Abstract
A surveillance study on antibiotic resistance of enterococcal isolates (n = 730) was carried out in North Rhine-Westphalia, Germany, in 1997. Resistance rates to ampicillin (7.4%), high-level gentamicin (15.0%), high-level streptomycin (27.9%), ciprofloxacin (37.9%), vancomycin (1.5%), and teicoplanin (1.5%) were determined. All vancomycin-resistant enterococci (VRE) carried the vanA gene. SmaI and ApaI macrorestriction patterns indicated an intra- and interhospital spread of VRE.
Enterococci have emerged in recent years as pathogens in association with serious nosocomial infections in spite of their low level of virulence (6, 15, 16). Antibiotic resistance, especially vancomycin resistance, has become a major problem in enterococcal infections. Vancomycin-resistant Enterococcus faecium was first isolated in Germany from peritoneal fluid in 1987 (13). In 1997 about 12% of enterococcal isolates and up to 16% in intensive care units (ICUs) in U.S. hospitals were vancomycin-resistant enterococci (VRE) (8). Moreover, VRE have been isolated from animals and environmental sources (1, 10, 11, 12), which might increase the number of potential reservoirs for infections.
In the present investigation 22 microbiological laboratories were asked to collect up to 100 consecutive enterococcal isolates prospectively between April and July 1997. The participating laboratories represented about 75% of all medical microbiological institutions situated in North Rhine-Westphalia, the German federal state with the highest number of inhabitants (17.1 million). One isolate per patient was allowed. Only strains with clinical significance, such as isolates from normally sterile body sites (blood and cerebrospinal fluid), isolates from wounds or urine (bacteriuria, ≥105 CFU/ml), and lower respiratory tract isolates from sputum or bronchealveolar lavage fluid, were included in the study. Identification criteria for enterococci were that they be catalase-negative gram-positive cocci, bile-esculin and pyrrolidonylarylamidase positive, and that they grow in 6.5% NaCl broth and possess the Lancefield group D antigen. Enterococci were identified to the species level by using the API STREP ID32 rapid gallery (bioMérieux, Nürtingen, Germany). Strains with unacceptable identification profiles were further characterized with the API 20 STREP gallery (bioMérieux), by motility testing, and according to the pigment production ability as described by Facklam et al. (7) and Devriese et al. (4). Susceptibility was determined by the agar dilution method recommended by the National Committee for Clinical Laboratory Standards (17). The following antimicrobials were tested: ampicillin (Sigma, Düsseldorf, Germany), penicillin G (Grünenthal, Aachen, Germany), erythromycin (Hoechst Marion Roussel, Romainville, France), clindamycin (Hoechst Marion Roussel), ciprofloxacin (Bayer, Leverkusen, Germany), gentamicin (Synopharm, Barsbuettel, Germany), streptomycin (Grünenthal, Stolberg, Germany), teicoplanin (Hoechst Marion Roussel), quinupristin-dalfopristin (Rhône Poulenc Rorer, Cologne, Germany), and vancomycin (Eli Lilly, Bad Homburg, Germany). Cefinase disks (BBL, Cockeysville, Md.) were used for screening for the presence of β-lactamase. Enterococcus faecalis ATCC 29212 (American Type Culture Collection, Rockville, Md.) was used as a reference strain.
A total of 25 vancomycin-resistant isolates were chosen for further molecular characterization. These included 11 strains from the present study, 10 isolates from a study of stool specimens at Aachen University Hospital (17a), and 4 isolates responsible for serious infection outbreaks at hospitals in other regions of Germany. All glycopeptide-resistant enterococci were analyzed for the presence of the vanA and vanB resistance genes by PCR (14).
For the amplification of vanA, the modified primers originally published by Clark et al. (2) and Dutka-Malen et al. (5) with the sequences 5′ CAT GAA TAG AAT AAA AGT TGC AAT AC 3′ (positions 1 to 26) and 5′ CCC CTT TAA CGC TAA TAC GAT CAA 3′ (positions 1029 to 1006) were selected; for the detection of vanB, the primers described by Miele et al. (14) with the sequences 5′ CCC GAA TTT CAA ATG ATT GAA AA 3′ (positions 440 to 462) and 5′ CGG CAT CCT CCT GCA AAA (positions 896 to 879) were chosen. To confirm the expression of vanA, the VanA ligase which is inducible by glycopeptides was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the whole cell protein extract according to the method of Klare et al. (9).
Macrorestriction analysis (MRA) was performed by using the restriction endonuclease SmaI (Boehringer) for digestion of the genomic DNA as described previously (3, 10, 12). Selected VRE from the present study and representative VRE originating from outbreaks (UW 400, UW 805, UW 901, UW 931, and UW 1505) were additionally typed with the MRA method by cleavage of the DNA with the restriction endonuclease ApaI.
Macrorestriction patterns of bacterial isolates differing in no more than three DNA bands were classified as indistinguishable or closely related as recommended by Tenover et al. (18).
Plasmid isolation was performed as described by Woodford et al. (21) and modified by Werner et al. (20). Nondigested and EcoRI-digested plasmids were electrophoresed through 0.8% agarose gels. Subsequently the resolved EcoRI-digested plasmid DNA was blotted onto a positively charged nylon membrane (Boehringer, Mannheim, Germany) by electrotransfer (blot chamber TE-22; Hoefer Scientific Instruments, San Francisco, Calif.). A digoxigenin-labeled probe for the vanA gene of E. faecium BM4147 was done by PCR according to the method of Werner et al. (20).
A total of 730 strains were collected: 312 (42.8%) were isolated from the urogenital tract, 168 (23.0%) from wounds, 79 (10.8%) from the respiratory tract, and 68 (9.3%) from blood. Furthermore, 39 (5.3%) isolates were catheter or drainage specimens, 4 (0.5%) were isolated from the central nervous system, and 59 (8.1%) were isolated from various additional sources.
E. faecalis was the leading enterococcal species (n = 648 [88.8%]), followed by E. faecium (n = 72 [9.9%]), Enterococcus avium (n = 3), Enterococcus casseliflavus (n = 3), Enterococcus hirae (n = 3), and Enterococcus durans (n = 1). The susceptibility patterns of all strains to the most important antibiotics are presented in Table 1.
TABLE 1.
Resistance patterns of 730 enterococcal strains isolated in North Rhine-Westphaliaa
| Antibiotic | No. (%) of resistant strains
|
||
|---|---|---|---|
| E. faecalis(n = 648) | E. faecium(n = 72) | Totalb(n = 730) | |
| Vancomycin | 3 (0.5) | 8 (11.1) | 11 (1.5) |
| Teicoplanin | 3 (0.5) | 7 (9.7) | 11 (1.5) |
| Ampicillin | 6 (0.9) | 48 (66.7) | 54 (7.4) |
| Ciprofloxacin | 218 (33.6) | 59 (81.9) | 277 (37.9) |
| HLG | 96 (14.8) | 13 (18.1) | 109 (15.0) |
| HLS | 166 (25.6) | 38 (52.8) | 204 (27.9) |
Resistance is defined according to National Committee for Clinical Laboratory Standards MIC breakpoints: vancomycin, ≥32 mg/liter; teicoplanin, ≥32 mg/liter; ampicillin, ≥16 mg/liter; ciprofloxacin, ≥4 mg/liter; HLG (high-level gentamicin), >500 mg/liter; HLS (high-level streptomycin), >2,000 mg/liter.
Includes 10 strains of other enterococcal species.
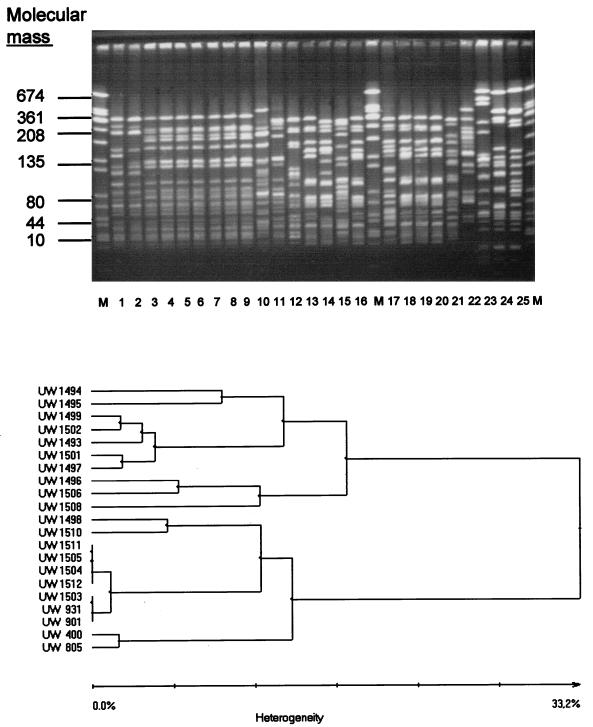
A total of 25 VRE were subjected to genotyping by SmaI MRA; special attention was given to 21 vanA-positive E. faecium isolates (Table 2). Vancomycin-resistant E. faecium strains isolated from nosocomial infections in a hospital in city L in North Rhine-Westphalia (strains UW 1503, UW 1504, UW 1505, UW 1511, and UW 1512), and those from nosocomial vancomycin-resistant E. faecium outbreaks in other German hospitals (hospital D1 in Hesse [isolate UW 901] and hospital G in Lower Saxony [strain UW 931]) showed identical patterns by SmaI MRA (Fig. 1) but differed from the strains from hospital N in Bavaria (strains UW 400 and UW 805). These data were also confirmed in MRA by ApaI digests (Fig. 2).
TABLE 2.
Characteristics of 25 vanA-positive high-level resistant enterococcia isolated from infected and colonized hospital patients and from fecal samples of outpatients
| Species | Strain (lane number in Fig. 1) | Origin of strain
|
Resistance patternb
|
SmaI MRA pattern | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Town (state) | Ward | Material | AMP | GEN | STR | CIP | Q-D | |||
| E. faecium | UW 400 (1) | N (Bavaria) | ICU | URTc | R | R | R | R | S | I |
| UW 805 (2) | N (Bavaria) | Nephrology | Blood | R | R | R | R | S | Ia | |
| UW 901 (3) | D1 (Hesse) | Dialysis unit | Wound | R | S | S | R | S | II | |
| UW 931 (4) | G (Lower Saxony) | ICU | BALc | R | S | S | R | S | II | |
| UW 1503 (5) | L (North Rhine-Westphalia) | n.d.c | Urine | R | S | S | R | S | II | |
| UW 1504 (6) | L (North Rhine-Westphalia) | Medicine | Bile | R | S | S | R | S | II | |
| UW 1505 (7) | L (North Rhine-Westphalia) | Surgery | Wound | R | S | S | R | S | II | |
| UW 1511 (8) | L (North Rhine-Westphalia) | Surgery | Wound | S | S | S | S | S | II | |
| UW 1512 (9) | L (North Rhine-Westphalia) | Surgery | Urine | R | S | S | R | S | II | |
| UW 1508 (10) | D2 (North Rhine-Westphalia) | Pediatrics | Urine | R | S | S | R | S | V | |
| UW 1510 (11) | A (North Rhine-Westphalia) | Medicine | Urine | R | S | R | R | S | III | |
| UW 1506 (12) | A (North Rhine-Westphalia) | Medicine | Wound | S | S | R | I | S | VI | |
| UW 1497 (13) | A (North Rhine-Westphalia) | Community | Stool | I | S | R | R | S | VIII | |
| UW 1495 (14) | A (North Rhine-Westphalia) | Community | Stool | I | S | R | I | S | IX | |
| UW 1498 (15) | A (North Rhine-Westphalia) | Community | Stool | I | S | R | R | S | IV | |
| UW 1501 (16) | A (North Rhine-Westphalia) | Community | Stool | I | S | S | I | S | VIII | |
| UW 1496 (17) | A (North Rhine-Westphalia) | Community | Stool | S | S | S | R | S | VII | |
| UW 1502 (18) | A (North Rhine-Westphalia) | Community | Stool | S | S | R | I | S | VIIIa | |
| UW 1499 (19) | A (North Rhine-Westphalia) | Community | Stool | S | S | S | I | S | VIIIb | |
| UW 1493 (20) | A (North Rhine-Westphalia) | Community | Stool | S | S | S | I | S | VIIIc | |
| UW 1494 (21) | A (North Rhine-Westphalia) | Community | Stool | S | S | S | R | S | X | |
| E. durans | UW 1492 (22) | A (North Rhine-Westphalia) | Community | Stool | S | S | S | S | S | n.d.c |
| E. faecalis | UW 1507 (23) | E (North Rhine-Westphalia) | Surgery | Blood | S | S | S | I | R | 1 |
| UW 1513 (24) | A (North Rhine-Westphalia) | Medicine | Wound | S | S | S | I | R | 2 | |
| UW 1509 (25) | D2 (North Rhine-Westphalia) | Medicine | Urine | S | S | S | R | R | 3 | |
According to the MICs, all enterococcal strains included were resistant to penicillin (≥16 mg/liter), erythromycin A (≥8 mg/liter), clindamycin (≥4 mg/liter), vancomycin (≥32 mg/liter), and teicoplanin (≥32 mg/liter). In all VRE the vanA gene and VanA protein were detectable.
Susceptibility (S), resistance (R), and intermediate (I) sensitivity were defined according to MIC breakpoints from the National Committee for Clinical Laboratory Standards (agar dilution method): ampicillin (AMP), ≤8 mg/liter (≥16 mg/liter); gentamicin (GEN), ≤500 mg/liter (>500 mg/liter); streptomycin (STR), ≤2,000 mg/liter (>2,000 mg/liter); ciprofloxacin (CIP), ≤1 mg/liter (≥4 mg/liter); and quinupristin-dalfopristin (Q-D), ≤1 mg/liter (≥4 mg/liter).
URT, upper respiratory tract; BAL, bronchoalveolar lavage fluid; n.d., no data.
FIG. 1.
Macrorestriction patterns (SmaI-digested genomic DNA) for 25 isolates of vanA-positive, high-level glycopeptide-resistant enterococci from clinical material from hospitalized patients and from fecal samples from outpatients (top panel). For the 21 vanA-positive E. faecium isolates in this group, the derived dendrogram (see the work of Claus et al. [3]) is presented (bottom panel). For the assignment of strains to lanes and for further characteristics of the strains, see Table 2. Lanes 1 to 21 are the results for the E. faecium isolates, lane 22 is the result for an E. durans strain, and lanes 23 to 25 are the results for E. faecalis strains. Lanes M, molecular size markers (strain S. aureus NCTC 8325).
FIG. 2.
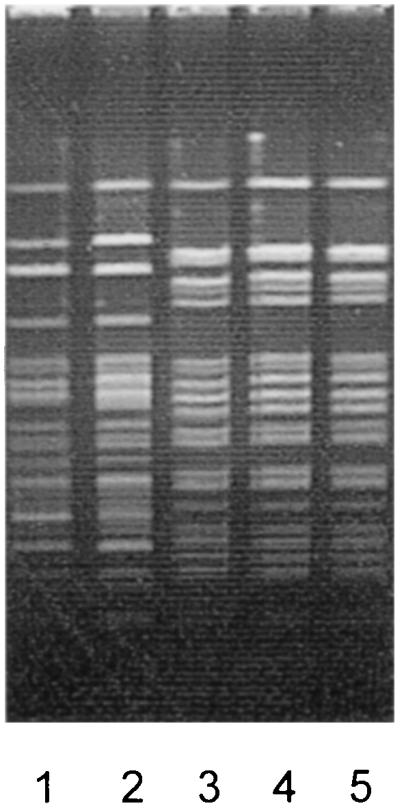
Macrorestriction patterns for ApaI-digested genomic DNA of vanA-positive E. faecium outbreak strains originating from different hospitals in Germany. Lane 1, UW 400, city N (Bavaria); lane 2, UW 805, city N (Bavaria); lane 3, UW 901, city D1 (Hesse); lane 4, UW 931, city G (Lower Saxony); lane 5, UW 1505, city L (North Rhine-Westphalia).
An interhospital spread of VRE was assumed. Therefore, these strains were further analyzed for their plasmid profiles and restriction endonuclease cleavage patterns of plasmids. These examinations showed that the two VRE isolates from hospitals in city N (UW 400 and UW 805) had only one large plasmid carrying vanA, in contrast to the other strains (UW 901, UW 931, and UW 1505) which had two plasmids each. The larger ones carried the vanA gene (hybridization results not shown). According to the EcoRI cleavage patterns the large plasmids of strains UW 400 and UW 805 were only distantly related. The EcoRI-cleaved plasmids of the VRE from a hospital in city G (strain UW 931) and a hospital in city L (strain UW 1505) were related but not identical. The appropriate plasmid pattern of the isolate from hospital D1 (strain UW 901) showed no homogeneity with any of the other patterns.
In summary, vancomycin resistance among enterococcal isolates seems not to be a major problem in Germany at the present time; this confirms data from a recently published multicenter study performed in southern Germany, which reported 12 VRE among 2,046 enterococcal isolates (19).
Data based on the analysis of SmaI and ApaI macrorestriction patterns suggest an interhospital spread of VRE. Additional analysis of plasmid profiles and restriction endonuclease cleavage patterns of plasmids indicates, however, that VRE exhibiting the same macrorestriction pattern are not necessarily totally identical. Nevertheless, the possibility cannot be excluded that there may be particular clones of E. faecium adapted to the hospital environment, which are supraregionally disseminated and have subsequently acquired different plasmids carrying the vanA gene cluster. The prevalence of VRE in clinical and animal sources should be carefully monitored in the future. A rational approach to the use of antibiotics is urgently warranted, in order to preserve the favorable situation regarding the low rate of enterococcal resistance to glycopeptides in Germany.
Acknowledgments
We thank M. Lemperle, M. Breuer-Wherle, I. Seyfarth, D. Badstübner, C. Konstabel, and I. Moonen for excellent technical assistance. We thank the following colleagues for providing the enterococcal isolates and clinical information: K.-D. Berg and R. Berndt (Eschweiler); P. Felgner and A. Kuhlencord (Paderborn); U. Hadding and G. Zysk (Düsseldorf); H.-J. Hagedorn (Herford); A. Höher and V. Knop-Hammad (Wuppertal); H. Kehren and B. Beckers (Mönchengladbach); G. Horpacsy and R. N. Schöngen (Leverkusen); R. Merten (Cologne); B. Neuhaus and G. Ahlemeyer (Münster); G. Peters and R. Gross (Münster); F. Pranada (Dortmund); G. Pulverer and H. Seifert (Cologne); P. Stolle and P. Nemes (Düsseldorf); U. Jäger (Münster); D. Winterhoff and W. Treder (Münster); C. H. Wirsing von König (Krefeld); R. Ansorg and E. N. Schmid (Essen); I. Scharmann (Bochum); W. Opferkuch and D. Kleiber-Imbeck (Bochum); and P. Courvalin (Paris).
REFERENCES
- 1.Bates J, Jordens J Z, Griffiths D T. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–514. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 2.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claus H, Cuny C, Pasemann B, Witte W. A database system for fragment patterns of genomic DNA of Staphylococcus aureus. Zentralbl Bakteriol. 1996;287:105–116. doi: 10.1016/s0934-8840(98)80154-0. [DOI] [PubMed] [Google Scholar]
- 4.Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 5.Dutka-Malen S, Molinas C, Arthur M, Courvalin P. The VANA glycopeptide resistance protein is related to d-alanyl-d-alanine ligase cell wall biosynthesis enzymes. Mol Gen Genet. 1990;224:364–372. doi: 10.1007/BF00262430. [DOI] [PubMed] [Google Scholar]
- 6.Eliopoulos G M. Increasing problems in the therapy of enterococcal infections. Eur J Clin Microbiol Infect Dis. 1993;12:409–412. doi: 10.1007/BF01967433. [DOI] [PubMed] [Google Scholar]
- 7.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarvis W R. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Prevention of the spread of vancomycin-resistant Enterococcus: what should we be telling hospitals to do?, abstr. S-146; p. 399. [Google Scholar]
- 9.Klare I, Collatz E, Al-Obeid S, Wagner J, Rodloff A C, Witte W. Glycopeptidresistenz bei Enterococcus faecium aus Besiedlungen und Infektionen von Patienten aus Intensivstationen Berliner Kliniken und einem Transplantationszentrum. Z Antimicrob Antineoplast Chemother. 1992;10:45–53. [Google Scholar]
- 10.Klare I, Heier H, Claus H, Böhme G, Marin S, Seltmann G, Hakenbeck R, Atanassova V, Witte W. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist. 1995;1:265–272. doi: 10.1089/mdr.1995.1.265. [DOI] [PubMed] [Google Scholar]
- 11.Klare I, Heier H, Claus H, Reissbrodt R, Witte W. VanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol Lett. 1995;125:165–171. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 12.Klare I, Heier H, Claus H, Witte W. Environmental strains of Enterococcus faecium with inducible high-level resistance to glycopeptides. FEMS Microbiol Lett. 1993;106:23–30. doi: 10.1111/j.1574-6968.1993.tb05930.x. [DOI] [PubMed] [Google Scholar]
- 13.Lütticken R, Kunstmann G. Vancomycin-resistant Streptococcaceae. Zentralbl Bakteriol Hyg A. 1988;267:379–382. doi: 10.1016/s0176-6724(88)80054-3. [DOI] [PubMed] [Google Scholar]
- 14.Miele A, Bandera M, Goldstein B P. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in vanA isolates. Antimicrob Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moellering R C., Jr Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992;14:1173–1176. doi: 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 16.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. 4th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 17a.Reinert, R. R., et al. Unpublished data.
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallrauch C, Elsner E, Milatovic D, Cremer J, Braveny I. Antibiotikaresistenz der Enterokokken in Deutschland. Med Klin. 1997;8:464–468. doi: 10.1007/BF03044913. [DOI] [PubMed] [Google Scholar]
- 20.Werner G, Klare I, Witte W. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol Lett. 1997;155:55–61. doi: 10.1111/j.1574-6968.1997.tb12685.x. [DOI] [PubMed] [Google Scholar]
- 21.Woodford N, Morrison D, Cookson B, George R C. Comparison of high-level gentamicin-resistant Enterococcus faecium isolates from different continents. Antimicrob Agents Chemother. 1993;37:681–684. doi: 10.1128/aac.37.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]