Abstract
Nicotine is inactivated by the polymorphic CYP2A6 enzyme to cotinine and then to 3′hydroxycotinine. The Nicotine Metabolite Ratio (NMR; 3′hydroxycotinine/cotinine) is a heritable nicotine metabolism biomarker, varies with sex and ancestry, and influences smoking cessation and disease risk. We conducted sex-stratified genome-wide association studies of the NMR in European American (EA) and African American (AA) smokers (NCT01314001, NCT00666978). In EA females (n = 389) and males (n = 541), one significant (P < 5e−8) chromosome 19 locus was found (top variant: rs56113850, CYP2A6 (intronic), for C vs. T: females: beta = 0.67, P = 7.5e−22, 21.8% variation explained; males: beta = 0.75, P = 1.2e−37, 26.1% variation explained). In AA females (n = 503) and males (n = 352), the top variant was found on chromosome 19 but differed by sex (females: rs11878604, CYP2A6 (~ 16 kb 3′), for C vs. T: beta = − 0.71, P = 6.6e−26, 16.2% variation explained; males: rs3865454, CYP2A6 (~ 7 kb 3′), for G vs. T: beta = 0.64, P = 1.9e−19, 18.9% variation explained). In AA females, a significant region was found on chromosome 12 (top variant: rs12425845: P = 5.0e−9, TMEM132C (~ 1 Mb 5′), 6.1% variation explained) which was not significant in AA males. In AA males, significant regions were found on chromosomes 6 (top variant: rs9379805: P = 4.8e−9, SLC17A2 (~ 8 kb 5′), 8.0% variation explained) and 16 (top variant: rs77368288: P = 3.5e−8, ZNF469 (~ 92 kb 5′), 7.1% variation explained) which were not significant in AA females. Further investigation of these associations outside of chromosome 19 is required, as they did not replicate. Understanding how sex and ancestry influence nicotine metabolism genetics may improve personalized approaches for smoking cessation and risk prediction for tobacco-related diseases.
Subject terms: Genetics, Neuroscience, Biomarkers
Introduction
Cigarette smoking remains a major public health concern1. Smoking behaviors, including the response to pharmacotherapy, differ between men and women. For instance, varenicline is more efficacious compared to nicotine patch or bupropion for women, whereas the treatment efficacy for all three is similar for men2. Smoking cessation is influenced by genetic variation in CYP2A6, coding for CYP2A6 which inactivates nicotine to cotinine, and metabolizes cotinine to 3′hydroxycotinine3,4. The nicotine metabolite ratio (NMR; 3′hydroxycotinine/cotinine ratio) is a biomarker of CYP2A6 activity, with higher NMR reflecting faster nicotine clearance5. CYP2A6 is induced by estrogen6, thus post-pubertal females who are pre-menopausal have faster nicotine clearance7 and higher NMR8 compared to males. African American (AA) smokers have lower NMR compared to European American (EA) smokers, due in part to AA having a higher prevalence of CYP2A6 decreased or loss-of-function variants9 and potentially also due to a higher prevalence of mentholated cigarette smoking8 compared to EA. Menthol can modestly inhibit CYP2A6 activity in vitro10 and nicotine clearance in vivo11, and smokers of mentholated cigarettes have lower NMRs on average8. Thus, sex, ancestry, and potentially mentholated cigarette smoking can alter CYP2A6 activity and the NMR.
The NMR has been associated with treatment response in bupropion and nicotine patch trials12–14. In the Pharmacogenetics of Nicotine Addiction Treatment (PNAT)-2 clinical trial (NCT01314001) where smokers were prospectively randomized by pre-treatment NMR, those with higher NMR had superior quit rates on varenicline (vs. nicotine patch), whereas those with lower NMR had similar quit rates15. Higher (vs. lower) CYP2A6 activity (measured by the NMR or CYP2A6 genetics) is also associated with higher exposure to tobacco-specific nitrosamines, volatile organic compounds, polycyclic aromatic hydrocarbons, and inflammatory markers16 and higher risk for tobacco-related diseases such as lung cancer17–19. CYP2A6 variation also alters the metabolism of chemotherapeutic agents (e.g. tegafur20 and letrozole21). In contrast to genomic assessments, the NMR can be reliably obtained only in current tobacco users22. An increased understanding of the genetics of the NMR, and if they differ by sex and ancestry, will enhance application in oncology and in epidemiologic studies of disease risk where non- and former-smokers are included.
Heritability estimates for the NMR range from 60 to 80%23,24. The vast majority of genome-wide significant (GWS) variants found in genome-wide association studies (GWASs) of the NMR in smokers were found in the CYP2A6 region of chromosome 1924–28; these GWASs all controlled for sex. Controlling for sex can confound the discovery and interpretation of genetic signals if the variant displays a sex-specific effect, or has a large but opposing direction of effect in males and females29. Sex differences in genetic effects on the expression and activity of CYP3A4, another CYP enzyme, have been reported30. Our primary aim was to identify sex differences in the genetic influences on CYP2A6 activity, measured by the NMR, and contrast these differences between AA and EA smokers. Compared to EA, far less is known regarding the biological underpinning of traits in minority populations including AA which have historically comprised < 5% of GWAS samples31. Our prior GWAS of the NMR (controlling for sex) in AA smokers identified several novel GWS SNPs in comparison to those found in EA smokers26. Here we performed sex-stratified GWASs of the NMR separately in EA and AA smokers.
Methods
Study participants
PNAT2 (NCT01314001) and Kick-it-at-Swope (KIS)-3 (NCT00666978) clinical trials
Genetic and biomarker data from PNAT2 and KIS3 clinical trial participants who provided written informed consent for DNA collection and release of de-identified information to investigators were used for this analysis, as previously described26. The clinical trial procedures were approved by IRBs at the University of Toronto and all clinical sites15,32 and were carried out in accordance with the Declaration of Helsinki. PNAT2 participants were EA and AA adult smokers (≥ 10 cigarettes/day) recruited at four clinical sites (University of Pennsylvania, University of Toronto/CAMH, SUNY Buffalo, and MD Anderson Cancer Center) and were randomized to placebo, nicotine patch, or varenicline based on their NMR at intake (for details see Ref.15). KIS3 participants were AA adult smokers from the Kansas City, Missouri area screened for participation in a clinical trial evaluating bupropion efficacy in AA light smokers (≤ 10 cigarettes/day) (for details see Ref.32).
Genome-wide SNP genotyping, quality control procedures, and imputation
Genome-wide genotyping was conducted at the Centre for Applied Genomics (Hospital for Sick Children; Toronto, Canada) using the Illumina HumanOmniExpressExome-8 v1.2 array and an iSelect custom add-on comprising 2688 variants associated with nicotine metabolism and/or smoking behaviours26. Quality control (QC) procedures implemented prior to imputation included removing samples with discordant sex, call rates < 98%, relatedness PI_HAT > 0.185, and heterozygosity > 3SD outside the mean. Variants with call rates < 98%, Hardy–Weinberg Equilibrium P < 1e−6, and minor allele frequencies (MAF) < 1% were excluded. Full details are described elsewhere26. EA and AA ancestry was determined via multidimensional scaling in combination with data and visualization from HapMap 3 (as described26). Genetic ancestries were highly concordant (> 95%) with self-reported ancestries. The number of individuals and variants available for phasing and imputation was: 390 female and 545 male PNAT2 EAs, 667,810 variants; 255 female and 251 male PNAT2 AAs, 733,629 variants; and 304 female and 154 male KIS3 AAs, 742,493 variants. The Michigan Imputation Server33 imputed genotypes using ShapeIT v2.r79034 and the HRC version r1.1 and 1000G Phase 3 v5 reference panels for EA and AA, respectively. Post-imputation variants with info (i.e. imputation quality) scores > 0.60 and MAF > 1% were analyzed. A total of 7,602,147 and 7,599,895 variants were analyzed in EA females and EA males, respectively. A total of 17,199,451 and 17,379,949 variants were meta-analyzed in AA females and AA males, respectively.
Phenotype: the nicotine metabolite ratio
As described elsewhere26, cotinine and 3′hydroxycotinine were measured using LC–MS/MS in PNAT2 and KIS3 participant blood samples collected at baseline during ad libitum smoking. To ensure reliability of NMR results, participants with cotinine < 10 ng/ml, suggestive of non-daily smoking, were excluded26. Seven PNAT2 participants (n = 5 EA and n = 2 AA) were excluded from further analyses due to missing and outlying (> 4 SD from the mean) NMR data, while n = 8 AA participants from KIS3 were excluded due to low cotinine. To correct for positive skew, the NMR (3′hydroxycotinine/cotinine) was rank-transformed using the rntransform function (https://rdrr.io/cran/GenABEL/man/rntransform.html) in the GenABEL package in RStudio.
Sex- and ancestry-stratified GWAS of the NMR
Frequentist additive models in SNPTEST (version 2.5.2)35 identified genetic associations with rank-transformed NMR separately in PNAT2 EA females, PNAT2 EA males, PNAT2 AA females, PNAT2 AA males, KIS3 AA females, and KIS3 AA males. As is convention in SNPTEST, the outcome measure was mean centered and scaled to have a variance of 1 for analysis. Chromosomes 1–22 were analyzed. Genotype uncertainty was controlled for by using the “-method expected” option26. Principal components 1–4, calculated separately in PNAT2 EA, PNAT2 AA, and KIS3 AA, were included in all models to adjust for population substructure. In previous genomic investigations of the NMR in these datasets, controlling for the first 2 versus 4 PCs26, or up to 10 PCs36, did not substantially alter the results. Potential covariates were evaluated based on a priori variables associated with rank NMR in PNAT28, which was the larger of the two trials and included both EA and AA. In EA, the main model included age (beta = 0.12; P = 0.0003), BMI (beta = − 0.10; P = 0.003), and alcohol use (# of drinks/week: beta = 0.06; P = 0.08) as covariates. In AA, the main model included age (beta = 0.12; P = 0.01), BMI (beta = − 0.10; P = 0.03), and mentholated cigarette use (beta = − 0.10; P = 0.047) as covariates. The final sample size for the main analysis was: n = 389 female and n = 541 male PNAT2 EA smokers, n = 205 female and n = 201 male PNAT2 AA smokers, and n = 298 female and n = 151 male KIS3 AA smokers. In a secondary analysis in EA, we further controlled for mentholated cigarette use. In a secondary analysis in KIS3 AA, where alcohol use (drinks/week) was significantly associated with NMR (beta = 0.16; P = 0.001), we further controlled for alcohol use. In PNAT2 AA, n = 98 participants were missing menthol data26, and were excluded from the main analysis; thus, we also performed a secondary analysis in all AA participants, without controlling for menthol. In KIS3 AA, n = 1 participant was missing BMI data, and was excluded from all analyses.
QQ plots are shown in Supplementary Fig. S1. Results from the PNAT2 AA females and KIS3 AA females were meta-analyzed, as were the results from the PNAT2 AA males and KIS3 AA males, using a fixed-effects model, info score threshold of 0.6, and META (version 1.7) software37.
To detect genome-wide association of individual SNPs explaining ≥ 10% of rank-transformed NMR variance, power was calculated to be 74%, 83%, 97%, and 98%, respectively, for sample sizes of n = 352, n = 389, n = 503, and n = 541. Notably, there was ≥ 80% power to detect SNPs explaining > 7% of rank-transformed NMR variation for n > 500. Power calculations were performed using Quanto version 1.2.438 and considered an additive model, two-sided significance level of 5e-8, and 5% MAF.
Conditional analysis, proportion of NMR variation explained, and sex × genotype interaction testing
To identify putative independent GWAS signals, conditional analyses were run in SNPTEST (version 2.5.2)35 for chromosomes that contained > 1 GWS variant, as previously described26. Analyses were performed separately for each chromosome, sex, and ancestry. For the AA sample, sex-stratified conditional analyses were performed in PNAT2 AA and KIS3 AA, followed by within sex meta-analysis. In conditional analysis, after controlling for the top variant, the next variant with the smallest P-value is controlled for along with the top independent signal in a second round of conditional analyses. The procedure is repeated until no additional significant (at P < 5e−8) signals emerge26.
In separate linear regression models examining the influence of genotype (coded additively: 0 vs. 1 vs. 2 copies of the effect allele) on rank-transformed NMR, we calculated the proportion of variation explained by each SNP by squaring the part correlation coefficient and multiplying by 100, after controlling for principal components 1–4 and NMR covariates (age, BMI, and alcohol use in EA; age, BMI, and mentholated cigarette smoking in AA). Analyses were conducted using SPSS version 23 (IBM, Armonk, New York, USA).
In AA, where sex-specific signals were observed, linear regression modeling examined the effect of the sex × genotype interaction term on rank-transformed NMR after controlling for NMR covariates (age, BMI, and menthol smoking) and main effects (sex and genotype, coded additively). Separate models were run for rs12425845, rs9379805, and rs77368288. Analyses were conducted using SPSS version 23 (IBM, Armonk, New York, USA).
Testing for replication of sex-specific genetic influences on the NMR in AA smokers
To determine whether the sex-specific signals on chromosome 12 (in AA females) and on chromosomes 6 and 16 (in AA males) replicated in an external sample, AA smokers with NMR data from the Quit-2-Live (NCT01836276)39 (n = 108 females and n = 108 males) and KIS240 (n = 334 females and n = 146 males) trials were analyzed. We genotyped samples for rs12426751 on chromosome 12, rs9295678 on chromosome 6, and rs77368288 on chromosome 16 using validated TaqMan SNP genotyping assays according to the manufacturer’s protocol (Applied Biosystems, Waltham, Massachusetts, USA). The rs12426751 and rs9295678 SNPs were selected since they are in high linkage disequilibrium (r2 = 0.95 in 1000G phase 3 AFR sample) with the sentinel SNPs (rs12425845 in chromosome 12 and rs9379805 in chromosome 6, respectively) and were directly genotyped by the array. For chromosome 16, the top SNP (rs77368288) was tested for replication.
The NMR was measured in KIS2 and Quit-2-Live at baseline during ad libitum smoking39,41. Linear regression models tested for association between genotype (coded additively) and rank-transformed NMR. Covariates included age, BMI, and mentholated cigarette smoking, which had similar distributions in the replication datasets (KIS2 and Quit-2-Live) as in the discovery datasets (PNAT2 and KIS3). Analyses were conducted using SPSS version 23 (IBM, Armonk, New York, USA). Assuming an additive model, alpha level of 0.05 for replication, and a similar MAF and effect size as found for the top chromosome 12, 6, and 16 SNPs in the discovery set, the power to detect an association in the replication set was > 0.99. Power was calculated using Quanto version 1.2.438.
eQTL and meQTL analysis
The GTEx Portal (https://www.gtexportal.org)42 and meQTL Database (http://www.mqtldb.org/)43 were used to determine whether the GWS variants were expression quantitative trait loci (eQTL) and/or methylation quantitative trait loci (meQTL), respectively. In GTEx, significant eQTL (using data release V8) were determined following cis-eQTL mapping using FastQTL, as described (https://www.gtexportal.org/home/documentationPage). For the meQTL analysis, the MatrixEQTL database and all time-points were selected, and the trans meQTL distance was set at > 1 Mb; i.e. variants found > 1 Mb away from the altered CpG position were defined as trans meQTL.
Results
Sex- and ancestry-stratified analysis of the NMR
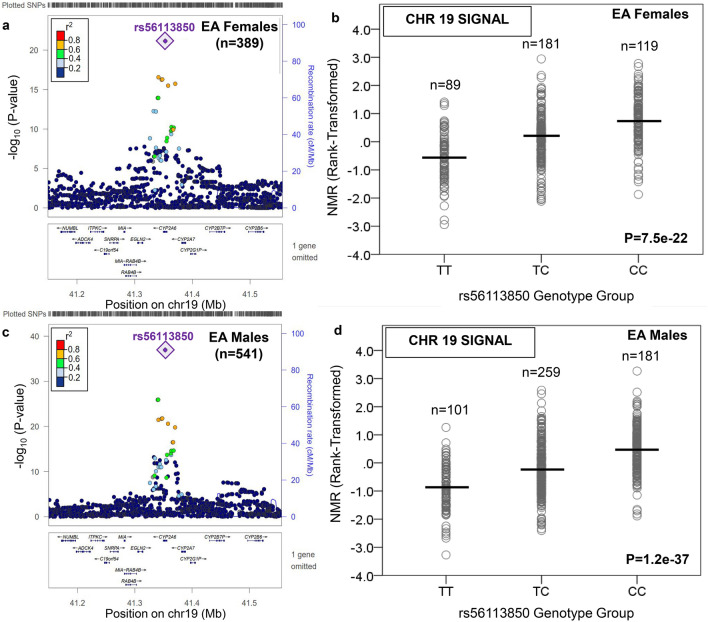
Participant characteristics are provided in Supplementary Table S1. The GWAS results for the main and secondary analyses are found in Supplementary Table S2. The calculated genomic inflation factors (ranging from 1.003 to 1.010) indicated little evidence of inflation (Supplementary Fig. S1). In EA females and males, n = 30 and n = 54 genome-wide significant (GWS) variants (P < 5e−8) were found, respectively; all were located on chromosome 19 (Supplementary Table S2). In each sex, there was one conditionally independent signal: the top variant was rs56113850 (for C vs. T: beta = 0.67, P = 7.5e−22, MAF = 47.8% in females; beta = 0.75, P = 1.2e−37, MAF = 44.9% in males) (Table 1, Fig. 1, and Supplementary Fig. S2). The rs56113850 variant is found in intron 4 of CYP2A6 and accounted for 21.8% and 26.1% of rank-transformed NMR variation in EA females and EA males, respectively. The rs56113850 variant was also significant in AA females and AA males (Supplementary Table S2).
Table 1.
Independent variants from each genome-wide significant locus in EA and AA smokers.
| Sample | Chr | Top SNP from region | Imputation Info (Quality) Score | MAF (%) | Effect Allele; Ref. Allele | Beta (SE); P-value | Gene location |
|---|---|---|---|---|---|---|---|
| EA females (n = 389) | 19 | rs56113850 | 0.92 in PNAT2 | 47.8 | C; T | 0.67 (0.07); 7.5e−22 | CYP2A6 (intron) |
| EA males (n = 541) | 19 | rs56113850 | 0.91 in PNAT2 | 44.9 | C; T | 0.75 (0.05); 1.2e−37 | CYP2A6 (intron) |
| AA females (n = 503) | 19 | rs11878604 | 0.98 in PNAT2, 0.93 in KIS3 | 22.8 | C; T |
− 0.71 (0.07); 6.6e−26c − 0.69 (0.11); 5.5e−10 in PNAT2 − 0.73 (0.09); 5.3e−15 in KIS3 |
CYP2A6 (16 kb 3′) |
| 19 | 19:41352257d | 0.90 in PNAT2, 0.92 in KIS3 | 31.9 | G; GT |
− 0.69 (0.07); 2.1e−25a,c − 0.68 (0.11); 2.9e−9 in PNAT2 − 0.69 (0.08); 2.8e−15 in KIS3 |
CYP2A6 (intron) | |
| 12 | rs12425845 | 1.0 in PNAT2, 0.99 in KIS3 | 8.4 | C; T |
0.64 (0.11); 5.0e−9c 0.73 (0.18); 7.1e−5 in PNAT2 0.59 (0.14); 2.9e−5 in KIS3 |
TMEM132C (1 Mb 5′) | |
| AA males (n = 352) | 19 | rs3865454 | 0.98 in PNAT2, 0.98 in KIS3 | 34.3 | G; T |
0.64 (0.07); 1.9e−19b 0.67 (0.09); 1.9e−11 in PNAT2 0.60 (0.11); 1.5e−7 in KIS3 |
CYP2A6 (7 kb 3′) |
| 6 | rs9379805 | 1.0 in PNAT2, 0.99 in KIS3 | 33.6 | A; T |
− 0.44 (0.08); 4.8e−9b − 0.34 (0.10); 8.3e−4 in PNAT2 − 0.58 (0.12); 1.6e−6 in KIS3 |
SLC17A2 (8 kb 5′) | |
| 16 | rs77368288 | 0.81 in PNAT2, 0.73 in KIS3 | 6.5 | C;T |
− 0.89 (0.16); 3.5e−8b − 0.96 (0.18); 4.6e−7 in PNAT2 − 0.65 (0.34); 5.4e−2 in KIS3 |
ZNF469 (92 kb 5′) |
Novel loci associated with the nicotine metabolite ratio (rank-transformed for analysis) were found in AA females and AA males and are indicated in italics.
EA European American, AA African American, Chr chromosome, info info (i.e. quality) score from imputation, MAF minor allele frequency, SE standard error.
Conditional analysis was used to determine the number of independent signals for each chromosome containing genome-wide significant variants.
aAfter conditioning on rs11878604, the beta (SE) for 19:41352257 was − 0.45 (0.08), and the P-value was 8.3e−9.
bBeta, SE, and P-value is reported for the meta-analyzed PNAT2 and KIS3 AA males.
cBeta, SE, and P-value is reported for the meta-analyzed PNAT2 and KIS3 AA females.
dNo rsID is available for this variant in dbSNP.
Figure 1.
The top signal associated with the Nicotine Metabolite Ratio in European ancestry females and males was found on chromosome 19 in the region containing CYP2A6. The nicotine metabolite ratio was rank-transformed for analysis. Regional plots, generated using LocusZoom44 (freely available at locuszoom.org), show the top overall SNP in European American (EA) females (rs56113850) (a). The influence of rs56113850 on the Nicotine Metabolite Ratio (NMR) in EA females is shown in (b). The rs56113850 SNP was also the top SNP in EA males (c), and the influence of rs56113850 on the NMR in EA males is shown in d). Linkage disequilibrium patterns in (a) and (c) are based upon the hg19/1000 Genomes November 2014 release European reference population. The plots in (b) and (d) were created using SPSS version 23 (can be purchased from IBM, Armonk, New York, USA). The black horizontal line represents the mean rank-transformed NMR in each group, and the P-value is from the GWAS after adjusting for population substructure and NMR covariates.
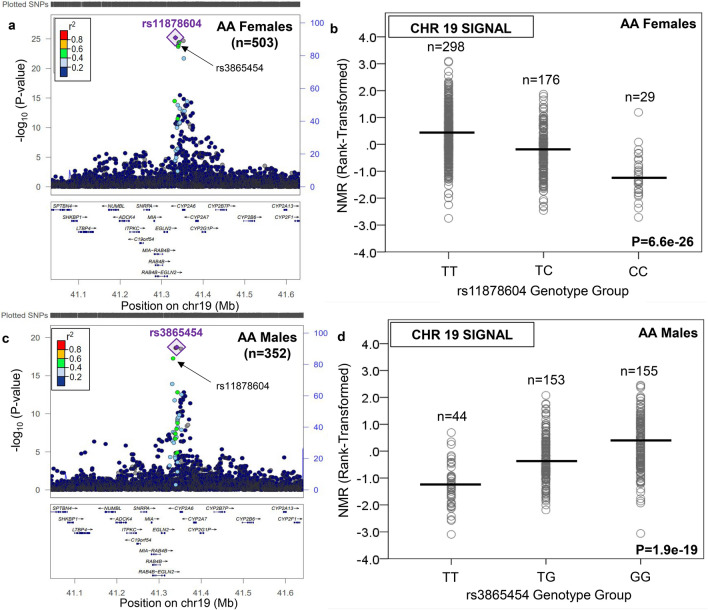
In AA females and males, there were n = 72 and n = 55 GWS variants, respectively (Supplementary Table S2). As for EA, the top overall SNP was found on chromosome 19 in AA. In AA females, the top SNP was rs11878604 (for C vs. T: beta = − 0.71, P = 6.6e−26, MAF = 22.8%) (Table 1, Fig. 2, and Supplementary Fig. S3), located 16 kb 3′ of CYP2A6, which explained 16.2% of rank-transformed NMR variation. The rs11878604 variant was the seventh top variant and had a similar beta in AA males, and was also significant in EA males (Supplementary Table S2). There was a second conditionally independent locus on chromosome 19 in AA females, with the top variant found at position 41352257 (for G vs. GT: beta = − 0.69, P = 2.2e−25, MAF = 31.9%) located in an intron of CYP2A6 (Table 1); this variant explained 3.6% of rank-transformed NMR variation after controlling for rs11878604, and was the sixth top variant and had a similar beta in AA males (Supplementary Table S2). In AA males, a single independent locus on chromosome 19 was observed. The top overall SNP was rs3865454 (for G vs. T: beta = 0.64, P = 1.9e−19, MAF = 34.3%) (Table 1, Fig. 2, and Supplementary Fig. S3), located 7 kb 3′ of CYP2A6, which explained 18.9% of rank-transformed NMR variation. The rs3865454 variant was the fourth top variant and had a similar beta in AA females (Supplementary Table S2). The betas for the top variants (rs11878604, variant at position 41352257, and rs3865454) were similar between PNAT2 AA and KIS3 AA (Supplementary Table S2). Of note, the top chromosome 19 SNPs (i.e. rs56113850 in EA males and EA females, rs11878604 in AA females, and rs3865454 in AA males) are in low-moderate linkage disequilibrium (LD; r2 = 0.13–0.51) in AA and EA populations (Supplementary Fig. S4).
Figure 2.
The top signal associated with the Nicotine Metabolite Ratio in African ancestry females and males was found on chromosome 19 in the region containing CYP2A6. The nicotine metabolite ratio was rank-transformed for analysis. Regional plots were generated using LocusZoom44 (freely available at locuszoom.org). In African American (AA) females, the top variant was rs11878604 (a), which was also significant in AA males. The influence of rs11878604 on the Nicotine Metabolite Ratio (NMR) in AA females is shown in (b). In AA males, the top variant was rs3865454 (c), which was also significant in AA females. The influence of rs3865454 on the NMR in AA males is shown in (d). Note: in AA females, there was a second independent signal on chromosome 19: the top variant was an indel at position 41352257 (P = 8.3e−9, after conditioning on rs11878604); this variant was also GWS in AA males. Linkage disequilibrium patterns in (a) and (c) are based upon the hg19/1000 Genomes November 2014 release African reference population. The plots in (b) and (d) were created using SPSS version 23 (can be purchased from IBM, Armonk, New York, USA). The black horizontal line represents the mean rank-transformed NMR in each group, and the P-value is from the GWAS after adjusting for population substructure and NMR covariates.
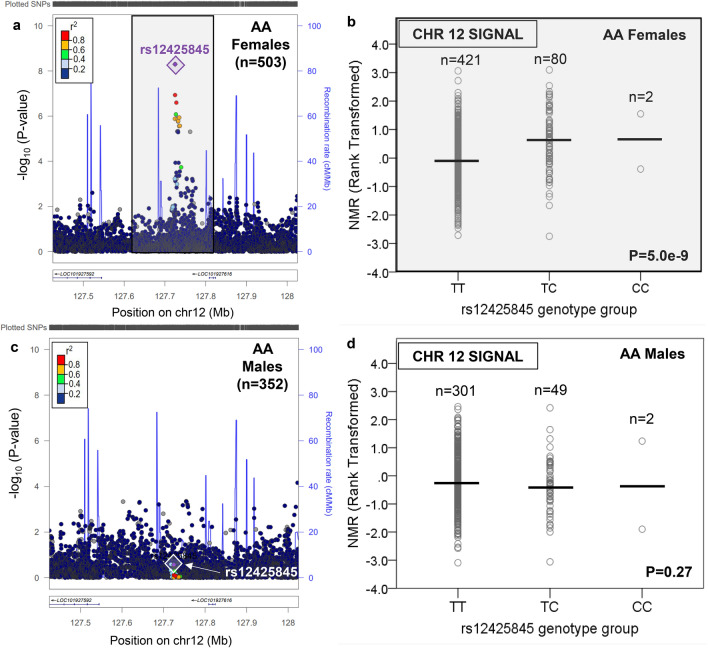
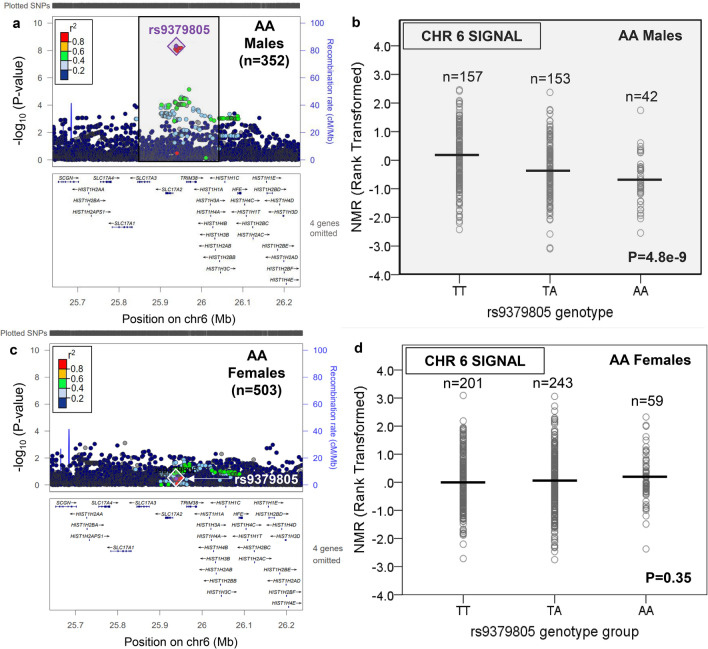
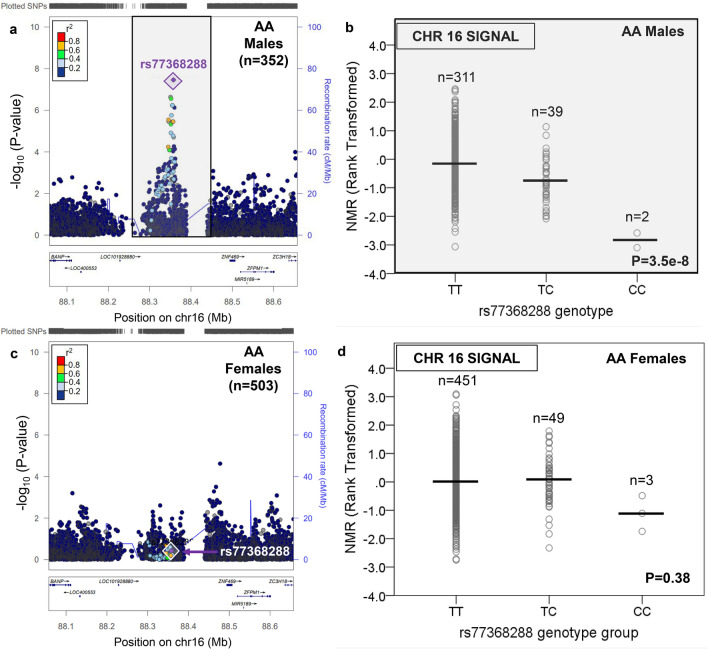
In contrast to EA, there were novel GWS loci found outside of chromosome 19 in AA (Supplementary Table S2). In AA females, one additional locus was found, on chromosome 12 (Table 1, Fig. 3, and Supplementary Fig. S5). The chromosome 12 signal comprised one GWS SNP, rs12425845 (for C vs. T: beta = 0.64, P = 5.0e−9, MAF = 8.4%), located ~ 1 MB 5′ of TMEM132C, which explained 6.1% of rank-transformed NMR variation. This chromosome 12 locus was not GWS in AA males (Fig. 3). In AA males, two additional loci were found outside of chromosome 19, on chromosome 6 (six GWS SNPs representing one independent signal) and chromosome 16 (one GWS SNP) (Table 1 and Figs. 4, 5 and Supplementary Fig. S5). The top chromosome 6 GWS SNP was rs9379805 (for A vs. T: beta = − 0.44, P = 4.8e−9, MAF = 33.6%), located ~ 8 kb 5′ of SLC17A2, which explained 8.0% of rank-transformed NMR variation. The lone chromosome 16 GWS SNP, rs77368288 (for C vs. T: beta = − 0.89, P = 3.5e−8, MAF = 6.5%), was found ~ 93 kb 5′ of ZNF469 and explained 7.1% of rank-transformed NMR variation. Neither the chromosome 6 nor the chromosome 16 loci were GWS in AA females (Figs. 4, 5). In AA, a significant sex × SNP interaction effect on rank-transformed NMR was observed for all three novel loci outside of chromosome 19: rs12425845 (P = 0.00002), rs9379805 (P = 0.000002), and rs77368288 (P = 0.001). Notably, variation in MRGPRX4, previously associated with menthol smoking in AA45, was not associated with rank-transformed NMR in either AA males or AA females (Supplementary Fig. S6).
Figure 3.
There was a chromosome 12 signal associated with the Nicotine Metabolite Ratio in African American females which was not significant in African American males. The nicotine metabolite ratio was rank-transformed for analysis. A regional plot, generated using LocusZoom44 (freely available at locuszoom.org), shows the top overall chromosome 12 variant (rs12425845) in AA females (a); the influence of rs12425845 on the Nicotine Metabolite Ratio (NMR) in AA females is shown in (b). The chromosome 12 locus containing rs12425845 was not significant in AA males (c); the lack of influence of rs12425845 on the NMR in AA males is shown in (d). Linkage disequilibrium patterns in (a) and (c) are based upon the hg19/1000 Genomes November 2014 release African reference population. The plots in (b) and (d) were created using SPSS version 23 (can be purchased from IBM, Armonk, New York, USA). The black horizontal line represents the mean rank-transformed NMR in each group, and the P-value is from the GWAS after adjusting for population substructure and NMR covariates.
Figure 4.
There was a chromosome 6 signal associated with the Nicotine Metabolite Ratio in African American males which was not significant in African American females. The nicotine metabolite ratio was rank-transformed for analysis. A regional plot, generated using LocusZoom44 (freely available at locuszoom.org), shows the top overall chromosome 6 variant (rs9379805) in AA males (a); the influence of rs9379805 on the Nicotine Metabolite Ratio (NMR) in AA males is shown in (b). The chromosome 6 locus containing rs9379805 was not significant in AA females (c); the lack of influence of rs9379805 on the NMR in AA females is shown in (d). Linkage disequilibrium patterns in (a) and (c) are based upon the hg19/1000 Genomes November 2014 release African reference population. The plots in (b) and (d) were created using SPSS version 23 (can be purchased from IBM, Armonk, New York, USA). The black horizontal line represents the mean rank-transformed NMR in each group, and the P-value is from the GWAS after adjusting for population substructure and NMR covariates.
Figure 5.
There was a chromosome 16 signal associated with the Nicotine Metabolite Ratio in African American males which was not significant in African American females. The nicotine metabolite ratio was rank-transformed for analysis. A regional plot, generated using LocusZoom LocusZoom44 (freely available at locuszoom.org), shows the top overall chromosome 16 variant (rs77368288) in AA males (a); the influence of rs77368288 on the Nicotine Metabolite Ratio (NMR) in AA males is shown in (b). The chromosome 16 locus containing rs77368288 was not significant in AA females (c); the lack of influence of rs77368288 on the NMR in AA females is shown in (d). Linkage disequilibrium patterns in (a) and (c) are based upon the hg19/1000 Genomes November 2014 release African reference population. The plots in (b) and (d) were created using SPSS version 23 (can be purchased from IBM, Armonk, New York, USA). The black horizontal line represents the mean rank-transformed NMR in each group, and the P-value is from the GWAS after adjusting for population substructure and NMR covariates.
In all secondary analyses, we observed similar effect sizes (betas) to those found in the main analysis with few substantial differences (Supplementary Table S2). Overall, the betas and P-values from the secondary analyses were highly correlated with the betas and P-values, respectively, from the main analysis (Supplementary Table S3).
Replication analysis
None of the signals from chromosome 12 (in AA females), chromosome 6 (in AA males), or chromosome 16 (in AA males) were significantly associated with the NMR in external clinical trials (i.e. KIS2 and Quit-2-Live), however the direction of effect for the chromosome 12 and 16 signals seemed consistent (Supplementary Fig. S7). Of note, the chromosome 12 and 6 signals were associated with the NMR in both PNAT2 (P = 7.1e−5 and 8.2e−4, respectively) and KIS3 (P = 2.9e−5 and P = 1.6e−6, respectively) (Table 1 and Supplementary Table S2). The chromosome 19 locus that we identified is consistent with prior GWASs of the NMR24–28.
eQTL and meQTL analyses
Most of the GWS variants associated with the NMR were significant eQTL and/or meQTL (Supplementary Table S2). In EA, the top SNP (rs56113850) was significantly associated with altered expression of CYP2A6, CYP2A7, EGLN2, RAB4B, CYP2G1P, CYP2T1P, and CTC-490E21.11, as previously noted27. In EA males, additional GWS variants mapping to CYP2A6 and CYP2A7 were associated with altered CYP2A6 expression in GTEx in the adrenal gland, which were not GWS in females (Supplementary Table S2).
In AA females, the top chromosome 19 SNP (rs11878604) was a significant eQTL for CYP2A6 in the adrenal gland, as well as for CYP2G1P, but was not a significant meQTL (Supplementary Table S2). In AA males, the top chromosome 19 SNP (rs3865454) was significantly associated with altered expression of CYP2A7, CYP2T1P, C19orf54, and CTC-490E21.11. However, rs3865454 was not a significant cis or trans meQTL. There were also several GWS SNPs in chromosome 19 in AA that were significant eQTL for CYP2A6 in subcutaneous adipose tissue.
The GWS SNP from chromosome 12 found in AA females (rs12425845) was a significant trans meQTL influencing methylation at a CpG site on chromosome 12 within TMEM132A but was not a significant eQTL. The top chromosome 6 SNP found in AA males (rs9379805), was not found in GTEx, however other SNPs from this locus were significant eQTL for several targets including three drug transporters (i.e. SLC17A1, SLC17A2, and SLC17A3) in adrenal gland and/or testis (Supplementary Table S2). Moreover, the GWS SNPs from the chromosome 6 locus were all cis meQTL. The GWS SNP from chromosome 16 in AA males (rs77368288) was a significant eQTL for APRT involved in the nucleotide salvage pathway.
Discussion
This is the first genome-wide investigation of sex differences in the genetic influences on the NMR, a sexually dimorphic nicotine metabolism biomarker5,6. Smoking behaviours, cessation outcomes, and tobacco-related disease risk, which are each associated with the NMR15,17,46, also differ between men and women2,47–49. We further stratified our sex-based analyses by genetic ancestry, due to differences in population substructure as well as in the NMR; AA have lower NMR on average compared to EA8. Moreover, smoking cessation outcomes39 and lung cancer risk50 differ between EA and AA.
In all four analytic sub-groups, the top overall variant was found on chromosome 19 and mapped to the CYP2A6 gene coding for the main metabolic enzyme for nicotine. In EA females and EA males, a single GWS locus was found on chromosome 19: the top variant was rs56113850 (intron 4 of CYP2A6). No other chromosomes contained GWS loci. The rs56113850 variant was the top SNP in all prior GWASs of the NMR in EA smokers that controlled for sex24,25,27,28 and was also significant in AA. Chromosome 19 also contained the top SNP in AA females (rs11878604, located 16 kb 3′ of CYP2A6) and AA males (rs3865454; 7 kb 3′ of CYP2A6). The rs11878604 and rs3865454 variants are in moderate LD (r2 = 0.48) in AA populations, and the rs11878604 variant was found in our prior NMR GWAS in AA (controlled for sex)26. The rs11878604 variant was also GWS in EA males, while rs3865454 was not significant in EA. The difference in top variant between EA and AA is likely due in part to distinct LD patterns and variant frequencies. In AA, rs28399454, which defines the known reduced function CYP2A6*17 allele51, was also GWS; rs28399454 is in low LD with rs11878604 (r2 = 0.28) and rs3865454 (r2 = 0.21). In contrast to EA, significant and novel loci were found outside of chromosome 19 in AA; these loci differed between AA females (chromosome 12) and AA males (chromosomes 6 and 16). In AA, the additional, and sex-specific, loci in our current study underscore the potential importance of performing sex-based analyses to better understand the biology of sexually dimorphic traits29 such as the NMR.
In the GWAS catalog (https://www.ebi.ac.uk/gwas/; accessed June 30, 2021), there are no reported associations for rs12425845 (chromosome 12), rs9379805 (chromosome 6), or rs77368288 (chromosome 16) with any traits. Only rs9379805 was found within 10 kb of a gene (i.e. SLC17A2). The SLC17A1-4 transporters are thought to transport organic anions, however their substrates and functional relevance are only partly understood52. Whether cotinine and/or 3′hydroxycotinine are substrates of the SLC17 transporter family is not known. The mechanism explaining the association between each of these non-chromosome 19 regions and the NMR remains to be determined but may involve regulatory regions, as an example.
Our study had several limitations, including the lack of association of the NMR with the sex-specific signals on chromosomes 12, 6, and 16 in an external sample, despite being sufficiently powered. In the discovery cohort, the chromosome 12 and 16 signals comprised a single GWS SNP, although other SNPs with low P-values in LD with the GWS SNP were found in the region. The inclusion criteria and recruitment strategy also differed between trials. However, in AA, the betas for the top SNPs were similar between PNAT2 and KIS3 despite differences in baseline smoking level. Further testing in external datasets is required to determine the robustness of the sex-specific findings. In our analyses controlling for menthol, many P-values were less significant than in models not controlling for menthol, however this could be due to the smaller sample size as the betas were highly correlated. Both GTEx and mQTLdb contain data from predominantly European (85% and 100%, respectively) samples43, limiting their generalizability to AA. In GTEx, 67% of the donors are male and sex differences in transcript levels have recently been identified53. Future studies could assess tissue expression levels from male and female donors separately. Moreover, all of the adults contributing samples to the mQTLdb are female, limiting generalizability to males; we therefore included all time-points in the mQTLdb so that multiple sexes could be considered. Finally, we examined the autosomes in this paper; examination of the X chromosome, which requires unique quality control procedures and statistical modeling considerations54, is underway. Strengths of our study include the sex-based analyses and the examination of multiple ancestries. Our sub-group analyses were well-powered for common variants (MAF ≥ 5%) each explaining at least 7–10% of rank-transformed NMR variance. The top SNPs each explained a relatively large portion of the variance in the NMR, consistent with previous NMR GWASs24,26–28. Future sex-based ancestry-specific analyses of the NMR, including genome-wide testing for sex-by-genotype interactions, in larger samples will determine the robustness of our findings, reveal new loci, and may suggest utility of sex-based genetic risk scoring approaches for the NMR.
In conclusion, we showed potential sex differences in the genetic influences on the NMR in AA, but not EA smokers. Identifying the causal SNPs and the mechanism behind these sex-based genetic influences will provide further biological insight into the NMR, which differs by sex8 and can only be measured in current tobacco users22. A greater understanding of how sex and ancestry influence nicotine metabolism biomarker genetics may improve genomics-based risk prediction modelling for tobacco-related illnesses and enhance personalized treatment approaches for tobacco dependence.
Supplementary Information
Acknowledgements
The authors acknowledge Drs. Andy Zhu for designing the custom array add-on, Jennifer Ware for quality control analyses, and Qian Zhou for genotyping. Computations were performed on the CAMH Specialized Computing Cluster, funded by the Canada Foundation for Innovation Research Hospital Fund. This work was funded by CIHR Catalyst (T2023973) and Project (PJY-159710) grants, NIH Grants PGRN DA020830, CA091912, DA031815, and R35 CA197461; and a Canada Research Chair in Pharmacogenomics (Tyndale).
Author contributions
M.J.C., J.K., and R.F.T. were responsible for the study concept and analysis plan. L.S.C., N.L.N., J.S.A., N.L.B., C.L., and R.F.T. oversaw the design of the original studies. M.J.C. performed data analysis. All authors contributed to the interpretation of the findings. M.J.C. and R.F.T. drafted the manuscript and revised it. All authors reviewed and approved the manuscript.
Data availability
Due to the type of informed consents given by study participants, individual level data cannot be made publicly available. GWAS summary statistics and scripts used for analyses are available upon request.
Competing interests
R. F. Tyndale has consulted for Quinn Emanuel and Ethismos. Dr. Benowitz has consulted with Achieve Life Sciences and Pfizer, companies that market or are developing smoking cessation medications, and has been a paid expert witness in litigation against tobacco companies. The other authors declare no conflicts of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-98883-z.
References
- 1.Jha P, Peto R. Global effects of smoking, of quitting, and of taxing tobacco. N. Engl. J. Med. 2014;370(1):60–68. doi: 10.1056/NEJMra1308383. [DOI] [PubMed] [Google Scholar]
- 2.Smith PH, Weinberger AH, Zhang J, Emme E, Mazure CM, McKee SA. Sex differences in smoking cessation pharmacotherapy comparative efficacy: A network meta-analysis. Nicotine Tob. Res. 2017;19(3):273–281. doi: 10.1093/ntr/ntw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J. Pharmacol. Exp. Ther. 1996;277(2):1010–1015. [PubMed] [Google Scholar]
- 4.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab. Dispos. 1996;24(11):1212–1217. [PubMed] [Google Scholar]
- 5.Dempsey D, Tutka P, Jacob P, 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin. Pharmacol. Ther. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, et al. Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab. Dispos. 2007;35(10):1935–1941. doi: 10.1124/dmd.107.016568. [DOI] [PubMed] [Google Scholar]
- 7.Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd Female sex and oral contraceptive use accelerate nicotine metabolism. Clin. Pharmacol. Ther. 2006;79(5):480–488. doi: 10.1016/j.clpt.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Chenoweth MJ, Novalen M, Hawk LW, Jr, Schnoll RA, George TP, Cinciripini PM, et al. Known and novel sources of variability in the nicotine metabolite ratio in a large sample of treatment-seeking smokers. Cancer Epidemiol. Biomark. Prev. 2014;23(9):1773–1782. doi: 10.1158/1055-9965.EPI-14-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, et al. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clin. Pharmacol. Ther. 2006;80(3):282–297. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 10.MacDougall JM, Fandrick K, Zhang X, Serafin SV, Cashman JR. Inhibition of human liver microsomal (S)-nicotine oxidation by (−)-menthol and analogues. Chem. Res. Toxicol. 2003;16(8):988–993. doi: 10.1021/tx0340551. [DOI] [PubMed] [Google Scholar]
- 11.Benowitz NL, Herrera B, Jacob P., 3rd Mentholated cigarette smoking inhibits nicotine metabolism. J. Pharmacol. Exp. Ther. 2004;310(3):1208–1215. doi: 10.1124/jpet.104.066902. [DOI] [PubMed] [Google Scholar]
- 12.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward personalized therapy for smoking cessation: A randomized placebo-controlled trial of bupropion. Clin. Pharmacol. Ther. 2008;84(3):320–325. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 13.Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin. Pharmacol. Ther. 2006;79(6):600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: A validation study. Pharmacol. Biochem. Behav. 2009;92(1):6–11. doi: 10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerman C, Schnoll RA, Hawk LW, Jr, Cinciripini P, George TP, Wileyto EP, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: A randomised, double-blind placebo-controlled trial. Lancet Respir. Med. 2015;3(2):131–138. doi: 10.1016/S2213-2600(14)70294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll DM, Murphy SE, Benowitz NL, Strasser AA, Kotlyar M, Carmella SG, et al. Relationships between the nicotine metabolite ratio and a panel of exposure and effect biomarkers: findings from two studies of U.S. commercial cigarette smokers. Cancer Epidemiol. Biomark. Prev. 2020;29(4):871–879. doi: 10.1158/1055-9965.EPI-19-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SL, Murphy SE, Wilkens LR, Stram DO, Hecht SS, Le Marchand L. Association of CYP2A6 activity with lung cancer incidence in smokers: The multiethnic cohort study. PLoS One. 2017;12(5):e0178435. doi: 10.1371/journal.pone.0178435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017;49(7):1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai J, Lv J, Zhu M, Wang Y, Qin N, Ma H, et al. Identification of risk loci and a polygenic risk score for lung cancer: A large-scale prospective cohort study in Chinese populations. Lancet Respir. Med. 2019;7(10):881–891. doi: 10.1016/S2213-2600(19)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamiya I, Yoshisue K, Ishii Y, Yamada H, Chiba M. Effect of CYP2A6 genetic polymorphism on the metabolic conversion of Tegafur to 5-fluorouracil and its enantioselectivity. Drug Metab. Dispos. 2014;42(9):1485–1492. doi: 10.1124/dmd.114.058008. [DOI] [PubMed] [Google Scholar]
- 21.Murai K, Yamazaki H, Nakagawa K, Kawai R, Kamataki T. Deactivation of anti-cancer drug letrozole to a carbinol metabolite by polymorphic cytochrome P450 2A6 in human liver microsomes. Xenobiotica. 2009;39(11):795–802. doi: 10.3109/00498250903171395. [DOI] [PubMed] [Google Scholar]
- 22.Lea RA, Dickson S, Benowitz NL. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: Prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J. Anal. Toxicol. 2006;30(6):386–389. doi: 10.1093/jat/30.6.386. [DOI] [PubMed] [Google Scholar]
- 23.Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. Genetic and environmental influences on the ratio of 3'hydroxycotinine to cotinine in plasma and urine. Pharmacogenet. Genom. 2009;19(5):388–398. doi: 10.1097/FPC.0b013e32832a404f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loukola A, Buchwald J, Gupta R, Palviainen T, Hallfors J, Tikkanen E, et al. A genome-wide association study of a biomarker of nicotine metabolism. PLoS Genet. 2015;11(9):e1005498. doi: 10.1371/journal.pgen.1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel YM, Park SL, Han Y, Wilkens LR, Bickeboller H, Rosenberger A, et al. Novel association of genetic markers affecting CYP2A6 activity and lung cancer risk. Cancer Res. 2016;76(19):5768–5776. doi: 10.1158/0008-5472.CAN-16-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chenoweth MJ, Ware JJ, Zhu AZX, Cole CB, Cox LS, Nollen N, et al. Genome-wide association study of a nicotine metabolism biomarker in African American smokers: Impact of chromosome 19 genetic influences. Addiction. 2018;113(3):509–523. doi: 10.1111/add.14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchwald J, Chenoweth MJ, Palviainen T, Zhu G, Benner C, Gordon S, et al. Genome-wide association meta-analysis of nicotine metabolism and cigarette consumption measures in smokers of European descent. Mol. Psychiatry. 2020;26(6):2212–2223. doi: 10.1038/s41380-020-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baurley JW, Edlund CK, Pardamean CI, Conti DV, Krasnow R, Javitz HS, et al. Genome-wide association of the laboratory-based nicotine metabolite ratio in three ancestries. Nicotine Tob. Res. 2016;18(9):1837–1844. doi: 10.1093/ntr/ntw117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powers MS, Smith PH, McKee SA, Ehringer MA. From sexless to sexy: Why it is time for human genetics to consider and report analyses of sex. Biol. Sex Differ. 2017;8:15. doi: 10.1186/s13293-017-0136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schirmer M, Rosenberger A, Klein K, Kulle B, Toliat MR, Nurnberg P, et al. Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics. 2007;8(5):443–453. doi: 10.2217/14622416.8.5.443. [DOI] [PubMed] [Google Scholar]
- 31.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox LS, Nollen NL, Mayo MS, Choi WS, Faseru B, Benowitz NL, et al. Bupropion for smoking cessation in African American light smokers: A randomized controlled trial. J. Natl. Cancer Inst. 2012;104(4):290–298. doi: 10.1093/jnci/djr513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 35.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 36.El-Boraie A, Chenoweth MJ, Pouget JG, Benowitz NL, Fukunaga K, Mushiroda T, et al. Transferability of ancestry-specific and cross-ancestry CYP2A6 activity genetic risk scores in African and European populations. Clin. Pharmacol. Ther. 2020;110(4):975–985. doi: 10.1002/cpt.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat. Med. 2002;21(1):35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 39.Nollen NL, Mayo MS, Cox LS, Benowitz NL, Tyndale RF, Ellerbeck EF, et al. Factors that explain differences in abstinence between black and white smokers: A prospective intervention study. Jnci-J. Natl. Cancer I. 2019;111(10):1078–1087. doi: 10.1093/jnci/djz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahluwalia JS, Okuyemi K, Nollen N, Choi WS, Kaur H, Pulvers K, et al. The effects of nicotine gum and counseling among African American light smokers: A 2 x 2 factorial design. Addiction. 2006;101(6):883–891. doi: 10.1111/j.1360-0443.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 41.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin. Pharmacol. Ther. 2009;85(6):635–643. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;31(17):61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozlitina J, Risso D, Lansu K, Olsen RHJ, Sainz E, Luiselli D, et al. An African-specific haplotype in MRGPRX4 is associated with menthol cigarette smoking. PLoS Genet. 2019;15(2):e1007916. doi: 10.1371/journal.pgen.1007916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanner JA, Tyndale RF. Variation in CYP2A6 activity and personalized medicine. J. Pers. Med. 2017;7(4):18. doi: 10.3390/jpm7040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huxley RR, Woodward M. Cigarette smoking as a risk factor for coronary heart disease in women compared with men: A systematic review and meta-analysis of prospective cohort studies. Lancet. 2011;378(9799):1297–1305. doi: 10.1016/S0140-6736(11)60781-2. [DOI] [PubMed] [Google Scholar]
- 48.Hymowitz N, Cummings KM, Hyland A, Lynn WR, Pechacek TF, Hartwell TD. Predictors of smoking cessation in a cohort of adult smokers followed for five years. Tob. Control. 1997;6(Suppl 2):S57–62. doi: 10.1136/tc.6.suppl_2.S57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perkins KA, Donny E, Caggiula AR. Sex differences in nicotine effects and self-administration: Review of human and animal evidence. Nicotine Tob. Res. 1999;1:301–315. doi: 10.1080/14622299050011431. [DOI] [PubMed] [Google Scholar]
- 50.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N. Engl. J. Med. 2006;354(4):333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 51.Fukami T, Nakajima M, Yoshida R, Tsuchiya Y, Fujiki Y, Katoh M, et al. A novel polymorphism of human CYP2A6 gene CYP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clin. Pharmacol. Ther. 2004;76(6):519–527. doi: 10.1016/j.clpt.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Reimer RJ. SLC17: A functionally diverse family of organic anion transporters. Mol. Aspects Med. 2013;34(2–3):350–359. doi: 10.1016/j.mam.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kassam I, Wu Y, Yang J, Visscher PM, McRae AF. Tissue-specific sex differences in human gene expression. Hum. Mol. Genet. 2019;28(17):2976–2986. doi: 10.1093/hmg/ddz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang D, Gao F, Slavney A, Ma L, Waldman YY, Sams AJ, et al. Accounting for eXentricities: Analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS One. 2014;9(12):e113684. doi: 10.1371/journal.pone.0113684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to the type of informed consents given by study participants, individual level data cannot be made publicly available. GWAS summary statistics and scripts used for analyses are available upon request.