Summary
Linalool is one of the common flavour‐related volatiles across the plant kingdom and plays an essential role in determining consumer liking of plant foods. Although great process has been made in identifying terpene synthase (TPS) genes associated with linalool synthesis, much less is known about regulation of this pathway. We initiated study by identifying PpTPS3 encoding protein catalysing enantiomer (S)‐(+)‐linalool synthesis, which is a major linalool component (˜70%) observed in ripe peach fruit. Overexpression of PpTPS3 led to linalool accumulation, while virus‐induced gene silencing of PpTPS3 led to a 66.5% reduction in linalool content in peach fruit. We next identified transcription factor (TF) PpbHLH1 directly binds to E‐box (CACATG) in the PpTPS3 promoter and activates its expression based on yeast one‐hybrid assay and EMSA analysis. Significantly positive correlation was also observed between PpbHLH1 expression and linalool production across peach cultivars. Peach fruit accumulated more linalool after overexpressing PpbHLH1 in peach fruit and reduced approximately 54.4% linalool production after silencing this TF. DNA methylation analysis showed increased PpTPS3 expression was associated with decreased 5 mC level in its promoter during peach fruit ripening, but no reverse pattern was observed for PpbHLH1. Arabidopsis and tomato fruits transgenic for peach PpbHLH1 synthesize and accumulate higher levels of linalool compared with wild‐type controls. Taken together, these results would greatly facilitate efforts to enhance linalool production and thus improve flavour of fruits.
Keywords: fruit aroma, linalool, transcription factor, epigenetics
Introduction
It has been well established that volatiles, together with sugars and acids, make large contributions to fruit flavour. Lacking a characteristic or distinctive flavour is a major source of consumer dissatisfaction. Compared to content of sugars and acids at mg/g level, volatiles are presented at ng/g in fruits. Therefore, small changes in volatile content have potential to affect fruit flavour quality and there is a renewed interest to regulate fruit flavour‐related volatiles (Klee and Tieman, 2018).
As a popular economic crop in the world, peach (Prunus persica L. Batsch) is a model plant of Rosaceae family (Zhu et al., 2012). More than 100 volatile chemicals have been identified in peach fruit, in which linalool is a key odorant that affects fruit aroma and consumer preference (Eduardo et al., 2010; Tian et al., 2017). Linalool, a linear monoterpene alcohol, is widely detected across 200 plant species. Due to the stereo‐centre with hydroxyl at C‐3, linalool has two enantiomers found in nature. R‐(−)‐linalool has a woody or lavender‐like aroma, while S‐(+)‐linalool possesses sweet and floral odour (Bonnländer et al., 2006). S‐(+)‐linalool is present in seed of coriander (Coriandrum sativum L.), flowers of Clarkia breweri and sweet orange (Citrus sinensis Osbeck), fruit of strawberry (Fragaria × ananassa Duch) and peach. R‐(−)‐linalool is a major constituent of sweet basil (Ocimum basilicum L.), lavender (Lavandula officinalis L.) and bergamot orange (Citrus aurantium L.) (Ravid et al., 1985, 1997). Different neural responses of people to each enantiomeric form of linalool were observed, and therefore, each enantiomer was classified as possessing a distinct odour (Raguso, 2016).
The biosynthesis pathway of linalool has been well characterized in plants. Terpene synthases (TPSs) are the vital terminal enzymes catalysing the formation of monoterpene linalool using geranyl diphosphate (GPP) as substrate (Vranová et al., 2012). TPSs associated with linalool synthesis have been isolated from multiple plant species, including Arabidopsis (Chen et al., 2003), tobacco (He et al., 2019), cotton (Huang et al., 2017), rose (Magnard et al., 2018), snapdragon flowers (Nagegowda et al., 2008), apple (Nieuwenhuizen et al., 2013), grape (Martin et al., 2010), strawberry (Aharoni et al., 2004) and tomato (Falara et al., 2011). However, transcriptional regulation of linalool metabolism remains largely unclear. In flowers of monocotyledonous plant Freesia hybrida, transcription factor (TF) FhMYB21L2 activated transcription of linalool synthesis gene FhTPS1 through binding to the MYBCORE sites in the promoter (Yang et al., 2020). In contrast, TF bHLH‐FhMYC2 represses the transcriptional activation function of FhMYB21L2 (Yang et al., 2020). Extra experiments are required to further elucidate the function of these TFs in regulating linalool synthesis, including overexpressing or silencing engineering evidence. Besides, candidate TFs related to SlTPS5 transcription were screened from tomato based on correlation analysis using a transcriptome database of stem trichome (Spyropoulou et al., 2014). Tomato SlTPS5 was related to linalool formation (Falara et al., 2011). Although many genes related to linalool synthesis have been characterized, no TFs have been identified to regulate linalool synthesis in fruits. Therefore, progress in identification of upstream TFs controlling volatile production lags far behind compared with fruit colour‐related chemicals such as anthocyanin.
Changes in DNA methylation are essential for fruit development and ripening (reviewed by Tang et al., 2020), and for regulating synthesis of flavour volatiles (Zhang et al., 2020; Zhang et al., 2016). For tomato fruit in response to postharvest cold storage, volatile loss is associated with reduced expression of synthesis‐related genes and increased DNA methylation levels in their promoters (Zhang et al., 2016). Tomatoes are harvested at mature or early ripe stage with high firmness suitable for transportation followed by exogenous ethylene to accelerate ripening. This postharvest handling caused global changes in DNA methylation status and expression of genes related to synthesis of flavour volatiles such as aldehydes, alcohols and esters (Zhang et al., 2020). As an important chemical related to fruit flavour, epigenetic regulation of linalool during fruit ripening remains unclear.
In the present study, RNA‐sequencing results revealed that PpTPS3 had the highest transcript abundance and correlated positively with linalool accumulation in peach fruit. Function of PpTPS3 in regulating linalool synthesis was elucidated based on both in vitro evidence and in vivo evidence. We next identified that transcription factor PpbHLH1 activated PpTPS3 expression via directly binding to the promoter. Correlation between gene expression and linalool content across natural cultivars was used to validate function of PpTPS3 and PpbHLH1 in linalool formation. In peach fruit, transient overexpressing and silencing strategies were applied to show their regulatory role in linalool synthesis. Moreover, PpbHLH1 was stably overexpressed in model plant Arabidopsis and tomato fruits to further clarify their function in linalool production. Besides, whole‐genome bisulphite sequencing was performed to analyse changes in cytosine methylation of linalool synthesis gene PpTPS3 and PpbHLH1 during peach fruit ripening. Taken together, we show a model of linalool regulation in peach fruit.
Results
Expression of PpTPS3 correlates with linalool content during peach fruit ripening
Peach fruits at five developmental stages were sampled (Figure 1a) for volatile terpene production analysis using GC‐MS. A total of 20 volatile terpenoids were identified in ripening peach fruit (Table S1), in which linalool was the predominant chemical (Figure 1b). Linalool production accumulated as peach fruit ripen and peaked at S5 stage, reaching up to 580.11 ng/g fresh weight and accounting for 75% of all terpenes detected in ripe peach fruit. Chiral analysis showed that approximately 70% of total linalool in ripe peach fruit was presented as S‐(+)‐linalool (Figure 2a). Previous study revealed that linalool is a major contributor to peach fruit aroma and flavour quality (Tian et al., 2017); therefore, we plan to identify genes associated with linalool synthesis and then to further investigate their regulatory mechanism.
Figure 1.
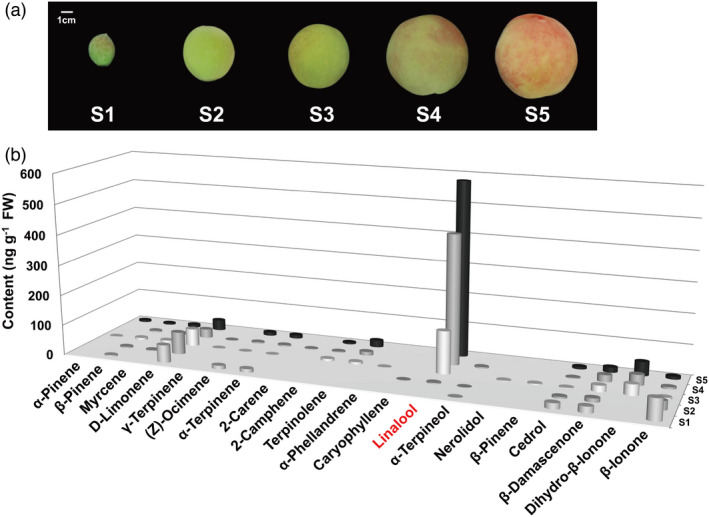
Changes in volatiles during peach fruit development and ripening. (a) Photo of peach fruit cv Hujingmilu at different stages: S1 (34 days after bloom, DAB), S2 (71 DAB), S3 (94 DAB), S4 (108 DAB) and S5 (111 DAB). Bars = 1 cm. (b) Volatile content of peach fruit. Averaged data from three biological replicates are shown. Standard errors are presented in Table S1.
Figure 2.
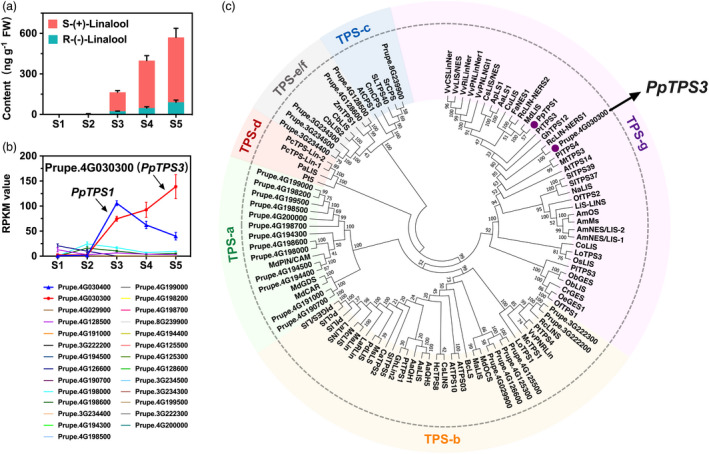
Characterization of PpTPS3 related to linalool production. (a) Content of linalool during peach fruit development and ripening. The linalool enantiomers were distinguished using chiral GC–MS analysis. (b) Expression pattern of peach TPS genes during peach development and ripening. (c) Phylogenetic analysis of plant TPSs based on deduced amino acid sequence. Accession numbers for these TPSs are given in Table S2.
To screen peach TPS genes associated with linalool synthesis, a total of 38 TPS genes displaying the HMM structure of the N‐terminal conserved domain (Pfam Accession Number PF01397) and the C‐terminal conserved domain (Pfam Accession Number PF03936) were identified (Figure S1; Kumar et al., 2018; Martin et al., 2010). Transcript levels of these PpTPS genes at five developmental stages were investigated using RNA‐Seq, and 27 members expressed in peach fruit were observed (Figure 2b). Among these PpTPS genes, PpTPS3 (Prupe.4G030300) and PpTPS1 (Prupe.4G030400) had the remarkable high transcript levels during peach fruit ripening. Our previous study reported that PpTPS1 was associated with biosynthesis of linalool, whose contents were reduced by UV‐B irradiation of peach fruit after harvest (Liu et al., 2017). In the present study, Pearson’s correlation analysis revealed a positive and significant correlation between transcript levels of PpTPS3 and linalool contents (R 2 = 0.902, P < 0.05), indicating a potential role of this gene in linalool synthesis during peach fruit ripening. Moreover, phylogenetic tree showed that PpTPS3 belongs to the TPS‐g subfamily like other linalool synthases (Figure 2c; Table S2). As shown in Figure 2a, (S)‐(+)‐linalool is a major contribution to linalool in ripe peach fruit. These results prompted us to investigate whether PpTPS3 is another candidate gene associated with linalool synthesis, particularly for (S)‐(+)‐linalool formation during peach fruit ripening.
Effect of overexpressing or silencing PpTPS3 on linalool production
Full‐length ORF was cloned and overexpressed in Escherichia coli to obtain recombinant PpTPS3 protein (Figure S2). Enzymatic activity analysis showed that PpTPS3 converted GPP to linalool (98.3% of total products) (Figure 3a). Chiral GC‐MS analysis revealed that S‐(+)‐linalool was major products produced by PpTPS3, accounting for approximately 95% of total linalool (Figure 3a).
Figure 3.
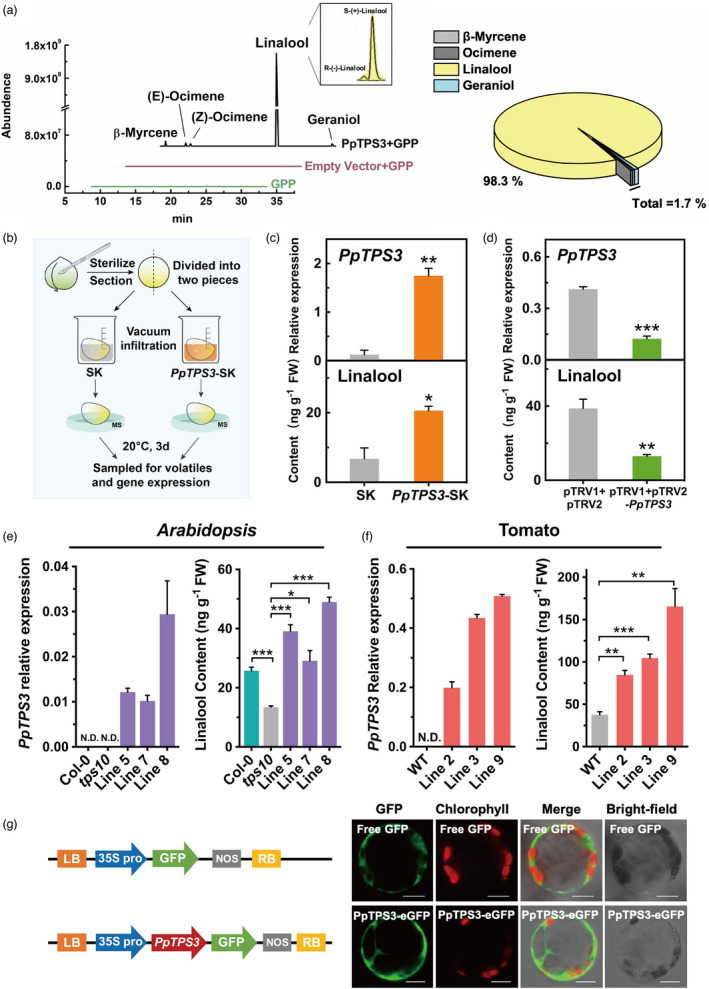
PpTPS3 catalyses linalool synthesis both in vitro and in vivo. (a) Enzymatic activity assay of PpTPS3 protein towards geranyl pyrophosphate (GPP) as substrate in vitro. (b) Schematic diagram for gene transient expression in peach fruits. (c) Transient overexpression of PpTPS3 increases linalool content in peach fruits. Empty SK vector was used as a control. (d) Silencing PpTPS3 by VIGS decreases linalool content in peach fruits. Empty pTRV1 + pTRV2 vector was used as a control. Phytoene desaturase (PDS) gene was used as a reporter gene for VIGS (Figure S3). (e) Overexpressing PpTPS3 induces linalool synthesis in Arabidopsis. tps10 mutant was used as a control. (f) Increased linalool content in tomato fruits after overexpressing PpTPS3. Wild‐type (WT) fruits were used as controls. (g) Subcellular localization of PpTPS3. Bars = 20 µm. Data are presented as mean ± standard error from three independent biological replicates. Significant differences are indicated with asterisks above the bars (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). N.D., not detected.
To our best knowledge, no transgenic peach fruits have been reported. Therefore, a homologous transient overexpression (Liu et al., 2017; Wu et al., 2019) and virus‐induced gene silencing (VIGS) system (Zhao et al., 2020) were used to validate the function of PpTPS3. For transient overexpression experiments, peach fruit was infiltrated with Agrobacterium harbouring PpTPS3‐pGreen‐SK construct or pGreen‐SK vector as control, and then, gene expression and volatile were detected after 3 d of infiltration (Figure 3b). Compared to the control, content of linalool was increased by 3.06‐fold after overexpressing PpTPS3 (Figure 3c). To confirm efficiency of VIGS system, phytoene desaturase (PpPDS) was used as a positive marker (Figure S3). As expected, peach fruits infiltrated with pTRV1+pTRV2‐PpTPS3 resulted in a significant reduction (˜66.5%) in linalool content relative to controls (infiltrated with pTRV1+pTRV2) (Figure 3d). No significant reduction was observed for other volatiles produced by PpTPS3 protein in vitro. These results indicated that PpTPS3 is involved in linalool synthesis in peach fruit.
To further test the role of PpTPS3 in linalool formation, heterologous stable transgenic experiments were performed in Arabidopsis and tomato plants. Homozygous Arabidopsis tps10 (SALK_041114C) mutant (Ginglinger et al., 2013) was used for overexpressing peach PpTPS3. Compared to tps10 mutants, linalool contents were increased by 2–3 times in three independent transgenic plants (Figure 3e). Linalool accumulation in 35S::PpTPS3/tps10 transgenic lines complemented linalool defect in the tps10 mutant, even exceeded the level of wild‐type Col‐0 (Figure 3e). No significant changes in transcript levels were observed for AtTPS10 and AtTPS14, which were associated with linalool formation, in transgenic Arabidopsis plants (Figure S4). These results revealed that the overexpression of PpTPS3 could restore synthesis of linalool in Arabidopsis tps10 mutant.
For transgenic tomatoes overexpressing PpTPS3, fruit at red ripe stage produced up to approximately 4.4‐fold higher contents of linalool than wild‐type controls (Figure 3f). Moreover, overexpressing PpTPS3 also induced accumulation of linalool in transgenic tomato leaves (Figure S5a, b). Subcellular localization of PpTPS3 was performed in Nicotiana benthamiana protoplasts. The result indicated that PpTPS3 was located in the cytoplasm (Figure 3g). Taken together, results mentioned above indicated that PpTPS3 is involved in biosynthesis of linalool in peach fruit based on both in vitro and in planta experiments.
Transcription factor PpbHLH1 activates PpTPS3 expression via direct binding to its promoter
Having identified the gene PpTPS3 responsible for linalool synthesis, we next investigated the regulation of its synthesis in peach fruit. Formation of volatile terpenes could be regulated by transcription factors (TFs) based on previous studies (Table S3). Therefore, homologues of these transcription factors were cloned from peach (Table S3). Besides, TFs whose expression positively correlated with PpTPS3 in fruit (R > 0.5) during peach fruit ripening were also cloned for promoter activation analysis (Table S4). A total of 58 TFs, including bHLH, bZIP, ERF, MADS, MYB, NAC and WRKY families, were selected for dual‐luciferase assay. Among these TFs, PpbHLH1 (Prupe.8G157500) had the strongest induction of PpTPS3, exhibiting approximately fourfold induction (Figure 4a). Although induction effect was also observed for PpbHLH2 (Prupe.6G211900) and PpbHLH3 (Prupe.6G212000), the highest transcript levels were detected for PpbHLH1 during peach fruit ripening (Figure S6a). Notably, PpbHLH1 showed no transcriptional activation for PpTPS1, which was another gene associated with linalool formation in peach fruit (Liu et al., 2017; Figure S6b). Therefore, PpbHLH1 was selected as a candidate to investigate whether this TF could regulate synthesis of linalool in peach fruit.
Figure 4.
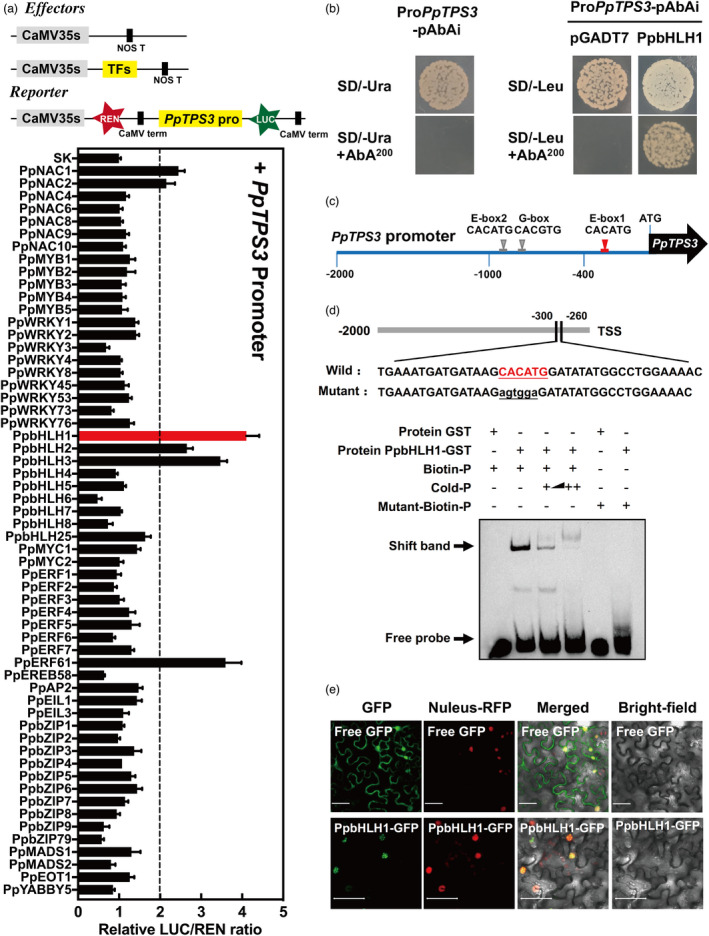
PpbHLH1 activates PpTPS3 and binds to its promoter. (a) Regulatory effects of transcription factors on the promoter of PpTPS3. Means and standard errors were calculated from six replicates. (b) Yeast one‐hybrid analysis of PpbHLH1 binding to the PpTPS3 promoter. Autoactivation was tested on SD‐Ura in the presence of AbA. AD‐empty and pAbAi‐PpTPS3 were used as negative controls. (c) G‐box (CACGTG) and E‐boxes (CACATG) of bHLH protein‐binding sites in the PpTPS3 promoter. (d) EMSA of 3’ boitin‐labelled dsDNA probes with the PpbHLH1‐binding protein. Presence or absence of specific probes is marked by symbol + or ‐. The mutated nucleotides in probe are indicated in red lowercase letters. (e) Subcellular localization of PpbHLH1 in Nicotiana benthamiana leaves. GFP, GFP channel; nucleus–RFP, transgenic tobacco plants with red fluorescence in the nucleus; merge, merged image of the GFP and nucleus–RFP channels; bright‐field, light microscopy image; bars = 50 µm.
A Y1H assay showed that PpbHLH1 could directly bind to the PpTPS3 promoter (Figure 4b). Sequence analysis of the PpTPS3 promoter (2000 bp) revealed that there are three bHLH binding sites, that is two E‐boxes (CACATG) and one G‐box (CACGTG) (Figure 4c). So, we designed three probes with 3’ biotin labelling based on these binding sites (Figure S7a), and the binding specificity of PpbHLH1 to the PpTPS3 promoter was confirmed by EMSA. Recombinant GST‐PpbHLH1 protein is shown in Figure S7b. EMSA result showed that PpbHLH1 only binds to the E‐box binding site closest to the PpTPS3 transcription start site (TSS) (Figure S7c). When the predicted binding sites were mutated, binding was eliminated (Figure 4d). With the increase in the cold probe as competitor, binding decreased. As expected, PpbHLH1 was localized in the nucleus (Figure 4e). Taken together, PpbHLH1 activated the expression of PpTPS3 through direct binding to the E‐box element in the PpTPS3 promoter.
Expression of PpbHLH1 correlates with linalool contents across peach cultivars
To further investigate the regulatory effects of PpbHLH1 on linalool, natural variation of gene expression and volatile contents were determined in peach cultivars (Figure 5a). The correlation between transcript level and linalool content was analysed using linear regression method. The results showed that PpTPS3 expression strongly positively correlated with linalool content (R 2 = 0.92, P < 0.001) (Figure 5b) and S‐(+)‐linalool content (R 2 = 0.93, P < 0.001; Figure S8a). PpbHLH1 expression significantly positively correlated with PpTPS3 expression (R 2 = 0.78, P < 0.01), linalool content (R 2 = 0.60, P < 0.05; Figure 5b) and S‐(+)‐linalool content (R 2 = 0.54, P < 0.05; Figure S8b) among the different cultivars. Taken together, these results indicate that the expression of PpbHLH1 is positively correlated with linalool accumulation, making PpbHLH1 a good candidate TF for regulating linalool biosynthesis in peach fruit.
Figure 5.
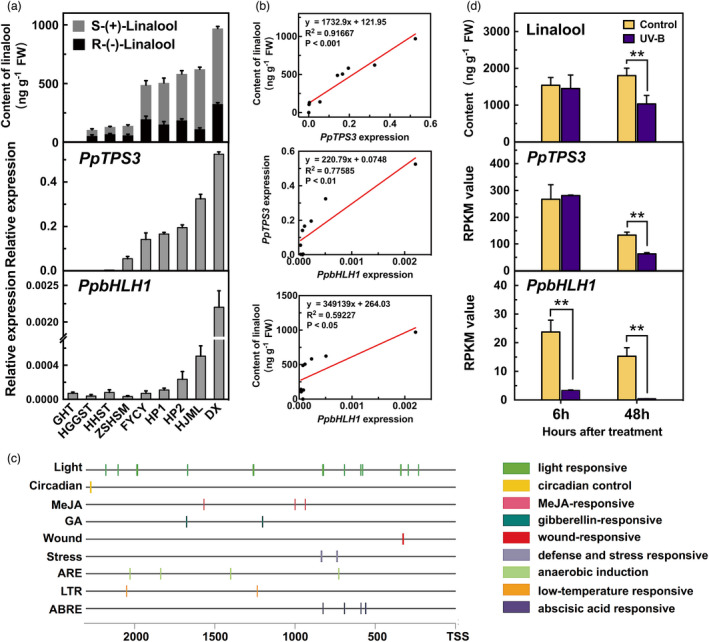
Correlation between transcript levels of PpbHLH1, PpTPS3 and contents of linalool in peach fruit. (a) Content of linalool and expression levels of PpTPS3 and PpbHLH1 across peach cultivars. (b) Correlation analysis between content of linalool and expression levels of PpTPS3 and PpbHLH1 across peach cultivars. (c) Analysis of cis‐acting elements of PpbHLH1 2kb promoter sequence based on the PlantCARE database. (d) Effect of UV‐B light treatment on content of linalool and expression levels of PpTPS3 and PpbHLH1 in peach fruit. Relative expression levels were determined using RT‐qPCR. Data are presented as mean ± standard error from three independent biological replicates. Significant differences are indicated with asterisks above the bars (**, P < 0.01).
Cis‐acting element analysis of PpbHLH1 promoter and its expression in response to UV‐B light
Synthesis of fruit volatiles is regulated by environmental factors (Liu et al., 2017; Wu et al., 2019; Zhang et al., 2016). Therefore, 2000 bp of the PpbHLH1 promoter sequence was submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify cis‐acting elements related to environmental stress (Higo et al., 1999). Notably, the greatest numbers of response elements were related to ‘light responsiveness’ in PpbHLH1 promoter (Figure 5c), and the specific motif sequences are shown in Table S5. Our previous study revealed a regulatory role of UV‐B light in linalool formation (Liu et al., 2017). Here, we found that transcript level of PpbHLH1 was significantly reduced after 6 h of UV‐B irradiation, followed by a decrease in PpTPS3 expression and reduced content of linalool compared with the control (Figure 5d). Content of linalool was reduced by 60% in peach fruit after 48 h of UV‐B irradiation. Our results supported a model that UV‐B inhibited PpbHLH1 expression, which in turn reduced PpTPS3 gene transcription and linalool synthesis in peach fruit.
Effect of overexpressing or silencing PpbHLH1 on linalool production in peach fruit
To further determine the role of PpbHLH1 on linalool synthesis, the TF was overexpressed or silenced in peach fruits using our published methods (Liu et al., 2017; Zhao et al., 2020). PpbHLH1‐pGreen‐SK vector was constructed for transient overexpression of PpbHLH1 in peach fruit. Compared with the empty vector, overexpression of PpbHLH1 promoted PpTPS3 expression, accompanying by significant accumulation of linalool (Figure 6a). Chiral analysis showed that the increased linalool is mainly contributed by S‐(+)‐linalool (Figure 6b). VIGS significantly reduced PpbHLH1 expression, accompanying by 53% reduction in PpTPS3 transcript level and 54% decline of linalool production, respectively (Figure 6c). Taken together, these results indicate that PpbHLH1 is involved in linalool production in peach fruit.
Figure 6.
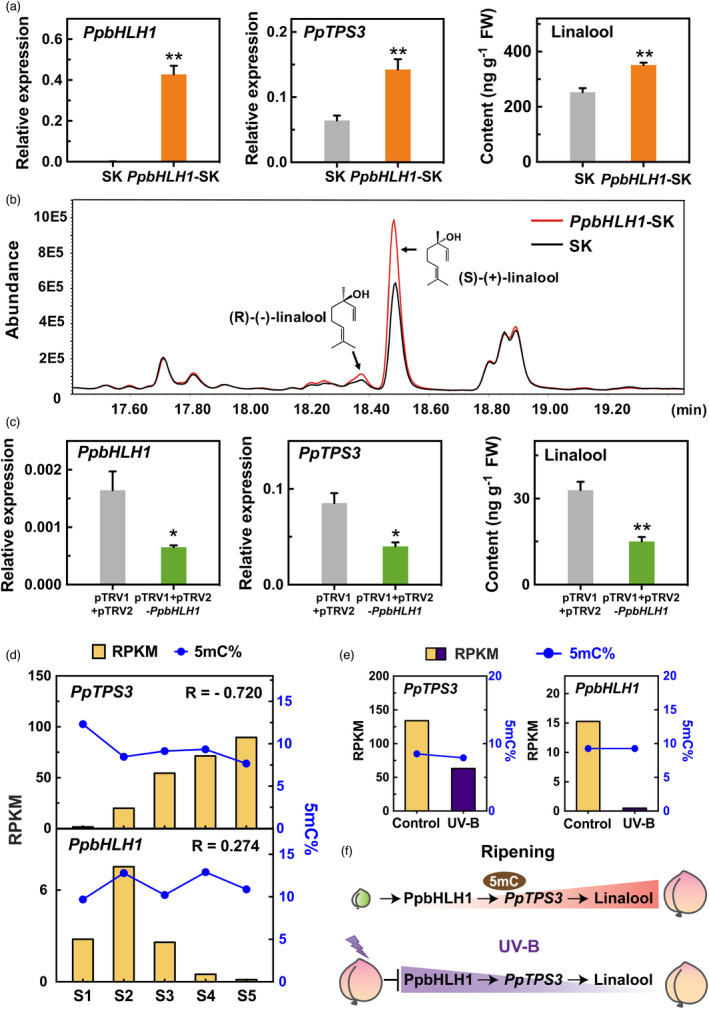
Regulatory effects of PpbHLH1 on linalool synthesis in peach fruit. (a) Overexpressing PpbHLH1 induces expression of PpTPS3 and linalool accumulation in peach fruits. Empty SK vector was used as a control. Significant differences are indicated with asterisks above the bars (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). (b) Chiral GC‐MS analysis of linalool enantiomers in transiently overexpressed peach fruits. (c) Silencing PpbHLH1 by VIGS decreases PpTPS3 expression and linalool content in peach fruit. Empty pTRV1 + pTRV2 vector was used as a control. (d) Changes in transcript levels and DNA methylation levels of PpTPS3 and PpbHLH1 during peach fruit ripening. (e) Effects of UV‐B irradiation on transcript levels and DNA methylation levels of PpTPS3 and PpbHLH1 in peach fruit. (f) A proposed regulatory model of linalool synthesis in peach fruit.
Changes in DNA methylation levels of linalool synthesis‐related genes during peach fruit ripening and in response to UV‐B irradiation
Epigenetic regulation is associated with fruit development and ripening (Tang et al., 2020), and stress responses (Zhang et al., 2016). To further investigate whether transcript levels of PpTPS3 and PpbHLH1 are associated with changes in DNA methylation, whole‐genome bisulphite sequencing of peach fruit at different development stages and under UV‐B treatment was performed. The average clean reads were 45.45 million, and the bisulphite conversion rates were >99.48% (Table S6). Mapping the clean reads showed that, on average 29.88 million reads (65.57%), covering >91.70% of the genome, were mapped to the peach reference genome (Tables S6 and S7). Sequencing depth was >25‐fold coverage per DNA strand (Table S7). The coverage and depth of sequencing in our study are comparable with those of published fruit methylomes (Cheng et al., 2018; Huang et al., 2019; Xu et al., 2018; Zhu et al., 2020). A decrease in DNA methylation levels was observed for PpTPS3 promoters throughout peach fruit development and ripening (Figure 6d). This declined DNA methylation was opposite of increased transcript levels of PpTPS3. Regarding PpbHLH1, no similar inverse pattern was observed between transcripts and cytosine methylation levels during peach fruit development and ripening (Figure 6d). For peach fruit irradiated with UV‐B light, although transcript levels of PpTPS3 and PpbHLH1 were significantly reduced, no remarkable increase in DNA methylation levels was observed (Figure 6e). Taken together, these results indicated that decreased DNA methylation is associated with increased PpTPS3 expression, which regulates linalool synthesis during peach fruit ripening (Figure 6f).
Engineering of PpbHLH1 induced linalool synthesis in tobacco, Arabidopsis and tomato fruit
To further confirm the function of PpbHLH1, it was overexpressed in other plant species. For tobacco, leaves were infiltrated with Agrobacterium harbouring PpbHLH1‐pGreen‐SK construct, and pGreen‐SK vector was used as control. Up‐regulating PpbHLH1 expression increased transcript of tobacco linalool synthase (NtLIS, XP_016510950) by sevenfold, accompanying by significant accumulation of linalool (Figure 7a). Compared to wild‐type Arabidopsis plants, stable overexpressing PpbHLH1 significantly enhanced transcript levels of AtTPS10 and induced contents of linalool in three independent T2 transgenic lines (Figure 7b). Besides, changes in linalool contents of T1 transgenic tomatoes were also analysed. For all three lines of transgenic tomato fruits, overexpressing PpbHLH1 increased SlTPS5 expression up to eightfold and produced up to fivefold higher contents of linalool compared with the wild‐type fruits, respectively (Figure 7c). These results supported that overexpressing peach PpbHLH1 enhanced linalool production in planta.
Figure 7.
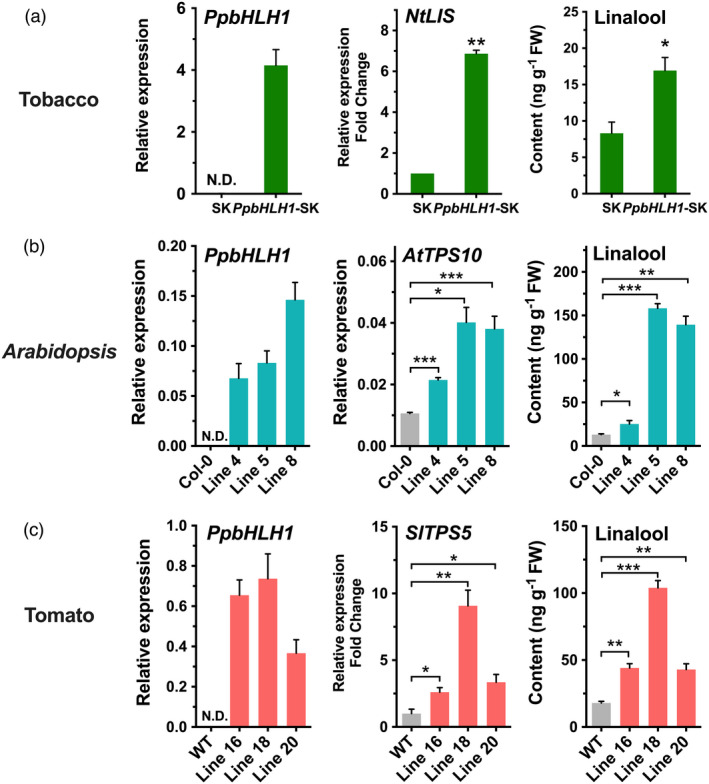
Overexpression of PpbHLH1 increases content of linalool in planta. (a) Transient overexpressing PpbHLH1 induces linalool accumulation in tobacco (Nicotiana benthamiana). (b) Transgenic overexpressing PpbHLH1 causes linalool accumulation in Arabidopsis. Wild‐type Arabidopsis Col‐0 was used as control. (c) Transgenic tomato fruits with overexpressing PpTPS3 produces higher content of linalool than wild‐type controls. Relative expression levels were determined using RT‐qPCR. Data are presented as mean ± standard error from three independent biological replicates. Significant differences are indicated with asterisks above the bars (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). N.D., not detected.
Discussion
Fruit flavour is determined by a complex mixture of sugars, acids and volatiles including linalool. Monoterpene linalool contributes to a sweet, floral note and is present in many edible fruit species, including guava, plum, pineapple, sweet orange and peach (Lewinsohn et al., 2001). Sensory analysis revealed that linalool contributed to consumers' preference of peach fruit (Liu et al., 2017; Tian et al., 2017). Besides its contribution to flavour, linalool is also an attractant of pollinators and acts as a defence against pathogens and herbivores (Raguso, 2016). Accumulation of linalool induced strong resistance to citrus canker in sweet orange (Shimada et al., 2017). Despite its important role during fruit ripening, regulation of linalool synthesis remains largely unclear.
PpTPS3 is a novel synthase regulating linalool synthesis in peach fruit
We initiated study with detecting linalool content during peach fruit ripening and found that linalool accumulated almost exclusively as the enantiomer S‐(+)‐linalool. The final step for linalool biosynthesis is catalysed by terpene synthase. Among multiple peach TPS genes, transcript levels of PpTPS1 peaked at 94 DAB followed by decline during fruit ripening, whereas PpTPS3 expression increased throughout fruit development. Here, we showed that PpTPS3 has higher specificity to produce linalool (98.3%) than PpTPS1 (65.1%, Liu et al., 2017). Moreover, chiral analysis showed a greater enrichment of S‐(+)‐linalool (95%) than (R)‐(−)‐linalool (5%) catalysed by PpTPS3 protein in vitro. For PpTPS1, equal ratios of (R)‐(−)‐linalool and (S)‐(+)‐linalool were detected for catalysed products (Liu et al., 2017). We further validated PpTPS3 function based on correlationship across peach cultivars, where transcript levels of PpTPS3 correlate with content of S‐(+)‐linalool (R 2 = 0.925, P < 0.001). In addition, overexpressing PpTPS3 resulted in significant accumulation, whereas silencing PpTPS3 led to significant reduction in linalool in peach fruit. Heterologous transgenic tomato fruit further verified the biological function of PpTPS3 in linalool production. These results indicated that PpTPS3 is a novel TPS gene family member and is associated with S‐(+)‐linalool synthesis in peach fruit based on both in vitro evidence and in vivo evidence.
A striking feature found in subcellular localization was the cytoplasm‐located PpTPS3. Sequence analysis showed no transit signal peptide of PpTPS3, supporting that its location in the cytoplasm (Figure S9). The biosynthesis of terpenoids depends on the substrate availability of TPS. In general, monoterpene linalool is synthesized in plastids using GPP as precursor. Lots of linalool synthesis‐related TPSs had a signal peptide in the N‐terminus protein, which guide enzymes into plastids to synthesis linalool (Bohlmann et al., 1998), including AmNES/LIS‐2 in snapdragon flowers (Nagegowda et al., 2008), AtTPS10 and AtTPS14 in Arabidopsis (Ginglinger et al., 2013) and FhTPS1 in Freesia × hybrida (Gao et al., 2018). However, studies suggested that GPP, FPP and their precursors, IPP and DMAPP, can communicate between plastids and cytoplasm through undetermined transporters (Gutensohn et al., 2013; Vranová et al., 2013). The increase in GPP content in plastids through transgene in tomato led to the metabolic flux of GPP towards cytoplasm, which can be used for monoterpene synthesis (Gutensohn et al., 2013). Isozymes of GGPPS were found localized in mitochondria, chloroplasts and endoplasmic reticulum in Arabidopsis, and this allows TPSs to synthesize monoterpenes using GPP in the cytoplasm (Okada et al., 2000; Thabet et al., 2012). CoLIS was found to be mainly responsible for the synthesis of S‐(+)‐linalool without transit signal peptide in Cinnamomum osmophloeum (Lin et al., 2013). This provides a new perspective for us to further understand that substrate availability and subcellular localization jointly determine TPS function in planta.
Enhanced linalool level by genetic engineering PpTPS3 upstream regulator PpbHLH1
Recently, TFs that regulate TPS expression have been identified in plants (Chuang et al., 2018; Yang et al., 2020). However, the direct effects of these TFs on linalool synthesis remain undetermined. In this study, peach PpMYB1, PpMYC2, PpWRKY73 and PpEOT1 with the highest homology to FhMYB21L1, SlMYC1, SlWRKY73 and SlEOT1, respectively, failed to activate the PpTPS3. To identify TFs associated with linalool synthesis, homologous TFs associated with terpene (monoterpenes, sesquiterpenes and biterpenes) formation were cloned. Moreover, TFs whose expression positively correlated with linalool production during peach fruit ripening were also cloned for screening. After testing 58 TFs, the highest induction of transcription was observed with PpbHLH1. Phylogenetic tree constructed using the sequences of bHLH proteins from both peach and Arabidopsis with some bHLH proteins, which regulate terpenoid formation, showed that PpbHLH1 belongs to the IVa subfamily (Figure S10). A direct effect of PpbHLH1 on expression of the PpTPS3 and production of linalool was detected in peach fruit with overexpressing and silencing PpbHLH1. Moreover, regulatory role of PpbHLH1 on linalool production was further validated based on significant positive correlations between gene expression and linalool contents across peach cultivars. Besides, a striking feature observed in PpbHLH1 promoter sequence is the enrichment of light responsiveness. Considering the effect of UV‐B light on peach fruit linalool production in our previous study (Liu et al., 2017), we here showed that the inhibition of PpbHLH1 expression was associated with linalool reduction after UV‐B irradiation. These results support the conclusion that PpbHLH1 transcriptionally activates the PpTPS3 gene and formation of volatile linalool in peach fruit.
Having identifying PpTPS3 and PpbHLH1 TF, we next tested whether these genes could enhance the levels of linalool in other plants through genetic engineering. Moreover, these genetic engineering experiments could further validate the function of PpTPS3 and PpbHLH1 in linalool formation. Indeed, early attempts to engineer plant linalool production mainly focused on TPS genes. For instance, heterologous overexpressing linalool synthase gene LIS found in Clarkia breweri induced accumulation of linalool in tomato fruits (Lewinsohn et al., 2001). Transgenic sweet orange plants overexpressing a linalool synthase gene CuSTS3‐1 generated higher levels of linalool in leaves (Shimada et al., 2017). Again, our transgenic plants produced significantly higher contents of linalool after engineering peach PpTPS3 in both Arabidopsis and tomato fruits. To our best knowledge, no reports have been published in plants engineering linalool production using TFs. Here, the contents of linalool were induced up to 8 and 3 times after overexpressing peach PpbHLH1 in Arabidopsis and tomato fruits, respectively. Although changed aroma of transgenic tomato fruit could be detected by the human nose, we have not carried out panel tests by tasting. Therefore, the consumers’ preference for the transgenic tomato fruit remains unknown.
We previously revealed that linalool is present in fruit as both free and glycosidic bound forms (Wu et al., 2019). Besides accumulation of free linalool detected in transgenic plants, significantly higher contents of the linalyl‐β‐d‐glucoside were also observed after the overexpression of PpTPS3 (Figure S11a). In PpbHLH1 transgenic tomato fruits, significant increase in the contents of linalyl‐β‐d‐glucoside was observed only in line 18, which has the highest content of free linalool (Figure S11b, Figure 7c). Similar accumulation of glycosylated‐bound linalool was also observed in engineered petunia flower (Lücker et al., 2001) and Arabidopsis (Aharoni et al., 2003) through heterologous overexpressing linalool synthases gene. Besides its contribution to aroma and flavour, linalool is also involved in plant defence. For instance, increased linalool confers strong resistance to citrus canker caused by Xanthormonas cirti subsp. Citri (Shimada et al., 2017). Genetic engineering of linalool metabolism in Arabidopsis and tomato may induce the plant’s defence mechanisms, although extra experiments are required.
Notably, a cytotoxic effect of linalool was observed based on increased mitochondrial reactive oxygen species (ROS) and apoptotic cell death caused by high contents of linalool (Cheng et al., 2017; Gu et al., 2010). These results suggested that high dosages of linalool are toxic to plant cells. Converting free linalool into nonvolatile glycosylated linalool is likely to be a strategy for detoxification in plant (Raguso and Pichersky, 1999). Glycosylation of linalool is catalysed by UGTs, which transfer sugar molecules to linalool (Wu et al., 2019). A large part of linalool produced in plants is converted to corresponding conjugated or derivatives, which are stored in tissues instead of releasing (Aharoni et al., 2003; Boachon et al., 2015). Besides, significant accumulation of cis‐linalool oxide was detected in transgenic tomato fruit after overexpressing PpbHLH1 (Figure S11c). Volatile linalool could be further oxidized into nonvolatile defensive chemicals, catalysing by cytochrome P450 enzymes (CYPs) (Boachon et al., 2019). Therefore, plants precisely regulate metabolism of linalool after long‐term evolution, including TPS‐catalysed biosynthesis, UGT catalysed glycosylation and CYP‐mediated oxidation. Correspondingly, plants regulate linalool content, making fruit attractive to seed‐dispersing animals, while reducing potential toxic effects of free linalool.
Peach fruit linalool synthesis is regulated not only by TF bHLH
Besides bHLH IVa subfamily, we also found three TFs, PpNAC1, PpNAC2 and PpERF61, with activation effect on PpTPS3 transcription (Figure 4a). It has been reported that PpNAC1 and PpNAC2 were associated with anthocyanin biosynthesis in peach (Zhou et al., 2015), suggesting that these two NACs play an important role in regulating peach secondary metabolism. Moreover, homologous NAC proteins in kiwifruit activate AaTPS1 transcription and thereby regulate monoterpene biosynthesis (Nieuwenhuizen et al., 2015). Transcript levels of PpERF61 were positively related to PpTPS3 expression during peach fruit ripening (R > 0.5), implying it might be an activator of linalool biosynthesis. Volatile linalool is a major contributor to flavour, therefore plays an important role for fruit consumption and help seed dispersal. Considering the important role of linalool has in plants and fruits, it seems reasonable to speculate that transcription of PpTPS3 may be regulated by multiple TFs in peach.
DNA methylation is one of the epigenetic modifications that play a critical role in fruit development and ripening (Tang et al., 2020). Although early DNA methylation studies were limited to model fruit tomato (Zhong et al., 2013), the increasing number of crop genomes has facilitated the DNA methylation studies in fruit crops such as orange (Huang et al., 2019), strawberry (Cheng et al., 2018), apple (Xu et al., 2018) and peach fruit (Zhu et al., 2020). In tomato fruit, changes in DNA methylation levels in the promoters of genes are associated with the production of flavour‐related chemicals including volatiles (Zhang et al., 2016, 2020). Here, decreased DNA methylation was observed during peach fruit ripening, accompanying by increased PpTPS3 expression and high accumulated linalool contents. High DNA methylation may reduce PpTPS3 expression and produce trace linalool to protect immature seeds of peach fruit. For ripe peach fruit, reduced DNA methylation may contribute to PpTPS3 expression and linalool accumulation, therefore making ripe fruit more attractive to consumers. To our best knowledge, this is the first study reporting fruit linalool synthesis is related to DNA methylation. It is possible that epigenetic regulation methylation is an added level of insurance for regulating linalool production during peach fruit ripening.
Conclusions
Accumulation of linalool during fruit ripening is important for fruit consumption and for seed dispersal. Here, we cloned and characterized a novel gene PpTPS3 encoding a protein capable of converting GPP to acyclic monoterpene linalool based on both in vitro and in vivo experiments. TF PpbHLH1 activates PpTPS3 transcription via binding to its promoter. Overexpressing PpbHLH1 enhances linalool, and silencing this gene reduces linalool content in peach fruit. Metabolic engineering of linalool production was generated by overexpression of PpbHLH1 in multiple plants such as Arabidopsis and tomato fruits. Moreover, our results showed that linalool accumulation is associated with DNA methylation of synthesis gene. Our study showed that linalool production is regulated not only by TFs, but also by epigenetic modifications during fruit ripening. Knowledge of how volatile pathways are regulated would greatly facilitate efforts to improve fruit flavour quality.
Methods
Plant materials and treatment
Peach (Prunus persica L. Batsch cv. Hujingmilu) fruits were obtained from the Melting Peach Research Institute of Fenghua, Zhejiang Province, China. They were harvested at five developmental stages, S1 (first rapid growth phase, 34 days after bloom, DAB), S2 (stone hardening, 71 DAB), S3 (second rapid growth phase, 94 DAB), S4 (mature stage, 108 DAB) and S5 (ripening stage, 111 DAB) (Wu et al., 2019). Peach fruits were exposed to 1.5 w/m2 UV‐B irradiation for 6 h and 48 h at 20 °C (Liu et al., 2017). Moreover, fruits of nine natural peach cultivars (Prunus persica) were sampled at the ripening stage, namely ‘Guanghetao’ (GHT), ‘Honggengansutao’ (HGGST), ‘Honghuashantao’ (HHST), ‘Zaoshanghaishuimi’ (ZSHSM), ‘Fuyangchiyue’ (FYCY), ‘Hongpan1’ (HP1), ‘Hongpan2’ (HP2), ‘Hujingmilu’ (HJML) and ‘Danxia’ (DX). Three biological replicates with five fruits each were sampled, frozen in liquid nitrogen and stored at −80 °C for further use.
Volatiles analysis by GC‐MS
Volatiles were analysed according to our previous study (Wu et al., 2019). Frozen peach fruit (5 g), peel (1 g), tomato fruit (5 g) and leaf (1 g), Arabidopsis (1 g) and tobacco leaf (1 g) were ground into powder under liquid nitrogen and transferred to vials containing 200 mm ethylenediaminetetraacetic acid (EDTA) and 20% CaCl2 solution. Before the vials were sealed, 30 μL 2‐octanol (0.8 mg/mL) was added as internal standard. A fibre coated with 65 μm of polydimethylsiloxane and divinylbenzene (PDMS−DVB) (Supeclo Co., Bellefonte, PA) was used for volatile collection. The vials were placed in the tray of a solid‐phase microextraction (SPME) autosampler (Combi PAL, CTC Analytics, Agilent Technologies), coupled to an Agilent 7890N gas chromatograph and an Agilent 5975C mass spectrometer. Volatiles were separated on a DB‐WAX column (30 m × 0.25 mm i.d. × 0.25 μm film thickness; J & W Scientific, Folsom, CA). Carrier gas helium rate was 1.0 mL/min. The temperature programme started at 40 °C and was increased by 3 °C/min to 100 °C and then to 245 °C at 5 °C/min. The column effluent was ionized by electron ionization at an energy of 70 eV with a transfer temperature of 250 °C and a source temperature of 230 °C. Volatiles were identified by comparing their electron ionization mass spectra with the NIST Mass Spectral Library (NIST‐08) and the retention time of authentic standards. Quantification of volatiles was performed using the peak area of the internal standard as a reference based on total ion chromatogram. Extraction and hydrolysis of glycosylated volatiles were performed according to Wu et al. (2019).
Chiral GC‐MS analysis
Chiral GC‐MS analysis was performed using a 7890A gas chromatograph coupled to an Agilent 5975C mass spectrometer. Helium was used as carrier gas. The enantiomers of linalool were separated with a β‐Dex™ 325 capillary column (30 m × 0.25 mm × 0.25 μm; Supelco Co.) according to methods described by Liu et al. (2017). The initial oven temperature was 35 °C, held for 1 min, increased to 118 °C by 5 °C/min, then increased to 118.4 °C and followed by a further increase to 230 °C by 5 °C/min. Authentic standards (Sigma‐Aldrich) were used for identification of linalool enantiomers.
Heterologous expression in Escherichia coli and enzymatic activity assay
Recombinant protein purification was performed following the method described previously (Wu et al., 2019). The full‐length cDNA of the PpTPS3 gene was inserted into the pET6 × HN expression vector (Clontech, Mountain View, CA) using the primers in Table S9. After sequence validation, pET vector was transformed into Escherichia coli BL21 (DE3) pLysS (Promega, Madison, WI). Recombinant protein was purified by the HisTALON™ gravity column (Clontech) following the manufacturer’s instructions and replaced with Tris‐HCl buffer (100 mm Tris, 2 mm DTT, pH 7.5) by the PD‐10 Desalting Column (GE Healthcare). SDS‐PAGE was performed, and the protein was visualized by Coomassie Brilliant Blue. Enzymatic activity assay was carried out according to Liu et al. (2017). Enzyme assay buffer B (50 mm HEPES, 7.5 mm MgCl2, 100 mm KCl, 5 mm dithiothreitol and 10% (v/v) glycerol, pH 7.0) used in the reaction with geranyl pyrophosphate (GPP) as substrate. The reaction was performed at 4‐mL sealed glass bottle, 200 µL of purified protein, 10 µg of reaction substrate and 800 µL of enzyme reaction buffer were added to 1 mL of the reaction system, and after adding 5 μL (0.2 mg/mL) of 2‐octanol as an internal standard, the reaction was performed at 42 °C for 15 min. The protein expressed by the empty vector and the substrate were heated separately as a negative control. The product catalysed by the enzyme was collected by SPME before GC‐MS analysis.
Gene expression analysis
Total RNA was extracted according to the protocol described by Zhang et al. (2006), and libraries for high‐throughput Illumina strand‐specific RNA‐seq were prepared as described previously (Zhang et al., 2016). RNA‐seq raw data used for the present study can be found in the National Center for Biotechnology Information (NCBI) Short Read Archive database. Accession Numbers PRJNA576753 and SRP103523 are for peach samples at different development and ripening stages and under UV‐B treatment, respectively. For quantitative reverse‐transcription PCR (RT‐qPCR) analysis, PrimeScript RT Reagent Kit with gDNA Eraser (Takara) was used to synthesize the first‐strand cDNA, and then, experiments were performed using CFX96 instrument (Bio‐Rad, Hercules, CA) and SsoFast EvaGreen Supermix (Bio‐Rad). Each RT‐qPCR analysis contains three replicates. Oligonucleotide primers used for RT‐qPCR analysis are listed in Table S9.
Dual‐luciferase assays
According to the previous protocol (Zhang et al., 2018), full‐length cDNAs of transcription factors were cloned into pGreen II 0029 62‐SK vector, and the promoter of PpTPS3 was cloned into pGreen II 0800‐LUC vector using the primers listed in Tables S4, S8 and S9. The above constructs were transformed into Agrobacterium tumefaciens GV3101::pSoup using the Gene Pulser Xcell™ Electroporation Systems (Bio‐Rad, Hercules, CA). To obtain the activity of a specific transcription factor on the PpTPS3 promoter, the Agrobacterium culture mixtures were comprised of 1 mL TFs and 100 μL promoter for transient expression in Nicotiana benthamiana leaves. The ratio of enzyme activities of firefly luciferase (LUC) and Renilla luciferase (REN) was measured using a Modulus Luminometer (Promega, Madison, WI) on the third day after infiltration. Enzyme activities of LUC and REN were assayed using dual‐luciferase assay reagents (Promega). The LUC/REN value of the empty vector SK on the promoter was set as 1, as a calibrator. For each transcription factor–promoter interaction, at least three independent experiments were performed, with six replicates in each experiment.
Yeast one‐hybrid assay
The sequence of the PpTPS3 promoter was cloned into the pAbAi vector, and the construct was integrated into the genome of the Y1HGold yeast strain. The background aureobasidin A resistance (AbAr) expression of Y1HGold PpTPS3‐pAbAi strain was tested on selective synthetic dextrose medium (SD) uracil. Then, the full length of PpbHLH1 was cloned into the pGADT7 vector for identification. After determining the minimal inhibitory concentration of AbA for the bait strains, the AD prey vectors were transformed into the bait strain and screened on an SD/‐Leu/AbA plate. Autoactivation and transcription factor–promoter interaction analysis were conducted according to the Matchmaker® Gold Yeast One‐Hybrid Library Screening System (Clontech) manufacturer’s protocol. All transformations and screenings were performed at least three times. Primers used in this assay are listed in Table S9.
Electrophoretic mobility shift assay (EMSA)
The coding sequence of PpbHLH1 was cloned into the pGEX‐4T‐1 vector using the primers listed in Table S9 and then was expressed in Escherichia coli BL21. Expression and purification of the recombinant protein were performed according to GST‐tag Protein Purification Kit (Beyotime) manufacturer’s instructions. The EMSA was conducted using the LightShift® Chemiluminescent EMSA Kit (Thermo Fisher Scientific). The details of the EMSA are provided by Li et al. (2017). The double‐stranded probes were made by annealing separately synthesized strands, with 3' biotin labelling. The probes used for EMSAs are listed in Table S9.
Gene transient overexpression in peach fruit
Transient overexpression in peach fruit was performed according to our previous study (Liu et al., 2017). Constructs were electroporated into Agrobacterium tumefaciens GV3101 and activated at 28 °C until the OD600 reached 0.8–1.0. After disinfecting the surface of the peach fruit, two flesh cubes (1 cm thick) were taken from opposite sides of each fruit. Each piece was cut into two halves on average, and A. tumefaciens carrying the PpTPS3 or PpbHLH1 construct and A. tumefaciens carrying empty construct were infiltrated under a −70 kPa vacuum, respectively. After vacuum infiltration, flesh cubes were rinsed three times with sterile water and cultured on Murashige and Skoog (MS) medium in a growth chamber (20 °C, RH 85%) for 3 day. Flesh cubes were then sampled for GC‐MS analysis. Transient expression treatments were repeated three times with five fruits each.
TRV‐based virus‐induced gene silencing (VIGS) in peach
Virus‐induced gene silencing was performed according to previous study (Zhao et al., 2020). Agrobacterium cells expressing pTRV1 and pTRV2 or pTRV2‐PpPDS/PpTPS3/PpbHLH1 constructs were cultured to an OD600 of 0.5–0.8, and then, Agrobacterium cells were collected by centrifugation and resuspended in 1/2 volume of MES infiltration buffer (200 mm acetosyringone, 10 mm MgCl2, 10 mm MES; pH 5.6). Agrobacterium with pTRV1 and pTRV2 constructs were mixed in a ratio of 1:1 and infiltrated using syringes into the flesh of peach fruit on the trees. Three biological replicates with ten fruits each were sampled for analysis.
Stable overexpression in tomato fruit
Full‐length cDNAs of PpTPS3 and PpbHLH1 were cloned into pBI121 vector or pBIN19‐E8, respectively, using the primers listed in Table S9. A. tumefaciens‐mediated tomato (cv MicroTom) transformation was performed according to Wang et al. (2005). The identified T1 generation of transgenic and wild‐type (WT) tomato plants were grown in a greenhouse (25 °C, 16‐h light/8‐h darkness). Tomato fruits at red ripe stage (breaker + 7 days) were frozen in liquid nitrogen and stored at −80 °C for analysis. Three tomato plants were selected from each line as three biological replicates, and each replicate contained five fruits.
Stable transformation of Arabidopsis
Four‐week‐old seedlings of Arabidopsis plants were transformed according to the floral dip method (Clough and Bent, 1998). PpTPS3‐pBI121 constructs were used to transform tps10 mutant in the Columbia‐0 genetic background, and PpbHLH1‐pBI121 was transformed in the wild‐type (WT) background. The homozygous Arabidopsis mutant (SALK_041114C) was purchased from the Arabidopsis Information Resource (TAIR). Transgenic seeds were selected on 1/2 Murashige and Skoog medium with 50 mg/L kanamycin, and the 1 month of whole plants (T2 and WT) was harvested and analysed.
DNA methylation analysis
DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used for genomic DNA extraction from peach fruit samples. DNA quality was monitored by agarose gel electrophoresis and a ratio of A260/A280. Genomic DNA with bisulphite conversion rates higher than 99.5% was used for Illumina DNA sequencing. A HiSeq 2500 was used for DNA sequencing according to Illumina protocols. Methylome data at single‐base resolution were achieved, and occurrence of DNA methylation was analysed according to our previous study (Zhang et al., 2016).
Subcellular localization analysis
The recombined GFP vectors were constructed using primers listed in Table S9 and were electroporated into A. tumefaciens GV3101 for transient expression in tobacco (N. benthamiana) leaves. After 48 h, leaves were detached for analysis using a confocal laser scanning microscope (LSM 780; Carl Zeiss, Oberkochen, Germany). Images were processed with the LSM Image Browser (Carl Zeiss). Protoplast extraction was performed using the plant protoplast preparation and transformation kit (RTU4052) (Real‐Times, China) following the manufacturer’s protocol.
Statistics
Figures were produced using GraphPad Prism 7.0 (GraphPad Software; San Diego, CA). The two‐sample significance test was calculated using unpaired Student’s t test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) (SPSS 19.0; SPSS Inc., Chicago, IL). MetaboAnalyst 4.0 was used for correlation analysis in the present study.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Author contributions
B.Z. conceived the research plans; C.W. and H.L. performed most of the experiments and analyses with help from X.C. and M.Z.; C.W. and B.Z. wrote the article; and B.Z., X.L. and K.C. reviewed and edited the manuscript. All authors have read and approved to the published version of the manuscript.
Supporting information
Figure S1 Alignment of amino acid sequences of 38 peach PpTPSs.
Figure S2 SDS‐PAGE analysis of PpTPS3 protein.
Figure S3 Silencing PpPDS (Prupe.1G174100) gene in peach fruit.
Figure S4 Expression of AtTPS10 and AtTPS14 in PpTPS3 transgenic Arabidopsis.
Figure S5 Changes in content of free linalool in PpTPS3 transgenic tomato leaves.
Figure S6 Expression profiles of three bHLH genes and regulatory effect of PpbHLH1 on the promoter of PpTPS1.
Figure S7 PpbHLH1 binds to PpTPS3 promoter.
Figure S8 Correlation between transcript and S‐(+)‐Linalool content in different peach cultivars.
Figure S9 Alignment of amino acid sequence of PpTPS3 with closely related sequences from TPS‐g subfamily.
Figure S10 Phylogenetic tree constructed using the sequences of bHLH proteins from both peach and Arabidopsis with some bHLH proteins which regulate linalool or terpene formation in other species.
Figure S11 Changes in content of linalyl‐β‐d‐glucoside and linalool oxide in PpTPS3 and PpbHLH1 transgenic tomato fruits.
Table S1 Content of volatiles in peach fruit during development and ripening.
Table S2 Accession numbers for plant TPSs used for sequence analysis in Figure 2.
Table S3 Peach homologues of transcription factors that regulate terpenoid formation.
Table S4 Primers of TFs which have expression correlation with PpTPS3 during peach fruit ripening.
Table S5 The light response‐related cis‐acting elements of PpbHLH1 promoter.
Table S6 Statistic of whole‐genome bisulfite sequencing reads from peach fruit at different development stages and under UV‐B treatment.
Table S7 Statistic of sequencing depth and covering in peach fruit at different development stages and under UV‐B treatment.
Table S8 Primers of TFs which homologues of transcription factors that regulate terpenoid formation.
Table S9 Other primer sequences used in the present study.
Acknowledgements
This research was supported by the National Key R & D Program of China (2016YFD0400101), the National Natural Science Foundation of China (31972379) and the Fundamental Research Funds for the Zhejiang Provincial Universities (2021XZZX026).
Wei, C. , Liu, H. , Cao, X. , Zhang, M. , Li, X. , Chen, K. and Zhang, B. (2021) Synthesis of flavour‐related linalool is regulated by PpbHLH1 and associated with changes in DNA methylation during peach fruit ripening. Plant Biotechnol J, 10.1111/pbi.13638
References
- Aharoni, A. , Giri, A.P. , Deuerlein, S. , Griepink, F. , de Kogel, W.J. , Verstappen, F.W. , Verhoeven, H.A. et al. (2003) Terpenoid metabolism in wild‐type and transgenic Arabidopsis plants. Plant Cell, 15, 2866–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A. , Giri, A. , Verstappen, F.W.A. , Bertea, C.M. , Sevenier, R. , Sun, Z. , Jongsma, M.A. et al. (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell, 16, 3110–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boachon, B. , Burdloff, Y. , Ruan, J.X. , Rojo, R. , Junker, R.R. , Vincent, B. , Nicolè, F. et al. (2019) A promiscuous CYP706A3 reduces terpene volatile emission from Arabidopsis flowers, affecting florivores and the floral microbiome. Plant Cell, 31, 2947–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boachon, B. , Junker, R.R. , Miesch, L. , Bassard, J.E. , Höfer, R. , Caillieaudeaux, R. , Seidel, D.E. et al. (2015) CYP76C1(Cytochrome P450)‐mediated linalool metabolism and the formation of volatile and soluble linalool oxides in Arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell, 27, 2972–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann, J. , Meyer‐Gauen, G. and Croteau, R. (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA, 95, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnländer, B. , Cappuccio, R. , Liverani, F.S. and Winterhalter, P. (2006) Analysis of enantiomeric linalool ratio in green and roasted coffee. Flavour Frag. J. 21, 637–641. [Google Scholar]
- Chen, F. , Tholl, D. , D’Auria, J.C. , Farooq, A. , Pichersky, E. and Gershenzon, J. (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell, 15, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J.F. , Niu, Q.F. , Zhang, B. , Chen, K.S. , Yang, R.H. , Zhu, J.K. , Zhang, Y.J. et al. (2018) Downregulation of RdDM during strawberry fruit ripening. Genome Biol. 19, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y.H. , Dai, C. and Zhang, J. (2017) SIRT3–SOD2–ROS pathway is involved in linalool‐induced glioma cell apoptotic death. Acta Biochim. Pol. 64, 343–350. [DOI] [PubMed] [Google Scholar]
- Chuang, Y.C. , Hung, Y.C. , Tsai, W.C. , Chen, W.H. and Chen, H.H. (2018) PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 69, 4363–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Eduardo, I. , Chietera, G. , Bassi, D. , Rossini, L. and Vecchietti, A. (2010) Identification of key odor volatile compounds in the essential oil of nine peach accessions. J. Sci. Food Agric. 90, 1146–1154. [DOI] [PubMed] [Google Scholar]
- Falara, V. , Akhtar, T.A. , Nguyen, T.T.H. , Spyropoulou, E.A. , Bleeker, P.M. , Schauvinhold, I. , Matsuba, Y. et al. (2011) The tomato terpene synthase gene family. Plant Physiol. 157, 770–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F. , Liu, B. , Li, M. , Gao, X.Y. , Fang, Q. , Liu, C. , Ding, H. et al. (2018) Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia × hybrida . J. Exp. Bot. 69, 4249–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginglinger, J.F. , Boachon, B. , Höfer, R. , Paetz, C. , Köllner, T.G. , Miesch, L. , Lugan, R. et al. (2013) Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell, 25, 4640–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. , Ting, Z. , Qiu, X. , Zhang, X.Z. , Gan, X.X. , Fang, Y.M. , Xu, X.H. et al. (2010) Linalool preferentially induces robust apoptosis of a variety of leukemia cells via upregulating p53 and cyclin‐dependent kinase inhibitors. Toxicology, 268, 19–24. [DOI] [PubMed] [Google Scholar]
- Gutensohn, M. , Orlova, I. , Nguyen, T.T.H. , Davidovich‐Rikanati, R. , Ferruzzi, M.G. , Sitrit, Y. , Lewinsohn, E. et al. (2013) Cytosolic monoterpene biosynthesis is supported by plastid‐generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J. 75, 351–363. [DOI] [PubMed] [Google Scholar]
- He, J. , Fandino, R.A. , Halitschke, R. , Luck, K. , Köllner, T.G. , Murdock, M.H. , Ray, R. et al. (2019) An unbiased approach elucidates variation in (S)‐(+)‐linalool, a context‐specific mediator of a tri‐trophic interaction in wild tobacco. Proc. Natl. Acad. Sci. USA, 116, 14651–14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Liu, R.E. , Niu, Q.F. , Tang, K. , Zhang, B. , Zhang, H. , Chen, K.S. et al. (2019) Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA, 116, 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X.Z. , Xiao, Y.T. , Köllner, T.G. , Jing, W.X. , Kou, J.F. , Chen, J.Y. , Liu, D.F. et al. (2017) The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 41, 261–274. [DOI] [PubMed] [Google Scholar]
- Klee, H.J. and Tieman, D.M. (2018) The genetics of fruit flavour preferences. Nat. Rev. Genet. 19, 347–356. [DOI] [PubMed] [Google Scholar]
- Kumar, Y. , Khan, F. , Rastogi, S. and Shasany, A.K. (2018) Genome‐wide detection of terpene synthase genes in holy basil (Ocimum sanctum L.). PLoS One, 13, e0207097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn, E. , Schalechet, F. , Wilkinson, J. , Matsui, K. , Tadmor, Y. , Nam, K.H. , Amar, O. et al. (2001) Enhanced levels of the aroma and flavor compound S‐linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 127, 1256–1265. [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Xu, Y.Y. , Shen, S.L. , Yin, X.R. , Klee, H. , Zhang, B. , Chen, K.S. et al. (2017) Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E‐geraniol in sweet orange fruit. J. Exp. Bot. 68, 4929–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.L. , Lee, Y.R. , Huang, W.K. , Chang, S.T. and Chu, F.H. (2013) Characterization of S‐(+)‐linalool synthase from several provenances of Cinnamomum osmophloeum . Tree Genet. Genomes, 10, 75–86. [Google Scholar]
- Liu, H.R. , Cao, X.M. , Liu, X.H. , Xin, R. , Wang, J.J. , Gao, J. , Wu, B.P. et al. (2017) UV‐B irradiation differentially regulates terpene synthases and terpene content of peach. Plant Cell Environ. 40, 2261–2275. [DOI] [PubMed] [Google Scholar]
- Lücker, J. , Bouwmeester, H.J. , Schwab, W. , Blaas, J. , van der Plas, L.H. and Verhoeven, H.A. (2001) Expression of Clarkia S‐linalool synthase in transgenic petunia plants results in the accumulation of S‐linalyl‐beta‐d‐glucopyranoside. Plant J. 27, 315–324. [DOI] [PubMed] [Google Scholar]
- Magnard, J.‐L. , Bony, A.R. , Bettini, F. , Campanaro, A. , Blerot, B. , Baudino, S. and Jullien, F. (2018) Linalool and linalool nerolidol synthases in roses, several genes for little scent. Plant Physiol. Biochem. 127, 74–87. [DOI] [PubMed] [Google Scholar]
- Martin, D.M. , Aubourg, S. , Schouwey, M.B. , Daviet, L. , Schalk, M. , Toub, O. , Lund, S.T. et al. (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 10, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagegowda, D.A. , Gutensohn, M. , Wilkerson, C.G. and Dudareva, N. (2008) Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 55, 224–239. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen, N.J. , Chen, X. , Wang, M.Y. , Matich, A.J. , Perez, R.L. , Allan, A.C. , Green, S.A. et al. (2015) Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE‐INSENSITIVE3‐like transcription factors. Plant Physiol. 167, 1243–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen, N.J. , Green, S.A. , Chen, X.Y. , Bailleul, E.J.D. , Matich, A.J. , Wang, M.Y. and Atkinson, R.G. (2013) Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol. 161, 787–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, K. , Saito, T. , Nakagawa, T. , Kawamukai, M. and Kamiya, Y. (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 122, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguso, R.A. (2016) More lessons from linalool: insights gained from a ubiquitous floral volatile. Curr. Opin. Plant Biol. 32, 31–36. [DOI] [PubMed] [Google Scholar]
- Raguso, R. and Pichersky, E. (1999) A day in the life of a linalool molecule: chemical communication in a plant–pollinator system. Part 1: linalool biosynthesis in flowering plants. Plant Spec. Biol. 14, 95–120. [Google Scholar]
- Ravid, U. , Putievsky, E. , Weinstein, V. and Ikan, R. (1985) Determination of the enantiomeric composition of natural flavouring agents by 1H‐NMR spectroscopy. In Essential Oils and Aromatic Plants ( Baerheim Svendsen, A. and Scheffer, J.J.C. , eds.), pp. 135–138. The Netherlands: Springer. [Google Scholar]
- Ravid, U. , Putievsky, E. , Katzir, I. and Lewinsohn, E. (1997) Enantiomeric composition of linalool in the essential oils of Ocimum species and in commercial basil oils. Flavour Frag. J. 12, 293–296. [Google Scholar]
- Shimada, T. , Endo, T. , Rodríguez, A. , Fujii, H. , Goto, S. , Matsuura, T. , Hojo, Y. et al. (2017) Ectopic accumulation of linalool confers resistance to Xanthomonas citri subsp. citri in transgenic sweet orange plants. Tree Physiol. 37, 654–664. [DOI] [PubMed] [Google Scholar]
- Spyropoulou, E.A. , Haring, M.A. and Schuurink, R.C. (2014) RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 15, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, D.G. , Gallusci, P. and Lang, Z.B. (2020) Fruit development and epigenetic modifications. New Phytol. 228, 839–844. [DOI] [PubMed] [Google Scholar]
- Thabet, I. , Guirimand, G. , Guihur, A. , Lanoue, A. , Courdavault, V. , Papon, N. , Bouzid, S. et al. (2012) Characterization and subcellular localization of geranylgeranyl diphosphate synthase from Catharanthus roseus . Mol. Biol. Rep. 39, 3235–3243. [DOI] [PubMed] [Google Scholar]
- Tian, H.L. , Wang, P. , Zhan, P. , Yan, H.Y. , Zhou, W.J. and Zhang, F. (2017) Effects of β‐glucosidase on the aroma characteristics of flat peach juice as assessed by descriptive sensory analysis and gas chromatography and compared by partial least squares regression. LWT‐Food Sci. Technol. 82, 113–120. [Google Scholar]
- Vranová, E. , Coman, D. and Gruissem, W. (2012) Structure and dynamics of the isoprenoid pathway network. Mol. Plant. 5, 318–333. [DOI] [PubMed] [Google Scholar]
- Vranová, E. , Coman, D. and Gruissem, W. (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Jones, B. , Li, Z. , Frasse, P. , Delalande, C. , Regad, F. , Chaabouni, S. et al. (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell, 17, 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B.P. , Cao, X.M. , Liu, H.R. , Zhu, C.Q. , Klee, H.J. , Zhang, B. and Chen, K.S. (2019) UDP‐glucosyltransferase PpUGT85A2 controls volatile glycosylation in peach. J. Exp. Bot. 70, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.D. , Zhou, S.S. , Gong, X.Q. , Song, Y. , van Nocker, S. , Ma, F.W. and Guan, Q.M. (2018) Single‐base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol. J. 16, 672–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z.Z. , Li, Y.Q. , Gao, F.Z. , Jin, W. , Li, S.Y. , Kimani, S. , Yang, S. et al. (2020) MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana . J. Exp. Bot. 71, 4140–4158. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Chen, K.S. , Bowen, J. , Allan, A. , Espley, R. , Karunairetnam, S. and Ferguson, I. (2006) Differential expression within the LOX gene family in ripening kiwifruit. J. Exp. Bot. 57, 3825–3836. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Tieman, D.M. , Jiao, C. , Xu, Y.M. , Chen, K.S. , Fei, Z.J. , Giovannoni, J.J. et al. (2016) Chilling‐induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. USA, 113, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Duan, W.Y. , Chen, K.S. and Zhang, B. (2020) Transcriptome and methylome analysis reveals effects of ripening on and off the vine on flavor quality of tomato fruit. Postharvest Biol. Technol. 162, 111096. [Google Scholar]
- Zhang, Y.Y. , Yin, X.R. , Xiao, Y.W. , Zhang, Z.Y. , Li, S.J. , Liu, X.F. , Zhang, B. et al. (2018) An ETHYLENE RESPONSE FACTOR‐MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE Expression in Strawberry. Plant Physiol. 178, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Dong, W.Q. , Zhu, Y.C. , Allan, A.C. , Wang, K.L. and Xu, C.J. (2020) PpGST1, an anthocyanin‐related glutathione S‐transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 18, 1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, S.L. , Fei, Z.J. , Chen, Y.R. , Zheng, Y. , Huang, M.Y. , Vrebalov, J. , McQuinn, R. et al. (2013) Single‐base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Lin‐Wang, K. , Wang, H.L. , Gu, C. , Dare, A.P. , Espley, R.V. , He, H.P. et al. (2015) Molecular genetics of blood‐fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 82, 105–121. [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Xia, R. , Zhao, B.Y. , An, Y.Q. , Dardick, C.D. , Callahan, A.M. and Liu, Z.R. (2012) Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 12, 149–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y.C. , Zhang, B. , Allan, A.C. , Lin‐Wang, K. , Zhao, Y. , Wang, K. , Chen, K.S. et al. (2020) DNA demethylation is involved in the regulation of temperature‐dependent anthocyanin accumulation in peach. Plant J. 102, 965–976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignment of amino acid sequences of 38 peach PpTPSs.
Figure S2 SDS‐PAGE analysis of PpTPS3 protein.
Figure S3 Silencing PpPDS (Prupe.1G174100) gene in peach fruit.
Figure S4 Expression of AtTPS10 and AtTPS14 in PpTPS3 transgenic Arabidopsis.
Figure S5 Changes in content of free linalool in PpTPS3 transgenic tomato leaves.
Figure S6 Expression profiles of three bHLH genes and regulatory effect of PpbHLH1 on the promoter of PpTPS1.
Figure S7 PpbHLH1 binds to PpTPS3 promoter.
Figure S8 Correlation between transcript and S‐(+)‐Linalool content in different peach cultivars.
Figure S9 Alignment of amino acid sequence of PpTPS3 with closely related sequences from TPS‐g subfamily.
Figure S10 Phylogenetic tree constructed using the sequences of bHLH proteins from both peach and Arabidopsis with some bHLH proteins which regulate linalool or terpene formation in other species.
Figure S11 Changes in content of linalyl‐β‐d‐glucoside and linalool oxide in PpTPS3 and PpbHLH1 transgenic tomato fruits.
Table S1 Content of volatiles in peach fruit during development and ripening.
Table S2 Accession numbers for plant TPSs used for sequence analysis in Figure 2.
Table S3 Peach homologues of transcription factors that regulate terpenoid formation.
Table S4 Primers of TFs which have expression correlation with PpTPS3 during peach fruit ripening.
Table S5 The light response‐related cis‐acting elements of PpbHLH1 promoter.
Table S6 Statistic of whole‐genome bisulfite sequencing reads from peach fruit at different development stages and under UV‐B treatment.
Table S7 Statistic of sequencing depth and covering in peach fruit at different development stages and under UV‐B treatment.
Table S8 Primers of TFs which homologues of transcription factors that regulate terpenoid formation.
Table S9 Other primer sequences used in the present study.


