Abstract
Background
The aim of the study was to determine if periodontitis, which often causes transient bacteraemia, associates with viable bacteria in standard blood donations.
Materials and methods
This was a cross-sectional study of 60 self-reported medically healthy blood donors aged over 50 years. According to standard procedures, whole blood was separated by fractionation into plasma, buffy-coat, and red blood cell (RBC)-fractions. The buffy-coat was screened for bacterial contamination using BacT/ALERT. Samples from plasma and RBC-fractions were incubated anaerobically and aerobically at 37°C for 7 days on trypticase soy blood agar (TSA). For identification, colony polymerase chain reaction was performed using primers targeting 16S rDNA.
Results
From 62% of the donors with periodontitis, bacterial growth was observed on at least 1 out of 4 plates inoculated with plasma or RBCs, whereas only 13% of plates inoculated with plasma or RBCs from periodontally healthy controls yielded bacterial growth (relative risk 6.4, 95% CI: 2.1; 19.5; p=0.0011). None of the donors tested positive for bacterial contamination using BacT/ALERT. Cutibacterium acnes was found in 31% of the donations from donors with periodontitis and in 10% of the donations from periodontally healthy donors. In addition, Staphylococcus species, Bacillus mycoides, Aggregatibacter aphrophilus, and Corynebacterium kroppenstedtii were detected.
Discussion
Periodontitis increased the risk of bacterial contamination of blood products. Contaminating bacteria are often associated with the RBC-fraction. As the BacT/ALERT test is generally performed on platelet products, routine screening fails to detect many occurrences of viable bacteria in the RBC-fraction.
Keywords: transfusion, infection, periodontal disease, periodontitis, bacteraemia
INTRODUCTION
Over 100 million blood units are transfused annually, making blood transfusion one of the most common procedures in hospitals1,2. Infections resulting from the introduction of bacteria into a patient through blood transfusion are known as transfusion-transmitted infections (TTIs)3, and remain a leading cause of post-transfusion mortality and morbidity4,5. Infectious complications to blood transfusion include sepsis, pneumonia, abscesses, wound infection, meningitis, haemolysis, empyema, urinary tract infection, and fever6. Such infections may be partly accounted for by an inhibitory effect of the transfusion per se on the immune system7–9, but another cause might be unrecognised bacterial contamination of the transfused blood units, as we have previously suggested10.
Bacteria in donor blood may derive from infections of the donor or from contamination during venipuncture. Previous studies showed that daily activities such as chewing and tooth brushing facilitate translocation of bacteria into the blood stream in patients with periodontitis11–14. Periodontitis is a multifactorial inflammatory disease in the tooth supporting tissues, triggered by bacteria forming a biofilm on the tooth surface15. The periodontal biofilm is complex, and often includes gram-negative anaerobic bacteria16. In most countries periodontitis affects more than half of the population over the age of 5017. In a recent study of 148 German blood donors with a mean age of 53.3 years, 74% were diagnosed with periodontitis18. Periodontitis causes inflammatory breakdown of tooth supporting tissues, as well as deepening and ulceration of periodontal pockets through which bacteria may gain access to the blood stream11,14,15,19. Though a standard quarantine of 24 hours after undergoing dental procedures applies, periodontitis is currently not an exclusion criterion for blood donation. Using direct culturing, we have previously shown a surprisingly high incidence of bacteria from both plasma and the red blood cell (RBC)-fractions from donors aged ≥50 years, with a total proportion of 60% donating bacterially contaminated blood10. This may explain the discrepancy between the relatively high rates of post-transfusion infections compared to the low rates of bacterial contamination detected by the BacT/ALERT test. The latter method is based on colourimetric detection of CO2 produced if growing micro-organisms are introduced into the culture medium from the sample. Our previous study had not been designed to find any explanation for the contamination, including a possible association of periodontitis with donation of bacterially-contaminated blood10.
Conventional tests for bacterial contamination of donor blood are based on sampling from the platelet-fraction20. This is due to the general perception that platelets are more likely to be associated with bacterial contamination than other blood components due to the fact that storage at room temperature facilitates bacterial growth21. However, sampling from plasma or platelets does not reveal bacteria adhering to RBCs, which may constitute a reservoir of blood-borne bacteria22. Opsonisation of bacteria with complement enables bacteria to adhere to RBCs via complement receptor 1 (CR1, CD35), a phenomenon referred to as immune adherence22–24.
We hypothesize that blood from donors with periodontitis has a higher incidence of viable bacteria than blood from donors without periodontitis. We further hypothesize that those viable bacteria identified by direct culture are missed by standard screening protocols for bacterial contamination of donor blood.
Therefore, the aims of the study were:
to determine the prevalence of periodontitis in a cohort of blood donors aged >50 years;
to identify viable bacteria in standard blood-pack units;
to determine the incidence of viable bacteria in donors with and without periodontitis;
to determine whether standard screening procedures identify viable bacteria with the same incidence as direct culture.
MATERIALS AND METHODS
Sample size
This was a cross-sectional study. Sample size was estimated using a two-sided power analysis with μ(0)=40, μ(1)=10, ∑=50, α=0.005 and a power of 95%. The total sample size required for the study was 60 consecutive donors.
Ethics
The study and consent procedures were approved by The Ethics Committee for The Capital Region of Denmark (#H-16049514). All donors attended the Zealand Regional Blood Bank, Zealand University Hospital, Roskilde, Denmark, between June 11th 2018 and January 22nd 2019. All donors gave informed written consent prior to blood donation.
Blood specimen collection
A total of 60 self-reportedly healthy donors aged ≥50 years and weighing >50 kilos were recruited. To avoid any risk of bias, periodontal status was unknown at the time of the blood collection. Prior to venipuncture of the antecubital vein, topical disinfection was performed using swabs containing 82% ethanol and 0.5% chlorhexidine for 30 s, followed by 30 s drying time in accordance with World Health Organization (WHO) guidelines2. The first 30–50 mL blood was directed into a pre-sample bag to minimise the risk of contamination from the skin plug following insertion of the needle. The following 450 mL of blood was collected into the triple blood-pack unit containing citrate-phosphate-dextrose solution (CPD) (MacoPharma, Tourcoing, France). The tube connecting the needle with the pre-sample bag and the 450 mL triple blood-pack units was welded off. The pre-sample bag was used to draw samples for standard analysis as required (viral screening, blood type). All blood donations were given a unique number, to allow subsequent blood cultivation to be blinded. The triple blood-pack units were transported and stored at room temperature until fractionation either the same day or the following morning at the Department of Clinical Immunology, Zealand University Hospital, Næstved, Denmark. Finally, using a sterile technique, 10 mL from each buffy-coat was transferred to a BacT/ALERT flask for aerobic growth for 7 days using the Biomerieux system (Biomerieux SA, Marcy l'Etoile, France).
To further investigate bacterial presence in the final products, the bottom hoses on the plasma and RBC blood pack units were disinfected twice with 85% alcohol in a laminar flow hood after which they were cut with sterile scissors. The first 30 mL blood/plasma were discarded to minimise risk of contamination from cutting the hose. The next 15 mL were poured directly into a sterile 15 mL tube.
Isolation of viable bacteria from blood
0.5 mL of plasma and 0.5 mL RBC-suspension were plated out separately under sterile conditions on duplicate sets of trypticase soy blood agar (TSA) plates containing 5 mg/L hemin and 50 μg/L vitamin K, and incubated at 37°C under anaerobic conditions in the presence of 10% CO2, 10% H2, and 80% N2, and aerobically in the presence of 5% CO2, respectively. The resulting four cultures per donor were included in further analyses of bacterial growth. Another 0.5 mL from the heparin vacutainer was handled similarly and incubated on TSA plates anaerobically or aerobically to test if it could predict bacterial contamination in any of the blood fractions from the actual blood donation bag. All four plates per donor were incubated for 7 days at 37°C.
Detection of colony forming units
All plates were visually examined for colonies after 7 days of incubation. If positive, the number of colonies on each plate was counted and the plate was photographed. Colonies were then individually transferred to fresh plates to obtain monocultures for identification of species. The re-plated colonies were incubated for 4 days using the same growth conditions.
Colony polymerase chain reaction and 16S rRNA gene sequence analysis
For identification of bacteria, colony polymerase chain reaction (PCR) was performed using primers targeting the bacterial 16S rRNA gene, as described by Bosshard et al.25. Colony PCR was performed at the Costerton Biofilm Center, Department of International Health, Immunology and Microbiology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark. Bacterial 16S rRNA gene sequences were compared with taxon sequences in the extended Human Oral Microbiome Database (eHOMD), the National Center for Biotechnology Information (NCBI) database, and the Ribosomal Database Project, with a special focus on designated type strains.
Periodontal examination
All participants underwent a periodontal examination using a headlamp, a mirror and a periodontal probe (PCP 15; Hu-Friedy, Chicago, IL, USA) immediately after the blood donation. Periodontal conditions were assessed with periodontal pocket probing depth (PPD) and clinical attachment level (CAL) at six measurement points for each tooth (mesio-buccal, buccal, disto-buccal, mesio-oral, oral and disto-oral) with the same millimetre-scaled periodontal probe (PCP 15; Hu-Friedy). Based on these parameters, the periodontal disease severity according to AAP/EFP criteria26 was classified in accordance with the criteria for periodontitis, stage II–IV, where stage I is the mildest and stage IV the most severe27. Number of teeth and condition of the oral mucosa were also recorded. Moreover, relevant anamnestic information regarding smoking, tooth brushing habits, last visit to the dentist, and cause of missing teeth were recorded. Periodontal examinations and diagnostics were performed by a trained Doctor of Dental Surgery (DDS; CD).
Radiologic examination
After the blood specimen and periodontal examination, all participants were asked to attend the Department of Radiology, Zealand University Hospital, Roskilde, Denmark, within 1 month for an orthopantomography of the teeth and jaws to be taken by a trained radiographer. Description and diagnostics were performed by a trained DDS (CD).
Statistical analysis
Relative risk was determined using GraphPad Prism software version 8 (GraphPad Software, San Diego, CA, USA).
RESULTS
Twenty-nine of the 60 (48%) blood donors with a mean age of 57.9 years suffered from periodontitis, whereas the remaining 31 donors with a mean age of 54.8 years were periodontally healthy. There was an even distribution of males (n=29) and females (n=31) within the two groups.
Donors with periodontitis had, on average, 28.1 teeth and 13.3 decayed, missing, and filled permanent teeth (DMFT), whereas the periodontally healthy donors had 28.8 teeth and 11.8 DMFT. Five donors with periodontitis were smokers, 10 were previous smokers and 14 were never-smokers (Table I). Only one periodontally healthy donor smoked, 14 were previous smokers, and 16 were never-smokers (Table I). There were no statistically significant differences between the donors with periodontitis and those without in terms of age, gender distribution, number of teeth, and DMFT (Table I).
Table 1.
Population characteristics
| Age in years* (SD) | Female | Male | N. of teeth* | DMFT* | Smokers | Previous smoker | Never- smoker | |
|---|---|---|---|---|---|---|---|---|
| Periodontitis | 57.9 (5.2) | 12 | 17 | 28.1 | 13.3 | 5 | 10 | 14 |
| Healthy | 54.8 (5.3) | 19 | 12 | 28.8 | 11.8 | 1 | 14 | 16 |
SD: standard deviation; DMFT: decayed, missing, filled teeth.
Mean values.
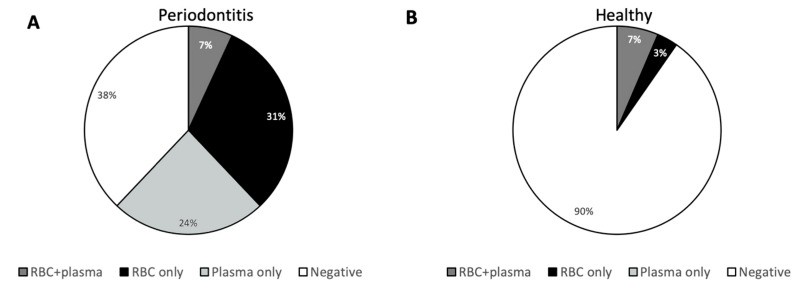
We inoculated TSA plates with plasma and RBCs, respectively, from each donor and incubated the plates under both aerobic and anaerobic conditions for 7 days. For 62% of the donors with periodontitis, bacterial growth was observed on at least one of these four plates (Figure 1A). They were distributed as 31% with growth only on plates inoculated with RBCs, 24% with growth only on plates inoculated with plasma, and 7% with growth on plates inoculated with RBCs and plasma (Figure 1A).
Figure 1. Frequency of viable bacteria in blood from donors.
Freshly drawn blood from 60 blood donors was fractioned into RBCs, buffy-coat and plasma. Plasma and RBCs were plated on trypticase soy blood agar (TSA) plates under aerobic or anaerobic conditions. Blood products were defined as positive if at least 1 colony was observed on at least one of the TSA plates. Shown are the frequencies of donors for whom bacteria were found in the RBC-fraction only, in the plasma-fraction only, in both fractions, or in none of the fractions. Donors with periodontitis (n=29) are shown in (A) and periodontally healthy donors (n=31) are shown in (B).
Among periodontally healthy donors, growth was only observed in 10%, of which 3% had growth only on plates with RBCs and 7% had growth on both RBCs and plasma (Figure 1B). All three periodontally healthy donors, who had bacterial growth, also had apical radiolucency visible on at least one tooth on the orthopantomogram, which is likely to be caused by an infected root canal.
The relative risk for having viable bacteria in the blood was 6.4 (95% CI: 2.1; 19.5) in donors with periodontitis compared to periodontally healthy donors.
None of the donors tested positive for bacterial contamination by the BacT/ALERT screening of each buffy-coat.
Among the 62% of donors with periodontitis for whom bacterial growth was demonstrated, 38% showed growth of more than one colony type (Figure 2).
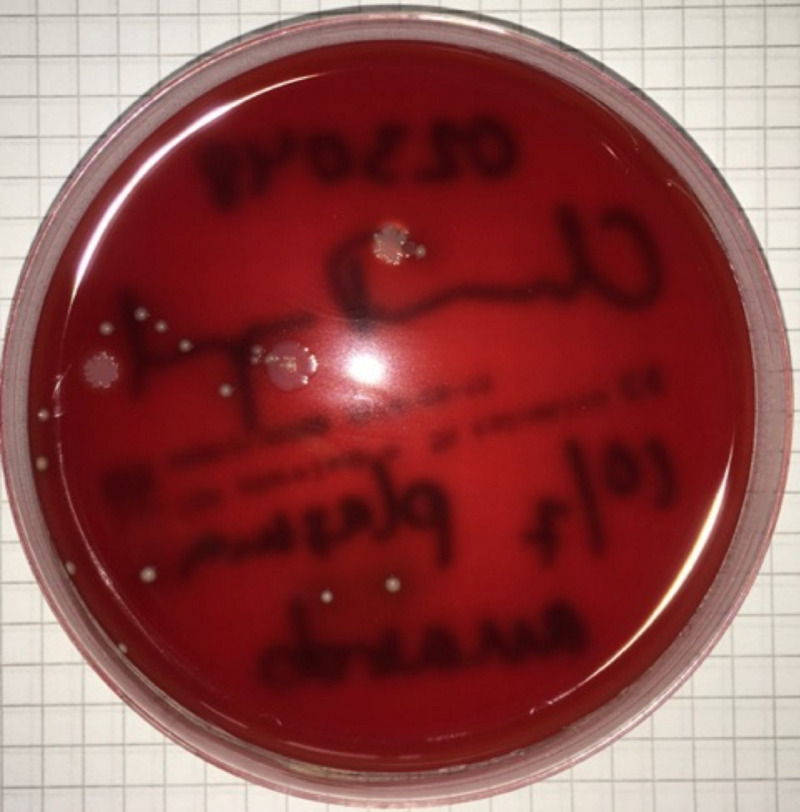
Figure 2. Example of a growth-positive plate.
Plasma from a donor with periodontitis plated on a trypticase soy blood agar (TSA) plate under anaerobic conditions for 7 days. Bacterial growth was identified as colonies of Staphylococcus epidermidis and Cutibacterium acnes.
Cutibacterium acnes was isolated from 20% of all participants, thus being the species most frequently identified (Table II). The bacterium was found in 31% of the donations from periodontitis-donors and in 10% of the donations from periodontally healthy donors (p=0.0544). Staphylococcus caprae/capitis was identified in 10% of the donations from periodontitis-donors and in 6% of the donations from periodontally healthy donors (p=0.6658), whereas S. epidermidis was identified in 7% of donors with periodontitis and 3% of the periodontally healthy donors (p=0.6059). Staphylococcus hominis, Staphylococcus lugdunensis and Bacillus mycoides were all identified in 7% of blood from donors with periodontitis and was not identified in blood from healthy donors. Aggregatibacter aphrophilus was identified in one donor with periodontitis. The majority of bacteria identified in the present study were facultatively anaerobic species.
Table II.
Bacterial species identified*
| Donors with periodontitis (n=18) | Periodontally healthy donors (n=3) | |
|---|---|---|
|
| ||
| Cutibacterium acnes | 9 | 3 |
| Staphylococcus caprae/capitis | 3 | 2 |
| Staphylococcus epidermidis | 2 | 1 |
| Staphylococcus lugdunensis | 2 | |
| Staphylococcus hominis | 2 | |
| Bacillus mycoides | 2 | |
| Corynebacterium kroppenstedtii | 1 | |
| Aggregatibacter aphrophilus | 1 | |
| Not identified | 1 | |
Bacteria were identified by comparing 16S rDNA sequence from the isolate taxons from the Expanded Human Oral Microbiome Database (eHOMD), the National Center for Biotechnology Information (NCBI) database, and the Ribosomal Project Database. All species were identified with minimum 99.1% confidence. Growth of the given species shown as number out of 60 blood donations.
DISCUSSION
Nosocomial-infections following transfusion with RBCs are known to occur at rates of up to 10.6–12.7%28, which are much higher than the rate of positive findings in conventional bacterial screening systems based on either pH-testing18, detection of CO2 29,30, or swirling of platelet concentrates31. It is to be noted that such testing is routinely performed only on the platelets and only under aerobic conditions28. In this study, we tested the plasma and RBC-fractions of donor blood for content of viable bacteria by cultivation on TSA plates under both aerobic and anaerobic conditions.
We confirm our previous finding of viable bacteria being frequently present in blood donations from donors aged ≥50 years. Also, our results confirm that bacteria can be found in the RBC-fraction as well as in the plasma fraction10. Our previous study did not consider the periodontal status of the donors, but here we show that patients with periodontitis have a relative risk of 6.4 for donating bacterium-contaminated blood compared to periodontally healthy donors. As expected17, around half of the blood donors had periodontitis, and all of these were aged >50 years.
None of the blood donations that yielded growth of bacteria, tested positive by bacterial screening using BacT/ALERT. Soeterboek et al. also found bacterial contamination of the RBC-fraction, but this was only based on BacT/ALERT-testing32. Accordingly, they reported considerably fewer cases of bacterial contamination than we do here, i.e., in 1% of RBC-products and 0.5% of the total blood products tested32. Likewise, Kunishima et al. found bacterial contamination of only 0.18% RBC concentrates based on cultivation in bottles of thioglycollate and soybean casein digest broth media33, in contrast to the direct cultures employed in the present study. In the French BACTHEM study, which included patients with transfusion-related adverse events, such as fever, chills, drop in blood pressure, shock, isolated dyspnoea, malaise, anxiety and digestive distress, 77% of samples yielded growth of bacteria by direct blood agar culture34. The species identified in the BACTHEM study were Gram-negative rods in 46% of contaminated blood portions, Gram-positive cocci in 28%, and Gram-positive rods in 21%34.
In agreement with the report by Kunishima et al.33, the most frequently isolated species in this study was Cutibacterium acnes (previously “Propionibacterium acnes”35), In addition, several coagulase-negative Staphylococcus species were frequently isolated. All of these species are typically associated with human skin, but are also inhabitants of the periodontium36,37. Only Aggregatibacter aphrophilus, which was recovered from one sample, is exclusively present in the human oral cavity and support the oral origin38. Theoretically, the majority of the other bacteria detected might have been introduced into the blood specimens at the collecting stage, although great effort was made to avoid this, including thorough disinfection of the skin and discarding the first 30 mL of collected blood. However, this explanation is not in agreement with the significantly higher frequency of bacterial contamination of blood from donors with periodontitis than periodontally healthy donors. Interestingly, it has been suggested that healthy individuals have a natural blood microbiome including bacterial phyla such as Proteobacteria, followed by Actinobacteria, Firmicutes, and Bacteroidetes39–41. Our findings suggest that the oral cavity is an important source of this microbiome, particularly in individuals with a jeopardised mucosal barrier due to periodontal inflammation. However, a limitation of this study is that we did not analyse the oral microbiome of the donors and thus cannot confirm that the bacterial species isolated from the blood are also present in the oral cavity or on the skin of the donors.
The question then arises as to why prominent periodontal bacteria such as Porphyromonas gingivalis, Fusobacterium and Prevotella spp.16 were not found in this study. One explanation may be that these bacteria are killed by the immune system in the whole blood preparations during overnight storage, while tolerating commensal bacteria such as staphylococci and C. acnes. Bacteria associated with periodontitis are mainly Gram-negative anaerobic bacteria, which are potent activators of leukocytes.
Although viable bacteria were considerably more frequent in blood donations from donors with periodontitis than in blood from periodontally healthy donors, 3 of the 31 donations from the latter did contain viable bacteria. Interestingly, however, all three periodontally healthy donors with bacterial growth also had apical radiolucency on at least one tooth. This suggests that inflammation due to infected root canals, such as periapical periodontitis, mediates translocation of bacteria into the circulation42.
The majority of the bacterial species identified in this study, including C. acnes and Staphylococcus spp., have previously been associated with nosocomial infections such as sepsis, endocarditis, brain abscesses, pneumonia, meningitis, urinary tract and wound infections43–47. Notably, TTI with Staphylococcus epidermidis has been reported to cause potentially fatal sepsis48,49.
We have previously shown a false-positivity rate of 8% using the method applied here10. Twenty-nine percent of the donors who tested positive had bacterial growth on more than one of the four plates, suggesting that at least those portions were not false-positive46. Moreover, 48% of the donations that tested positive for bacterial contamination presented with more than one colony, which also argues against a high frequency of false-positive findings.
In conclusion, 48% of blood donors in this cohort (mean age 57.9 years) suffered from periodontitis. Using direct cultivation of both plasma and the RBC-fraction, the relative risk for having viable bacteria in the blood was 6.4 (95% CI: 2.1; 19.5) in donors with periodontitis compared to periodontally healthy donors. None of the donors tested positive for bacterial contamination using BacT/ALERT screening of each buffy-coat. This suggests that blood donors with periodontitis are considerably more likely to donate blood contaminated with bacteria, and that routine screening of platelets using BacT/ALERT may not be sufficient to detect contaminations that may cause TTI.
ACKNOWLEDGEMENTS
We thank Professor Carsten Thomsen and Chief Physician Karina Vinum for taking the orthopantomographies at the Department of Radiology, Zealand University Hospital, Roskilde, Denmark. We thank Professor Tim Tolker- Nielsen for supplying reagents at the Department of Immunology and Microbiology, Costerton Biofilm Center, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
Footnotes
FUNDING
This study was supported by the Danish Dental Association.
CONTRIBUTIONS
CHN and PH contributed equally to this work.
CD, PH, SGS and CHN conceived and designed the experiments. CD, SGS and MN performed the experiments. CD, MN, MK, SGS, PH and CHN analysed the data. CD, PH, SGS and CHN wrote the paper.
The Authors declare no conflicts of interest.
Commented by DOI 10.2450/2021.0140-21
REFERENCES
- 1.Chassé M, McIntyre L, English SW, et al. Effect of blood donor characteristics on transfusion outcomes: a systematic review and meta-analysis. Transf Med Rev. 2016;30:69–80. doi: 10.1016/j.tmrv.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Blood safety and availability. WHO Glob Database Blood Saf. 2014. [Accessed on 23/09/2020]. Available at: http://www.who.int/mediacentre/factsheets/fs279/en.
- 3.Kuehnert MJ, Roth VR, Haley NR, et al. Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion. 2000;41:1493–9. doi: 10.1046/j.1537-2995.2001.41121493.x. [DOI] [PubMed] [Google Scholar]
- 4.Perel P, Clayton T, Altman DG, et al. Red blood cell transfusion and mortality in trauma patients: risk-stratified analysis of an observational study. PLoS Med. 2014;11:e1001664. doi: 10.1371/journal.pmed.1001664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–13. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Offner PJ, Moore EE, Biffl WL, et al. Increased rate of infection associated with transfusion of old blood after severe injury. Arch Surg. 2002;137:711–7. doi: 10.1001/archsurg.137.6.711. [DOI] [PubMed] [Google Scholar]
- 7.Hill GE, Frawley WH, Griffith KE, et al. Allogeneic blood transfusion increases the risk of postoperative bacterial infection: a meta-analysis. J Trauma. 2003;54:908–14. doi: 10.1097/01.TA.0000022460.21283.53. [DOI] [PubMed] [Google Scholar]
- 8.Rogers MA, Blumberg N, Saint S, et al. Hospital variation in transfusion and infection after cardiac surgery: a cohort study. BMC Med. 2009;7:37. doi: 10.1186/1741-7015-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers MA, Blumberg N, Saint SK, et al. Allogeneic blood transfusions explain increased mortality in women after coronary artery bypass graft surgery. Am Heart J. 2006;152:1028–34. doi: 10.1016/j.ahj.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Damgaard C, Magnussen K, Enevold C, et al. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS One. 2015;10:e0120826. doi: 10.1371/journal.pone.0120826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forner L, Larsen T, Kilian M, et al. Incidence of bacteremia after chewing, tooth brushing and scaling in individuals with periodontal inflammation. J Clin Periodontol. 2006;33:401–7. doi: 10.1111/j.1600-051X.2006.00924.x. [DOI] [PubMed] [Google Scholar]
- 12.Parahitiyawa NB, Jin LJ, Leung WK, et al. microbiology of odontogenic bacteremia: beyond endocarditis. Clin Microbiol Rev. 2009;22:386. doi: 10.1128/CMR.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horliana AC, Chambrone L, Foz AM, et al. Dissemination of periodontal pathogens in the bloodstream after periodontal procedures: a systematic review. PLoS One. 2014;9:e98271. doi: 10.1371/journal.pone.0098271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomás I, Diz P, Tobías A, et al. Periodontal health status and bacteraemia from daily oral activities: systematic review/meta-analysis. J Clin Periodontol. 2012;39:213–28. doi: 10.1111/j.1600-051X.2011.01784.x. [DOI] [PubMed] [Google Scholar]
- 15.Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol 2000. 2013;62:203–17. doi: 10.1111/j.1600-0757.2012.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 17.Eke PI, Thornton-Evans GO, Wei L, Borgnakke WS, et al. Periodontitis in US adults: National Health and Nutrition Examination Survey 2009–2014. J Am Dental Assoc. 2018;149:576–88. doi: 10.1016/j.adaj.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmalz G, Hübscher AE, Angermann H, et al. High prevalence of periodontitis in blood donors and the possibility of questionnaire-based screening - results of a cross-sectional study. Transfus Med. 2019;29:394–400. doi: 10.1111/tme.12633. [DOI] [PubMed] [Google Scholar]
- 19.Klausen SS, Hervig T, Seghatchian J, et al. Bacterial contamination of blood components: Norwegian strategies in identifying donors with higher risk of inducing septic transfusion reactions in recipients. Transfus Apher Sci. 2014;51:87–102. doi: 10.1016/j.transci.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Benjamin RJ, McDonald CP. The international experience of bacterial screen testing of platelet components with an automated microbial detection system: a need for consensus testing and reporting guidelines. Transfus Med Rev. 2014;28:61–7. doi: 10.1016/j.tmrv.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Hillyer CD, Josephson CD, Blajchman MA, et al. Bacterial contamination of blood components: risks, strategies, and regulation: joint ASH and AABB educational session in transfusion medicine. Hematology Am Soc Hematol Educ Program. 2003;1:575–89. doi: 10.1182/asheducation-2003.1.575. [DOI] [PubMed] [Google Scholar]
- 22.Belstrøm D, Damgaard C, Holmstrup P, et al. The atherogenic bacterium Porphyromonas gingivalis evades circulation phagocytes by adhering to erythrocytes. Infect Immun. 2011;17:1559–65. doi: 10.1128/IAI.01036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson RA., Jr The immune-adherence phenomenon; an immunologically specific reaction between microorganisms and erythrocytes leading to enhanced phagocytosis. Science. 1953;118:733–7. doi: 10.1126/science.118.3077.733. [DOI] [PubMed] [Google Scholar]
- 24.Ng YC, Schifferli JA, Walport MJ. Immune complexes and erythrocyte CR1 (complement receptor type 1): effect of CR1 numbers on binding and release reactions. Clin Exp Immunol. 1988;71:481–5. [PMC free article] [PubMed] [Google Scholar]
- 25.Bosshard PP, Abels S, Altwegg M, et al. Comparison of coventional and molecular methods for identification of aerobic catalase-negative Gram-positive cocci in the clinical laboratory. J Clin Microbiol. 2004;42:2065–73. doi: 10.1128/JCM.42.5.2065-2073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caton JG, Armitage G, Berglundh T, et al. A new classification scheme for periodontal and peri-implant diseases and conditions - Introduction and key changes from the 1999 classification. J Clin Periodontol. 2018;45(Suppl 20):S1–8. doi: 10.1111/jcpe.12935. [DOI] [PubMed] [Google Scholar]
- 27.Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Clin Periodontol. 2018;45(Suppl 20):S162–70. doi: 10.1111/jcpe.12946. [DOI] [PubMed] [Google Scholar]
- 28.Rhode JM, Dimcheff DE, Blumberg N, et al. Health care associated infection after red blood cell transfusion: a systematic review and meta-analysis. JAMA. 2014;311:1317–26. doi: 10.1001/jama.2014.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walther-Wenke G. Incidence of bacterial transmission and transfusion reactions by blood components. Clin Chem Lab Med. 2008;46:919–25. doi: 10.1515/CCLM.2008.151. [DOI] [PubMed] [Google Scholar]
- 30.Brecher ME, Jacobs MR, Katz LM, et al. Survey of methods used to detect bacterial contamination of platelet products in the United States in 2011. Transfusion. 2013;53:911–8. doi: 10.1111/trf.12148. [DOI] [PubMed] [Google Scholar]
- 31.Montag T. Strategies of bacteria screening in cellular blood components. Clin Chem Lab Med. 2008;46:926–32. doi: 10.1515/CCLM.2008.176. [DOI] [PubMed] [Google Scholar]
- 32.Soeterboek AM, Welle FH, Marcelis JH, et al. Sterility testing of blood products in 1994/1995 by three cooperating blood banks in The Netherlands. Vox Sang. 1997;72:61–2. doi: 10.1046/j.1423-0410.1997.00061.x. [DOI] [PubMed] [Google Scholar]
- 33.Kunishima S, Inoue C, Kamiya T, et al. Presence of Propionibacterium acnes in blood components. Transfusion. 2001;41:1126–9. doi: 10.1046/j.1537-2995.2001.41091126.x. [DOI] [PubMed] [Google Scholar]
- 34.Perez P, Salmi LR, Follea G, et al. the French Haemovigilance Network. Determinants of transfusion-associated bacterial contamination: results of the French BACTHEM Case-Control Study. Transfusion. 2001;41:862–72. doi: 10.1046/j.1537-2995.2001.41070862.x. [DOI] [PubMed] [Google Scholar]
- 35.Scholz CFP, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol. 2016;66:4422–32. doi: 10.1099/ijsem.0.001367. [DOI] [PubMed] [Google Scholar]
- 36.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niazi SA, Clarke D, Do T, et al. Propionibacterium acnes and Staphylococcus epidermidis isolated from refractory endodontic lesions are opportunistic pathogens. J Clin Microbiol. 2010;48:3859–69. doi: 10.1128/JCM.01326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nørskov-Lauritsen N, Kilian M. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int J Syst Evol Microbiol. 2006;56:2135–46. doi: 10.1099/ijs.0.64207-0. [DOI] [PubMed] [Google Scholar]
- 39.Gosiewski T, Ludwig-Galezowska A, Huminska K, et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method-the observation of DNAemia. Eur J Clin Microbiol Infect Dis. 2017;36:329–36. doi: 10.1007/s10096-016-2805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Païssé S, Valle C, Servant F, et al. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56:1138–47. doi: 10.1111/trf.13477. [DOI] [PubMed] [Google Scholar]
- 41.Whittle E, Leonard MO, Harrison R, et al. Multi-method characterization of the human circulating microbiome. Front Microbiol. 2019;9:3266. doi: 10.3389/fmicb.2018.03266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debelian GJ, Olsen I, Tronstad L. Anaerobic bacteremia and fungemia in patients undergoing endodontic therapy: an overview. Ann Periodontol. 1998;3:281–7. doi: 10.1902/annals.1998.3.1.281. [DOI] [PubMed] [Google Scholar]
- 43.Otto M. Staphylococcus epidermidis-the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–67. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regalado NG, Martin G, Antony SJ. Acinetobacter lwoffii: bacteremia associated with acute gastroenteritis. Travel Med Infect Dis. 2009;7:316–7. doi: 10.1016/j.tmaid.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Seifert H, Kaltheuner M, Perdreau-Remington F. Micrococcus luteus endocarditis: case report and review of the literature. Zentralbl Bakteriol. 1995;282:431–5. doi: 10.1016/s0934-8840(11)80715-2. [DOI] [PubMed] [Google Scholar]
- 46.Hall KK, Lyman JA. Updated review of blood culture contamination. Clin Microbiol Rev. 2006;19:788–802. doi: 10.1128/CMR.00062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry A, Lambert P. Propionibacterium acnes: infection beyond the skin. Expert Rev Anti Infect Ther. 2011;9:1149–56. doi: 10.1586/eri.11.137. [DOI] [PubMed] [Google Scholar]
- 48.Muder RR, Yee YC, Rihs JD, et al. Staphylococcus epidermidis bacteremia from transfusion of contaminated platelets: application of bacterial DNA analysis. Transfusion. 1992;32:771–4. doi: 10.1046/j.1537-2995.1992.32893032109.x. [DOI] [PubMed] [Google Scholar]
- 49.Goldman M, Delage G. A fatal case of transfusion-transmitted Staphylococcus epidermidis sepsis. Transfusion. 2001;41:1075–6. doi: 10.1046/j.1537-2995.2001.41081075.x. [DOI] [PubMed] [Google Scholar]