Highlights
-
•
Pyroptosis genes were differentially expressed between HNSCC and normal tissue.
-
•
Developed a model for prognostic prediction of HNSCC based on pyroptosis genes.
-
•
The risk-score model was proved to be promising and reliable in practical patients.
Key words: HNSCC, Prognosis, Pyroptosis, Carcinoma, Prediction
Abstract
We aimed at establishing a risk – score model using pyroptosis-related genes to predict the prognosis of patients with head and neck squamous cell carcinoma (HNSCC). A total of 33 pyroptosis-related genes were selected. We then evaluated the data of 502 HNSCC patients and 44 normal patients from TCGA database. Gene expression was then profiled to detect differentially expressed genes (DEGs). Using the univariate, the least absolute shrinkage and selection operator (LASSO) Cox regression analyses, we generated a risk – score model. Tissue samples from neoplastic and normal sites of 44 HNSCC patients were collected. qRT-PCR were employed to analyze the mRNA level of the samples. Kaplan-Meier method was used to evaluate the overall survival rate (OS). Enrichment analysis was performed to elucidate the underlying mechanism of HNSCC patient's differentially survival status from the perspective of tumor immunology. 17 genes were categorized as DEGs. GSDME, IL-6, CASP8, CASP6, NLRP1 and NLRP6 were used to establish the risk – score model. Each patient's risk score in the TCGA cohort was calculated using the risk – score formula. The risk score was able to independently predict the OS of the HNSCC patients (P = 0.02). The OS analysis showed that the risk score model (P < 0.0001) was more reliable than single gene, a phenomenon verified by practical patient cohort. Additionally, enrichment analysis indicated more active immune activities in low-risk group than high-risk group. In conclusion, our risk – score model has provided novel strategy for the prediction of HNSCC patients’ prognosis.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is one of the most common malignancies of head and neck, with a propensity for high recurrence and low survival rates. HNSCC accounts for an estimated incidence of more than 880,000 cases and more than 450,000 deaths [1].
Despite significant improvement in the treatment of HNSCC, such as functional surgery, induction chemotherapy, and adjuvant radiotherapy in the past three decades, the estimated 5-year survival rate of patients with HNSCC remains low (50 – 55%) [2,3] Thus, there is an urgent need for development of more effective molecules, which could help in the diagnosis and prediction of prognosis in patients with HNSCC.
Apoptosis and necrosis are the two main types of cell death that help in maintaining cellular homeostasis. However, recent reports have shown existence of pyroptosis, a cell death form that is triggered by proinflammatory signals [4,5]. Gasdermin family molecules such as GSDMA, GSDMB, GSDMC, GSDMD, GSDME and DFNB59, are key effector molecules that mediate the occurrence of pyroptosis. For instance, GSDMD-mediated pyroptosis requires the activation of caspase – 1 and caspase-4/5/11 molecules. The activated caspase – 1/4/5/11 split the GSDMD into the N-terminal that has perforation effect and the C-terminal that has self -inhibition effect. The N-terminal then binds to membrane lipids, phosphatidylinositol and cardiolipin, leading to localization into cell membrane pores. Cellular gasdermin family proteins have been shown to form 10 to 20 nm pores in the cell membrane, and then cytoplasmic contents such as interleukin (IL) – 1B and IL – 18 are recruited to release inflammatory cells. This cascade of events triggers inflammatory responses causing cell pyroptosis [6], [7], [8]. Furthermore, GSDME – mediated pyroptosis relies on caspase-3 [9].
Pyroptosis was initially shown to be key in combating infection [10]. However, recent reports have demonstrated that pyroptosis could also play an important role in the development of tumors. For instance, Akino et al. reported that about 52% of 89 gastric cancer tissue samples showed abnormal methylation in the GSDME promoter [11]. Besides, Kim et al. treated three colorectal cancer cell lines: HCT116, HT29 and DLD-1 with demethylation inhibitor 5-Azad C and showed upregulation of GSDME gene expression. Furthermore, there was increased methylation of the GSDME gene promoter in colorectal and breast tumor tissues, compared to normal colorectal tissues [12,13].
Whereas pyroptosis plays an important role in oncogenesis and anticancer processes, there is limited data on the significance of pyroptosis in HNSCC lesions. Here we performed a systematic study to determine the expression profile of pyroptosis-related genes in normal and HNSCC tissues. We then developed a novel risk-score model to predict the prognosis of HNSCC patients.
Material and methods
Database and patients
We obtained RNA sequencing (RNA-seq) data of 502 HNSCC patients and 44 normal human head and neck samples as well as their corresponding clinical features from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/repository). Fresh HNSCC and normal tissues from 44 human patients (collected postoperatively from April 2010 to October 2014), the same cohort with our previous study, were collected by the Department of Oral and Maxillofacial Head and Neck Oncology, Shanghai Ninth People's Hospital [14]. During operations, frozen sections of the tissue at the surgical margins were examined at the time when the expanded resection was finished. If the pathological results were positive, the expanded resection was continued until the surgical margins in all directions were negative, and the normal tissue were resected from surrounding area. Patients were diagnosed through pathological examination by pathologists at Department of Pathology, Shanghai Ninth People's Hospital. All cases were clinically staged according to the 8th Edition TNM Classification for Head and Neck Cancer developed by the Union for International Cancer Control (UICC) and the American Joint Committee on Cancer (AJCC) [15]. The clinical patients’ data was shown in Table S1. All of the 44 patients with HNSCC were followed up by telephone survey until October 2019. This study was approved by the Human Research Ethics Committee of the Ninth People's Hospital, Shanghai JiaoTong University School of Medicine (Shanghai, China). Given the retrospective nature of this study, informed consent was not available.
Identification of differentially expressed pyroptosis-related genes
We screend and extracted 33 pyroptosis-related genes from previous studies (Table S2) [16], [17], [18], [19]. We then normalized the expression data in TCGA database to fragment per kilobase million (FPKM) values prior to comparison. The “limma” package was used to identify the DEGs with |log2FC| ≥ 0.5 and FDR < 0.05.
Establishment of the prognostic risk-model
To assess the prognostic value of the pyroptosis-related genes, we employed univariate Cox regression analysis to evaluate the correlation between each gene and survival status in the TCGA cohort. To prevent omissions, we set 0.05 as the cut-off P-value, and then identified 17 survival-related genes for subsequent analysis. The least absolute shrinkage and selection operator (LASSO) Cox regression model (R package “glmnet”) was then utilized to narrow down the candidate genes and develop the prognostic model. A total of six genes and their coefficients were retained through multivariate Cox regression analysis, and then the penalty parameter (λ) was decided by minimum criteria. In addition, we calcuated the risk score after centralization and standardization (applying the “scale” function in R) of the TCGA expression data, using the risk score formula(X: coefficients, Y: gene expression level):
The HNSCC patients in the TCGA cohort were divided into low- and high-risk subgroups according to the median risk score, and then the overall survival (OS) time was compared between the two subgroups via Kaplan–Meier analysis. Principal component analysis (PCA) based on the 6-gene model was performed by the “prcomp” function in the“stats” R package. The “survival”, “survminer” and “timeROC” R packages were used to perform a 5-year ROC curve analysis.
Independent prognostic analysis of the risk score
We extracted the clinical data of 502 HNSCC patients in the TCGA cohort. We then analyses the variables against the risk score in our regression model. Univariate and multivariate Cox regression models were employed for the analysis.
Total RNA extraction and reverse transcription
The clinical specimens of 44 patients with HNSCC were collected during surgeries and the specimens were immersed into the RNAlater Solution (Invitrogen, USA) immediately and then used for RNA extraction or stored at −80 °C. Total RNA was extracted from the fresh tissues using TRIzol Reagent (Invitrogen) and cDNA was synthesized from 10 μg of total mRNA by using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following manufacturers’ instructions.
qRT-PCR analysis and RNA sequencing
qRT-PCR was performed by using FastStart Universal SYBR Green Master Mix (Roche) and QuantStudio™ 6 Flex (Applied Biosystems). Primers used for the qRT-PCR were showed in Table S3. We performed RNA-seq analysis using the NovelBrain Cloud Analysis Platform, China. In brief, after total RNA was extracted, the cDNA libraries were constructed for each pooled RNA sample using the VAHTSTM Total RNA-seq (H/M/R). The gene expression level was determined by the FPKM method.
Immune function analysis between the low- and high-risk groups
The HNSCC patients in the TCGA cohort were stratified into two subgroups according to the median risk score. The “gsva” package was utilized to perform single-sample gene set enrichment analysis (ssGSEA) and then calculate the scores of infiltrating immune cells to evaluate the activity of immune-related pathways.
Statistical analysis
Single-factor analysis of variance was used to compare the gene expression between the normal tissues and HNSCC tissues, while the Pearson chi‐square test was used to compare the categorical variables. To compare the OS of the patients between subgroups, we employed the Kaplan – Meier method with a two – sided log – rank test. We used univariate and multivariate Cox regression models to assess the independent prognostic value of the risk – score model. In addition, we used the Mann–Whitney test to compare immune cell infiltration and immune pathway activation between the two groups. All statistical analyses were performed in R software (v4.0.2) and Graphpad Prism 8.0. A P < 0.05 was considered statistically significant.
Results
Identification of DEGs between normal and HNSCC tissues
The 33 pyroptosis-related gene expression levels from 44 normal and 502 HNSCC tissues were compared in The Cancer Genome Atlas (TCGA), and a total of 17 DEGs were identified (all |log2FC| ≥ 0.5 and FDR < 0.05). The analysis showed that 3 genes (ELANE, IL-18, IL-6) were downregulated while 14 genes (GSDMD, PLCG1, IL1B, GSDME, PYCARD, NLRP1, CASP8, IL18, IL6, NLRP6, NLRC4, AIM2, GSDMB and CASP6) were enriched in the tumor group (Fig. S1). In addition, we profiled the RNA expression of these genes as shown in heatmaps (Fig. 1A). To further explore the interactions among the pyroptosis-related genes, we conducted a correlation network analysis with all the pyroptosis-related genes (Fig. 1B).
Fig. 1.
Expressions of the 33 pyroptosis-related genes and their interaction. (A) Heatmap (green: low expression level; red: high expression level) of the pyroptosis-related genes between the normal (N, brilliant blue) and the tumor tissues (T, red). (B) The correlation network of the pyroptosis-related genes (red line: positive correlation; blue line: negative correlation. The depth of the colours reflects the strength of the relevance).
Development of a prognostic gene risk-model in the TCGA cohort
A total of 502 HNSCC samples were matched with the corresponding patients who had complete survival data. Univariate Cox regression analysis was used for primary screening of the survival-related genes. Out of the 7 genes (GSDME, IL-6, IL-1B, NLRP1, CASP8, NLRP6, and CASP6) that had a P < 0.05, 5 genes (GSDME, IL-6, IL-1B, CASP8 and CASP6) were associated with increased cancer risk with HRs >1, while the other 2 genes (NLRP1and NLRP6) were protective genes with HRs <1 (Fig. 2A). In addition, the LASSO Cox regression analysis yielded 6 genes that were used to construct a 6 – gene model according to the optimum λ value (Fig. 2B, C). The risk score was calculated as follows: risk score = (9.95×10−4* expression level of GSDME) + (2.96×10−5* expression level of IL6) + (−2.67×10−5* expression level of NLRP1) + (2.65×10−4* expression level of CASP8) + (−0.11* expression level of NLRP6) + (2.75×10−3* expression level of CASP6).
Fig. 2.
Construction of risk score model in the TCGA cohort. (A) Univariate Cox regression analysis of OS for each pyroptosis-related gene, and 7 genes with P < 0.05. (B) LASSO regression of the 7 OS-related genes. (C) Cross-validation for tuning the parameter selection in the LASSO regression. (D) Distribution of patients based on the risk score. (E) PCA plot for HNSCCs based on the risk score. (F) The survival status for each patient (low-risk population: on the left side of the dotted line; high-risk population: on the right side of the dotted line).
Based on the median score, the HNSCC patients were randomly and equally divided into low- and high-risk subgroups (Fig. 2D). PCA showed efficient discrimination of patients with different risks into two clusters (Fig. 2E). Furthermore, t-distributed stochastic neighbor embedding (tSNE) was performed to validate the PCA result (Fig. S2). Patients in the high-risk group had a higher death rate and shorter survival compared to those in the low-risk group (Fig. 2F).
Survival analysis based on risk-score model in comparison with single gene
HNSCC patients were equally and randomly divided into low- and high - expression subgroups on the expression profile of the six pivotal genes to perform single gene survival analysis. However, there is only difference in OS between the low-and high-expression groups based on NLRP1 (P = 0.022, Fig. 3C), all of other genes were nonsignificant (Fig. 3A, B, G-I). It is worth noting that there was a remarkable difference between low-and high-risk groups in survival analysis (P < 0.0001, Fig. 3M). ROC analysis was applied to evaluate the sensitivity and specificity of the prognostic model, and we found that the area under the ROC curve (AUC) was 0.869 for 1-year, 0.759 for 3- year, and 0.757 for 5-year survival (Fig. 3N). But the result of the single genes’ ROC analysis fell short of our expectation when compared with the result of risk-score, which demonstrated that our risk-score model yielded better performances on the survial analysis of TCGA cohort (Fig 3 D-F, J-L).
Fig. 3.
OS analysis and ROC analysis based on each single gene and risk score. (A) Kaplan-Meier curve based on GSDME. (B) Kaplan-Meier curve based on IL-6. (C) Kaplan-Meier curve based on NLRP1. (D) ROC analysis based on GSDME. (E) ROC analysis based on IL-6. (F) ROC analysis based on NLRP1. (G) Kaplan-Meier curve based on CASP8 (H) Kaplan-Meier curve based on NLRP6 (I) Kaplan-Meier curve based on CASP6 (J) ROC analysis based on CASP8 (K) ROC analysis based on NLRP6 (L) ROC analysis based on CASP6 (M) Kaplan-Meier curve based on risk score. (N) ROC analysis based on risk score.
Independent prognostic value of the risk-model
Next, we used univariate and multivariate Cox regression analyses to evaluate whether the risk scores derived from the gene model could serve as an independent prognostic marker for prognosis in HNSCC patients. The univariate Cox regression analysis indicated that the T stage, patient's age, use of radiotherapy and risk score affect the HNSCC patients’ survival time (all P < 0.05, Table 1). Through the multivariate analysis, we showed that the risk score could serve as an independent prognostic marker in patients with HNSCC (P = 0.02, Table 1).
Table 1.
Univariate and multivariate analyses of clinical characteristics for overall survival in patients with HNSCC (TCGA cohort).
| Characteristics | Amount (%) | Univariate P 1value | HR2 (95% CI3) | Multivariate P 1value | HR2 (95% CI3) |
|---|---|---|---|---|---|
| Age | 0.02* | 0.31 | |||
| ≤60 | 246 (49.0%) | 1.352 | 1.252 | ||
| > 60 | 256 (51.0%) | (1.056 – 1.639) | (0.956 –1.639) | ||
| Gender | 0.066 | ||||
| Male | 368 (73.3%) | 0.764 | |||
| Female | 134 (26.7%) | (0.574 – 1.018) | |||
| Smoking history | 0.618 | ||||
| Present | 381 (77.4%) | 1.089 | |||
| Absent | 111 (22.6%) | (0.778 – 1.525) | |||
| Alcohol history | 0.733 | ||||
| Present | 333 (67.8%) | 0.952 | |||
| Absent | 158 (32.2%) | (0.716 – 1.265) | |||
| T stage | 0.037* | 0.12 | |||
| T1& T2 | 319 (63.5%) | 1.245 | 1.245 | ||
| T3& T4 | 183 (36.5%) | (1.132 – 1.361) | (0.932 –1.661) | ||
| Clinical stage | 0.238 | ||||
| StageⅠ&Ⅱ | 388 (77.3%) | 1.217 | |||
| StageⅢ&Ⅳ | 114 (22.7%) | (0.878 – 1.688) | |||
| Radiation therapy | 0.002** | 0.03* | |||
| Present | 288 (57.4%) | 0.613 | 0.672 | ||
| Absent | 214 (42.6%) | (0.452 – 0.831) | (0.483 –0.840) | ||
| Risk score | 0.002** | 0.02* | |||
| Low | 251 (50%) | 1.275 | 1.525 | ||
| High | 251 (50%) | (1.118 – 1.358) | (1.278 –1.712) |
All statistical tests were 2-sided. 2HR, Hazard ratio. 3CI, confdence interval.*= P < 0.05; **= P < 0.01.
Expression difference of pivotal genes in practical patients
We performed the qRT-PCR to detect the mRNA level of six fundamental genes of our risk-score model in each patients’ fresh HNSCC tissue and normal tissue. The expression difference of GSDME, IL-6, NLRP1, CASP 8, NLRP6, CASP6 between neoplastic and normal tissue in practical patients cohort were in accordance with that of TCGA cohort. (Fig. 4A-D, F: P < 0.001, Fig. 4E: P < 0.01).
Fig. 4.
qRT – PCR analyses of six fundamental genes between HNSCC and normal tissue. (A) Relative mRNA level of GSDME. (B) Relative mRNA level of IL-6. (C) Relative mRNA level of NLRP1. (D) Relative mRNA level of CASP8. (E) Relative mRNA level of NLRP6. (F) Relative mRNA level of CASP6. ** = P < 0.01, *** = P < 0.001.
Validation of the risk-model in practical application
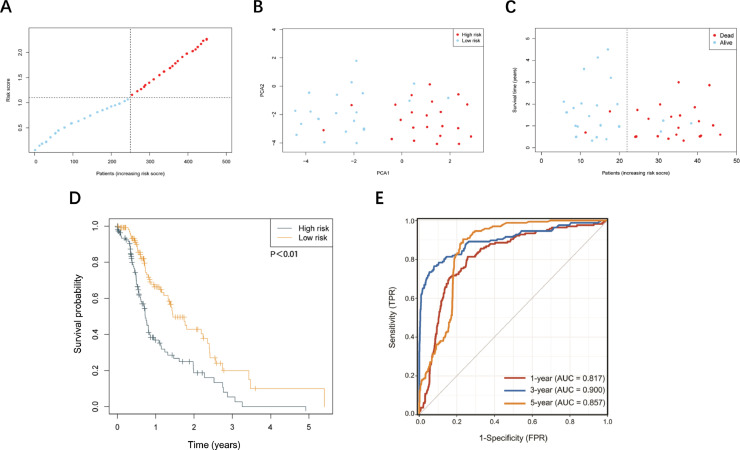
Next, we applied the RNA-seq to patient's HNSCC tissue in order to obtain each patient's gene expression level. We applied this part of data to the risk score formula to get each patient's risk score. 44 HNSCC patients were divided into two groups according to median score (Fig. 5A). Finally, we connected the risk score with patients’ prognostic status in order to see if the risk-model is applicable in practical use. Patients from low-risk group enjoyed longer survival time and better prognosis than those from high-risk group (Fig. 5B-D). Furthermore, in practical patient cohort, our risk-score model was a promising method for predicting HNSCC patients’ prognosis (Fig. 5E).
Fig. 5.
Validation of risk score model in the practical patient cohort. (A) Distribution of patients based on the risk score. (B) PCA plot for HNSCCs based on the risk score. (C) The survival status for each patient (low-risk population: on the left side of the dotted line; high-risk population: on the right side of the dotted line). (D) Kaplan – Meier curve based on risk score. (E) ROC analysis based on risk score.
Comparison of the immune activity between the subgroups
Given that pyroptosis plays a critical role in the immune processes in human cells, especially in the tumor microenvironment, we employed single-sample gene set enrichment analysis (ssGSEA) to compare the enrichment scores of 16 types of immune cells and the activity of 13 immune-related pathways between the low and high-risk groups in TCGA cohort [20], [21], [22]. As shown in Fig. 6A, the high-risk subgroup had lower infiltration of immune cells, especially macrophages, neutrophils or T helper (Th) cells compared to the low-risk subgroup. In addition, patients from the low-risk subgroup had significantly higher activation of chemotactic cytokines receptors (CCR) pathway, human leukocyte antigen (HLA) pathway and typeⅡFN reponse pathway compared to patients in high-risk subgroup (Fig. 6B).
Fig. 6.
Comparison of the ssGSEA scores for immune cells and immune pathways. (A) Comparison of the enrichment scores of 16 types of immune cells between low- (blue box) and high-risk (red box) group in the TCGA cohort. (B) Comparison of the enrichment scores of 13 immune-related pathways between low- (blue box) and high-risk (red box) group in the TCGA cohort. P values were showed as: ns not significant. *P = < 0.05; **P = < 0.01; ***P = < 0.001.
Discussion
Given the low survival rate as a result of local recurrence and lymphatic metastasis, head and neck squamous cell carcinoma (HNSCC) presents great treatment challenges [23]. Besides, there is no standard diagnostic or prognostic tools in oncology. We previously demonstrated that NUDT1, a hydrolase, could independently predict survival of patients with oral squamous cell carcinoma [3]. Despite the promising data, prediction of prognosis using a single gene or factor could be unreliable. Here, we developed a novel model for the prediction of prognosis using risk score formula generated from multiple genes.
Pyroptosis, a form of pattern of cell death that is triggered by proinflammatory signals maintains homeostasis, eliminates abnormal cells and controls the human immune system [10]. In this study, we collected 33 pyroptosis – realated genes, and then screened for differentially expressed genes (DEGs) in the HNSCC and normal tissues. Through the univariate and LASSO Cox regression analysis, 6 of the DEGs (GSDME, IL-6, NLRP1, CASP8, NLRP6, and CASP6) were shown to be pivotal in generation of a risk-score model. Just as mentioned above, single prognostic factor was vulnerable to external influences, multifactor prognostic model may cover this shortage. In present study, only NLRP1 met the requirement to evaluate HNSCC patients’ survival status independently, nevertheless, its distinguishing ability of prediction model was not eligible by ROC analyzing (Fig. 3C, F). Our risk score model based on several pyroptosis – realated genes demonstrated more reliability compared to the use of a single factor both in the TCGA cohort and practical patients cohort.
In present study, NLRP6 possessed the largest absolute value of coefficient among the six fundamental genes of the risk score formula, not only suggesting NLRP6 could be served as the tumor suppressing gene in HNSCC, but indicating that the expression level of NLRP6 in HNSCC patients affects the final score predominantly. Our findings were in accordance with previous study that NLRP6 exerts as a tumor suppressor by interaction with GRP78 and mediating its degradation in gastric cancer [24]. Moreover, NLRP6 deficiency has been implicated in the aggravation of chemical-induced colitis and subsequent tumorigenesis [25]. An interesting study by Normand et al. reported that NLRP6 performs essential functions in the regulation of tissue repair necessary for protection against chemically induced colorectal carcinogenesis. This study showed that NLRP6 deficiency led to dysregulated colonocyte proliferation and migration, thereby facilitating tumor formation [26]. Together, these observations established NLRP6 as a negative regulator of colorectal cancer and suggest that modulation of NLRP6 may be a promising therapeutic alternative for its treatment. Although there were not enough reports shed light on the specific function of NLRP6 in tumorigenesis of HNSCC, it has provided new angle to discover the correlation between cell pyroptosis and HNSCC based on NLRP6 functions in our further studies.
Furthermore, the strong inflammatory effect of cell pyroptosis suggests that it may be dominant in the regulation of tumor immune microenvironment [27], [28], [29], [30]. Thus, we performed enrichment analysis to evaluate whether the low-risk patients were associated with better survival due to higher respone of the immune system. Interestingly, on the macro level, low – risk patients were shown to recruit more immune cells and trigger higher activation of inmmune pathways compared to the high – risk patients. This phenomenon was associated with the fact that macrophages and T helper (Th) cells (Th1 cells, not Th2 cells) are essential in the HNSCC microenvironment in the regulation of the inflammatory reactions caused by pyroptosis (Fig. 6A). Monocyte chemotactic protein-1 (MCP-1, also CCL2), which is secreted by tumor cells and tumor microenvironment, can bind to receptor CCR2 and recruit CCR2+Ly6chigh monocytes. The latter is thought to be precursor cells of tumor-associated macrophages (TAMs) [31]. CCL2 can not only recruit monocytes, but initiate the transformation from CCR2+Ly6chigh monocytes to TAMs and the polarization process of TAMs [32]. There are two main types of polarization of TAMS in the tumor microenvironement under the influence of chemokines: M1 and M2 macrophages. M1 macrophages are involved in the classic activation of the Th1 cell responses and are able to suppress tumor development by releasing NO (nitric oxide), ROS (reactive oxygen species) and other tumor killing factors [33,34]. On the other hand, M2 macrophages are involved in alternative activation pathway in the Th2 cell response. The M2 macrophages secrete interleukin-10 (IL-10), transforming growth factor-β (TGF-β), prostaglandin-E2 (PGE2), vascular endothelial growth factor (VEGF) and matrix metalloproteinases (MMPs) which facilitate tumor genesis and angiogenesis [35,36]. Thus, the evidences account for the high level of macrophages, T helper (Th) cells, especially Th1 cells and CCR pathway activation in the low – risk subgroup (Fig. 6B).
Finally, by putting forward our pyroptosis-related prognostic model, we hope it could aid in determining HNSCC treatment regimens and allocating theraputic resources appropriately.
Conclusions
Taken together, our findings demonstrate that there is differential expression of pyroptosis – related genes between the normal and HNSCC tissues. The differentially expressed pyroptosis – related genes were used to successfully generate a risk – score model that could independently predict prognosis in HNSCC patients. Since, there was a strong correlation between our risk – score model and cellular immune activities, there is need to study underlying mechnisms of pyroptosis in tumor immunology.
Declaration of Competing Interest
The authors have declared no conflicts of interest.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (No. 81871458), the Health Clinical Research Project of Shanghai Municipal Health Commission (No. 202040328), Fundamental research program funding of Ninth People's Hospital affiliated to Shanghai Jiao Tong university School of Medicine (No. JYZZ076), Clinical Research Program of Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine (No. JYLJ201801, JYLJ201911) and the China Postdoctoral Science Foundation (No. 2017M611585).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101233.
Contributor Information
Xitao Yang, Email: xitao123456@126.com.
Lixin Su, Email: sulixin1975@126.com.
Xindong Fan, Email: fanxindong@aliyun.com.
Appendix. Supplementary materials
References
- 1.Sung H. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Bray F. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Shen Y. NUDT1: a potential independent predictor for the prognosis of patients with oral squamous cell carcinoma. J. Oral Pathol. Med. 2020;49:210–218. doi: 10.1111/jop.12974. [DOI] [PubMed] [Google Scholar]
- 4.Tonnus W. The pathological features of regulated necrosis. J. Pathol. 2019;247:697–707. doi: 10.1002/path.5248. [DOI] [PubMed] [Google Scholar]
- 5.Cookson B.T., Brennan M.A. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9:113–114. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- 6.Ding J. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 7.Feng S., Fox D., Man S.M. Mechanisms of gasdermin family members in inflammasome signaling and cell death. J. Mol. Biol. 2018;430:3068–3080. doi: 10.1016/j.jmb.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Ding J., Shao F. SnapShot: the Noncanonical Inflammasome. Cell. 2017;168 doi: 10.1016/j.cell.2017.01.008. 544-544 e541. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 10.Shi J., Gao W., Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 2017;42:245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Akino K. Identification of DFNA5 as a target of epigenetic inactivation in gastric cancer. Cancer Sci. 2007;98:88–95. doi: 10.1111/j.1349-7006.2006.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M.S. Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene. 2008;27:3624–3634. doi: 10.1038/sj.onc.1211021. [DOI] [PubMed] [Google Scholar]
- 13.Fujikane T. Genomic screening for genes upregulated by demethylation revealed novel targets of epigenetic silencing in breast cancer. Breast Cancer Res. Treat. 2010;122:699–710. doi: 10.1007/s10549-009-0600-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L.M. Epigenetic regulation of VENTXP1 suppresses tumor proliferation via miR-205-5p/ANKRD2/NF-kB signaling in head and neck squamous cell carcinoma. Cell Death. Dis. 2020;11:838. doi: 10.1038/s41419-020-03057-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S.H., O'Sullivan B. Overview of the 8th Edition TNM Classification for Head and Neck Cancer. Curr. Treat. Options Oncol. 2017;18:40. doi: 10.1007/s11864-017-0484-y. [DOI] [PubMed] [Google Scholar]
- 16.Karki R., Kanneganti T.D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer. 2019;19:197–214. doi: 10.1038/s41568-019-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia X. The role of pyroptosis in cancer: pro-cancer or pro-"host"? Cell Death. Dis. 2019;10:650. doi: 10.1038/s41419-019-1883-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B., Yin Q. AIM2 inflammasome activation and regulation: a structural perspective. J. Struct. Biol. 2017;200:279–282. doi: 10.1016/j.jsb.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Man S.M., Kanneganti T.D. Regulation of inflammasome activation. Immunol. Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupaul-Chicoine J. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. ImmunityImmunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Wei Q. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab. Invest. 2014;94:52–62. doi: 10.1038/labinvest.2013.126. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen I., Rayamajhi M., Miao E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017;17:151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskovitz J., Moy J., Ferris R.L. Immunotherapy for head and neck squamous cell carcinoma. Curr. Oncol. Rep. 2018;20:22. doi: 10.1007/s11912-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X. NLRP6 suppresses gastric cancer growth via GRP78 ubiquitination. Exp. Cell Res. 2020;395 doi: 10.1016/j.yexcr.2020.112177. [DOI] [PubMed] [Google Scholar]
- 25.Chen G.Y., Liu M., Wang F., Bertin J., Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Normand S. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature. 2020;579:421–426. doi: 10.1038/s41586-020-2079-1. [DOI] [PubMed] [Google Scholar]
- 28.Xi G. GSDMD is required for effector CD8(+) T cell responses to lung cancer cells. Int. Immunopharmacol. 2019;74 doi: 10.1016/j.intimp.2019.105713. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368 doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–420. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian B.Z. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z. CCL2/CCR2 axis promotes the progression of salivary adenoid cystic carcinoma via recruiting and reprogramming the tumor-associated macrophages. Front. Oncol. 2019;9:231. doi: 10.3389/fonc.2019.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray P.J. Macrophage polarization. Annu. Rev. Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 34.Shapouri-Moghaddam A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 35.Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 36.Lurier E.B. Transcriptome analysis of IL-10-stimulated (M2c) macrophages by next-generation sequencing. Immunobiology. 2017;222:847–856. doi: 10.1016/j.imbio.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.