Abstract
Endemic seasonal coronaviruses cause morbidity and mortality in a subset of patients, but no specific treatment is available. Molnupiravir is a promising pipeline antiviral drug for treating SARS-CoV-2 infection potentially by targeting RNA-dependent RNA polymerase (RdRp). This study aims to evaluate the potential of repurposing molnupiravir for treating seasonal human coronavirus (HCoV) infections. Molecular docking revealed that the active form of molnupiravir, β-D-N4-hydroxycytidine (NHC), has similar binding affinity to RdRp of SARS-CoV-2 and seasonal HCoV-NL63, HCoV-OC43 and HCoV-229E. In cell culture models, treatment of molnupiravir effectively inhibited viral replication and production of infectious viruses of the three seasonal coronaviruses. A time-of-drug-addition experiment indicates the specificity of molnupiravir in inhibiting viral components. Furthermore, combining molnupiravir with the protease inhibitor GC376 resulted in enhanced antiviral activity. Our findings highlight the great potential of repurposing molnupiravir for treating seasonal coronavirus infected patients.
Keywords: Molnupiravir, Seasonal coronavirus, RdRp, Drug repurposing
Coronaviruses constitute a large family of single-stranded positive-sense RNA viruses infecting mammals and birds. There are currently seven types of coronaviruses known to infect humans, although all of them are thought to have originated in animals. The three highly pathogenic members—MERS-CoV, SARS-CoV-1 and SARS-CoV-2—can cause severe acute respiratory diseases. The four endemic seasonal human coronaviruses (HCoV)—NL63, OC43, 229E and HKU1—usually but not exclusively cause mild and self-limiting respiratory tract infections (Ma et al., 2020).
Endemic seasonal HCoVs have been neglected by both public and research communities. Globally, the four seasonal HCoVs contribute to 5% of the several billion upper respiratory infections each year (Li et al., 2020b). Systematic analysis of reported clinical studies estimated that about 25% of patients infected with seasonal HCoVs will actually develop pneumonia potentially resulting in serious complications (Li et al., 2020a), although such estimation likely has bias due to limited data available. Importantly, fatality has been reported usually in vulnerable populations but also in healthy individuals (Veiga et al., 2021). The clinical burden of seasonal HCoVs infection is clearly undeniable, but there is no therapeutic option available.
Based on the close genetic relationship among different coronaviruses (Ma et al., 2020), we hypothesize the feasibility of repurposing anti-SARS-CoV-2 agents for treating seasonal HCoVs infections. The ribonucleoside analog β-D-N4-hydroxycytidine (NHC) has broad-spectrum antiviral activity against various RNA viruses, including hepatitis C virus, Ebola virus and influenza viruses (Reynard et al., 2015; Stuyver et al., 2003; Toots et al., 2019). Molnupiravir (EIDD-2801 or MK-4482), the pro-drug of NHC, has been shown to potently inhibit the replication of SARS-CoV-2 in human airway cell culture and animal models (Cox et al., 2021; Wahl et al., 2021). Importantly, recent results of a Phase 2a trial has demonstrated that as the first oral, direct-acting antiviral, molnupiravir is highly effective in reducing nasopharyngeal SARS-CoV-2 infectious virus and viral RNA levels, and has a favorable safety and tolerability profile (Fischer et al., 2021). It thus represents as one of the most promising pipeline antiviral drug candidates for treating COVID-19, the disease caused by SARS-CoV-2 infection. This study aims to evaluate the potential antiviral activity of molnupiravir against three seasonal HCoVs using molecular docking and cell culture models. Because of the unavailability of HCoV-HKU1 cell culture system, it was excluded in this study.
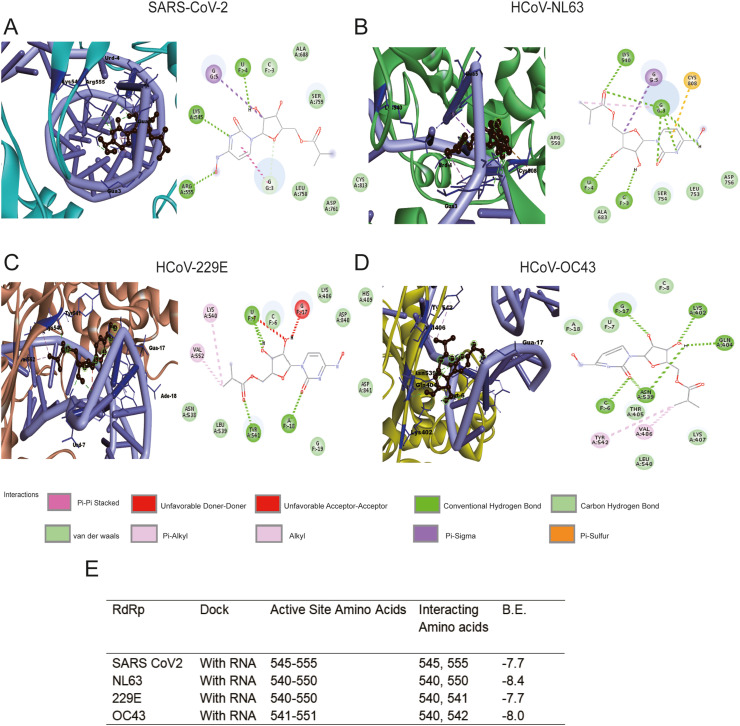
One possible antiviral mechanism of molnupiravir is to introduce lethal mutagenesis during viral RNA replication (Sheahan et al., 2020). Thus, it is intuitive to expect direct interactions between NHC (the active form of molnupiravir) and coronavirus RdRp. RdRp is virtually encoded by all RNA viruses and can be targeted with a high degree of selectivity. As a key virus-encoded enzyme in the viral replication cycle, RdRp plays an important role in transcribing mRNA from genome templates and acts as a replicase to copy genomic RNA. Given the structural similarity among RdRps and the conservation of their structural elements, it has become one of the best targets for developing broad-spectrum antiviral agents. To map such potential interactions, we retrieved the experimentally solved crystal structure of SARS-CoV-2 RdRp, and successfully modelled the RdRp structures of HCoV-NL63, HCoV-OC43 and HCoV-229E (see details in supplementary methods). Through preliminary blind docking, a grid was defined around the active regions from 540 to 555 of the four RdRp structures (Fig. S1, Fig. 1 and Fig. S2). Site-specific docking of this region consistently indicated interactions between NHC with the four RdRps (Fig. S1). As RdRp catalyzes the replication of viral RNA, it is important to further confirm the binding affinity of NHC on a polymerase-RNA complex. As the crystal structure of polymerase-RNA complex of HCoV-NL63, HCoV-229E and HCoV-OC43 is not available, the polymerase was docked with RNA in the HDOCK server. The resulting conformation was downloaded and further docked with NHC (the active form of molnupiravir) (Fig. 1). As the crystal structure of polymerase-RNA complex of SARS-CoV-2 (PDB 7C2K) is available, this polymerase was first docked with RNA in crystal structure (Fig. S2A). To be consistent, this was also performed in the HDOCK server (Fig. 1A). Overall, the binding scores of NHC with polymerase-RNA complex compared to polymerase alone are substantially higher, suggesting that the drug has higher affinity towards the polymerase-RNA complex (Fig. S1E, Fig. 1E and Fig. S2B). Importantly, NHC was found to form several conventional hydrogen bonds with the residues of all the RdRps, along with other electrostatic interactions such as van der Walls (Fig. S1, Fig. 1 and Fig. S2). These results suggest that NHC has comparable binding affinity towards the RdRp of SARS-CoV-2 and seasonal coronaviruses. This encouraged us to further assess the antiviral activity in cell culture models of the three seasonal coronaviruses.
Fig. 1.
Site specific binding mode of NHC to coronavirus polymerase-RNA complex. The active form of molnupiravir (β-D-N4-hydroxycytidine triphosphate; NHC), binding to the RdRp-RNA complex (atom color ribbons) of SARS-CoV-2 (A), HCoV-NL63 (B), HCoV-229E (C) and HCoV-OC43 (D), is depicted as surface representation. H-bond donor (purple) and acceptor (green) interactions are depicted. (E) Summary of binding mode and affinity index (B.E.). HCoV: human coronavirus. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
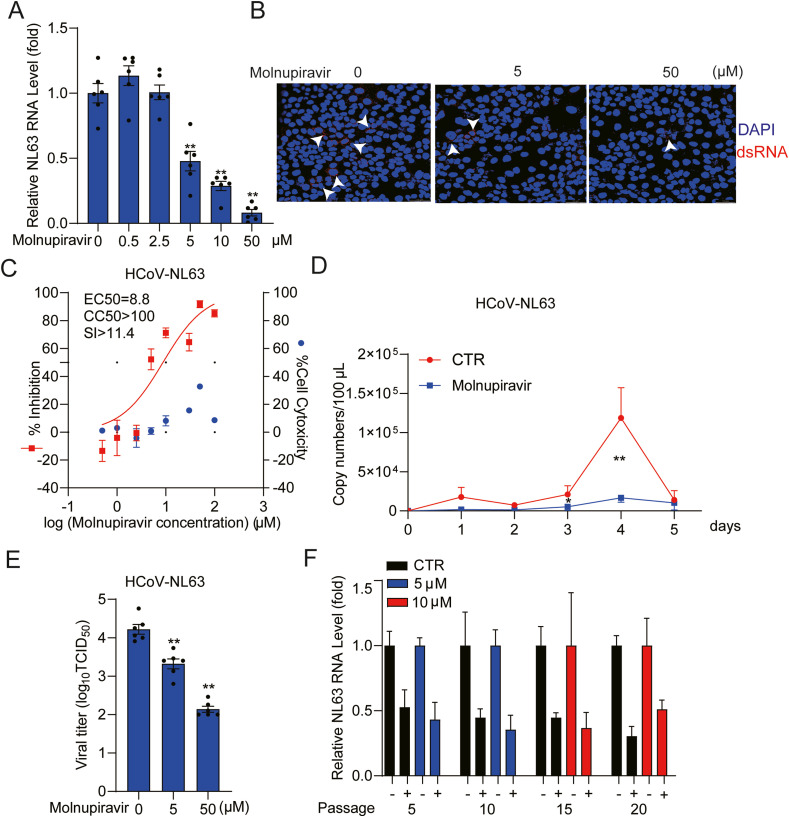
Interestingly, HCoV-NL63 is the only member of seasonal HCoVs that utilizes angiotensin converting enzyme 2 (ACE2) as its receptor for viral entry (Hofmann et al., 2005), similar to SARS-CoV-1 and SARS-CoV-2. It was first isolated from a 7-month-old child suffering from bronchiolitis and conjunctivitis in the Netherlands (van der Hoek et al., 2004). We tested a series of concentrations (0.5–100 μM) of molnupiravir in different cell models infected with HCoV-NL63 (Fig. 2 A, Fig. S3A and Fig. S3B). Molnupiravir treatment inhibited viral replication in a dose-dependent manner in all tested cell models. For example, treatment with 50 μM molnupiravir for 48 h decreased intracellular virus RNA level by 91.8 ± 2.5% (mean ± SEM, n = 6, p< 0.0001) in Caco2 cells. This inhibitory effect was further confirmed by immunofluorescent staining of dsRNA, an intermediate of genomic viral RNA replication (Fig. 2B). Of note, high dose treatment with molnupiravir had moderate effects on cell viability (Fig. S4A, Fig. S4B and Fig. S4C). The half maximum effective concentration (EC50) of molnupiravir against HCoV-NL63 replication was 8.8 μM and the half maximum cytotoxic concentration (CC50) was above 100 μM, which resulted in a selective index (SI) above 10, indicating a substantial therapeutic window (Fig. 2C). We monitored the dynamic effects on virus production in a consecutive 5-day course by treatment with 5 μM molnupiravir in Caco2 cells, and found significant inhibition of viral RNA release into culture supernatant (Fig. 2D and Fig. S5A). Especially on day 4, there was 83.5 ± 4.7% (mean ± SEM, n = 6, P < 0.0001) reduction of viral RNA release. By harvesting Caco2 cells and culture supernatant at 48 h post-treatment, we performed 50% Tissue Culture Infective Dose (TCID50) assay to determine the titers of viruses treated with molnupiravir. Consistently, the titers of produced HCoV-NL63 with infectivity were significantly reduced by different concentrations of molnupiravir treatment (Fig. 2E). Treatment with 50 μM molnupiravir resulted in a 98.8 ± 0.5% (mean ± SEM, n = 6, P < 0.0001) reduction of viral titer.
Fig. 2.
Antiviral effects of molnupiravir against HCoV-NL63. (A) Dose-dependent inhibition of HCoV-NL63 replication in Caco2 cell line by molnupiravir treatment. Intracellular viral RNA quantified by qRT-PCR was normalized to housekeeping gene GAPDH and presented relative to the control (CTR) (set as 1) (n = 6). (B) Immunofluorescence microscopy analysis of dsRNA (red), the intermediate of replicating HCoV-NL63 genomic RNA, upon treatment of different concentrations of molnupiravir in Caco2 cell line. Nuclei were visualized by DAPI (blue). (C) Caco2 cells were infected with HCoV-NL63 at an MOI of 0.1 in the treatment of different concentrations of molnupiravir for 48 h. Viral yield in the cell supernatant was quantified by qRT-PCR. Cytotoxicity was determined by MTT assay. The half maximum effective concentration (EC50) and the half maximum cytotoxic concentration (CC50) were calculated based on the model Y = 100/(1 + 10^(LogEC50-X)) using GraphPad Prism 8.0.2 software. The left and right Y-axis of the graphs represent mean % inhibition of virus yield and cytotoxicity of the drugs, respectively. (n = 6–16). (D) Caco2 cells were infected with HCoV-NL63 at the MOI of 0.5, and then untreated or treated with 5 μM molnupiravir for 5 days. Supernatant was collected every day to quantify secreted viruses by qRT-PCR, calculated as genomic copy numbers (n = 6). Standard curve for calculation of genomic copy numbers is included in Supplementary Fig. S3A. (E) Caco2 cells were infected with 0.5 MOI HCoV-NL63, and then untreated or treated with 5 or 50 μM molnupiravir for 48 h. Virus titers from different groups were determined by TCID50 assay (n = 6). (F) HCoV-NL63 was serially passaged in Caco2 cells exposed to no molnupiravir (as control) or increasing concentrations of molnupiravir for 20 passages. 5 μM molnupiravir was used in passage 1–10, which was increased to 10 μM at the subsequent passages. The effect of molnupiravir (5 μM) on HCoV-NL63 harvested at passage 5, 10, 15 and 20 was quantified using qRT-PCR. Data represent as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. HCoV: human coronavirus. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Importantly, serially passaging of HCoV-NL63 in the presence of escalating concentrations (5 μM for passage 1–10 and 10 μM for passage 11–20) of molnupiravir maintained the sensitivity to the treatment (Fig. 2F). Our results are in accordance with several previous studies that molnupiravir exposure does not easily develop resistance in several viral infection models, including influenza virus, Venezuelan equine encephalitis virus, mouse hepatitis virus (beta-coronavirus) and MERS-CoV (Agostini et al., 2019; Toots et al., 2019; Urakova et al., 2018). All these studies failed to induce viral escape by dose escalation, suggesting that molnupiravir has a high barrier against viral resistance development.
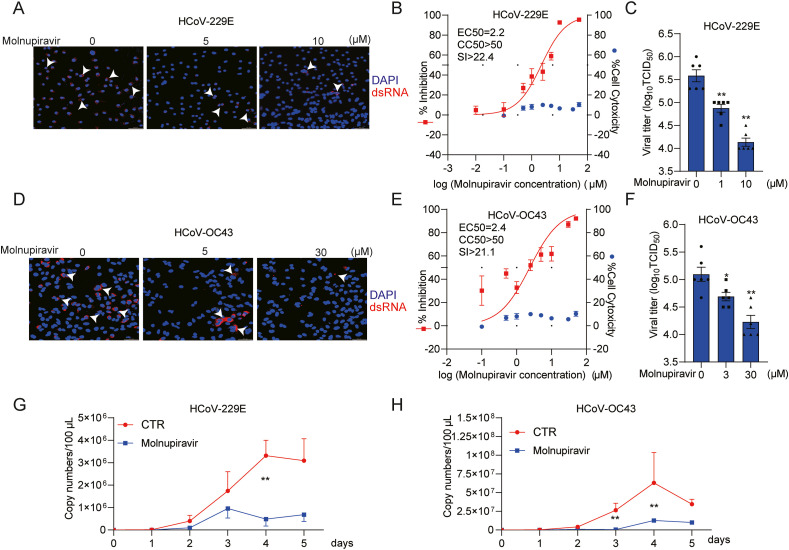
To evaluate whether molnupiravir has pan-coronavirus antiviral activity, we tested another two seasonal HCoVs, HCoV-229E and HCoV-OC43, in human lung A549 cell model. Immunofluorescent staining of viral dsRNA showed reduction of the number of infected cells by molnupiravir treatment (Fig. 3 A and Fig. 3D). The EC50 value against HCoV-229E was 2.2 μM with CC50 value above 50 μM. The EC50 value against HCoV-OC43 was 2.4 μM and CC50 was above 50 μM. The selective indexes were above 20 for both viruses (Fig. 3B and E, Fig. S3C, Fig. S3D and Fig. S4D). TCID50 assay demonstrated significant reduction of viral titers of produced infectious viruses by molnupiravir (Fig. 3C and F). Treatment with 10 or 30 μM molnupiravir resulted in nearly 90% reduction of HCoV-229E and HCoV-OC43 viral titers respectively. Molnupiravir (5 μM) significantly inhibited the release of viral RNA into supernatant in a consecutive 5-day course (Fig. 3G and H, Fig. S5B and Fig. S5C). Similar to HCoV-NL63, the inhibitory effect was most prominent on day 4, and for example, the level of secreted 229E viral RNA was reduced by 87.6 ± 6.6% (mean ± SEM, n = 6, P < 0.0001). Of note, the production of all three HCoVs was dramatically decreased by day 5. One of the possible explanations could be the cytopathogenic effect caused by coronavirus at the late stage of infection in cell culture.
Fig. 3.
Antiviral effects of molnupiravir against HCoV-229E and HCoV-OC43 infection. (A) and (D) Immunofluorescence microscope analysis of dsRNA (red) upon treatment with different concentrations of molnupiravir in A549 cell line. Nuclei were visualized by DAPI (blue). (B) and (E) A549 cells were infected HCoV-229E or HCoV-OC43 at an MOI of 0.1 in the treatment of different concentrations of molnupiravir for 48 h. The viral yield in the cell supernatant was then quantified by qRT-PCR. Cytotoxicity was determined by MTT assay. The half maximum effective concentration (EC50) and the half maximum cytotoxic concentration (CC50) were calculated based on the model Y = 100/(1 + 10^(LogEC50-X)) using GraphPad Prism 8.0.2 software. The left and right Y-axis of the graphs represent mean % inhibition of virus yield and cytotoxicity of the drugs, respectively. (n = 6–16). (C) and (F) The supernatant and cells of each well under molnupiravir treatment was harvested after freezing and thawing for three times. Virus titers from different groups were determined by TCID50 assay (n = 6). (G) and (H) A549 cells were infected with HCoV-229E or HCoV-OC43 at the MOI of 0.1, and then untreated or treated with 5 μM molnupiravir for 5 days. Supernatant was collected every day to quantify secreted viruses by qRT-PCR, calculated as genomic copy numbers (n = 6). Standard curve for calculation of genomic copy numbers is included in Supplementary Figs. S3B and S3C. Data represent as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. HCoV: human coronavirus. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
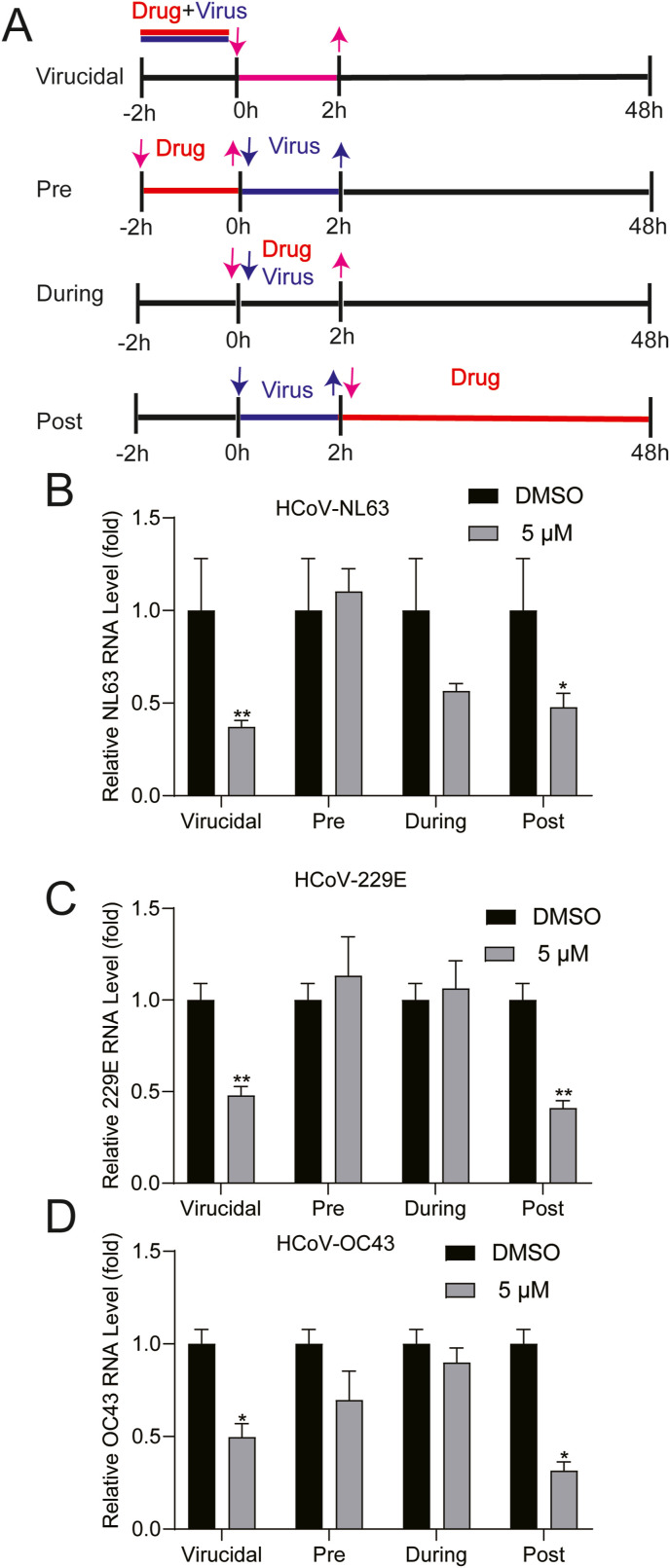
To explore which step(s) of the viral lifecycle is blocked by molnupiravir, we performed a time-of-drug-addition experiment (Daelemans et al., 2011) (Fig. 4 A). Pre-treatment and treatment during virus inoculation had minor effects on the three coronaviruses. Surprisingly, virucidal treatment for 2 h resulted in comparable antiviral potency when compared to post-infection treatment for 48 h (Fig. 4B, C and D). For example, virucidal treatment (5 μM) resulted in reduction of NL63 viral RNA by 62.8 ± 3.5% (mean ± SEM, n = 6, P < 0.0001), whereas post-infection treatment resulted in 52.2 ± 7.5% (mean ± SEM, n = 6, P < 0.0001) inhibition. It is clear that potent antiviral activity by post-infection treatment is attributed to inhibiting viral replication through targeting RdRp. However, the mechanism why virucidal treatment exerted potent antiviral effects remains unclear. It is interesting to be further explored.
Fig. 4.
Time-of-addition analysis of the antiviral activity of molnupiravir. (A) Schematic illustration of the time-of-addition experiment. (B), (C) and (D) Caco2 or A549 cells were infected with HCoV-NL63, HCoV-229E or HCoV-OC43 at an MOI of 0.1 for 2 h (0–2 h) respectively. 5 μM molnupiravir was introduced at different time points, designated as virucidal, pretreatment (pre), during treatment (during) or post-treatment (post). The inhibitory effect of molnupiravir in each group was determined by qRT-PCR. Data represent as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. HCoV: human coronavirus.
Combination treatment is often used to enhance antiviral efficacy and to avoid drug resistance development in clinical applications. Intuitively, combining agents with distinct antiviral mechanisms would be more likely to exert synergistic effects. GC376 is a viral protease inhibitor, which is currently at clinical development phase for treating COVID-19 (Fu et al., 2020). We evaluated the combined antiviral effects of molnupiravir with GC376 in cell culture models and calculated by Synergy Finder based on mathematical modeling (Ianevski et al., 2020). A moderate additive effect was observed for all three coronaviruses (Fig. S6). The tested concentrations of the compounds had minimal cytotoxicity on host cells (Fig. S7).
In summary, we have demonstrated that molnupiravir is a potent inhibitor of HCoVs based on molecular docking with viral RdRp and testing in cell culture models. Seasonal coronaviruses are imposing an undeniable clinical burden in special populations with association of fatalities in some cases (Veiga et al., 2021). We believe that repurposing anti-SARS-CoV-2 drugs is a viable option for expeditiously developing therapeutics for seasonal coronavirus infected patients. Currently, remdesivir, an RdRp inhibitor, is the only FDA-approved antiviral for treating hospitalized COVID-19 patients. However, whether remdesivir actually has meaningful clinical benefits remains widely questioned (Consortium et al., 2021). Another issue with remdesivir is that it is intravenously administered, which limits its wide application in particular for unhospitalized patients. In this respect, molnupiravir has clear advantages, as it is orally active with potent antiviral activity consistently shown in preclinical SARS-CoV-2 models. In addition, our results in line with previous studies (Agostini et al., 2019; Toots et al., 2019; Urakova et al., 2018) suggest molnupiravir has a high barrier to the development of drug resistance variants. Combination with other antivirals, such as GC376, may further enhance antiviral potency and prevent drug resistance development. If the upcoming large clinical trials would indeed prove that molnupiravir is highly effective for treating COVID-19 patients, we would strongly recommend its repurposing for treating patients with severe seasonal coronavirus infection.
CRediT authorship contribution statement
Yining Wang: Conceptualization, Vitro experiments, Methodology, Investigation, Writing – original draft, Writing – review & editing. Pengfei Li: Vitro experiments, Methodology, Data curation, Writing – review & editing. Kundan Solanki: Molecular docking, Methodology, Investigation, Writing – original draft, Writing – review & editing. Yang Li: Vitro experiments, Methodology, Data curation, Writing – review & editing. Zhongren Ma: Writing – review & editing. Maikel P. Peppelenbosch: Conceptualization, Supervision, Writing – review & editing. Mirza S. Baig: Conceptualization, Molecular docking, Methodology, Resources, Supervision, Writing – review & editing. Qiuwei Pan: Conceptualization, Methodology, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors thank Dr. Lia van der Hoek (Amsterdam UMC location AMC, University of Amsterdam, the Netherlands) for providing the stock of human coronavirus NL63.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.09.009.
Funding
This study is supported by a VIDI grant (No. 91719300) from the Netherlands Organisation for Scientific Research (NWO) to Q. Pan, and the China Scholarship Council for funding PhD fellowships to Yining Wang (No.201903250082), Pengfei Li (No. 201808370170), Yang Li (No. 201703250073).
Ethical approval
Not required.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Andres E.L., Bluemling G.R., Lockwood M.A., Sheahan T.P., Sims A.C., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G.R., Baric R.S., Denison M.R. Small-molecule antiviral beta-d-N (4)-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93 doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium W.H.O.S.T., Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernandez Garcia C., Kieny M.P., Malekzadeh R., Murthy S., Reddy K.S., Roses Periago M., Abi Hanna P., Ader F., Al-Bader A.M., Alhasawi A., Allum E., Alotaibi A., Alvarez-Moreno C.A., Appadoo S., Asiri A., Aukrust P., Barratt-Due A., Bellani S., Branca M., Cappel-Porter H.B.C., Cerrato N., Chow T.S., Como N., Eustace J., Garcia P.J., Godbole S., Gotuzzo E., Griskevicius L., Hamra R., Hassan M., Hassany M., Hutton D., Irmansyah I., Jancoriene L., Kirwan J., Kumar S., Lennon P., Lopardo G., Lydon P., Magrini N., Maguire T., Manevska S., Manuel O., McGinty S., Medina M.T., Mesa Rubio M.L., Miranda-Montoya M.C., Nel J., Nunes E.P., Perola M., Portoles A., Rasmin M.R., Raza A., Rees H., Reges P.P.S., Rogers C.A., Salami K., Salvadori M.I., Sinani N., Sterne J.A.C., Stevanovikj M., Tacconelli E., Tikkinen K.A.O., Trelle S., Zaid H., Rottingen J.A., Swaminathan S. Repurposed antiviral drugs for covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daelemans D., Pauwels R., De Clercq E., Pannecouque C. A time-of-drug addition approach to target identification of antiviral compounds. Nat. Protoc. 2011;6:925–933. doi: 10.1038/nprot.2011.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Eron J.J., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R., Duke E.R., Azizad M.M., Borroto-Esoda K., Wohl D.A., Loftis A.J., Alabanza P., Lipansky F., Painter W.P. Molnupiravir, an oral antiviral treatment for COVID-19. medRxiv. 2021 doi: 10.1101/2021.06.17.21258639. [DOI] [Google Scholar]
- Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski A., Giri A.K., Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ikram A., Peppelenbosch M.P., Ma Z., Pan Q. Systematically mapping clinical features of infections with classical endemic human coronaviruses. Clin. Infect. Dis. 2020;73(3):554–555. doi: 10.1093/cid/ciaa1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Liu J., Ma Z., Bramer W.M., Peppelenbosch M.P., Pan Q. Estimating global epidemiology of low-pathogenic human coronaviruses in relation to the COVID-19 context. J. Infect. Dis. 2020;222:695–696. doi: 10.1093/infdis/jiaa321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Li P., Ji Y., Ikram A., Pan Q. Cross-reactivity towards SARS-CoV-2: the potential role of low-pathogenic human coronaviruses. Lancet Microbe. 2020;1:e151. doi: 10.1016/S2666-5247(20)30098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard O., Nguyen X.N., Alazard-Dany N., Barateau V., Cimarelli A., Volchkov V.E. Identification of a new ribonucleoside inhibitor of Ebola virus replication. Viruses. 2015;7:6233–6240. doi: 10.3390/v7122934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver L.J., Whitaker T., McBrayer T.R., Hernandez-Santiago B.I., Lostia S., Tharnish P.M., Ramesh M., Chu C.K., Jordan R., Shi J., Rachakonda S., Watanabe K.A., Otto M.J., Schinazi R.F. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 2003;47:244–254. doi: 10.1128/AAC.47.1.244-254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots M., Yoon J.J., Cox R.M., Hart M., Sticher Z.M., Makhsous N., Plesker R., Barrena A.H., Reddy P.G., Mitchell D.G., Shean R.C., Bluemling G.R., Kolykhalov A.A., Greninger A.L., Natchus M.G., Painter G.R., Plemper R.K. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakova N., Kuznetsova V., Crossman D.K., Sokratian A., Guthrie D.B., Kolykhalov A.A., Lockwood M.A., Natchus M.G., Crowley M.R., Painter G.R., Frolova E.I., Frolov I. beta-d-N (4)-hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018;92 doi: 10.1128/JVI.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga A., Martins L.G., Riediger I., Mazetto A., Debur M.D.C., Gregianini T.S. More than just a common cold: endemic coronaviruses OC43, HKU1, NL63, and 229E associated with severe acute respiratory infection and fatality cases among healthy adults. J. Med. Virol. 2021;93:1002–1007. doi: 10.1002/jmv.26362. [DOI] [PubMed] [Google Scholar]
- Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, Liu H., Madden V.J., Krzystek H.M., De C., White K.K., Gully K., Schafer A., Zaman T., Leist S.R., Grant P.O., Bluemling G.R., Kolykhalov A.A., Natchus M.G., Askin F.B., Painter G., Browne E.P., Jones C.D., Pickles R.J., Baric R.S., Garcia J.V. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.