Abstract
Campylobacter jejuni produces a toxin called cytolethal distending toxin (CDT). Knowledge of the prevalence and homogeneity of Campylobacter sp. cdt genes is incomplete. In this work, we identified four PCR primer pairs that collectively amplified cdt genes in all of the C. jejuni and Campylobacter coli strains tested. Restriction analyses of the cdt PCR products showed clear differences between the cdt genes of these two species, yet there were few heterogeneities noted between members of the same species. Consequently, it may be possible to speciate C. jejuni and C. coli isolates on the basis of restriction patterns within their cdt genes.
Campylobacteriosis is one of the most common bacterial enteritises in many industrialized countries, including the United States. The most common Campylobacter species implicated as causes of Campylobacter-induced enteritis are Campylobacter jejuni and Campylobacter coli; C. jejuni may account for as much as 95% of all Campylobacter isolates from diarrheal cases in the United States (11, 17). The clinical spectrum of Campylobacter enteritis ranges from a watery, nonbloody, noninflammatory diarrhea to a severe inflammatory diarrhea with abdominal pain and fever (3, 13, 30).
Several potential virulence determinants have been proposed for C. jejuni and C. coli, including motility, adherence, invasive capabilities, and toxin production (12, 30, 31). The best characterized of the toxins attributed to Campylobacter spp. is cytolethal distending toxin (CDT), a toxic activity first shown by Johnson and Lior (10) to be produced by several Campylobacter spp. CDT causes progressive cellular distention and, ultimately, death in Chinese hamster ovary (CHO), Vero, HEp-2, and HeLa cells (10). The C. jejuni cdt genes have been cloned and sequenced (24), and Whitehouse et al. (32) have recently shown that C. jejuni CDT causes sensitive cells to become blocked in the G2 phase of their cell cycle, indicating that CDT has a novel mechanism of action for a bacterial toxin. While the role of CDT in disease has not been defined, Okuda et al. (19) have shown that a CDT produced by Shigella dysenteriae is capable of causing diarrhea in a suckling mouse model.
Cdt genes have also been found in some Escherichia coli isolates (4, 7, 9, 20, 23, 25), in some Shigella sp. isolates (8, 18, 19), in Haemophilus ducreyi (5), and in Actinobacillus actinomycetemcomitans (29). In all cases there are three adjacent or slightly overlapping genes, cdtA, cdtB, and cdtC, all of whose expression is apparently required for activity (19, 23, 24). The predicted amino acid sequences for the three Cdt proteins indicate that considerable amino acid sequence divergence has occurred, particularly within the CdtA and CdtC sequences. For example, the predicted amino acid sequences of the CdtA proteins from E. coli 9142-88 and C. jejuni 81-176 are only 34% similar (21% identical and 13% conserved amino acids [23]). In addition, Cdt proteins from isolates from the same species can be substantially divergent, since the predicted amino acid sequences of the CdtA proteins from E. coli 9142-88 and E6468/62 are only 53% similar (37% identical and 17% conserved amino acids [23, 25]). Given this sequence divergence that is seen both within the same species and among different species, it seemed likely that the best methods for screening for cdt genes in many Campylobacter isolates would be a PCR method that would not depend on extensive regions of DNA homology. Consequently, we decided to design and test PCR primers for use in the detection of cdt genes in C. jejuni and C. coli strains. The development of useful cdt gene PCR primers will allow investigators to quickly evaluate whether Campylobacter sp. isolates contain cdt genes, thereby furthering our knowledge about the prevalence of these genes in isolates from a variety of sources.
Bacterial strains and media.
C. jejuni 81-176 has been described previously (1, 14). C. jejuni 84-142, 85-452, 79-193, 85-360, G13, 84-19, and D133 and C. coli 43473, D730, D2593, 78-64, D2594, D115, and D126 and their relevant CDT characteristics were described by Pickett et al. (24). C. jejuni 79-101 was isolated from a human stool (2), and C. jejuni Lior 19 was isolated from a chicken (16). C. jejuni AED973, AED974, and AEB9710 and C. coli AEB971, AEB979, AEB9713b, and AED9715 were isolated during this work from chicken carcasses by the method described by Hunt and Abeyta (6). Species identification of the chicken isolates and the low-CDT-producing C. jejuni strains was performed by using both API-Campy (Bio-Merieux, Marcy l’Etoile, France) and a species-specific PCR method which uses two sets of primers that amplify unique regions of either the C. jejuni or C. coli genome (27, 28). In addition, a PCR method for detection of the C. jejuni hippuricase gene was used as an additional speciation method for strain AED974 (15), since this strain produced an atypical API-Campy result. A 735-bp product was amplified from strain AED974 by using the hippuricase primers, indicating that AED974 is a C. jejuni strain. Campylobacter species were grown on brucella agar as previously described (22). When necessary, selective antibiotics for Campylobacter were added to the following final concentrations: for cephalothin, 15 μg/ml; for vancomycin, 10 μg/ml; and for trimethoprim, 5 μg/ml.
PCR.
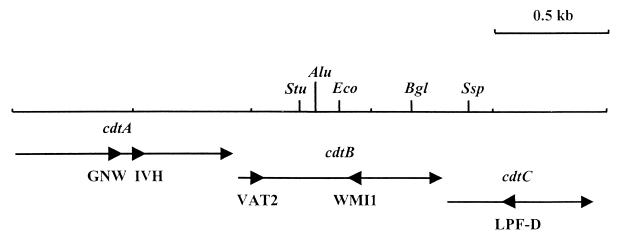
Total bacterial cell DNA was isolated from Campylobacter strains either by the method described by Silhavy et al. (26) or by using the QIAamp tissue kit according to the manufacturer’s specifications (Qiagen, Santa Clarita, Calif.). PCR reagents were obtained from Perkin-Elmer (Norwalk, Conn.), and all primers were purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). Chromosomal DNA from C. jejuni and C. coli isolates was used as the template in PCRs with different primer pairs as follows: VAT2 and WMI1, VAT2 and LPF-D, GNW and WMI1, or IVH and WMI1. VAT2 and WMI1 (Fig. 1) were described by Pickett et al. (24) and are both based on highly conserved regions of the cdtB genes of E. coli 9142-88 and E6468/62. LPF-D (Fig. 1) is based on a conserved amino acid sequence (LPFGYVQ) in the C. jejuni 81-176 and C. coli D730 cdtC genes and has the sequence 5′-(AGT)AA(CT)TG(ACGT)AC(AGT)TA(ACGT)CC(AGT)AA(ACGT)GG-3′ (21, 24). GNW and IVH (Fig. 1) are based on conserved regions of the C. jejuni 81-176 and C. coli D730 cdtA genes, which encode the amino acid sequences GNWIWGY and IVHYPCD, respectively (21, 24). The nucleotide sequence of GNW is 5′-GG(ACGT)AA(CT)TGGAT(ACT)TGGGG(ACGT)TA-3′, and that of IVH is 5′-AT(ACT)GT(ACGT)CA(CT)TA(CT)CC(ACGT)TG(CT)GA-3′. All PCRs contained 0.2 mM (each) dATP, dCTP, dGTP, and TTP, 1.5 mM MgCl2, 1× Taq DNA polymerase buffer, 0.25 μM (each) primer, 0.25 μg of template DNA, and 2.5 U of Taq polymerase. Parameters for all reactions were 30 cycles at 94°C for 1 min, 42°C for 2 min, and 72°C for 3 min. Restriction endonucleases were used in accordance with the specifications of the supplier (New England Biolabs, Boston, Mass.).
FIG. 1.
Partial restriction map of the C. jejuni 81-176 cdt genes and location of the PCR primers used in this study. Arrows indicate the direction of transcription of the three cdt genes. Large arrowheads indicate the location and priming direction of primers. Restriction endonuclease abbreviations: Stu, StuI; Alu, AluI; Eco, EcoRI; Bgl, BglII; Ssp, SspI.
Toxin assay.
The HeLa cell assays for detection of CDT activity and determination of CDT titers were performed as described by Pickett et al. (23, 24). Each strain was assayed in at least three independent assays. Titers are expressed as the geometric means ± the standard deviations of the means.
Production of CDT.
The C. jejuni and C. coli strains used in this study were deliberately selected, because they were isolated in diverse geographical locations from both humans and animal species and because they had a variety of CDT titers in the HeLa assay. Twelve of the thirteen C. jejuni isolates produced active CDT, and 10 of these produced CDT titers greater than 100. The exceptions were strains D133, 84-19, and AED974, which had mean CDT titers of 11 ± 19, 22 ± 16, and 0, respectively. All of the C. coli isolates examined had CDT titers lower than 5, except 78-64 which had a titer of 18 ± 19. C. coli lysates do not appear to contain much CDT activity when assayed on HeLa cells (24). It is not clear if this low activity results from poor expression of the C. coli cdt genes, if the C. coli CDT has poor specific activity, or if this CDT is simply not very active on HeLa cells.
Detection and characterization of cdt genes.
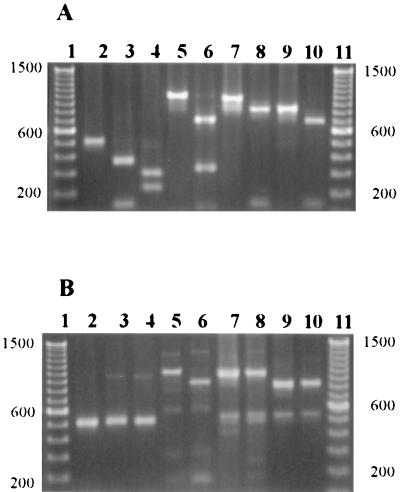
The degenerative primers VAT2 and WMI1 (Fig. 1) were tested for their ability to amplify a portion of the cdtB gene in 13 C. jejuni and 11 C. coli isolates. A single product of the expected size (approximately 0.5 kb) was observed for 11 of the 13 C. jejuni strains and for all 11 of the C. coli strains (Table 1; Fig. 2A and B, lane 2). The restriction endonuclease (RE) fragment patterns produced by EcoRI digestion of these 0.5-kb PCR products were examined. Sequence data from C. jejuni 81-176 and C. coli D730 indicated that this enzyme would cut the amplified portion of at least some C. jejuni strains’ cdtB genes but perhaps not the C. coli cdtB gene (21, 24). The 0.5-kb PCR products of all 11 C. jejuni strains were cut once with EcoRI, yielding fragments with approximate sizes of 0.37 and 0.15 kb (Table 1; Fig. 2A, lane 3), which were close to the expected sizes of 0.36 and 0.14 kb. None of the C. coli VAT2-WMI1 PCR products were cut with EcoRI (Table 1; Fig. 2B, lane 3). Restriction patterns of the VAT2-WMI1 PCR products of the 11 C. jejuni strains were also digested with two additional endonucleases, StuI and AluI (Table 1). StuI cut all of the C. jejuni strains’ VAT2-WMI1 products once, resulting in fragments with apparent sizes of 0.3 and 0.22 kb (Fig. 2A, lane 4). None of the C. coli strains’ PCR products were cut with StuI (Fig. 2B, lane 4). Nucleotide sequence data predicted that AluI would cut C. jejuni VAT2-WMI1 products to yield a fragment of 0.28 kb and five smaller fragments. We saw the larger fragment in AluI cuts of the 11 C. jejuni strains tested, but the smaller fragments were not resolved on our agarose gels (data not shown). Sequence data from C. coli strain D730 (21) indicated that AluI would cut this PCR product four times to produce fragments of approximately 0.21, 0.11, 0.10, 0.05, and 0.01 kb. We were able to consistently see the 0.21-kb fragment in AluI cuts of the VAT2-WMI1 product from the four C. coli strains tested, as well as a fragment, likely a doublet, at about 0.11 kb, but the other bands were not resolved on our gels (data not shown).
TABLE 1.
Restriction analysis of cdt PCR products
| Strain | Primer pair and enzyme(s)a
|
||||||
|---|---|---|---|---|---|---|---|
| VAT2-WMI1
|
VAT2–LPF-D
|
GNW-WMI1, EcoRI | IVH-WMI1, EcoRI | ||||
| EcoRI | StuI | AluI | BglII | SspI | |||
| C. jejuni | |||||||
| 81-176 | + | + | + | + | + | + | + |
| 84-142 | + | + | + | + | + | + | + |
| 85-452 | + | + | + | + | + | + | + |
| 79-193 | + | + | + | + | + | + | + |
| 85-360 | + | + | + | + | + | + | + |
| G13 | + | + | + | + | + | + | + |
| D133 | + | + | + | − | + | + | + |
| 79-101 | + | + | + | + | + | + | + |
| Lior19 | + | + | + | + | + | + | + |
| AED973 | + | + | + | + | + | + | + |
| AED974 | NP | NP | NP | − | + | NP | NP |
| AEB9710 | + | + | + | + | + | + | + |
| 84-19 | NP | NP | NP | − | + | NP | NP |
| C. coli | |||||||
| 43473 | − | − | + | + | ND | − | − |
| D730 | − | − | + | + | ND | − | − |
| D2593 | − | − | + | + | ND | − | − |
| 78-64 | − | − | + | + | ND | − | − |
| D2594 | − | − | + | + | ND | − | − |
| D115 | − | − | + | + | ND | − | − |
| D126 | − | − | + | + | ND | − | − |
| AEB971 | − | − | + | + | ND | − | − |
| AEB979 | − | − | + | + | ND | − | − |
| AEB9713b | − | − | + | + | ND | − | − |
| AED9715 | − | − | + | + | ND | − | − |
Symbols: +, PCR product was cut by the indicated restriction endonuclease; −, PCR product was not cut by the indicated restriction endonuclease. NP, no product produced; ND, not determined.
FIG. 2.
PCR products obtained with the cdt primers and template DNA from representative C. jejuni and C. coli strains. (A) C. jejuni PCR products. (B) C. coli PCR products. Lanes 1 and 11, DNA size standard (100-bp ladder); lane 2, uncut VAT2-WMI1 PCR product; lane 3, EcoRI-cut VAT2-WMI1 PCR product; lane 4, StuI-cut VAT2-WMI1 PCR product; lane 5, uncut VAT2–LPF-D PCR product; lane 6, BglII-cut VAT2–LPF-D PCR product; lane 7, uncut GNW-WMI1 PCR product; lane 8, EcoRI-cut GNW-WMI1 PCR product; lane 9, uncut IVH-WMI1 PCR product; lane 10, EcoRI-cut IVH-WMI1 PCR product. DNA fragment standard sizes, in base pairs, are indicated to the sides of the figure.
We next used the cdtA-based degenerative primers GNW and IVH paired with the downstream cdtB primer WMI1 (Fig. 1) in order to see how these primers compared with VAT2-WMI1 in their ability to amplify cdt genes from these C. jejuni and C. coli isolates. The predicted PCR product sizes for the GNW-WMI1 and IVH-WMI1 primer pairs are 0.96 and 0.82 kb, respectively. Appropriately sized PCR products for both GNW-WMI1 and IVH-WMI1 primer pairs were amplified from 11 of 13 C. jejuni strains (Table 1; Fig. 2A, lanes 7 and 9). These two primer pairs failed to amplify a product from the same C. jejuni strains, 84-19 and AED974, for which no product was obtained with VAT2-WMI1 (Table 1). It should be noted that these are two of the three C. jejuni strains that produced little or no detectable CDT. Both of these primer pairs amplified cdt sequences from all 11 C. coli strains tested (Table 1; Fig. 2B, lanes 7 and 9). The 0.96-kb GNW-WMI1 PCR product from the C. jejuni strains was cut once with EcoRI, yielding 0.85- and 0.15-kb fragments; predicted sizes were 0.82 and 0.14 kb (Fig. 2A, lanes 8). Similarly, the 0.82-kb IVH-WMI1 PCR product from the C. jejuni strains was cut once with EcoRI, producing 0.7- and 0.15-kb fragments, close to the expected 0.68- and 0.14-kb fragments (Fig. 2A, lane 10). As expected, neither the GNW-WMI1 nor the IVH-WMI1 PCR products from any C. coli strain were cut with EcoRI (Table 1; Fig. 2B, lanes 8 and 10, respectively).
None of the three primer pairs already discussed, all of which use WMI1 as the downstream primer, successfully amplified cdt genes from two C. jejuni strains. We therefore decided to test a fourth PCR primer pair which did not include the WMI1 primer. The degenerative primer, LPF-D, which is based on a conserved region of the cdtC genes of C. jejuni 81-176 and C. coli D730, was paired with VAT2 (Fig. 1). These primers successfully produced a PCR product of approximately 1.05 kb from all 13 C. jejuni strains (Table 1; Fig. 2A, lane 5). Nucleotide sequence data from C. jejuni 81-176 suggested that the C. jejuni VAT2–LPF-D products would likely be cut once by BglII and SspI to produce fragments of approximately 0.7 and 0.3 kb and 0.9 and 0.1 kb, respectively. Ten of the thirteen C. jejuni VAT2–LPF-D PCR products were cut once with BglII, producing 0.75- and 0.33-kb fragments (Fig. 2A, lane 6). The remaining three C. jejuni strains’ (D133, AED974, and 84-19) PCR products were not cut with BglII. However, all 13 C. jejuni products were cut with SspI, yielding fragments with apparent sizes of 0.95 and 0.12 kb, with the exception of strain 84-19, for which the fragments were 0.65 and 0.42 kb (data not shown). Since strains AED974 and 84-19 had not produced PCR products from the other primer pairs, we tested the ability of the VAT2–LPF-D products from these strains to hybridize to the VAT2-WMI1 PCR product amplified from C. jejuni 81-176 (24). The results confirmed that the VAT2–LPF-D PCR products from these two strains were indeed amplified regions of their cdt genes (data not shown). The VAT2–LPF-D primer pair also successfully amplified sequences from all of the test C. coli strains (Table 1; Fig. 2B, lane 5). The C. coli VAT2–LPF-D products were cut once by BglII, generating products of approximately 0.9 and 0.2 kb (Table 1; Fig. 2B, lane 6). The C. coli products were not tested for SspI digestion, since nucleotide sequence data suggested that no SspI site would be present.
In summary, we tested several different PCR primer pairs for their ability to amplify cdt genes from a variety of C. jejuni and C. coli isolates. The four primer pairs described here all appear to be useful for detection of cdt sequences in isolates from these Campylobacter species. The ability of these primers to amplify cdt sequences from both C. jejuni and C. coli should be useful, since DNA-DNA hybridization between the cdt genes of these species is poor (21, 24).
Overall, our results suggest that the cdt gene sequences are relatively conserved within species boundaries. Our restriction analysis of the cdt genes’ PCR products amplified from different strains of the same species revealed only occasional differences (Table 1). Supporting this conclusion is work done in our laboratory to sequence the cdt genes from an additional C. jejuni strain and from two C. coli strains (21). The sequence of the cdt genes from the second C. jejuni strain differs from the sequence of those from 81-176 at only a few positions. The cdt sequences from the two C. coli strains are nearly identical, in contrast to what has been found for isolates of E. coli, in which the cdt genes from three different strains exhibit numerous dissimilarities (20, 23, 25). Thus, while it is clear that sequence variation can be tolerated within cdt genes, and that the encoded variant Cdt proteins can still be functional, the cdt sequences of C. jejuni or C. coli isolates may be more conserved.
The only two C. jejuni strains for which all of these primers did not amplify cdt sequences are the two low- or non-CDT-producing strains, 84-19 and AED974. The lack of success with the WMI1-containing primer pairs suggests that at least a portion of the cdtB gene in these two strains is changed in one or more critical ways. Our previous hybridization results (24) indicate that the cdt genes from strain 84-19 may differ from the cdt genes of strain 81-176 at many positions, since both the size of the ClaI fragment from strain 84-19 to which a 81-176 cdtB probe hybridized and the strength of the hybridization were atypical. In any case, these two strains are clearly unusual C. jejuni strains; the typical C. jejuni strain appears to have cdt sequences that are readily amplified by any of the primers described here.
Hybridization studies by Pickett et al. (24) suggested that the cdtB genes of C. jejuni and C. coli had diverged. We expected, therefore, to find differences between the cdt genes of C. jejuni and C. coli. Analysis of the restriction profiles of various cdt PCR products verified at least some sequence divergence between the cdt sequences of these two closely related species. Our results also indicate that C. jejuni and C. coli cdt genes can be distinguished simply by EcoRI digestion of any of the PCR products analyzed in this work, since the EcoRI site present in the C. jejuni cdtB gene is not present in the C. coli cdtB gene. In addition, StuI appears to be an alternative restriction enzyme for easy differentiation of C. jejuni and C. coli cdt gene sequences.
Acknowledgments
Thanks to Daniel Cottle for technical assistance and Ozhan Eyigor for assistance with the figures. Thanks to Richard Meinersmann for providing some of the strains used in this work.
This work was supported in part by the National Institutes of Health Public Health Services grant AI/DK 41477 to C.L.P. A.E. was supported by a fellowship from the Turkish Higher Education Council and Uludag University, Bursa, Turkey.
REFERENCES
- 1.Black R E, Levine M M, Clements M L, Hughes T P, Blaser M J. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- 2.Blaser, M. J. Personal communication.
- 3.Butzler J P, Skirrow M B. Campylobacter enteritis. Clin Gastroenterol. 1979;8:737–765. [PubMed] [Google Scholar]
- 4.Comayras C, Tasca C, Peres S Y, Ducommun B, Oswald E, De Rycke J. Escherichia coli cytolethal distending toxin blocks the HeLa cell cycle at the G2/M transition by preventing cdc2 protein kinase dephosphorylation and activation. Infect Immun. 1997;65:5088–5095. doi: 10.1128/iai.65.12.5088-5095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cope L D, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson R S, Jr, Lagergard T, Radolf J D, Hansen E J. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt J M, Abeyta C. Isolation of Campylobacter species from food and water. In: Tomlison L A, editor. FDA bacteriological analytical manual. 8th ed. Baltimore, Md: AOAC International; 1995. pp. 7.01–7.27. [Google Scholar]
- 7.Johnson W M, Lior H. Response of Chinese hamster ovary cells to a cytolethal distending toxin (CDT) of Escherichia coli and possible interpretation as heat-labile (LT) enterotoxin. FEMS Microbiol Lett. 1987;43:19–23. [Google Scholar]
- 8.Johnson W M, Lior H. Production of Shiga toxin and a cytolethal distending toxin (CLDT) by serogroups of Shigella spp. FEMS Microbiol Lett. 1987;48:235–238. [Google Scholar]
- 9.Johnson W M, Lior H. A new heat-labile cytolethal distending toxin (CLDT) produced by Escherichia coli isolates from clinical material. Microb Pathog. 1988;4:103–113. doi: 10.1016/0882-4010(88)90052-6. [DOI] [PubMed] [Google Scholar]
- 10.Johnson W M, Lior H. A new heat-labile cytolethal distending toxin (CLDT) produced by Campylobacter spp. Microb Pathog. 1988;4:115–126. doi: 10.1016/0882-4010(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 11.Karmali M A, Penner J L, Fleming P C, Williams A, Hennessy J N. The serotype and biotype distribution of clinical isolates of Campylobacter jejuni and Campylobacter coli over a three-year period. J Infect Dis. 1983;147:243–246. doi: 10.1093/infdis/147.2.243. [DOI] [PubMed] [Google Scholar]
- 12.Ketley J M. Virulence of Campylobacter species: a molecular genetic approach. J Med Microbiol. 1995;42:312–317. doi: 10.1099/00222615-42-5-312. [DOI] [PubMed] [Google Scholar]
- 13.Ketley J M. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143:5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 14.Korlath J A, Osterholm M T, Judy L A, Forfang J C, Robinson R A. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- 15.Linton D, Lawson A J, Owen R J, Stanley J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol. 1997;35:2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lior H, Woodward D L, Edgar J A, Laroche L J, Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982;15:761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Advisory Committee on Microbiological Criteria for Foods. Campylobacter jejuni/coli. Dairy Food Environ Sanit. 1995;15:133–153. [Google Scholar]
- 18.Okuda J, Kurazono H, Takeda Y. Distribution of the cytolethal distending toxin A gene (cdtA): among species of Shigella and Vibrio, and cloning and sequencing of the cdt gene from Shigella dysenteriae. Microb Pathog. 1995;18:167–172. doi: 10.1016/s0882-4010(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 19.Okuda J, Fukumoto M, Takeda Y, Nishibuchi M. Examination of diarrheagenicity of cytolethal distending toxin: suckling mouse response to the products of the cdtABC genes in Shigella dysenteriae. Infect Immun. 1997;65:428–433. doi: 10.1128/iai.65.2.428-433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peres S Y, Marches O, Daigle F, Nougayrede J P, Herault F, Tasca C, De Rycke J, Oswald E. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol Microbiol. 1997;24:1095–1107. doi: 10.1046/j.1365-2958.1997.4181785.x. [DOI] [PubMed] [Google Scholar]
- 21.Pickett, C. L. Unpublished data.
- 22.Pickett C L, Auffeberg T, Pesci E C, Sheen V L, Jusuf S S D. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect Immun. 1992;60:3872–3877. doi: 10.1128/iai.60.9.3872-3877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickett C L, Cottle D L, Pesci E C, Bikah G. Cloning, sequencing, and expression of the Escherichia coli cytolethal distending toxin genes. Infect Immun. 1994;62:1046–1051. doi: 10.1128/iai.62.3.1046-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickett C L, Pesci E C, Cottle D L, Russell G, Erdem A N, Zeytin H. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB genes. Infect Immun. 1996;64:2070–2078. doi: 10.1128/iai.64.6.2070-2078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott D A, Kaper J B. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect Immun. 1994;62:244–251. doi: 10.1128/iai.62.1.244-251.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silhavy R J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. p. 137. [Google Scholar]
- 27.Stonnet V, Guesdon J-L. Campylobacter jejuni: specific oligonucleotides and DNA probes for use in polymerase chain reaction-based diagnosis. FEMS Immunol Med Microbiol. 1993;7:337–344. doi: 10.1111/j.1574-695X.1993.tb00415.x. [DOI] [PubMed] [Google Scholar]
- 28.Stonnet V, Sicinschi L, Megraud F, Guesdon J L. Rapid detection of Campylobacter jejuni and Campylobacter coli isolated from clinical specimens using the polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1995;14:355–360. doi: 10.1007/BF02116533. [DOI] [PubMed] [Google Scholar]
- 29.Sugai M, Kawamota T, Peres S Y, Ueno Y, Komatsuzawa H, Fujiwara T, Kurihara H, Suginaka H, Oswald E. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect Immun. 1998;66:5008–5019. doi: 10.1128/iai.66.10.5008-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker R I, Caldwell M B, Lee E C, Guerry P, Trust T J, Ruiz-Palacios G M. Pathophysiology of Campylobacter enteritis. Microbiol Rev. 1986;50:81–94. doi: 10.1128/mr.50.1.81-94.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wassenaar T M. Toxin production by Campylobacter spp. Clin Microbiol Rev. 1997;10:466–476. doi: 10.1128/cmr.10.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehouse C A, Balbo P B, Pesci E C, Cottle D L, Mirabito P M, Pickett C L. Campylobacter jejuni cytolethal distending toxin causes a G2-phase cell cycle block. Infect Immun. 1998;66:1934–1940. doi: 10.1128/iai.66.5.1934-1940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]