Abstract
Bridging integrator-1 (BIN1) is a family of banana-shaped molecules implicated in cell membrane tubulation. To understand the curvature sensitivity and functional roles of BIN1 splicing isoforms, we engineered vertical nanobars on a cell culture substrate to create high and low curvatures. When expressed individually, BIN1 isoforms with phosphoinositide-binding motifs (pBIN1) appeared preferentially at high-curvature nanobar ends, agreeing well with their membrane tubulation in cardiomyocytes. In contrast, the ubiquitous BIN1 isoform without phosphoinositide-binding motif (uBIN1) exhibited no affinity to membranes around nanobars but accumulated along Z-lines in cardiomyocytes. Importantly, in pBIN1-uBIN1 coexpression, pBIN1 recruited uBIN1 to high-curvature membranes at nanobar ends, and uBIN1 attached the otherwise messy pBIN1 tubules to Z-lines. The complementary cooperation of BIN1 isoforms (comboBIN1) represents a novel mechanism of T-tubule formation along Z-lines in cardiomyocytes. Dysregulation of BIN1 splicing, e.g., during myocardial infarction, underlied T-tubule disorganization, and correction of uBIN1/pBIN1 stoichiometry rescued T-tubule morphology in heart disease.
Keywords: nanopillar array, BIN1, splicing isoforms, T-tubule, muscle contraction, heart disease
Graphical Abstract
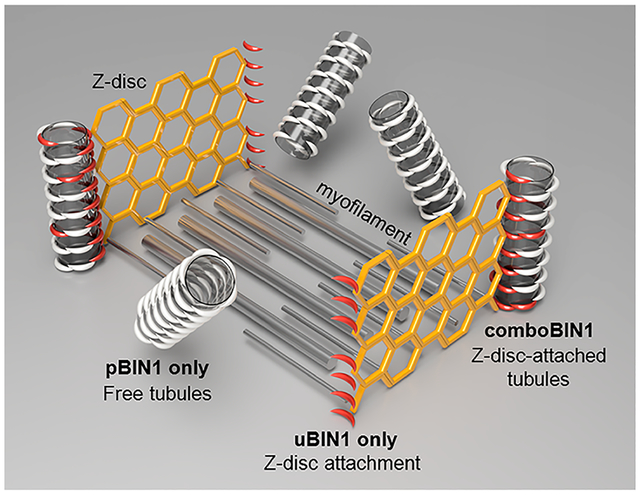
Bridging integrator 1 (BIN1, also known as amphiphysin-2) is a protein family containing a ~25 nm long, banana-shaped Bin/Amphiphysin/Rvs (BAR) domain for membrane-curvature sensing, a C-terminal Src homology 3 (SH3) domain for protein–protein interaction, and a variable central region that diversifies their functions.1,2 Alternative splicing of 20 BIN1 exons results in at least 10 isoforms with distinct functions and tissue specificity, some of which are involved in membrane tubulation in a variety of cells.3–9 In skeletal muscle, BIN1 is implicated in T-tubules (TTs) morphogenesis.8–11 TTs are invagination of the cell membrane with 50–200 nm cross sections.12–14 By forming junctions with the sarcoplasmic reticulum (SR), the major intracellular Ca2+ store, TTs play an essential role in controlling the Ca2+ signaling and contraction in striated muscles.12,14–21 The BIN1 splicing isoform with the phosphoinositide-binding (PI) motif encoded by exon 11 (pBIN1, mainly Bin1 + 11, with a small component of Bin1 + 11 + 17) has been found essential for the generation and maintenance of TTs,1,8–11 while the function of the ubiquitously expressed BIN1 isoform without the PI motif1,8 (uBIN1, mainly Bin1, with a small component of Bin1 + 17) is not known.
In heart muscle, pBIN1, the only BIN1 isoform linked to TT formation in skeletal muscle, has not been found in cardiomyocytes.3,8 Although uBIN1 is expressed, its function is not clear. Currently, the only BIN1 isoform with characterized function is the cardiac-specific BIN1 (Bin1 + 13 + 17, or cBIN1), which creates microdomains on TTs rather than forming a TT network.3,13,22 Therefore, how TTs are formed in cardiomyocytes and whether BIN1 isoforms are involved in cardiac TT formation are open but important questions. In the present study, we utilized the nanobar array technology to characterize the nanocurvature sensing properties and interactions of major BIN1 isoforms expressed in cardiomyocytes and revealed a novel mechanism of cardiac TT formation along Z-lines.
We first profiled the expression of BIN1 splice variants in rat ventricular cardiomyocytes using primers flanking the BIN1 cDNA region between exons 10 and 18. The fragments were cloned into pEASY-T1 plasmids. After sequencing of 200 randomly picked clones, we found 6 BIN1 variants, namely, Bin1, Bin1 + 17, Bin1 + 11, Bin1 + 11 + 17, Bin1 + 13, and Bin1 + 13 + 17 (Figure 1a). Exon 11 encodes the PI motif with only 15 amino acid residues and has not been detected previously in heart muscle.3 We therefore confirmed the expression of pBIN1 in rat cardiomyocytes by the Western blot assay using an antibody against the PI motif (Figure 1b). Moreover, immuno-electron microscopy using pre-embedded immunogold labeling specifically recognized TT membranes in the close vicinity of Z-lines (Figure 1c). These experiments indicated that pBIN1 was expressed natively on TTs in rat heart cells.
Figure 1.
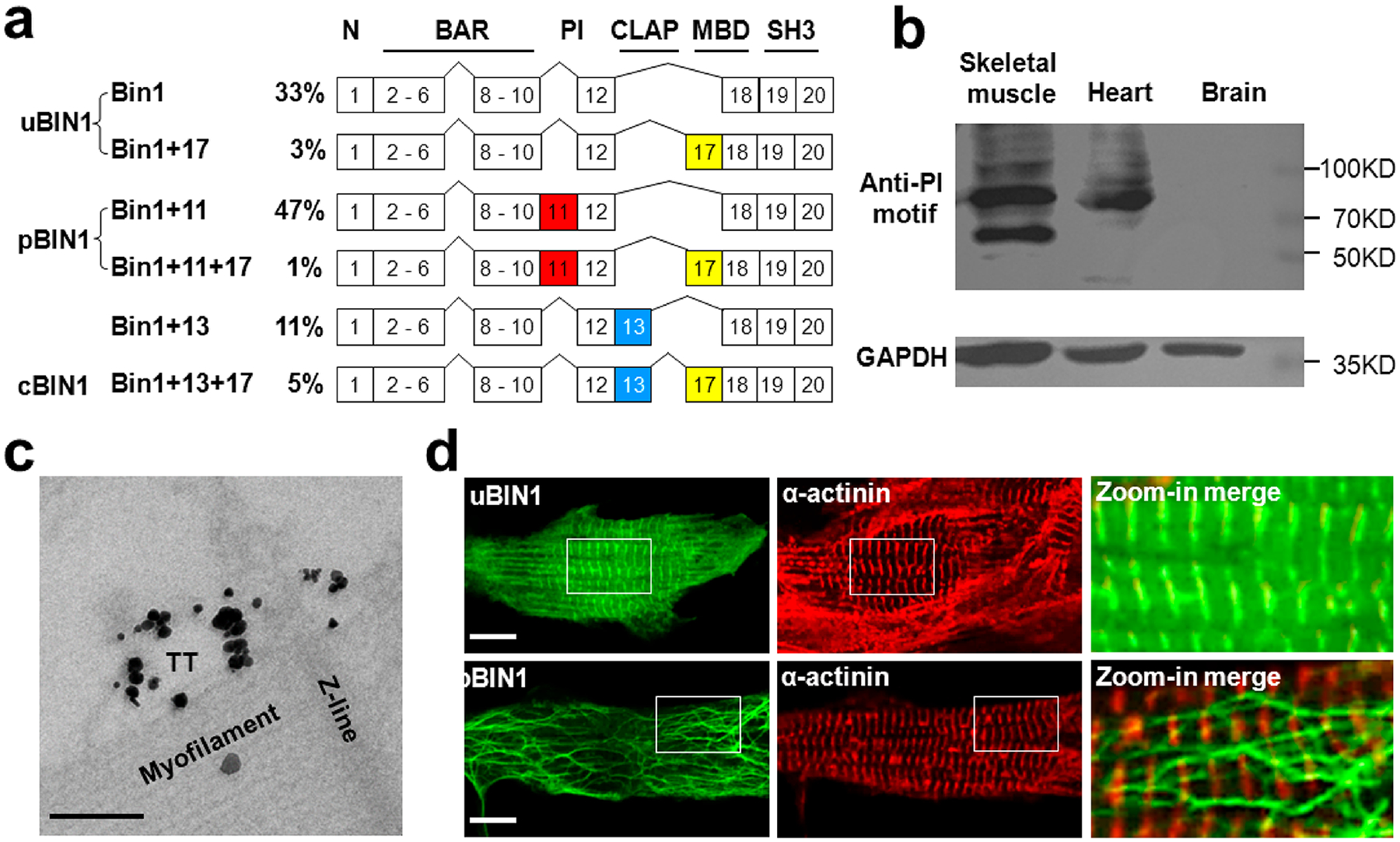
Expression of BIN1 isoforms in rat neonatal cardiomyocytes. (a) Relative expression (in percentage) of BIN1 mRNA splicing (top) and the 6 isoforms identified in cardiomyocytes. N,N-terminal amphipathic helix domain; BAR, BIN1-Amphiphysin-Rvs167 domain; PI, phosphoinositide-binding domain; CLAP, clathrin-associated protein-binding domain; MBD, Myc-binding domain; SH3, Src homology 3 domain. (b) Western blot assay of pBIN1 in rat brain, heart, and skeletal muscle samples. (c) Immuno-EM image of adult rat cardiomyocyte labeled for pBIN1. The gold particle signals were enhanced by HQ SILVER (Nanoprobes). Scale bar, 200 nm. (d) Representative fluorescence images of rat neonatal cardiomyocytes transfected with plasmids of uBIN1-eGFP (upper) and pBIN1-eGFP (lower). The panels from left to right are the fluorescence of eGFP, immunofluorescence of α-actinin, and the zoom-in merged images of the white square areas, respectively. Scale bar, 10 μm.
To explore the possible roles of BIN1 isoforms in cardiac TT formation, we expressed eGFP-tagged uBIN1 and pBIN1 (without exon 17, Figure 1a), in neonatal rat cardiomyocytes, in which native TTs were not formed. We found that the expression of uBIN1 alone did not promote membrane tubulation (Figure 1d upper). uBIN1 signals were highly colocalized with the immunofluorescence of α-actinin (Figure 1d), indicating that the uBIN1 proteins were attached on Z-lines. In contrast, the expression of pBIN1 resulted in a blossom of branched tubules without colocalization with α-actinin (Figure 1d lower). Ultrastructural quantification confirmed that membrane tubules were significantly increased in pBIN1-expressed cardiomyocytes, but not in uBIN1-expressed cells (Figure S1).
uBIN1 and pBIN1 contain the same curvature-sensing BAR domains,1,8 but why only the pBIN1 expression induced membrane tubules? To date, the curvature-sensing capacity of proteins has been primarily studied using in vitro systems.23–25 However, given the complex intracellular protein environment, these properties need to be validated in live cells. Recently, we have developed patterned nanostructures with well-defined curvatures to characterize the curvature-sensing properties of proteins.26–31 Therefore, to understand the functional difference between uBIN1 and pBIN1, we engineered a SiO2 nanobar platform to culture the BIN1 isoform-expressed cells (Figure 2a). In this platform, each nanobar structure locally induced two different membrane curvatures: high curvature at the two ends and zero curvature in the middle (Figure 2b; Figure S2). As shown previously,26,27,32 the plasma membrane of cultured cells wrapped evenly around the bars, provide local membrane models with distinct curvatures.
Figure 2.
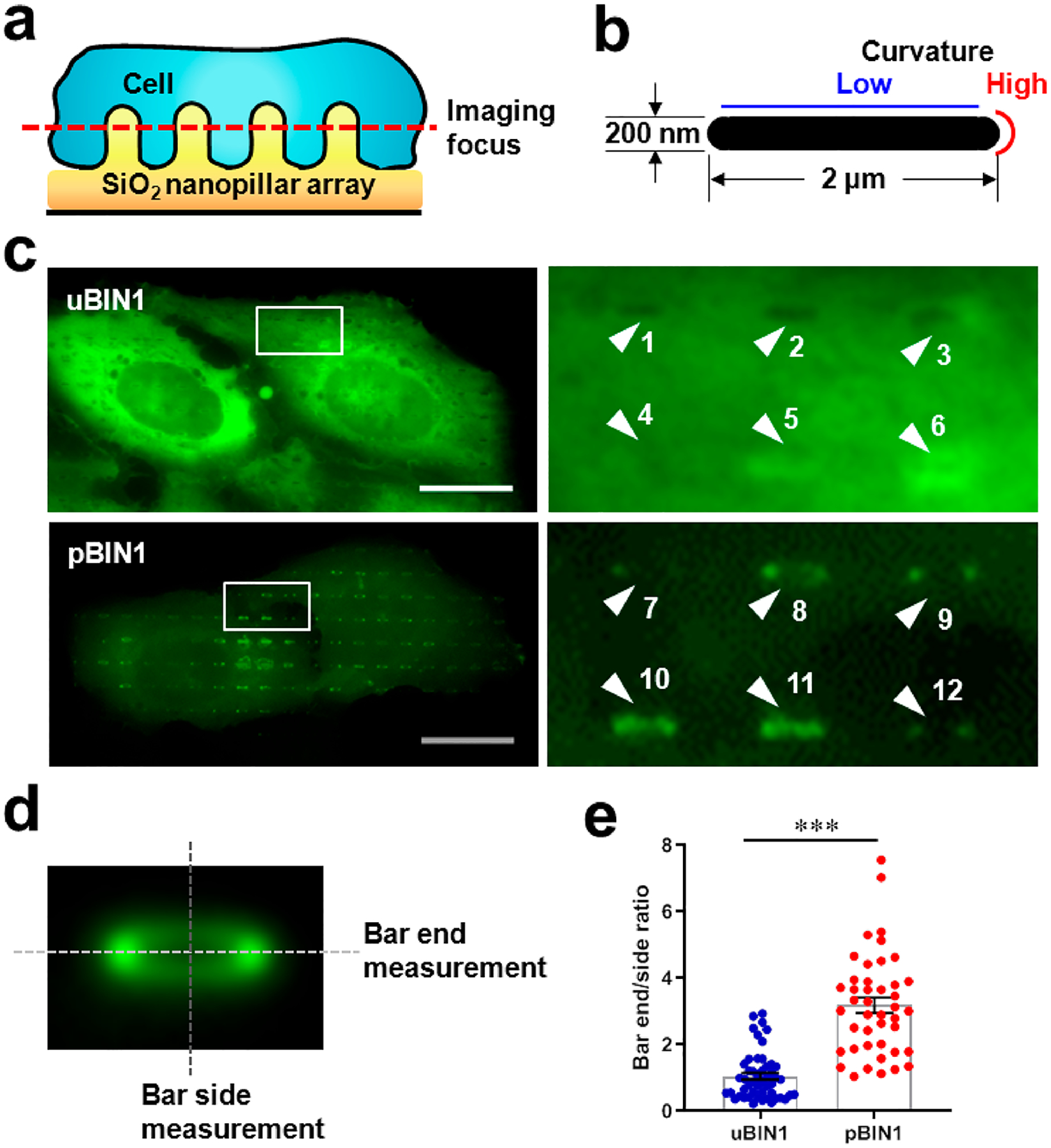
Nanobar experiments to test BIN1 isoforms for curvature sensing. (a) Schematic diagram of nanobar platform. (b) Cross-section of a nanobar illustrating low and high curvatures. (c) Representative fluorescence images of U2OS cells cultured on gelatin-coated, bar-shaped SiO2 nanobar arrays and transfected with eGFP-tagged uBIN1 (upper) and pBIN1 (lower). Left panels were zoom-in images of the white square areas. Numbered arrowheads point to the nanobar signals. (d) Averaged images of 760 nanobars. The dashed lines denote the measurement of the peak fluorescence intensities at bar-ends and bar-sides, respectively. (e) Comparison of bar-end/bar-side intensity ratios. Scale bar, 5 μm. N = 1663 and 1300 nanobars (only 50 plotted) for eGFP-tagged uBIN1 and pBIN1 groups, respectively, ***P < 0.001 vs eGFP-tagged uBIN1 group by t-test.
To test the curvature-sensing and membrane-binding capacities of BIN1 isoforms on the nanobar platform, we expressed uBIN1 and pBIN1 in U2OS cells, which do not have membrane tubules and native BIN1 proteins. When eGFP-tagged uBIN1 was expressed, the fluorescent signals distributed evenly in the cytosol (Figure 2c upper) with little accumulation around nanobars (e.g., Figure 2c Arrows 1–3), indicating that uBIN1 was not bound to the plasma membrane in these areas. Occasionally, uBIN1 fluorescence signals were slightly higher around nanobars (e.g., Arrows 5–6), but their distribution exhibited no curvature preference between bar-ends and bar-sides.
In contrast, when pBIN1 was expressed, the fluorescence signals around nanobars were robustly stronger than the cytosolic background (Figure 2c bottom). The averaged images of over 760 nanobars clearly demonstrated that a ring of pBIN1 fluorescence surrounded nanobars (Figure 2d), indicating that pBIN1 had membrane-binding capability. For most nanobars, the fluorescence in the two bar-end areas was much stronger than that in the bar-side areas (Figure 2c Arrows 7–9 and 12). A few nanobars display fluorescence at both bar-ends and bar-sides (Figure 2c Arrows 10–11). To quantify the curvature preference, we measured the ratio between the peak fluorescence intensity at bar-ends and that at bar-sides (illustrated in Figure 2d). We found that the bar end/side ratio in pBIN1-expressed cells was significantly higher than in cells expressed with uBIN1 (Figure 2e), indicating pBIN1, but not uBIN1 had curvature-sensing capacity.
The only difference between uBIN1 and pBIN1 is the PI motif encoded by exon 11. In order to confirm the specific role of PI motif in making the difference in curvature-sensing, we went on to examine uBIN1 and pBIN1 with the Myc-binding (MB) domains encoded by exon 17 (Bin1 + 17 and Bin1 + 11 + 17). We found that U2OS cells expressed with the uBIN1 with an MB domain shared the same pattern as the uBIN1 without an MB domain, in terms of the evenly distributed florescence in the cytosol (Figure 3a upper), indicating that the additional MB domain did not alter the membrane-phobic property of uBIN1. Again, cells expressed with the pBIN1 with an MB domain exhibited brighter fluorescence around nanobars (Figure 3a lower) and significantly higher bar end/side ratio (Figure 3b) than cells expressed with the uBIN1 with an MB domain, indicating that the gain of curvature-sensing function was attributable specifically to the PI motif encoded by exon 11.
Figure 3.
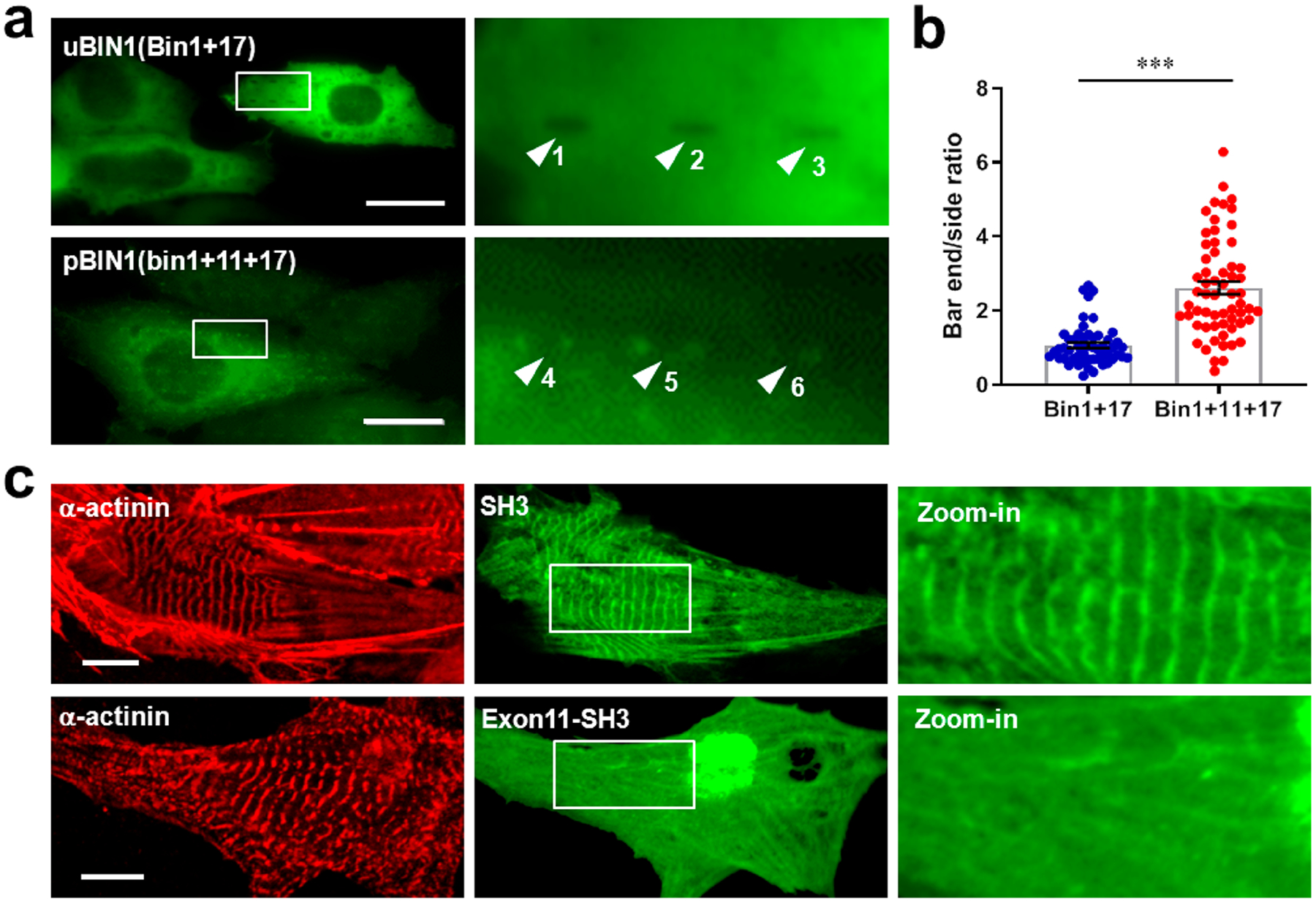
The specificity of PI domain in determining BIN1 isoform properties. (a) Representative fluorescence images of U2OS cells cultured on gelatin-coated, bar-shaped SiO2 nanobar arrays and transfected with eGFP-tagged Bin1 + 17 (upper) and Bin1 + 11 + 17 (lower). White arrows indicate different typical fluorescent signals on the nanobar platform from the two groups. Scale bar, 20 μm. (b) Comparison of bar-end/bar-side intensity ratios. N = 1520 and 1860 nanobars (only 50 plotted) for eGFP-tagged Bin1 + 17 and Bin1 + 11 + 17 groups, respectively, ***P < 0.001 vs eGFP-tagged Bin1 + 17 group. (c) Representative confocal images of rat neonatal cardiomyocytes transfected with SH3-eGFP (upper) and exon 11-SH3-eGFP (lower). The panels from left to right are the immunofluorescence of α-actinin, fluorescence of eGFP, and zoom-in images of eGFP of the square areas in the middle panels, respectively. Scale bar, 10 μm.
In the PI motif, 9 of the 15 amino acid residuals are positively charged.33 Based on previous reports, although the positively changed residues render the PIP2-binding capacity, they may neutralize the electrostatic effect of the SH3 domain.33,34 Indeed, expression of eGFP-tagged SH3 domain alone displayed a Z-line pattern in neonatal cardiomyocytes (Figure 3c upper). However, expression of PI motif-linked SH3 no longer showed a Z-line pattern (Figure 3c lower), confirming the effect of PI-SH3 interdomain neutralization in differentiating uBIN1 and pBIN1 functions.
In light of the above experiments, it is intriguing to know whether pBIN1 is able to interact with uBIN1 and recruits uBIN1 to plasma membrane. The application of the nanobar platform provided a unique opportunity to test this possibility. We therefore cotransfected eGFP-tagged uBIN1 and RFP-tagged pBIN1 (without MB domains) in nanobar-cultured U2OS cells. We observed that, when uBIN1 and pBIN1 were expressed simultaneously in a same cell, the spatial pattern of uBIN1 fluorescence was converted from the otherwise membrane-phobic distribution (Figure 2c upper) to a membrane-preferred distribution (Figure 4a–d). Comparison between eGFP and RFP signals showed that uBIN1 and pBIN1 exhibited quite similar distribution around each nanobar. In most bar areas, both uBIN1 and pBIN1 signals exhibited bar-end biased patterns (Figure 4b). When pBIN1 signals extended to bar-sides, uBIN1 signals were also bright at bar-sides (Figure 4c); when pBIN1 signals were weak, uBIN1 signals showed dark holes (Figure 4d Arrows 1, 2, 4, and 5) similar to those when uBIN1 was expressed alone (Figure 2c upper). The correlation between uBIN1 and pBIN1 signals was not due to spectral overlap, because in cells expressing pBIN1 only, the RFP signal did not cause any detectable signal in the eGFP channel (white arrowheads in Figure 4e), and vice versa (yellow arrowhead in Figure 4e; Figure S3). We also coexpressed eGFP protein with RFP-tagged pBIN1 (Figure 4f). Despite the bright signal outlining the bars in the RFP channel, the bar areas in the eGFP channel still appeared as black holes (e.g., arrows in Figure 4f). The Pearson’s correlation coefficient between eGFP and RFP channels was below zero in cells coexpressed with eGFP and pBIN1, while that in cells coexpressed with uBIN1 and pBIN1 was 0.66 (Figure 4g). In the eGFP channel, while the bar-end/bar-side ratio in cells coexpressed with eGFP and uBIN1 or expressed with uBIN1 only remained around 1 (no preference), that in uBIN1-pBIN1-coexpressed cells was significantly higher (bar-end preferred. Figure 4h), indicating that uBIN1, but not eGFP, was recruited preferentially to high-curvature regions in a pBIN1-dependent manner.
Figure 4.
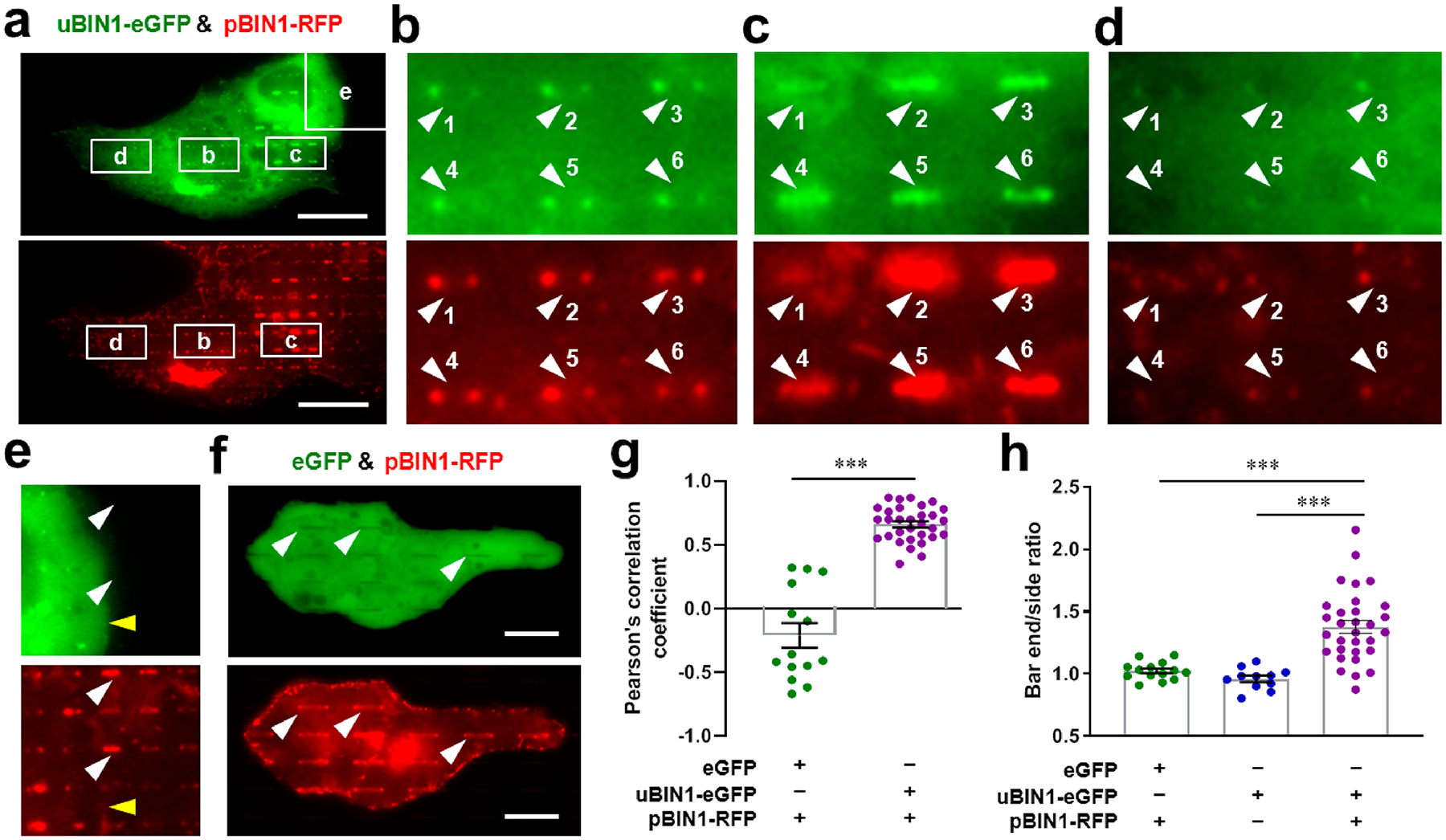
Nanobar experiments to test the interaction between uBIN1 and pBIN1. (a) Representative images of U2OS cells cultured on nanobar arrays and cotransfected with uBIN1-eGFP (upper) and pBIN1-RFP (lower). Scale bar, 20 μm. (b)–(e) The zoom-in images of square areas marked in (a). The numbered white arrowheads point to pBIN1 signals around nanobars (lower) and corresponding positions in the eGFP channel (upper). The yellow arrowheads in (e) denote the border of strong signals in the eGFP channel (upper), which did not cause an background difference in the RFP channel (lower). (f) Representative images of U2OS cells cotransfected with eGFP (upper) and pBIN1-RFP (lower). (g) Pearson’s correlation coefficient between eGFP and RFP channels in cells coexpressed with eGFP and coexpressed with uBIN1 and pBIN1. (h) Comparison of bar-end/bar-side intensity ratios among cells coexpressed with eGFP and pBIN1-RFP, expressed with uBIN1-eGFP only and coexpressed with uBIN1-eGFP and pBIN1-RFP. ***P < 0.001 vs eGFP-tagged uBIN1 group by t-test.
The finding of uBIN1 recruitment by pBIN1 suggested an intriguing hypothesis that TTs may be formed based on an intermolecular integration of uBIN1 and pBIN1 (“combo-BIN1”). In mature cardiomyocytes, TTs are formed along Z-lines. However, the tubules formed by pBIN1 alone did not attached to Z-lines (Figure 1d lower). Although uBIN1 was attached to Z-lines (Figure 1d upper), it was not capable of tubule generation (Figure S1). Therefore, comboBIN1 may provide a perfect solution for TT formation along Z-lines.
To test the comboBIN1 hypothesis directly, we coexpressed eGFP-tagged uBIN1 and RFP-tagged pBIN1 in neonatal rat cardiomyocytes (Figure 5a). We found that the tubules formed under this condition became fewer but better organized. In intracellular areas with regularly spaced α-actinin or Z-line signals, pBIN1 and uBIN1 signals colocalized with each other forming short tubules along Z-lines (white arrowheads in Figure 5a). Even for pBIN1 signals not occurring as typical tubular structures, they were attached on Z-lines as bud-like structures (yellow arrowheads in Figure 5a). This finding demonstrated that comboBIN1 indeed enabled Z-line-based TT formation.
Figure 5.
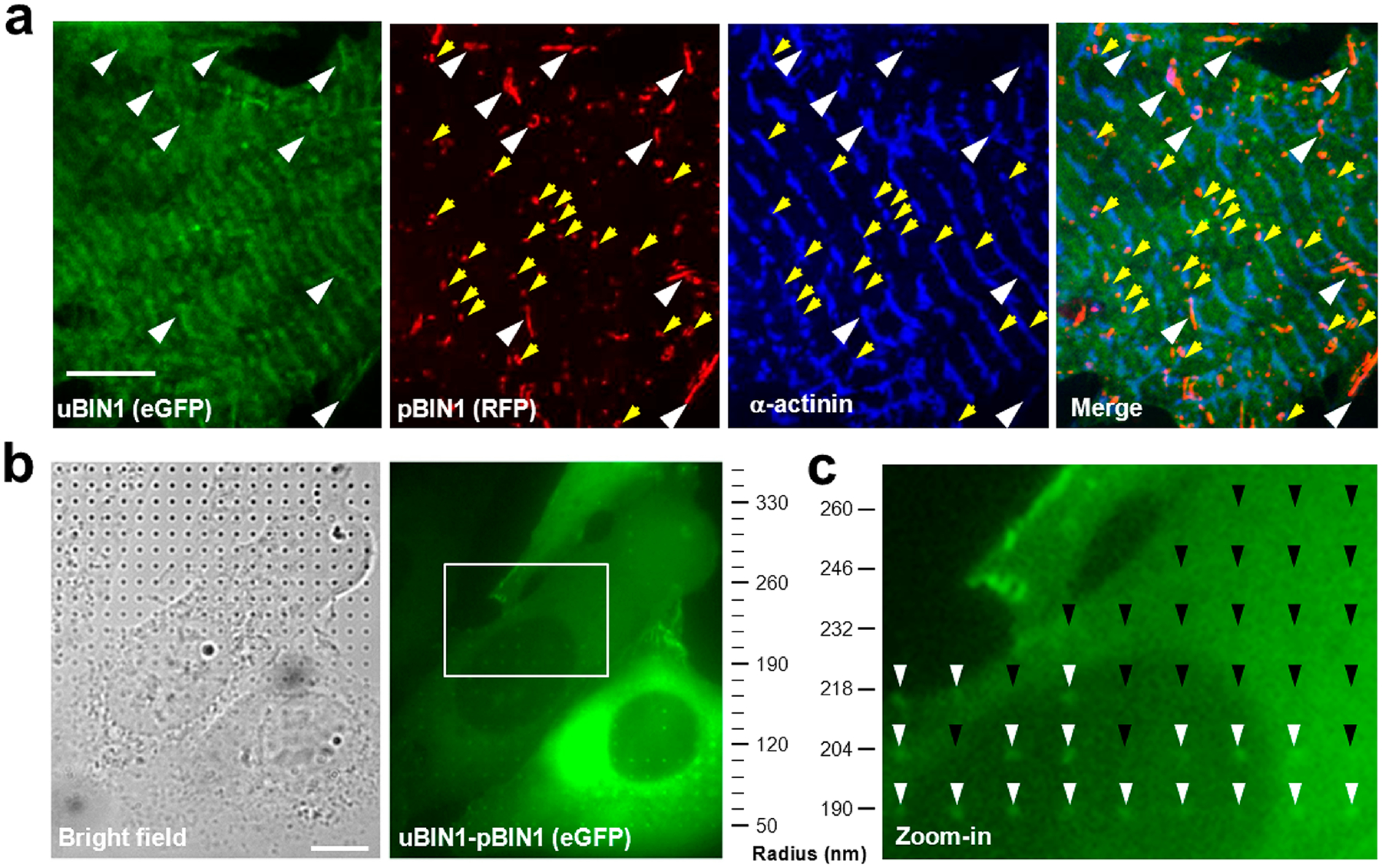
ComboBIN1 enabled Z-line-based tubule formation with nanocurvature preference. (a) Structured illumination microscopy images of rat neonatal cardiomyocytes cotransfected with uBIN1-eGFP and pBIN1-RFP. White arrowheads indicate comboBIN1-induced tubules along Z-lines, and yellow arrowheads denote tubule buds in the vicinity of Z-lines. Scale bar, 5 μm. (b). Typical images of bright field (left) and eGFP fluorescence (right) in cells expressed with eGFP-pBIN1-uBIN1 (linked ComboBIN1, right). The cells were cultured on a nanopillar platform with a gradient of pillar radii from 50 to 500 nm (up to 350 nm displayed). Scale bar, 10 μm. (c) The zoom-in image of the square area in (b). The white and black arrowheads denote the presence and absence of eGFP signals at the nanopillar positions.
In cardiomyocytes, the radius of TTs usually ranges around 50–200 nm.12,13 The comboBIN1-mediated TT formation requires that their curvature preference matches the TT morphology. To examine the curvature preference of comboBIN1, we utilized a SiO2 nanopillar array with a gradient of pillar radii from 50 to 500 nm with a 14 nm increment.26,27 To avoid the variation of local stoichiometry caused by inhomogeneity of comboBIN1 components, uBIN1 and pBIN1 were connected with a flexible linker, fixing their stoichiometry at 1:1, roughly reflecting their relative mRNA abundance in rat heart cells (Figure 1a). When the linked comboBIN1 was expressed in U2OS cells couture on gradient nanopillar array (Figure 5b left), we found that the comboBIN1 signals occurred mainly in the 50–200 nm radii range (Figure 5b right). Beyond 200 nm, the chance for comboBIN1 signals to occur around nanopillars, even within a same cell, decayed sharply to zero (white arrow heads in Figure 5c). These results indicated that the curvature preference of comboBIN1 well matched the requirement of TT formation.
In heart diseases with defective E–C coupling, such as ischemic cardiomyopathy and heart failure, TTs are degraded.15–20 To determine whether abnormal TT remodeling involves an altered stoichiometry of comboBIN1, we analyzed BIN1 expression in a rat myocardial infarction (MI) model created by ligation of the left anterior descending coronary artery, which displayed a defective TT network (Figure S4a) as reported previously. Real-time RT-PCR assay revealed decreased uBIN1 expression but increased pBIN1 expression in the MI group (Figures S4b,c). It has been reported that the splicing regulator muscleblind-like-1 (MBNL1) binds to BIN1 pre-mRNA and promotes splicing inclusion of exon 11.8 We found that Mbnl1 expression was upregulated in the MI group (Figure S4d), agreeing well with the increased pBIN1/uBIN1 ratio. Given the Z-line anchoring property of uBIN1, we inferred that the decrease of uBIN1 expression would compromise the attachment of pBIN1 encompassed TTs along Z-lines, which would thus explain at least partially the irregularity of TTs in the MI group.
To test the above hypothesis, we created an inducible rat model for heart-specific uBIN1 (Bin1) overexpression (BOE, Figure 6a). Two weeks after the tamoxifen treatment (Figure 6b), eGFP-tagged uBIN1 expression was induced in adult BOE rat heart cells without altering native BIN1 expression (Figures 6c and d). The striated florescence in BOE cardiomyocytes indicated the Z-line-biased positioning of overexpressed eGFP-tagged uBIN1 (Figure 6e upper right). We then generated the MI model in control and BOE rats and analyzed the morphology of TTs labeled with CellMask Deep Red (Figure 6e lower panels) 1 week after the MI surgery. Quantification of the regularity of TTs showed that the spatial frequency power corresponding to inter-Z-line space was significantly higher in the BOE group than in the control group (Figure 6f). This result supported the idea that higher uBIN1/pBIN1 ratio would promote the Z-line positioning of TTs.
Figure 6.
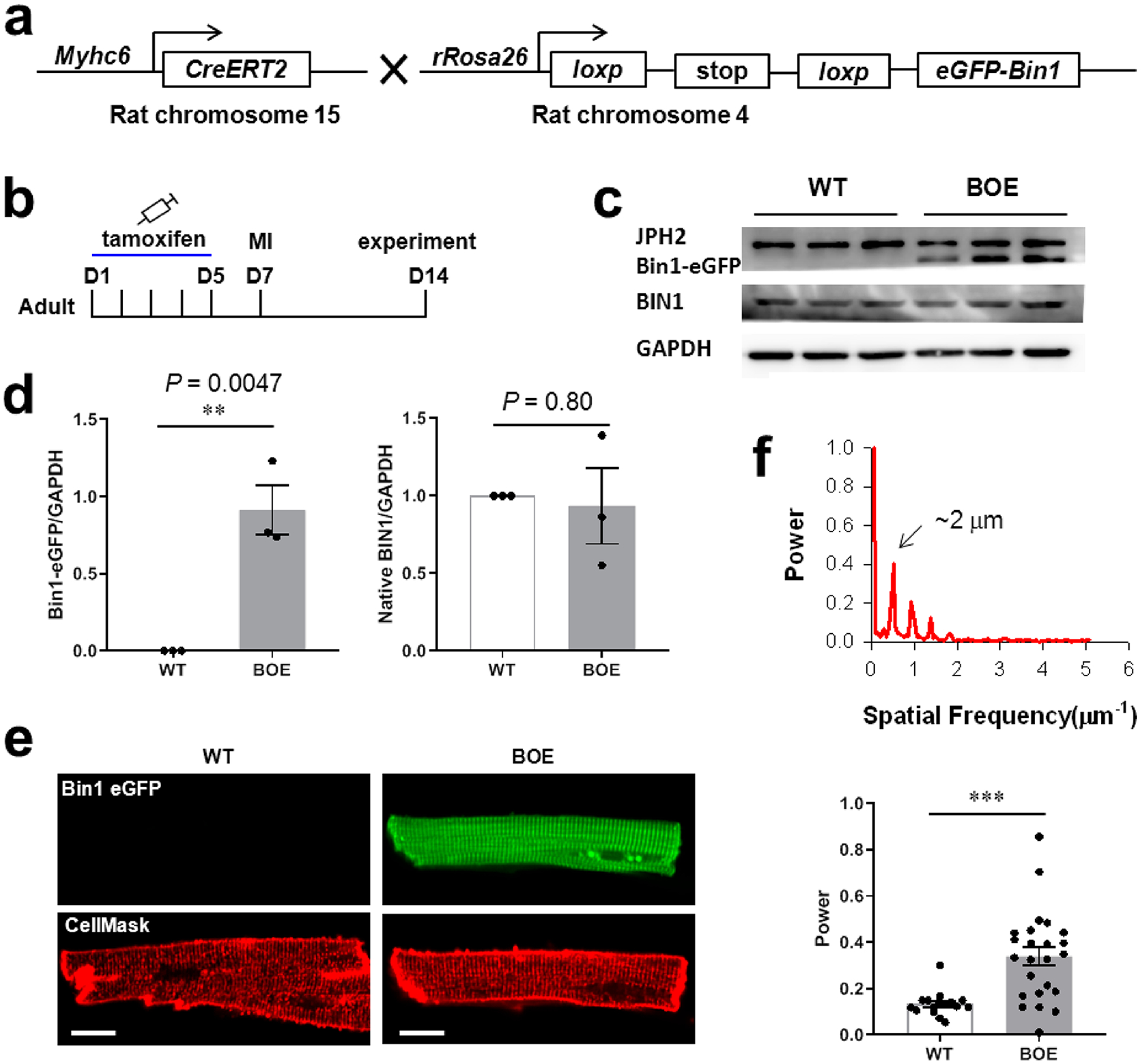
Overexpression of uBIN1 (BOE) in vivo promoted Z-line positioning of TTs in rat MI model. (a) Schematic illustration of α-MHCCreERT2 (Cre) rats and rROSA26loxp-stop-loxp-eGFP-Bin1 rats, the cross of which produces inducible cardiac-specific BOE rats. (b) Time schedule of tamoxifen injection, MI model generation, and the functional experiments. (c) Typical Western blot images of samples from WT and BOE groups. (d) Comparison of overexpressed uBIN1 (Bin1-eGFP, left) and native total BIN1 (right) protein levels between WT and BOE groups. N = 3, **P < 0.01 vs sham group by t-test. (e) Typical images of eGFP fluorescence (upper) and CellMask Deep Red fluorescence (lower) of the cardiomyocytes form the border zone after MI surgery in WT and BOE rats. Scale bar, 10 μm. (f) Quantification of the spatial frequency by fast Fourier transformation of the cardiomyocytes stained by the CellMask Deep Red dye. The upper panel shows an example of fast Fourier transformation, and the lower panel shows the comparison between WT and BOE groups. N = 16 and 24 cardiomyocytes for WT and BOE groups, respectively. ***P < 0.001 vs WT group by t-test.
As members of the BAR domain superfamily, BIN1 isoforms play essential roles in the formation of different high-curvature membrane structures in a large variety of cell types.1,3–9 In cardiomyocytes, 4 BIN1 splicing isoforms, including Bin1, Bin1 + 17, Bin1 + 13, and Bin1 + 13 + 17, have been reported.3,13,22 pBIN1 was considered as the skeletal muscle-specific isoform because it has not been previously found in other tissues, including mouse myocardium.3,13 In the present study, we have for the first time detected the expression of pBIN1 in heart cells. Our experiments provided 3 lines of evidence supporting its expression in rat (but not mouse) hearts: (1) We cloned pBIN1 in rat myocardium using primers flanking the BIN1 cDNA region between exons 10–18 and identified exon 11 in 48%. (2) Using a specific antibody against the PI domain, we detected pBIN1 proteins in rat myocardium. (3) Immuno-electron microscopy showed that pBIN1 was a native component of the TTs in rat cardiomyocytes.
Using the nanobar arrays technology, we proved that pBIN1, but not uBIN1, was a curvature-sensing and membrane-binding molecule. When expressed in neonatal cardiomyocytes, pBIN1, but not uBIN1, promoted the generation of membrane tubules. Although the cardiac specific cBIN1 is found to create membrane folds on TTs, it does not mediate tubulation.3 Therefore, our finding of the curvature-sensing and tubule-generating properties of pBIN1 provides the first candidate mechanism for the generation of membrane tubules in heart cells.
Although uBIN1 is expressed in both skeletal and cardiac muscles, its function was not known. Here, our study provided the following new information: (1) When expressed alone, pBIN1 (but not uBIN1) was able to organize the formation of free membrane tubules with a certain curvature, while uBIN1 (but not pBIN1) was attached on Z-lines without membrane association. (2) The nanobar/nanopillar experiments established that pBIN1 was able to recruits uBIN1 to high-curvature membrane regions. (3) Coexpression of uBIN1 and pBIN1 generated premature tubules anchored to Z-lines in neonatal cardiomyocytes, preventing the random tubulation by pBIN1. (4) Decreased uBIN1/pBIN1 ratio is associated with decreased TT positioning along Z-lines under MI conditions. (5) Overexpression of uBIN1 to increase the uBIN1/pBIN1 ratio improved the regularity of TTs along Z-lines.
On the basis of these evidence, we proposed a “comboBIN1” model (Figure S5) to explain the roles of uBIN1 and pBIN1 in Z-line-based tubule generation: In cardiomyocytes expressing both uBIN1 and pBIN1, while pBIN1 promotes tubule generation, uBIN1, due to its Z-line tethering, anchors the tubules to Z-lines presumably via uBIN1-pBIN1 interaction. This model provided a novel mechanism of TT formation and suggested new strategies for treating related heart diseases.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Drs. TingTing Hong and Robin Shaw in Cedars-Sinai Medical Center and UCLA for helps and insightful comments, and Drs. Ying-Chun Hu, Chun-Yan Shan and Xue-Mei Hao in the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with electron microscopy and optical imaging. This study was supported by the State Research and Development Program (2016YFA0500401); the National Natural Science Foundation of China (91854209, 31630035, 31971116, and 31327901); the National Institute of Health (NIH 1R01GM128142 and R01GM125737); and the Packard Fellowship for Science and Engineering (to B.C.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c01957.
Figure S1, tubules induced by pBIN1 but not by uBIN1; Figure S2, morphology of engineered 3D nanostructures; Figure S3, independence between RFP and eGFP fluorescent channels; Figure S4, altered expression of BIN1 splice variants in rat MI models; Figure S5, comboBIN1 model for the roles of uBIN1 and pBIN1 cooperation in tubule generation along Z-lines; detailed description of the animals, nanostructures fabrication, cardiac-specific BOE rats, induction of myocardial infarction (MI), cardiomyocytes preparation, molecular cloning, quantitative RT-PCR analysis of BIN1 isoforms, Western blot analysis, membrane labeling and imaging, immunofluorescence imaging, immnuno-electron microscopy, transmission electron microscopy (TEM) imaging and analysis, BIN1 curvature-sensing assay and data statistics (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.nanolett.0c01957
The authors declare no competing financial interest.
Contributor Information
Lin-Lin Li, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China; National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing 100190, China.
Qian-Jin Guo, State Key Laboratory of Membrane Biology, Institute of Molecular Medicine, Peking University, Beijing 100871, China.
Hsin-Ya Lou, Department of Chemistry, Stanford University, Stanford, California 94305, United States.
Jing-Hui Liang, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China.
Yang Yang, Department of Chemistry, Stanford University, Stanford, California 94305, United States.
Xin Xing, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China.
Hong-Tao Li, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China.
Jing Han, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China.
Shan Shen, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China.
Hui Li, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China.
Haihong Ye, Department of Medical Genetics and Developmental Biology, School of Basic Medical Sciences, Beijing Key Laboratory of Neural Regeneration and Repair, Capital Medical University, Beijing 100069, China.
Hao Di Wu, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China.
Bianxiao Cui, Department of Chemistry, Stanford University, Stanford, California 94305, United States;.
Shi-Qiang Wang, State Key Laboratory of Membrane Biology, College of Life Sciences, Peking University, Beijing 100871, China;.
REFERENCES
- (1).Lee E; Marcucci M; Daniell L; Pypaert M; Weisz OA; Ochoa GC; Farsad K; Wenk MR; De Camilli P Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 2002, 297 (5584), 1193–1196. [DOI] [PubMed] [Google Scholar]
- (2).Bhatia VK; Madsen KL; Bolinger PY; Kunding A; Hedegard P; Gether U; Stamou D Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009, 28 (21), 3303–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hong T; Yang H; Zhang SS; Cho HC; Kalashnikova M; Sun B; Zhang H; Bhargava A; Grabe M; Olgin J; Gorelik J; Marban E; Jan LY; Shaw RM Cardiac BIN1 folds T-tubule membrane, controlling ion flux and limiting arrhythmia. Nat. Med 2014, 20 (6), 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wechsler-Reya R; Sakamuro D; Zhang J; Duhadaway J; Prendergast GC Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J. Biol. Chem 1997, 272 (50), 31453–31458. [DOI] [PubMed] [Google Scholar]
- (5).De La Mata A; Tajada S; O’Dwyer S; Matsumoto C; Dixon RE; Hariharan N; Moreno CM; Santana LF BIN1 Induces the Formation of T-Tubules and Adult-Like Ca2+ Release Units in Developing Cardiomyocytes. Stem Cells 2019, 37 (1), 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gowrisankaran S; Wang Z; Morgan DG; Milosevic I; Mim C Cells Control BIN1-Mediated Membrane Tubulation by Altering the Membrane Charge. J. Mol. Biol 2020, 432 (4), 1235–1250. [DOI] [PubMed] [Google Scholar]
- (7).Andrew RJ; De Rossi P; Nguyen P; Kowalski HR; Recupero AJ; Guerbette T; Krause SV; Rice RC; Laury-Kleintop L; Wagner SL; Thinakaran G Reduction of the expression of the late-onset Alzheimer’s disease (AD) risk-factor BIN1 does not affect amyloid pathology in an AD mouse model. J. Biol. Chem 2019, 294 (12), 4477–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Fugier C; Klein AF; Hammer C; Vassilopoulos S; Ivarsson Y; Toussaint A; Tosch V; Vignaud A; Ferry A; Messaddeq N; Kokunai Y; Tsuburaya R; de la Grange P; Dembele D; Francois V; Precigout G; Boulade-Ladame C; Hummel MC; Lopez de Munain A; Sergeant N; Laquerriere A; Thibault C; Deryckere F; Auboeuf D; Garcia L; Zimmermann P; Udd B; Schoser B; Takahashi MP; Nishino I; Bassez G; Laporte J; Furling D; Charlet-Berguerand N Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med 2011, 17 (6), 720–725. [DOI] [PubMed] [Google Scholar]
- (9).Nicot AS; Toussaint A; Tosch V; Kretz C; Wallgren-Pettersson C; Iwarsson E; Kingston H; Garnier JM; Biancalana V; Oldfors A; Mandel JL; Laporte J Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat. Genet 2007, 39 (9), 1134–9. [DOI] [PubMed] [Google Scholar]
- (10).Lionello VM; Nicot AS; Sartori M; Kretz C; Kessler P; Buono S; Djerroud S; Messaddeq N; Koebel P; Prokic I; Herault Y; Romero NB; Laporte J; Cowling BS Amphiphysin 2 modulation rescues myotubular myopathy and prevents focal adhesion defects in mice. Sci. Transl. Med 2019, 11 (484), eaav1866. [DOI] [PubMed] [Google Scholar]
- (11).Cowling BS; Prokic I; Tasfaout H; Rabai A; Humbert F; Rinaldi B; Nicot AS; Kretz C; Friant S; Roux A; Laporte J Amphiphysin (BIN1) negatively regulates dynamin 2 for normal muscle maturation. J. Clin. Invest 2017, 127 (12), 4477–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Brette F; Orchard C T-tubule function in mammalian cardiac myocytes. Circ. Res 2003, 92 (11), 1182–1192. [DOI] [PubMed] [Google Scholar]
- (13).Hong T; Shaw RM Cardiac T-Tubule Microanatomy and Function. Physiol. Rev 2017, 97 (1), 227–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pinali C; Bennett H; Davenport JB; Trafford AW; Kitmitto A Three-dimensional reconstruction of cardiac sarcoplasmic reticulum reveals a continuous network linking transversetubules: this organization is perturbed in heart failure. Circ. Res 2013, 113 (11), 1219–1230. [DOI] [PubMed] [Google Scholar]
- (15).Guo A; Zhang C; Wei S; Chen B; Song LS Emerging mechanisms of T-tubule remodelling in heart failure. Cardiovasc. Res 2013, 98 (2), 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Feng Y; Zhang Y; Xiao H AMPK and cardiac remodeling. Sci. China: Life Sci 2018, 61 (1), 14–23. [DOI] [PubMed] [Google Scholar]
- (17).Zhang C; Chen B; Guo A; Zhu Y; Miller JD; Gao S; Yuan C; Kutschke W; Zimmerman K; Weiss RM; Wehrens XH; Hong J; Johnson FL; Santana LF; Anderson ME; Song LS Microtubule-mediated defects in junctophilin-2 trafficking contribute to myocyte transverse-tubule remodeling and Ca2+ handling dysfunction in heart failure. Circulation 2014, 129 (17), 1742–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Du X Post-infarct cardiac injury, protection and repair: roles of non-cardiomyocyte multicellular and acellular components. Sci. China: Life Sci 2018, 61 (3), 266–276. [DOI] [PubMed] [Google Scholar]
- (19).Zhang HB; Li RC; Xu M; Xu SM; Lai YS; Wu HD; Xie XJ; Gao W; Ye H; Zhang YY; Meng X; Wang SQ Ultrastructural uncoupling between T-tubules and sarcoplasmic reticulum in human heart failure. Cardiovasc. Res 2013, 98 (2), 269–276. [DOI] [PubMed] [Google Scholar]
- (20).Zhang XX; Liu HY; Gao J; Zhu M; Wang YP; Jiang CT; Xu M Metabolic disorder in the progression of heart failure. Sci. China: Life Sci 2019, 62 (9), 1153–1167. [DOI] [PubMed] [Google Scholar]
- (21).Pinali C; Malik N; Davenport JB; Allan LJ; Murfitt L; Iqbal MM; Boyett MR; Wright EJ; Walker R; Zhang Y; Dobryznski H; Holt CM; Kitmitto A, Post-Myocardial infarction T-tubules form enlarged branched structures with dysregulation of junctophilin-2 and bridging integrator 1 (BIN-1). J. Am. Heart Assoc 2017, 6 (5). DOI: 10.1161/JAHA.116.004834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Xu B; Fu Y; Liu Y; Agvanian S; Wirka RC; Baum R; Zhou K; Shaw RM; Hong T The ESCRT-III pathway facilitates cardiomyocyte release of cBIN1-containing microparticles. PLoS Biol. 2017, 15 (8), No. e2002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Daum B; Auerswald A; Gruber T; Hause G; Balbach J; Kuhlbrandt W; Meister A Supramolecular organization of the human N-BAR domain in shaping the sarcolemma membrane. J. Struct. Biol 2016, 194 (3), 375–382. [DOI] [PubMed] [Google Scholar]
- (24).Heinrich MC; Capraro BR; Tian A; Isas JM; Langen R; Baumgart T Quantifying Membrane Curvature Generation of Drosophila Amphiphysin N-BAR Domains. J. Phys. Chem. Lett 2010, 1 (23), 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Simunovic M; Evergren E; Golushko I; Prevost C; Renard HF; Johannes L; McMahon HT; Lorman V; Voth GA; Bassereau P How curvature-generating proteins build scaffolds on membrane nanotubes. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (40), 11226–11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhao W; Hanson L; Lou HY; Akamatsu M; Chowdary PD; Santoro F; Marks JR; Grassart A; Drubin DG; Cui Y; Cui B Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nat. Nanotechnol 2017, 12 (8), 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Santoro F; Zhao W; Joubert LM; Duan L; Schnitker J; van de Burgt Y; Lou HY; Liu B; Salleo A; Cui L; Cui Y; Cui B Revealing the Cell-Material Interface with Nanometer Resolution by Focused Ion Beam/Scanning Electron Microscopy. ACS Nano 2017, 11 (8), 8320–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li X; Matino L; Zhang W; Klausen L; McGuire AF; Lubrano C; Zhao W; Santoro F; Cui B A nanostructure platform for live-cell manipulation of membrane curvature. Nat. Protoc 2019, 14 (6), 1772–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lou HY; Zhao W; Li X; Duan L; Powers A; Akamatsu M; Santoro F; McGuire AF; Cui Y; Drubin DG; Cui B Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proc. Natl. Acad. Sci. U. S. A 2019, 116 (46), 23143–23151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hanson L; Lin ZC; Xie C; Cui Y; Cui B Characterization of the cell-nanopillar interface by transmission electron microscopy. Nano Lett. 2012, 12 (11), 5815–5820. [DOI] [PubMed] [Google Scholar]
- (31).Xie C; Hanson L; Xie W; Lin Z; Cui B; Cui Y Noninvasive neuron pinning with nanopillar arrays. Nano Lett. 2010, 10 (10), 4020–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Galic M; Jeong S; Tsai FC; Joubert LM; Wu YI; Hahn KM; Cui Y; Meyer T External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nat. Cell Biol 2012, 14 (8), 874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kojima C; Hashimoto A; Yabuta I; Hirose M; Hashimoto S; Kanaho Y; Sumimoto H; Ikegami T; Sabe H Regulation of Bin1 SH3 domain binding by phosphoinositides. EMBO J. 2004, 23 (22), 4413–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wu T; Baumgart T BIN1 membrane curvature sensing and generation show autoinhibition regulated by downstream ligands and PI(4,5)P2. Biochemistry 2014, 53 (46), 7297–7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


