Abstract
Tumor‐associated macrophages (TAMs), one of the most common cell components in the tumor microenvironment, have been reported as key contributors to cancer‐related inflammation and enhanced metastatic progression of tumors. To explore the underlying mechanism of TAM‐induced tumor progression, TAMs were isolated from colorectal cancer patients, and the functional interaction with colorectal cancer cells was analyzed. Our study found that coculture of TAMs contributed to a glycolytic state in colorectal cancer, which promoted the stem‐like phenotypes and invasion of tumor cells. TAMs produced the cytokine transforming growth factor‐β to support hypoxia‐inducible factor 1α (HIF1α) expression, thereby upregulating Tribbles pseudokinase 3 (TRIB3) in tumor cells. Elevated expression of TRIB3 resulted in activation of the β‐catenin/Wnt signaling pathway, which eventually enhanced the stem‐like phenotypes and cell invasion in colorectal cancer. Our findings provided evidence that TAMs promoted colorectal cancer progression in a HIF1α/TRIB3‐dependent manner, and blockade of HIF1α signals efficiently improved the outcome of chemotherapy, describing an innovative approach for colorectal cancer treatment.
Keywords: colorectal cancer, HIF1α, TAM, TGF‐β, TRIB3
Our findings provided evidence that tumor‐associated macrophages promoted colorectal progression in a HIF1α/TRIB3‐dependent manner, and blockade of HIF1α signals efficiently improved the outcome of chemotherapy, describing an innovative approach for colorectal cancer treatment.
1. INTRODUCTION
Tumor cells are characterized by a variety of genic alterations, including the activation of oncogenes and restraint of tumor suppressors. Specific autogenous mutations or microenvironment‐derived cytokines, which are involved in the enzyme activity or activation of prosurvival signaling pathways and contribute to the alterations of protein expression and tumor progression, are commonly observed in tumor tissues.1, 2 One such adaptation is aberrant glycolysis, in which tumor cells display enhanced glycometabolism by preferential conversion of pyruvate to lactate instead of entering the tricarboxylic acid cycle. Increasing evidence has indicated that enhanced glycolysis is involved in sustained tumor progression in several tumor types.3 The enhanced glycolysis in tumor cells leads to an accumulation of lactate, thereby resulting in the upregulation of hypoxia‐inducible factor 1α (HIF1α) through suppressing the HIF degradation. Subsequently, HIF1α could further promote tumor progression by facilitating angiogenesis, upregulating prosurvival signaling pathways, and mediating the activation of protumor transcription factors.4, 5 Additionally, HIF1α is prone to participate in immune suppressive effects through regulating the expression of programmed cell death‐ligand 1 and CD73, resulting in the antiinflammatory features in the tumor microenvironment.6
Tumor development is not exclusively dependent on the genetic alterations in cancer cell populations, but is also affected by interaction with the ECM and other subpopulations of cell types in the tumor microenvironment.7 The tumor microenvironment is composed of the ECM, soluble cytokines, mesenchymal cells, or infiltrating lymphocytes, including CD4 T cells, myeloid‐derived suppressor cells, macrophages, and neutrophils. Among those immune cell subpopulations, macrophages constitute one of the most abundant cell types in the tumor microenvironment, which show both pro‐ and antitumor characteristics. Activated M1‐like macrophages participate in pro‐inflammatory responses, which are involved in Toll‐like receptor signaling activation and T cell‐dependent injury effects on tumor cells.8 In contrast, M2‐like macrophages display antiinflammatory features, which are tightly correlated with immune suppression and tumor progression in various tumor types. Tumor‐associated macrophages (TAMs), usually sharing similar phenotypes with M2‐like macrophages, are reported to participate in tumor progression by secreting cytokines to suppress the immune responses of tumor‐infiltrating lymphocytes and promote tumor cell invasion or distant metastasis.9 Although the protumor effects of TAMs have been reported in several tumor types, the underlying mechanism of TAMs and metabolism‐induced tumor progression remains controversial.
In this study, we aimed to further explore the role of TAMs in colorectal cancer development. Our findings suggested that TAMs secreted transforming growth factor‐β (TGF‐β) to affect cell metabolism, resulting in the activation of the HIF1α/Tribbles pseudokinase 3 (TRIB3) signaling pathway, therefore leading to crucial impact to rewiring tumor progression. Furthermore, suppression of HIF1α signaling contributed to improved anticancer effects, revealing a novel strategy for clinical colorectal cancer treatment.
2. MATERIALS AND METHODS
2.1. Cell culture and reagents
Human colorectal cancer HCT116 cells and murine colorectal cancer CT26 cells were purchased from the Cell Bank of Chinese Academy of Sciences. All cell lines were cultured in RPMI‐1640 (Gibco) supplemented with 10% FBS (Gibco). Transforming growth factor‐β1 was purchased from Thermo Fisher Scientific. Anti‐human TGF‐β1 neutralizing Ab was purchased from Sigma. The HIF1α inhibitor GN44028 and β‐catenin/Wnt inhibitor PNU‐74654 were purchased from MCM. 5‐Fluorouracil (5‐FU) and oxaliplatin (OXA) were purchased from Sangon Biotech.
2.2. Tumor tissue collection and culture of TAMs
Colorectal tumor samples were obtained after surgery at Tianjin Medical University Cancer Institute and Hospital (No. LLSP2019‐016). All samples were reviewed by a pathologist according to the WHO classification. The samples were divided into metastatic and nonmetastatic groups according to follow‐up visits. All sample collection and processing were undertaken according to the Declaration of Helsinki and ethical approval was obtained from the Committee of Tianjin Medical University Cancer Institute and Hospital. For TAM isolation and culture, tumor tissues from patients were cut into pieces as small as possible after surgery. The tissues were digested by DMEM culture medium containing ACCUMAX (Sigma) at 37℃ with 5% CO2 in an incubator for 2 hours, followed by filtration (40 μm; Thermo Fisher Scientific). The cell population was stained by primary Ab to CD68 (eBioscience) for TAM isolation and analysis. The isolated TAMs were seeded into a 6‐well plate containing 2 mL DMEM culture medium with 10% FBS at 37℃. Tumor‐associated macrophages were cultured for a maximum of 7 days.
2.3. Cell proliferation analysis
Cell proliferation was detected using a CCK‐8 kit (Solarbio). Briefly, 2 × 103 HCT116 cells cocultured with TAMs or not were seeded into 96‐well culture plates. CCK‐8 solution (20 μL) was added to the 96 wells at determined time points. After incubation at 37℃ of 2 hours, absorbance was measured at 450 nm on a microplate reader (Bio‐Rad). Each experiment was carried out independently three times.
2.4. Colony formation and tumorigenesis analysis
For the colony formation assay, CT26 or HCT116 cells (500 cells per well) were seeded into 6‐well plates and cultured at 37℃ for 10 days. The colonies were then fixed using 4% paraformaldehyde and stained with crystal violet. Colonies were photographed and counted. Each experiment was repeated independently in triplicate. For tumorigenesis analysis, 105 HCT116 cells (50 μL PBS) were subcutaneously injected into NOD‐SCID mice and 2 × 104 CT26 cells (50 μL PBS) were subcutaneously injected into Balb/C mice (n = 10 per group). After 30 days, the tumor‐bearing mice were counted in each group. Each experiment was carried out independently three times.
2.5. Transwell analysis
For the cell invasion assay, HCT116 and CT26 cells (5 × 104 cells) were seeded in the upper Transwell chamber (8 μm; Corning). The bottom chamber was filled with 0.5 mL medium containing 20% FBS. After 24 hours, cells were fixed with 4% paraformaldehyde and then stained with 0.05% crystal violet. The cell numbers were counted in 10 fields. Each experiment was repeated independently in triplicate.
2.6. Immunohistochemical and immunofluorescence staining
The paraffin sections of tumor tissues from colorectal cancer were obtained from Tianjin Medical University Cancer Institute and Hospital. The sections were then deparaffinized and rehydrated in alcohol and water. Antigen retrieval was undertaken in sodium citrate buffer for 5 minutes at 100℃. The sections were then incubated with anti‐CD68 (1:300; Abcam), anti‐TGF‐β1 (1:100, Ab215715; Abcam), anti‐Wnt3A (1:200, Ab219412; Abcam), anti‐TRIB3 (1:500, Ab137526; Abcam), and anti‐HIF1α (1:300, Ab179483; Abcam) primary Abs at 4℃ overnight. For immunofluorescent staining, the samples were incubated with goat anti‐rabbit secondary Abs (1:1000; Thermo Fisher Scientific) and the nuclei were stained with DAPI (Solarbio). Images were obtained using a laser scanning confocal microscope (Leica). For immunohistochemical staining, samples were incubated with an ABC HRP Kit (Thermo Fisher Scientific) and counterstained with hematoxylin (Solarbio). The intensity of protein expression was calculated by ImageJ 2.0 (immunofluorescence) and Image‐Pro Plus 7.0 (immunohistochemistry) software. The mean of protein expression intensity in 30 fields was determined in each sample. Fifteen samples from 15 colorectal cancer patients were included in each group.
2.7. Western blot analysis
The protein lysates from CT26 and HCT116 cells were separated by SDS‐PAGE and then transferred to a PVDF membrane (Millipore). The membrane was incubated with the primary Abs against anti‐HIF1α (1:1000, Ab179483; Abcam), anti‐TRIB3 (1:1000, Ab137526; Abcam), anti‐β‐catenin (1:1000, Ab32572; Abcam), anti‐Wnt3A (1:1000, Ab219412; Abcam), and anti‐β‐actin (1:1000, Ab8226; Abcam), followed by incubation with an HRP‐conjugated secondary Ab (1:1000; Abcam).
2.8. RNA interference
For RNA interference, human TRIB3, HIF1α, and Wnt3A siRNA were purchased from Ruibo. All siRNA were transfected at a concentration of 20 μmol/L using Lipofectamine RNAiMAX (Thermo Fisher Scientific). The silencing efficiency of all siRNA was greater than 60% in HCT116 cells. Independent experiments were carried out in triplicate.
2.9. Immunoassays
For lactic acid or glucose measurement, 5 × 105 pretreated tumor cells were seeded in 6‐well plates for 24 hours. The RPMI‐1640 culture medium was replaced with 2 mL fresh medium; after 24 hours, the supernatant was collected and glucose and lactic acid concentrations were measured using a Glucose Assay Kit (Abcam) and Lactic Acid Test Kit (Sigma).
For TGF‐β measurement, 105 macrophages were collected and cultured for the same length of indicated time, and TGF‐β production of cells was measured according to the instructions of a TGF‐β1 human ELISA kit (Thermo Fisher Scientific).
2.10. Animal protocols
Female Balb/C mice (6‐8 weeks old) and NOD‐SCID mice (6‐8 weeks old) were purchased from Huafukang. All mice were housed in a specific pathogen‐free facility. All animal experiments were carried out according to the guidelines approved by the Institute Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. For the subcutaneous colorectal cancer model, 106 HCT116 cells (50 μL PBS) were subcutaneously injected into NOD‐SCID mice and 2 × 105 CT26 cells (50 μL PBS) were subcutaneously injected into Balb/C mice. After 14 days, mice were treated with PBS, 5‐FU (25 mg/kg), OXA (10 mg/kg), or GN44028 (5 mg/kg) by tail vein injection twice a week. The tumor volumes of mice were recorded every day (n = 6). Survival was recorded on a daily basis (n = 6). The calculation of tumor volume used the formula: tumor volume = length × width2/2.
2.11. Statistical analysis
Each experiment was undertaken independently at least three times. Results are presented as the mean ± SEM and the statistical significance was analyzed using GraphPad 6.0 software. Statistical significance between groups was calculated by Student’s t test for two groups or by one‐way ANOVA for more than two groups. The survival rates were determined by Kaplan‐Meier survival analysis (*P < .05; **P < .01).
3. RESULTS
3.1. Tumor‐associated macrophages promoted stem‐like phenotypes and metastasis in colorectal cancer
Previous reports have shown that colorectal cancer with strengthened migratory and invasive characteristics exhibited typical genomic expression profiles, including oncogenic signatures or subset genes with the capability to drive stem‐like phenotypes.10 Here, our aim was to explore the role of the tumor microenvironment and, more specifically, the role of TAMs in driving colorectal cancer progression. To do this, we isolated colorectal tumor tissues from patients, which were divided into nonmetastatic and metastatic groups. Immunohistochemical staining of CD68, a marker of human macrophages, was carried out and enriched TAM distribution was observed in tumor tissues from metastatic colorectal cancer patients (Figure 1A). Subsequently, flow cytometry screening was used to isolate CD68+ macrophages. Macrophages and colorectal cancer HCT116 cells were cocultured for 72 hours in a Transwell inset/24‐well system (Figure 1B). After coculturing with TAMs, the tumor cells showed enhanced capabilities of colony formation (Figure 1C) and tumorigenicity (Figure 1D), suggesting acquired stem‐like phenotypes of colorectal cancer cells after coculture. As previously mentioned, cancer stem cells might possess enhanced capabilities of proliferation and migration and, consequently, promote sustained tumor growth and distant metastasis in vivo. To assess the influence of TAMs, the proliferation and invasion of TAM‐cocultured HCT116 cells were examined in vitro. Intriguingly, no obvious difference was observed in the proliferation of TAM‐cocultured HCT116 cells (Figure 1E), whereas a significantly increased number of migrating cells was found in the TAMs coculture group compared with the control group (Figure 1F), indicating that TAMs could promote metastasis of colorectal cancer. These findings suggested a direct relationship between the subtype of TAMs and colorectal cancer development. Moreover, it should be addressed that TAMs have the capability to promote stem‐like phenotypes and invasion of tumor cells, instead of regulating colorectal cancer cell proliferation directly.
FIGURE 1.
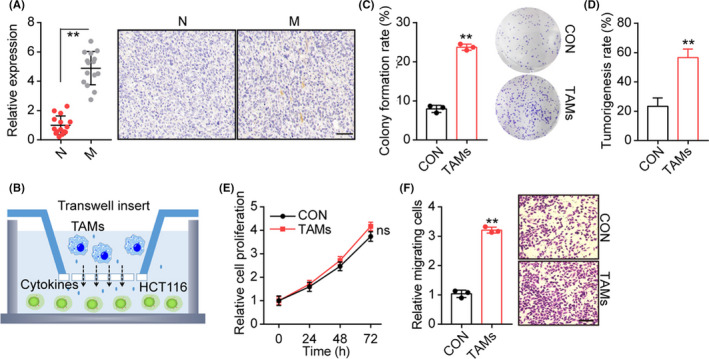
Tumor‐associated macrophages (TAMs) promoted stem‐like phenotypes and cancer metastasis. A, Immunohistochemical staining of CD68 in tumor tissues from metastasis (M) and nonmetastasis (N) colorectal cancer patients. The TAMs percentages were calculated by Image‐Pro Plus 6.0 (n = 15 per group). Scale bar, 100 μm. B, Schematic diagram of macrophages and HCT116 cell coculture system. TAMs were seeded in the upper inset (3 μm) and HCT116 cells were cultured in the bottom of the 24‐well system (TAMs : HCT116, 1:5). HCT116 cells were cocultured with TAMs for 72 h. C, Colony formation rates of HCT116 cocultured with TAMs or not. D, Tumorigenesis capability of HCT116 cells in NOD‐SCID mice was examined after TAMs coculture or not. E, Relative cell proliferation of HCT116 cells cocultured with TAMs or not. F, Relative migrating cell numbers of HCT116 cocultured with TAMs or not, using Transwell analysis. Scale bar, 80 μm. **P < .01. ns, no significant difference
3.2. Tumor‐associated macrophages secreted TGF‐β to facilitate colorectal cancer development
To clarify the mechanism underlying TAM‐induced tumor progression, we further examined the cytokines produced by patient‐derived TAMs. Compelling findings showed that CD206+ M2 macrophages expressed high amounts of TGF‐β, a major cytokine involved in the cancer metastasis.11 Thereby, we sought to assess whether elevated TGF‐β was produced by TAMs to regulate colorectal cancer progression. Notably, significantly increased TGF‐β secretion was observed in the culture medium of TAMs compared to blood‐derived macrophages (Figure 2A). In addition, expression of TGF‐β was increased in tumor tissues from metastatic patients (Figure 2B), suggesting that TGF‐β might be involved in TAM‐associated colorectal cancer progression. We further treated HCT116 cells with TAMs combining TGF‐β neutralizing Abs. Consistent with our hypothesis, TGF‐β neutralizing Ab treatment suppressed the colony formation (Figure S1A) and cell migration (Figure S1B) induced by TAMs, whereas free neutralizing Ab treatment had no impact on HCT116 cells. To further explore the role of TGF‐β in colorectal cancer progression, TGF‐β was added to the culture medium of colorectal cancer cell lines HCT116 and CT26. After 72 hours of culture, HCT116 and CT26 cells revealed enhanced capability of colony formation (Figure 2C) and tumorigenicity (Figure 2D). Increased migrating cells in TGF‐β treated HCT116 and CT26 cells was also observed (Figure 2E). Collectively, these results suggested that TAMs promoted colorectal cancer progression through secretion of TGF‐β.
FIGURE 2.
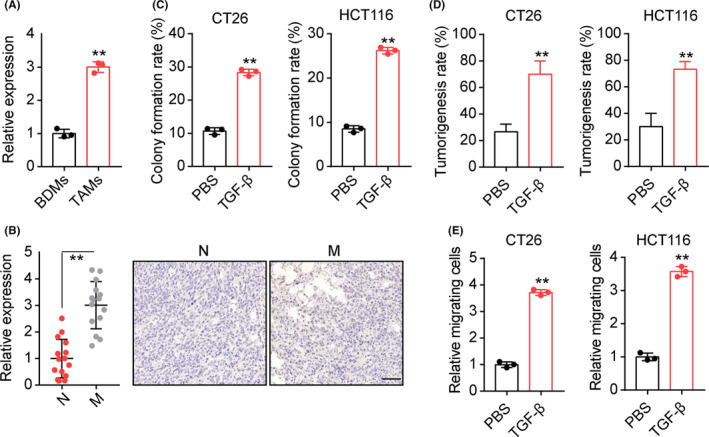
Tumor‐associated macrophages (TAMs) secreted transforming growth factor‐β (TGF‐β) to facilitate colorectal cancer development. A, TGF‐β concentration was detected in the culture medium of TAMs and blood‐derived macrophages (BDMs) (105 macrophages in 1 mL culture medium for 48 h). B, Immunohistochemical staining of TGF‐β in tumor tissues from metastasis (M) and nonmetastasis (N) colorectal cancer patients. Expression intensity was calculated by Image‐Pro Plus 6.0 (n = 15 per group). Scale bar, 100 μm. C, Colony formation rates of CT26/HCT116 cells treated with PBS or TGF‐β (100 ng/mL). D, Tumorigenesis capability of CT26/HCT116 cells in NOD‐SCID mice was examined after PBS or TGF‐β (100 ng/mL) treatment. E, Numbers of relative migrating CT26/HCT116 cells treated with PBS or TGF‐β (100 ng/mL). **P < .01
3.3. Transforming growth factor‐β upregulated HIF1α to promote TRIB3 expression in colorectal cancer
We next investigated the underlying mechanism by which TGF‐β promotes colorectal cancer development. Several lines of evidence suggested that TGF‐β could promote glycolysis of tumor cells, resulting in the accumulation of succinate and HIF1α signal activation.12 Indeed, enhanced glucose consumption and lactic acid production was found in TAM‐cocultured or TGF‐β treated HCT116 cells (Figure S2A,B). We also observed enhanced expression of HIF1α in TAM‐cocultured HCT116 cells (Figure 3A), and TGF‐β treatment significantly upregulated the expression of HIF1α in HCT116 and CT26 cells (Figure 3B). Repressing the HIF1α signaling by inhibitor GN44028 abrogated the TGF‐β‐induced colony formation (Figure 3C) and tumorigenicity (Figure 3D) induced by TGF‐β. Meanwhile, blockade of HIF1α suppressed the TGF‐β associated cells invasion in HCT116 and CT26 cells (Figure 3E). Similar results were observed in HIF1α silenced HCT116 cells (Figure S2C,D), indicating that TGF‐β promoted colorectal cancer progression in a HIF1α‐dependent manner.
FIGURE 3.
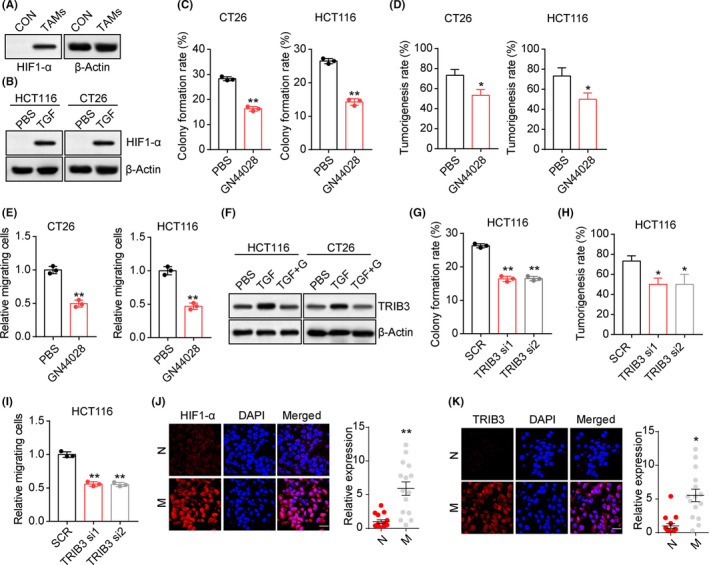
Transforming growth factor‐β (TGF‐β) upregulated hypoxia‐induble factor 1α (HIF1α) to promote Tribbles pseudokinase 3 (TRIB3) expression. A, Western blotting of HIF1α in HCT116 cells cocultured with tumor‐associated macrophages (TAMs) or not (CON). B, Western blotting of HIF1α in HCT116/CT26 cells treated with PBS or TGF‐β (100 ng/mL). C, Colony formation rates of TGF‐β (100 ng/mL) cultured CT26/HCT116 cells treated with PBS or GN44028 (20 nmol/L). D, Tumorigenesis capability of TGF‐β (100 ng/mL) cultured CT26/HCT116 cells treated with PBS or GN44028 (20 nmol/L). E, Numbers of relative migrating TGF‐β (100 ng/mL) cultured CT26/HCT116 cells treated with PBS or GN44028 (20 nmol/L). F, Western blotting of TRIB3 in CT26/HCT116 cells treated with TGF‐β (100 ng/mL) combined with PBS or GN44028 (20 nmol/L) (TGF+G). G, Colony formation rates of TGF‐β (100 ng/mL) cultured HCT116 cells treated with scramble RNA (SCR) or TRIB3 siRNA. H, Tumorigenesis capability of TGF‐β (100 ng/mL) cultured HCT116 cells treated with scramble RNA or TRIB3 siRNA. I, Numbers of relative migrating TGF‐β (100 ng/mL) cultured HCT116 cells treated with SCR or TRIB3 siRNA. J, Immunofluorescence staining of HIF1α in tumor tissues from metastasis (M) and nonmetastasis (N) colorectal cancer patients. Intensity of HIF1α expression was determined in 30 tumor tissues. Scale bar, 30 μm. K, Immunofluorescence staining of TRIB3 in tumor tissues from M and N colorectal cancer patients. Intensity of TIRIB3 expression was determined in 30 tumor tissues. Scale bar, 30 μm. *P < .05; **P < .01
Compelling findings provided evidence to suggest that TRIB3 denoted a poor prognosis in breast cancer and was associated with hypoxia response.13, 14 Intriguingly, we showed that TRIB3 was further upregulated in TGF‐β treated HCT116/CT26 cells. We hypothesized that TRIB3, a known tumor stemness regulator, might serve as the downstream regulator of HIF1α and, consequently, contributed to colorectal cancer progression. Upon suppression of HIF1α by GN44028, the upregulation of TRIB3 induced by TGF‐β was retarded in HCT116 and CT26 cells (Figure 3F). Furthermore, we found that silence of TRIB3 by siRNA efficiently suppressed colony formation (Figure 3G) and tumorigenicity (Figure 3H). Additionally, TRIB3 siRNA weakened the capability of cell invasion (Figure 3I), indicating that TGF‐β mediated colorectal cancer development through HIF1α/TRIB3 signaling. Consistently, the expression of HIF1α (Figure 3J) and TRIB3 (Figure 3K) was significantly elevated in tumor tissues from metastatic patients compared to the nonmetastatic group. Thus, those results suggested that TGF‐β upregulated HIF1α/TRIB3 expression to promote colorectal cancer development.
3.4. β‐Catenin/Wnt signaling activation promoted by TRIB3
Previous reports have suggested that TRIB3 could interact with β‐catenin and transcription factor 4 to activate the Wnt‐associated prosurvival signaling pathways.15 Herein, we further examined the activation of the β‐catenin/Wnt signaling pathway in TGF‐β treated colorectal cancer cells. As shown in Figure 4A, elevated expression of β‐catenin/Wnt was observed in TGF‐β treated HCT116/CT26 cells, and blockade of HIF1α by GN44028 or TRIB3 silence abrogated the TGF‐β‐induced β‐catenin/Wnt signal activation. To further explore the role of β‐catenin/Wnt signals in colorectal cancer, PNU‐74654, a β‐catenin/Wnt inhibitor, was added to the culture medium of HCT116/CT26 cells. Consistently, blockade of β‐catenin/Wnt signals suppressed the colony formation (Figure 4B) and tumorigenicity (Figure 4C) induced by TGF‐β. Additionally, suppression of β‐catenin/Wnt signals retarded invasion of HCT116 and CT26 cells (Figures 4D and S3), indicating that TGF‐β mediated the activation of Wnt signaling in colorectal cancer. Elevated expression of Wnt signals was also found in tumor tissues from metastatic patients compared to the nonmetastatic group (Figure 4E). Together, these results suggested that TGF‐β facilitated HIF1α/TRIB3/β‐catenin/Wnt signaling pathway activation to promote colorectal cancer progression.
FIGURE 4.
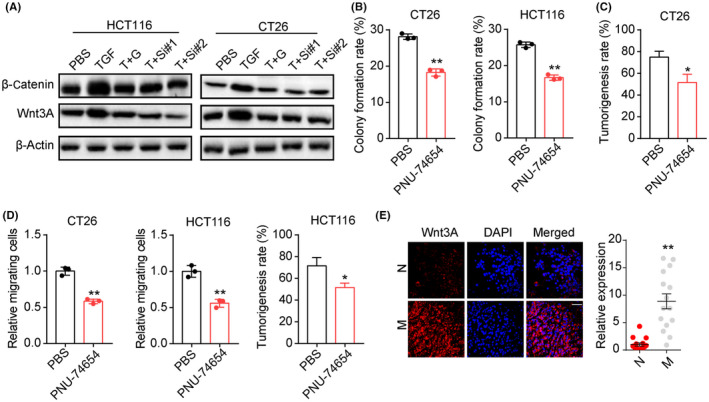
Tribbles pseudokinase 3 (TRIB3) promoted β‐catenin/Wnt signaling. A, Western blotting of β‐catenin and Wnt3A in HCT116/CT26 cells treated with PBS, transforming growth factor‐β (TGF‐β, 100 ng/mL), TGF‐β (100 ng/mL) combined with GN44028 (20 nmol/L) (T+G), or TGF‐β (100 ng/mL) combined with TRIB3 siRNA (T+Si#1 and T+S#2). B, Colony formation rates of TGF‐β (100 ng/mL) cultured CT26/HCT116 cells treated with PBS or PNU‐74654 (50 μmol/L). C, Tumorigenesis capability of TGF‐β (100 ng/mL) cultured CT26/HCT116 cells treated with PBS or PNU‐74654 (50 μmol/L). D, Relative migrating cell numbers of TGF‐β (100 ng/mL) cultured CT26/HCT116 treated with PBS or PNU‐74654 (50 μmol/L). E, Immunofluorescence staining of Wnt3A in tumor tissues from metastasis (M) and nonmetastasis (N) colorectal cancer patients. Intensity of β‐catenin/Wnt expression was determined in 30 patients. Scale bar, 50 μm. *P < .05. **P < .01
3.5. Blockade of HIF1α inhibited tumor growth and lung metastasis of colorectal cancer
Given the importance of HIF1α signals in colorectal cancer development described above, we examined whether suppression of HIF1α could improve the outcome of chemotherapy. Tumors arising from the subcutaneous implication of CT26 and HCT116 cells in mice was established for analysis of anticancer effects. Using this model, subcutaneous CT26 and HCT116 tumors were established in mice, then treated with chemotherapeutic 5‐FU, OXA, and HIF1α inhibitor GN44028. Notably, GN44028 treatment resulted in the suppression of tumor growth, and strengthened the anticancer effects of 5‐FU (Figure 5A,B) and OXA (Figure 5C,D) in HCT116 mice. The similar anticancer effects of GN44028 were observed in CT26‐bearing mice treated with 5‐FU (Figure 5E,F). Next, we collected lung tissues from CT26‐bearing mice to assess the pulmonary cancer metastasis. Both 5‐FU and GN44028 treatment contributed to reduced pulmonary nodules in tumor‐bearing mice, and enhanced metastasis‐suppressive effects were found in the combination group (Figure 5G). On the basis of our results, we suggested that suppression of HIF1α signals by GN44028 could efficiently improve the outcome of chemotherapy and impair tumor metastasis in colorectal cancer.
FIGURE 5.
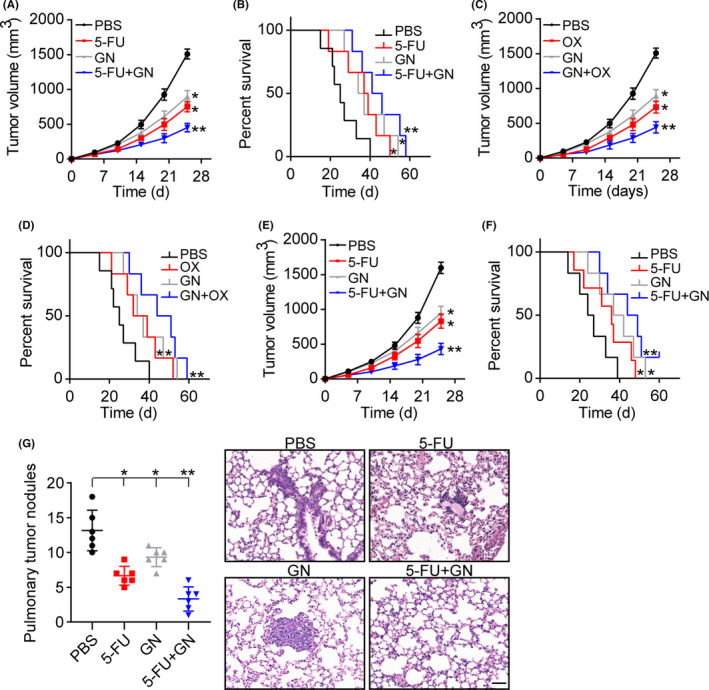
Blockade of hypoxia‐induble factor 1α (HIF1α) inhibited tumor growth and lung metastasis. A, Tumor volume of HCT116‐bearing mice treated with PBS, 5‐fluorouracil (5‐FU), GN44028 (GN), or 5‐FU combined with GN44028. B, Overall survival of HCT116‐bearing mice treated with PBS, 5‐FU, GN, or 5‐FU+GN. C, Tumor volume of HCT116‐bearing mice treated with PBS, oxaliplatin (OX), GN, or GN+OX. D, Overall survival of HCT116‐bearing mice treated with PBS, OX, GN, or GN+OX. E, Tumor volume of CT26‐bearing mice treated with PBS, 5‐FU, GN, or 5‐FU+GN. F, Overall survival of CT26‐bearing mice treated with PBS, 5‐FU, GN, or 5‐FU+GN. G, H&E staining of lung tissues and metastatic pulmonary nodule numbers in CT26‐bearing mice treated with PBS, 5‐FU, GN, or 5‐FU+GN. Scale bar, 100 μm. *P < .05. **P < .01
4. DISCUSSION
Both intrinsic and extrinsic stressors, including the tumor microenvironment and oncogene mutations, could promote the activation of prosurvival signaling pathways to strengthen cancer cell survival. Tumor‐associated macrophages serve as the common immune cell subpopulation in the tumor microenvironment, and are involved in multiple tumor progression and have been recognized as crucial determinant in cancer development.16, 17, 18 However, the molecular mechanism by which TAMs sustain tumor progression remain poorly understood. In our study, we showed that enriched TAMs in tumor tissues from metastatic colorectal cancer patients and the distribution of TAMs is positively associated with the poor prognosis of patients. Those TAMs isolated from tumor tissues could significantly promote the capability of tumorigenesis and invasion of colorectal cancer cells in vitro and in vivo. Our results suggested that the TGF‐β produced by TAMs could activate the HIF1α/TRIB3/β‐catenin/Wnt signals to mediate the TAM‐associated tumor progression. Importantly, interrupting HIF1α signaling by inhibitor GN44028 weakened the tumor‐initiating capability and improved the anticancer effects of chemotherapy, which describe an innovative strategy in colorectal cancer treatment.
Tumor‐associated macrophages in the tumor microenvironment have been reported to participate in the progression of multiple tumors and serve as potential prognostic indicators in breast cancer treatment.19 In fact, the cross‐talk between tumor cells and stromal/immune cells has been recognized as a crucial determinant for cancer development.20 For instance, interleukin‐10 derived from TAMs has been proven to facilitate tumor cell invasion and drug resistance in several tumor types.21 The chemokine (C‐C motif) ligand 5 produced by TAMs could promote prostate stem cancer cells and tumor metastasis through activating β‐catenin/STAT3 signaling.19 By contrast, lactic acid produced by tumor cells, as a result of glycolysis, could be taken up by neighboring TAMs, resulting in M2‐like polarization of macrophages and immunosuppressive phenotypes in the tumor microenvironment.22 The phenomenon suggested a loop between the TAMs and tumor cells.23 Here, our study further reported that TAMs could secret TGF‐β to promote glycolysis of tumor cells, resulting in the activation of downstream HIF1α/TRIB3 signaling activation and tumor development in colorectal cancer. As described previously, TGF‐β is a cytokine typically produced by antiinflammatory macrophages to promote the EMT process of tumor cells.24 Our study described the role of TGF‐β in stem‐like phenotypes and distant metastasis of colorectal cancer. In addition, we correlated TGF‐β with HIF1α signaling and determined the underlying mechanism of TGF‐β‐associated tumor progression.
Glycolytic metabolites in vivo, such as lactate or alanine accumulation, facilitates the stabilization of HIF1α in tumor cells.25 Additionally, HIF1α served as an oncogenic transcription factor to efficiently promote tumorigenesis by enhancing angiogenesis and prosurvival signaling activation.26 More importantly, HIF1α has been reported to regulate the expression of immune checkpoint proteins, which are expressed in several tumor types and correlated with prognosis in patients.27 TRIB3 served as a stress sensor in response to diverse stress factors, including inflammation, hypoxia, and metabolism alteration in tumor cells.28 Compelling reports provided evidence that TRIB3 expression contributed to tumor progression in various tumor types.29, 30 Yu et al13 reported that TRIB3 activates the AKT1‐FOXO1‐SOX2 axis to promote breast cancer stem cells, resulting in poor prognosis in patients. Increasing evidence also implied that expression of TRIB3 correlated with the activation of Wnt prosurvival signal activation in glioma.31 Our study confirmed the role of HIF1α/TRIB3 in TAM‐induced colorectal cancer progression. The HIF1α signals could be upregulated through TGF‐β‐induced glycolytic metabolism in colorectal cancer cells, subsequently resulting in downstream TRIB3 and β‐catenin/Wnt signaling pathway activation. In fact, current studies have provided evidence that patients with colorectal cancer frequently displayed mutations in Wnt/β‐catenin‐associated molecules,32, 33 resulting in a poor prognosis in the clinic. In our study, we further confirmed the role of Wnt/β‐catenin signaling in promoting colorectal cancer, and described the underlying mechanism of aberrant Wnt/β‐catenin signaling activation in tumor cells. This further highlights the role of Wnt/β‐catenin signaling in clinical cancer therapy, and correlates with TAMs and Wnt/β‐catenin signaling, describing novel insight in tumor immunology. More importantly, blockade of HIF1α signals interrupted the activation of downstream prosurvival signaling, resulting in superior anticancer effects and prolonged survival time in our animal model, which described novel insights into colorectal cancer therapy.
In summary, our study indicated that enriched TAMs link TGF‐β to mediate colorectal progression, which is dependent on HIF1α/TRIB3/β‐catenin/Wnt signaling. This work provided proof‐of‐concept for targeting colorectal cancer by inhibiting HIF1α signals.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Fig S1‐3
ACKNOWLEDGMENT
We are grateful of all participants in Tianjin Medical University Cancer Institute and Hospital. This study was supported by the China Health Promotion Foundation, with the Supporting Program No. XM_2020_031_0295_01.
Liu C, Zhang W, Wang J, Si T, Xing W. Tumor‐associated macrophage‐derived transforming growth factor‐β promotes colorectal cancer progression through HIF1‐TRIB3 signaling. Cancer Sci. 2021;112:4198–4207. 10.1111/cas.15101
REFERENCES
- 1.Vitale I, Manic G, Coussens L, Kroemer G, Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30(1):36‐50. [DOI] [PubMed] [Google Scholar]
- 2.Arneth B. Tumor microenvironment. Medicina. 2020;56(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Xu Z, Duan H, et al. Tumor‐associated macrophage interleukin‐β promotes glycerol‐3‐phosphate dehydrogenase activation, glycolysis and tumorigenesis in glioma cells. Cancer Sci. 2020;111(6):1979‐1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kierans S, Taylor C. Regulation of glycolysis by the hypoxia‐inducible factor (HIF): implications for cellular physiology. J Physiol. 2021;599(1):23‐37. [DOI] [PubMed] [Google Scholar]
- 5.Feng J, Li J, Wu L, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge Y, Yoon S, Jang H, Jeong J, Lee Y. Decursin promotes HIF‐1α proteasomal degradation and immune responses in hypoxic tumour microenvironment. Phytomedicine. 2020;78: 153318. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro Franco P, Rodrigues A, de Menezes L, Pacheco MM. Tumor microenvironment components: allies of cancer progression. Pathol Res Pract. 2020;216(1):152729. [DOI] [PubMed] [Google Scholar]
- 8.Rhee I. Diverse macrophages polarization in tumor microenvironment. Arch Pharm Res. 2016;39(11):1588‐1596. [DOI] [PubMed] [Google Scholar]
- 9.Wu K, Lin K, Li X, et al. Redefining tumor‐associated macrophage subpopulations and functions in the tumor microenvironment. Front Immunol. 2020;11:1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang A, Qian Y, Ye Z, et al. Cancer‐associated fibroblasts promote M2 polarization of macrophages in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6(2):463‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua W, Ten Dijke P, Kostidis S, Giera M, Hornsveld M. TGFβ‐induced metabolic reprogramming during epithelial‐to‐mesenchymal transition in cancer. Cell Mol Life Sci. 2020;77(11):2103‐2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J‐M, Sun W, Wang Z‐H, et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat Commun. 2019;10(1):5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wennemers M, Bussink J, Grebenchtchikov N, Sweep F, Span P. TRIB3 protein denotes a good prognosis in breast cancer patients and is associated with hypoxia sensitivity. Radiother Oncol. 2011;101(1):198‐202. [DOI] [PubMed] [Google Scholar]
- 15.Hua F, Shang S, Yang Y‐W, et al. TRIB3 interacts with β‐catenin and TCF4 to increase stem cell features of colorectal cancer stem cells and tumorigenesis. Gastroenterology. 2019;156(3):708‐721.e715. [DOI] [PubMed] [Google Scholar]
- 16.Yahaya M, Lila M, Ismail S, Zainol M, Afizan N. Tumour‐associated macrophages (TAMs) in colon cancer and how to reeducate them. J Immunol Res. 2019;2019:2368249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi J, Gyamfi J, Jang H, Koo J. The role of tumor‐associated macrophage in breast cancer biology. Histol Histopathol. 2018;33(2):133‐145. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Wu T, Zheng B, Chen L. Individualized precision treatment: targeting TAM in HCC. Cancer Lett. 2019;458:86‐91. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Ji H, Niu X, et al. Tumor‐associated macrophages secrete CC‐chemokine ligand 2 and induce tamoxifen resistance by activating PI3K/Akt/mTOR in breast cancer. Cancer Sci. 2020;111(1):47‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothlin C, Carrera‐Silva E, Bosurgi L, Ghosh S. TAM receptor signaling in immune homeostasis. Annu Rev Immunol. 2015;33:355‐391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson M, Cheever A, White S, Thompson R, Wynn T. IL‐10 blocks the development of resistance to re‐infection with Schistosoma mansoni . PLoS Pathog. 2011;7(8):e1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan T, Chen S, Chen XI, et al. M2‐TAM subsets altered by lactic acid promote T‐cell apoptosis through the PD‐L1/PD‐1 pathway. Oncol Rep. 2020;44(5):1885‐1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y, Xu J, Lan H. Tumor‐associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol. 2019;12(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morikawa M, Derynck R, Miyazono K. TGF‐β and the TGF‐β family: context‐dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8(5):a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi L, Wang R, Huang G, et al. HIF1alpha‐dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367‐1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones R, Dorsett K, Hjelmeland A, Bellis S. The ST6Gal‐I sialyltransferase protects tumor cells against hypoxia by enhancing HIF‐1α signaling. J Biol Chem. 2018;293(15):5659‐5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniel S, Sullivan K, Labadie K, Pillarisetty V. Hypoxia as a barrier to immunotherapy in pancreatic adenocarcinoma. Clin Transl Med. 2019;8(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin R, Wu I, Hong J, et al. Capsaicin‐induced TRIB3 upregulation promotes apoptosis in cancer cells. Cancer Manag Res. 2018;10:4237‐4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J‐J, Zhou D‐D, Yang X‐X, et al. TRIB3‐EGFR interaction promotes lung cancer progression and defines a therapeutic target. Nat Commun. 2020;11(1):3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma BO, Zhang H, Wang YU, et al. Corosolic acid, a natural triterpenoid, induces ER stress‐dependent apoptosis in human castration resistant prostate cancer cells via activation of IRE‐1/JNK, PERK/CHOP and TRIB3. J Exp Clin Cancer Res. 2018;37(1):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, Li L, Chen L, Gao Y, Chen X, Cao Y. TRIB3 confers glioma cell stemness via interacting with β‐catenin. Environ Toxicol. 2020;35(6):697‐706. [DOI] [PubMed] [Google Scholar]
- 32.Sebio A, Kahn M, Lenz H. The potential of targeting Wnt/β‐catenin in colon cancer. Expert Opin Ther Targets. 2014;18(6):611‐615. [DOI] [PubMed] [Google Scholar]
- 33.Chen G, Gao C, Gao X, et al. Wnt/β‐Catenin pathway activation mediates adaptive resistance to BRAF inhibition in colorectal cancer. Mol Cancer Ther. 2018;17(4):806‐813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐3


