Abstract
Differential expression of cell adhesion molecules in neuronal populations is one of the many mechanisms promoting the formation of functional neural circuits in the developing nervous system. The IgLON family consists of five cell surface immunoglobulin proteins that have been associated with various developmental disorders, such as autism spectrum disorder, schizophrenia, and major depressive disorder. However, there is still limited and fragmented information about their patterns of expression in certain regions of the developing nervous system and how their expression contributes to their function. Utilizing an in situ hybridization approach, we have analyzed the spatiotemporal expression of all IgLON family members in the developing mouse brain, spinal cord, eye, olfactory epithelium, and vomeronasal organ. At one prenatal (E16) and two postnatal (P0 and P15) ages, we show that each IgLON displays distinct expression patterns in the olfactory system, cerebral cortex, midbrain, cerebellum, spinal cord, and eye, indicating that they likely contribute to the wiring of specific neuronal circuitry. These analyses will inform future functional studies aimed at identifying additional roles for these proteins in nervous system development.
Subject terms: Cellular neuroscience, Development of the nervous system, Neural circuits, Olfactory system, Peripheral nervous system, Developmental biology, Neuroscience
Introduction
The developing nervous system relies on many mechanisms to form functioning neural circuits. One such mechanism is cell–cell or axon–axon interaction which relies on adhesion between cell surface molecules belonging to the large family of cell adhesion molecules (CAMs). By allowing for adhesive interactions, CAMs modulate a wide array of processes during neuronal development, including neurogenesis, fasciculation and guidance of axons, and synapse formation and selectivity1–4. The selective expression of different families of CAMs, or of individual members of specific subfamilies, in populations of neurons is especially important during the development of sensory neural maps. For example, differential expression of members of the Teneurin and Kirrel family of CAMs have been implicated in the formation of the olfactory maps in Drosophila and mice, respectively5–10.
The IgLON family of cell surface proteins belongs to the large immunoglobulin family of CAMs. To date, five IgLON family members have been identified: opioid-binding cell adhesion molecule (OBCAM/IgLON1), neurotrimin (Ntm/IgLON2), limbic-system associated membrane protein (LAMP/LSAMP/IgLON3), neuronal growth regulator 1 (NEGR1/Kilon/IgLON4), and IgLON511–14. IgLONs are composed of three C2-like Ig domains and are tethered to the surface of the membrane by a glycosylphosphatidylinositol anchor15. Each Ig domain can be glycosylated, but glycosylation sites vary between IgLON family members16. The organization of IgLON genes also varies across family members. While a dual-promoter structure has been identified for IgLON1-3 genes, leading to expression of two different isoforms, a single promoter regulates the expression of IgLON4 and IgLON517–21 On the extracellular surface of the plasma membrane, IgLONs form homo- and heterodimers that facilitate adhesion22–26. IgLONs have been implicated in the regulation of several neurodevelopmental processes including neurite outgrowth, axonal fasciculation, and synaptogenesis27,28. IgLON4 can promote neurite outgrowth and attract hippocampal neuron axons, and its ablation reduces hippocampal neurite outgrowth29,30 Hippocampal neurons deficient for IgLON2 showed premature sprouting and increased elongation, suggesting IgLON2 may have an inhibitory role, whereas IgLON3 likely promotes neurite initiation31. Conversely, an inhibitory effect of IgLON3 on neurite outgrowth was observed in dorsal root ganglia neurons32. Proper fasciculation of dopaminergic afferent fibers and guidance to the lateral habenula has been shown to require IgLON3 expression on lateral habenula efferent projections33,34. Furthermore, shedding of IgLONs from the cell surface by metalloproteinases promotes neurite outgrowth in both cortical and dorsal root ganglion neurons35,36. Evidence for a role of IgLONs in modulating synapse development include the observation that expression of IgLON1 or IgLON3 in rat hippocampal neuron cultures leads to increased synapse numbers, while overexpression of IgLON4 reduced synapse numbers37. While IgLONs are conserved across deuterostomes, protostomes such as Drosophila have evolved a pair of binding partners called DIPs and DPRs, whose combinatorial expression contributes to the sorting and targeting of olfactory receptor neuron axons38.
IgLONs have been linked to various developmental disorders. Mice with an IgLON2 deletion show decreased performance in the active avoidance task, suggesting a deficit in emotional learning39. Polymorphisms in the IGLON3 gene have been associated with major depressive disorder, panic disorder, and schizophrenia40–42. Interestingly, mice bearing a deletion in the IgLON3 gene showed reduced anxiety in novel environments and lowered sensitivity to stressors43. Genome-wide association studies have linked IGLON4 with increased risk of major depressive disorder44. As well, placental DNA methylation studies associated IGLON4 with increased body mass index (BMI) and neurodevelopment defects in children45. Mouse knock-out models of IgLON4 showed increased adiposity and behavioral deficits associated with neuropsychiatric disorders46–48. IgLON5, the least characterized family member, was first identified in patients with anti-IgLON5 disorder that present a variety of symptoms, including altered sleep, gait abnormalities, bulbar dysfunction, and chorea13,49,50.
Despite IgLONs’ roles in key neurodevelopmental events, there is fragmented information available in the literature about their expression in certain regions of the developing nervous system17,21,22,32,51–53. We utilized an in situ hybridization approach to create a spatiotemporal map of IgLON mRNA expression in the murine nervous system. We have analyzed one prenatal and two postnatal ages for the expression of the five IgLON family members. We demonstrate that during late prenatal and postnatal development, IgLONs are differentially expressed in specific regions of the nervous system, including the olfactory system and the developing eye. Our study provides data to inform future functional studies into the role of these adhesion molecules in the development of specific structures of the nervous system.
Results
Specificity of the cRNA probes
To examine the patterns of expression of IgLON family members in the developing nervous system, we incubated sections of embryonic day (E) 16 embryos, postnatal day (P) 0, and P15 brains with sense and antisense cRNA probes for IgLON mRNAs. cRNA probes were designed to detect expression of both the 1a and 1b transcripts for IgLON1, IgLON2, and IgLON3. Representative sections showing the specific patterns of expression of IgLON1–5 are shown in Fig. 1, and no signal was observed on sections incubated with sense probes throughout our studies (as exemplified in Figs. 4, 6, and 8). To assess the potential cross-reactivity of the cRNA probes among IgLON family members, we reasoned that the observation of a region detected by a single cRNA probe would provide evidence that cRNA probes for other family members do not cross-react with this family member. For three of the family members, IgLON2, IgLON3, and IgLON4, we were able to detect specific structures that were positive for a single IgLON family cRNA probe. For example, the glomerular cell layer of the olfactory bulb (Fig. 3F) only expressed IgLON2 at P15, the olfactory epithelium only expressed IgLON3 at E16 (Fig. 2G), and the apical ventricular zone of the cortex was only positive for IgLON4 at E16 (Fig. 4J). Cross-reactivity was also assessed by identifying specific brain regions that were positive for one IgLON and negative for another. For example, the expression of IgLON2, 3, and 5 in the eye, and of IgLON4 in the entorhinal cortex, was not detected with the IgLON1 cRNA probe, indicating that it does not bind to these mRNAs (Figs. S2D and S3B, C, E). Overall, we were able to find specific examples of brain areas that were positive for one IgLON and negative for another to cover most combinations of possible cross-reactivity between the probes as summarized in Table 1, demonstrating the specificity of the cRNA probes used.
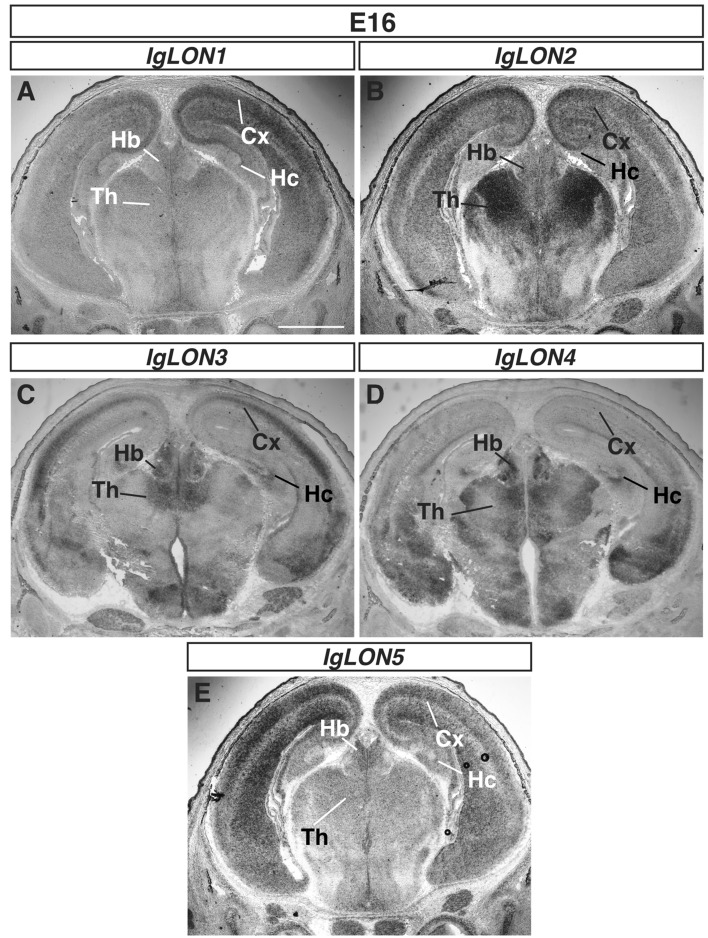
Figure 1.
mRNA expression of IgLON family members in the developing brain. In situ hybridization of coronal sections of brains from E16 embryos with antisense cRNA probes specific for IgLON1 (A), IgLON2 (B), IgLON3 (C), IgLON4 (D), and IgLON5 (E) transcripts. IgLON1–5 are widely expressed in the brain but show differential expression in certain regions, such as the cortex and thalamus. IgLON-1, 2, 3 and 5, are expressed in the developing cortex (Cx), while IgLON1–5 are expressed in the habenula (Hb) and various nuclei of thalamus (Th) (A, B, C, D, E). Scale bar = 1 mm.
Figure 4.
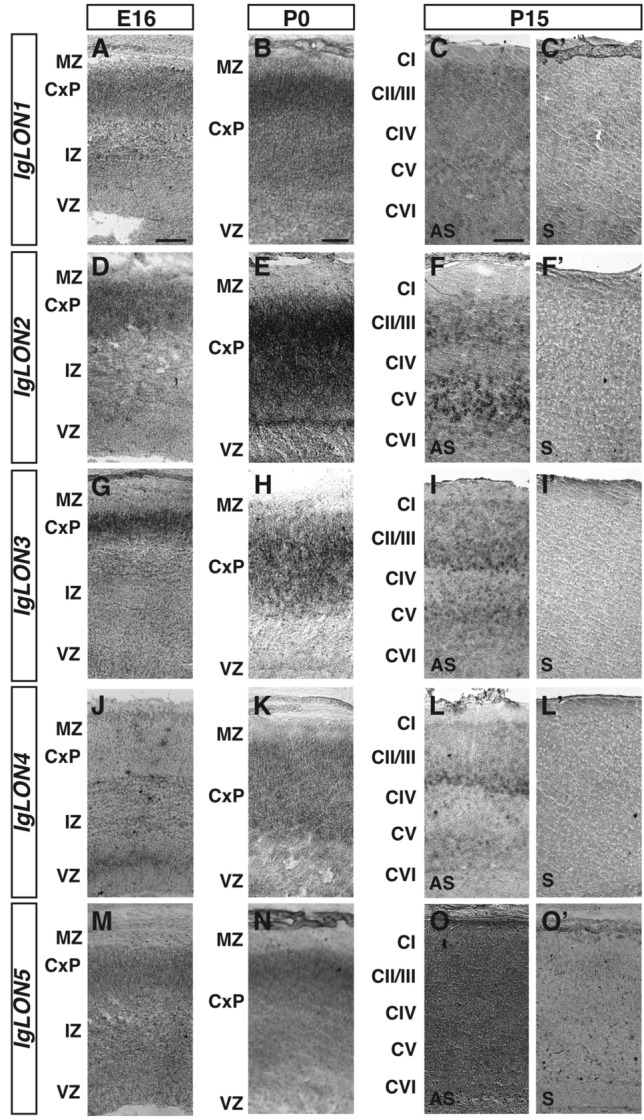
IgLONs mRNA expression during development of the cerebral cortex. In situ hybridization of coronal sections of cerebral cortex from E16, P0, and P15 mice with antisense (A–O) or sense (C′–O′) cRNA probes for IgLON genes. IgLON1 was expressed in the cortical plate (CxP) at E16 and P0 (A, B). At P15, IgLON1 mRNA was observed in cortical layers II/III (CII/CIII) and V–VI (CV–VI) (C). IgLON2 mRNA was detected in the CxP at E16 and upregulated in the CxP at P0 (D, E). At P15, IgLON2 is expressed in layers CII/III and CV–VI of the cortex (F). IgLON3 expression was detected in the CxP at E16 (G) and P0 (H), and in layers CII/III and CV-VI at P15 (I). IgLON4 mRNA expression is restricted to the apical region of the ventricular zone (VZ) at E16 and at lower levels in the intermediate zone (IZ) (J). At P0, IgLON4 expression was detected in the CxP (K). Expression at P15 was observed in layers CII/III and CV–VI (L). IgLON5 was expressed at low levels in the CxP at E16 and P0 (M, N). At P15, IgLON5 mRNA was observed at low levels throughout the cortex (O). Scale bars = 125 µm (A, D, G, J, M), 250 µm (B, E, H, K, N), and 500 µm (C, C′, F, F′, I, I′, L, L′, O, O′).
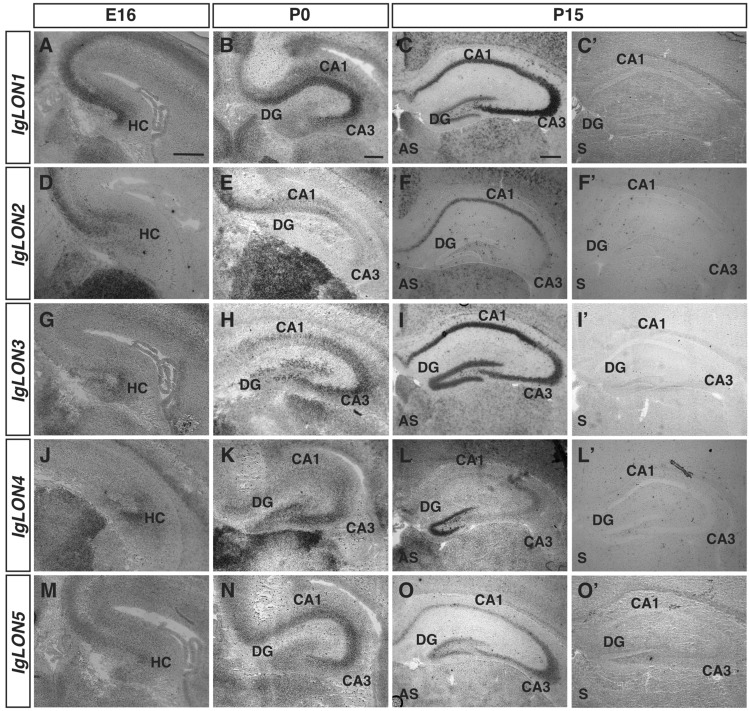
Figure 6.
IgLON mRNA expression in the developing hippocampal region. In situ hybridization of coronal sections of the hippocampal region from E16, P0, and P15 mice with antisense (A–O) or sense (C′–O′) cRNA probes for IgLON transcripts. IgLON1–5 are expressed in the developing hippocampal formation (HC) at E16 (A, D, G, J, M). IgLON1 mRNA is observed in the CA1, CA3, and dentate gyrus (DG) of the hippocampus at P0 and P15 (B, C). IgLON2 expression is restricted to the CA1 region of the hippocampus at P0 and P15 (E, F). IgLON3 is uniformly expressed in both the CA1 and CA3 regions, as well as in the DG, during postnatal development (H, I), whereas IgLON4 is expressed in the DG, CA3, and presumptive CA2 regions at P0 and P15 (K, L). IgLON5 expression peaks at P0 in the CA1 and CA3 regions but dissipates by P15 (N, O). Scale bars = 250 µm (A, B, D, E, G, H, J, K, M, N), and 500 µm (C, C′, F, F′, I, I′, L, L′, O, O′).
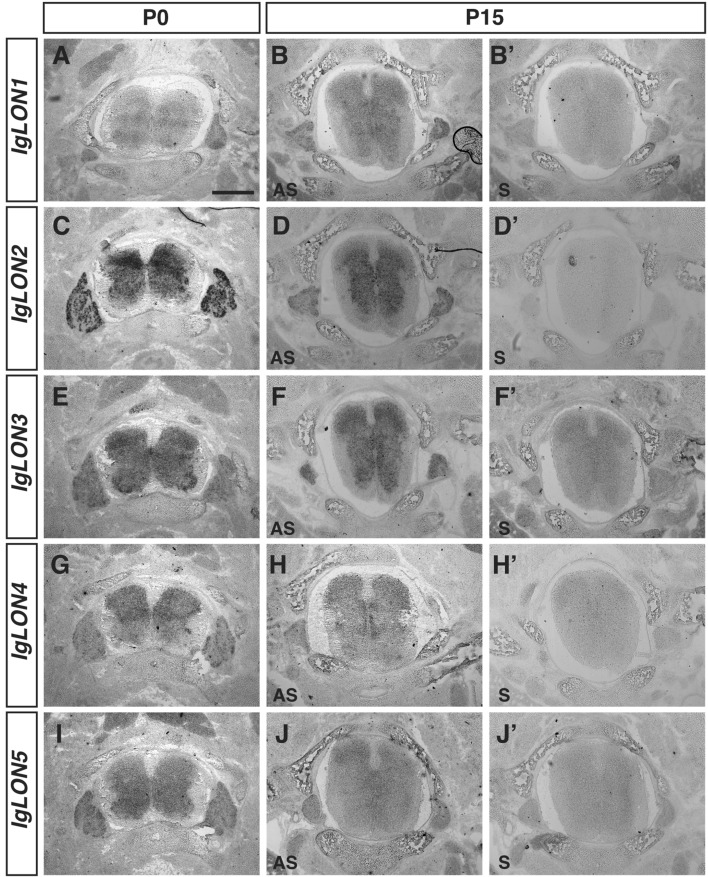
Figure 8.
IgLONs mRNA expression during spinal cord development. In situ hybridization of coronal sections of the spinal cord from P0 and P15 mice with antisense (A–J) or sense (B′–J′) cRNA probes for IgLON transcripts. IgLON1, IgLON2, IgLON3, IgLON4, and IgLON5 mRNAs are detected at various levels in the gray matter of the spinal cord at P0 and P15 (A–J). At P0, higher levels of expression of IgLON2 and IgLON4 are observed in the dorsal region of the spinal cord (C, G) while higher levels of IgLON3 and IgLON5 are detected in the ventral part of the spinal cord at P0 (I). All IgLON family members are expressed in sensory neurons within the dorsal root ganglia at P0 (A, C, E, G, I) while only IgLON1, IgLON2, and IgLON3 mRNAs could be detected in these structures at P15 (B, D, F). Scale bar = 1 mm.
Figure 3.
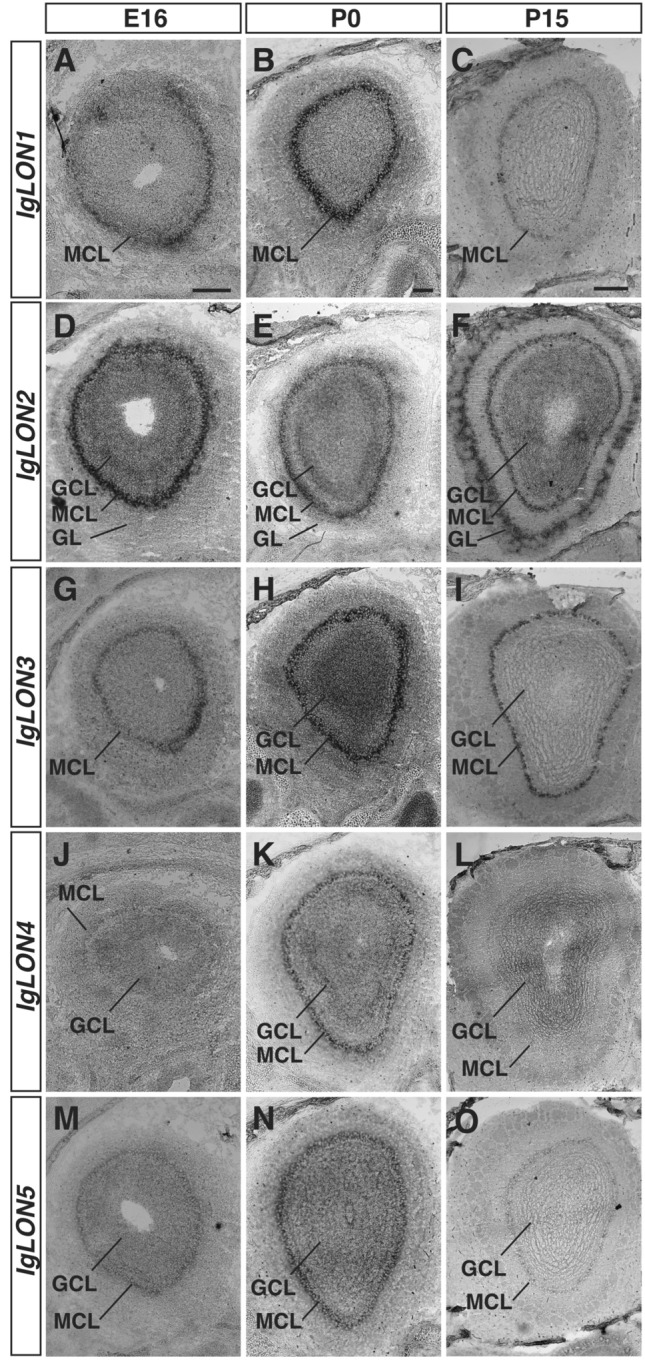
IgLONs mRNA expression in the developing olfactory bulb (OB). In situ hybridization of coronal sections of OB from embryonic day (E) 16 and postnatal day (P) 0 and 15 mice with antisense cRNA probes for IgLON transcripts. IgLON1 mRNA is detected in the mitral cell layer (MCL) of the OB at E16 and is maintained at P0 and P15 (A–C). IgLON2 mRNA expression is observed in the MCL and in the granule cell layer (GCL) of the OB at E16, P0, and P15 (D–F). By P15, IgLON2 expression is also detected in the glomerular cell layer (GL) (F). IgLON3 is expressed in the MCL at E16, P0, and P15, and could also be detected in the GCL at P0 (G–I). IgLON4 mRNA expression is observed at low levels in the GCL and in the MCL at E16 (J). Its expression in these regions is increased at P0 but is downregulated in the MCL at P15 (K, L). IgLON5 mRNA is detected in the MCL and GCL at E16 (E) and P0 (N) and at lower levels at P15 (O). Scale bars = 200 µm (A, B, D, E, G, H, J, K, M, N) and 500 µm (C, F, I, L, O).
Figure 2.
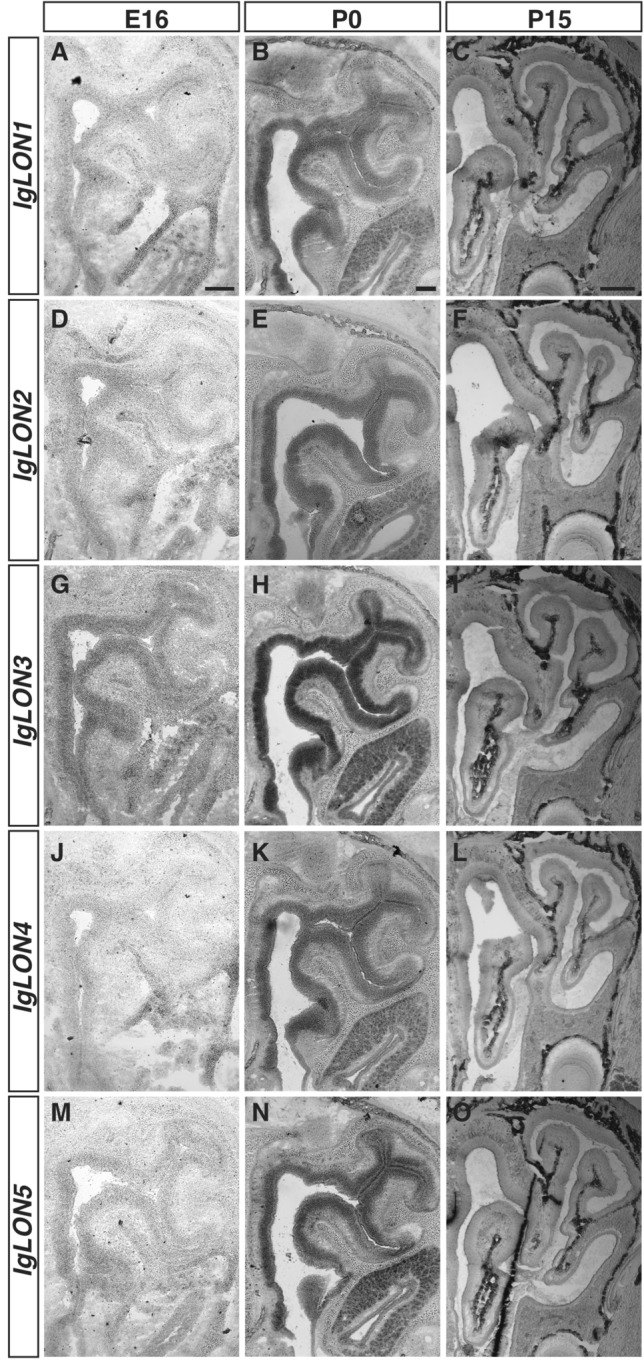
IgLONs mRNA expression in the developing olfactory epithelium (OE). In situ hybridization of coronal sections of OE from E16, P0, and 15 mice with antisense cRNA probes for IgLON transcripts. IgLON3 mRNA is detected in the OE at E16 (G), while other IgLON family members could not be detected in the OE at that age (A, D, J, M). By P0 all IgLONs are expressed throughout the OE (B, E, H, K, N) but their expression is downregulated by P15 (C, F, I, L, O). Scale bars = 200 µm (A, B, D, E, G, H, J, K, M, N) and 500 µm (C, F, I, L, O).
Table 1.
Summary of IgLON gene expression in the developing murine nervous system.
| IgLON | E16 | P0 | P15 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| I. Telencephalon | |||||||||||||||
| Eye | |||||||||||||||
| Neuroblastic layer | − | + + | + + + | − | − | ||||||||||
| Inner granular layer | − | + + | + + + | − | + + + | ||||||||||
| Olfactory system | |||||||||||||||
| Vomeronasal organ | − | + | + + | − | + + | + + + | + + + | + + + | − | + + + | − | − | − | − | − |
| Olfactory epithelium | − | − | + + | − | − | + + | + + | + + + | + + | + + | − | − | − | − | − |
| Glomerular layer | − | − | − | − | − | − | + | − | − | − | − | + + + | − | − | − |
| Mitral cell layer | + + + | + + + | + + + | − | + + | + + + | + + + | + + + | + + | + + | + + | + + + | + + + | − | + |
| Granular layer | − | + + | − | − | − | − | + + | + + + | + | + | − | + + | − | + + | + |
| Cerebral cortex | |||||||||||||||
| Cortical plate | + + | + + + | + + + | − | + + | + + | + + + | + + + | + + | + + | |||||
| Intermediate zone | + | − | − | + | − | ||||||||||
| Ventricular zone | − | − | − | + + | − | − | − | − | − | − | |||||
| CI | − | − | − | − | + | ||||||||||
| CII–CIII | + + | + + | + + + | + + | + | ||||||||||
| CIV–CVI | + | + + + | + + + | + | + | ||||||||||
| Entorhinal cortex | − | + | + + + | + + + | + | ||||||||||
| Hippocampal formation | + + | + + | + + | + + | + | ||||||||||
| CA1 | + + + | + + | + + + | − | + + | + + | + + + | + + + | − | + + | |||||
| CA3 | + + + | − | + + + | + + | + + | + + + | − | + + + | + | + + | |||||
| Dentate gyrus | + | − | + + | + + + | + | + + | − | + + + | + + + | + | |||||
| II. Diencephalon | |||||||||||||||
| Thalamus | + + | + + + | + + | + + + | + | + + | + + + | + + | + | + | + | + + | + + | + | + |
| Habenula | + + | + + | + + | + + + | + | + + | + + | + + | + | + | + + | + + | + + | + | + |
| III. Rhombencephalon | |||||||||||||||
| Cerebellum | |||||||||||||||
| Molecular layer | − | − | − | − | − | − | − | − | − | − | |||||
| Purkinje layer | + | + + + | + | − | + | + + | + + + | + | − | + | |||||
| Inner granular layer | − | + | + + | − | − | − | + | + + | + + | + + | |||||
| IV. Other | |||||||||||||||
| Spinal cord | + | + + + | + + + | + + | + + | + | + + + | + + + | + + | + | |||||
| Dorsal root ganglion | + | + + + | + + + | + | + + | + | + + + | + + + | + | + | |||||
Relative gene expression: − , not detected; + , low expression; + + , medium expression; + + + , high expression, E, embryonic; P, postnatal day.
IgLON expression in the olfactory systems
Olfactory sensory neurons (OSNs) located in the olfactory epithelium (OE) are responsible for detecting odorants. OSNs project their axons to the olfactory bulb (OB) where they coalesce into neuropil structures termed glomeruli. In these structures, OSN axons form synapses with second order neurons, the mitral/tufted cells that send projections to pyramidal cells in the olfactory cortex. In contrast, vomeronasal sensory neurons (VSNs) located in the vomeronasal organ detect chemosignals that modulate social and sexual behaviour in mice and project their axons to the accessory olfactory bulb54. We examined the expression of IgLONs in the olfactory systems at E16, when OSN and VSN axons are projecting to their targets, P0 when glomeruli start to form, and P15 when glomeruli are refined55,56. While at E16, we could only detect IgLON3 (Fig. 2A, D, G, J, M) in the OE, all IgLON family members were expressed uniformly throughout the OE by P0 (Fig. 2B, E, H, K, N). Interestingly, the levels of IgLON family members in the OE appear to decrease beyond detectable levels by P15 (Fig. 2C, F, I, L, O). In the OB (Fig. 3), IgLON1, IgLON3, and IgLON5 mRNA were mainly detected in the mitral cell layer (MCL), with IgLON3 mRNA also in the granule cell layer (GCL), which contains cells implicated in lateral inhibition of the mitral cells. While IgLON1 and IgLON3 MCL expression was maintained from E16 to P15 (Fig. 3A–C, G–I respectively), IgLON5 expression decreased by P15 (Fig. 3M–O). IgLON2 mRNA was detected in the MCL and GCL at all ages examined (Fig. 3D–F). It was also expressed in periglomerular cells present in the glomerular layer (GL) at P15 (Fig. 3F). IgLON4 was detected in the MCL and GCL at E16 and P0, but only in the GCL at P15 (Fig. 3J–L). As observed in the OE, most IgLONs are expressed in the vomeronasal organ (VNO) by P0, except for IgLON4, and their expression decreases by post-natal day 15 (Fig. S1A–O). At E16, IgLON3 and IgLON5 mRNA were detected more basally in the VNO, but both genes were expressed throughout the VNO by P0 (Fig. S1G, H, M, N). IgLON1 and IgLON2 also showed uniform expression in the VNO at P0 but could not be detected at P15 (Fig. S1B, E, C, F).
IgLON expression in the cortex
Cells from the ventricular zone and ganglionic eminences migrate during embryogenesis towards the cortical plate and develop into cortical neurons and interneurons that form the layered cortex. By E16, proliferative cells in the ventricular zone (VZ) have begun to migrate out to the cortical plate (CxP). By birth, cortical layer formation starts to emerge and further development leads to the fully formed cortex containing six neuronal layers. These layers are composed of neurons with different morphological features that serve various functions and that project their axons to a wide variety of targets, including the thalamus and the contralateral cortex57–59. Several IgLONs are expressed in the developing somatosensory cortex and some of their expression is maintained postnatally. At E16, IgLON1, IgLON2, IgLON3, and IgLON5 were detected in the cortical plate (CxP). While IgLON1, IgLON2, and IgLON5 were expressed throughout the CxP, IgLON3 expression was higher in the apical region of the CxP (Fig. 4A, D, G, M). In contrast, IgLON4 expression was mainly observed in the ventricular zone (VZ), where dividing cells are located, as well as at low levels in the intermediate zone60 (Fig. 4J). The expression of several IgLON family members was maintained as development proceeds into early post-natal stages. At P0, all family members were detected in the cortical plate. While IgLON1 expression appeared higher in the apical region of the CxP, IgLON2, IgLON3, IgLON4, and IgLON5 were detected throughout the CxP (Fig. 4B, E, H, K, N). At P15, all IgLON family members could be detected in the cortex (Fig. 4C, F, I, L, O). IgLON1 was weakly detected in cortical layers II-III (CII-CIII) and V-VI (CV-VI), while IgLON2, IgLON3, and IgLON4 mRNA were detected at high levels in layers CII-CIII and CV-VI, and IgLON5 showed low levels of expression throughout the cortex (Fig. 4C, F, I, L, O).
IgLON expression in the diencephalon
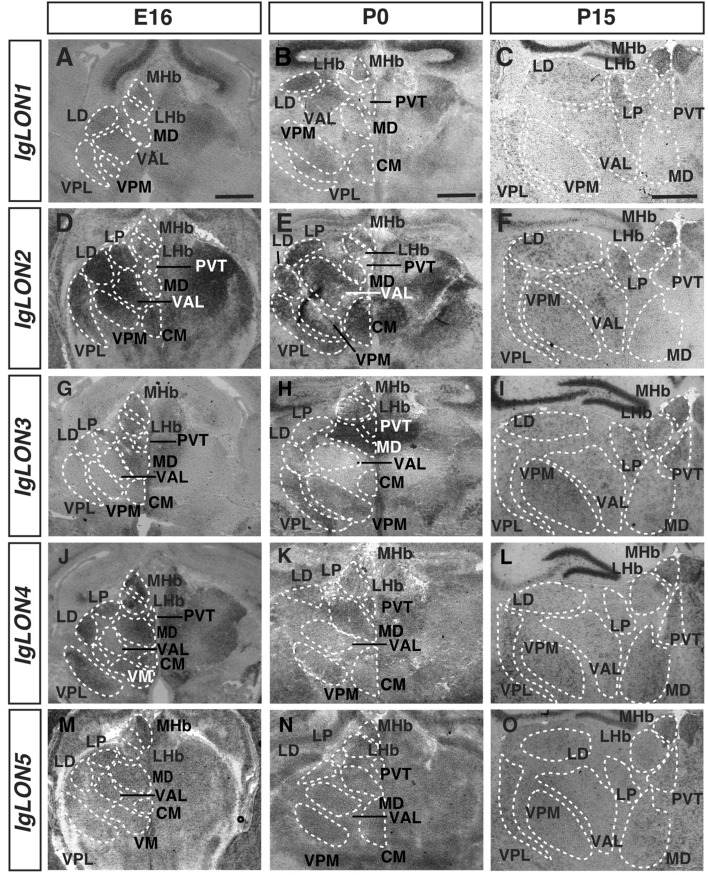
The thalamus is the central hub for sensory input, and largely acts as a relay station to the cerebral cortex61. The thalamic complex is composed of various thalamic nuclei, the habenula, and the hypothalamus62. The habenula is divided into medial (MHb) and lateral (LHb) regions, and functions mainly in relaying input received from the basal ganglia and forebrain to monoaminergic brain structures such as the ventral tegmental area63. The modality-specific nuclei in the thalamus are largely responsible for relaying sensory information to the cortex, while hypothalamic nuclei function in innate survival behaviors such as feeding and fighting64–66. IgLON1 mRNA expression was detected in the MHb, the ventral anterolateral thalamic nucleus (VAL), and the laterodorsal thalamic nucleus (LD) at E16, and P15 (Fig. 5A, C). At P0, IgLON1 could be detected in these nuclei as well as in the central posteromedial thalamic nucleus (VPM) and central medial thalamic nucleus (CM) (Fig. 5B). IgLON2 showed expression at E16 and P0 in the LHb, MHb, LD, lateroposterior thalamic nucleus (LP), VAL, VPM, CM, the ventral posterolateral thalamic nucleus (VPL) and medial dorsal thalamic nucleus (MD) (Fig. 5D, E). The expression of IgLON2 was maintained in these regions at P15 (Fig. 5F). At all ages analyzed, IgLON3 mRNA was detected in the LHb, MHb, MD, and paraventricular thalamic nucleus (PVT) (Fig. 5G–I). At P0 and P15, some IgLON3 expression was also detected in the LD, VPM, VPL, and LP (Fig. 5H, I). IgLON4 mRNA expression was extensive at E16; mRNA was detected in the MHb, LHb, PVT, LP, LD, MD, CM, VPL, and ventromedial thalamic nucleus (VM) (Fig. 5J). IgLON4 was also detected in all the above nuclei at ages P0 and P15 (Fig. 5K, L). IgLON5 mRNA was observed in the MHb at all ages examined. Low levels of expression were also detected in the LHb and in most thalamic nuclei (Fig. 5M–O).
Figure 5.
IgLONs mRNA expression in the developing thalamus. In situ hybridization of coronal sections of the thalamus from E16, P0, and 15 mice with antisense cRNA probes for IgLON transcripts. IgLON1 mRNA was detected in the medial habenula (MHb), laterodorsal thalamic nucleus (LD), and ventral anterolateral thalamic nucleus (VAL) at all ages examined. IgLON1 mRNA was also detected in additional nuclei, such as the ventral posteromedial thalamic nucleus (VPM) and ventral posterolateral thalamic nucleus (VPL) at P0 and P15, respectively (A–C). IgLON2 showed expression in the lateral habenula (LHb), the lateroposterior thalamic nucleus (LP), LD, VAL, VPM, VPL, and the mediodorsal thalamic nucleus (MD) at all ages analyzed (D–F). At E16 and P0, IgLON2 could also be detected in the centromedial thalamic nucleus (CM), and expression in the PVT was observed at P15 (D–F). IgLON3 was detected in the MHb, LHb, PVT, and MD (G–I). At P15, expression could also be detected in the LD, VPM, and VPL (I). At E16, IgLON4 showed expression in the LHb, MHb, PVT, LP, LD, MD, CM, VPL, and the ventromedial thalamic nucleus (VM) (J). IgLON4 was also observed in the majority of these nuclei at P0 and P15 (K, L). IgLON5 mRNA expression was visible in the MHb at all ages analyzed. Low levels of expression were also detected in most thalamic nuclei. (M–O). All scale bars = 500 µm.
IgLON expression in the hippocampus
The hippocampus functions in memory formation and spatial navigation. The hippocampal formation receives input from the entorhinal cortex to the granule cells of the dentate gyrus (DG). The DG cell axons project to CA3 pyramidal neurons as a bundle known as the mossy fiber pathway. The CA3 pyramidal cell axons in turn project to CA1 pyramidal neurons via the Schaffer collateral pathway67,68. Signals are then either relayed back to the entorhinal cortex or to the cerebral cortex. Our analysis of the expression patterns of IgLONs in the hippocampal formation revealed that all members of this family are expressed in various subregions. At E16, when it remains difficult to identify some subregions of the hippocampal formation, IgLON1 to 5 mRNA could be detected (Fig. 6A, D, G, J, M). At P0, IgLON1 was detected in the CA1-CA3 regions and in the DG (Fig. 6B). This expression was maintained at P15, although higher levels of expression was observed in the CA3 at that age (Fig. 6C). IgLON2 was detected in the CA1 region at both P0 and P15 (Fig. 6E, F). IgLON3 was expressed in the CA1–CA3 regions and in the DG at these ages (Fig. 6H, I). IgLON4 was expressed in the DG, CA3, and presumptive CA2 region of the hippocampus at P0 and P15 (Fig. 6K, L). IgLON5 was detected in the DG and in the CA1-CA3 regions at P0 and P15 (Fig. 6N, O). IgLON2, IgLON3, IgLON4, and IgLON5 were also expressed at varying levels in the entorhinal cortex, which is a major source of inputs to the dentate gyrus of the hippocampus (Fig. S2B–E)69.
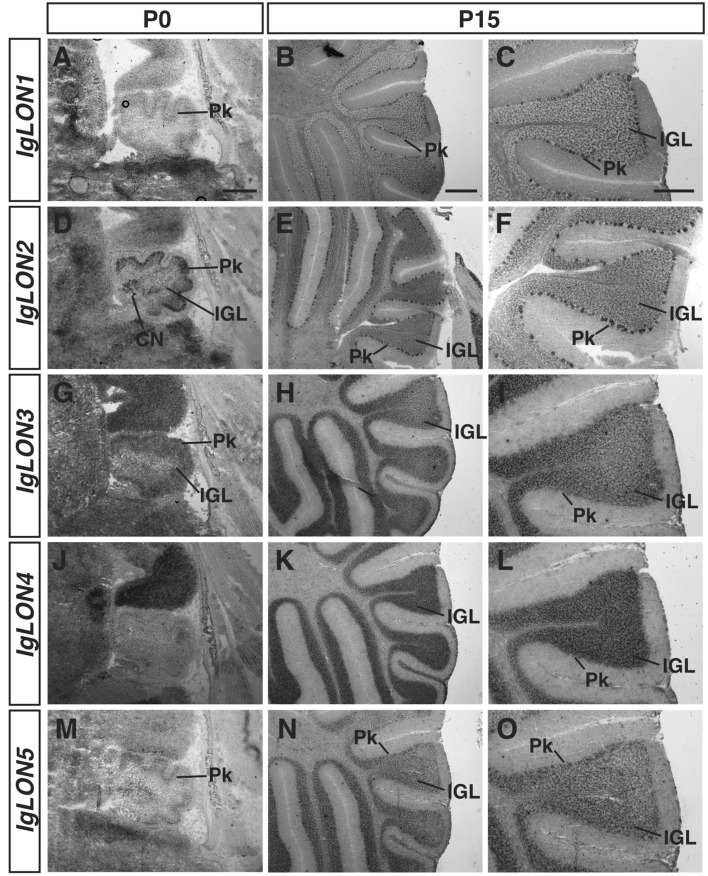
IgLON expression in the cerebellum
The cerebellum is important for controlling motor coordination70. The dense layer of excitatory granule cells receives input from various regions of the brain and project to Purkinje cells, which in turn mainly project to cerebellar nuclei. The different cellular and molecular layers of the developing cerebellum begin to be distinguishable around age P0 and are well-defined by age P15 (Fig. 7). IgLON1 mRNA was detectable in the Purkinje layer (Pk) at P0 and P15 (Fig. 7A–C). IgLON2 mRNA was detected in both the inner granular layer and in the Pk at P0 and P15 (Fig. 7D–F). IgLON3 showed strong expression in the inner granular layer (IGL) and weak expression in the Pk (Fig. 7G–I). IgLON4 mRNA could be detected at low levels in the Purkinje layer at P0 and P15, and its expression increased in the inner granular layer by P15 (Fig. 7J–L). IgLON5 was expressed in the Pk at P0 and in the Pk and IGL at P15 (Fig. 7M–O).
Figure 7.
IgLONs mRNA expression in the cerebellum. In situ hybridization of sagittal sections from P0 and P15 mouse brains with antisense cRNA probes for IgLON transcripts. IgLON1 was expressed at low levels in the Purkinje cell layer (Pk) at P0 and P15 (A–C). IgLON2 showed expression in the Pk and in the internal granule layer (IGL) at P0 and P15 (D–F). IgLON3 mRNA could be detected in the IGL and at low levels in the Pk at P0 and P15 (G–I). IgLON4 showed weak expression in the Pk at P0 (J) but was restricted to the IGL by P15 (K, L). IgLON5 mRNA expression was detected in the Pk layer at P0 and in both Pk and IGL layers at P15 (M–O). Scale bars = 200 µm (A, C, D, F, G, I, J, L, M, O) and 500 µm (B, E, H, K, N).
IgLON expression in the spinal cord and dorsal root ganglia
In the developing spinal cord, IgLONs are expressed in the grey matter and in the dorsal root ganglia (DRG) containing sensory neurons. IgLON1, IgLON3, and IgLON5 showed widespread expression in the gray matter at P0 and P15, and in DRGs at both ages (Fig. 8A, B, E, F, I, J). While IgLON3 is expressed throughout the grey matter, higher levels of expression are observed in the ventral part of the spinal cord containing motor neurons at E16 (Fig. 8E). IgLON2 was also expressed in the grey matter but lower levels of expression were observed in the most dorsal lamina at P0 (Fig. 8D). IgLON4 mRNA was enriched in the dorsal part of the spinal cord at both P0 and P15 (Fig. 8G, H) where sensory interneuron cell bodies are located (Fig. 8G, H). All IgLON family members were also detected in the cell bodies of DRG neurons. While IgLON1, IgLON2, IgLON3, and IgLON5 appeared to be expressed in the majority of DRG neurons, IgLON4 expression was detected in a subset of DRG neurons. Whether IgLON4 is expressed in a subset of DRG neurons with a specific sensory modality remains to be established.
IgLON expression in the developing eye
At E16, the retina is composed of a developing inner nuclear layer (INL), a retinal ganglion cell layer (GCL), and a proliferative neuroblastic cell layer (NBL) in which photoreceptors and horizontal cells will form (Kaufman, 1992). IgLON1 and IgLON4 mRNA was not detected in any eye structure at age E16 (Fig. S3A, D). IgLON2 and IgLON3 showed expression in the GCL, presumptive INL, and in the NBL (Fig. S3B, C). In addition, IgLON3 was highly expressed in the lens of the eye (Fig. S3C). IgLON5 only showed expression in the GCL and presumptive INL (Fig. S3E).
Discussion
Cell adhesion molecules modulate a wide variety of processes during neuronal circuit development and maintenance. Although IgLON family members have been implicated in the regulation of some of these processes, including axonal growth and synapse formation, our understanding of their role in the development of neuronal circuitry in specific regions of the brain remains limited. To begin to gain insight into the roles of IgLONs in the development of specific regions of the nervous system, we have performed a detailed analysis of their patterns of expression in the murine nervous system. Our results show that IgLON family members are expressed in distinct patterns of expression and that their expression is temporally regulated during development. In multiple regions of the brain, several IgLON family members are expressed in a complementary fashion, suggesting a functional diversity or neuronal subtype-specific roles for the different IgLONs. Furthermore, in addition to being high during embryogenesis, IgLON expression is maintained postnatally in several structures analyzed, suggesting they also contribute to processes that take place later in development, including synapse formation and refinement.
During formation of olfactory glomerular maps, the differential expression of cell adhesion molecules on olfactory and vomeronasal receptor neurons contributes to the coalescence of subsets of axons expressing the same olfactory or vomeronasal receptors into specific glomeruli of the olfactory and accessory olfactory bulbs, respectively71. Although members of the Kirrel family of cell adhesion molecules have been shown to play an important role in this process, it is likely that additional cell adhesion molecules also modulate axonal coalescence during olfactory map formation. In Drosophila, the IgLON homologs, DIPs, are differentially expressed in olfactory receptor neurons and have been proposed to serve as tags to promote coalescence of axons expressing the same olfactory receptor during formation of glomeruli in the mushroom body72,73. Our examination of the patterns of expression of IgLONs in the mouse olfactory and vomeronasal epithelia revealed that most family members are highly expressed in olfactory and vomeronasal sensory neurons when axons are projecting and innervating their targets. However, we did not observe differential expression of IgLON family members among these neurons, suggesting they are unlikely to act as recognition tags for these projecting axons.
In contrast to the olfactory epithelium, IgLON family members are differentially expressed in specific regions of several brain structures, such as the thalamus. It remains possible that the differential expression of IgLONs in the thalamus serves to modulate the fasciculation and segregation of different populations of projecting axons. In keeping with this possibility, proper fasciculation of dopaminergic afferent fibers and guidance to the lateral habenula requires IgLON3 expression on lateral habenula efferent projections33. IgLONs are also differentially expressed in regions of the hippocampus. While IgLON2 expression appears restricted to the CA1–CA2 region, IgLON1 is more highly expressed in the CA3 region than in the CA1–CA2 region, and IgLON4 is more highly expressed in the dentate gyrus. In vitro, expression of IgLON1 or IgLON3 in hippocampal neurons increased synapse numbers while expression of IgLON4 reduced synapse numbers37. The differential expression of IgLONs in the hippocampus may therefore contribute to synapse specificity by promoting or inhibiting synapse formation between specific populations of neurons in the hippocampus. The expression of IgLONs may also regulate axonal growth in this system as cell surface cleavage of IgLON4 in cultures of primary neurons can promote neurite outgrowth30,74,75.
The patterns of expression we observe mostly correlate with previously observed expression profiles in specific regions of the brain described in the literature17,21,22,32,51,52,76,77. However, while we observed expression of IgLON3/Lsamp in the developing cortex at E16, we did not detect its expression in the ventricular zone as previously reported for Lsamp 1b20. In contrast to our in situ hybridization approach, Philips et al. examined the expression of Lsamp 1b using a very sensitive Xgal enzymatic reaction on brain sections from mice carrying an insertion of a LacZ gene cassette in-frame with the Lsamp 1b promoter. It is therefore likely that our inability to detect IgLON3 expression in the ventricular zone is due to a lower sensitivity of the in situ hybridization approach we have used. Alternatively, it is possible that the Lsamp 1b transcript is expressed in the ventricle at earlier time-points during development and downregulated by E16, but that residual β-galactosidase protein is still present at that age in the Lsamp 1b-lacZ mouse, leading to an Xgal-positive signal in this structure.
In conclusion, our survey of IgLON expression shows that IgLONs are widely expressed in the developing nervous system, with partially overlapping, yet distinct, patterns of expression. These analyses will permit the future development of in vivo assays to address the function of these proteins in the development and maintenance of specific regions of the nervous system using available and newly developed genetically-modified mouse models.
Methods
Animals and ethics
E16 mouse embryos were obtained from timed-pregnant females of the CD1 strain purchased from Charles River. P0 and P15 CD1 mouse pups were obtained from Charles River. The date of vaginal plug was considered E0, and the day of birth was considered P0. Heads and embryos were cryoprotected with Tissue-Tek O.C.T. compound (Miles, Elkhart,IN) and flash-frozen in dry ice cooled 2-methylbutane. Tissue was stored at − 80 °C until cryosectioning. All animal procedures used were approved by the Neuro Animal Care Committee and McGill University, in accordance with the guidelines of the Canadian Council on Animal Care. Data and methods are reported in accordance with the ARRIVE guidelines78.
Riboprobe synthesis
For each IgLON member, digoxigenin-labeled cRNA probes with either antisense or sense orientation were prepared by in vitro transcription using DIG labelling mix (Sigma,USA). Probe sequences were PCR-amplified from cDNAs prepared from mouse brain cDNA using primers designed based on sequences from the Allen Brain Atlas and Gene Paint Database (see Supplemental Material for probe sequences) and cloned into a pBluescript vector. cRNA probes were designed to detect expression of both the 1a and 1b transcripts for IgLON1, IgLON2, and IgLON3. Following in vitro transcription, the riboprobes were partially hydrolyzed in a 10 mM DTT and 200 mM NaHCO3/Na2CO3 solution (pH 11) for 25 min at 60 °C79. Hydrolysis was stopped by addition of 100 mM acetic acid, and cRNA probes were precipitated by addition of 1/10th volume 4 M LiCl and ethanol. Precipitated cRNA fragments were resuspended in diethylpyrocarbonate (DEPC)-treated water and stored at − 80 °C.
In situ hybridization
In situ hybridization was performed as previously described80. 20 µm thick sections of fresh-frozen heads or embryos were thaw-mounted onto Fisherbrand Superfrost Plus microscope slides (Fisher Scientific, Hampton, NH) and allowed to dry for 1 h. Sections were incubated in diethyl pyrocarbonate (DEPC)-treated 4% paraformaldehyde in 0.1 M phosphate-buffered isotonic saline (PBS; pH 7.4) for 20 min and washed 3 times in 1 × PBS for 5 min each, followed by acetylation in 0.25% acetic anhydride in 1% triethanolamine solution for 10 min. Sections were washed in 1X PBS and 1X standard saline citrate (SSC; pH 7.4) and pre-hybridized in hybridization solution (50% formamide, 5 × Denhardt's solution, 5 × SSC, 200 µg/ml baker’s yeast tRNA) in a humidified chamber at 60 °C for 2 h to overnight. Slides were then incubated overnight at 60 °C in hybridization solution containing approximately 100 ng/ml of IgLON cRNA probes.
The following day, slides were washed in 5 × SSC at 60 °C for 5 min and 2 × SSC at 60 °C for 1 min. The slides were then incubated in 0.2 × SSC in 50% formamide solution at 60 °C for 30 min, followed by a 5-min wash in Tris-buffered saline (TBS; 100 mM Tris-HC1, pH 7.5, 150 mM NaCl) and a 1-h block in 1% blocking reagent (Sigma) in TBS. After a 5-min wash in TBS, slides were incubated with anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (1:3000) for 3 h, washed in TBS, and subjected to a color reaction overnight at room temperature. Slides were coverslip mounted with Mowiol 4-88 (Calbiochem, San Diego, CA) and sections examined on a Zeiss Axio imager upright microscope. Each in situ hybridization experiment was repeated a minimum of three times on a minimum of three different samples to confirm the patterns of expression observed.
Supplementary Information
Acknowledgements
We thank members of the Cloutier lab for helpful discussions and technical advice. This work was supported by the Canadian Institutes for Health Research and the Natural Sciences and Engineering Research Council of Canada.
Author contributions
S.F. performed the experiments. S.F., R.R., and J.-F.C. designed the experiments, wrote, and edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-97768-5.
References
- 1.Arikkath J, Reichardt LF. Cadherins and catenins at synapses: Roles in synaptogenesis and synaptic plasticity. Trends Neurosci. 2008;31:487–494. doi: 10.1016/j.tins.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Vactor D. Adhesion and signalling in axonal fasciculation. Curr. Opin. Neurobiol. 1998;8:80–86. doi: 10.1016/S0959-4388(98)80011-1. [DOI] [PubMed] [Google Scholar]
- 3.Pollerberg GE, Thelen K, Theiss MO, Hochlehnert BC. The role of cell adhesion molecules for navigating axons: Density matters. Mech. Dev. 2013;130:359–372. doi: 10.1016/j.mod.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Dodd J, Jessell TM. Axon guidance and the patterning of neuronal projections in vertebrates. Science. 1988;80(242):692–699. doi: 10.1126/science.3055291. [DOI] [PubMed] [Google Scholar]
- 5.Mosca TJ, Hong W, Dani VS, Favaloro V, Luo L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 2012;484:237–241. doi: 10.1038/nature10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young TR, Leamey CA. Teneurins: Important regulators of neural circuitry. Int. J. Biochem. Cell Biol. 2009;41:990–993. doi: 10.1016/j.biocel.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Hong W, Mosca TJ, Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484:201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prince JEA, Brignall AC, Cutforth T, Shen K, Cloutier J-F. Kirrel3 is required for the coalescence of vomeronasal sensory neuron axons into glomeruli and for male-male aggression. Development. 2013;140:2398–2408. doi: 10.1242/dev.087262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaddadi N, et al. Kirrel2 is differentially required in populations of olfactory sensory neurons for the targeting of axons in the olfactory bulb. Development. 2019;146:dev173310. doi: 10.1242/dev.173310. [DOI] [PubMed] [Google Scholar]
- 10.Brignall AC, et al. Loss of Kirrel family members alters glomerular structure and synapse numbers in the accessory olfactory bulb. Brain Struct. Funct. 2018;223:307–319. doi: 10.1007/s00429-017-1485-0. [DOI] [PubMed] [Google Scholar]
- 11.Rosen CL, Lisanti MP, Salzer JL. Expression of unique sets of GPI-linked proteins by different primary neurons in vitro. J. Cell Biol. 1992;117:617–627. doi: 10.1083/jcb.117.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Struyk AF, et al. Cloning of neurotrimin defines a new subfamily of differentially expressed neural cell adhesion molecules. J. Neurosci. 1995;15:2141–2156. doi: 10.1523/JNEUROSCI.15-03-02141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabater L, et al. A novel NREM and REM parasomnia with sleep breathing disorder associated with antibodies against IgLON5: A case series, pathological features, and characterization of the antigen. Lancet Neurol. 2014;131016:575–586. doi: 10.1016/S1474-4422(14)70051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schofield PR, et al. Molecular characterization of a new immunoglobulin superfamily protein with potential roles in opioid binding and cell contact. EMBO J. 1989;8:489–495. doi: 10.1002/j.1460-2075.1989.tb03402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karagogeos D. Neural Gpi-anchored cell adhesion molecules. Front. Biosci. 2003;8:1304–1320. doi: 10.2741/1214. [DOI] [PubMed] [Google Scholar]
- 16.Itoh S, et al. Glycosylation analysis of IgLON family proteins in rat brain by liquid chromatography and multiple-stage mass spectrometry. Biochemistry. 2008;47:10132–10154. doi: 10.1021/bi8009778. [DOI] [PubMed] [Google Scholar]
- 17.Vanaveski T, et al. Promoter-specific expression and genomic structure of Iglon family genes in mouse. Front. Neurosci. 2017;11:1–15. doi: 10.3389/fnins.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinla I, et al. Gene expression patterns and environmental enrichment-induced effects in the hippocampi of mice suggest importance of Lsamp in plasticity. Front. Neurosci. 2015;9:1–9. doi: 10.3389/fnins.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karis K, et al. Altered expression profile of IgLON family of neural cell adhesion molecules in the dorsolateral prefrontal cortex of schizophrenic patients. Front. Mol. Neurosci. 2018;11:1–12. doi: 10.3389/fnmol.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philips MA, et al. Lsamp is implicated in the regulation of emotional and social behavior by use of alternative promoters in the brain. Brain Struct. Funct. 2015;220:1381–1393. doi: 10.1007/s00429-014-0732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagomäe T, Singh K, Philips M, Jayaram M, Seppa K. Alternative promoter use governs the expression of IgLON cell adhesion molecules in histogenetic fields of the embryonic mouse brain. Int. J. Mol. Sci. 2021;22:6955. doi: 10.3390/ijms22136955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodge AP, Howard MR, McNamee CJ, Moss DJ. Co-localisation, heterophilic interactions and regulated expression of IgLON family proteins in the chick nervous system. Mol. Brain Res. 2000;82:84–94. doi: 10.1016/S0169-328X(00)00184-4. [DOI] [PubMed] [Google Scholar]
- 23.Reed J. Diglons are heterodimeric proteins composed of IgLON subunits, and Diglon-CO inhibits neurite outgrowth from cerebellar granule cells. J. Cell Sci. 2004;117:3961–3973. doi: 10.1242/jcs.01261. [DOI] [PubMed] [Google Scholar]
- 24.Akeel M, McNamee CJ, Youssef S, Moss D. DIgLONs inhibit initiation of neurite outgrowth from forebrain neurons via an IgLON-containing receptor complex. Brain Res. 2011;1374:27–35. doi: 10.1016/j.brainres.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 25.McNamee CJ, Reed JE, Howard MR, Lodge AP, Moss DJ. Promotion of neuronal cell adhesion by members of the IgLON family occurs in the absence of either support or modification of neurite outgrowth. J. Neurochem. 2002;80:941–948. doi: 10.1046/j.0022-3042.2002.00798.x. [DOI] [PubMed] [Google Scholar]
- 26.Mcnamee CJ, Youssef S, Moss D. IgLONs form heterodimeric complexes on forebrain neurons. Cell Biochem. Funct. 2011;29:114–119. doi: 10.1002/cbf.1730. [DOI] [PubMed] [Google Scholar]
- 27.Gil OD, Zanazzi G, Struyk AF, Salzer JL. Neurotrimin mediates bifunctional effects on neurite outgrowth via homophilic and heterophilic interactions. J. Neurosci. 1998;18:9312–9325. doi: 10.1523/JNEUROSCI.18-22-09312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada M, et al. Synaptic adhesion molecule OBCAM; synaptogenesis and dynamic internalization. Brain Res. 2007;1165:5–14. doi: 10.1016/j.brainres.2007.04.062. [DOI] [PubMed] [Google Scholar]
- 29.Schäfer M, Bräuer AU, Savaskan NE, Rathjen FG, Brümmendorf T. Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Mol. Cell. Neurosci. 2005;29:580–590. doi: 10.1016/j.mcn.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Singh K, et al. Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-019-41991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh K, et al. The combined impact of IgLON family proteins Lsamp and Neurotrimin on developing neurons and behavioral profiles in mouse. Brain Res. Bull. 2018;140:5–18. doi: 10.1016/j.brainresbull.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Gil OD, et al. Complementary expression and heterophilic interactions between IgLON family members neurotrimin and LAMP. J. Neurobiol. 2002;51:190–204. doi: 10.1002/neu.10050. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt ERE, et al. Subdomain-mediated axon–axon signaling and chemoattraction cooperate to regulate afferent innervation of the lateral habenula. Neuron. 2014;83:372–387. doi: 10.1016/j.neuron.2014.05.036. [DOI] [PubMed] [Google Scholar]
- 34.Zhukareva V, Levitt P. The limbic system-associated membrane protein (LAMP) selectively mediates interactions with specific central neuron populations. Development. 1995;121:1161–1172. doi: 10.1242/dev.121.4.1161. [DOI] [PubMed] [Google Scholar]
- 35.Sanz R, Ferraro GB, Fournier AE. IgLON cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. J. Biol. Chem. 2015;290:4330–4342. doi: 10.1074/jbc.M114.628438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz RL, Ferraro GB, Girouard MP, Fournier AE. Ectodomain shedding of limbic system-associated membrane protein (LSAMP) by ADAM Metallopeptidases promotes neurite outgrowth in DRG neurons. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hashimoto T, Maekawa S, Miyata S. IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem. Funct. 2008;27:496–498. doi: 10.1002/cbf.1600. [DOI] [PubMed] [Google Scholar]
- 38.Cheng S, et al. Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence. Proc. Natl. Acad. Sci. 2019;116:201818631. doi: 10.1073/pnas.1818631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazitov T, Bregin A, Philips MA, Innos J, Vasar E. Deficit in emotional learning in neurotrimin knockout mice. Behav. Brain Res. 2017;317:311–318. doi: 10.1016/j.bbr.2016.09.064. [DOI] [PubMed] [Google Scholar]
- 40.Koido K, et al. Associations between LSAMP gene polymorphisms and major depressive disorder and panic disorder. Transl. Psychiatr. 2012;2:e152. doi: 10.1038/tp.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koido, et al. Associations between polymorphisms of LSAMP gene and schizophrenia. Psychiatr. Res. 2014;215:797–798. doi: 10.1016/j.psychres.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Innos J, et al. Lower anxiety and a decrease in agonistic behaviour in Lsamp-deficient mice. Behav. Brain Res. 2011;217:21–31. doi: 10.1016/j.bbr.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 43.Innos J, et al. Deletion of the Lsamp gene lowers sensitivity to stressful environmental manipulations in mice. Behav. Brain Res. 2012;228:74–81. doi: 10.1016/j.bbr.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, et al. Integrating genome-wide association study and expression quantitative trait loci data identifies NEGR1 as a causal risk gene of major depression disorder. J. Affect. Disord. 2020;265:679–686. doi: 10.1016/j.jad.2019.11.116. [DOI] [PubMed] [Google Scholar]
- 45.Breton E, et al. Placental NEGR1 DNA methylation is associated with BMI and neurodevelopment in preschool-age children. Epigenetics. 2020;15:323–335. doi: 10.1080/15592294.2019.1666653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh K, et al. Neuronal growth and behavioral alterations in mice deficient for the psychiatric disease-associated Negr1 gene. Front. Mol. Neurosci. 2018;11:1–14. doi: 10.3389/fnmol.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noh K, et al. Negr1 controls adult hippocampal neurogenesis and affective behaviors. Mol. Psychiatr. 2019;1:1189–1205. doi: 10.1038/s41380-018-0347-3. [DOI] [PubMed] [Google Scholar]
- 48.Joo Y, Kim H, Lee S, Lee S. Neuronal growth regulator 1-deficient mice show increased adiposity and decreased muscle mass. Int. J. Obes. 2019;43:1769–1782. doi: 10.1038/s41366-019-0376-2. [DOI] [PubMed] [Google Scholar]
- 49.Sabater L, Planagumà J, Dalmau J, Graus F. Cellular investigations with human antibodies associated with the anti-IgLON5 syndrome. J. Neuroinflamm. 2016;13:1–12. doi: 10.1186/s12974-016-0689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaig C, et al. HLA and microtubule-associated protein tau H1 haplotype associations in anti-IgLON5 disease. Neurol. Neuroimmunol. Neuroinflamm. 2019;6:1–7. doi: 10.1212/NXI.0000000000000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bräuer AU, et al. IG-molecule kilon shows differential expression pattern from LAMP in the developing and adult rat hippocampus. Hippocampus. 2000;10:632–644. doi: 10.1002/1098-1063(2000)10:6<632::AID-HIPO1001>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 52.Pimenta AF, Fischer I, Levitt P. cDNA cloning and structural analysis of the human limbic-system-associated membrane protein (LAMP) Gene. 1996;170:189–195. doi: 10.1016/0378-1119(96)84698-1. [DOI] [PubMed] [Google Scholar]
- 53.Miyata S, Taguchi K, Maekawa S. Dendrite-associated opioid-binding cell adhesion molecule localizes at neurosecretory granules in the hypothalamic magnocellular neurons. Neuroscience. 2003;122:169–181. doi: 10.1016/S0306-4522(03)00609-2. [DOI] [PubMed] [Google Scholar]
- 54.Brignall AC, Cloutier JF. Neural map formation and sensory coding in the vomeronasal system. Cell. Mol. Life Sci. 2015;72:4697–4709. doi: 10.1007/s00018-015-2029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katreddi RR, Forni PE. Mechanisms underlying pre- and postnatal development of the vomeronasal organ. Cell. Mol. Life Sci. 2021;78:5069–5082. doi: 10.1007/s00018-021-03829-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kam JWK, Raja R, Cloutier JF. Cellular and molecular mechanisms regulating embryonic neurogenesis in the rodent olfactory epithelium. Int. J. Dev. Neurosci. 2014;37:76–86. doi: 10.1016/j.ijdevneu.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 57.Price DJ, et al. The development of cortical connections. Eur. J. Neurosci. 2006;23:910–920. doi: 10.1111/j.1460-9568.2006.04620.x. [DOI] [PubMed] [Google Scholar]
- 58.Canty AJ, Murphy M. Molecular mechanisms of axon guidance in the developing corticospinal tract. Prog. Neurobiol. 2008;85:214–235. doi: 10.1016/j.pneurobio.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 60.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Constantinople CM, Bruno RM. Deep cortical layers are activated directly by thalamus (supplementary materials) Science. 2013;340:1591–1594. doi: 10.1126/science.1236425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagalski A, et al. Molecular anatomy of the thalamic complex and the underlying transcription factors. Brain Struct. Funct. 2016;221:2493–2510. doi: 10.1007/s00429-015-1052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallace ML, et al. Anatomical and single-cell transcriptional profiling of the murine habenular complex. Elife. 2020;9:1–22. doi: 10.7554/eLife.51271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El-Boustani S, et al. Anatomically and functionally distinct thalamocortical inputs to primary and secondary mouse whisker somatosensory cortices. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-17087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sternson SM. Hypothalamic survival circuits: Blueprints for purposive behaviors. Neuron. 2013;77:810–824. doi: 10.1016/j.neuron.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lo L, et al. Connectional architecture of a mouse hypothalamic circuit node controlling social behavior. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7503–7512. doi: 10.1073/pnas.1817503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eichenbaum H. The hippocampus and mechanisms of declarative memory. Behav. Brain Res. 1999;103:123–133. doi: 10.1016/S0166-4328(99)00044-3. [DOI] [PubMed] [Google Scholar]
- 68.Zemla R, Basu J. Hippocampal function in rodents. Curr. Opin. Neurobiol. 2017;43:187–197. doi: 10.1016/j.conb.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Groen T, Miettinen P, Kadish I. The entorhinal cortex of the mouse: Organization of the projection to the hippocampal formation. Hippocampus. 2003;13:133–149. doi: 10.1002/hipo.10037. [DOI] [PubMed] [Google Scholar]
- 70.Sgaier SK, et al. Morphogenetic and cellular movements that shape the mouse cerebellum: Insights from genetic fate mapping. Neuron. 2005;45:27–40. doi: 10.1016/j.neuron.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 71.Cho JH, Prince JEA, Cloutier JF. Axon guidance events in the wiring of the mammalian olfactory system. Mol. Neurobiol. 2009;39:1–9. doi: 10.1007/s12035-008-8047-7. [DOI] [PubMed] [Google Scholar]
- 72.Cheng S, et al. Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila. Elife. 2019;8:1–27. doi: 10.7554/eLife.41028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones CD, et al. Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. PLOS Genet. 2018;14:e1007560. doi: 10.1371/journal.pgen.1007560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pischedda F, Piccoli G. The IgLON family member negr1 promotes neuronal arborization acting as soluble factor via FGFR2. Front. Mol. Neurosci. 2016;8:1–12. doi: 10.3389/fnmol.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szczurkowska J, et al. NEGR1 and FGFR2 cooperatively regulate cortical development and core behaviours related to autism disorders in mice. Brain. 2018;141:2772–2794. doi: 10.1093/brain/awy190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pimenta AF, Reinoso BS, Levitt P. Expression of the mRNAs encoding the limbic system-associated membrane protein (LAMP): II. Fetal rat brain. J. Comput. Neurol. 1996;375:289–302. doi: 10.1002/(SICI)1096-9861(19961111)375:2<289::AID-CNE8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 77.Pimenta AF, Reinoso BS, Levitt P. Expression of the mRNAs encoding the limbic system-associated membrane protein (LAMP): I. Adult rat brain. J. Comput. Neurol. 1996;375:274–288. doi: 10.1002/(SICI)1096-9861(19961111)375:2<289::AID-CNE8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 78.du Sert NP, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020;18:e3000410. doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: In situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 80.Beaubien F, Cloutier JF. Differential expression of Slitrk family members in the mouse nervous system. Dev. Dyn. 2009;238:3285–3296. doi: 10.1002/dvdy.22160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.