Abstract
Introduction
Prognostic biomarkers and novel therapeutic approaches have been slow to emerge in the treatment of head and neck squamous cell carcinoma (HNSCC). In this study, an HNSCC patient cohort is created and performance of putative prognostic biomarkers investigated in a population-validated setting. The overall goal is to develop a novel way to combine biomarker analyses with population-level clinical data on HNSCC patients and thus to improve the carryover of biomarkers into clinical practice.
Materials and methods
To avoid selection biases in retrospective study design, all HNSCC patients were identified and corresponding clinical data were collected from the Southwest Finland geographical area. A particular emphasis was laid on avoiding potential biases in sample selection for immunohistochemical staining analyses. Staining results were evaluated for potential prognostic resolution.
Results
After comprehensive evaluation, the patient cohort was found to be representative of the background population in terms of clinical characteristics such as patient age and TNM stage distribution. A negligible drop-out of 1.3% (6/476) was observed during the first follow-up year. By immunohistochemical analysis, the role of previously implicated HNSCC biomarkers (p53, EGFR, p16, CIP2A, Oct4, MET, and NDFIP1) was investigated.
Discussion
Our exceptionally representative patient material supports the use of population validation to improve the applicability of results to real-life situations. The failure of the putative prognostic biomarkers emphasizes the need for controlling bias in retrospective studies, especially in the heterogenous tumor environment of HNSCC. The resolution of simple prognostic examination is unlikely to be sufficient to identify biomarkers for clinical practice of HNSCC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00405-021-06650-7.
Keywords: HNSCC, Population-validation, Prognosis, Biomarkers
Introduction
Head and neck squamous cell carcinomas (HNSCC) compose a behaviorally diverse field of cancers united by their common localization to the head and neck regions [1, 2]. Clinical problems such as early metastatic behavior and serial recurrences due to field cancerization are frequently encountered. Especially intriguing phenomena are the unexpected aggressiveness of small tumors and, in a favorable way, the surprising treatment response of some large tumors. The current therapy stratification of HNSCC is based on the overall state of the patient and clinical observations about the tumor [3, 4].
The site and extent of the tumor do not, however, have a decisive effect on patient prognosis [5, 6]. Attempts to explain clinical diversity of HNSCC by genetical and molecular analysis have thus far proven unsuccessful, leaving the determination of patient prognosis uncertain. A multitude of biomarkers has been suggested, with little success in translating findings to clinical practice [7]. The enthusiastically awaited inclusion of p16/HPV in the staging of oropharyngeal HNSCC has not met all expectations [8]. Some reasons to lack of success may be found in the uneven inclusion of patients to especially small retrospective patient cohorts, bias in inclusion criteria, and poor definition of clinical questions to be tackled [7, 9].
Northern European healthcare system offers an intriguing prospect for unbiased patient sampling, because cancer patients in need of oncological treatment are referred to regional tertiary centers independent of insurance or socioeconomic status of the patients. In addition, based on EUROCARE-5 data, the results of head and neck cancer treatment in Nordic countries and especially in Finland are remarkably superior to other regions in Europe [10].
In this study, a population-based cohort of all new HNSCC patients treated between 2005 and 2010 in Southwest Finland region, covering one sixth of Finland’s population, was collected. This cohort of HNSCC patients corresponds to the real-life patient succession treated at our institute. Tumor samples were retrieved, sampling bias analyzed, and a panel of immunohistochemical biomarkers analyzed.
Thus, we re-evaluated the real-life capability of a panel of immunohistochemical biomarkers to prognosticate patient 5-year overall survival (OS), when identified clinical prognostic variables are taken into account. All of these biomarkers have previously been reported to function as prognostic markers in HNSCC. The biomarkers included loss of tumor suppressor p53 expression associated with p53 mutations, that are the most often encountered mutations in HNSCC associated with metastatic behavior and radio resistance [11]. EGFR overexpression has been the focus of intense study in HNSCC, as EGFR inhibitors are available [12]. p16 has a clinical application as oropharyngeal cancer prognosticator [13, 14]. CIP2A is an mTOR and MYC-associated inhibitor of tumor suppressor protein phosphatase 2A [15]. MET and Oct4 are associated with a stemness phenotype [16, 17] and NDFIP1 was listed in the top three unfavorable HNSCC biomarker in Protein Atlas database [18].
Materials and methods
Primary HNSCC patient cohort
The HNSCC patient cohort was formed by identifying and including all patients treated for new HNSCC in Turku University Hospital (TUH) region in 2005–2010. Tumors were staged according to TNM criteria applicable at the time of diagnosis. Treatment protocols were decided in a multidisciplinary Tumor Board for head and neck cancer. OS was defined from end-of-treatment to end-of-follow-up or death. Age-standardized OS were calculated using International Cancer Survival Standards for weighting.
The usage of human tissue samples was approved by the Finnish national authority for medicolegal affairs (V/39706/2019), regional ethics committee of University of Turku (51/1803/2017) and Auria biobank scientific board (AB19-6863). Patient formalin-fixed, paraffin-embedded (FFPE) samples were acquired from pathology archives through Auria Biobank. Final TMA blocks of duplicate 0.6 mm cores were made in TMA Grand Master (3D Histech) according to annotations on scanned HE slides. Samples of normal liver were included in each block for orientation.
Immunohistochemistry (IHC)
FFPE blocks were cut into 6 um sections. CIP2A IHC was carried out after protocol optimization in Ventana BenchMark XT staining automate (Ventana Medical Systems, Inc) using mouse monoclonal anti-CIP2A antibody (1:25, 2G10-3B5, sc-80659, SantaCruz). p16, p53, and EGFR IHC were carried out in Ventana in clinical pathology laboratory. Oct4 IHC was performed as previously described with anti-Oct4 antibody sc-5279 (1:200 mouse monoclonal, Santa Cruz Biotechnology) [17]. NDFIP1 immunohistochemistry was carried out with anti-NDFIP1 antibody HPA009682 (1:1000 rabbit polyclonal, Atlas Antibodies). MET stainings were performed as previously reported [16].
Immunohistochemical stainings were analyzed by two authors independently, and differences were discussed until consensus was reached. p53 staining was analyzed using the established 3-tier system. Cytoplasmic/membraneous EGFR, MET, and CIP2A expression were scored semiquantitatively based on intensity of the staining on a scale of 1–3. Nuclear Oct4 was scored positive, when a subpopulation of strong positive nuclei was present. p16 immunostaining was regarded positive, when at least 70% of cells demonstrated strong nuclear and cytoplasmic staining intensity. Nuclear NDFIP1 staining was regarded positive when strong, uniform nuclear staining was present. For all statistical analyses, dichotomous cutoffs were applied.
Statistical analysis
Patient data and staining results were entered into SPSS 24 software (SPSS, IBM). For Cox hazards models, the proportionality of hazards was testing using log-minus-log plotting and plotting Schoenfeld residuals against survival time, when appropriate. For all multivariable analysis, stepwise approach with backward LR method was applied, if not otherwise indicated, with p value limits for inclusion and exclusion at 0.05 and 0.10, respectively. For Kaplan–Meier survival estimation, significance was analyzed using log-rank method. To test prognostic potential of biomarkers, their combinations and their interactions, Cox regression was used by first entering the prognostic clinicopathological variables and in another block the biomarker combinations. p values of less than 0.05 were considered significant.
Results
Southwest Finland regional cohort corresponds with Nordic EUROCARE-5 population
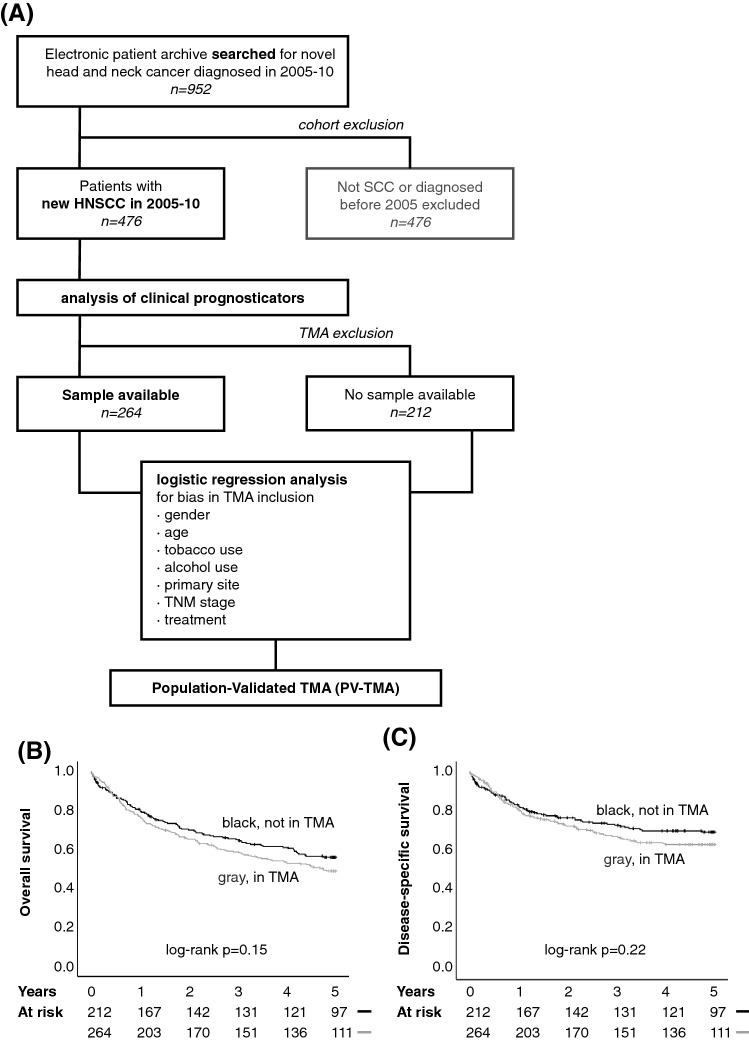
An electronic database screen was made to include all HNSCC patients treated in Southwest Finland region during years 2005–2010 (Fig. 1a). Altogether 952 patients’ records were accessed. After initial evaluation, the final cohort included 476 patients diagnosed and treated for new HNSCC tumor (Table 1). Two-hundred and thirty-two patients (49%) were diagnosed with early stage HNSCC, 164 patients (34%) had nodal metastasis at presentation, and five patients (1.1%) were diagnosed with distant metastasis. Only 1.3% (6/476) of patients were lost during the first year of follow-up.
Fig. 1.
a Principle of the population-validated TMA. First, a background population was screened for comprehensive inclusion of all patients treated for HNSCC in Southwest Finland during the time period of 2005–2010. This background population was used to assess clinical prognostic factors. All available samples were included in TMA. The representativeness of the TMA was analyzed with logistic regression analysis for multiple variables. After the representativeness was confirmed, the TMA is considered a population-validated TMA (PV-TMA). b Overall survival, and c disease-specific survival of the patients included in PV-TMA was slightly lower than of patients not included in PV-TMA. In multivariable analysis, there was no difference in survival
Table 1.
Clinicopathological variables of the patient cohort. Univariate (left panels) and multivariable (right panels) survival analysis of HNSCC cohort
| Total | Survival effect | Total | Survival effect | |||||
|---|---|---|---|---|---|---|---|---|
| n | % | HR (95% CI) | p | n | % | HR (95% CI) | p | |
| Gender | ||||||||
| Male | 325 | 68 | 1.03 (0.78–1.37) | 0.84 | 325 | 68 | not included | – |
| Female | 151 | 32 | 1 | – | 151 | 32 | ||
| Age at diagnosis | ||||||||
| < 65 | 236 | 50 | 1.02 (1.01–1.03) /year | < 0.001 | 236 | 50 | 1.04 (1.02–1.05) / yr | < 0.001 |
| > 65 | 240 | 50 | – | 240 | 50 | – | ||
| Smoking status | ||||||||
| Current smoker | 202 | 42 | 1.31 (0.99–1.74) | 0.063 | 202 | 42 | 1.19 (0.80–1.78) | 0.39 |
| Former smoker | 73 | 15 | 0.93 (0.61–1.41) | 0.73 | 73 | 15 | 0.89 (0.57–1.39) | 0.61 |
| Non-smoker | 201 | 42 | 1 | – | 201 | 42 | 1 | – |
| Alcohol consumption | ||||||||
| Yes | 139 | 29 | 0.69 (0.52–0.91) | 0.008 | 139 | 29 | 1.45 (1.02–2.06) | 0.037 |
| No | 337 | 71 | 1 | – | 337 | 71 | 1 | – |
| Primary tumor site | ||||||||
| Oral cavity | 226 | 47 | 1 | – | 226 | 47 | 1 | – |
| Oropharynx | 89 | 19 | 0.86 (0.59–1.26) | 0.86 | 89 | 19 | 0.69 (0.46–1.05) | 0.086 |
| Larynx | 105 | 22 | 1.24 (0.90–1.73) | 0.19 | 105 | 22 | 1.03 (0.71–1.49) | 0.88 |
| Hypopharynx | 20 | 4 | 2.65 (1.51–4.63) | 0.001 | 20 | 4 | 1.61 (0.88–2.96) | 0.13 |
| Other | 36 | 8 | 1.08 (0.65–1.78) | 0.77 | 36 | 8 | 1.15 (0.67–1.97) | 0.61 |
| T class | ||||||||
| T0-2 | 311 | 65 | 0.32 (0.24–0.41) | < 0.001 | 311 | 65 | 0.27 (0.17–0.44) | < 0.001 |
| T3-4 | 165 | 35 | 1 | – | 165 | 35 | 1 | – |
| N class | ||||||||
| N0 | 312 | 66 | 0.67 (0.51–0.88) | 0.003 | 312 | 66 | 0.54 (0.36–0.78) | 0.001 |
| N + | 164 | 34 | 1 | – | 164 | 34 | 1 | – |
| Stage | ||||||||
| 0–II | 232 | 49 | 0.46 (0.35–0.60) | < 0.001 | 232 | 49 | 1.41 (0.77–2.58) | 0.26 |
| III–IV | 244 | 51 | 1 | – | 244 | 51 | 1 | – |
| Recidive in 5 years | ||||||||
| Yes | 137 | 29 | 5.34 (3.92–7.27) | < 0.001 | 137 | 29 | not included | – |
| No | 289 | 61 | 1 | – | 289 | 61 | – | – |
| No curative treatment | 49 | 10 | 30.07 (20.06–45.08) | < 0.001 | 49 | 10 | – | – |
| Living at 5 years | ||||||||
| Yes | 253 | 53 | – | – | 253 | 53 | not included | – |
| No, died of HNSCC | 150 | 32 | – | – | 150 | 32 | – | – |
| No, died of other cause | 73 | 15 | – | – | 73 | 15 | – | – |
| Surgical treatment | ||||||||
| No surgery | 141 | 30 | 1 | – | 141 | 30 | 1 | – |
| Local operation | 282 | 59 | 0.59 (0.45–0.76) | < 0.001 | 282 | 59 | 0.74 (0.55–0.98) | 0.038 |
| Neck dissection | 173 | 36 | 0.86 (0.66–1.14) | 0.29 | 173 | 36 | 0.73 (0.53–1.00) | 0.049 |
| Treatment type | ||||||||
| Surgery only | 172 | 36 | 1 | – | 172 | 36 | 1 | – |
| RT only | 51 | 11 | 2.71 (1.78–4.12) | < 0.001 | 51 | 11 | 2.12 (1.26–3.57) | 0.005 |
| CRT only | 75 | 16 | 1.57 (1.05–2.36) | 0.028 | 75 | 16 | 0.81 (0.46–1.43) | 0.47 |
| RT + surgery | 46 | 10 | 1.97 (1.24–3.11) | 0.004 | 46 | 10 | 1.27 (0.77–2.07) | 0.40 |
| CRT + surgery | 116 | 24 | 1.16 (0.80–1.70) | 0.43 | 116 | 24 | 0.74 (0.44–1.23) | 0.25 |
| No treatment | 15 | 3 | 15.75 (8.82–28.17) | < 0.001 | 15 | 3 | 5.80 (2.96–11.38) | < 0.001 |
Results from Cox proportional hazards model regression. In multivariable modeling, treatment effects were analyzed by entering the clinical prognostic variables (separated by a horizontal line.)
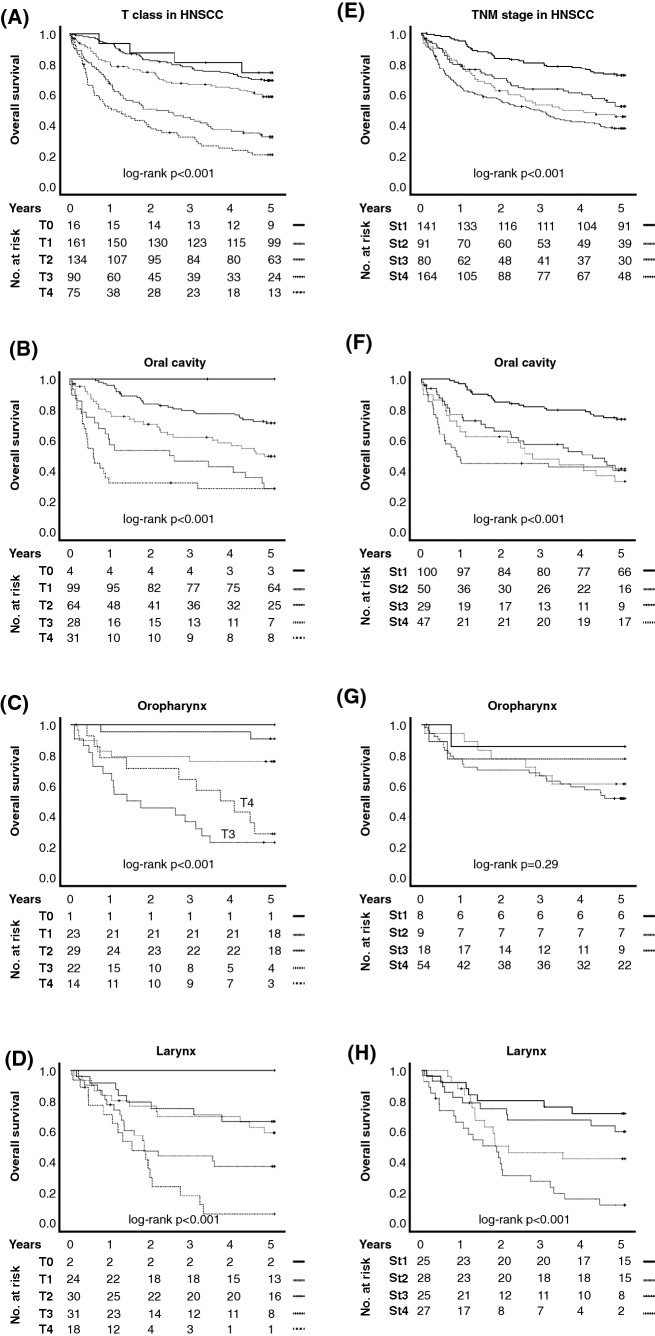
OS was influenced by previously acknowledged risk factors: patient age, advanced T class, nodal positivity, and alcohol use (Table 1). Interestingly, T class proved to be a superior prognosticator than TNM stage in all major subsites of HNSCC (Fig. 2a–h and Table 1). However, inadequate prognostic resolution between T1 and T2 as well as T3 and T4, respectively, was noted, especially in laryngeal cancer (Fig. 2d). Thus, for multivariable analysis, T class was divided dichotomously in T0-2 vs T3-4, providing a highly significant prognostic stratification (Table 1; HR 0.27, 95% CI 0.17–0.44, p < 0.001). While the primary tumor site had no decisive impact on patient OS, inclusion of primary tumor site in the following multivariable models was deemed appropriate.
Fig. 2.
Overall survival was highly affected by tumor T class in both a HNSCC overall and the three main subsites, b oral cavity, c oropharynx, and d larynx. e–h TNM stage was an inferior prognosticator as compared to tumor T class in HNSCC overall and the three main subsites, especially in oropharynx, where the prognostic resolution was virtually non-existent. In oral cancer, TNM stage offered minimal prognostic resolution between stage 2 and stage 3
One-hundred and seventy-two patients (36%) were given only surgical treatment (Table 1). Ninety-seven and 191 patients were treated with radiotherapy or chemoradiotherapy, respectively. Fifteen patients were offered no treatment. In a multivariable model fitting age at diagnosis, primary tumor site, T class, nodal status and alcohol consumption, no treatment type proved clearly superior with regard to OS impact, although surgical treatment was associated with a statistically significant improvement in prognosis.
Survival data were compared to results of EUROCARE-5 study (summarized in Table 2). In comparison to general Finnish, Northern European, and whole European average head and neck cancer patient survival, the observed survival rates in Southwest Finland region were higher especially in elderly patients and hypopharyngeal cancer.
Table 2.
Survival rates in TUH HNSCC patient cohort compared with Eurocare-5 data for Northern Europe
| Oral cavity | Larynx | Oropharynx | Hypopharynx | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HNSCC | Eurocare | HNSCC | Eurocare | HNSCC | Eurocare | HNSCC | Eurocare | HNSCC | Eurocare | |
| OAS 5-years | 56% | – | 48% | – | 70% | – | 30% | – | 53% | – |
| DSS 5-years | 71% | – | 69% | – | 65% | – | 40% | – | 68% | – |
| ICSS 5-year observed survival rate | 58% | 43% | 50% | 52% | 57% | 41% | 36% | 17% | 53% | 41% |
| ICSS 5-year relative survival rate | – | 50% | – | 62% | – | 46% | – | 19% | – | 46% |
Eurocare-5 data accessed at https://w3.iss.it/site/EU5Results/
Construction of representative population-validated tissue microarray (PV-TMA)
Altogether 264 patients’ tumor samples were available for TMA (Fig. 1a). A thorough analysis of TMA construction biases was carried out (Table 3). Compared to clinical data of the background population, HNSCC patients treated in Southwest Finland region in 2005–2010, the established PV-TMA was shown to be representative in terms of age distribution, tobacco and alcohol exposure and especially TNM class, whereas uneven site distribution was observed.
Table 3.
Univariate (left panels) and multivariable (right panels) analysis of TMA inclusion bias
| Total | TMA patients | TMA inclusion | TMA patients | TMA inclusion | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | OR (95% CI) | p | n | % | OR (95% CI) | p | |
| Gender | ||||||||||
| Male | 325 | 68 | 164 | 62 | 0.52 (0.35–0.78) | 0.001 | 164 | 62 | 0.56 (0.36–0.88) | 0.011 |
| Female | 151 | 32 | 100 | 38 | 1 | – | 100 | 38 | 1 | – |
| Age at diagnosis | ||||||||||
| < 65 | 236 | 50 | 137 | 52 | 1.23 (0.86–1.77) | 0.26 | 137 | 52 | Not included | |
| > 65 | 240 | 50 | 127 | 48 | 1 | – | 127 | 48 | ||
| Smoker | ||||||||||
| > 20 pack years | 225 | 47 | 115 | 44 | 0.72 (0.50–1.03) | 0.071 | 115 | 44 | NS | |
| < 20 pack years | 251 | 53 | 149 | 56 | 1 | – | 149 | 56 | ||
| Alcohol consumption | ||||||||||
| Yes | 139 | 29 | 78 | 30 | 0.96 (0.65–1.43) | 0.85 | 78 | 30 | Not included | |
| No | 337 | 71 | 186 | 70 | 1 | – | 186 | 70 | ||
| Primary tumor site | ||||||||||
| Oral cavity | 226 | 47 | 137 | 52 | 1 | – | 137 | 52 | 1 | – |
| Oropharynx | 89 | 19 | 64 | 24 | 1.66 (0.98–2.84) | 0.062 | 64 | 24 | 2.34 (1.21–4.54) | 0.012 |
| Larynx | 105 | 22 | 35 | 13 | 0.33 (0.20–0.53) | < 0.001 | 35 | 13 | 0.68 (0.37–1.24) | 0.21 |
| Hypopharynx | 20 | 4 | 11 | 4 | 0.79 (0.32–1.99) | 0.62 | 11 | 4 | 1.64 (0.58–4.63) | 0.35 |
| Other | 36 | 8 | 17 | 6 | 0.58 (0.29–1.18) | 0.13 | 17 | 6 | 0.93 (0.42–2.03) | 0.85 |
| T class | ||||||||||
| T0-2 | 311 | 65 | 173 | 66 | 1.02 (0.70–1.49) | 0.92 | 173 | 66 | Not included | |
| T3-4 | 165 | 35 | 91 | 34 | 1 | – | 91 | 34 | ||
| N class | ||||||||||
| N0 | 312 | 66 | 157 | 59 | 0.54 (0.37–0.80) | 0.002 | 157 | 59 | NS | |
| N + | 164 | 34 | 107 | 41 | 1 | – | 107 | 41 | ||
| Stage | ||||||||||
| 0–II | 232 | 49 | 118 | 45 | 0.70 (0.48–1.00) | 0.049 | 118 | 45 | NS | |
| III–IV | 244 | 51 | 146 | 55 | 1 | – | 146 | 55 | ||
| Recidive in 5 yrs | ||||||||||
| Yes | 137 | 29 | 84 | 32 | 1.40 (0.93–2.12) | 0.11 | 84 | 32 | Not Included | |
| No | 289 | 61 | 152 | 58 | 1 | – | 152 | 58 | ||
| No curative treatment | 49 | 10 | 28 | 11 | 1.20 (0.65–2.21) | 0.56 | 28 | 11 | ||
| Living at 5 years | ||||||||||
| Yes | 253 | 53 | 131 | 50 | 0.72 (0.48–1.08) | 0.11 | 131 | 50 | Not Included | |
| No, died of HNSCC | 150 | 32 | 90 | 34 | 1 | – | 90 | 34 | ||
| No, died of other cause | 73 | 15 | 43 | 16 | 0.96 (0.54–1.69) | 0.88 | 43 | 16 | ||
| Surgical treatment | ||||||||||
| No surgery | 141 | 30 | 0 | 1 | – | 55 | 21 | 1 | – | |
| Local operation | 282 | 59 | 174 | 66 | 1.88 (1.30–2.73) | 0.001 | 174 | 66 | 1.75 (1.05–2.94) | 0.033 |
| Neck dissection | 173 | 36 | 125 | 47 | 3.10 (2.07–4.63) | < 0.001 | 125 | 47 | 2.30 (1.49–3.56) | < 0,001 |
| Treatment type | ||||||||||
| Surgery only | 172 | 36 | 92 | 35 | 1 | – | 92 | 35 | NS | |
| RT only | 51 | 11 | 20 | 8 | 0.56 (0.30–1.06) | 0.075 | 20 | 8 | ||
| CRT only | 75 | 16 | 30 | 11 | 0.58 (0.33–1.01) | 0.052 | 30 | 11 | ||
| RT + surgery | 46 | 10 | 34 | 13 | 2.46 (1.20–5.08) | 0.015 | 34 | 13 | ||
| CRT + surgery | 116 | 24 | 82 | 31 | 2.10 (1.27–3.46) | 0.004 | 82 | 31 | ||
| no treatment | 15 | 3 | 5 | 2 | 0.44 (0.14–1.33) | 0.14 | 5 | 2 | ||
Results from logistic regression modeling
Importantly, TMA inclusion was not a significant predictor of 5-year OS or disease-specific survival in neither univariate analysis nor in multivariable survival model fitting for established clinical risk factors (Fig. 1b, c). In conclusion, the PV-TMA constructed for this work can be considered to be well representative of HNSCC patients treated in the region of Southwest Finland in 2005–2010.
Analysis of representative HNSCC patient TMA demonstrates poor performance of putative biomarkers for prognostication
Using this exceptionally representative PV-TMA material, we analyzed the prognostication capability of multiple biomarkers—p53, EGFR, p16, CIP2A, MET, Oct4, and NDFIP1—previously shown to function as prognostic markers in HNSCC (Fig. 3). The prognostic information of CIP2A and p16 reached significance in univariate analysis (Fig. 3i, o, respectively). However, regardless of the hypothesis-based selection of the candidate biomarkers and their previous association with poor prognosis in HNSCC, none of the biomarkers showed significant prognostic value in multivariable analysis using PV-TMA material (Table 4).
Fig. 3.
Representative immunohistochemical stains and prognostic trends (estimates using Kaplan–Meier method and log-rank method for significance) of the investigated biomarkers in HNSCC. a–c p53, d–f EGFR, g–i CIP2A, j-l Oct4, m–o p16, p–r NDFIP1, s–u MET
Table 4.
Prognostic performance of investigated biomarker staining intensities
| Total | 5-yr survival | Survival analysis | ||||
|---|---|---|---|---|---|---|
| n | % | alive, n | % | HR | p | |
| p53 | ||||||
| Absent | 73 | 29 | 34 | 47 | 0.91 (0.62–1.36) | 0.65 |
| wt or high | 176 | 71 | 89 | 51 | 1 | – |
| EGFR | ||||||
| Low-moderate | 190 | 78 | 93 | 49 | 1.27 (0.82–1.97) | 0.29 |
| Strong | 53 | 22 | 26 | 49 | 1 | – |
| CIP2A | ||||||
| Low-moderate | 150 | 66 | 79 | 53 | 1.20 (0.81–1.76) | 0.37 |
| High | 78 | 34 | 30 | 38 | 1 | – |
| Oct4 | ||||||
| Negative | 101 | 39 | 56 | 55 | 0.73 (0.50–1.07) | 0.11 |
| Positive | 160 | 61 | 75 | 47 | 1 | – |
| p16 | ||||||
| Negative | 181 | 81 | 81 | 45 | 0.91 (0.68–1.22) | 0.54 |
| Positive | 43 | 19 | 26 | 60 | 1 | – |
| NDFIP1 | ||||||
| Negative | 137 | 60 | 63 | 46 | 1.18 (0.81–1.73) | 0.40 |
| Positive | 90 | 40 | 46 | 51 | 1 | – |
| cMET | ||||||
| Low | 151 | 66 | 76 | 50 | 0.80 (0.55–1.15) | 0.22 |
| Moderate-high | 79 | 34 | 38 | 48 | 1 | – |
Staining distributions, survival rates, and results of multivariable survival analysis (Cox proportional hazards model controlling for age, T class, nodal status and alcohol use)
Further, the possible prognostication value of the biomarkers for oral cavity, oropharyngeal, or laryngeal cancer patients was further investigated using a multivariable model entering the above identified clinical prognosticators. None of the investigated biomarkers provided statistically significant prognostic information in the three main subsites of HNSCC (Supplementary Table 1). Furthermore, no combination or interaction of the investigated biomarkers could not provide significant prognostic potential in multivariable survival regression, when clinical prognostic variables were included in the models (data not shown).
Discussion
Our study demonstrates, that in a non-biased HNSCC patient population treated with optimal results, the putative biomarkers failed to offer significant prognostic information. In order to improve retrospective as well as future prospective studies, a population-based analysis should be mandatory to appreciate the potential biases in patient selection. Further, the recent failures of significant prospective drug trials in HNSCC [19–21] suggest that optimization of retrospective studies is an underappreciated step in discovery of biomarkers for patient treatment stratification.
This study emphasizes the need for thorough exploration of inclusion bias, since some exclusion of patients due to loss of samples and inadequate sample size is unavoidable. In our patient cohort, this is achieved by analysis of the population giving rise to the TMA cohort, the Southwest Finland HNSCC patients from 2005 to 2010. The statistical analysis reveals that our PV-TMA is an exceptionally representative and unbiased study environment for retrospective analysis of biomarkers. Population-validation approach thus improves the robustness and reliability of data analysis.
High risk of bias is present in patient inclusion to both retrospective and prospective cohorts [22, 23]. Inclusion biases include unequal recruitment of patients with different socioeconomic status or limited insurance coverage, supposedly having a poor prognosis, and on the other hand patients with small tumors with good prognosis. Moreover, variance in the given cancer treatments between different hospitals, and between individual clinicians can also be a confounding factor in the analysis of treatment outcomes. Clinical validation of our patient cohort is made possible by the referral system in Northern Europe, leading to an unbiased, institutional patient population, which serves as a representative cross-section of the regional population. Thus, this dataset represents the real-life patient succession observed in the clinic and is, in this respect, superior to recruited prospective cohorts. Furthermore, loss to follow-up is virtually non-existent due to the Nordic public health care system and electronic databases.
Particularly good head and neck cancer treatment results in Nordic countries increases the interest of this dataset [10]. Interestingly, in our regional data, the Southwest Finland patient prognosis was even better than in Finnish EUROCARE-5 data. This may be due to more wide-spread use of cisplatin radiosensitization and, most importantly, the long-standing multi-disciplinary tumor board practice, guaranteeing optimized protocols, meticulous treatment planning, and impartial response monitoring. Of special clinical interest is also the superior prognostic resolution afforded by T class in comparison to complete TNM stage. However, the observed 34% survival rate of T1-2 patients provides rationale for biomarker-based prognostication.
Particularly interesting are our results when putative biomarkers with auspicious publication history for prognostication of HNSCC were tested in PV-TMA. Importantly, we failed to recognize significant prognostic factors, when clinical prognosticators were taken into account, either in the patient material as a whole or in any major subsite. Surprisingly, no combination or interaction of biomarkers proved useful in prognostication of our patient material. More complex statistical analysis used in previous studies to create prognostic biomarker panels [24, 25] could not be applied in this study, concentrating in an unselected patient population. Despite the disappointing failure of the biomarkers, our approach highlights the value of unbiased cross-sectional regional control of patient inclusion in biomarker discovery.
Immunohistochemistry for p16 is the only clinically approved biomarker for HNSCC and is applied in oropharyngeal cancer staging. In our study, p16 was a surprisingly poor prognosticator of HNSCC patients’ OS, in contrast to earlier reports [14, 26, 27]. Whether this is attributable to better overall prognosis of HPV-negative patients or the widespread use of cisplatin radiosensitization, remains an intriguing question. The failure of recent p16 deintensification trials seems, however, to demonstrate a need for better understanding of the role of p16 in both radio- and chemoradioresistance [8, 20]. Thus, our finding cautions against p16-based deintensification with regard to current treatment guidelines in Finland.
The main strength of this study is the impartial inclusion of all HNSCC patients treated in our regional referral center. Thus, the patient cohort is representative of the real-life population encountered in the routine clinical practice, increasing the applicability of our results to clinical decision-making. Despite the crucial representativeness of our patient cohort, there are weaknesses in this study as well. The patient number remains relatively low, especially in site-specific analysis. Further, the patient numbers do not readily allow for more complex statistical approaches, such as multivariable analysis of biomarker combinations and more detailed analysis of staining cut-offs, including integration of data on subcellular localization changes.
In conclusion, we demonstrate the value of population-validation methodology for retrospective biomarker studies, and wish to emphasize the need for population level evaluation for inclusion biases. Impartial cancer patient selection, comprehensive patient registers available for researchers, and exceptionally good cancer treatment outcomes demonstrate optimal possibilities for retrospective analysis of biomarkers. Similar approach should be applied for the design of future prospective trials in molecularly diverse cancers.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr Oliver von Ahsen for his permission in using MET staining data in this paper. We thank the Auria Biobank for their kind collaboration in preparing the TMA. Dr. Eliisa Löyttyniemi from the Biostatistics Department at University of Turku is acknowledgment for her most pertinent remarks on the statistical methods of this paper.
Funding
Open access funding provided by University of Turku (UTU) including Turku University Central Hospital. This research was funded by the Finnish Medical Foundation, Sigrid Jusélius Foundation, the Finnish ORL-HNS Research Foundation, and State Research Funding.
Compliance with ethical standards
Conflict of Interest
No potential conflicts of interest were disclosed by the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chow LQM. Head and Neck Cancer. N Engl J Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Alsahafi E, Begg K, Amelio I, et al. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis. 2019;10:540. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammerman PS, Hayes DN, Grandis JR. Therapeutic insights from genomic studies of head and neck squamous cell carcinomas. Cancer Discov. 2015;5:239–244. doi: 10.1158/2159-8290.CD-14-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koch WM, Ridge JA, Forastiere A, Manola J. Comparison of clinical and pathological staging in head and neck squamous cell carcinoma: results from Intergroup Study ECOG 4393/RTOG 9614. Arch Otolaryngol Head Neck Surg. 2009;135:851–858. doi: 10.1001/archoto.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma SJ, Wagner S, Reder HSF, et al. The 8th edition JCC/UICC TNM staging for p16-positive oropharyngeal carcinoma: is there space for improvement? Eu rArch Oto-rhino-laryngol. 2018;275:3087–3091. doi: 10.1007/s00405-018-5156-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim KY, McShane LM, Conley BA. Designing biomarker studies for head and neck cancer. Head Neck. 2014;36:1069–1075. doi: 10.1002/hed.23444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chera BS, Amdur RJ. Current status and future directions of treatment deintensification in human papilloma virus-associated oropharyngeal squamous cell carcinoma. Semin Radiat Oncol. 2018;28:27–34. doi: 10.1016/j.semradonc.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Noor AM, Holmberg L, Gillett C, Grigoriadis A. Big Data: the challenge for small research groups in the era of cancer genomics. Br J Cancer. 2015;113:1405–1412. doi: 10.1038/bjc.2015.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatta G, Botta L, Sánchez MJ, et al. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. Eur J Cancer (Oxford, England: 1990) 2015;51:2130–2143. doi: 10.1016/j.ejca.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 11.Servomaa K, Kiuru A, Grénman R, et al. p53 mutations associated with increased sensitivity to ionizing radiation in human head and neck cancer cell lines. Cell Prolif. 1996;29:219–230. doi: 10.1046/j.1365-2184.1996.01009.x. [DOI] [PubMed] [Google Scholar]
- 12.Bossi P, Resteghini C, Paielli N, et al. Prognostic and predictive value of EGFR in head and neck squamous cell carcinoma. Oncotarget. 2016;7:74362–74379. doi: 10.18632/oncotarget.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jouhi L, Hagström J, Atula T, Mäkitie A. Is p16 an adequate surrogate for human papillomavirus status determination? Curr Opi Otolaryngol Head Neck Surg. 2017;25:108–112. doi: 10.1097/MOO.0000000000000341. [DOI] [PubMed] [Google Scholar]
- 14.Chung CH, Zhang Q, Kong CS, et al. p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J Clin Oncol. 2014;32:3930–3938. doi: 10.1200/JCO.2013.54.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khanna A, Pimanda JE, Westermarck J. Cancerous inhibitor of protein phosphatase 2A, an emerging human oncoprotein and a potential cancer therapy target. Can Res. 2013;73:6548–6553. doi: 10.1158/0008-5472.CAN-13-1994. [DOI] [PubMed] [Google Scholar]
- 16.Khan M, Khaznadar SS, Routila J, et al. Hepatocyte Growth Factor Receptor overexpression predicts reduced survival but its targeting is not effective in unselected HNSCC patients. Head Neck. 2020 doi: 10.1002/hed.26049.10.1002/hed.26049. [DOI] [PubMed] [Google Scholar]
- 17.Ventelä S, Sittig E, Mannermaa L, et al. CIP2A is an Oct4 target gene involved in head and neck squamous cell cancer oncogenicity and radioresistance. Oncotarget. 2015;6:144–158. doi: 10.18632/oncotarget.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science (New York, NY) 2017;357:eaan507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 19.Burtness B, Harrington KJ, Greil R, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet (London, England) 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. [DOI] [PubMed] [Google Scholar]
- 20.Mehanna H, Robinson M, Hartley A, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet (London, England) 2019;393:51–60. doi: 10.1016/S0140-6736(18)32752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klinghammer K, Gauler T, Dietz A, et al. Cetuximab, fluorouracil and cisplatin with or without docetaxel for patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck (CeFCiD): an open-label phase II randomised trial (AIO/IAG-KHT trial 1108) Eur J Cancer (Oxford, England: 1990) 2019;122:53–60. doi: 10.1016/j.ejca.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Kyzas PA, Loizou KT, Ioannidis JPA. Selective reporting biases in cancer prognostic factor studies. JNCI J Natl Cancer Inst. 2005;97:1043–1055. doi: 10.1093/jnci/dji184. [DOI] [PubMed] [Google Scholar]
- 23.Virk JS, Ingle M, Podesta CM, et al. Survival outcomes for head and neck cancer patients with N3 cervical nodal metastases. Clin Otolaryngol. 2020;45:342–349. doi: 10.1111/coa.13501. [DOI] [PubMed] [Google Scholar]
- 24.Marioni G, Blandamura S, Lionello M, et al. Indications for postoperative radiotherapy in laryngeal carcinoma: A panel of tumor tissue markers for predicting locoregional recurrence in surgically treated carcinoma. A pilot study. Head Neck. 2014;36:1534–1540. doi: 10.1002/hed.23493. [DOI] [PubMed] [Google Scholar]
- 25.Franz L, Tealdo G, Contro G, et al. Biological tumor markers (maspin, CD105, nm23-H1) and disease relapse in laryngeal cancer: cluster analysis. Head Neck. 2020;42:2129–2136. doi: 10.1002/hed.26152. [DOI] [PubMed] [Google Scholar]
- 26.Bryant AK, Sojourner EJ, Vitzthum LK, et al. Prognostic role of p16 in nonoropharyngeal head and neck cancer. J Natl Cancer Inst. 2018;110:1393–1399. doi: 10.1093/jnci/djy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Xia X, Gross N, et al. Prognostic implications of human papillomavirus status and p16 expression in laryngeal squamous cell carcinoma. Head Neck. 2019;41:4151–4163. doi: 10.1002/hed.25961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.