐specific PD‐1‐mediated antitumor immune responses via cross‐presentation of tumor antigen. Cancer Immunol Res. 2015;3:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lugade AA, Moran JP, Gerber SA, Rose RC, Frelinger JG, Lord EM. Local radiation therapy of B16 melanoma tumors increases the generation of tumor antigen‐specific effector cells that traffic to the tumor. J Immunol. 2005;174:7516–7523. [DOI] [PubMed] [Google Scholar]
- 3.Gong J, Le TQ, Massarelli E, Hendifar AE, Tuli R. Radiation therapy and PD‐1/PD‐L1 blockade: the clinical development of an evolving anticancer combination. J Immunother Cancer. 2018;6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu X, Cao Y, Ju X, et al. Personalized designs of adjuvant radiotherapy for pancreatic cancer based on molecular profiles. Cancer Sci. 2021;112(1):287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonia SJ, Villegas A, Daniel D, et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med. 2018;379:2342–2350. [DOI] [PubMed] [Google Scholar]
- 6.Shaverdian N, Lisberg AE, Bornazyan K, et al. Previous radiotherapy and the clinical activity and toxicity of pembrolizumab in the treatment of non‐small‐cell lung cancer: a secondary analysis of the KEYNOTE‐001 phase 1 trial. Lancet Oncol. 2017;18:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dovedi SJ, Adlard AL, Lipowska‐Bhalla G, et al. Acquired resistance to fractionated radiotherapy can be overcome by concurrent PD‐L1 blockade. Cancer Res. 2014;74:5458–5468. [DOI] [PubMed] [Google Scholar]
- 8.Deng L, Liang H, Burnette B, et al. Irradiation and anti‐PD‐L1 treatment synergistically promote antitumor immunity in mice. J Clin Invest. 2014;124:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato H, Niimi A, Yasuhara T, et al. DNA double‐strand break repair pathway regulates PD‐L1 expression in cancer cells. Nat Commun. 2017;8:1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasof GM, Goyal L, White E. Btf, a novel death‐promoting transcriptional repressor that interacts with Bcl‐2‐related proteins. Mol Cell Biol. 1999;19:4390–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPherson JP, Sarras H, Lemmers B, et al. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ. 2009;16:331–339. [DOI] [PubMed] [Google Scholar]
- 12.Sarras H, Alizadeh AS, McPherson JP. In search of a function for BCLAF1. ScientificWorldJournal. 2010;10:1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Li X, Cheng Y, et al. BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat Commun. 2014;5:4581. [DOI] [PubMed] [Google Scholar]
- 15.Lee YY, Yu YB, Gunawardena HP, Xie L, Chen X. BCLAF1 is a radiation‐induced H2AX‐interacting partner involved in gammaH2AX‐mediated regulation of apoptosis and DNA repair. Cell Death Dis. 2012;3:e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savage KI, Gorski JJ, Barros EM, et al. Identification of a BRCA1‐mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell. 2014;54:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Wen Y, Tian Y, et al. Heat shock protein 90alpha‐dependent B‐cell‐2‐associated transcription factor 1 promotes hepatocellular carcinoma proliferation by regulating MYC proto‐oncogene c‐MYC mRNA stability. Hepatology. 2019;69:1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohigashi Y, Sho M, Yamada Y, et al. Clinical significance of programmed death‐1 ligand‐1 and programmed death‐1 ligand‐2 expression in human esophageal cancer. Clin Cancer Res. 2005;11:2947–2953. [DOI] [PubMed] [Google Scholar]
- 19.Dundas SR, Lawrie LC, Rooney PH, Murray GI. Mortalin is over‐expressed by colorectal adenocarcinomas and correlates with poor survival. J Pathol. 2005;205:74–81. [DOI] [PubMed] [Google Scholar]
- 20.Brown GT, Cash B, Alnabulsi A, Samuel LM, Murray GI. The expression and prognostic significance of bcl‐2‐associated transcription factor 1 in rectal cancer following neoadjuvant therapy. Histopathology. 2016;68:556–566. [DOI] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP‐1 and SHP‐2 associate with immunoreceptor tyrosine‐based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol. 2004;173:945–954. [DOI] [PubMed] [Google Scholar]
- 23.Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD‐1 immunoreceptor inhibits B cell receptor‐mediated signaling by recruiting src homology 2‐domain‐containing tyrosine phosphatase 2 to phosphotyrosine. Proc Natl Acad Sci U S A. 2001;98:13866–13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 25.Burr ML, Sparbier CE, Chan YC, et al. CMTM6 maintains the expression of PD‐L1 and regulates anti‐tumour immunity. Nature. 2017;549:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mezzadra R, Sun C, Jae LT, et al. Identification of CMTM6 and CMTM4 as PD‐L1 protein regulators. Nature. 2017;549:106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vohhodina J, Barros EM, Savage AL, et al. The RNA processing factors THRAP3 and BCLAF1 promote the DNA damage response through selective mRNA splicing and nuclear export. Nucleic Acids Res. 2017;45:12816–12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen B, Tan M, Mu X, et al. Upregulated SMYD3 promotes bladder cancer progression by targeting BCLAF1 and activating autophagy. Tumour Biol. 2016;37:7371–7381. [DOI] [PubMed] [Google Scholar]
- 29.Wen Y, Zhou X, Lu M, et al. Bclaf1 promotes angiogenesis by regulating HIF‐1alpha transcription in hepatocellular carcinoma. Oncogene. 2019;38:1845–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4


