Abstract
We report here the identification and characterization of a novel Vav family member, Vav-3. Signaling experiments demonstrate that Vav-3 participates in pathways activated by protein tyrosine kinases. Vav-3 promotes the exchange of nucleotides on RhoA, on RhoG and, to a lesser extent, on Rac-1. During this reaction, Vav-3 binds physically to the nucleotide-free states of those GTPases. These functions are stimulated by tyrosine phosphorylation in wild-type Vav-3 and become constitutively activated upon deletion of the entire calponin-homology region. Expression of truncated versions of Vav-3 leads to drastic actin relocalization and to the induction of stress fibers, lamellipodia, and membrane ruffles. Moreover, expression of Vav-3 alters cytokinesis, resulting in the formation of binucleated cells. All of these responses need only the expression of the central region of Vav-3 encompassing the Dbl homology (DH), pleckstrin homology (PH), and zinc finger (ZF) domains but do not require the presence of the C-terminal SH3-SH2-SH3 regions. Studies conducted with Vav-3 proteins containing loss-of-function mutations in the DH, PH, and ZF regions indicate that only the DH and ZF regions are essential for Vav-3 biological activity. Finally, we show that one of the functions of the Vav-3 ZF region is to work coordinately with the catalytic DH region to promote both the binding to GTP-hydrolases and their GDP-GTP nucleotide exchange. These results highlight the role of Vav-3 in signaling and cytoskeletal pathways and identify a novel functional cross-talk between the DH and ZF domains of Vav proteins that is imperative for the binding to, and activation of, Rho GTP-binding proteins.
Rho and Rac family members participate in coordinated cellular responses to extracellular stimuli (11, 28). Their action is important in promoting the formation of cytoskeletal structures, the activation of serine/threonine kinase cascades, and the induction of gene expression (11, 28). Rho proteins are regulated by the binding of guanosine nucleotides (1). In quiescent cells, these GTPases are bound to GDP molecules and are in an inactive state. Stimulation of cells via a number of extracellular stimuli leads to the exchange of GDP by GTP, a transition that allows the acquisition of a conformation optimal for the binding to their effector molecules (1). Because the intrinsic GDP-GTP exchange rate of these GTPases is low under physiological conditions, the activation of these proteins during signal transduction requires the participation of enzymes generically known as guanosine nucleotide exchange factors (GEFs) (1). To date, two families of Rho GEFs have been identified. The first group is composed of Rho GDP dissociation stimulators, a family of proteins distantly related to the Cdc25 homology regions present in Ras GEFs (1). The second group comprises an extensive number of enzymes containing Dbl homology (DH) domains with catalytic activity exclusively directed towards Rho and Rac GTPases (4).
Although Rho GEFs have been extensively characterized biochemically and oncogenically, little information is available regarding the mechanism by which they become activated during signal transduction. To date, the best example for the participation of a DH-containing protein in receptor-mediated cell signaling is perhaps the product of the vav proto-oncogene, a protein preferentially expressed in the hematopoietic system (2). In addition to the DH and Pleckstrin homology (PH) regions commonly found in Rho and Rac GEFs, Vav contains other structural motifs, including a calponin homology (CH) region, an acidic (Ac) motif, a zinc finger (ZF) domain, two SH3 regions, and one SH2 domain (2). Vav becomes tyrosine-phosphorylated during the signaling of many membrane receptors, and binds to a number of cytoplasmic molecules via its SH2 and SH3 domains (2). Recently, biochemical experiments have demonstrated that the phosphorylation of Vav on tyrosine residues leads to the activation of its GDP/GTP exchange activity towards Rac-1 in vitro (7). In agreement with such observations, it has been shown that several elements of the Rac-1 pathway, including Rac-1 itself and JNK, are activated in vivo by wild type Vav protein upon tyrosine phosphorylation (7, 26). Deletion of vav via gene targeting leads to decreased proliferation of prothymocytes (31), to defective positive and negative selection of immature T cells (15, 27) and to ineffective functional responses of mature T and B cells (25, 31). In T lymphocytes, this phenotype is linked to abnormal actin clustering upon receptor engagement (10, 14). Vav appears to provide therefore a direct connection between membrane receptors and Rac-1, a pathway that is essential for the generation of proper mitogenic, developmental, and antigenic responses.
Vav was long considered to have unique structural and signaling properties. However, this initial view has changed recently after the identification of Vav-2, a Vav-related GEF whose activity is modulated by tyrosine phosphorylation (13, 22, 23). Vav and Vav-2 differ in several biological properties. For example, Vav-2 displays a ubiquitous pattern of expression during embryonic and adult mouse stages (22). Moreover, the stable expression of the vav-2 oncogene in rodent fibroblasts elicits a morphological phenotype that is different from that induced by the vav oncogene (22, 23). Perhaps more importantly, it has been shown that Vav-2 targets preferentially RhoG and RhoA subfamily members, whereas Vav does so on RhoG and Rac subfamily members (7, 23). These observations indicate that the two Vav family members use overlapping, but not identical, signal transduction pathways. Whether Vav and Vav-2 utilize common membrane receptors for activation and bind to similar spectra of intracellular signaling molecules remains to be determined.
The discovery of Vav-2 suggested that the Vav family was likely to be more complex and that additional members of the family remain to be discovered. The presence of a Caenorhabditis elegans gene encoding another Vav-related protein (30), gave us the final impetus to look for the possibility of new Vav members. This search resulted in the identification of Vav-3, a third representative of the Vav family present in humans. We present here the functional characterization of this novel member of the Vav family. Our results demonstrate that Vav-3 participates in signal transduction processes and in the activation of several members of the Rho family, an activity that, when deregulated, leads to marked cytoskeletal changes and to alterations in the process of cell division. Finally, we provide evidence regarding the subcellular localization of Vav-3 and the identification of the structural domains that are important for the interaction with, and activation of, GTP-binding proteins. These results expand the spectrum of known Vav family members and provide information about a new mechanism that is essential for the catalytic activity of Vav family members.
MATERIALS AND METHODS
Cloning of VAV-3 cDNAs.
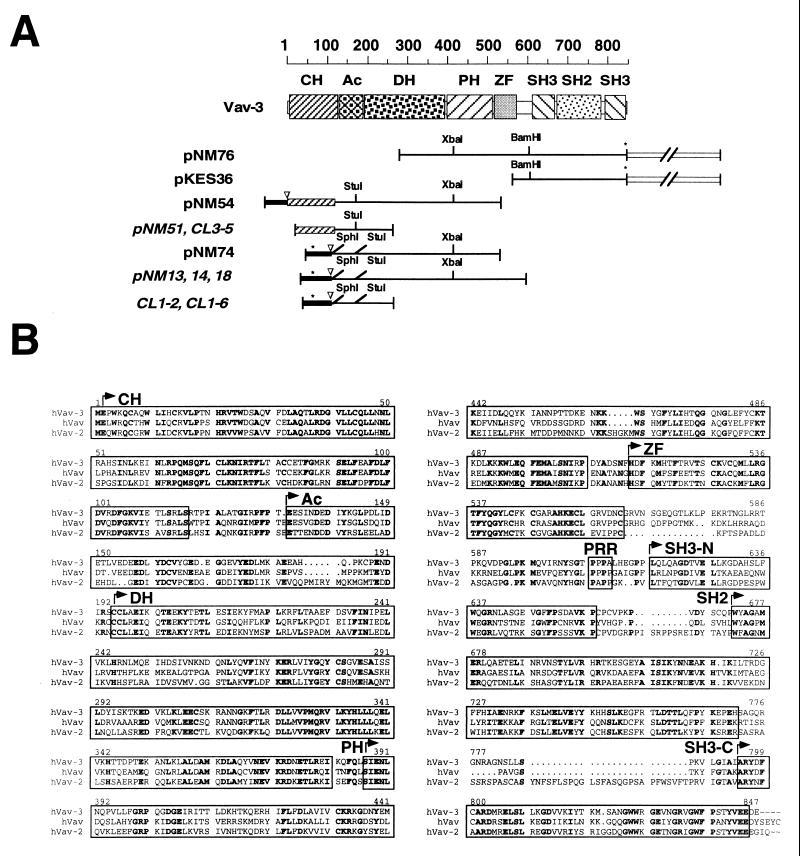
A scheme of all cDNA fragments isolated to clone the human VAV-3 cDNA is depicted in Fig. 1A. After obtaining the EST cDNA containing the VAV-3 3′ from the American Tissue Culture Collection, the cDNA fragment was used to screen a human placenta cDNA library (Stratagene). More than 60 cDNA clones were obtained, all of them containing truncated versions of the VAV-3 cDNA, as determined by PCR analysis. One of these incomplete cDNA clones was excised from the phage DNA by using a helper phage, resulting in the generation of the pKES36 plasmid (Fig. 1A). After sequencing the 5′ end of the pKES36 cDNA, we used a set of two nested oligonucleotides (VAV-3#1 and VAV-3#2) to obtain the missing 5′ end of the VAV-3 cDNA by using the rapid amplification of cDNA ends (RACE) technique. This approach resulted in the isolation of three new VAV-3 cDNA fragments (pNM13, pNM14, and pNM18) each derived from independent PCR reactions of placental cDNAs (Fig. 1A). Since sequence analysis indicated that these cDNAs contained an unusual N terminus for Vav family proteins (the Vav-3β isoform, see Results), another set of nested oligonucleotides (VAV-3-19RACE and VAV-3-20RACE) was used to isolate the 5′ cDNA sequences expected to be homologous to the other known members of the Vav family (referred to as Vav-3α hereafter). The RACE fragments amplified with these oligonucleotides were cloned into the pCR2.1 vector (Invitrogen) and screened by digestion with SphI, a restriction enzyme that cuts at the ATG position of the VAV-3β cDNA (Fig. 1A). Clones lacking SphI sites were then sequenced, resulting in the identification of two cDNA clones (pNM51 and clone #3-5) that encoded part of the Vav-3 CH domain but still lacking the initiator methionine (Fig. 1A). Since the PCR amplification of a new placental cDNA sample yielded fragments only corresponding to the VAV-3β cDNA end (i.e., clones #1-6 and #1-2), we decided to use the cDNA clone contained in pNM51 to screen a different human placental cDNA library (Clontech). This screening yielded two additional VAV-3 cDNAs, one containing the 5′ end of VAV-3α (pNM54) and the other containing the 5′ end of the VAV-3β isoform (pNM74) (Fig. 1A). The cDNA fragment of pNM54 was then used to re-screen the λgt11 placenta cDNA library to make sure that all VAV-3 cDNA sequences were derived from cDNA libraries. This approach resulted in the isolation of a truncated VAV-3 cDNA (pNM76) overlapping with the 3′ end of the pNM54 cDNA but extending farther downstream into the 3′ untranslated region (UTR) (Fig. 1A). To generate the full-length VAV-3β cDNA (pNM77), a 3.1-kbp XbaI fragment was liberated from pNM76 and ligated into an XbaI fragment of pNM74 containing pBluescript SK and the 5′ end of the VAV-3β cDNA (Fig. 1A). To generate the full-length VAV-3α cDNA (pNM81), we amplified by PCR the 5′ end of this cDNA by using oligonucleotides VAV-3α-ATF-F (forward) and VAV-3-14R (reverse) and the pNM54 plasmid as the DNA template. After digestion with SphI (contained in the forward plasmid) and StuI (present in the VAV-3 cDNA) (Fig. 1A), the fragment was gel purified and ligated into a 7.6-kbp SphI-StuI fragment of pNM78 containing pMEX (see below) vector sequences and VAV-3 cDNA sequences located downstream of the StuI site. Sequence analysis indicated that no mutations were created during this amplification step. Automatic nucleotide sequence analysis of the VAV-3 cDNAs was performed on the cDNA fragments contained in plasmids pNM13, pNM14, pNM18, pNM54, pNM74, and pNM76 by using overlapping oligonucleotides that annealed to both strands of the VAV-3 cDNA clones. The sequences of the oligonucleotides used in these studies are available upon request.
FIG. 1.
(A) Schematic representation of the VAV-3 cDNAs isolated in this study. Clones obtained from the screening of cDNA libraries are shown in regular typeface. Those obtained from RACE amplifications are shown in italics. The 5′ UTRs are shown as closed boxes. The 3′ UTRs are shown as open boxes and not in scale. The 5′ ends encoding the Vav-3 CH domain are shown as shaded boxes. Stop codons are indicated by asterisks. Start codons are indicated by inverted triangles. Important restriction sites for cloning purposes are indicated. (B) Alignment of the amino acid sequences of the three members of the Vav family in Homo sapiens. Identical residues present in the three proteins are shown on boldface. The individual structural domains are boxed. PRR, proline-rich region.
Antibodies.
Monoclonal antibodies to FLAG, hexahistidines, and Shc were obtained from Kodak, Sigma, and Transduction Laboratories, respectively. Polyclonal antibodies against glutathione S-transferase (GST), Dbl, and green fluorescent protein (GFP) were obtained from Santa Cruz Biotechnology (GST and Dbl) and Clontech (GFP). A monoclonal antibody to the maltose binding protein was obtained from Sigma. A rabbit polyclonal antibody to Vav-3 was developed with a synthetic peptide corresponding to sequences 201 to 216 of human Vav-3 as antigen (Research Genetics, Inc.). Lack of cross-reactivity among these antibodies was demonstrated by immunoprecipitation and immunoblot analysis with Vav, Vav-2, and Vav-3 proteins purified from Sf9 cells (19).
mRNA expression studies.
The VAV-3 cDNA fragment contained in pNM54 (Fig. 1A) and a 300-bp fragment of the ubiquitin cDNA (Clontech) were labeled with [α-32P]dCTP by using a random hexamer primer technique (Amersham) and hybridized overnight at 65°C with the filters according to the manufacturer’s instructions.
Insect cell and bacterial expression vectors.
Baculovirus was generated from pFastBac-derivatives (Gibco-BRL) encoding oncogenic Dbl (pNM12) and truncated Vav-3 (residues 144 to 847) protein (pNM85). To generate the baculovirus encoding the polyhistidine-tagged Vav-3 Ac+DH domains, pNM85 was digested with XbaI (Fig. 1A), gel purified to eliminate 3′ sequences, and religated to generate the pNM92 plasmid. To generate the baculovirus encoding the 6xHis-tagged Ac+DH+PH domains of Vav-3, pNM85 was digested with XhoI, gel purified to eliminate the 3′ sequences, and religated to generate the pNM120 plasmid. To make the baculovirus encoding the polyhistidine tagged version of wild-type Vav-3, a 0.5-kbp cDNA fragment encoding the Vav-3 N terminus was generated by PCR from pNM81 and, after digestion with EcoRI and StuI, ligated into EcoRI-StuI-linearized pNM85. 6xHis–Vav-3 (144 to 847) harboring mutations in the PH (W493L) (pNM93) and ZF (C527S) (pNM94) were generated from pNM85 by using the Quikchange site-directed mutagenesis kit (Stratagene). All pFastBac constructs were then recombined in E. coli with the baculovirus DNA by using a helper phage; successful recombinants were identified by lacZ gene inactivation and PCR and used to generate baculovirus particles in Spodoptera frugiperda (Sf9) cells exactly as recommended by the supplier (Gibco-BRL). Rho proteins were induced in bacteria by using pGEX derivatives (23). To express the Vav-3 ZF region (residues 506 to 584) in bacteria, a 0.5 kbp cDNA fragment was amplified from pNM77 by PCR, digested with KpnI and EcoRI, and ligated into KpnI-EcoRI-linearized pMAL-c (New England Biolabs) or pTrc-HisC (Invitrogen) to generate the plasmids pNM128 and pNM125, respectively. After expression, these proteins were purified by using amylose (New England Biolabs) or nickel (Qiagen) beads according to the specifications of each commercial supplier.
Mammalian expression vectors.
For focus formation assays in rodent fibroblasts, all genes were cloned in pMEX, a vector capable of driving high levels of protein expression via its mouse mammary tumor virus long terminal repeat. Plasmids were those encoding the full length Vav-3 protein (pNM81) and the truncated Vav-3 proteins lacking residues 1 to 144 (pNM78) or 1 to 184 (pNM80). Other pMEX-derivatives containing vav and vav-2 sequences were previously described (23).
For transient expression experiments in mammalian cells, a truncated version of Vav-3 lacking both the N-terminal (residues 1 to 144) and SH3-SH2-SH3 domains (residues 607 to 847) was expressed using a pcDNAHisA vector (pNM88, Invitrogen). EGFP-tagged proteins were generated as follows: Vav-3 (Δ1–144+Δ607–847) was liberated from the pNM84 plasmid by digestion with StuI and BamHI and cloned into the pEGFP-C3 vector (Clontech) to generate pNM99. Vav-3 (Δ1–144+Δ607–847) with mutations in either PH (W493L) (pNM101) or ZF (C527S) (pNM102) domains were generated from the cDNA fragments contained in pNM93 and pNM94, respectively (see above). Vav-3 (Δ1–144+Δ607–847) with a mutation in the DH domain (L211Q) (pNM127) was generated by site-directed mutagenesis from pNM99. To generate EGFP–Vav-3 (Δ1–144) (pNM100), a BamHI VAV-3 cDNA fragment was obtained from pNM78 and ligated to BamHI-linearized pNM99. pNM100 was also used for site directed mutagenesis to generate vectors encoding Vav-3 (Δ1–144) proteins with mutations in either the DH (L211Q) (pNM126) or the ZF (C527S) (pNM122). Constructs for vav, vav-2, DBL, lckY505F, and rho genes were previously described (23). The pSRαNeo-FLAGLbc plasmid was provided by D. Toksoz (Tufts University School of Medicine, Boston, Mass.). The rabbit rsc oncogene was provided by A. Pellicer (New York University).
Protein purification, in vitro GDP-GTP exchange, and GTPase assays.
Purification of Vav-3 proteins from baculovirus-infected Sf9 cells, purification of GTPases from Escherichia coli cells, [35S]GTP-γS incorporation, [3H]GDP release, and GTP hydrolysis assays were performed as described earlier (23). GST-tagged Lck and untagged Hck proteins purified from Sf9 cells were gifts from J. Fragnoli (Bristol-Myers Squibb Pharmaceutical Research Institute, Princeton, N.J.) and T. Miller (Department of Physiology, State University of New York, Stony Brook, N.Y.), respectively. In vitro kinase reactions were performed as previously indicated (7). Dbl total cellular lysates were obtained from baculovirus-infected Sf9 cells as indicated earlier (33). GST-GTPases were normalized for activity and concentration by using [35S]GTP-binding experiments.
In vitro binding experiments.
GST-GTPases (75 pmol) were stripped of nucleotides by incubating them for 10 min at room temperature in binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2.5 ng of bovine serum albumin per ml, 10 mM EDTA, 0.1% Triton X-100, 1 mM dithiothreitol, 10% glycerol) supplemented with a cocktail of protease inhibitors (Cømplete; Roche Molecular Biochemicals). After these incubations, the buffer was supplemented with 50 mM MgCl2 plus either GDP (450 μM), GTP (450 μM), or no nucleotides and then incubated 30 min longer. At this moment, 30 μl of glutathione beads (Pharmacia/LKB) and either the Vav-3 proteins (55 pmol) or the Dbl-containing Sf9 lysates (50 μl) were added to the GTPase solutions. After an incubation of 3 h at 4°C, the beads were washed three times with binding buffer plus 50 mM MgCl2, boiled in the presence of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and analyzed by immunoblot.
Cell stimulation, transfection assays, and immunofluorescence techniques.
Jurkat cells were cultured in RPMI medium supplemented with 10% fetal calf serum (HyClone) and antibiotics. For stimulation, exponentially growing cell cultures were harvested, washed once with RPMI and resuspended in RPMI at 25 × 106 cell · ml−1. Cells were stimulated with 10 μg of anti-CD3 antibodies (Dako) per ml for the indicated periods of time. After stimulation, cells were lysed by the addition of 5× lysis buffer (50 mM Tris-HCl [pH 8.0], 5% Triton X-100, 750 mM NaCl) supplemented with 50 mM sodium fluoride, 0.5 mM sodium orthovanadate and protease inhibitors (Cømplete). In the case of COS-1 cells, transfected cells were cultured for 48 h in 10-cm plates and then starved in serum-free Dulbecco modified Eagle medium overnight. Cells were then either left unstimulated or stimulated for the indicated periods of time with EGF (0.25 μg · ml−1) and disrupted in 1 ml of 1× lysis buffer. Cell lysates were centrifuged for 30 min at 11,000 × g and supernatants were immunoprecipitated by using the appropriate antibodies. Immunoprecipitation and immunoblot analysis were performed as previously described (3). Focus formation, transient-transfection, and immunofluorescence techniques were performed as indicated earlier (23).
RESULTS
Isolation of the human VAV-3 gene.
To look for new members of this protein family, we searched GenBank databases with small stretches of the primary structure of Vav and Vav-2 to identify any possible Vav-related sequences present in available expressed sequence tag (EST) libraries. This search yielded one EST cDNA clone (GenBank entry R56753) that was found to encode half of an SH2 domain and a complete SH3 domain, both of them highly homologous to the corresponding domains of Vav and Vav-2. High homology was also found in the linker region connecting the SH2 domain and the most C-terminal SH3 region of the known Vav proteins, further suggesting that this EST clone encoded a new member of the Vav family rather than an unrelated SH2-SH3 protein. We then cloned the full-length cDNA of this protein with a combination of standard cDNA library screening and RACE techniques (see Materials and Methods) (Fig. 1). Sequence analysis of this cDNA (accession number [AN] AF118887) confirmed that it encoded a novel 847-amino-acid polypeptide (97.8 kDa, pI 6.65) with overall sequence similarities of 69.4 and 66.1% with the human Vav and Vav-2 proteins, respectively. This new Vav family member was designated Vav-3.
During the final characterization of our VAV-3 gene product, another VAV-3 sequence was posted in the GenBank (AN AF067817). This VAV-3 cDNA clone encodes a protein with two amino acid changes when compared to our cDNA isolate, one change being located in the CH domain (K107E) and the other in the DH region (T298S). The lysine at position 107 present in our cDNA isolate is conserved in Vav and Vav-2 proteins. Instead, the residues located at position 298 are more variable in Vav family members. Minor discrepancies are also observed between both isolates in the 3′ noncoding region (19). It is plausible that those sequence discrepancies could represent allelic variations of the human VAV-3 gene. In addition to those cDNAs, at least 11 human and 1 mouse (AN AA517102) EST clones containing fragments of the 3′ end of the VAV-3 cDNA were also found in gene databases.
In addition to this full-length protein, the screening of cDNA libraries by both PCR and probe hybridization frequently rendered additional cDNAs with a different 5′ end (AN AF118886) (Fig. 1A; see also Materials and Methods). All those cDNAs encode a truncated protein (referred to hereafter as Vav-3β) in which the first 107 amino acid residues of Vav-3 are replaced by a shorter open reading frame of 13 residues (Fig. 1A). However, we have found no evidence for the expression of this putative Vav-3 isoform in vivo, suggesting that this alternative form probably represents an incomplete spliced form of the VAV-3 hnRNA present in some commercial cDNA libraries.
Vav-3 is expressed broadly and participates in signal transduction pathways.
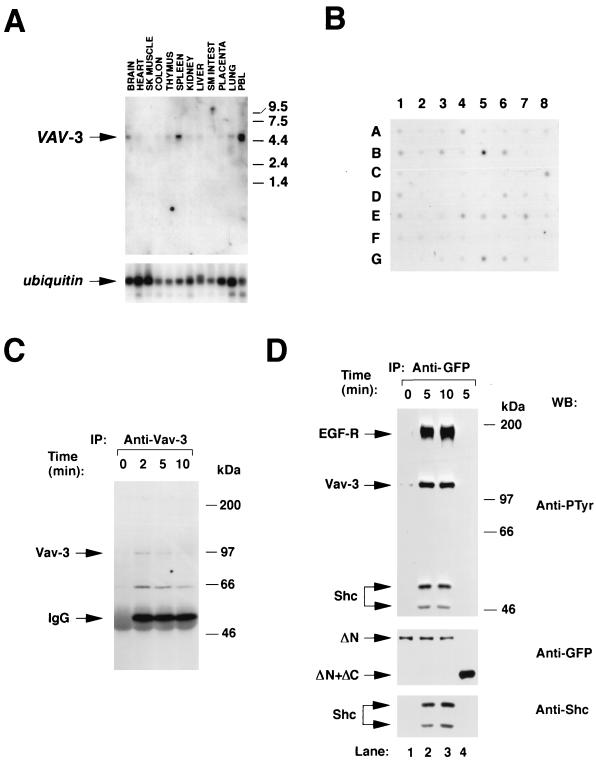
To determine the distribution of VAV-3 transcripts, we performed Northern hybridization analysis of polyadenylated mRNA from various human tissues by using a 32P-labeled probe containing the 5′ end of the human VAV-3 cDNA. We detected a single 5-kb transcript that was highly expressed in peripheral blood lymphocytes, spleen, and brain (Fig. 2A, upper panel). Lower, but detectable, levels of VAV-3 mRNA were detected in thymus, heart, kidney, liver, placenta, and lung (Fig. 2A, upper panel). Rehybridization of the same filter with a 32P-labeled ubiquitin probe demonstrated the integrity of all these RNA samples (Fig. 2A, lower panel). To analyze further the pattern of VAV-3 gene expression, we conducted hybridization experiments by using poly(A) mRNAs derived from 50 different human tissues. In addition to the places where VAV-3 mRNAs had been detected in our Northern blot experiments, VAV-3 messages were found in several regions of the nervous system (cerebellum, occipital lobe, substantia nigra, thalamus and subthalamus), in hematopoietic tissues (lymph nodes), and in other tissues such as the testis, thyroid gland, and stomach (Fig. 2B). In addition, high levels of expression of the VAV-3 gene were observed in embryonic hematopoietic tissues (Fig. 2B). Taken together, these results indicate that VAV-3 is widely expressed in human tissues.
FIG. 2.
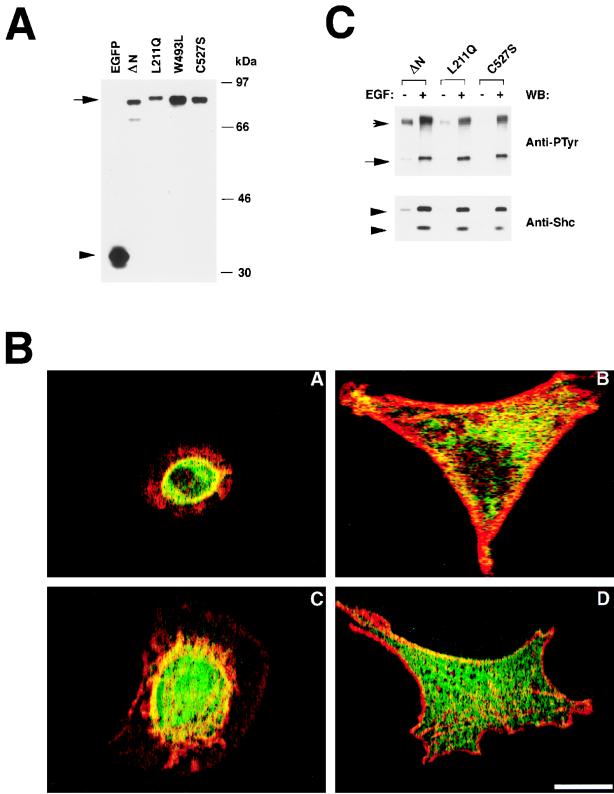
(A) Distribution of VAV-3 transcripts in adult human tissues. A filter containing 2 μg of poly(A) mRNA isolated from the indicated tissues (MTN blot; Clontech) was hybridized to either a VAV-3 (upper panel) or a ubiquitin (lower panel) probe as described in Materials and Methods. The hybridized filter was exposed for 48 h (upper panel) and 1 h (lower panel). The migration of the VAV-3 and ubiquitin mRNAs is indicated by arrows. The migration of molecular weight markers is indicated on the right. (B) Dot blot analysis of the expression of the VAV-3 gene in adult and embryonic human tissues. The hybridized filter (Human RNA Master blot; Clontech) contained poly(A) mRNA from the following sources: A1 (whole brain, 257 ng), A2 (amigdala, 135 ng), A3 (caudate nucleus, 161 ng), A4 (cerebellum, 109 ng), A5 (cerebral cortex, 99 ng), A6 (frontal lobe, 89 ng), A7 (hippocampus, 118 ng), A8 (medulla oblongata, 124 ng), B1 (occipital lobe, 207 ng), B2 (putamen, 202 ng), B3 (substantia nigra, 132 ng), B4 (temporal lobe, 117 ng), B5 (thalamus, 125 ng), B6 (subthalamic nucleus, 127 ng), B7 (spinal cord, 169 ng), C1 (heart, 410 ng), C2 (aorta, 106 ng), C3 (skeletal muscle, 210 ng), C4 (colon, 177 ng), C5 (bladder, 100 ng), C6 (uterus, 114 ng), C7 (prostate, 190 ng), C8 (stomach, 293 ng), D1 (testis, 213 ng), D2 (ovary, 205 ng), D3 (pancreas, 514 ng), D4 (pituitary gland, 331 ng), D5 (adrenal gland, 349 ng), D6 (thyroid gland, 169 ng), D7 (salivary gland, 393 ng), D8 (mammary gland, 93 ng), E1 (kidney, 213 ng), E2 (liver, 429 ng), E3 (small intestine, 275 ng), E4 (spleen, 175 ng), E5 (thymus, 209 ng), E6 (peripheral blood lymphocytes, 94 ng), E7 (lymph node, 164 ng), E8 (bone marrow, 147 ng), F1 (appendix, 152 ng), F2 (lung, 243 ng), F3 (trachea, 235 ng), F4 (placenta, 361 ng), G1 (fetal brain, 137 ng), G2 (fetal heart, 135 ng), G3 (fetal kidney, 210 ng), G4 (fetal liver, 243 ng), G5 (fetal spleen, 149 ng), G6 (fetal thymus, 104 ng), and G7 (fetal lung, 245 ng). The filter was exposed for 1 week. (C) Phosphorylation of Vav-3 in Jurkat cells. Cells were stimulated for the indicated periods of time, lysed, and immunoprecipitated with antibodies specific for Vav-3. Immunocomplexes were then subjected to anti-phosphotyrosine immunoblot analysis. The mobility of Vav-3 and the immunoglobulin G heavy chain is indicated by arrows. (D) Phosphorylation of Vav-3 proteins in COS-1 cells. Transfected cells were made quiescent and then stimulated for the indicated periods of time with EGF. Proteins were then immunoprecipitated with anti-GFP antibodies and immunoblotted with anti-PTyr antibodies (upper panels). The filter was then blotted with anti-GFP (middle panel) and, after being stripped with SDS at 50°C for 30 min, incubated with anti-Shc antibodies (lower panel). The position of the EGF-R, Vav-3, and Shc isoforms is indicated by arrows. Lanes 1 to 3 contain immunoprecipitated EGFP–Vav-3 (Δ1–144) (ΔN). Lane 4 contains immunoprecipitated EGFP–Vav-3 (Δ1–144+Δ607–847) (ΔN+ΔC). (C and D) The mobility of the coelectrophoresed molecular weight markers is indicated on the right of each panel. IP, immunoprecipitation; WB, Western blot.
To verify whether Vav-3 participates in signal transduction pathways, we first studied the phosphorylation state of the endogenous Vav-3 present in Jurkat cells, a T-cell line that expresses the three known members of the Vav family (19). As shown in Fig. 2C, Vav-3 displays low levels of tyrosine phosphorylation in nonstimulated cells but becomes rapidly phosphorylated after cross-linking of the T-cell receptor with anti-CD3 antibodies. Maximal levels of phosphorylation were detected after 2 min of stimulation and decreased gradually at 5 and 10 min (Fig. 2C). The peak of phosphorylation of Vav-3 matched those of Vav and Vav-2 in the same cell type (19).
We next evaluated the behavior of Vav-3 during the signaling of the EGF-R present in COS-1 cells. Since immunoprecipitation experiments indicated that this cell line contains only Vav-2 (19), we expressed ectopically an enhanced green fluorescent protein (EGFP) fusion protein containing residues 144 to 847 of Vav-3. This protein contains all but one (Y141) of the tyrosine residues present in Vav-3 (Fig. 1B). As shown in Fig. 2D, quiescent COS-1 cells responded to the addition of EGF with rapid tyrosine phosphorylation of EGFP–Vav-3 (Δ1–144). This phosphorylation was due to an increased number of phosphorylated residues, because immunoblot analysis confirmed that the protein levels of this protein did not change during stimulation (Fig. 2D, middle panel). EGF-stimulation of COS-1 cells resulted also in the physical association of Vav-3 with the autophosphorylated EGF-R and with two phosphoproteins of 48 and 52 kDa (Fig. 2D, upper panel). Immunoblot analysis demonstrated that these proteins corresponded to the two small isoforms of the endogenous Shc protein present in COS-1 cells (Fig. 2D, lower panel). Phosphorylation of Vav-3 and its physical association with both the EGF-R and Shc were dependent on an intact SH3-SH2-SH3 region, because a truncated version of Vav-3 (Δ1–144+Δ607–847) lacking the C-terminal region was inactive in all these responses (Fig. 2D, lane 4). Immunoblot analysis showed that this truncated protein was expressed at even higher levels that the larger construct (Fig. 2D, middle panel, lane 4). These results indicate that Vav-3 participates in signal transduction processes activated by receptors with either intrinsic (EGF-R) or associated (TCR) protein tyrosine kinase activity.
Vav-3 acts biochemically as a GEF for Rho GTPases.
We investigated next the biochemical specificity of Vav-3 towards members of the Rho-Rac family of GTP hydrolases. To this end, we first generated a baculovirus capable of expressing a polyhistidine-tagged version of human Vav-3 (residues 144 to 847) after infection of Sf9 cells. According to previous results with Vav-2 (23), we expected that this protein would catalyze the exchange of nucleotides in a phosphorylation-independent manner. After infection of Sf9 cells, the induced protein was purified by chromatography onto a nickel resin, a method that allows the efficient purification of Vav-3 (Δ1–144) free of other protein contaminants (Fig. 3A). Next, we purified several representative members of the Rho family as GST fusion proteins to be used as substrates by using a standard bacterial expression system (Fig. 3B). After purification, the activity of these proteins was demonstrated by testing their ability to hydrolyze [α-32P]GTP into [α-32P]GDP (Fig. 3C).
FIG. 3.
(A) Coomassie staining of a aliquots of purified Vav-3 (Δ1–144) from Sf9 cells. (B) Aliquots of the purified GST-GTPases from E. coli. (C) GTPase activity of purified Rho proteins estimated as described in Materials and Methods. The mobility of GTP and GDP molecules is indicated. (D) Exchange activity of Vav-3 (Δ1–144) with [3H]GDP release assays. [3H]GDP-loaded GTPases (15 pmol) were incubated with unlabeled GTP either in the absence (black columns) or in the presence (gray columns) of purified Vav-3 (Δ1–144) protein (30 pmol) (left panel). After 45 min at room temperature, aliquots from each incubation were taken in duplicates, and the exchange obtained under each experimental condition was determined by a filter immobilization assay. A similar incubation was performed for 30 min with lysates from either mock (black columns)- or Dbl (light columns)-infected Sf9 cells (right panel). (E) Exchange activity of Vav-3 (Δ1–144) by [35S]GTP-γS incorporation assays. The indicated GDP-loaded GTPases (4 pmol) were incubated with [35S]GTP-γS in the absence (black columns) or presence (gray columns) of purified Vav-3 (Δ1–144) (1.8 pmol). Exchange values were determined as in panel A. In panels D and E, values represent the mean and standard deviation (SD) of three independent determinations each performed in duplicate.
The enzyme activity of Vav-3 (Δ1–144) was then determined by measuring its ability to induce the exchange of nucleotides on representative members of the Rho family. Using [3H]GDP-releasing assays, we found that saturating concentrations of Vav-3 could promote strong nucleotide exchange on RhoA and RhoG and, to a lesser extent, on Rac-1 (Fig. 3D, left panel). In contrast, Vav-3 (Δ1–144) was inactive on the Cdc42 GTPase (Fig. 3D, left panel). To verify that this GTPase was active in this assay, the same preparations of Cdc42 were subjected to exchange reactions in the presence of insect cell lysates containing the human Dbl oncoprotein. As expected (33), Dbl elicited a strong exchange activity on RhoA, RhoG, and Cdc42 (Fig. 3D, right panel), confirming that the lack of activity of Vav-3 (Δ1–144) towards Cdc42 was not due to the use of an inactive GTPase. To further demonstrate that the activity of Vav-3 (Δ1–144) represents a bona fide exchange reaction, we also analyzed the nucleotide exchange induced by substoichiometric amounts of Vav-3 (Δ1–144) by using [35S]GTP-γS incorporation assays. As shown in Fig. 3E, Vav-3 (Δ1–144) was highly active on RhoA and RhoG and showed much lower activity on Rac-1. Again, no detectable activity was observed when Vav-3 (Δ1–144) was incubated with GDP-loaded Cdc42 (Fig. 3E). These results identify Vav-3 as a GEF with substrate specificity toward RhoG, RhoA and, to a lesser extent, Rac-1.
Next, we compared the activities of Vav-3 (Δ1–144) and wild-type Vav-3. To this end, 6xHis–Vav-3 was purified from Sf9 cells (Fig. 4A) and subjected to GDP/GTP exchange assays with RhoA as substrate. Unlike Vav (Δ1–144), it was found that wild-type Vav-3 did not promote exchange activity on RhoA even after long incubation times (Fig. 4B). The phosphorylation of Vav-3 by Lck stimulated its latent exchange activity, leading to the efficient turnover of nucleotides on RhoA (Fig. 4B). In contrast, the activity of Vav-3 (Δ1–144) was similar before and after treatment with Lck (Fig. 4B). Further experiments indicated that phosphorylated Vav-3 was also active on RhoG (19) and, when at high concentrations, on Rac-1 (Fig. 4C). However, like Vav-3 (Δ1–144), wild-type Vav-3 was inactive on Cdc42 (Fig. 4C). Immunoblot analysis indicated that Vav-3 and Vav (Δ1–144) were phosphorylated at similar levels in Sf9 cells (Fig. 4D) and after incubation with Lck (19), indicating that the deregulated activity of this truncated protein was not due to its hyperphosphorylation on tyrosine residues. These results indicate that wild type Vav-3 is a phosphorylation-dependent GEF for RhoA, RhoG and, to a lower extent, Rac-1. Deletion of the N-terminal residues generates a truncated protein with identical substrate specificity but with phosphorylation-independent activity.
FIG. 4.
(A) Coomassie staining of aliquots of wild-type Vav-3 purified from Sf9 cells. (B) GDP-loaded RhoA (4 pmol) was incubated with [35S]GTP-γS in the presence of 2.0 pmol of either nonphosphorylated Vav-3 (□), nonphosphorylated Vav-3 (Δ1–144) (○), phosphorylated Vav-3 (■), or phosphorylated Vav-3 (Δ1–144) (●). As a negative control, RhoA was also incubated with GST-Lck (◊). Exchange values were determined at the indicated time points as indicated in Fig. 3D. (C) Exchange activity of Vav-3 on Rho/Rac family members by using either 3:1 (left panel) or 1:3 (right panel) molar ratios of GEF-GTPase. The indicated GDP-loaded GTPases were incubated for 45 min at room temperature with [35S]GTP-γS in the presence of GST-Lck (open boxes), nonphosphorylated Vav-3 (gray boxes), or Lck-phosphorylated Vav-3 (closed boxes). Exchange activities were determined as in Fig. 3D. (D) Phosphorylation levels of Vav-3 proteins. Purified wild-type Vav-3 (WT) and Vav-3 (Δ1–144) were separated electrophoretically and subjected to immunoblot analysis with anti-hexahistidine (upper panel) and anti-phosphotyrosine (lower panel) antibodies.
Vav-3 associates with its GTPase substrates in a nucleotide-dependent manner.
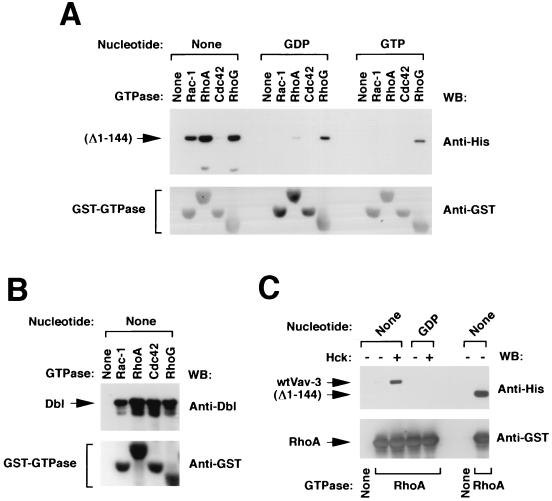
To further define the functional relationship between Vav-3 and Rho family GTPases, we tested the ability of Vav-3 (Δ1–144) and wild-type Vav-3 to interact physically with Rho family proteins in vitro. In good agreement with our previous exchange reactions, we found that Vav-3 (Δ1–144) could interact with both RhoA and RhoG and, more weakly, with Rac-1 (Fig. 5A). The binding affinity of Vav-3 (Δ1–144) was maximal when these GTPases were in the nucleotide-free state (Fig. 5A) and was independent on tyrosine phosphorylation (19). No significant association was observed between Vav-3 (Δ1–144) and Cdc42 (Fig. 5A). However, this GTPase did bind to Dbl (Fig. 5B), confirming again that Cdc42 is functional in our assays. Wild-type Vav-3 showed also binding to nucleotide-free RhoA, but only after being phosphorylated by tyrosine kinases (Fig. 5C). These results indicate that Vav-3 proteins appear to catalyze the exchange of nucleotides in members of the Rho family via the stabilization of the nucleotide-free forms of its substrates.
FIG. 5.
(A) Binding of Vav-3 (Δ1–144) to Rho family proteins. Vav-3 (Δ1–144) was incubated with the indicated GTPases as described in Materials and Methods. Bound proteins were separated by SDS-PAGE and subjected to immunoblot analysis by using either anti-hexahistidine residues (upper panel) or anti-GST (lower panel) antibodies. The type of guanosine nucleotide bound to the GTPases used in this study is indicated at the top. (B) Binding of Dbl to Rho proteins. Total cellular lysates from baculovirus-infected Sf9 cells were incubated with the indicated GTPases and processed as indicated above. Blots were incubated with anti-Dbl antibodies and then with anti-GST antibodies to visualize the appropriate proteins. (C) Binding of wild type Vav-3 to RhoA. Wild-type Vav-3 and Vav-3 (Δ1–144) were incubated in kinase buffer alone (−) or with Hck (+) for 15 min and then incubated with either beads alone (None) or coated with GST-RhoA in the indicated nucleotide states. Binding of Vav-3 proteins to RhoA in each experimental condition was determined by immunoblot analysis with anti-hexahistidine antibodies. The mobility of Vav-3 and Vav-3 (Δ1–144) is indicated by arrows. The nucleotide state of each RhoA protein is indicated at the top. In panels A to C the blotting antibody is indicated on the right.
Vav-3 is not transforming but induces morphological change in NIH 3T3 cells.
Expression of truncated forms of either Vav or Vav-2 in NIH 3T3 cells leads to high levels of morphological transformation (2). To verify whether this is a property common to all Vav family members, we tested the ability of Vav-3 to induce cellular transformation in this rodent cell line. To this end, we generated a collection of mammalian expression vectors containing either the full-length or truncated versions of Vav-3 (Table 1). To our surprise, all constructs containing VAV-3 cDNAs failed to induce transformation either alone or in combination with vectors encoding LckY505F, a protein tyrosine kinase that activates the GDP-GTP exchange activity of wild-type Vav-3 (Table 1). In contrast, the oncogenic versions of vav and vav-2 yielded high levels of morphological transformation in the same transfection experiments (Table 1). As expected, cotransfection of either the vav or the vav-2 proto-oncogene with LckY505F generated high levels of morphological transformation (Table 1). Thus, these results indicate that Vav-3 is not transforming when overexpressed in rodent fibroblasts.
TABLE 1.
Transforming activity of Vav family proteinsa
| Plasmid | Description | Amount (ng/10-cm plate) | Foci |
|---|---|---|---|
| pNM81 | Wild-type Vav-3 | 1,000 | 0 |
| pNM79 | Vav-3 (Δ1–144) | 1,000 | 0 |
| pNM80 | Vav-3 (Δ1–184) | 1,000 | 0 |
| AatII-pNM81 | Linearized plasmid | 1,000 | 0 |
| AatII-pNM80 | Linearized plasmid | 1,000 | 0 |
| pJC11 | Wild-type Vav | 500 | 0 |
| pKES12 | Vav (Δ1–187) | 100 | TMTCb |
| pXRB138 | Wild-type Vav-2 | 500 | 0 |
| pXRB141 | Vav-2 (Δ1–183) | 300 | TMTC |
| pXEM-LckY505F | LckY505F | 800 | 0 |
| pNM81 + pXEM-LckY505F | Wild-type Vav-3+ LckY505F | 500 + 800 | 0 |
| pJC11 + pXEM-LckY505F | Wild-type Vav+ LckY505F | 500 + 800 | 203 |
| pXRB139 + pXEM-LckY505F | Wild-type Vav-2+ LckY505F | 500 + 800 | 1,344 |
Transforming activity was determined using focus formation assays. For plasmid linearization, DNAs were digested with AatII (present in the plasmid backbone), extracted with phenol/chloroform, precipitated thrice with ethanol, and resuspended in sterile water.
TMTC, too many to count.
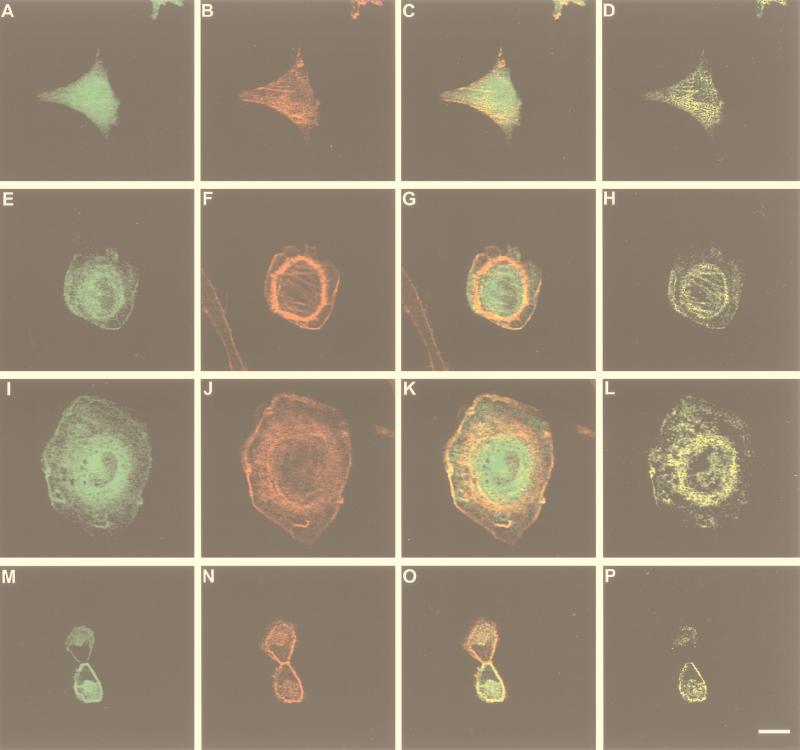
The lack of transforming activity of Vav-3 led us to measure the biological activity of this protein by using an alternative experimental approach. Since Rho proteins have an implication in the organization of the actin cytoskeleton (11, 28), we studied next whether Vav-3 had some role in regulating the distribution of F-actin in rodent fibroblasts. To this end, we transfected transiently NIH 3T3 cells with mammalian expression vectors encoding EGFP–Vav-3 (Δ1–144) and EGFP–Vav-3 (Δ1–144+Δ607–847). This last protein contains only the Ac, DH, PH, and ZF domains of Vav-3. According to our previous biochemical assays (Fig. 3 and 5), we expected that these proteins could induce constitutive signals due to their phosphorylation-independent GDP-GTP exchange activity. Twenty four hours post-transfection, cells were fixed, incubated with rhodamine-labeled phalloidin to visualize the actin network, and subjected to confocal microscopy analysis. We found that the expression of either of these two Vav-3 proteins led to extensive lamellipodia, membrane ruffling, and the formation of an actomyosin ring (Fig. 6F and J). In addition, thin bundles of stress fibers were observed in the central region of Vav-3-expressing cells (Fig. 6F and J). We also detected a tendency of Vav-3-expressing cells to round up and loose adherence to the substrate (19), a phenotype that was only observed upon the overexpression of RhoA subfamily members (23). The changes elicited by Vav-3 were similar to those induced by Vav and Vav-2 oncoproteins (23) but not to other GEFs such as the Rho-specific Lbc oncoprotein (34), the Ral-specific Rsc (8), and the Ras-specific RasGRF (1, 19). Expression of the EGFP protein alone did not induce any detectable morphological change in NIH 3T3 cells (Fig. 6B). A similar morphological phenotype was observed when these two Vav-3 proteins were expressed in the absence of EGFP tags (19). Untagged wild-type Vav-3 induced also morphological change, but only when coexpressed with activated protein tyrosine kinases such as LckY505F (19).
FIG. 6.
Biological effect and subcellular distribution of Vav-3 in transiently transfected NIH 3T3 cells. Cells expressing EGFP (A to D), EGFP–Vav-3 (Δ144–606) (E to H), or EGFP–Vav-3 (Δ1–144+Δ607–847) (I to P) were fixed, stained with phalloidin-rhodamine, and analyzed by confocal microscopy by using filters for fluorescein (A, E, I, and M) or rhodamine (B, F, J, and N). Panels C, G, K, and O show the overlap of both images with the areas of coexpression shown in yellow. Panels D, H, L, and P show only the areas of overlap between Vav-3 and F-actin. The scale bar in panel P is equivalent to 20 μm.
The presence of the EGFP tag in these proteins allowed us to study their subcellular distributions in NIH 3T3 cells. EGFP–Vav-3 (Δ1–144) and EGFP–Vav-3 (Δ1–144+Δ607–847) appeared localized mostly around, although excluded from, the nucleus of non mitotic NIH 3T3 cells (Fig. 6E and I). Vav-3 signals were also detected along the stress fibers, actomyosin ring, membrane ruffles, and plasma membrane (Fig. 6E and I). Vav-3 colocalized with F-actin in all these structures (Fig. 6G, H, K, and L). In general, this colocalization was not total, because the codistribution of Vav and actin was limited to a narrow region of overlap in those cellular structures, with regions typically showing a region of Vav-3–actin colocalization and an adjacent region displaying F-actin alone (Fig. 6G and K). In mitotic cells, Vav-3 was found again excluded from the nucleus and located both in the cell body and in the cleavage region of the daughter cells (Fig. 6M). Vav-3 partially colocalized with F-actin in these cells (Fig. 6O and P). For instance, in the cleavage furrow, a region of F-actin alone was surrounded by an adjacent layer of Vav-3–F-actin coexpression and a third layer containing only Vav-3 (Fig. 6O and P and data not shown). These results underscore the connection between Vav-3 and the actin cytoskeleton. Moreover, the similar distribution of these two Vav-3 proteins suggests that the central Vav-3 region (residues 144 to 606) contains all of the structural information required for biological activity and proper subcellular localization.
Expression of Vav-3 induces abnormal cell division.
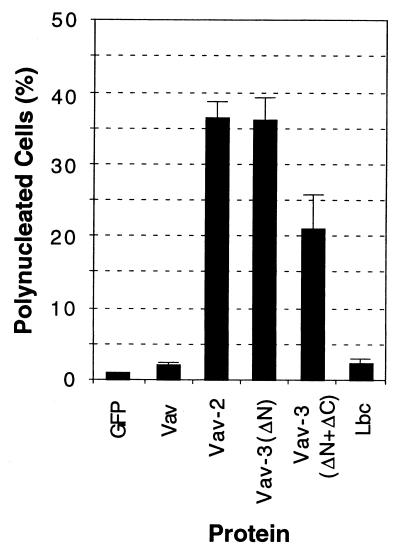
During the transfection experiments, it became apparent that a significant proportion of VAV-3 expressing cells were binucleated. To analyze this in more detail, we performed transient transfections with EGFP–Vav-3 proteins, Vav, Vav-2, and Lbc. Transfected cells were cultured for 28 h, fixed, stained with Hoechst 33258 to visualize the nuclei, and counted. As shown in Fig. 7, EGFP–Vav-3 (Δ1–144) and, to a lesser extent, EGFP–Vav-3 (Δ1–144+Δ607–847), both induced a significant increase in the number of binucleated cells present in the cultures. This phenotype was independent of the EGFP tag, since similar results were obtained with untagged Vav-3 proteins (19). The Vav-2 oncoprotein induced a similar phenotype that EGFP-Vav-3 (Δ1–144) (Fig. 7). In contrast, expression of the vav and LBC oncogenes resulted in marginal numbers of polynucleated cells when compared with cells expressing EGFP protein alone (Fig. 7). No multinucleated cells were detected after expression of the wild-type versions of Vav-3, Vav, or Vav-2 (19). Thus, the expression of deregulated Vav-3 proteins induces cytokinesis defects that lead to the generation of polynucleated cells.
FIG. 7.
Vav-3 induces polynucleated cells. NIH 3T3 cells were transiently transfected with constructs encoding EGFP–Vav-3 (Δ1–144) (Vav-ΔN) and EGFP–Vav-3 (Δ1–144+Δ607–847) (Vav-3ΔN+ΔC) (2 μg each) or with the pEGFP-C1 vector alone (0.25 μg) or in combination with vav-, vav-2-, and LBC-containing plasmids (3 μg each). After 28 h, cells were fixed and then stained with Hoechst 33258 (Sigma), and the number of nuclei present in EGFP-containing cells was scored. Values represent the mean and SD of three independent determinations in which more than 180 cells were counted.
The DH and ZF domains, but not the PH domain, are required for Vav-3 biological activity.
The previous results indicated that the DH-PH-ZF domains of Vav-3 contain all the functional information required for morphological change, alterations in cytokinesis, and subcellular localization. To investigate the specific task of these domains in Vav-3 function, we investigated the effect generated by point mutations in the Vav-3 DH, PH, and ZF regions in the biological activity of this protein in vivo. To this end, we used expression plasmids encoding EGFP fused to Vav-3 (Δ1–144+Δ607–847) proteins containing inactive DH (L211Q), PH (W493L), or ZF (C527S) regions. These mutations were previously shown to inactivate the function of the Vav DH and ZF (5, 6) and the PH of other Rho and Rac GEFs (29). All of these plasmids were capable of expressing the expected proteins, as determined by immunoblot analysis with anti-EGFP antibodies of total cellular lysates obtained from transfected COS-1 cells (Fig. 8A). When these plasmids were introduced into NIH 3T3 cells, we found that the Vav-3 protein lacking a functional PH region was as active as the wild-type version, as assessed by its ability to induce both F-actin reorganization and morphological change in the transfected cells (Fig. 8B, panels A and C). In contrast, the Vav-3 proteins harboring defective DH or ZF regions were totally inactive in both processes (Fig. 8B, panels B and D). Similar results were obtained when these mutants were constructed in the EGFP-Vav (Δ1–144) background (19). To demonstrate that these proteins were active in other biological processes, we determined their participation in the signal transduction pathway activated by EGF. EGFP–Vav-3 (Δ1–144+L211Q) and EGFP–Vav-3 (Δ1–144+C527S) proteins became phosphorylated after treatment of quiescent cells with EGF and associated with both the EGF-R and Shc (Fig. 8C). This result indicates that the DH and ZF mutations specifically impair the activation of GTPase pathways in vivo but do not affect the overall structure of these proteins.
FIG. 8.
(A) Expression of the EGFP–Vav-3 proteins in COS-1 cells. Total cellular lysates from COS-1 cells expressing either EGFP or the indicated EGFP–Vav-3 (Δ1–144+Δ607–847) proteins were subjected to immunoblot analysis with anti-EGFP antibodies. The mobility of Vav-3 proteins is indicated by an arrow. The position of nonchimeric EGFP is marked by an arrowhead. ΔN refers to the Vav-3 (Δ1–144+Δ607–847) protein. (B) Biological activity of Vav-3 proteins. Cells expressing either EGFP–Vav-3 (Δ1–144+Δ607–847) (panel A), EGFP–Vav-3 (Δ1–144+Δ607–847+L211Q) (panel B), EGFP–Vav-3 (Δ1–144+Δ607–847+W293L) (panel C), or EGFP-Vav-3 (Δ1–144+Δ607–847+C527S) (panel D) were fixed, stained with phalloidin-rhodamine, and analyzed by confocal microscopy by using filters for fluorescein (EGFP, green) and rhodamine (actin, red) as indicated in Fig. 6. The panels show the overlap of both images, with the areas of coexpression shown in yellow. In each case, a representative cell of each transfection is shown. Similar results were obtained in three independent transfections. The scale bar in panel D is equivalent to 20 μm. (C) Participation of Vav-3 mutant proteins in the EGF-activated pathway. EGFP–Vav-3 (Δ1–144) (ΔN), Vav-3 (Δ1–144+L211Q) (L211Q), and Vav-3 (Δ1–144+C527S) (C527S) were immunoprecipitated from quiescent (−) and EGF-stimulated (+) COS-1 cells and then tested for phosphorylation (upper panel) and binding to the EGF-R (upper panel) and Shc (lower panel) by using appropriate antibodies. The position of the EGF-R and Vav proteins is indicated by arrows. The position of Shc isoforms is indicated by arrowheads. The blotting antibody is indicated on the right.
Vav-3 requires both the DH and the ZF domains for the activation of GTPases.
While the absence of activity of Vav-3 L211Q mutants was consistent with the catalytic role of this structural domain, the experiments described above could not distinguish whether the inactivity of the Vav-3 C527S mutants was due to effects in cis or trans. To get further insight into the role of the PH and ZF domains to the function of Vav-3, we decided to investigate the relative contribution of those domains to the biochemical activity of Vav-3 using in vitro assays. To this end, we generated baculoviruses capable of inducing the expression of several mutants of Vav-3 in insect cells. These proteins included the Vav-3 Ac+DH domain, the Vav-3 Ac+DH+PH, and two N-terminally truncated Vav-3 proteins (Δ1–144) harboring mutations in either the PH or the ZF domain (Fig. 9A). After purification from Sf9 cells (Fig. 9B), these proteins were tested for exchange activity by using GST-RhoA as substrate. As shown in Fig. 9C, a Vav-3 protein with a defective PH domain was capable of inducing an exchange of nucleotides with kinetics identical to those induced by the Vav-3 (Δ1–144) protein. This indicates that the PH region of Vav-3 does not affect the catalytic properties of this protein either positively or negatively. By contrast, a Vav-3 protein containing a point mutation (C527S) in the ZF region was defective in such activity (Fig. 9C, left panel). Similar low activities were obtained when the Vav-3 Ac+DH domain or the Vav-3 Ac+DH+PH regions were used, indicating that the lack of activity of C527S is not due to an artifactual effect of the point mutation in the Vav-3 structure (Fig. 9C, left panel).
FIG. 9.
(A) Schematic representation of the proteins used in this study. The polyhistidine tag is depicted as an open box (not in scale). (B) Vav-3 proteins purified from Sf9 cells. (C) (Left panel). Exchange activity of Vav-3 proteins. Equal molar amounts of each Vav-3 protein were incubated with GDP-loaded RhoA in the presence of free [35S]GTP-γS. After 45 min, aliquots of each reaction were taken in duplicate and the exchange determined as indicated in Fig. 3. Similar results were obtained in two independent experiments, each performed in duplicate. (Right panel) Kinetics of GDP-GTP exchange of RhoA either alone (○) or with Vav-3 (Δ1–144) (□) and Vav-3 (Δ1–144+W293L) (▵). (D) Binding of the indicated Vav-3 proteins to RhoA (upper panel) and immunoblot analysis of the same filter with anti-GST antibodies (lower panel). Experiments were conducted as indicated in Fig. 5. The mobility of Vav-3 (DN) and GST-RhoA is indicated by arrows. The blotting antibody is indicated on the right.
We also evaluated the ability of those Vav-3 proteins to bind to RhoA in vitro. As shown in Fig. 9D (upper panel), both the wild-type and the Vav-3 PH mutant protein bound to nucleotide-free RhoA at comparable levels. Loading of RhoA with GDP reduced the binding of both Vav-3 proteins, confirming the specificity of those interactions (Fig. 9D, upper panel). Instead, the Vav-3 Ac+DH domains, the Ac+DH+PH, and Vav-3 (Δ1–144+C527S) showed no physical association with RhoA (Fig. 9D, upper panel). Immunoblot analysis with anti-GST antibodies confirmed that comparable amounts of GST-RhoA were present in these incubations (Fig. 9D, lower panel). Based on these results, we conclude that the DH and the ZF region of Vav-3 act coordinately to assure optimal binding to, and exchange of nucleotides of, Rho proteins. In contrast, the PH region appears to be dispensable for both the biological and biochemical activities of Vav-3 protein.
The ZF region contributes to the interaction of Vav-3 with GTPases.
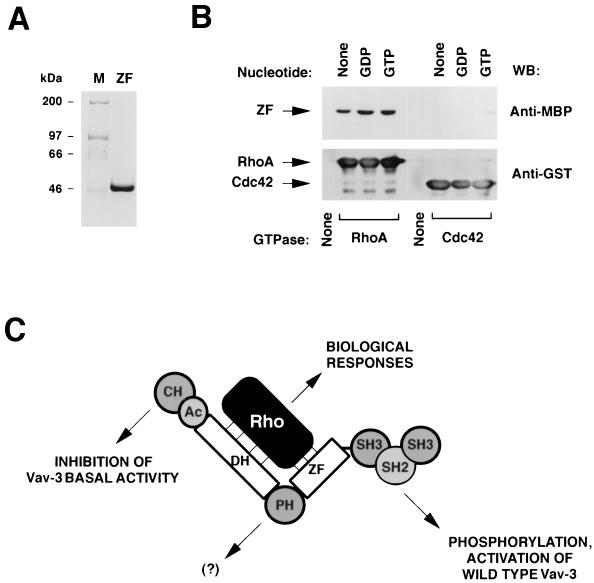
To further address the role of the Vav-3 ZF in the regulation of the activity of this GEF, we investigated whether this isolated domain could interact with RhoA. To this end, we purified a maltose-binding protein (MBP) containing the Vav-3 ZF region (residues 506 to 584) from E. coli (Fig. 10A). The purified protein was then incubated with GST-GTPases, and its binding to them was measured by anti-MBP immunoblot analysis. As shown in Fig. 10B (upper panel), the MBP-Vav-3ZF associated physically with RhoA in the absence of nucleotides and, with slightly better affinity, with its GDP- and GTP-bound forms. In contrast, the MBP–Vav-3 ZF protein did not associate with Cdc42 or with the glutathione beads used to immobilize the GTPases (Fig. 10B, upper panel), confirming the specificity of the Vav-3 ZF–RhoA interaction. Reblotting of the same filter with anti-GST antibodies demonstrated that RhoA and Cdc42 proteins were present at comparable levels in these incubations (Fig. 10B, lower panel). Additional binding experiments confirmed that this ZF region could also associate with Rac-1 and RhoG (19). Binding of the Vav-3 ZF to RhoA was also detected when a polyhistidine-tagged version of this region was used in these experiments (19). These results suggest that the Vav-3 ZF region affects the biochemical activity of Vav-3 by, at least in part, contributing to its interaction with the GTPase substrates.
FIG. 10.
(A) Coomassie blue staining of an aliquot of the MBP–Vav-3 ZF protein purified from E. coli cells. (B) The MBP–Vav-3 ZF protein was incubated with the indicated GST-GTPases as described in Materials and Methods. As negative control, MBP–Vav-3 ZF was incubated with beads alone (None). Bound proteins were separated by SDS-PAGE and subjected to immunoblot analysis by using either anti-MBP (upper panel) or anti-GST (lower panel) antibodies. The GTPases used in the study and their nucleotide state are indicated at the bottom and top, respectively. The mobility of the Vav-3 ZF (upper panel) and GTPases (lower panel) is indicated by arrows. The blotting antibody is indicated on the right. (C) Our proposed model for Vav-3 activity.
DISCUSSION
In this study, we describe the identification and functional characterization of VAV-3, a new member of the VAV gene family of guanosine nucleotide exchange factors. The product of the human VAV-3 gene is a 98-kDa protein which shares a high degree of homology with the products of the VAV and VAV-2 genes. Experiments conducted with both endogenous and ectopically expressed Vav-3 demonstrate its participation in the signal transduction processes. Moreover, the analysis of the enzyme specificity of Vav-3 indicates that it catalyzes preferentially the exchange of guanosine nucleotides on RhoA and RhoG and, to a lower extent, on the Rac-1 GTPase. Vav-3 binds also physically to those GTPases when they are in the nucleotide-free state, a result previously shown for most Rho and Ras GEFs (12). By analogy to those proteins, we can infer that Vav-3 will catalyze GDP-GTP exchange by stabilizing the nucleotide-free state of its substrates.
The role of Vav-3 as an activator of Rho GTPases is further substantiated by transient transfections in rodent fibroblasts. We have shown that truncated Vav-3 proteins (Δ1–144 and Δ1–144+Δ607–847) are capable of inducing a rapid reorganization of the actin cytoskeleton, leading to membrane ruffling, lamellipodia, and the formation of thin bundles of stress fibers in NIH 3T3 cells. In COS-1 cells, the expression of Vav-3 leads also to the formation of extensive membrane ruffling, a phenotype similar to that observed upon overexpression of the other Vav family proteins, Rac-1Q61L, and RhoGQ61L (19). Given the similarity of the morphologies induced by Rac-1 and RhoG, it is not possible at present to discriminate whether such lamellipodia and membrane ruffling formation represents the activation of Rac-1, RhoG, or both. However, since Vav-3 works only at substoichiometric concentrations on RhoA and RhoG, we favor the idea that those responses are mediated mainly through the stimulation of RhoG.
The expression of truncated versions of Vav-3 induces also marked alterations in the process of cell division, leading to the generation of binucleated cells in about 35% of all Vav-3-expressing cells. This phenotype is shared with Vav-2, but it is not observed upon expression of Vav and other Rho GEFs such as Lbc. The rapid induction of this phenotype (28 h) suggests that this response is the consequence of the direct unregulated activity of Vav-3 and Vav-2 rather than to an epistatic event occurring upon the long-term constitutive activation of the Vav-2/Vav-3 pathways. Interestingly, the examination of Vav-3-expressing cells shows at least two different types of alterations. In the majority of cases, the two new nuclei separate effectively into the daughter cells but the septum fails to form, leading to lack of partition of the cells. In a minority of cases, it has been observed that one of the nuclei of binucleated cells undergoes mitosis, while the second one remains resting. Division of those cells leads to the generation of a new cell inheriting one of the new nucleus, while the second daughter cell inherits the other newly formed nucleus and the undivided one (19). Thus, multinucleation of Vav-3-expressing cells appears to be derived from alterations in both the cytokinetic machinery and from the asynchronous DNA synthesis in the nuclei of already binucleated cells. A recent report has shown that a RhoA downstream target, citron, can induce polynucleation in HeLa cells due to abnormal cytokinesis (18). It is tempting to speculate that the biological effects of Vav-3 and Vav-2 could be mediated, at least in part, by activation of this RhoA-regulated serine and threonine kinase.
Despite the similar activity of Vav-3 and Vav-2 both in vitro and in vivo, we were unable to detect any transforming activity of this new gene in standard focus formation assays. The explanation for this result is not straightforward to us. Due to the lack of transformation of the initial VAV-3 cDNA obtained by PCR (see Materials and Methods), we recloned the full length VAV-3 cDNA using standard screening procedures to avoid the possibility of a recurrent mutation in all our PCR clones. However, the library-derived clones showed the same primary structure and still lacked transforming activity, even when truncations that hyperactivated the oncogenicity of Vav and Vav-2 (23) were generated in the VAV-3 cDNA. VAV-3 was also inactive when linearized plasmids were used in the transfections, a method that traditionally enhances the transforming activity of oncogene-harboring plasmids by up to 200-fold, including those for vav (19). Species differences can also be ruled out, because the human VAV gene is as transforming as its rodent counterpart (5). It is possible, therefore, that the lack of VAV-3 oncogenic activity could derive from a different behavior of this gene in vivo, such as distinct subcellular localization or to lower levels of activity of Vav-3 in vivo. More work will be necessary to fully understand the lack of transformation potential of this new Vav family member.
The study of Vav-3 has also allowed us to gain a better understanding on the intramolecular interactions that regulate the activity of this GDP-GTP exchange factor. Thus, we have shown that the biochemical activity of wild-type Vav-3 is strictly dependent on tyrosine phosphorylation. Since these assays are done with purified proteins, this result indicates that this posttranslational modification induces an intramolecular effect that activates the latent biochemical activity of Vav. In this regard, we have shown that this effect is mediated, at least in part, by an increase in the binding avidity of Vav-3 toward its substrates. However, since Vav-3 activity is always associated with both the physical interaction to the GTPases and catalysis of GDP-GTP exchange, it is impossible at this point to determine whether phosphorylation plays only a role in the former process or whether, in addition, it also participates in regulating the catalytic activity of Vav-3 towards the bound substrate. Mutagenesis experiments capable of dissociating these two functional points in the Vav-3–Rho relationship will help to answer this important question. By using an N-terminally truncated Vav-3 protein (Δ1–144), we have also demonstrated that these Vav-3 mutant proteins are constitutively active due to the lack of dependency of tyrosine phosphorylation for the activation of Rho proteins. This is not due to higher levels of tyrosine phosphorylation, because this truncated protein displays comparable levels of phosphorylation that the wild type protein when purified from Sf9 cells and after phosphorylation by Hck. In good agreement with these in vitro observations, we have shown that Vav-3 proteins lacking the N terminus can induce effective morphological change and cell multinucleation even in the absence of the C-terminal SH3-SH2-SH3 domains, a region essential for the tyrosine phosphorylation of Vav-3 and for its interaction with protein tyrosine kinases and other cytoplasmic phosphoproteins. This constitutive activation appears to be another intramolecular effect mediated by the missing CH region, since it can be induced in vitro by using highly purified preparations of Vav-3. Taken together, these results are consistent with the idea that the main function of the Vav-3 SH3-SH2-SH3 is to mediate the phosphorylation of the wild-type protein, thereby eliminating the inhibitory function of the CH region on other structural domains of Vav-3 (Fig. 10C).
Since the DH, PH, and ZF domains appear to drive all the biological and biochemical activities of Vav-3, we decided to investigate the individual contributions of the DH, PH, and ZF regions for both the enzyme and biological activities of Vav-3. Not surprisingly, we found that a point mutation affecting the DH region (L211Q) inhibits the biological activity of Vav-3. This finding is in good agreement with our previous results with Vav showing that the mutation in the same residue (L213Q) disrupts its ability to promote JNK-1 activation and Rac-1 GDP-GTP exchange in vivo (6, 7). This inactivation is not due to a major alteration in the folding of Vav-3 because this mutant protein works well in other functions such as the EGF-dependent binding to the EGF-R and Shc.
Further analysis of the DH-PH-ZF region has shown that the integrity of the Vav-3 ZF domain is also essential for the function of this protein. Indeed, we have shown that a point mutation affecting a conserved cysteine residue (C527S) of the Vav-3 ZF totally eliminates the activity of this protein in vivo and in vitro. Several independent observations indicate that this inhibition reflects a truly functional role of the Vav-3 ZF. First, we have shown that this C527S mutant does participate in signal transduction pathways, becoming phosphorylated on tyrosine residues after treatment of quiescent COS-1 cells with EGF. Second, this mutant protein can also interact physically with EGF-R and the p48/p52Shc isoforms in a ligand-dependent manner. These two properties are similar to the ones seen for the intact Vav-3 protein when expressed in the same cell background. Third, we have demonstrated that different Vav-3 deletion mutants lacking the entire ZF region are also inactive in vitro. Thus, the inactivation of the Vav-3 C527S mutant protein cannot be attributed to artifactual causes, such as the formation of new intramolecular covalent bonds between cysteines of the disrupted ZF and cysteine residues located elsewhere in the molecule.
Although we do not know as yet how the ZF works with the DH domain to promote binding and catalysis of the GTPases, we believe that our in vitro results are consistent with a dual functional role for this domain. On one hand, our observations demonstrating that the isolated Vav-3 ZF can interact physically with RhoA in vitro suggest that this domain contributes to the Vav-3–GTPase interaction by establishing points of contact with specific regions of Rho proteins (Fig. 10C). These regions may not be conserved in all GTPases because the Vav-3 ZF binds to the Vav-3 substrates but not to Cdc42. On the other hand, the fact that the Vav-3 ZF region has a completely different specificity for the guanosine nucleotide state of the GTPases than do wild-type Vav-3 and Vav-3 (Δ1–144) is not consistent with this region being the main determinant of the stable interaction of Vav-3 with its substrates. Accordingly, we propose a model in which the ZF also makes contributions to the overall structure of the DH-PH-ZF cassette which are essential for the biochemical action of the catalytic DH domain (Fig. 10C). Such function may be analogous to the contribution of the PH domain of Trio to the catalytic efficiency of its DH domain (17). Studies aimed at solving the crystal structure of Vav-3 will allow determination of the specific contributions of the ZF region to the effector functions of Vav-3.
Finally, our experiments have shown that a mutation that completely disrupts the structure of the PH domain (W493L) has no detectable consequences for biochemical and biological activity of Vav-3. In agreement with these results, recent experiments indicate that the Vav-3 W493L mutant protein is also active in the activation of another Rac-1/RhoG downstream element, JNK (19). To date, only Vav-3 and the Lbc exchange factor (21) appear to be completely independent of their respective PH domains for the induction of their in vivo effects. The PH domain of Dbl is also dispensable for the catalytic activity in vitro, but it is essential for its effects in vivo, presumably by facilitating the interaction of this GEF with the cytoskeleton (35). Recent structural studies have shown that the N-terminal region of the PH domain of Sos-1 folds into the C terminus of the DH region, leading to the inhibition of the basal exchange activity of this catalytic domain (24, 32). This inhibitory effect appears to be eliminated by the binding of phospholipids to the Sos-1 PH region (20). These results indicate that some PH domains may influence the activity of Rho and Rac GEFs via intramolecular interactions and through the binding to intracellular second messengers. Since our experiments have not included mutations in the N-terminal region of the Vav-3 PH domain, we cannot rule out at the present time whether the Vav-3 PH domain has regulatory functions similar to those described for Sos-1. However, two observations argue indirectly against such possibility. First, the N-terminal regions of the PH domains of Sos-1 and Vav-2 have very low amino acid sequence similarity and differ significantly in length. This suggests that the interactions that take place between the Sos-1 DH and PH domains probably cannot occur in the case of Vav-3. Second, previous nuclear magnetic resonance and crystal structure studies have shown that the residue targeted in our mutagenesis experiments (W493) establishes hydrogen bonds and water-mediated interactions with the N-terminal β1 region of the same PH region (9, 16). The W493L mutation created in Vav-3 is expected therefore to cause major conformational changes in the entire N-terminal region of its PH domain. In this regard, our kinetic analysis showing that the Vav-3 (W493L) protein shows catalytic activity identical to that of the wild type version further suggests that this domain does not play either positive or negative roles in the GDP-GTP exchange activity of this protein. In any case, the definitive answer to the putative negative regulatory role of the Vav-3 PH region will require extensive mutagenesis of the residues located in the N-terminal region of this structural domain. Another interesting question that needs to be addressed is whether the model reported here for the Vav-3 PH region is conserved in the other members of the family. This is an important consideration because the PH regions are, along with the most proximal SH3 domain, one of the less-conserved regions among these proteins. It will be interesting therefore to compare the functional properties of the Vav and Vav-2 PH regions and to study whether the lack of transforming activity of Vav-3 is due to structural or functional differences among Vav family members in this structural domain. Future studies with point mutants of Vav and Vav-2 and with chimeric Vav-3/Vav and Vav-3/Vav-2 proteins will help to solve these two pending questions.
In summary, we have presented a comprehensive functional characterization of the VAV-3 gene product both biochemically and biologically and have provided new data regarding mechanistic aspects of Vav-3 GDP-GTP exchange regulation. It is likely that the isolation of VAV-3 will not represent the last step on the characterization of the Vav family. Although the presence of additional Vav-related proteins in mammals remains to be further addressed, the presence of Vav-related proteins in C. elegans has been already demonstrated (30). It is therefore plausible that, as for many other signaling molecules involved in developmental pathways, Vav proteins will be present in other animal models such as flies and nonmammalian vertebrates. The isolation of these genes will provide in the future important tools to study genetically the signal transduction pathway activated by these proteins and to understand the different mechanisms by which tyrosine kinase receptors couple their activation with the stimulation of Rho/Rac pathways.
ACKNOWLEDGMENTS
We would like to thank Todd Miller (Department of Physiology, State University of New York at Stony Brook) for his gift of purified Hck.
This work was made possible by a research grant from the U.S. National Cancer Institute to X.R.B. (CA7373501), who is a Sinsheimer Scholar for Cancer Research.
REFERENCES
- 1.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 2.Bustelo X R. The VAV family of signal transduction molecules. Crit Rev Oncog. 1996;7:65–88. doi: 10.1615/critrevoncog.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- 3.Bustelo X R, Suen K L, Michael W M, Dreyfuss G, Barbacid M. Association of the vav proto-oncogene product with poly(rC)-specific RNA-binding proteins. Mol Cell Biol. 1995;15:1324–1332. doi: 10.1128/mcb.15.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerione R A, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 5.Coppola J, Bryant S, Koda T, Conway D, Barbacid M. Mechanism of activation of the vav protooncogene. Cell Growth Differ. 1991;2:95–105. [PubMed] [Google Scholar]
- 6.Crespo P, Bustelo X R, Aaronson D S, Coso O A, Lopez-Barahona M, Barbacid M, Gutkind J S. Rac-1-dependent stimulation of the JNK/SAPK signaling pathway by Vav. Oncogene. 1996;13:455–460. [PubMed] [Google Scholar]
- 7.Crespo P, Schuebel K E, Ostrom A A, Gutkind J S, Bustelo X R. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 8.D’Adamo D R, Novick S, Kahn J M, Leonardi P, Pellicer A. rsc: a novel oncogene with structural and functional homology with the gene family of exchange factors for Ral. Oncogene. 1997;14:1295–1305. doi: 10.1038/sj.onc.1200950. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson K M, Lemmon M A, Schlessinger J, Sigler P B. Crystal structure at 2.2 Å resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 10.Fischer K D, Kong Y Y, Nishina H, Tedford K, Marengere L E, Kozieradzki I, Sasaki T, Starr M, Chan G, Gardener S, Nghiem M P, Bouchard D, Barbacid M, Bernstein A, Penninger J M. Vav is a regulator of cytoskeletal reorganization mediated by the T-cell receptor. Curr Biol. 1998;8:554–562. doi: 10.1016/s0960-9822(98)70224-6. [DOI] [PubMed] [Google Scholar]
- 11.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 12.Hart M, Powers S. Ras-Cdc25 and Rho-Dbl binding assays: complex formation in vitro. Methods Enzymol. 1995;255:129–135. doi: 10.1016/s0076-6879(95)55016-x. [DOI] [PubMed] [Google Scholar]
- 13.Henske E P, Short M P, Jozwiak S, Bovey C M, Ramlakhan S, Haines J L, Kwiatkowski D J. Identification of VAV2 on 9q34 and its exclusion as the tuberous sclerosis gene TSC1. Ann Hum Genet. 1995;59:25–37. doi: 10.1111/j.1469-1809.1995.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 14.Holsinger L J, Graef I A, Swat W, Chi T, Bautista D M, Davidson L, Lewis R S, Alt F W, Crabtree G R. Defects in actin-cap formation in Vav-deficient mice implicate an actin requirement for lymphocyte signal transduction. Curr Biol. 1998;8:563–572. doi: 10.1016/s0960-9822(98)70225-8. [DOI] [PubMed] [Google Scholar]
- 15.Kong Y Y, Fischer K D, Bachmann M F, Mariathasan S, Kozieradzki I, Nghiem M P, Bouchard D, Bernstein A, Ohashi P S, Penninger J M. Vav regulates peptide-specific apoptosis in thymocytes. J Exp Med. 1998;188:2099–2111. doi: 10.1084/jem.188.11.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemmon M A, Ferguson K M, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak E T, Meadows R P, Schkeryantz J M, Janowick D A, Harlan J E, Harris E A, Staunton D E, Fesik S W. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- 18.Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 19.Movilla, M., and X. R. Bustelo. Unpublished results.
- 20.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 21.Olson M F, Sterpetti P, Nagata K, Toksoz D, Hall A. Distinct roles for DH and PH domains in the Lbc oncogene. Oncogene. 1997;15:2827–2831. doi: 10.1038/sj.onc.1201594. [DOI] [PubMed] [Google Scholar]
- 22.Schuebel K E, Bustelo X R, Nielsen D A, Song B J, Barbacid M, Goldman D, Lee I J. Isolation and characterization of murine vav2, a member of the vav family of proto-oncogenes. Oncogene. 1996;13:363–371. [PubMed] [Google Scholar]
- 23.Schuebel K E, Movilla N, Rosa J L, Bustelo X R. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 1998;17:6608–6621. doi: 10.1093/emboj/17.22.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soisson S M, Nimnual A S, Uy M, Bar-Sagi D, Kuriyan J. Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell. 1998;95:259–268. doi: 10.1016/s0092-8674(00)81756-0. [DOI] [PubMed] [Google Scholar]
- 25.Tarakhovsky A, Turner M, Schaal S, Mee P J, Duddy L P, Rajewsky K, Tybulewicz V L. Defective antigen receptor-mediated proliferation of B and T cells in the absence of Vav. Nature. 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 26.Teramoto H, Salem P, Robbins K C, Bustelo X R, Gutkind J S. Tyrosine phosphorylation of the vav proto-oncogene product links FcɛRI to the Rac1-JNK pathway. J Biol Chem. 1997;272:10751–10755. doi: 10.1074/jbc.272.16.10751. [DOI] [PubMed] [Google Scholar]
- 27.Turner M, Mee P J, Walters A E, Quinn M E, Mellor A L, Zamoyska R, Tybulewicz V L. A requirement for the Rho-family GTP exchange factor Vav in positive and negative selection of thymocytes. Immunity. 1997;7:451–460. doi: 10.1016/s1074-7613(00)80367-2. [DOI] [PubMed] [Google Scholar]
- 28.Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 29.Whitehead I, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- 30.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R, Alt F W, Davidson L, Orkin S H, Swat W. Defective signalling through the T- and B-cell antigen receptors in lymphoid cells lacking the vav proto-oncogene. Nature. 1995;374:470–473. doi: 10.1038/374470a0. [DOI] [PubMed] [Google Scholar]
- 32.Zheng J, Chen R H, Corblan-Garcia S, Cahill S M, Bar-Sagi D, Cowburn D. The solution structure of the pleckstrin homology domain of human SOS1. A possible structural role for the sequential association of diffuse B cell lymphoma and pleckstrin homology domains. J Biol Chem. 1997;272:30340–30344. doi: 10.1074/jbc.272.48.30340. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y, Hart M J, Cerione R A. Guanine nucleotide exchange catalyzed by dbl oncogene product. Methods Enzymol. 1995;256:77–84. doi: 10.1016/0076-6879(95)56011-4. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Y, Olson M F, Hall A, Cerione R A, Toksoz D. Direct involvement of the small GTP-binding protein Rho in lbc oncogene function. J Biol Chem. 1995;270:9031–9034. doi: 10.1074/jbc.270.16.9031. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, Zangrilli D, Cerione R A, Eva A. The pleckstrin homology domain mediates transformation by oncogenic dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]