Summary
Hemp (Cannabis sativa L.) is an annual and typically dioecious crop. Due to the therapeutic potential for human diseases, phytocannabinoids as a medical therapy is getting more attention recently. Several candidate genes involved in cannabinoid biosynthesis have been elucidated using omics analysis. However, the gene function was not fully validated due to few reports of stable transformation for Cannabis tissues. In this study, we firstly report the successful generation of gene‐edited plants using an Agrobacterium‐mediated transformation method in C. sativa. DMG278 achieved the highest shoot induction rate, which was selected as the model strain for transformation. By overexpressing the cannabis developmental regulator chimera in the embryo hypocotyls of immature grains, the shoot regeneration efficiency was substantially increased. We used CRISPR/Cas9 technology to edit the phytoene desaturase gene and finally generated four edited cannabis seedlings with albino phenotype. Moreover, we propagated the transgenic plants and validated the stable integration of T‐DNA in cannabis genome.
Keywords: Agrobacterium‐mediated transformation, CRISPR, Cas9, Hemp, genome editing
Introduction
Cannabis sativa belongs to the Cannabaceae family and its ability to induce a relaxed state has been known for over 2700 years (Russo et al., 2008). Since the last decade, renewed interest in the medical effects of cannabinoids emanated (Galán‐Ávila et al., 2020). Although cannabis is best known for the psychoactive effects of D9‐tetrahydrocannabinol (THC), it contains various phytocannabinoids with the therapeutic potential for the treatment of multiple human diseases from complex neurological diseases to cancer, such as cannabidiol (CBD), cannabinol (CBN), cannabigerol (CBG), and cannabichromene (CBC). At present, the medical cannabis market is one of the fastest‐growing markets worldwide, which was worth $3.5 billion at retail prices in 2019. A growing market worth $20.2 billion is expected in the forecast period of 2020 to 2025 (Aliekperova et al., 2020). By 2020, medical cannabis and cannabinoids have been legalized in more than 50 countries, including China (YUNNAN and HEILONGJIANG), Australia, Germany, Israel, Canada and most of the US, etc.
Despite a lot of genes involved in cannabinoid biosynthesis had been elucidated (Liu et al., 2021), functions of these genes were not yet fully validated due to few reports of stable transformation for Cannabis tissues. Recently, the nanoparticle‐based method was used in the transient transformation of Cannabis, as an effective carrier to introduce foreign DNA fragments (Ahmed et al., 2021). Compared with Nicotiana benthamiana, cannabis seedlings and explants are less susceptible to be transformed with A. tumefaciens, another important tool of generating the genome‐edited plants that could be used for further gene function study (Sorokin et al., 2020). In addition, the regeneration of transgenic plantlets from transformed cells is a time‐consuming process and often limited by cannabis varieties (Chaohua et al., 2016; Galán‐Ávila et al., 2020).
Since conventional genetic transformation is a huge obstacle to C. sativa, we plan to reprogram plant meristems and manipulate the genome by coexpressing developmental regulators and genome‐editing components, which have been reported to be effective in several monocot and dicot plant species such as maize, wheat, rice, and sunflower (Debernardi et al., 2020; Kong et al., 2020; Lowe et al., 2018). When BABY BOOM (BBM) and WUSCHEL (WUS) were transformed into immature maize embryos, these embryos can germinate into plants without a callus phase (Lowe et al., 2018). Although the expression of BBM and WUS improved plant regeneration efficiency, these developmental regulators had to be excised from transgenic plants because of the negative pleiotropic effects, which restricted their application (Luo and Palmgren, 2020). Additional developmental genes are proposed to further improve the regeneration efficiency. In N. benthamiana, overexpression of the developmental regulators SHOOT MERISTEMLESS (STM) and ISOPENTENYL TRANSFERASE (IPT) were sufficient to induce the shoot organogenesis (Maher et al., 2020). It was demonstrated that the transcription factor GROWTH‐REGULATING FACTOR (GRF) could form a functional transcriptional complex with GRF‐INTERACTING FACTOR (GIF), which regulated the transition between stem cells and boost genetic transformation in various crop species (Debernardi et al., 2020; Kong et al., 2020). This GRF‐GIF duo performed well under various transformation protocols, and even antibiotic resistance markers were not required due to the high regeneration rate without exogenous cytokinin.
The research described in this paper is initiated to develop a stable Agrobacterium tumefaciens‐mediated transformation system for C. sativa. We estimated the regeneration efficiency within different explants, such as cotyledon, hypocotyl, true leaf, and immature embryo hypocotyl isolated from immature grains after anthesis. Then we cloned the cannabis candidate genes homologous to ZmWUS2, NbSTM, NbIPT, OsGRF4 and AtGIF1, which had been reported to stimulate somatic embryogenesis or organogenesis in other species (Duan et al., 2015; Maher et al., 2020; Shimano et al., 2018). We overexpressed the different combinations of these developmental regulators in the immature embryo hypocotyl, evaluated the effects on the shoot regeneration of different cannabis varieties, and further obtained stable C. sativa transgenic plants and seedlings with knocked out phytoene desaturase gene (CsPDS1), a common marker gene that can test genetic manipulation tools. The system we developed can be applied to functional genomic studies and release the real potential of genome editing in C. sativa.
Results
Effect of explant, variety, and developmental regulator on shoot organogenesis
To evaluate the regeneration efficiency of different explant types, seeds of hemp strain YUNMA7 were surface sterilized and cultured in the Murashige & Skoog (MS) medium. True leaves, cotyledons and hypocotyls were collected from these seedlings about 17 days later after the germination (Figure 1a‐c). In addition, the embryo hypocotyls of immature grains were collected from YUNMA7 spikes approximately 10 days, 15 days, 20 days and 25 days after anthesis (Figure 1d). All the explants were cultured in a callus induction medium. Over 20% of the immature embryo hypocotyls developed embryogenic calli within 5 days (Figure 1h), and the hypocotyls collected 15 days after anthesis (D15) produced more calli (at an average of 31.08%) compared to those collected earlier or later (Table 1). Throughout the 4‐week incubation, we observed the induction frequencies of only 5.97% in true leaves, 7.65% in cotyledons and 5.31% in hypocotyls (Table 1; Figure 1e‐g). After an additional 2 weeks, proliferating tissue was transferred to regeneration medium at 26 °C under continuous light (50 μM/m2/s) for 5 weeks. About 6.12% of the D15 calli produced shoots, and less than 3% of the calli developed proliferated shoots from the other three explants (Table 1; Figure 1i‐l).
Figure 1.
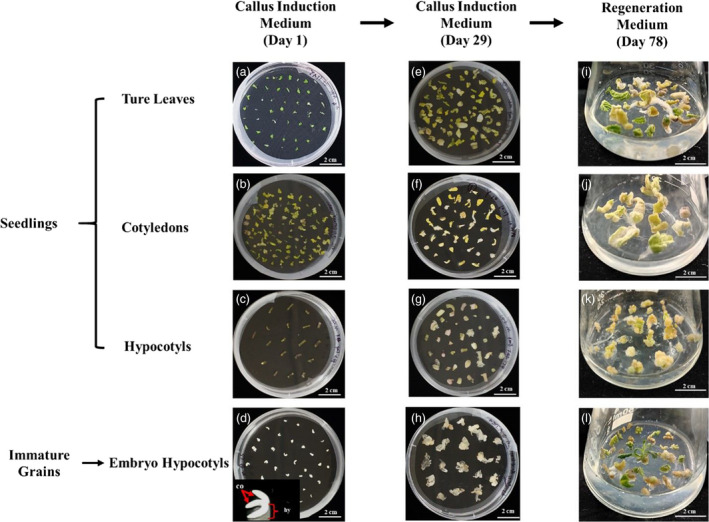
Callus induction and shoot regeneration from different explants of YUNMA7. The developmental stages are described as follows: (a, b and c) Isolated true leaves, cotyledons and hypocotyls from 17‐days‐old seedlings. (d) Embryo hypocotyls dissected from immature grains 15 days after anthesis; co: cotyledon; hy: hypocotyl. (e, f, g and h) Formation of embryogenic calli at the callus induction medium after 29 days of in vitro culture. (i, j, k and l) Development of shoots at the regeneration medium after 78 days of in vitro culture. Scale bars: 2 cm.
Table 1.
Effect of explant on callus induction and regeneration rate of YUNMA7 explants
| n | Responding explants at callus induction stage (%) | Significance | Responding explants at regeneration stage (%) | Significance* | ||
|---|---|---|---|---|---|---|
| Embryo hypocotyls of immature grains | 10 Days after anthesis | 1969 | 17.18 ± 5.28 | b | 5.71 ± 1.06 | ab |
| 15 Days after anthesis | 1561 | 31.08 ± 8.54 | a | 6.12 ± 1.51 | a | |
| 20 Days after anthesis | 1846 | 21.46 ± 6.81 | ab | 5.29 ± 0.85 | b | |
| 25 Days after anthesis | 1227 | 19.77 ± 2.15 | ab | 5.77 ± 1.16 | ab | |
| Seedlings | Ture leaves | 1828 | 5.97 ± 1.56 | c | 2.27 ± 1.29 | c |
| Cotyledons | 1757 | 7.65 ± 2.17 | c | 1.78 ± 0.76 | c | |
| Hypocotyls | 1413 | 5.31 ± 2.38 | c | 1.41 ± 0.47 | c |
Mean of responding explants is denoted as a percentage (±SE) of the total explants.
Different letters (a, b and c) among the levels of each factor indicate the significant differences between them (P < 0.05).
Genotype dependence is one of the major limitations of regeneration and Agrobacterium‐mediated transformation for Cannabis sativa. To increase the robustness of the protocol and evaluate the regeneration differences, we performed a regeneration assay for one hundred cannabis varieties obtained from the national germplasm bank of Institute of Bast Fiber Crops (IBFC), which were planted in the experimental station during the 2020 growing season. The immature grains were collected from spikes approximately 15 days after anthesis because D15 hypocotyls produced more shoots on average, compared with the other three groups (Table 1). After sterilization, their hypocotyls were isolated and incubated in the mediums to estimate the regeneration ability. Significant differences between varieties were observed among all the varieties estimated in this study, DMG278 achieved the highest shoot induction rate of 7.09%, which was selected as the model variety for the Agrobacterium‐mediated transformation (Figure 2). DMG278 was a hemp strains, which was the F2 line bred from the crossing of Red Cherry Berry and YUNMA7. Red Cherry Berry was an indica‐dominant hybrid originating from a cross between Skunk #1 and Californian Indica. YUNMA7 was a commercial hemp strain in China.
Figure 2.
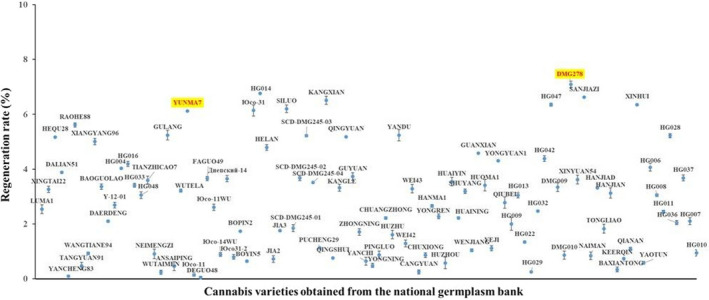
Effect of cannabis varieties on the shoot regeneration rate of immature embryo hypocotyls. For each variety, 90–130 hypocotyls were dissected from immature embryos, cultured in callus induction medium and then transferred to regeneration medium. The regeneration rate was calculated as the percentage of regenerated shoots /total calli for each variety, which were 7.09% for DMG278 and 6.12% for YUNMA7. The two varieties are coloured red and yellow.
To enhance and accelerate the shoot formation, we reprogrammed plant meristems by expressing developmental regulators with positive influences on shoot organogenesis (Debernardi et al., 2020; Kong et al., 2020; Lowe et al., 2018). To compare their effects on regeneration efficiency, we cloned the cDNA sequences of five cannabis candidate genes based on their homology to ZmWUS2, NbSTM, NbIPT, OsGRF4 and AtGIF1 (Figure S2). We designed ten candidate combinations, cloned their cDNA fragments into the pKSE401 binary vector and transformed into Agrobacterium strain AGL1 separately. After that, the D15 hypocotyls isolated from DMG278 immature grains were inoculated with Agrobacterium for 40 min and transferred to callus induction mediums with 100 mg/L Kanamycin. The average regeneration efficiencies for the CsGRF3–CsGIF1 chimera and four chimeras harbouring CsWUS4 were 1.7‐fold higher than for the empty vector control (Figure 3). The average regeneration efficiency of the separate CsSBH1, CsIPT3 and their chimera did not accelerate the shoot regeneration significantly compared to that for the control.
Figure 3.
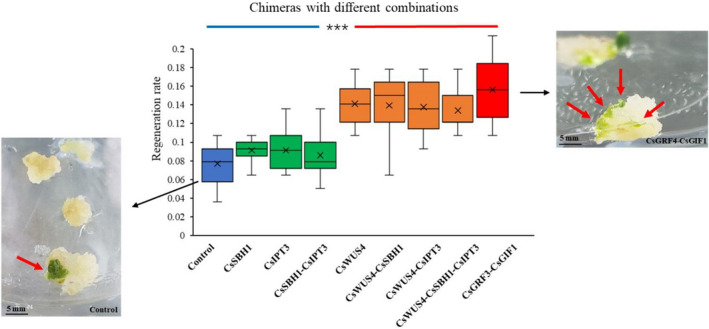
Effect of developmental regulators on shoot regeneration frequency. All the calli used in the transformation experiments were produced from the hypocotyls of immature DMG278 grains (15 days after anthesis). Box plots show the regeneration frequencies for different regulator combinations as well as the control (empty pKSE401 vector). The box is divided by the median with the range from the first to third quartile, and the range of whiskers from the minimum to the maximum values in each treatment (***P < 0.001). The growing green shoots are noted in the callus (red arrows). Control: pKSE401 empty vector.
Application of protoplast technology to estimate gene‐editing efficiency
Gene editing reagents single guide RNA (sgRNA) and Cas9 protein can be synthesized to form active ribonucleoprotein complexes in protoplast and mutagenize the target gene. Thus, a reliable protoplast isolation and transformation protocol can greatly benefit the determination of mutagenesis efficiency. The protoplast validations of targeting mutagenesis mediated by CRISPR/Cas9 were reported in several species, while this strategy remains a bottleneck to cannabis. Protoplast isolation and transfection were performed following Borgato et al. (2007) for Solanaceae since this protocol was successfully applied in several dicot plant species. However, the protoplast yield was much lower compared with descriptions in the paper. To improve the protoplast isolation efficiency, 10 g/L Cellulase RS Onozuka and 2 g/L Macerozyme enzymes were replaced with 12 g/L pectinase and 5 g/L cellulose R10 enzymes, which released more protoplasts in the digestion treatment.
Protoplast quality is important for successful transient transformation. To calculate the efficiency of transformation, pA7‐GFP plasmid was mixed with protoplasts and cultured in the dark at room temperature for 18 h. The number of GFP‐fluorescing cells was checked using a fluorescence microscope (Figure S1). The transfection efficiency in cannabis protoplasts was calculated as the percentage of GFP‐fluorescing cells /intact cells, which was 63% on average (the basic requirement for successful transient transformation was >40% for most plant species).
To demonstrate the genetic transformation system useful to generate genome‐edited plants, six candidates of target sites within CsPDS1 were selected using the online software CHOPCHOP (https://chopchop.cbu.uib.no/). Since the CsGRF3–CsGIF1 chimera resulted in higher regeneration efficiency than other combinations, each of the targeting portions was synthesized as a pair of reverse complementary oligonucleotides and assembled into the genome‐editing vector pG41sg harbouring CsGRF3–CsGIF1 chimera. The constructs expressing gRNA targeting the CsPDS1 gene were co‐transformed into protoplasts and incubated in the dark for 48 h. Protoplast DNA was isolated and the genomic regions of CsPDS1 gene were amplified by PCR and detect the mutations efficiency by deep sequencing (Figure 4). The mutagenesis efficiencies for guide3, guide4 and guide5 were higher compared with efficiencies for guide1, guide2 and guide6. Based on the editing results in protoplast, higher efficiency of targeted mutagenesis can be achieved in Exon 6 of CsPDS1 because the targeting sites of guide3, guide4 and guide5 were all located in this region.
Figure 4.
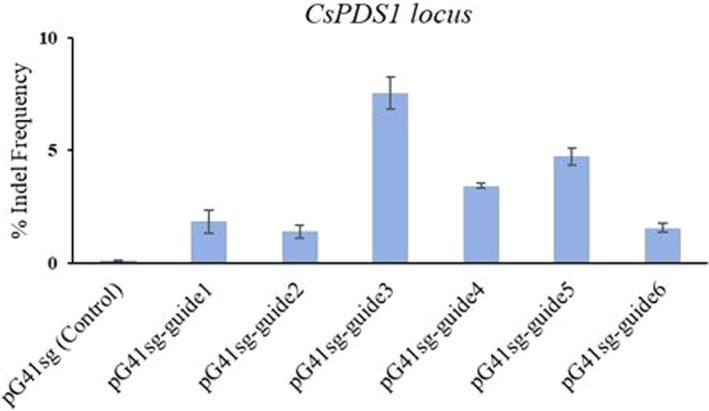
Indel efficiency of pG41sg at the six targeting sites of CsPDS1. The empty pG41sg vector was used as a control. The indel frequency was calculated as the percentage of successful mutations for each target sites, which were 7.55% for guide3, 3.43% for guide4 and 4.72% for guide5.
Characterization of the CsPDS1‐targeted editing in C. sativa transformants
The guide3 sgRNA fused to pG41sg was delivered to the immature embryo hypocotyls (D15) via infection by AGL1. We isolated about 3950 DMG278 hypocotyls for transformation. After a 4‐week incubation in a callus induction medium, a total of 1185 immature hypocotyls developed embryogenic calli. Then we transferred these calli to the regeneration medium containing 50 mg/L kanamycin. A total of 161 calli produced shoots after a 5‐week incubation under continuous light, and the regeneration efficiency of embryogenic calli was 13.59% for DMG278. Of these shoots, 83 had chimeric tissues with the upper shoot having green leaves, whereas the lower shoot displayed white and developmental abnormalities. Only four calli produced white seedlings, approximately 2.48% of the generated shoots and 0.101% of the dissected hypocotyls. All these shoots did not form full plants probably because of the CsPDS1 inactivation, and their vitality was compromised by a lack of chlorophyll (Figure 5a, b and c). Nevertheless, we extracted the genomic DNA from the four white shoots and chimeric shoots including both green and white leaves. Samples were sequenced to identify any insertions or deletions within the targeted region. There were six kinds of deletions observed in the samples (from 4 bp to 17 bp in length), which caused frameshift mutations in CsPDS1 (Figure 5d and e). The six kinds of deletions were found in both white shoots and chimeric shoots, and there was no significant difference in mutagenesis efficiencies between them.
Figure 5.
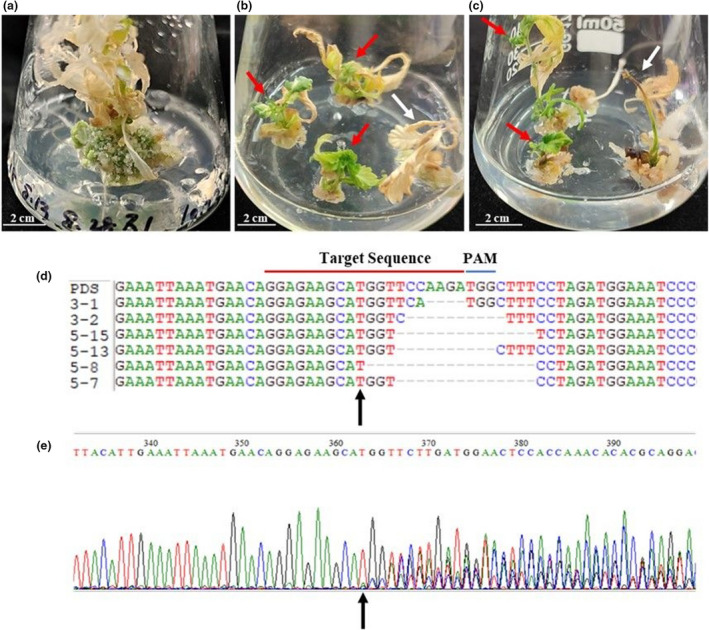
Representative images of gene‐edited shoots and sequencing results of indels at the desired place. (a) A gene‐edited shoot germinated from the callus that displayed photobleaching. (b and c) Chimeric tissues with the upper shoot green and lower shoot white (red arrows) as well as white shoots in developmental abnormalities (white arrows). (d) and (e) Sequencing results of indels at the desired place and their chromatograms. The black arrows indicate the mutations. The red line denotes the guide3‐sgRNA target sequence. The blue line denotes the protospacer adjacent motif (PAM) sequence serves as a binding signal for Cas9.
When estimating different developmental regulators on shoot organogenesis, pG41sg empty vector was delivered to the immature embryo hypocotyls via infection by AGL1. The first fully expanded leaf was sampled from each of the regenerated shoots after a 6‐week incubation and identified with the primers (AtU6‐F1/R1) specific to the T‐DNA sequence of pG41sg. We finally obtained one transgenic seedling G41‐1 carrying the T‐DNA fragment. To propagate more seedlings and confirm whether the pG41sg T‐DNA has been inserted into the G41‐1 genome stably, the G41‐1 stem was cut into pieces and incubated in the regeneration medium containing kanamycin for 6 weeks. There were five shoots germinated from the stem explants of G41‐1 (Figure 6a), which were transferred to the root‐induction medium and then planted in soil condition. The first fully expanded leaves were sampled every three weeks for the transgenic screening (Figure 6b). Meanwhile, eleven chimeric seedlings which contained mutagenesis at CsPDS1 were randomly selected and transferred to the soil after incubation in the root‐induction medium. Their first fully expanded leaves were sampled for screening too. In the second round of screening, the eleven chimeric plants were identified as non‐transgenic plants based on the PCR result (Figure 6c), which means these plants have lost the T‐DNA fragment in new leaves. However, the five shoots germinated from G41‐1 were identified as transgenic based on transgenic‐specific PCR results with two pairs of primers AtU6‐F1/R1 and CsCAS9F2/R2 (Figure 6d and e), which confirmed the stable T‐DNA insertion within the cannabis genome.
Figure 6.
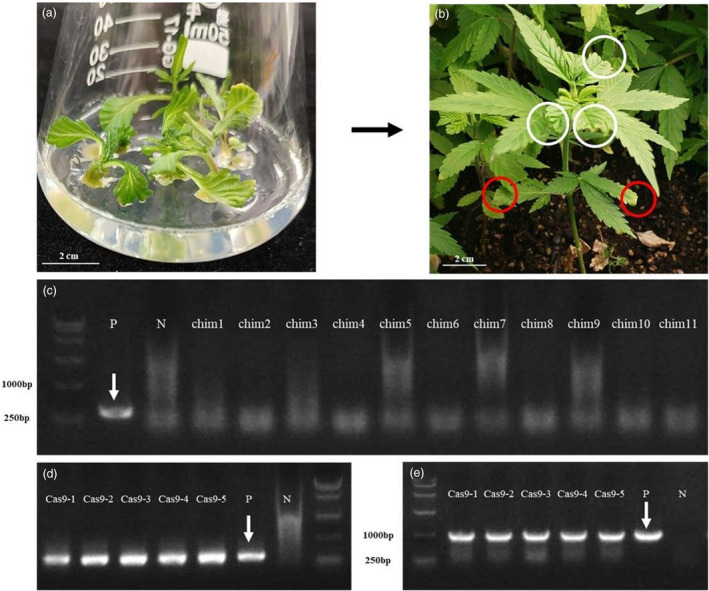
Transgenic cannabis seedlings and results of transgenic screening. (a) Shoots regenerated from stems of the transgenic seedling. In the estimation of developmental regulator effects on shoot organogenesis, we obtained one transgenic seedling G41‐1 carrying the pG41sg T‐DNA fragment. Then G41‐1 stem was cut into pieces and incubated in the regeneration medium containing kanamycin for 6 weeks. There are five shoots germinated from the stem explant. (b) A transgenic seedling regenerated from the G41‐1 stem. All the five shoots were transferred to soil after a 5‐week incubation in the root‐induction medium. The first fully expanded leaves were sampled every three weeks when growing in greenhouse. Red circle: sampling in the first round of screening; white circle: sampling in the second round of screening. (c) Transgenic‐specific PCR result of the chimeric plants containing mutagenesis at CsPDS1. Eleven chimeric seedlings (chim1‐11) were randomly selected and transferred to soil after incubation in the root‐induction medium. Since their first fully expanded leaves lost the T‐DNA fragment, these plants were identified as no transgenic with primers AtU6‐F1/R1 in the second round of screening. P: DNA sample of pG41sg, N: DNA sample of no transgenic plant; white arrows: specific PCR product. (d) and (e) Transgenic‐specific PCR results of the five seedlings regenerated from G41‐1. In the second round of screening, these plants (Cas9‐1 to Cas 9‐5) were identified as transgenic plants based on transgenic‐specific PCR results amplified with primers AtU6‐F1/R1 and CsCAS9F2/R2.
Discussion
Tissue culture has been widely used to advance plant research by transgenesis and gene‐editing technologies (Azeez and Busov, 2021; Li et al., 2020; Liu et al., 2019). Although successful regeneration of transformed seedlings has been reported in some species, it is still the bottleneck for the C. sativa to produce transgenic or gene‐edited plants. Only the cannabis hairy roots were verified to be susceptible to the infection with A. rhizogenes, which induced transformed root cultures in a short time (Wahby et al., 2013). Until recently, the transient GUS expression and PDS gene silencing were detected in cannabis tissues using agrobacterium or agroinfiltration mediated transformation (Deguchi et al., 2020; Sorokin et al., 2020). A nanoparticle‐based method was used to overexpress multiple transgenes in the transient transformation of Cannabis, as an effective carrier to introduce foreign DNA fragments simultaneously (Ahmed et al., 2021). Although transient expression analysis can be applied to study the gene function, stable transformation protocols are still the foundation to enhance cannabis phenotypes through genetic engineering.
Calli derived from mature plants or immature embryos were two main sources for Agrobacterium‐mediated transformation in plant species (Debernardi et al., 2020; Kong et al., 2020). Cannabis explants from seedlings or plants have already been demonstrated to be less susceptible to transformation, which can only be used for transient expression (Deguchi et al., 2020; Sorokin et al., 2020). Our focus turned to the calli developed from immature embryos, since embryogenic calli provided a large number of embryogenic competent cells that were extremely susceptible to transformation by Agrobacterium (Leelavathi et al., 2004). In our protocol, an additional step of 4‐week incubation in callus induction was applied to explants after infection treatment. To estimate the effect of explant types on embryogenic calli induction, the embryo hypocotyls of immature grains were collected approximately 10 days, 15 days, 20 days and 25 days after anthesis. True leaves, cotyledons and hypocotyls from seedlings were dissected and cultured as control since these explants showed high regeneration ability in the previous reports (Chaohua et al., 2016; Galán‐Ávila et al., 2020). In the pre‐experiment, we also estimated the callus induction rate of immature cotyledons, the regeneration rate was lower than the seedling explants, the relevant data was not mentioned here. After a 4‐week incubation, hypocotyls collected 15 days after anthesis (D15) produced more calli as well as more shoots compared to other explants.
Since the shoot regeneration efficiency also varied greatly among different cannabis varieties (Sorokin et al., 2020), we estimated the genotype effect of one hundred cannabis varieties obtained from the national germplasm bank. These varieties were planted in field conditions, and their D15 hypocotyls were dissected to compare the regeneration differences. In Figure 2, significant differences can be observed between the varieties evaluated. DMG278 achieved the highest shoot induction rate of 7.09%, which was higher than for YUNMA7 (6.12%). Based on the effect estimation of different explant types and cannabis varieties, D15 hypocotyls from DMG278 were selected as the model explant for the Agrobacterium‐mediated transformation.
For plant species recalcitrant to regeneration, it has been observed that overexpression of developmental regulators showed positive influences on shoot organogenesis (Debernardi et al., 2020; Kong et al., 2020; Lowe et al., 2018). To increase the regeneration efficiency of cannabis calli, we cloned the cannabis candidate genes homologous to ZmWUS2, NbSTM, NbIPT, OsGRF4 and AtGIF1, which were effective in several monocot and dicot plant species (Debernardi et al., 2020; Kong et al., 2020; Lowe et al., 2018), designed ten combinations and delivered these chimeras into D15 hypocotyls. The average regeneration efficiencies for the CsGRF3–CsGIF1 chimera and four chimeras harbouring CsWUS4 were 1.7‐fold higher than that for the empty vector control, while no significant differences were observed between CsSBH1, CsIPT3, CsSBH1‐CsIPT3 and control. The CsGRF3–CsGIF1 chimera performed the best among all the regulators, with the regeneration efficiency of 13.43% compared to 7.88% in the control. To create more editing events, we assessed the mutagenesis efficiency of the six targeted regions designed for CsPDS1 by transient expression in the cannabis protoplasts. The mutagenesis efficiency for guide3 was 7.55%, significantly higher than that of the other regions.
In this study, we increased the regeneration efficiency by optimizing the explant, variety, developmental regulator and gene‐editing tool. There were finally four calli generated white seedlings with CsPDS1 edited, which account for 2.48% of the generated shoots. Most of the transgenic shoots were chimeric tissues, that lost the T‐DNA fragment of pG41sg in new leaves when growing in the greenhouse two months later. Fortunately, we obtained one transgenic plant carrying the T‐DNA fragment when estimating the CsGRF3–CsGIF1 chimera effect on regeneration frequency. To confirm whether the pG41sg T‐DNA has been inserted into the plant genome stably, we propagated the transgenic plant by direct in vitro regeneration from the stem explants. Two pairs of primers were prepared identifying the transgenic plants, which were designed to amplify the regions near the two ends of the T‐DNA insertion. The T‐DNA was identified as integrating with the genome by these specific primers in five regenerated seedlings, which were planted in greenhouse. After 6 weeks, the T‐DNA can still be identified by PCR from the new leaves on the top, conforming to the stable T‐DNA insertion that can be transmitted into progeny.
In conclusion, this study constitutes the first report of successful gene editing as well as stable transformation in C. sativa. By optimizing the explant, variety, developmental regulator and gene‐editing tool, we developed a protocol for Agrobacterium‐mediated transformation in C. sativa. This method therefore can play an important role in functional genomics of cannabis genes and release the real potential of gene editing in the production of improved cannabis varieties.
Methods
Plant material and growth conditions
One hundred cannabis varieties were analyzed in this study, which were obtained from the materials stored in the national germplasm bank of Institute of Bast Fiber Crops (IBFC), Chinese Academy of Agriculture Sciences. Most of these varieties were collected from 21 provinces of China, and 19 commercial cultivars from US, Canada, Germany, France, Russia, Ukraine, Poland, Morocco, Netherlands, and Slovenia. Field experiments were conducted during the growing season in 2020 at the Changsha Agro‐Eco‐Experimental Station of the IBFC, located in the south‐central part of China (28°20’ N latitude, 112°69’ E longitude, 63 m elevation). All the varieties were randomly sown in the plots (24 m2 for each pot), with four replications. Immature grains were collected from spikes approximately 10 days, 15 days, 20 days and 25 days after anthesis, sterilized for 25 min in the solution of 2% (vol/vol) sodium hypochlorite followed by 5 min in 75% (vol/vol) ethanol, and washed three times with the sterilized water. Immature embryo hypocotyls (about 0.6 cm in length) were isolated with scalpels and collected for transformation. Mature seeds from hemp strain YUNMA7, a widely planted cultivar in YUNNAN (China), were surface sterilized and germinated in plastic petri dishes containing the autoclaved Murashige & Skoog (MS) medium. True leaves, cotyledons and hypocotyls were dissected from 17‐days‐old seedlings and employed as explants. The recipes of all the mediums mentioned in regeneration and transformation were listed in Table S1.
Agrobacterium strains and binary plasmids
Agrobacterium tumefaciens strain (AGL1) carrying a nopaline Ti plasmid pTiBo542DT was used in our study (Biomed ®, Beijing, China). The coding regions of CsWUS4, CsSBH1, CsIPT3, CsGRF3 and CsGIF1 were amplified using cDNA generated from YUNMA7 leaves (Figure S2 and Table S2). To develop the vectors coexpressing developmental regulators (DRs) and genome‐editing components, PCR fragments were cloned into pDONR vector with the MultiSite Gateway Pro Plus Kit (Invitrogen, Carlsbad, CA) and overlapped to generate the chimeras. The CaMV 35S promoter and NOS terminator for controlling the gene expression were cloned and assembled before and after the developmental regulators by In‐fusion (Takara Bio USA) into the pKSE401 binary vector from the laboratory of Dr. Chen (China Agricultural University), which contains a codon‐optimized Cas9 and gRNA scaffold for insertion of target sequence (Addgene, 6220). These modified pKSE401vectors were transformed into AGL1 to evaluate their effect on shoot regeneration of different cannabis cultivars. After validating the regeneration rate in different vector treatments, we assembled the gRNA construct targeting the coding region of CsPDS1 gene into the modified vector providing the best performance in shoot regeneration.
Cloning of CsPDS1 fragment and selection of CRISPR/Cas9 target sites
Genomic DNA was extracted from seedlings according to a modified CTAB method (Dvorak et al., 1988). Primers PDS‐F6 and PDS‐R6 were designed to amplify the fragments of CsPDS1 (Table S2). The PCR amplification was completed with TIANGEN DNA polymerase (Beijing, China) in a total volume of 50 μL, with the reaction conditions: 94 °C for 5 min; 35 cycles of 94 °C for 10 s, 58 °C for 10 s, and 72 °C for 50 s; and 72 °C for 10 min. The PCR products were purified with the Gel Extraction Kit (TIANGEN Scientific, China), and then analyzed by Sanger sequencing (Sangon Biotech, China). All possible CRISPR target sites within CsPDS1 were identified with the online software CHOPCHOP (https://chopchop.cbu.uib.no/). Six candidates of CRISPR target sites were selected based on the genomic location and potential off‐target scores. Each of the targeting portions was synthesized as a pair of reverse complementary oligonucleotides, assembled into the CsGRF3–CsGIF1–pKSE401 (pG41sg) vector by GoldenGate reaction assembly (NEB, Hitchin, UK). These constructs were transformed into Agrobacterium strain AGL1 separately.
Assessment of the mutagenesis efficiency
Protoplast isolation and transfection were performed following Borgato et al. (2007) for Solanaceae. The protoplasts were transferred into plates, which were precoated with 1% BSA solution. To calculate the efficiency of transformation, five micrograms of pA7‐GFP plasmid (Katrin Czempinski, Potsdam University, Germany) in 15 μL volume was mixed with 50 μL of protoplasts gently. Protoplasts were cultured in the dark at room temperature for 18 h. The GFP signal was estimated using the Leica DMI8 inverted fluorescent microscope with the excitation‐wavelength 488 nm. The pG41sg constructs expressing gRNA targeting the CsPDS1 gene were co‐transformed into protoplasts and incubated in the dark for 48 h. For DNA isolation, protoplasts were pooled by centrifugation and extracted using a Plant Genomic DNA Extraction Kit (TIANGEN Scientific, China). Genomic regions of CsPDS1 gene harbouring the gRNA targets were PCR amplified and detect the efficiency of mutation by deep sequencing which was performed following Kang et al. (2018).
Agrobacterium‐mediated plant transformation
Agrobacterium strain containing the DR‐modified vector was used to infect the explants. Single colony of AGL1 was selected and grown in YEP liquid media containing 50 mg/L kanamycin and 20 mg/L rifampicin in an incubator shaker 200 rpm overnight at 28°C. A. tumefaciens culture was centrifuged at 3500 g for 30 min at room temperature. The supernatant was removed and the pellet was resuspended using MS media with 7.5 mm MgCl2. The culture was centrifuged at 2000 x g and resuspended using liquid MS media with 5 mm MgSO4 (pH = 5.4) to reach the final OD600 = 0.1. The explants were infected for 40 min and then transferred to the MS medium. After 3 days of cultivation, the explants were rinsed in sterile water three times and transferred to the selection medium which was renewed every two weeks. Kanamycin‐resistant shoots were cut and transferred to the root‐induction medium.
Molecular analysis and detection of mutations
Two pairs of primers (AtU6‐F1/R1 and CsCAS9F2/R2) were used to identify transgenic plants containing the T‐DNA fragment fused into the genome (Table S2). The positive plants were deep sequenced to identify any insertions or deletions within the targeted region. The phenotypes of the transgenic plants were photographed and described in the results.
Data analysis
Factors evaluated in this study were explant, variety and developmental regulator. Each factor was analyzed using eight biological replicates at least, and each biological replicate consisted explants from 10 to 15 different seedlings of the same variety. To evaluate significant differences between factors, Kruskal–Wallis non‐parametric test and pairwise Wilcoxon test (P < 0.01) were used. Statistical analysis was carried out using the SPSS statistical package (version 16.0; SPSS Inc., Chicago, IL).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
X.Z. designed the experiments, analyzed the results. X.Z. and L.L. wrote the manuscript. G.X. and C. C. participated in most of the experiments under X.Z.’s direction. The project was conceived, planned and supervised by Q.T. and J.S. Other authors helped with the field work. All authors discussed the results and provided feedback on the manuscript.
Supporting information
Table S1 Recipes.
Table S2 Primers used in this study.
Figure S1 PEG‐mediated transformation of cannabis protoplasts.
Figure S2 cDNA sequences of the developmental regulators (CsWUS4, CsSBH1, CsIPT3, CsGRF3 and CsGIF1) and CsPDS1.
Acknowledgements
We appreciate Dr. Katrin Czempinski (Potsdam University, Germany) for providing the pA7‐GFP plasmid. This research was supported by grants from the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Science [grant number ASTIP‐IBFSC01] and Basic Research Foundation of IBFC.
Zhang, X. , Xu, G. , Cheng, C. , Lei, L. , Sun, J. , Xu, Y. , Deng, C. , Dai, Z. , Yang, Z. , Chen, X. , Liu, C. , Tang, Q. and Su, J. (2021) Establishment of an Agrobacterium‐mediated genetic transformation and CRISPR/Cas9‐mediated targeted mutagenesis in Hemp (Cannabis Sativa L.). Plant Biotechnol J, 10.1111/pbi.13611
†These authors contributed equally to this article.
Contributor Information
Qing Tang, Email: sujianguang@caas.cn.
Jianguang Su, Email: zhangxiaoyuyyy@gmail.com.
References
- Ahmed, S. , Gao, X. , Jahan, M.A. , Adams, M. , Wu, N. and Kovinich, N. (2021) Nanoparticle‐based genetic transformation of Cannabis sativa . J. Biotechnol. 326, 48–51. [DOI] [PubMed] [Google Scholar]
- Aliekperova, N. , Kosyachenko, К. and Kaniura, O. (2020) Perspectives on formation of medical cannabis market in Ukraine based on holistic approach. J. Cannabis Res. 2, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeez, A. and Busov, V. (2021) CRISPR/Cas9‐mediated single and biallelic knockout of poplar STERILE APETALA (PopSAP) leads to complete reproductive sterility. Plant Biotechnol. J. 19, 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgato, L. , Pisani, F. and Furini, A. (2007) Plant regeneration from leaf protoplasts of Solanum virginianum L. (Solanaceae). Plant Cell Tissue Organ Cult. 88, 247–252. [Google Scholar]
- Cheng, C. , Zang, G. , Zhao, L. , Gao, C. , Tang, Q. , Chen, J. , Guo, X. et al. (2016) A rapid shoot regeneration protocol from the cotyledons of hemp (Cannabis sativa L.). Ind. Crops Prod. 83, 61–65. [Google Scholar]
- Debernardi, J.M. , Tricoli, D.M. , Ercoli, M.F. , Hayta, S. , Ronald, P. , Palatnik, J.F. and Dubcovsky, J. (2020) A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi, M. , Bogush, D. , Weeden, H. , Spuhler, Z. , Potlakayala, S. , Kondo, T. , ZhanyuanJ, Z. et al. (2020) Establishment and optimization of a hemp (Cannabis sativa L.) agroinfiltration system for gene expression and silencing studies. Sci. Rep. 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, P. , Ni, S. , Wang, J. , Zhang, B. , Xu, R. , Wang, Y. , Chen, H. et al. (2015) Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants, 2, 1–5. [DOI] [PubMed] [Google Scholar]
- Dvorak, J. , McGuire, P.E. and Cassidy, B. (1988) Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome, 30, 680–689. [Google Scholar]
- Galán‐Ávila, A. , García‐Fortea, E. , Prohens, J. and Herraiz, F.J. (2020) Development of a direct in vitro plant regeneration protocol from Cannabis sativa L. seedling explants: developmental morphology of shoot regeneration and ploidy level of regenerated plants. Front. Plant Sci. 11, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.C. , Yun, J.Y. , Kim, S.T. , Shin, Y. , Ryu, J. , Choi, M. , Woo, J.W. et al. (2018) Precision genome engineering through adenine base editing in plants. Nat. Plants, 4, 427–431. [DOI] [PubMed] [Google Scholar]
- Kong, J. , Martin‐Ortigosa, S. , Finer, J. , Orchard, N. , Gunadi, A. , Batts, L.A. , Thakare, D. et al. (2020) Overexpression of the transcription factor GROWTH‐REGULATING FACTOR5 improves transformation of dicot and monocot species. Front. Plant Sci. 11, 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelavathi, S. , Sunnichan, V.G. , Kumria, R. , Vijaykanth, G.P. , Bhatnagar, R.K. and Reddy, V.S. (2004) A simple and rapid Agrobacterium‐mediated transformation protocol for cotton (Gossypium hirsutum L.): embryogenic calli as a source to generate large numbers of transgenic plants. Plant Cell Rep. 22, 465–470. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Wu, G. , Zhao, Y. , Wang, B. , Zhao, B. , Kong, D. and Wei, H. (2020) CRISPR/Cas9‐mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height. Plant Biotechnol. J. 18, 2520–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Wang, Y. , Xu, S. , Tang, X. , Zhao, J. , Yu, C. , Guo, H. et al. (2019) Efficient genetic transformation and CRISPR/Cas9‐mediated genome editing in Lemna aequinoctialis . Plant Biotechnol. J. 17, 2143–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Zhu, P. , Cai, S. , Haughn, G. and Page, J.E. (2021) Three novel transcription factors involved in cannabinoid biosynthesis in Cannabis sativa L. Plant Mol. Biol. 2021, 1–17. [DOI] [PubMed] [Google Scholar]
- Lowe, K. , La Rota, M. , Hoerster, G. , Hastings, C. , Wang, N. , Chamberlin, M. , Wu, E. et al. (2018) Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev. Biol. Plant, 54, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, G. and Palmgren, M. (2020) GRF‐GIF chimeras boost plant regeneration. Trends Plant Sci. 26, 201–204. [DOI] [PubMed] [Google Scholar]
- Maher, M.F. , Nasti, R.A. , Vollbrecht, M. , Starker, C.G. , Clark, M.D. and Voytas, D.F. (2020) Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, E.B. , Jiang, H.E. , Li, X. , Sutton, A. , Carboni, A. , Del Bianco, F. , Mandolino, G. et al. (2008) Phytochemical and genetic analyses of ancient cannabis from Central Asia. J. Exp. Bot. 59, 4171–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano, S. , Hibara, K.I. , Furuya, T. , Arimura, S.I. , Tsukaya, H. and Itoh, J.I. (2018) Conserved functional control, but distinct regulation, of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF‐INTERACTING FACTOR 1 orthologs. Development, 145, 7. [DOI] [PubMed] [Google Scholar]
- Sorokin, A. , Yadav, N.S. , Gaudet, D. and Kovalchuk, I. (2020) Transient expression of the β‐glucuronidase gene in Cannabis sativa varieties. Plant Signal Behav. 15, 1780037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahby, I. , Caba, J.M. and Ligero, F. (2013) Agrobacterium infection of hemp (Cannabis sativa L.): establishment of hairy root cultures. J. Plant Interact. 8, 312–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Recipes.
Table S2 Primers used in this study.
Figure S1 PEG‐mediated transformation of cannabis protoplasts.
Figure S2 cDNA sequences of the developmental regulators (CsWUS4, CsSBH1, CsIPT3, CsGRF3 and CsGIF1) and CsPDS1.


