Abstract
Background
The competitive endogenous RNA (ceRNA) mechanism has been discovered recently and regulating cancer-related gene expressions. The ceRNA network participates in multiple processes, such as cell proliferation and metastasis, and potentially drives the progression of cancer. In this study, we focus on the ceRNA networks of esophageal squamous cell carcinoma and discovered a novel biomarker panel for cancer prognosis.
Methods
RNA expression data of esophageal carcinoma from the TCGA database were achieved and constructed ceRNA network in esophageal carcinoma using R packages.
Results
Four miRNAs were discovered as the core of the ceRNA model, including miR-93, miR-191, miR-99b, and miR-3615. Moreover, we constructed a ceRNA network in esophageal carcinoma, which included 4 miRNAs and 6 lncRNAs. After ceRNA network modeling, we investigated six lncRNAs which could be taken together as a panel for prognosis prediction of esophageal cancer, including LINC02575, LINC01087, LINC01816, AL136162.1, AC012073.1, and AC117402.1. Finally, we tested the predictive power of the panel in all TCGA samples.
Conclusions
Our study discovered a new biomarker panel which may have potential values in the prediction of prognosis of esophageal carcinoma.
1. Introduction
Esophageal squamous cell carcinoma is one of the most frequently diagnosed malignancies among women worldwide. Despite the substantial research findings in investigating the intracellular mechanisms of esophageal cancer, it remains to be a significant cause of cancer death to women. One reason for this urgent situation is that we have not found effective methods for the early diagnosis and prognosis of esophageal cancer. Previous work has proved that some noncoding RNAs (NcRNAs), including circular RNA (circRNA), long noncoding RNAs (lncRNAs), and micro RNAs (miRNAs) [1], could interact with each other [2].
Based on the functions of NcRNAs, most researches focus on the functional NcRNAs, which also play essential roles in the ceRNA network [3], in brief, miRNAs bind with mRNAs via MREs, and the functions of mRNAs are thus inhibited by miRNAs, at the same if functional lncRNAs bind with the corresponding miRNAs, the functions of miRNAs are thus inhibited which indirectly release the mRNAs which were inhibited by miRNAs, and the interaction network are called ceRNETs [4]. Recently, more and more evidence have proved that the ceRNA network has key roles in the progression of cancer, and some ceRNAs could be further used to construct biomarkers for the prediction of patient prognosis [5, 6].
Esophageal cancer is one of the most aggressive cancer types and sixth most common cause of cancer death worldwide [7]. Esophageal cancer has two major subtypes: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). The incidence rate of ESCC is high in the Asian regions including Japan, China, and India [8, 9]. ESCC is one of the most deadly forms of human malignancy characterized by late metastasis, stage diagnosis, therapy resistance, and frequent recurrence [10]. Earlier diagnosing ESCC could improve treatment outcomes of patients as well as prolong patients' OS. ESCC has a close relation with aberrant expressions of genes, which makes clinical diagnosis more complicated to patients [11]. The ceRNA network could improve our insight into the progression of esophageal carcinoma, and it may help us to build a better biomarker panel for the diagnosis and prognosis of patients [12].
Numerous experimental studies have demonstrated ceRNA network had a crucial role in modulating the pathogenesis of human diseases. For example, lncRNA PART1 suppresses ESCC cell proliferation by regulating the miR-18a-5p/SOX6 signaling axis [13]. Linc00941 regulates ESCC via functioning as a competing endogenous RNA for miR-877-3p to modulate PMEPA1 expression [14]. Moreover, analysis of the ceRNA network has been utilized in biomarker construction, drug target discovery, and even drug resistance studies [15]. Based on the ceRNA network theory, Cantile et al. discovered that a single lncRNA, HOTAIR, was highly expressed in ESCC, which was the first breast cancer hallmark and a biomarker to predict the patient's OS [16]. Since a single lncRNA was not enough considering poor sensitivity and specificity in a statistic perspective [17], taking multiple dysregulated RNAs into evaluation of the prognosis of patients were gradually accepted by researchers.
Taken together, it has been generally approved that lncRNAs have great predict powers in the prediction of cancer stages and patients' prognosis. In this study, we discovered a ceRNA network by analyzing RNA expressions in esophageal squamous cell carcinoma. Through bioinformatic procedures, we further explored novel ESCC progression-related lncRNAs, then construct a prediction model, and evaluated the predictive power in TCGA samples.
2. Material and Methods
2.1. RNA-Seq Data Procession
The study was done in R environment, and RTCGAToolBox was used to download mRNA and miRNA database. GDC Data Portal was used to download the lncRNA database. The following R packages were selected for data filtration and analysis in this paper, including Hmisc, plyr, hash, ggplot2, R.utils, ggbiplot, pROC, gplots, survival, ggbiplot, edgeR, survminer, RColorBrewer, magrittr, GenomicDataCommons, rtracklayer, grid, corrplot, scales, and limma.
2.2. RNA-Seq Data Procession
ESCC RNA-seq data containing 161 tumor samples and 11 normal samples from the TCGA database were retrieved with the software R. All databases were opensource and free to use.
2.3. RNA Expression Analysis
All RNA expression profiles were divided into high and low expression groups with Edge R and limma as previously described by Anders and Huber [18]. GENCODE was used to analyze lncRNA expressions. In this study, log2 FC ≥ 1.5 and the FDR < 0.01 were set as threshold.
2.4. ceRNA Network Construction
MiRWalk (http://mirwalk.umm.uni-heidelberg.de/) was selected to screen miRNAs and corresponding target genes. The miRNA-lncRNA correlation > 0.45 and P < 0.05 was set to filter out powerful correlations [19]. Cytoscape 3.6.3 was used to construct the network model. Cluster Profiler was used to do GO and KEGG analysis followed by methods previously described [20].
A schematic view of this study is shown in Supplementary Figure 1 (Figure S1). In brief, the key procedures of this study could be listed as follows:
Download mRNA, miRNA, and lncRNA data from the TCGA portal
Data filtering and gather DERNAs
Analyze all DERNAs and gather information of all possible ceRNA network connections. In this study, all published or validated proof of ceRNA network is taken into account for the following analysis. MicroRNA survival analysis followed by correlation analysis generate all possible core miRNAs
Adopt unsupervised clustering strategy to further filter DElncRNAs to a proper amount to build ceRNA network
Training the lncRNA panel
3. Survival Analysis
We referred to a previously described method for the construction of CESC prognosis prediction model [21]. Survival package was selected to generate survival plots, which were then used to evaluate the prediction power of the biomarker panel. Cox regression statistic procedures were processed to test the predictive value of the panel. ROC and AUC curves were plotted with the Daim package to evaluate model specificity and sensitivity [22].
4. Results
4.1. Database Download and Clinical Features
All ESCC TCGA samples with level 2 RNA expression info were downloaded. The sample information was shown in Table S1. The transcriptome data of 161 ESCC samples and 11 normal samples were achieved. Log2 FC > 1.5 and P < 0.01 set as threshold. To help our understanding of the intracellular mechanisms which were involved in the development of tumor progression, KEGG analysis was also performed in the R-based ProfilerCluster package. The pathways in cancer (P < 0.01) were shown in Figure 1(a). Heatmap package was used to plot the heatmap with the top 1000 differentially expressed genes (Figure 1(b)), followed by PCA plots in Figure 1(c). Expression data were gathered in Table S2.
Figure 1.
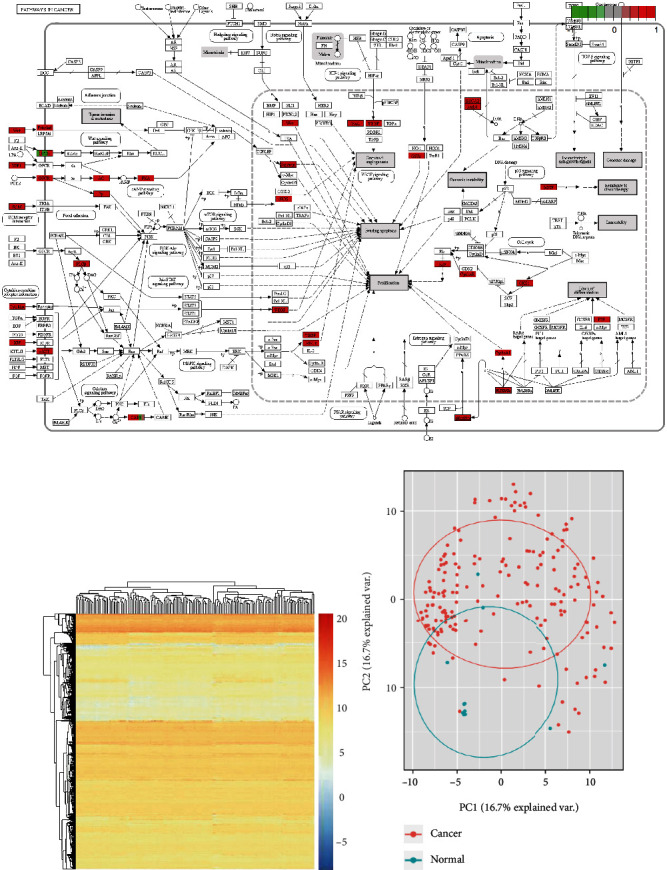
Transcriptome profiles in esophageal carcinoma. (a) Pathways of cancer. up or downregulated DEmRNAs were marked red or green, respectively. (b) The DEmRNAs were shown in heatmap. (c) PCA plot analysis of ESCA and normal samples based on DEmRNAs' expression.
4.2. lncRNA and miRNA Profiles
Log2 FC > 1.5 and P < 0.01 were set as threshold, as shown in Figure 2(a) (DEGs were shown in Table S3). Next, PCA plots were shown in Figure 2(b). Expression data were grouped into multiple categories based on clinical features to evaluate prognostic significance.
Figure 2.
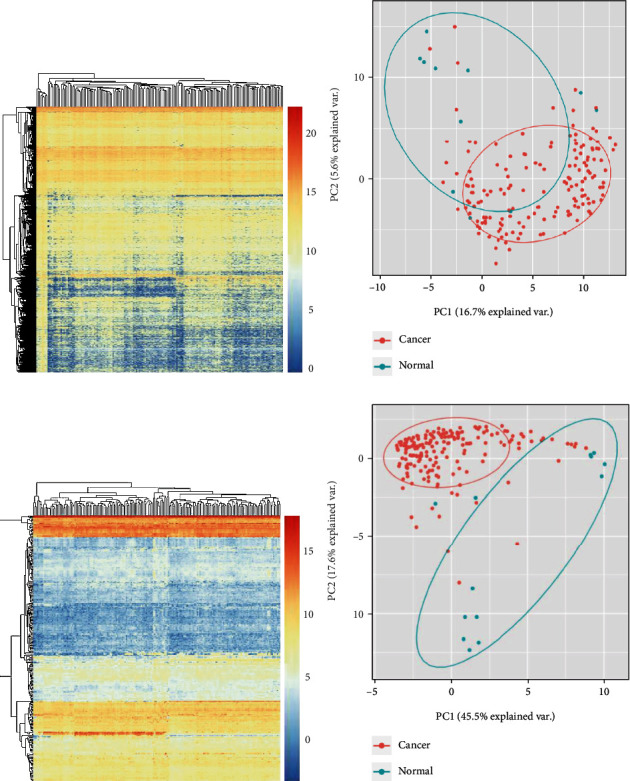
lncRNA and miRNA profiles in esophageal carcinoma. (a) The DElncRNAs were depicted in heatmap. (b) PCA plots of ESCA and normal samples based on DElncRNAs' expression. (c) The DEmiRNAs were shown in heatmap. (d) PCA plots of ESCA and normal samples based on DEmiRNAs' expression.
To generate ceRNA network, we further processed the miRNA expression data. Dysexpressed miRNAs (DEmiRNAs) were shown in Table S4. Heatmap was shown in Figure 2(c). The PCA plots were shown in Figure 2(d).
4.3. miRNA Survival Analysis
As miRNAs form the core of ceRNA network, survival plots were generated to filter out the miRNAs with clinical significance. Significant correlation among four miRNAs, namely, miR-93, miR-191, miR-99b, and miR-3615 and the patients' OS were found (P < 0.05). To be more specific, all the four miRNAs were positively correlated with patients' OS (Figures 3(a)–3(d)).
Figure 3.
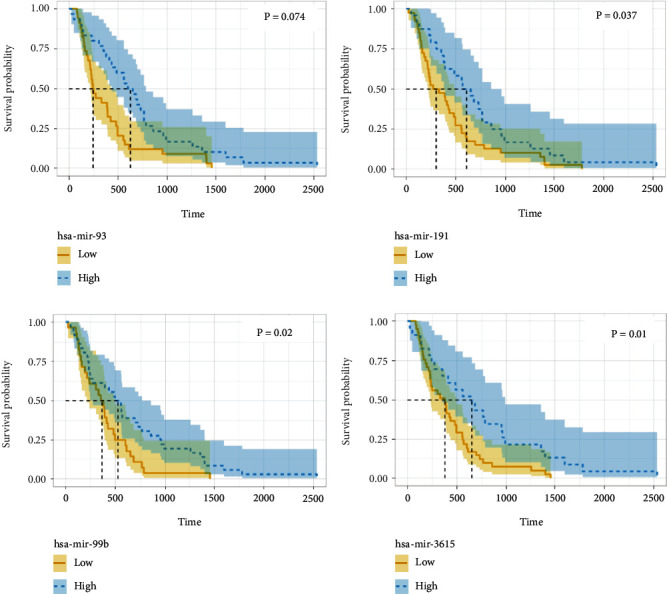
miRNA survival analysis in esophageal carcinoma. (a)–(d) The dysregulation of miR-93 (a), miR-191 (b), miR-99b (c), and miR-3615 (d) was correlated to overall survival in esophageal carcinoma.
4.4. Construction of ceRNA Network in Esophageal Carcinoma
In the first step, we tried to find a hubLncRNA panel without training the lncRNA data. But the result showed that the smallest panel we could find contained as many as 50 hubLncRNAs, which were not appropriate for future application. The initial ceRNA network was shown in Figure 4(a). Then, we trained the lncRNAs data by unsupervised hierarchical clustering strategy. After unsupervised clustering, the number of lncRNAs was limited to a proper amount, and we constructed an initial ceRNA network that directly yielded four key miRNAs (Figure 4(b)). For instance, two lncRNAs, namely, LINC02575 and LINC01087 could competitively bind with miR-191. Two lncRNAs, namely, LINC01816 and AL136162.1, could competitively bind with miR-3615. One lncRNA, AC012073.1, could competitively bind with miR-93. One lncRNA, AC117402.1, could competitively bind with miR-99b. Next, DElncRNAs and DERNAs were used to build a linear regression model. Results proved a strong correlation between miRNAs, lncRNAs, and mRNAs (Figure 4(c)).
Figure 4.
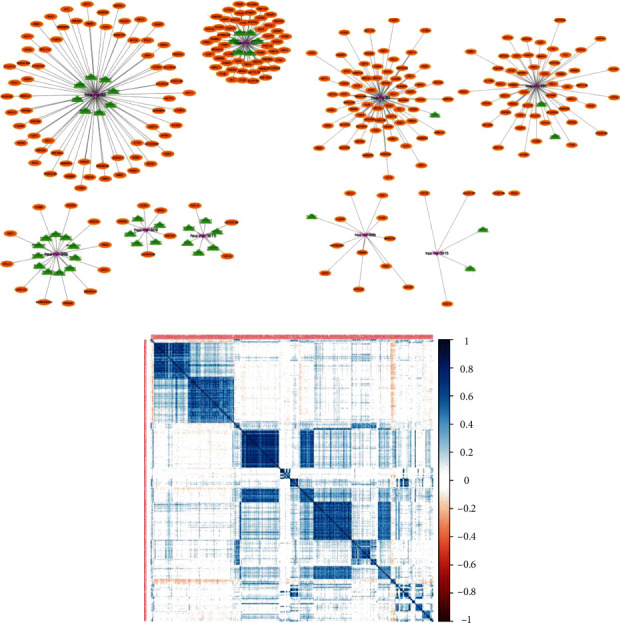
Construction of ceRNA network in esophageal squamous carcinoma. (a) the ceRNA network model. lncRNAs, miRNAs, and mRNAs are marked as green, brown, and purple, respectively. (b) After unsupervised clustering, we constructed an optimized lncRNA-miRNA-mRNA network in esophageal carcinoma. lncRNAs, miRNAs, and mRNAs were marked as green, purple, and brown, respectively. (c) miRNA-lncRNA network linear regression plots.
4.5. Generation of Prediction Model
Topological analysis was selected to discover the potential hub lncRNAs in the construction of the prognosis prediction model (Figures 5(a)–5(d)). Finally, six hub lncRNAs were filtered out: LINC02575, LINC01087, LINC01816, AL136162.1, AC012073.1, and AC117402.1, and the interaction network of the hublncRNAs is shown in Table S5.
Figure 5.
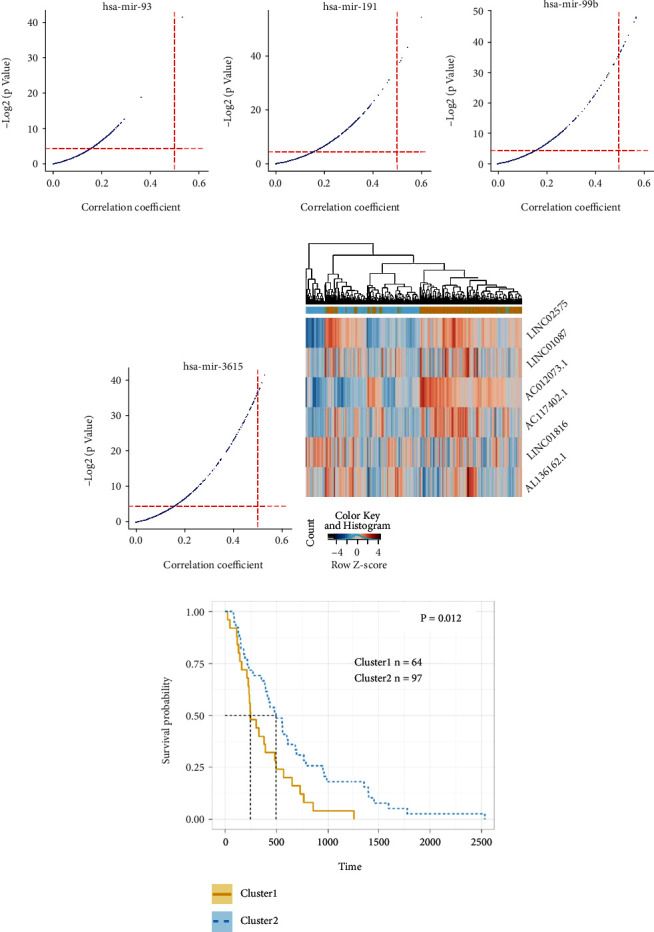
Statistical features of hublncRNAs in esophageal squamous carcinoma. (a)–(d) miRNA and lncRNA coefficiency plot analysis of miR-93 (a), miR-191 (b), miR-99b (c), and miR-3615 (d). (e) Heatmap plot analysis of six hub lncRNAs in ESCA and normal samples. (f) Survival plots of cluster 1 and cluster 2.
To clearly investigate the predictive power of the six hub genes, by unsupervised clustering strategy, all 161 patients were grouped into two clusters (Figure 5(e)). Survival analysis showed statistical significance (median OS, 494 vs. 247 days) (log-rank test P = 0.012; Figure 5(f)).
The survival data of the six HublncRNAs were taken to computing the patients' risk score which comes as below:
| (1) |
Then, we analyzed the prediction model with all sample data via Cox regression analysis (Table 1).
Table 1.
Univariate/multivariate Cox regression analysis in all patients.
| Variables | Univariate statistics | Multivariate statistics | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI of HR | P value | HR | 95% CI of HR | P value | |
| Patient age | 1 | 0.98-1 | 0.96 | 0.99 | 0.97-1 | 0.616708 |
| Stage | ||||||
| Stage T1 | 0.63 | 0.32-1.8 | 0.048 | 0.2 | 0.023-1.8 | 0.153204 |
| Stage T2 | 0.73 | 0.39-1.4 | 0.89 | 0.23 | 0.027-2 | 0.181557 |
| Stage T3 | 1.6 | 0.94-2.8 | 0.083 | 0.39 | 0.048-3.2 | 0.382688 |
| Stage T4 | 1.7 | 0.39-7 | 0.49 | 0.46 | 0.039-5.4 | 0.533969 |
| Risk_score | 0.55 | 0.32-0.95 | 0.031 | 0.52 | 0.28-0.97 | 0.0397278 |
Abbreviations: HR: hazard ratio; CI: confidence interval.
4.6. Clinical Prognostic Value of Biomarker Panel
Clinical relevances of all 161 samples were shown in Figures 6(a)–6(d). Six HublncRNAs were considered as key risk factors, including LINC02575, LINC01087, LINC01816, AL136162.1, AC012073.1, and AC117402.1. For example, the AUC value reached 0.686, which means a strong predictive power. When combined all together, the six lncRNAs constructed a powerful biomarker panel. For example, in male patients, the OS curves showed statistical significance (log-rank test P < 0.05, Figure 6(e)). AUC curve was shown in Figure 6(d). Furthermore, similar results could be found in stages I-II/III-IV as well (log-rank test P < 0.05, Figures 6(g)–6(h)).
Figure 6.
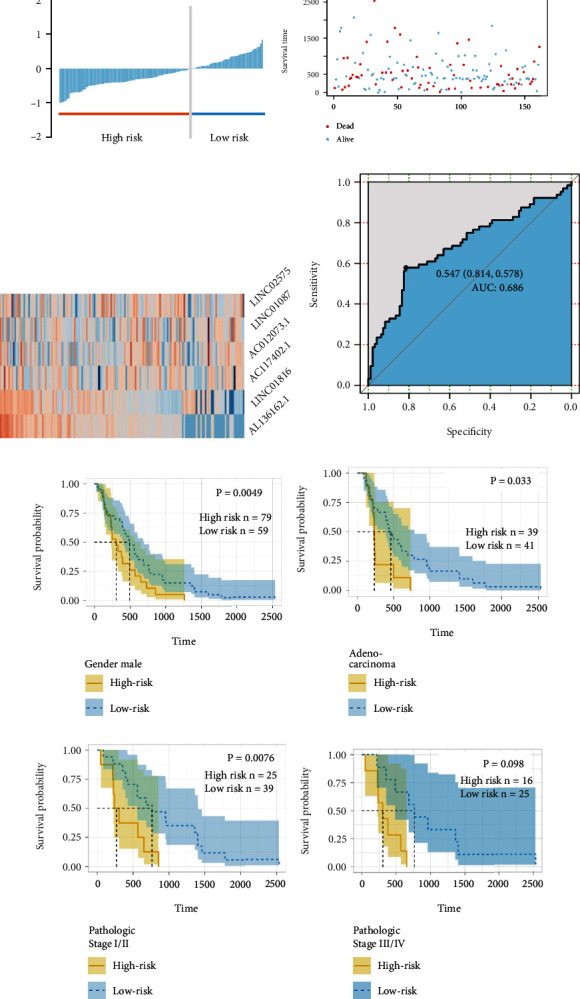
Clinical prognostic significance of biomarker panel. (a)–(c) Risk distribution plots of biomarker panel. Grey line indicated the cutoff value based on risk scores. (d) ROC plot analysis of biomarker panel in ESCC. (e) Overall survival plot analysis of high-risk and low-risk group in male esophageal cancer patients. (f) Overall survival plots analysis of high-risk and low-risk group in esophageal adenocarcinoma patients. (g) Overall survival plots analysis of high-risk and low-risk group in stage I/II esophageal cancer patients. (h) Overall survival plot analysis of high-risk and low-risk group in stage III/IV esophageal cancer patients.
5. Discussion
The ceRNA model is found on the basis of miRNA competitive binding. More and more evidence shows that these miRNAs and their competitive endogenous targets can form complex ceRNA networks [23].
Thanks to the free access to the comprehensive TCGA database, this study managed to develop an ESCC prognostic prediction biomarker panel. Our ceRNA network-based prediction model provided a deeper perspective into the intracellular mechanisms of ESCC.
As discussed in many studies, lncRNAs have great predict power in prognosis and diagnosis, which are sometimes even stronger than miRNAs and mRNAs [6, 17, 24, 25], some lncRNAs such as Fender have already been applied in practice. In esophageal cancer, several lncRNAs had been revealed to be related to the cancer prognosis and progression [26]. For example, overexpression of LEF1-AS1 predicts a poor prognosis in ESCC [26]. Li et al. reported an eight-lncRNA signature could predict the overall survival of ESCC [27]. Moreover, upregulation of HOTAIR was related to the poor prognosis of ESCC [28]. However, few studies on ceRNA have focused on predicting ESCC prognosis. Moreover, there are rare lncRNAs, mRNAs, or miRNAs related to ESCC could be treated as reliable biomarkers to detect ESCC and stratify the ESCC risk. Under this background and ceRNA network hypothesis, our study identified 95 dysexpressed lncRNAs, such as DIAPH3-AS1, NCBP2-AS1, ALG9-IT1, PRR7-AS1, SLC25A5-AS1, and TBL1XR1-AS1, which were strongly correlated with the progression of esophageal carcinoma. One lncRNA worth noting is LINC01087, it is the only lncRNA that has been reported to promote the cell division and metastasis of cancer [29]. As far as we know, the rest 5 lncRNAs are not reported to have prognostic value or molecular insights. One thing worth noting, at the beginning of this study, first, we tried to find a hub lncRNA panel without training the lncRNA data. But the result showed that the smallest panel we could find contained as many as 50 hub lncRNAs, which were not appropriate for future application. So we trained the lncRNA data by unsupervised hierarchical clustering strategy. Unsupervised clustering strategy has been adopted in many researches, for example, a multi-institutional study of the International Mesothelioma Panel from MESOPATH Reference Center proved that unsupervised clustering strategy is a powerful tool in achieving the predict accuracy of biomarker panel [30].
Given that any transcripts harbouring MREs could theoretically function as ceRNAs, as previously reported, they may represent in a widespread multiple forms of regulation in both pathology and physiology [31]. In this study, as shown in Figure 4(b), we found two lncRNAs, namely, LINC02575 and LINC01087, could competitively bind with miR-191. Two lncRNAs, namely, LINC01816 and AL136162.1, could competitively bind with miR-3615. One lncRNA, AC012073.1, could competitively bind with miR-93. One lncRNA, AC117402.1, could competitively bind with miR-99b. Among the core miRNAs found in this study, miR-191 has been previously reported in esophageal carcinoma which predicted poor prognosis and promoted cell proliferation and invasion [32]. Furthermore, miR-191 has been proved to be emerged as key participants of p53 signaling pathways in breast cancer [33]. miR-93 has been proved to promote cervical cancer progression by targeting THBS2/MMPS signal pathway [34]. miR-99b was previously reported to be a tumor suppressor by targeting IGF-1R in gastric cancer [35], while in this study, miR-99b is highly expressed in cancer patients and is supposed to be a tumor promoter. Moreover, miR-99b and miR-375 as a combination are found to be potential predictive response biomarker panel for preoperative chemoradiotherapy in rectal cancer [36]. miR-3615 was previously reported to be a potential oncogene in human cancer. For example, miR-3615 was overexpressed in liver cancer and related to the high TNM stage [37]. Although the ceRNA network we found in this study identifies many ESCC-related miRNAs, lncRNAs, and mRNAs, the correlation and the extent of ceRNA effects remain to be investigated by in vivo experiments. Recent studies have proved that by constitutive posttranscriptional regulations, binding free energy and repression feedbacks are pivotal factors for crosstalk between ceRNAs [38, 39], which means that there are many criteria for validating ceRNA networks, such as miRNAs and RBPs, kinetic parameters, the target ratio of miRNAs, the quantitative measurements of miRNAs, and the size and affinities of the competing targets [40, 41]. Moreover, we reported six lncRNAs which could be taken together as a panel for prognosis prediction of esophageal cancer, including LINC02575, LINC01087, LINC01816, AL136162.1, AC012073.1, and AC117402.1. Of note, several previous reports had implied the functional importance of these lncRNAs in cancers. For instance, LINC02575 was reported to promote the proliferation of laryngeal squamous cell carcinoma cells [42]. LINC01087 is highly expressed in breast cancer [43]. LINC01816 was found to promote the migration of thyroid carcinoma cells [44]. Therefore, our findings still need to be verified through in the future in vivo and in vitro experiments as well as clinical practice.
This study showed that the six lncRNAs together can be taken as a biomarker panel for the prognosis of esophageal cancer.
6. Conclusion
This study provides a novel biomarker for the prediction of prognosis to patients diagnosed with esophageal squamous carcinoma.
Acknowledgments
This study is supported by the China–Japan Friendship Hospital under grant number 2016-2-QN-20.
Data Availability
The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Authors' Contributions
CL designed the project. JZ proposed the research concept and wrote the manuscript. FX, GQ, and ZZ carried out the data search and downloading and literature collection. QM and YH conducted the bioinformatic analysis. HX constructed the graphic images and data charts and performed the statistical processing.
Supplementary Materials
Supplementary Table 1: the sample information was shown in Table S1. Supplementary Table 2: expression data of genes were gathered in Table S2. Supplementary Table 3: differently expressed lncRNAs were shown in Table S3. Supplementary Table 4: differently expressed miRNAs were shown in Table S4. Supplementary Table 5: the interaction network of between lncRNAs and miRNAs is shown in Table S5.
References
- 1.Consortium E. P. An integrated encyclopedia of DNA elements in the human genome. Nature . 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X., Yang L., Chen L. L. The biogenesis, functions, and challenges of circular RNAs. Molecular Cell . 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 3.Chan J. J., Tay Y. Noncoding RNA:RNA regulatory networks in cancer. International journal of molecular sciences . 2018;19(5):p. 1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdollahzadeh R., Daraei A., Mansoori Y., Sepahvand M., Amoli M. M., Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. Journal of Cellular Physiology . 2019;234(7):10080–10100. doi: 10.1002/jcp.27941. [DOI] [PubMed] [Google Scholar]
- 5.Do D., Bozdag S. Cancerin: a computational pipeline to infer cancer-associated ceRNA interaction networks. PLoS Computational Biology . 2018;14(7, article e1006318) doi: 10.1371/journal.pcbi.1006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L., Zhang Y., Fu Y., et al. Long non-coding RNA MALAT1 regulates BLCAP mRNA expression through binding to miR-339-5p and promotes poor prognosis in breast cancer. Bioscience reports . 2019;39(2) doi: 10.1042/BSR20181284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbas G., Krasna M. Overview of esophageal cancer. Annals of cardiothoracic surgery . 2017;6(2):131–136. doi: 10.21037/acs.2017.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abnet C. C., Arnold M., Wei W. Q. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology . 2018;154(2):360–373. doi: 10.1053/j.gastro.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang H., Fan J. H., Qiao Y. L. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biology & Medicine . 2017;14(1):33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichenbach Z. W., Murray M. G., Saxena R., et al. Clinical and translational advances in esophageal squamous cell carcinoma. Advances in Cancer Research . 2019;144:95–135. doi: 10.1016/bs.acr.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Mao S., Li Y., Lu Z., et al. Survival-associated alternative splicing signatures in esophageal carcinoma. Carcinogenesis . 2019;40(1):121–130. doi: 10.1093/carcin/bgy123. [DOI] [PubMed] [Google Scholar]
- 12.Bhan A., Soleimani M., Mandal S. S. Long noncoding RNA and cancer: a new paradigm. Cancer Research . 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., Zhang Q., Liu H., Wang N., Zhang X., Yang S. lncRNA PART1, manipulated by transcriptional factor FOXP2, suppresses proliferation and invasion in ESCC by regulating the miR‑18a‑5p/SOX6 signaling axis. Oncology Reports . 2021;45(3):1118–1132. doi: 10.3892/or.2021.7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Zhu H., Sun N., et al. Linc00941 regulates esophageal squamous cell carcinoma via functioning as a competing endogenous RNA for miR-877-3p to modulate PMEPA1 expression. Aging (Albany NY) . 2021;13(13):17830–17846. doi: 10.18632/aging.203286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong X., Hu S., Yuan Y., et al. Analysis of lncRNA, miRNA and mRNA-associated ceRNA networks and identification of potential drug targets for drug-resistant non-small cell lung cancer. Journal of Cancer . 2020;11(11):3357–3368. doi: 10.7150/jca.40729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantile M., Di Bonito M., Cerrone M., Collina F., De Laurentiis M., Botti G. Long non-coding RNA HOTAIR in breast cancer therapy. Cancers . 2020;12(5):p. 1197. doi: 10.3390/cancers12051197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sørensen K. P., Thomassen M., Tan Q., et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Research and Treatment . 2013;142(3):529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 18.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biology . 2010;11(10):p. R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sticht C., De La Torre C., Parveen A., Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One . 2018;13(10, article e0206239) doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merico D., Isserlin R., Stueker O., Emili A., Bader G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One . 2010;5(11, article e13984) doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Y., Zhang T., Qi L., et al. Integrated analysis of co-expression and ceRNA network identifies five lncRNAs as prognostic markers for breast cancer. Journal of Cellular and Molecular Medicine . 2019;23(12):8410–8419. doi: 10.1111/jcmm.14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontoura C. A., Castellani G., Mombach J. C. The R implementation of the CRAN package PATHChange, a tool to study genetic pathway alterations in transcriptomic data. Computers in Biology and Medicine . 2016;78:76–80. doi: 10.1016/j.compbiomed.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. A _ceRNA_ hypothesis: The Rosetta Stone of a hidden RNA language? Cell . 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Zhang W., Liu P., et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Oncotargets and Therapy . 2018;Volume 11:1403–1412. doi: 10.2147/OTT.S149511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y., Lu J., Zhou J., et al. Long non-coding RNA Loc554202 regulates proliferation and migration in breast cancer cells. Biochemical and Biophysical Research Communications . 2014;446(2):448–453. doi: 10.1016/j.bbrc.2014.02.144. [DOI] [PubMed] [Google Scholar]
- 26.Zong M. Z., Feng W. T., Du N., Yu X. J., Yu W. Y. Upregulation of long noncoding RNA LEF1-AS1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. European Review for Medical and Pharmacological Sciences . 2019;23(18):7929–7934. doi: 10.26355/eurrev_201909_19007. [DOI] [PubMed] [Google Scholar]
- 27.Li W., Liu J., Zhao H. Identification of a nomogram based on long non-coding RNA to improve prognosis prediction of esophageal squamous cell carcinoma. Aging (Albany NY) . 2020;12(2):1512–1526. doi: 10.18632/aging.102697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen F. J., Sun M., Li S. Q., et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Molecular Carcinogenesis . 2013;52(11):908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 29.Li X., Yan X., Wang F., et al. Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. Journal of Cellular and Molecular Medicine . 2019;23(4):2920–2932. doi: 10.1111/jcmm.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salle F. G., Le Stang N., Tirode F., et al. Comprehensive molecular and pathologic evaluation of transitional mesothelioma assisted by deep learning approach: a multi-institutional study of the international mesothelioma panel from the MESOPATH reference center. Journal of thoracic oncology . 2020;15(6):1037–1053. doi: 10.1016/j.jtho.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi X., Zhang D. H., Wu N., Xiao J. H., Wang X., Ma W. ceRNA in cancer: possible functions and clinical implications. Journal of Medical Genetics . 2015;52(10):710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 32.Gao X., Xie Z., Wang Z., Cheng K., Liang K., Song Z. Overexpression of miR-191 predicts poor prognosis and promotes proliferation and invasion in esophageal squamous cell carcinoma. Yonsei Medical Journal . 2017;58(6):1101–1110. doi: 10.3349/ymj.2017.58.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma S., Nagpal N., Ghosh P. C., Kulshreshtha R. P53-miR-191-SOX4regulatory loop affects apoptosis in breast cancer. RNA . 2017;23(8):1237–1246. doi: 10.1261/rna.060657.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X. Y., Han X. M., Zhao X. L., Cheng X. M., Zhang Y. MiR-93-5p promotes cervical cancer progression by targeting THBS2/MMPS signal pathway. European Review for Medical and Pharmacological Sciences . 2019;23(12):5113–5121. doi: 10.26355/eurrev_201906_18175. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z., Zhao Z., Yang Y., et al. miR-99b-5p and miR-203a-3p function as tumor suppressors by targeting IGF-1R in gastric cancer. Scientific Reports . 2018;8(1):p. 10119. doi: 10.1038/s41598-018-27583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campayo M., Navarro A., Benítez J. C., et al. miR-21, miR-99b and miR-375 combination as predictive response signature for preoperative chemoradiotherapy in rectal cancer. PLoS One . 2018;13(11, article e0206542) doi: 10.1371/journal.pone.0206542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan X., Zhang Y., Yu Z. Expression and clinical significance of miR-3615 in hepatocellular carcinoma. The Journal of International Medical Research . 2021;49(1, article 300060520981547) doi: 10.1177/0300060520981547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Figliuzzi M., Marinari E., De Martino A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophysical Journal . 2013;104(5):1203–1213. doi: 10.1016/j.bpj.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Figliuzzi M., De Martino A., Marinari E. RNA-based regulation: dynamics and response to perturbations of competing RNAs. Biophysical Journal . 2014;107(4):1011–1022. doi: 10.1016/j.bpj.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jens M., Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nature Reviews. Genetics . 2015;16(2):113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 41.Bosson A. D., Zamudio J. R., Sharp P. A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Molecular Cell . 2014;56(3):347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y., Yang D., Qin Y. Identifying prognostic lncRNAs based on a ceRNA regulatory network in laryngeal squamous cell carcinoma. BMC Cancer . 2021;21(1):p. 705. doi: 10.1186/s12885-021-08422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.She J. K., Fu D. N., Zhen D., Gong G. H., Zhang B. LINC01087 is highly expressed in breast Cancer and regulates the malignant behavior of cancer cells through miR-335-5p/Rock1. Oncotargets and Therapy . 2020;Volume 13:9771–9783. doi: 10.2147/OTT.S255994. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Zhao H., Zhu X., Luo Y., et al. LINC01816 promotes the migration, invasion and epithelial‑mesenchymal transition of thyroid carcinoma cells by sponging miR‑34c‑5p and regulating CRABP2 expression levels. Oncology reports . 2021;45(5) doi: 10.3892/or.2021.8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: the sample information was shown in Table S1. Supplementary Table 2: expression data of genes were gathered in Table S2. Supplementary Table 3: differently expressed lncRNAs were shown in Table S3. Supplementary Table 4: differently expressed miRNAs were shown in Table S4. Supplementary Table 5: the interaction network of between lncRNAs and miRNAs is shown in Table S5.
Data Availability Statement
The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.


