Abstract
RNase MRP is a ribonucleoprotein endoribonuclease that has been shown to have roles in both mitochondrial DNA replication and nuclear 5.8S rRNA processing. SNM1 encodes an essential 22.5-kDa protein that is a component of yeast RNase MRP. It is an RNA binding protein that binds the MRP RNA specifically. This 198-amino-acid protein can be divided into three structural regions: a potential leucine zipper near the amino terminus, a binuclear zinc cluster in the middle region, and a serine- and lysine-rich region near the carboxy terminus. We have performed PCR mutagenesis of the SNM1 gene to produce 17 mutants that have a conditional phenotype for growth at different temperatures. Yeast strains carrying any of these mutations as the only copy of snm1 display an rRNA processing defect identical to that in MRP RNA mutants. We have characterized these mutant proteins for RNase MRP function by examining 5.8S rRNA processing, MRP RNA binding in vivo, and the stability of the RNase MRP RNA. The results indicate two separate functional domains of the protein, one responsible for binding the MRP RNA and a second that promotes substrate cleavage. The Snm1 protein appears not to be required for the stability of the MRP RNA, but very low levels of the protein are required for processing of the 5.8S rRNA. Surprisingly, a large number of conditional mutations that resulted from nonsense and frameshift mutations throughout the coding regions were identified. The most severe of these was a frameshift at amino acid 7. These mutations were found to be undergoing translational suppression, resulting in a small amount of full-length Snm1 protein. This small amount of Snm1 protein was sufficient to maintain enough RNase MRP activity to support viability. Translational suppression was accomplished in two ways. First, CEN plasmid missegregation leads to plasmid amplification, which in turn leads to SNM1 mRNA overexpression. Translational suppression of a small amount of the superabundant SNM1 mRNA results in sufficient Snm1 protein to support viability. CEN plasmid missegregation is believed to be the result of a prolonged telophase arrest that has been recently identified in RNase MRP mutants. Either the SNM1 gene is inherently susceptible to translational suppression or extremely small amounts of Snm1 protein are sufficient to maintain essential levels of MRP activity.
RNase MRP was first identified in mouse cells as an endoribonuclease involved in mitochondrial DNA replication (4–6). It cleaves RNA transcripts from mitochondrial origins of replication in a manner consistent with the formation of the primer for the initiation of DNA synthesis (23–25, 60), hence, the name MRP, for mitochondrial RNA processing. Like the mammalian RNase MRP, the yeast enzyme cleaves an RNA substrate complementary to a yeast mitochondrial origin of replication at an exact site of RNA-to-DNA linkage (2, 52, 57). However, the majority of RNase MRP is localized in the nucleus (20, 26, 39), where it directly processes the rRNA precursor at the A3 site (7, 9, 28, 44). The loss of RNase MRP activity leads to the loss of the production of one of two different species of 5.8S rRNA (41, 44). Some question has arisen as to whether the loss of processing of the rRNAs is the essential function of RNase MRP (28, 32). Recent work has identified a late cell cycle arrest in RNase MRP mutants, and it is some function in the control of the cell cycle that may be responsible for the essentiality of RNase MRP (2a).
RNase MRP is a ribonucleoprotein complex with a single RNA component and several protein subunits (3, 8, 10, 29, 43, 45). The 340-nucleotide (nt) RNA component of RNase MRP is encoded by an essential nuclear gene, NME1 (43). RNase MRP shares many functional and structural similarities with another ribonucleoprotein complex, RNase P, which is involved in pre-tRNA processing (14, 37, 38, 46). Both ribonucleoproteins have sequence-specific endoribonuclease activity and have functions in both the mitochondria and the nucleus. The RNA components of both complexes can be folded into similar secondary structures (14, 46). In the yeast Saccharomyces cerevisiae, RNase P and RNase MRP share several protein components: Pop1p (100.5 kDa), Pop3p (22.6 kDa), Pop4p (32.8 kDa), Pop5p (19.6 kDa), Pop6p (18.2 kDa), Pop7p (15.8 kDa), Pop8p (15.5 kDa), and Rpp1p (32.2 kDa) (3, 8, 10, 29, 53). Pop1p, Pop3p, and Pop4p were identified by genetic screening. Rpp1p was identified as a protein subunit of RNase P by virtue of its homology with a human scleroderma autoimmune antigen, Rpp30, which copurifies with human RNase P. Yeast RNase P has been purified biochemically to apparent homogeneity. Four more proteins that purify with RNase P activity, Pop5p (19.6 kDa), Pop6p (18.2 kDa), Pop7p/Rpp2p (15.8 kDa), and Pop8p (15.5 kDa), have been demonstrated to be shared by RNase P and RNase MRP (3).
Snm1p, the only protein component unique to the RNase MRP complex (not shared with RNase P), was identified as a high-copy-number suppressor of a temperature-sensitive nme1 mutant (45). SNM1 at a high copy number can fully complement the rRNA-processing defect of this nme1 mutant at the nonpermissive temperature. Snm1p binds the RNase MRP RNA but not the RNase P RNA. The SNM1 gene encodes an essential protein of 198 amino acids with a predicted molecular mass of 22.5 kDa. The primary sequence reveals three possible structural motifs. The amino-terminal portion of the protein contains a series of leucines spaced 6 to 8 amino acids apart that could potentially form a coiled-coil domain. The central portion of the protein contains two cysteine clusters that may function in binding zinc ions. Similar cysteine clusters can be found in a binuclear cluster arrangement seen in DNA-binding proteins (31, 54). The carboxy-terminal portion of the protein is rich in serines and lysines. Of the last 63 amino acids, 15 are serines and 16 are lysines. There is no obvious RNA binding motif in Snm1p, although it was found to specifically bind the MRP RNA (45). In order to learn more about the function of this protein that is exclusive to the RNase MRP enzyme complex, we undertook mutagenesis screening of the SNM1 gene. Here we report the isolation and characterization of a number of temperature-conditional SNM1 mutants. These mutants have led us to find a previously undiscovered role for the yeast RNase MRP in chromosomal segregation.
MATERIALS AND METHODS
Strains and media.
Yeast media and genetic manipulations have been described elsewhere (48). The Escherichia coli strain used for cloning, DH5α, has the genotype psi80dlacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 λ− gyrA96 relA1 Δ(lacZYA-argF)U169 F−. Basic molecular biology techniques were performed as described elsewhere (42).
PCR mutagenesis of SNM1.
PCR mutagenesis was performed as described by Muhlrad et al. (34). Plasmid DNA carrying a full-length SNM1 gene was used as a template. The PCR mixtures (total reaction volume, 100 μl) contained 1 μg of DNA template (pMES193 [pBS containing SNM1]) (45); Vent DNA polymerase buffer (New England Biolabs, Beverly, Mass.); either 1.0 mM each dGTP, dCTP, and dTTP and 0.2 mM dATP or 1.0 mM each dATP, dCTP, and dTTP and 0.2 mM dGTP; 3 U of Deep VentR (exo−) DNA polymerase (New England Biolabs); 0.5 mM MnCl2; and 1 μg each of primers O-SNM-2 (5′-CTG CAG GCA AGG CGT ACA TGG CGT G-3′) and O-SNM-5 (5′-GAT GAG TAC TAC TAG TGG CTA-3′). Thirty cycles were completed at an annealing temperature of 40°C. PCR products of the correct size (∼845 bp) were purified from agarose gels by filter centrifugation.
Plasmid pTHR100 is a derivative of plasmid pMES194 (45). To remove BamHI and SpeI from the polylinker of pMES194, the plasmid was digested with SmaI and XbaI, and the ends were filled in with the Klenow fragment and ligated together to generate plasmid pTHR100. Plasmid pTHR100 (pRS315 containing LEU2, CEN, and SNM1) was used to remove from between the BamHI and NsiI sites a 696-bp fragment which contained 100% of the SNM1 open reading frame (45). The purified mutagenized snm1 PCR product and the gap-containing pTHR100 plasmid were cotransformed into the haploid strain THR200 (MATa ade2-1 leu2-3,112 his3-Δ200 trp1-Δ1 ura3-52 snm1-Δ1::HIS3 pTHR101 [URA3 CEN SNM1]) for in vivo gap repair of pTHR100 (11). Transformants were selected on SD plates without leucine (0.67% yeast nitrogen base containing ammonium sulfate, 2% dextrose, and 2% Bacto-Agar and supplemented with 20 mg each of adenine, uracil, and tryptophan per liter) and then transferred to SCD-5FOA plates (0.67% yeast nitrogen base containing ammonium sulfate, 2% dextrose, 0.5% Casamino Acids, and 2% Bacto-Agar and supplemented with 20 mg each of adenine, uracil, tryptophan, and 0.2% 5-fluoro-orotic acid [5-FOA] per liter). Two transfers to 5-FOA-containing plates were performed to ensure that the URA3-containing plasmid was lost (50). Transformants were then replicated to SCD plates without 5-FOA at 17, 24, 30, or 37°C to screen for mutants.
Growth testing of yeast mutants.
Mutant yeast strains were analyzed for temperature-conditional growth on SCD plates at 17, 24, 30, and 37°C. One hundred microliters of sterile water was distributed into the wells of a 96-well Falcon tissue culture plate. Two fresh colonies of each mutant yeast strain were mixed into the first-row wells. Tenfold serial dilutions of these cell mixtures were placed into the second-, third-, and fourth-row wells. A 48-pin Frogger (Dan-kar Corp.) was used to transfer diluted cell mixtures to SCD plates. The cells were grown at the temperatures specified for 2 days or at 17°C for 5 days.
Analysis of yeast RNA.
RNA was extracted as previously described (47). Approximately 10 μg of whole-cell RNA was separated on 6% acrylamide–7 M urea gels. The gels were stained with ethidium bromide to visualize the 5.8S rRNA, or Northern blot analysis was performed (44). The probes used were SNM1 (a 705-bp BamHI-NsiI fragment of the SNM1 gene), NME1 (a 1,042-bp EcoRI fragment), and SCR1 (a 736-bp EcoRI-SpeI fragment) (20, 21, 39). Probes were radiolabeled for hybridization with [α-32P]dCTP by use of the Prime-It Kit (Stratagene, Inc.). Radioactive blots were analyzed on a Molecular Dynamics PhosphorImager.
Preparation of yeast cell extracts.
Yeast cells were grown in 50 ml of liquid SCD at 30°C until they reached exponential growth (107 cells/ml). The cultures were then shifted to the nonpermissive temperature and grown for 6 h. The cells were collected, washed with 1 ml of buffer A (20 mM Tris-HCl [pH 8], 150 mM KCl, 5 mM EDTA, 0.1% Triton X-100, 10% glycerol, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]), pelleted in a microcentrifuge, and resuspended in 400 μl of buffer A. Two hundred milligrams of glass beads was added to the cell mixture, which was then vortexed for 15 min at 4°C and centrifuged at 16,000 × g for 2 min at 4°C. The supernatant was transferred to a clean tube and centrifuged at 16,000 × g for an additional 15 min. Three hundred microliters of the supernatant was collected, and the protein concentration was determined (30).
Immunoprecipitation by anti-Snm1p antibody.
Immunoprecipitation was performed as described before (45). Fifty microliters of protein A-Sepharose CL4B beads (200 mg/ml) was incubated with 3 μl of rabbit serum containing anti-Snm1p antibody for 1 h at 4°C, and the mixture was washed three times with buffer A. The bead volume was brought up to 1.25 ml with buffer A, and the mixture was incubated with 375 μg of yeast extract for 1 h at 4°C. The samples were washed four times with 1 ml of buffer A, the volume of each sample was brought up to 100 μl with buffer A, 100 μl of 5% sodium dodecyl sulfate (SDS)–10% β-mercaptoethanol was added, and the samples were heated to 95°C for 5 min. After the beads were removed by centrifugation, the RNA was extracted with phenol-chloroform equilibrated to pH 5.3, precipitated with ethanol, and examined by Northern analysis.
Affinity purification of anti-Snm1p antibody.
Snm1p was expressed in E. coli BL21(DE3) cells by use of a T7 promoter system as described before (45). After induction with isopropyl-β-d-thiogalactopyranoside, (IPTG), cells were pelleted and washed with lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM KCl, 5 mM EDTA, 1 mM DTT, 1 mM PMSF). Cells were then sonicated and washed with lysis buffer containing 0.5% Triton X-100. Inclusion bodies were pelleted, solubilized in 6 M guanidine chloride, and diluted in 50 mM Tris-HCl–0.5 mM DTT–10 μM ZnCl2; about 60% of Snm1p was soluble. Snm1p was concentrated by use of an Amicon concentrator with a molecular mass cutoff of 10,000 Da. Approximately 10 mg of Snm1p was coupled to 6 ml of Affi-Gel 10 (Bio-Rad Corp., Hercules, Calif.) according to the instructions of the manufacturer, and 25 ml of serum was loaded on an Snm1p column and washed with 25 column volumes of 10 mM Tris (pH 7.5) and then with 25 column volumes of 10 mM Tris (pH 7.5)–0.5 M NaCl. Antibody was eluted with 10 column volumes of 100 mM glycine (pH 2.5). The column was equilibrated with 10 mM Tris (pH 8.8), and antibody was eluted with 10 column volumes of 100 mM triethylamine (pH 11.5). The two eluates were combined and dialyzed overnight against phosphate-buffered saline (PBS). The antibody was precipitated with 50% ammonium sulfate, resuspended in 10 ml of PBS, and dialyzed against PBS.
Western blot analysis.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed as previously described (47). Forty micrograms of yeast protein extract was denatured in 2% SDS–5% β-mercaptoethanol for 5 min at 95°C and resolved on a 15% polyacrylamide gel. The separated proteins were electrophoretically transferred to a BA-S NC-supported nitrocellulose membrane (Schleicher & Schuell, Inc., Keene, N.H.) by use of a Bio-Rad semidry transfer cell. The membrane was stained for 10 min with Ponceau S solution (0.1% Ponceau S, 5% acetic acid) to ensure proper transfer. The membrane was blocked with 5% milk–TBST (20 mM Tris-Cl [pH 7.6], 150 mM NaCl, 0.1% Tween 20) for 1 h at 24°C, incubated with affinity-purified anti-Snm1p antibody (1:500 dilution in 5% milk–TBST) for 1 h, washed with TBST three times for 10 min each time, incubated with peroxidase–anti-rabbit antibody (1:10,000 dilution in TBST; Boehringer Mannheim Biochemicals, Indianapolis, Ind.) for 30 min, and washed with TBST three times for 10 min each time. The peroxidase–anti-rabbit antibody was detected with a Boehringer chemiluminescence Western blotting kit and exposed to film for 10 min and then again overnight.
Plasmid segregation assay.
A 3.6-kb BamHI fragment containing the ADE2 gene was cloned into the pRS316 and YCPlac33 vectors to make the reporter plasmids pTC185 and pTC186, respectively (15, 51). Both reporter plasmids were transformed into THR200-derived yeast strains carrying the appropriate SNM1 allele by selecting for the URA3 marker on the plasmids (11). snm1 mutant strains carrying an ADE2 reporter plasmid were grown at 30°C in SCD overnight and diluted in sterile water, and 500 cells of each yeast strain were plated onto 150-mm YPD (1% yeast extract, 2% peptone, 2% dextrose) plates. The plates were scanned by use of a Linotype Hell JADE2 scanner after 3 days of growth at 30°C, when the color was fully developed.
Tubulin immunofluorescence.
Cells were prepared for immunofluorescence as previously described (36). Rat monoclonal antitubulin antibody YOL1/34 and indocarbocyanine (Cy3)-conjugated rabbit anti-rat secondary antibody were obtained from Accurate Chemical Corp., Westbury, N.Y. Cells were viewed by use of a Ziess Axioskop microscope equipped with epifluorescence and Narmoski optics and a Zeiss Plan-Apochromat 100× objective. Images were captured in real time by use of a Diagnostics Instruments Spot Camera 2 directly linked to a Macintosh PowerMac G3 computer.
RESULTS
Isolation of conditional mutations in the SNM1 gene.
Random PCR mutagenesis of the SNM1 gene was performed to further define the functional regions of the Snm1 protein. Plasmid pTHR100 (pRS315 containing SNM1) was digested with restriction enzymes BamHI and NsiI to remove a 696-bp fragment containing the SNM1 coding region. At the same time, plasmid pMES193 (pBS containing SNM1) was used as a template to produce a linear fragment containing the SNM1 gene by use of PCR. The nucleotide concentrations in the PCR were made unequal to encourage mutations. The mutagenized snm1 PCR product was cotransformed with gap-containing plasmid pTHR100 into haploid yeast strain THR200, with selection for the LEU2 gene. Yeast cells will repair the gap-containing plasmid with the PCR product by recombining homologous regions found at the ends of the PCR product and the ends of the gap-containing plasmid. Yeast strain THR200 contains a chromosomal deletion of the SNM1 gene and a wild-type copy of SNM1 on a URA3-based CEN plasmid (YCp50) (40). Transformants were replicated to two successive 5-FOA plates and then replicated to SCD plates at various temperatures between 17 and 37°C.
Approximately 8,000 separate yeast transformants were screened for a growth phenotype due to the presence of the mutagenized snm1 gene. Strains that displayed either cold- or temperature-sensitive growth were selected. Growth phenotypes were verified by testing a second time on SCD plates at various temperatures. Two hundred possible mutants that showed defective growth phenotypes were selected.
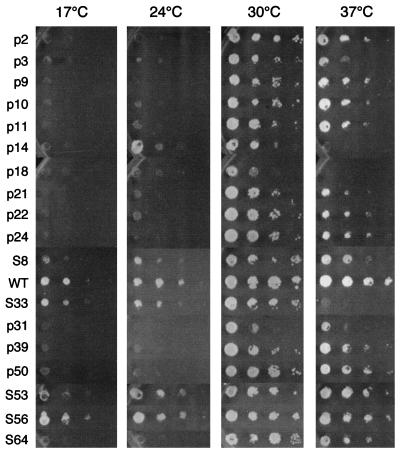
We then confirmed that the growth phenotypes seen were due to the presence of the pTHR100 plasmid repaired with the mutagenized snm1 gene. The plasmids from 180 of the original transformants were rescued into E. coli and retransformed back into THR200 to confirm that the initial growth phenotype seen was linked to the plasmid. Mutants were then more closely screened for temperature-conditional phenotypes. They were grown on dextrose-based medium plates at 17, 24, 30, or 37°C. Eighteen conditional plasmid-linked mutations of the snm1 gene were isolated in this way. Fourteen were cold sensitive (p2, p3, p9, p10, p11, p21, p22, p24, S8, p31, p39, p50, S53, and S64), 1 was temperature sensitive (S33), and 2 were both cold sensitive and temperature sensitive (p14 and p18) (Fig. 1). One showed slightly slower growth at elevated temperatures (S56).
FIG. 1.
Growth phenotypes of yeast strains carrying a mutant snm1 gene. Two colonies from each mutant yeast strain were mixed with 100 μl of sterile double-distilled H2O in the first-row wells of a 96-well Falcon tissue culture plate; 10-fold serial dilutions of these cell mixtures were placed in the second-, third-, and fourth-row wells. A 48-pin Frogger was used to transfer diluted cell mixtures to SCD plates. The cells were grown for 2 days at 24, 30, and 37°C and for 4 days at 17°C. SNM1 alleles are listed in Table 1. All SNM1 alleles are carried on LEU2 CEN vector pRS315 (51) in strain THR200. WT, wild type.
The snm1 insert in all 18 mutant plasmids was sequenced on both strands. Fifteen of the plasmids showed mutations within the snm1 coding region. Several transitions, transversions, insertions, and deletions were found. In addition to the formation of missense mutants, several frameshift and nonsense mutants were identified. Sequence changes identified in the mutants are listed in Table 1.
TABLE 1.
SNM1 alleles used in this study
| Designation(s)a | Mutation | Effectb | Extent of growth at the following temp (°C)c:
|
5.8Sa/5.8Sb ratiod | Stability of the MRP RNAe | IP resultf | |||
|---|---|---|---|---|---|---|---|---|---|
| 17 | 24 | 30 | 37 | ||||||
| Wild type | None | None | 3 | 3 | 4 | 4 | 10:1 | 4 | ++ |
| p3 | C250→T | Gln84→stop (TAA) | 0 | 0 | 3 | 2 | 1:1 | 4 | − |
| p9 | G91→A; A463→G | Ala31→Thr; Ser155→Gly | 0 | 0 | 3 | 3 | 1:1 | 4 | − |
| p21, p22, p39 | Missing an A between 294 and 300 | Frameshift at Lys100; changes Lys100 to Asn and adds 6 aa | 0 | 0 | 3 | 2 | 1:1 | 4 | − |
| p31 | T320→A | Leu107→stop (TAG) | 0 | 0 | 2 | 1 | 1:1 | 4 | − |
| p50 | T430→C; missing A288 | Tyr144→His; frameshift at Gly97; changes Gly97 to Asp and adds 9 aa | 0 | 0 | 4 | 3 | 1:1 | 4 | − |
| S8 | C153→T; G157→T | Pro52→Ser; Glu53→stop (TAA) | 1 | 2 | 3 | 2 | 1:1 | 4 | −/+ |
| S56 | C79→G; T206→G; G289→A | Leu27→Val; Val69→Gly; Gly97→Arg | 3 | 3 | 4 | 4 | 10:1 | 4 | − |
| p2 | Extra A between 514 and 515 | Frameshift at Leu172; changes Leu172 to Phe and adds 33 aa | 0 | 0 | 4 | 3 | 1:1 | 4 | −/+ |
| S53 | Missing A538 | Frameshift at Lys180; adds 11 aa | 1 | 3 | 4 | 3 | 1:1 | 4 | + |
| S64 | T425→C; extra A between 514 and 515 | Val142→Ala; frameshift at Leu172; changes Leu172 to Phe and adds 33 aa | 0 | 0 | 4 | 3 | 1:1 | 4 | + |
| p14 | G392→T; missing A24 | Ser131→Ile; frameshift at Lys8; changes Lys8 to Asn and adds 18 aa | 0 | 2 | 3 | 0 | 1:8 | 4 | − |
| p18 | A451→T | Lys151→stop (TAG) | 0 | 0 | 2 | 0 | 1:10 | 4 | +++ |
| S33 | A65→G; G199→A; G205→C | His22→Arg; Val67→Ile; Val69→Leu | 2 | 2 | 4 | 0 | 1:8 | 4 | − |
| p10, p11, p24 | None | None | 0 | 0 | 3 | 3 | 1:1 | 4 | − |
| 172 | T189→G; G190→C (site directed) | Cys64→Ala | 3 | 3 | 4 | 0 | 1:1 | 4 | − |
| 170 | T315→G; G316→C (site directed) | Cys106→Ala | 3 | 3 | 4 | 3 | 8:1 | 4 | + |
| p3 (YCp) | C250→T | Gln84→stop (TAA) | 0 | 0 | 3 | 2 | 1:1 | ND | ND |
| p18 (YCp) | A451→T | Lys151→stop (TAG) | 0 | 0 | 2 | 0 | 1:10 | ND | ND |
| p21 (YCp) | Missing an A between 294 and 300 | Frameshift at Lys100; changes Lys100 to Asn and adds 6 aa | 0 | 0 | 2 | 1 | 1:1 | ND | ND |
| p31 (YCp) | T320→A | Leu107→stop (TAG) | 0 | 0 | 2 | 1 | 1:1 | ND | ND |
| S8 (YCp) | C153→T; G157→T | Pro52→Ser; Glu53→stop (TAA) | 0 | 1 | 2 | 1 | 1:1 | ND | ND |
| I-p3 | C250→T | Gln84→stop (TAA) | 0 | 0 | 2 | 1 | 1:8 | ND | ND |
| I-p18 | A451→T | Lys151→stop (TAG) | 0 | 0 | 2 | 0 | 1:10 | ND | ND |
| I-S8 | C153→T; G157→T | Pro52→Ser; Glu53→stop (TAA) | 0 | 1 | 2 | 1 | 1:10 | ND | ND |
Mutations were maintained in yeasts in one of three ways. (i) They were placed in the LEU2 CEN vector pRS315 (51) and placed in strain THR200 as the sole copy of SNM1. (ii) They were placed in the LEU2 CEN vector YCplac111 (15) and placed in strain THR200 as the sole copy of SNM1 (YCp group). (iii) They were placed in the LEU2 integration vector YIplac128 and integrated into the ClaI site of the chromosomal LEU2 locus in THR200 (I group).
The effects of all the SNM1 mutants are listed. aa, amino acids.
The yeast strains were grown on dextrose at the indicated temperatures to check for temperature-dependent growth phenotypes. The extent of growth is described as a number ranging from 0 to 4, 4 being wild-type growth and 0 reflecting no growth.
Ratio of the two 5.8S rRNAs present in the mutant strains.
Scaled the same way as the growth phenotypes; ND, not determined.
Ability of the mutant SNM1 proteins to immunoprecipitate (IP) MRP RNA with anti-Snm1p antibodies. +, MRP RNA was precipitated, although at reduced levels compared to the wild-type level; ++, MRP RNA was precipitated at wild-type levels; +++, MRP RNA was precipitated at higher-than-wild-type levels; −/+, MRP RNA was precipitated at just-detectable levels; −, MRP RNA was not detected in the immunoprecipitation experiments; ND, not determined.
Two site-directed mutations were created to specifically change cysteines 64 and 106, which are believed to be involved in zinc binding (snm1-170 and snm1-172, respectively). The 170 mutant displayed no growth defect from 17 to 37°C, while the 172 mutant was temperature sensitive.
Defects in 5.8S rRNA processing in snm1 mutants.
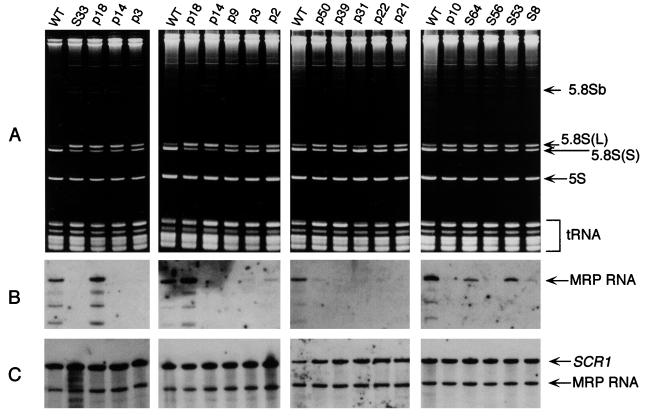
The severity of the mutations and the resulting growth phenotypes suggested that the defects in Snm1p affected the function of the RNase MRP enzyme complex. Although Snm1p has been shown to be an essential protein that is closely associated with the MRP RNA, it has not been directly shown to be required for the function of the MRP enzyme (45). Mutations in nme1, the gene for the MRP RNA, or in Pop1p, another protein component of yeast RNase MRP, lead to defective 5.8S rRNA processing (7, 29, 44). In addition, transcriptional depletion of mRNAs for other RNase MRP protein components leads to a similar phenotype (3, 8). Two species of 5.8S rRNA that differ in length by only 7 nucleotides exist in yeast (41). Normally, they are present at a ratio of 10:1, small to large. Loss of yeast RNase MRP components results in a loss of processing of the smaller 5.8S rRNA. To see if mutations in the snm1 gene produce the same result, mutants were screened for anomalies in 5.8S rRNA processing. The mutants were grown at 30°C until they reached the exponential phase and then were shifted to the nonpermissive temperature to determine the effect of growth at the nonpermissive temperature on 5.8S rRNA processing. Whole-cell RNA extracts from the mutants were analyzed on 6% acrylamide–7 M urea gels. Nearly all the mutants displayed some defect in 5.8S rRNA processing. The severity of the temperature-dependent growth seen in the mutants closely coincided with the severity of the defect seen in 5.8S rRNA processing (Fig. 2A). In addition, nearly all of the mutants displayed some loss of 5.8S rRNA processing at the permissive temperature (data not shown). These results demonstrate that Snm1p is required for the function of the RNase MRP enzyme in rRNA processing.
FIG. 2.
rRNA processing, RNase MRP RNA association, and Northern analysis of MRP RNA in snm1 mutant strains. Yeast strains were grown to 107 cells/ml at 30°C in YPD and then shifted to the nonpermissive temperature for 6 h. Cells were harvested, total RNA was isolated from one-half of the culture, and whole-cell protein extracts were made from the other half (see Materials and Methods). SNM1 alleles are listed in Table 1. All SNM1 alleles are carried on LEU2 CEN vector pRS315 (51) in strain THR200. (A) Ten micrograms of total RNA from the indicated strains was separated on 6% acrylamide–7 M urea gels and examined by staining with ethidium bromide. The locations of the relevant 5.8S(S), 5.8S(L), and 5.8Sb RNAs are shown. The ratios of the two 5.8S bands are provided in Table 1. WT, wild type. (B) Northern analysis of the RNase MRP RNA that coimmunoprecipitated with the anti-Snm1p antibody. Four hundred micrograms of protein extract from each mutant was incubated with polyclonal anti-Snm1p antibody. Antibody-protein RNA complexes were collected as described in Materials and Methods. RNA that coimmunoprecipitated with the antibody was separated on 6% acrylamide–7 M urea gels, and the MRP RNA was detected by hybridization with a 32P-labeled probe corresponding to a 1,042-bp EcoRI fragment containing the NME1 gene (43). (C) Northern analysis of the RNase MRP RNA in yeast snm1 mutant strains. Ten micrograms of whole-cell RNA isolated from yeast snm1 mutant strains was separated on 6% acrylamide–7 M urea gels and blotted to nylon membranes. The MRP RNA was visualized by hybridization with 32P-labeled probes corresponding to a 1,042-bp EcoRI fragment containing the NME1 gene (43) and a 736-bp EcoRI-SpeI fragment containing the SCR1 gene as a loading control (12).
The stability of the MRP RNA is not affected in snm1 mutants.
The RNA component of yeast RNase MRP is essential for the proper functioning of the enzyme and cellular viability. It is known that Snm1p is associated with the MRP RNA in vivo (45). It was postulated that mutations in Snm1p could compromise the stability of the RNA component, explaining the mutant phenotypes seen. To test this possibility, Northern analyses were performed on whole-cell RNAs isolated from the mutant yeast strains after they were shifted to nonpermissive temperatures. Whole-cell RNA extracts from the mutants were analyzed for the presence of the MRP RNA and SCR1 RNA, an abundant cytoplasmic RNA, used as a loading control (12). As indicated in Fig. 2C, MRP RNA stability was unaffected in all of the snm1 mutants. This result is in contrast to the rapid loss of MRP RNA stability caused by a mutation in the pop1 gene (29).
The association of Snm1p with the MRP RNA changes in the mutants.
Antibody against Snm1p has been previously shown to coimmunoprecipitate the MRP RNA. Coimmunoprecipitation experiments with mutant Snm1 proteins were performed to test if this interaction was maintained (45). Anti-Snm1p antibody was bound to protein A-Sepharose beads. Whole-cell mutant yeast extracts were incubated with the anti-Snm1p antibody attached to protein A-Sepharose beads and washed several times. The RNA was removed from the beads and analyzed by Northern analysis for the presence of both the MRP RNA and SCR1 RNA as a negative control. As shown in the Northern blot analyses of the coimmunoprecipitation experiments (Fig. 2B), wild-type controls were able to precipitate the MRP RNA by use of the anti-Snm1p antibody. However, many of the snm1 mutants failed to precipitate appreciable amounts of the MRP RNA, indicating that these mutants have a reduced MRP RNA association in vivo. Similar results were seen whether extracts were made from cells at either the permissive or the nonpermissive temperature (data not shown). Immunoprecipitations were repeated at least three times for each mutant, with similar results each time. In addition, degradation of nonprecipitated MRP RNA was never seen. Precipitated MRP RNA was undetectable in this assay for the p3, p9, p21, p22, p39, p31, p50, S56, p14, S33, 172, and p10 mutants. Mutants p2, S53, S8, S64, and 170 were able to precipitate some MRP RNA, although to a lesser extent than the wild type. The p18 mutant consistently precipitated more MRP RNA than the wild type. Unlike the processing of the 5.8S rRNA, there was no discernable correlation between Snm1p binding to the MRP RNA and the growth phenotype of the mutant. Many mutant strains, such as S56, grew quite well but still failed to bind any detectable RNase MRP RNA, while the p18 mutant appeared to bind more MRP RNA than the wild type yet grew slowly at most temperatures and had a severe rRNA-processing defect.
Essentiality testing of the SNM1 gene.
In the snm1 mutants isolated, several frameshift and nonsense mutations were found. Some of these mutations were so severe (p14 had a frameshift mutation at Lys8, p3 had a nonsense mutation at Gln84, and S8 had a nonsense mutation at Glu53) that we expected no functional Snm1p was being made. However, all of these mutants, regardless of the severity of the mutations, could grow at least moderately well, if not as well as the wild type, at 30°C, despite little or no detectable MRP RNA binding. This result suggests that Snm1p may not be essential for the viability of yeast cells at 30°C. This notion would be in conflict with a previous deletion analysis of the SNM1 gene (45) and suggested the possibility that some other gene is tightly linked to the SNM1 gene and that a loss of the function of the linked gene is responsible for the lack of viability observed for snm1 deletion strains. We tested this possibility by using three different approaches. First, we placed the SNM1 coding sequence (from ATG to TAG) in between an ACT1 promoter and terminator and transformed this construct into yeast strain THR200, which has a chromosomal deletion of the SNM1 gene and is maintained by a wild-type SNM1 gene on a URA3-based plasmid. After removal of the wild-type SNM1 plasmid by growth on 5-FOA plates, we found that the heterologous promoter construct could fully complement the chromosomal deletion of the SNM1 gene at all temperatures (data not shown). This result indicated that the SNM1 coding region was required for viability. Second, we combined three different snm1 nonsense and frameshift mutations (p14, S8, and p18) to make all of the possible double and triple mutations. All of these mutations were unable to complement the chromosomal deletion, demonstrating that Snm1p was required and eliminating the possibility that the SNM1 RNA transcript was required for cell viability. Indeed, this result suggested that the snm1 mutants were translationally suppressed. Third, we had previously constructed a carboxy-terminal hemagglutonin (3×HA)-tagged version of the SNM1 gene. This tagged version complemented an snm1 deletion, but with reduced protein levels. We combined this 3×HA-tagged version of SNM1 with snm1 nonsense and frameshift mutations (p3 and p14). Neither of these tagged mutations could complement a chromosomal snm1 deletion, indicating that translational readthrough to the carboxy-terminal end of Snm1p was required for complementation of these mutations.
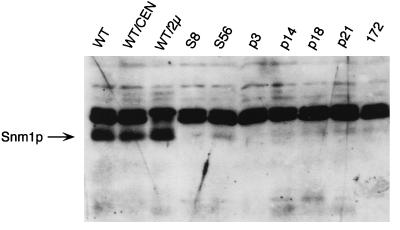
Reduction in Snm1p levels in snm1 mutants.
An affinity-purified anti-Snm1p antibody was used in the Western analysis to examine Snm1p levels in snm1 mutants (45). As shown in Fig. 3, this antibody readily detects Snm1p in yeast cell extracts. In addition, this antibody cross-reacts with another protein of approximately 27 kDa, providing a convenient loading control. A yeast strain with the SNM1 gene on a high-copy-number plasmid (WT/2μ) produces a signal with an intensity stronger than that of a wild-type single-copy SNM1 strain (wild type or WT/CEN), confirming that this band is Snm1p. In all the mutants shown (S8, p3, p14, p18, and p21), Snm1p levels are very low. However, after a long exposure, a full-length Snm1p band is detected for most of these nonsense and frameshift mutations. This result suggests that very little Snm1p is required for cell viability and that these nonsense and frameshift mutations are translationally suppressed, but at a very low rate. Surprisingly, very little p18 protein is detected despite increased immunoprecipitation of the MRP RNA from this mutant. A low-abundance band with the molecular weight expected for the truncated p18 protein can be reproducibly seen in the Western analysis. This low-abundance protein may have a higher affinity than the wild-type protein for the MRP RNA and/or polyclonal anti-Snm1 antibodies when in its native configuration. The results obtained with mutant p18 show that the last third of Snm1p is dispensable for MRP RNA binding but not for RNase activity.
FIG. 3.
Western analysis of snm1 mutants. Whole-cell protein extracts were made from yeast strains grown to 107 cells/ml at 30°C in SCD (see Materials and Methods). Forty micrograms of protein extracts was denatured in 2% SDS–5% β-mercaptoethanol for 5 min at 95°C. Proteins were separated by SDS-PAGE through a 15% polyacrylamide gel. Proteins were subsequently transferred to a nitrocellulose membrane and probed with an affinity-purified polyclonal anti-Snm1p antibody at a 1:500 dilution. The blot was probed with a secondary peroxidase–anti-rabbit antibody and visualized with the Boehringer chemiluminescence kit. The Snm1p band is indicated. The band above Snm1p is a nonspecific protein that serves as a convenient loading control. SNM1 alleles are listed in Table 1. SNM1 alleles WT/CEN, S8, S56, p3, p14, p18, p21, and 172 are carried on LEU2 CEN vector pRS315 (51) in strain THR200. The WT/2μ strain is identical to the other strains, except that the wild-type SNM1 gene is maintained on yeast 2μm plasmid YEp351 (18). WT is DBY2006, which contains a wild-type chromosomal SNM1 gene (43).
Other genes are not translationally suppressed in snm1 mutants.
Since Snm1p and the RNase MRP enzyme are involved in ribosome biogenesis, we suspected that defects in RNase MRP could lead to the production of ribosomes that have compromised translational fidelity (55). To test this possibility, we looked for cross suppression of the yeast nonsense mutations lys1-1 (UAA), trp1-1 (UAG), tyr7-1 (UAG), ilv1-1 (UAG), ade3-26 (UAG), and met8-1 (UAG) (27) both by snm1 mutations and by mutations in the MRP RNA (NME1) (44). We were unable to detect translational suppression of any of these mutations by snm1 or nme1 mutations (data not shown). This result indicated that the translational suppression was specific for the SNM1 gene and that there is no general compromise of translational fidelity in snm1 mutants.
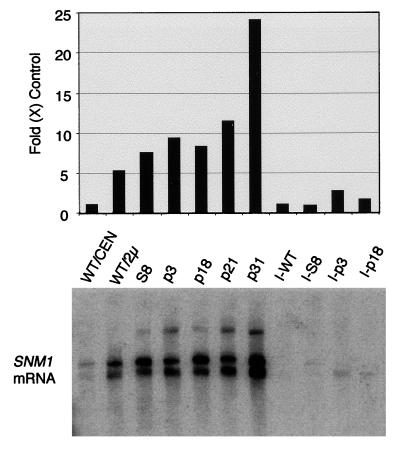
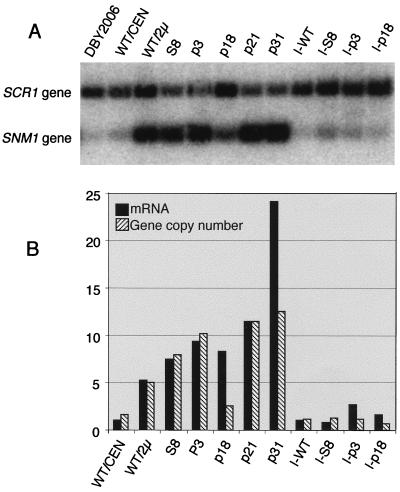
Increase in snm1 mRNA levels in snm1 mutants.
Since we unexpectedly found translational suppression of the snm1 mutants, we decided to perform Northern analysis on the suppressed snm1 mutants. As shown in Fig. 4, the 800-nucleotide SNM1 transcript was barely detectable in a wild-type yeast strain (WT/CEN). However, in most of the mutants examined, snm1 mRNA levels were increased between 7- and 25-fold, to levels even higher than the level of expression from a wild-type SNM1 2μm plasmid (WT/2μ). This result suggests that high snm1 mRNA levels in nonsense and frameshift mutants may lead to sufficient translational suppression to sustain cell viability.
FIG. 4.
Northern analysis of SNM1 mRNA. Total RNA was isolated from yeast snm1 mutant strains grown to 107 cells/ml at 30°C in SCD (see Materials and Methods). Twenty micrograms of total RNA per lane was loaded on a 6% acrylamide–7 M urea gel and blotted to a nylon membrane. The mRNA corresponding to transcripts from the SNM1 allele was detected by hybridization with a 32P-labeled probe corresponding to the 696-bp BamHI-NsiI fragment containing the SNM1 coding region. The lower panel shows the location of the SNM1 transcripts. The upper panel shows the signal corresponding to the SNM1 mRNA, measured with a PhosphorImager and normalized to the mRNA level in WT/CEN, which was assigned a level of 1. SNM1 alleles are listed in Table 1. The WT/CEN and mutant SNM1 genes (S8, p3, p18, p21, and p31) are carried on LEU2 CEN vector YCplac111 (15) in strain THR200. The WT/2μ strain is identical to the other strains, except that the wild-type SNM1 gene is maintained on yeast 2μm plasmid YEp351 (18). I, the given allele was integrated into the yeast chromosome at the LEU2 gene in THR200 (see Materials and Methods).
Plasmid specificity of snm1 mutants.
The mutant snm1 genes were maintained on a CEN LEU2 (pRS315) plasmid in all of the mutants (51). To examine if the mRNA level increase was plasmid specific, we transferred several of the snm1 mutations into plasmid YCplac111 and tested them for replacement of a wild-type SNM1 gene (15). All of the mutations still complemented a deletion strain at 30°C, except for the p14 mutation (frameshift at amino acid 8) (data not shown). We also placed several of the mutant snm1 genes into plasmid YIplac128 and integrated them into the chromosomal LEU2 gene. After integration, only the nonsense mutations were still able to complement the snm1 deletion at 30°C, and most of these nonsense mutants grew very slowly (only p18 grew at a rate comparable to that when maintained on a plasmid). None of the frameshift mutations were able to complement when integrated. These results were consistent with the placement of SNM1 on the plasmid playing a direct role in snm1 mRNA amplification and with past reports that frameshift mutations are more difficult to suppress than nonsense mutations (33).
Increase in plasmid copy number in snm1 mutants.
To test if the large increase in mRNA expression was caused by plasmid amplification, copy numbers of the snm1-carrying plasmids were examined by Southern analysis. The signal was normalized to the single chromosomal copy of SNM1 in strain DBY2006 and normalized between strains by use of the single chromosomal copy of the SCR1 gene. This strategy allowed us to estimate plasmid copy numbers in the various strains. All the mutants showed increased snm1 plasmid copy numbers compared to the wild type, ranging from 5 to 15 copies per cell (Fig. 5). The wild-type CEN plasmid was estimated at about 1.5 copies per cell, and the wild-type 2μm plasmid was estimated at about 6 copies. However, most of the plasmids containing mutant snm1 genes were highly amplified even when CEN was present. We were able to find no other reports in the literature of this type of CEN-containing plasmid amplification and suppression. These results suggest that mutations in the snm1 gene may cause plasmid amplification, either by directly causing overreplication or missegregation because of a defective RNase MRP enzyme or by selecting for yeast that missegregates or overreplicates or by both methods. The integrated snm1 mutations were all maintained at one copy per cell.
FIG. 5.
Southern analysis of SNM1 gene copy number in various mutants. (A) Genomic DNA was isolated from yeast snm1 mutant strains grown to 2 × 107 cells/ml at 30°C in SCD (see Materials and Methods). Ten micrograms of genomic DNA per lane was digested with EcoRI and SpeI overnight, separated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled SNM1 probe and SCR1 probe for standardization. The locations of the SNM1 and SCR1 genes are indicated. (B) Band intensity was measured with a PhosphorImager and standardized against the intensities of the SCR1 and DBY2006 SNM1 bands as single-copy controls to calculate copy number. Copy numbers were plotted against mRNA levels from Fig. 4. SNM1 alleles are listed in Table 1. The WT/CEN and mutant SNM1 genes (S8, p3, p18, p21, and p31) are carried on LEU2 CEN vector YCplac111 (15). The WT/2μ strain is identical to the other strains, except that the wild-type SNM1 gene is maintained on yeast 2μm plasmid YEp351 (18). I, the given allele was integrated into the yeast chromosome at the LEU2 gene in THR200 (see Materials and Methods).
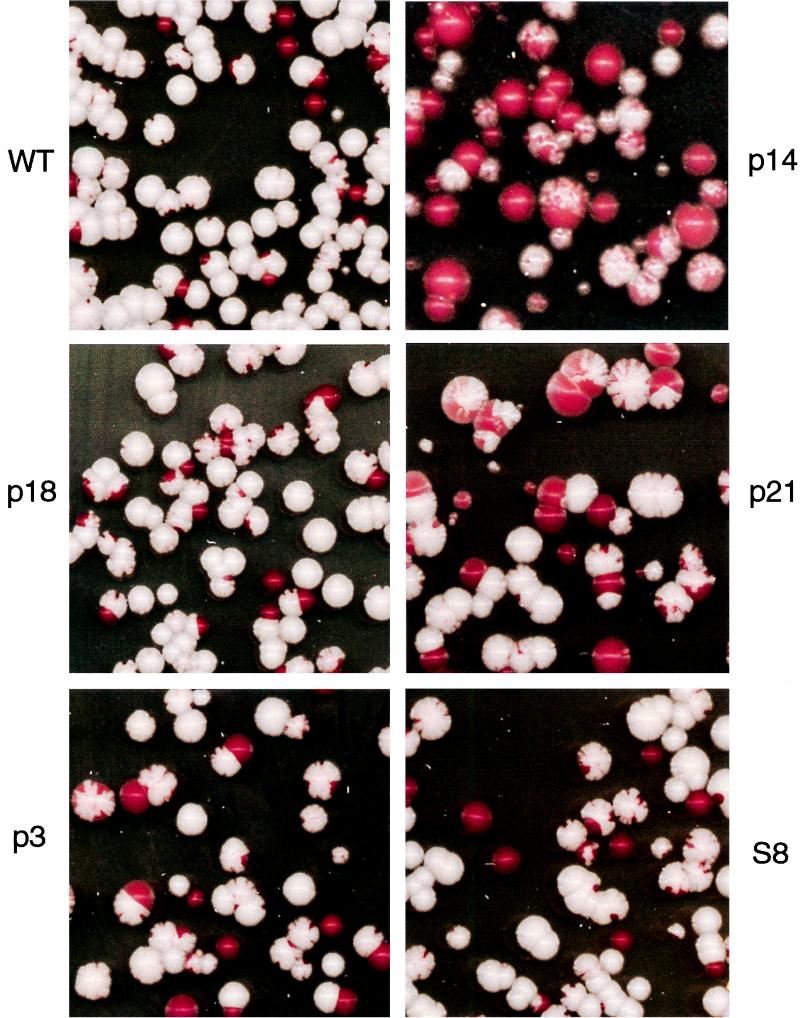
Plasmid missegregation in snm1 mutants.
If the plasmid amplification is directly due to selection to amplify the mutant snm1 gene on the plasmid, maintenance of a separate plasmid should be normal. To test plasmid segregation in snm1 mutant strains, we constructed reporter plasmid pTC185. It contains a wild-type ADE2 gene in the CEN URA3 vector pRS316 (51) that is independent of the mutant snm1 gene. All the snm1 mutant strains have an ade2-1 mutation on the chromosome and form red colonies on YPD plates. After having been transformed with plasmid pTC185 containing a wild-type ADE2 gene, they form white colonies. Upon loss of the plasmid, red sectors are formed in white colonies. If there is a segregation problem, more red sectors will be seen.
Yeast strains with the reporter plasmid were grown in nonselective media overnight. The cells were counted, diluted, plated at 500 cells per 150-mm petri dish, and grown at 30°C for several days. The results are shown in Fig. 6. Most wild-type cells are white, although a small number of red colonies are seen due to plasmid loss during nonselective growth. A very small number of sectoring colonies are found; most of these have fewer than three small sectors. In the snm1 mutant strains, we see a clear increase in plasmid missegregation. The p18, p3, and S8 mutants have more sectoring colonies than the wild type, and the sectoring colonies usually have more than three sectors, indicating more frequent missegregation during colony formation. The p21 strain has more sectoring colonies than p18, p3, and S8 strains, consistent with a higher plasmid copy number. The p14 strain, which grows slowly even at 30°C, has an extreme sectoring phenotype and an increase in the number of all red colonies. These results show that defects in Snm1p function can cause plasmid missegregation.
FIG. 6.
Plasmid segregation in snm1 mutants. The yeast snm1 mutant strains were transformed with a plasmid carrying a wild-type ADE2 gene as a reporter for plasmid maintenance. These mutants were grown at 30°C in SCD with uracil to remove selection for the plasmid for 18 h and were diluted in sterile water, and 500 cells of each yeast strain were plated on 150-mm petri dishes. Pictures were taken after 3 days of growth at 30°C, when the color was fully developed. Strain p14 is shown at twice the magnification, since it grows slower than the other strains at 30°C. SNM1 alleles are listed in Table 1. All SNM1 alleles are carried on LEU2 CEN vector pRS315 (51) in strain THR200. WT, wild type.
To confirm that missegregation was not specific for the pRS vectors, we also used the YCplac vectors, which have a CEN4-ARS1 cassette for plasmid maintenance (15); similar results were obtained (data not shown).
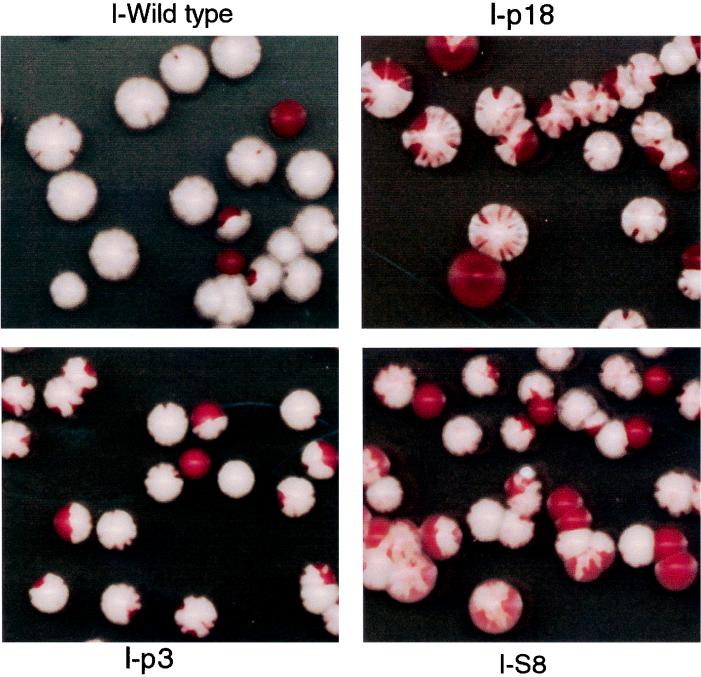
We wanted to rule out the possibility that the high-copy-number CEN maintenance in the mutant strains was not causing the missegregation of the independent ADE2-containing plasmid. We did this by examining plasmid segregation in strains with an integrated mutant SNM1 gene. These strains have no plasmids other than the introduced ADE2-containing plasmid and should have no selection for or against the presence of this plasmid. As shown in Fig. 7, strains with an integrated mutant SNM1 gene still have a large increase in plasmid instability compared to the wild-type strain. This result rules out the possibility that plasmid instability is caused by plasmid competition. We also tested an MRP RNA mutant in the segregation assay and obtained identical results, indicating that the missegregation phenotype is due to the loss of RNase MRP activity and is not SNM1 specific (data not shown).
FIG. 7.
Plasmid missegregation is independent of high-copy-number plasmid maintenance or a need to amplify the SNM1 gene. Yeast strains with snm1 mutations integrated into the LEU2 locus on chromosome III were transformed with a plasmid carrying a wild-type ADE2 gene as a reporter for plasmid maintenance. These mutants were grown at 30°C in SCD with uracil to remove selection for the plasmid for 18 h and were diluted in sterile water, and 500 cells of each yeast strain were plated on 150-mm petri dishes. Pictures were taken after 3 days of growth at 30°C, when the color was fully developed. SNM1 alleles are listed in Table 1.
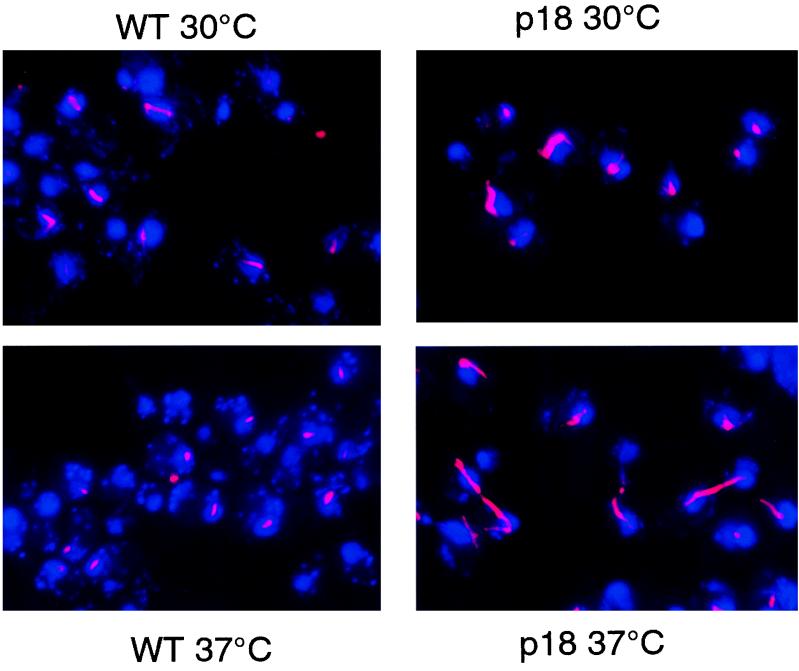
Recent results have identified a role for the RNase MRP enzyme in a late stage of the cell cycle (2a). RNase MRP RNA mutants showed a cell cycle arrest, with large-bud cells, a long mitotic spindle, and divided nuclei (M to G1 arrest). This phenotype is characteristic of the late-telophase arrest seen in the cdc15 class of mutants (16, 19, 56). Many of these mutants have also been found to have chromosome segregation problems. We tested an snm1 mutant for a similar arrest phenotype. Using the p18 mutant, we examined the mitotic spindle by using an antitubulin antibody and immunofluorescence when this mutant was grown at either the permissive or the nonpermissive temperature (21). As shown in Fig. 8, the p18 mutant displayed a clear telophase arrest when grown at the nonpermissive temperature for 3 h. In addition, even at the permissive temperature, we found cells that looked delayed at this stage of the cell cycle. The appearance of this defect in the p18 mutant (maintained by a low plasmid copy number) indicates that the arrest is not the result of a problem with segregating the many CEN plasmids found in some snm1 mutants. Other snm1 mutants, including 172 and p14, displayed an identical arrest phenotype. These data, along with a similar arrest phenotype in MRP RNA mutants (2a), indicate an RNase MRP-specific defect as opposed to an SNM1-specific or an SNM1-allele-specific defect.
FIG. 8.
Immunolocalization of mitotic spindles. Yeast strains were grown to 2 × 106 cells/ml at 30°C in YPD and either maintained at 30°C for an additional 3 h or shifted to 37°C for 3 h. Cells were fixed, and the mitotic spindle and DNA were localized as described in Materials and Methods. 4′,6-Diamidino-2-phenylindole (DAPI) staining of DNA is shown in blue, and Cy3 staining of tubulin is shown in red. The wild-type gene and the p18 mutation are carried on LEU2 CEN vector pRS315 (51) in strain THR200.
DISCUSSION
Generation of snm1 mutations.
Random PCR mutagenesis of the SNM1 gene yielded 18 mutants with various temperature-conditional growth phenotypes. DNA sequence analysis of the mutants revealed mutations located throughout the gene and ranging from missense mutations to frameshift and nonsense mutations. Because the SNM1 gene is essential, such serious frameshift and nonsense mutations were not expected. The phenotypes of most of the frameshift and nonsense mutants were very similar. They all displayed cold-sensitive growth (except for p18 and p14, which are also temperature sensitive), a decrease in 5.8S rRNA production, a decrease in Snm1p levels, and a decrease in Snm1p associated with MRP RNA (except for p18). There have been no other reports of cold-sensitive translational suppression, so the cold sensitivity may be the result of a reduction in binding of the MRP RNA by Snm1p at colder temperatures.
Translational suppressors of frameshift and nonsense mutations have been reported for yeast (33, 59). However, these extragenic suppressors usually lead to cross suppression of similar mutations in different genes, a finding which was not obtained in this study (27). Yeast can read through nonsense mutations in the absence of translational suppressors, but at a low rate (13). Nonsense mutations in the HSF1 gene were found to be subject to translational readthrough without second-site translational suppressors; however, the copy number of the HSF1 centromere plasmid in that study was not determined (22). It also has been demonstrated that the overexpression of nonsense HSF1 mutations on a 2μm plasmid can provide full viability, indicating that a high copy number is able to suppress at least some nonsense mutations (22). In this study, all the frameshift and nonsense mutations isolated conferred viability, suggesting that suppression does occur. Observation of the full-length readthrough product in Western analyses and the lethality of the mutations when combined with a carboxy-terminal 3×HA tag also advocate readthrough. After examining mRNA levels and plasmid copy numbers in the snm1 mutants, we found that nonsense and frameshift mutations in the snm1 gene are suppressed by amplification of the plasmid carrying the snm1 gene. In the mutants studied, plasmid copy number is amplified 5- to 15-fold, with a parallel increase in mRNA levels. At high mRNA levels, it is postulated that a small portion of the mRNA can be read through by the ribosomes, resulting in the translation of full-length products. These experiments indicate that very little full-length Snm1p is required for cell viability. A careful search of the literature revealed no other reports of translational suppression conferred by CEN plasmid amplification.
Function of Snm1p in RNase MRP.
We show for the first time that mutations in the snm1 gene can adversely effect RNase MRP activity, causing a loss of 5.8S rRNA processing. Since it has been previously established that Snm1p interacts with the MRP RNA and that it is a component of the RNase MRP enzyme complex, one role for Snm1p may be to stabilize the MRP RNA component. This possibility was explored by analyzing whole-cell RNA isolated from the snm1 mutants for the presence of the MRP RNA. Northern analyses of the RNA isolated from the snm1 mutants showed that the stability of the MRP RNA was unaffected in all of the mutants. The stability of the MRP RNA is crucial for maintaining proper RNase MRP activity and yeast cell viability. Depletion of other known protein components of the yeast RNase MRP enzyme complex, Pop1p, Pop4p, Pop5p, Pop6p, Pop7/Rpp2p, Pop8p, and Rpp1p, results in a loss of the stability of the RNA component (3, 8, 53). The only other known protein component that has no role in MRP RNA stability is the Pop3 protein, which is believed to be loosely associated with RNase MRP and tightly associated with RNase P (10). Since Snm1p is the only unique protein component of RNase MRP, we speculate that the function of Snm1p may involve substrate recognition and/or binding. The eight other protein components shared by RNase MRP and RNase P probably have functions in stabilizing the component RNAs, assembling, transporting, and localizing the ribonucleoprotein complexes, and potentially catalyzing the RNA cleavage reactions. Either Rpr2p, which is a unique component of RNase P, or Pop3p may play the counterpart role of Snm1p in RNase P.
Although not required for the stability of the MRP RNA in vivo, Snm1p appears to be required to produce a fully functional RNase MRP complex capable of processing rRNAs. To test if a direct interaction with the MRP RNA is required for function, mutants were tested in coimmunoprecipitation experiments with anti-Snm1p antibody to see if mutant Snm1p still associates with the MRP RNA. Almost all mutants showed reduced or undetectable levels of immunoprecipitable MRP RNA, implying that these mutants have levels of Snm1p in association with MRP RNA that are below the range of the detection system used in these experiments (approximately <2% the wild-type level). Western analysis showed that most snm1 mutants produced full-length Snm1p, but at a very low level. These low levels of Snm1p explain the low levels of MRP RNA coimmunoprecipitated with Snm1p. Interestingly, the p18 mutant could consistently coimmunoprecipitate more MRP RNA than the wild type. In addition, this is the only nonsense mutation that was unaffected by the carboxy-terminal 3×HA tag, indicating that the truncated version is active in this case. p18 has a Lys151 nonsense mutation that is predicted to produce a truncated protein (about two thirds the size of the full-length protein) without the lysine- and serine-rich tail. This protein appears to associate more tightly with the MRP RNA or has a higher affinity for the antibody than the full-length protein, therefore allowing more MRP RNA to be immunoprecipitated in this mutant. A higher antibody affinity would also necessitate the antibody binding better to the native protein than to the denatured protein, since Western analysis detected very little mutant p18 protein. The p18 mutant has a very strong rRNA-processing defect and similar growth defects when found on a plasmid or integrated into the chromosome. The immunoprecipitation results imply that Snm1p binding to the MRP RNA requires the zinc cluster half of the protein and that this interaction is not sufficient for the function of Snm1p. Indeed, point mutations that fell within the predicted zinc binding site failed to bind appreciable levels of the MRP RNA (170, S56, and S33), demonstrating the essentiality of this region for RNA binding. The 170 and S56 mutants, despite minimal MRP RNA binding, were still nearly wild type for both growth and rRNA processing. Conversely, mutations in the last third of the protein (p18, S53, S64, and p2) were all capable of MRP RNA binding but were deficient in RNase MRP activity, as measured by rRNA processing. These results suggest that Snm1p has two functionally independent domains, one domain (zinc cluster) being required for association with the MRP RNA and a second domain (lysine- and serine-rich tail) being required for the activity of the enzyme, potentially in binding the substrate or folding the substrate into a conformation that can be cleaved. The binding of Snm1p to the MRP RNA may not be essential for the function of the second domain in RNase MRP activity.
The essential function of RNase MRP.
RNase MRP was first found as an enzyme involved in mitochondria DNA replication (4) and subsequently has been found to have a direct role in rRNA processing (44). However, yeast cells without mitochondrial DNA are viable, and others have questioned whether the role of RNase MRP in rRNA processing is essential (1, 17). The RNA component and all the protein components of yeast RNase MRP identified so far are essential, indicating that the RNase MRP complex has a function which is essential for normal cell growth.
In these studies of the snm1 mutants, we found that plasmids carrying the nonsense and frameshift mutations were amplified and that this amplification is due to plasmid missegregation. Initially, we used the pRS set of vectors in our mutagenesis experiments (51); these vectors use a CEN6-ARS4 cassette for plasmid maintenance. To confirm that missegregation was not caused by the pRS vectors, we also used the YCplac set of vectors; these vectors use a CEN4-ARS1 cassette for plasmid maintenance. Similar results were obtained (data not shown) (15). The YCplac vectors were found to be more stable than the pRS vectors in both the wild type and various snm1 mutants; however, a higher rate of missegregation was always seen in the snm1 mutants. This was also the case when the mutations were integrated into the chromosome, indicating that the missegregation was not the result of a cell trying to maintain too many CEN plasmids and was independent of the need to amplify the SNM1-containing plasmid. We also performed plasmid segregation experiments on the nme1-P6 mutant (44, 45), which has a point mutation at position 122 in the MRP RNA. A high rate of missegregation was also observed in this mutant compared to the isogenic wild-type strain. These results indicate that defects in Snm1p resulting in plasmid missegregation are directly caused by a defect in RNase MRP function and not by direct selection for the plasmid.
Other experiments from our laboratory have identified a role for the RNase MRP enzyme late in the mitotic cell cycle, in the breakdown of the mitotic spindle and in movement out of mitosis (2a, 35). Yeast cells with RNase MRP defects arrest during telophase with large-bud cells, divided nuclei, and a long mitotic spindle. This is also clearly the case for snm1 mutants. The essential function of RNase MRP may be in cell division cycle control. The arrest phenotype may be the cause of the plasmid missegregation observed, since a prolonged period of anaphase or improper disassembly of the mitotic spindle could easily be expected to lead to improper segregation of CEN plasmids or even potentially of chromosomes. The exact role of RNase MRP in exit from mitosis is unknown but may be associated with the release of Cdc14 phosphatase from the nucleolus (49, 58).
ACKNOWLEDGMENTS
We thank M. Hale, B. A. Morisseau, P. Kane, and D. Amberg for comments and helpful discussions during the preparation of the manuscript.
This work was supported by grant NP-936 from the American Cancer Society.
REFERENCES
- 1.Allmang C, Henry Y, Morrissey J P, Wood H, Petfalski E, Tollervey D. Processing of the yeast pre-rRNA at sites A(2) and A(3) is linked. RNA. 1996;2:63–73. [PMC free article] [PubMed] [Google Scholar]
- 2.Baldacci G, Chérif-Zahar B, Bernardi G. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 1984;3:2115–2120. doi: 10.1002/j.1460-2075.1984.tb02099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Cerio, M., and M. E. Schmitt. Unpublished data.
- 3.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang D D, Clayton D A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 5.Chang D D, Clayton D A. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang D D, Clayton D A. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- 7.Chu S, Archer R H, Zengel J M, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;18:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu S, Zengel J M, Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton D A. A nuclear function for RNase MRP. Proc Natl Acad Sci USA. 1994;91:4615–4617. doi: 10.1073/pnas.91.11.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn B, Szauter P, Pardue M L, Szostak J W. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1984;39:191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- 12.Felici F, Cesareni G, Hughes J M X. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal growth. Mol Cell Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firoozan M, Grant C M, Duarte J A, Tuite M F. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast. 1991;7:173–183. doi: 10.1002/yea.320070211. [DOI] [PubMed] [Google Scholar]
- 14.Forster A C, Altman S. Similar cage-shaped structure for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 15.Gietz R, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 16.Hardy C F, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–6782. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill J H, Meyers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 19.Hogan E, Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson M R, Cao L G, Wang Y L, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilmartin J V, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopczynski J B, Raff A C, Bonner J J. Translational readthrough at nonsense mutations in the HSF1 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1992;234:369–378. doi: 10.1007/BF00538696. [DOI] [PubMed] [Google Scholar]
- 23.Lee D Y, Clayton D A. Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J Biol Chem. 1996;271:24262–24269. doi: 10.1074/jbc.271.39.24262. [DOI] [PubMed] [Google Scholar]
- 24.Lee D Y, Clayton D A. RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication. Genes Dev. 1997;11:582–592. doi: 10.1101/gad.11.5.582. [DOI] [PubMed] [Google Scholar]
- 25.Lee D Y, Clayton D A. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 26.Li K, Smagula C S, Parsons W J, Richardson J A, Gonzalez M, Hagler H K, Williams R S. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liebman S W, Sherman F, Stewart J W. Isolation and characterization of amber suppressors in yeast. Genetics. 1976;82:251–272. doi: 10.1093/genetics/82.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 29.Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 30.Markwell M A K, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 31.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 32.Morrissey J P, Tollervey D. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- 33.Mortimer R K, Gilmore R A. Suppressors and suppressible mutations in yeast. Adv Biol Med Phys. 1968;12:319–331. doi: 10.1016/b978-1-4831-9928-3.50017-5. [DOI] [PubMed] [Google Scholar]
- 34.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 35.Paluh J L, Clayton D A. A functional dominant mutation in Schizosaccharomyces pombe RNase MRP RNA affects nuclear RNA processing and requires the mitochondrial-associated nuclear mutation ptp1-1 for viability. EMBO J. 1996;15:4723–4733. [PMC free article] [PubMed] [Google Scholar]
- 36.Pringle J R, Preston R A, Adams E M, Sterns T, Drubin D G, Haarer B K, Jones E W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 37.Reddy R, Shimba S. –1996. Structural and functional similarities between MRP and RNase P. Mol Biol Rep. 1995;22:81–85. doi: 10.1007/BF00988710. [DOI] [PubMed] [Google Scholar]
- 38.Reilly T H, Schmitt M E. –1996. The yeast, Saccharomyces cerevisiae, RNase P/MRP ribonucleoprotein endoribonuclease family. Mol Biol Rep. 1995;22:87–93. doi: 10.1007/BF00988711. [DOI] [PubMed] [Google Scholar]
- 39.Reimer G, Raska I, Scheer V, Tan E M. Immunolocalization of 7-2 ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- 40.Rose M, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 41.Rubin G M. Three forms of the 5.8S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem. 1974;41:197–202. doi: 10.1111/j.1432-1033.1974.tb03260.x. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Schmitt M E, Clayton D A. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- 44.Schmitt M E, Clayton D A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt M E, Clayton D A. Isolation of a unique protein component of the yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain. Genes Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt M E, Bennett J L, Dairaghi D J, Clayton D A. Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. FASEB J. 1993;7:208–213. doi: 10.1096/fasebj.7.1.7678563. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt M E, Brown T A, Trumpower B L. A rapid, improved method for isolation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 49.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W S, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 50.Sikorski R S, Boeke J D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 51.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stohl L L, Clayton D A. Saccharomyces cerevisiae contains an RNase MRP that cleaves at a conserved mitochondrial RNA sequence implicated in replication priming. Mol Cell Biol. 1992;12:2561–2569. doi: 10.1128/mcb.12.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 1997;11:2414–2425. doi: 10.1101/gad.11.18.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaminathan K, Flynn P, Reece R J, Marmorstein R. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat Struct Biol. 1997;4:751–759. doi: 10.1038/nsb0997-751. [DOI] [PubMed] [Google Scholar]
- 55.Tavernarakis N, Alexandraki D, Liodis P, Tzamarias D, Thireos G. Gene overexpression reveals alternative mechanisms that induce GCN4 mRNA translation. Gene. 1996;179:271–277. doi: 10.1016/s0378-1119(96)00379-4. [DOI] [PubMed] [Google Scholar]
- 56.Toyn J H, Johnston L H. Spo12 is a limiting factor that interacts with the cell cycle protein kinases Dbf2 and Dbf20, which are involved in mitotic chromatid disjunction. Genetics. 1993;135:963–971. doi: 10.1093/genetics/135.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Dyck E, Clayton D A. Transcription-dependent DNA transactions in the mitochondrial genome of a yeast hypersuppressive petite mutant. Mol Cell Biol. 1998;18:2976–2985. doi: 10.1128/mcb.18.5.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visintin R, Hwang E, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 59.Wickner R B. Prions and RNA viruses of Saccharomyces cerevisiae. Annu Rev Genet. 1996;30:109–139. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 60.Xu B, Clayton D A. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol Cell Biol. 1995;15:580–589. doi: 10.1128/mcb.15.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]