Abstract
Epithelial-to-mesenchymal transition (EMT) is an essential mechanism for development and wound healing, but in cancer it also mediates the progression and spread of aggressive tumors while increasing therapeutic resistance. Adoption of a mesenchymal state is also associated with increased iron uptake, but the relationship between EMT and the key regulators of cellular iron metabolism remains undefined. In this regard, the human adrenal cortical carcinoma SW13 cell line represents an invaluable research model as HDAC inhibitor treatment can convert them from an epithelial-like (SW13-) cell type to a mesenchymal-like (SW13+) subtype. In this study we establish SW13 cells as a model for exploring the link between iron and EMT. Increased iron accumulation following HDAC inhibitor mediated EMT is associated with decreased expression of the iron export protein ferroportin, enhanced ROS production, and reduced expression of antioxidant response genes. As availability of redox active iron and loss of lipid peroxide repair capacity are hallmarks of ferroptosis, a form of iron-mediated cell death, we next examined whether HDAC inhibitor treatment could augment ferroptosis sensitivity. Indeed, HDAC inhibitor treatment synergistically increased cell death following induction of ferroptosis. The exact mechanisms by which HDAC inhibition facilitates cell death following ferroptosis induction requires further study. As several HDAC inhibitors are already in use clinically for the treatment of certain cancer types, the findings from these studies have immediate implications for improving iron-targeted chemotherapeutic strategies.
Keywords: Epithelial-to-mesenchymal transition (EMT), Iron, Ferroptosis, Histone deacetylase (HDAC) inhibitors
Highlights
-
•
HDAC inhibitor mediated EMT alters cellular iron metabolism and redox homeostasis.
-
•
Increased iron regulatory protein binding activity promotes iron accumulation in mesenchymal cells.
-
•
Altering cellular iron availability during HDAC inhibitor mediated EMT reduces cell viability.
-
•
HDAC inhibitor treatment increases cell death following ferroptosis induction.
Abbreviations:
- BAX
BCL2 associated X, apoptosis regulator
- BCL2
BCL2 apoptosis inhibitor
- CHAC1
ChaC glutathione specific gamma-glutamylcyclotransferase 1
- DFO
desferrioxamine
- EMT
epithelial-to-mesenchymal transition
- FER1
ferrostatin 1
- GAPDH
Glyceraldehyde-3-Phosphate Dehydrogenase
- GPX4
glutathione peroxidase 4
- HDAC
histone deacetylase
- IRP
iron regulatory protein
- MMP2
matrix metallopeptidase 2
- MMP9
matrix metallopeptidase 9
- PPIB
peptidylprolyl isomerase B
- ROS
reactive oxygen species
- SLC7A11
solute carrier family 7 member 11
- SMARCA2
SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin 2
- SNAI1
snail family transcriptional repressor 1
- SOD2
superoxide dismutase 2
- TGF-β1
transforming growth factor-β 1
- TP53
tumor suppressor p53
1. Introduction
The epithelial-to-mesenchymal-transition (EMT) is a fundamental transcriptional program that is critical for proper embryogenesis and wound healing, but EMT can become aberrantly activated during pathologic conditions such as cancer, ischemia, and chronic inflammation [[1], [2], [3]]. During EMT, epithelial cells undergo changes in gene expression resulting in the reorganization of the cytoskeleton, loss of cell-cell adhesion, and the acquisition of migratory and invasive properties as they transition to a mesenchymal state. Recent studies in the cancer biology field have identified iron as an important regulator of the EMT process by demonstrating that mesenchymal-like cancer stem cells have an increased dependence on intracellular iron [[4], [5], [6]]. Moreover, adoption of a mesenchymal state is associated with increased sensitivity to ferroptosis, a form non-apoptotic programmed cell death mediated by the iron-dependent accumulation of lipid reactive oxygen species (ROS) [7]. Yet, the mechanisms promoting mesenchymal cell iron acquisition and utilization are not understood.
In this regard, the human adrenal cortical carcinoma SW13 cell line may serve as an ideal research model for investigating the changing iron requirements of epithelial cells as they transition to a mesenchymal phenotype. Under standard growth conditions, the majority of SW13 cells exist as a rapidly growing, highly tumorigenic, epithelial-like subtype, that lacks the expression of the tumor suppressor protein SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin 2 (SMARCA2), and are termed SW13-. However, ∼2% of the cells in culture exist as a slow growing, highly invasive, mesenchymal-like subtype that expresses SMARCA2 and the mesenchymal markers vimentin and CD44 and are referred to as SW13+ cells [8]. Intriguingly, HDAC inhibition can induce a phenotype transition from the tumorigenic SW13- to the metastatic SW13+, and the conversion can be confirmed by morphological changes and induction of SMARCA2 expression [8,9].
In this study, we establish the epigenetically plastic SW13 cell line as a model to temporally control EMT initiation and explore the relationship between iron and EMT. Using this model, we found that neither iron chelation nor iron supplementation alters the progression of HDAC inhibitor mediated EMT in SW13 cells. However, increased iron accumulation and reduced antioxidative capacity in mesenchymal-like SW13+ cells do increase their sensitivity to alterations in iron availability and augments their cell death following induction of ferroptosis.
2. Materials and methods
2.1. Cell culture and treatment conditions
The SW13 cell line (ATCC CCL-105) is a primary small cell adrenal gland/cortex carcinoma cell line derived from a 55-year-old Caucasian human. For all experiments, cells were cultured in DMEM 1640 1X with l-glutamine (Corning #17-207-CV Manassas, VA) supplemented with 10% fetal bovine serum and 100 IU penicillin and 100 μg/ml streptomycin (Corning #30-002-Cl Manassas, VA) in a temperature and humidity-controlled incubator (37 °C, 95% humidity, 5% CO2).
For EMT induction, SW13 cells were treated with 2 nM Romidepsin (FK228, Selleckchem, Houston, TX, USA) for 48 h. To examine the influence of iron availability on EMT progression, SW13 cells were treated with 2 nM FK228 for 24 h and then co-treated with 50 μM desferrioxamine (DFO), an iron chelator, or 40 μM hemin, an iron-containing porphyrin, for another 24 h. To induce ferroptosis, cells were treated with either 1 μM or 5 μM erastin for 24 or 48 h. All experiments followed the described treatment dose and length unless mentioned otherwise in the methods session.
2.2. Morphologic and immunofluorescence analysis
To examine the effects of iron availability on the morphologic changes that occur with HDAC inhibitor-induced EMT, SW13 cells were seeded at 4000 cells/well into pre-collagen coated 8-well chamber slides (Ibidi, Fitchburg, WI, USA) and treated as described above. Following their respective treatments, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton-X, and blocked with 1% BSA before incubation with phalloidin conjugated to Alexafluor 488 (A12379, Invitrogen, Waltham, MA, USA) at 1:1000 or Vimentin (Cell Signaling, #5741) at 1:100 in 1% BSA for 1 h. Cells were washed three times with PBS and slides were mounted ProLong™ Gold Antifade Mountant with DAPI (Invitrogen, Waltham, MA, USA). Images were obtained using a BZ-X700 Fluorescent Microscope (Keyence, Osaka, Japan) at 495 nmexcitation/518 nmemission and 360 nmexcitation/460 nmemission for actin filaments and DAPI, respectively, with uniform exposure.
2.3. Intracellular iron assay
Total intracellular iron was measured from at least 1 × 106 control SW13 cells and SW13 cells that had been converted to a mesenchymal like subtype by treatment with 2 nM FK228 for 48 h using an Iron Assay kit (MilliporeSigma, Burlington, MA, USA). Following cell lysis, released iron was reduced and reacted with a chromagen to produce a colorimetric (593 nm) product proportional to the total iron present. Colorimetric changes were measured using a Biotek Synergy H1 (Biotek, Winooski, VT, USA) plate reader. Calculated total iron levels were then normalized to total cell number to account for the influence of HDAC inhibitor treatment effects on cell growth.
2.4. mRNA expression analysis
To assess changes in relative mRNA abundance following HDAC inhibitor-mediated conversion to a mesenchymal subtype, total RNA was extracted from treated cells using Trizol reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer's instructions. Following confirmation of RNA integrity by agarose gel electrophoresis, cDNA was synthesized using Superscript II (ThermoFisher, Waltham, MA, USA). Differences in mRNA expression levels were determined by real-time qPCR using SYBR green chemistry on a Bio-Rad CFX Opus 384 Real-Time PCR system (Bio-Rad, Hercules, CA, USA) and normalized relative to peptidylprolyl isomerase B (PPIB) abundance using the 2-ΔΔCt method [10]. Primer sequences for each mRNA of interest are listed in Supplementary Table 1.
2.5. Cell viability and proliferations assays
To examine the effects of iron availability on cell viability following HDAC inhibitor-induced EMT, SW13 cells were seeded at 4000 cells per well in a 96-well plate and cultured for 24 h before treatment with DMSO (control) or the previously described doses of FK228 and DFO or Hemin. Following a 30 min incubation with Prestoblue reagent (ThermoFisher, Waltham, MA, USA) differences in fluorescence intensity were read at 560 nmexcitation/570 nmemission using a Biotek Synergy H1 (Biotek, Winooski, VT, USA) plate reader. Differences in cell viability were normalized relative to the vehicle control group for each cell line.
To assess the impact of iron availability on proliferation following HDAC inhibitor induced EMT, SW13 cells were seeded into pre-collagen coated 8-well chamber slides (Ibidi, Fitchburg, WI, USA) at 4000 cells/well. Cells were treated with FK228 and DFO or hemin as described above and incubated with 10 μM Click-iT EdU reagent (ThermoFisher, Waltham, MA, USA). Prior to imaging, nuclei were stained with Hoeschst 33,342 stain diluted at 1:1000 for 10 min. Cells were then imaged live on a 4X objective using a BZ-×700 Fluorescent Microscope (Keyence, Okaka, Japan) at 361 nmexcitation/497 nmemission and 495 nmexcitation/519 nmemission wavelengths for Hoechst 33,342 and EdU, respectively. Low photobleach settings and exposure were held consistent throughout the imaging process. ImageJ software [11] was used to count numbers of green (EdU) and blue (Hoechst 33,342) nucleus, and percent proliferation was determined by the ratio between number of newly synthesized DNA (EdU) by the total nuclei (Hoechst 33,342).
2.6. Electrophoretic mobility shift assay (EMSA)
Cells were plated in a 6-well plate at 1 × 105 cells per well and cultured for 24 h before treatment with DMSO (control) or 2 nM FK228 for another 24 h. Cells were washed with 1X PBS and pelleted at 1000×g for 5 min at 4 °C. Cytosolic protein was collected by incubating cells in 2-vol cytosol buffer (1 mM HEPES, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 0.1 M PMSF, 10 μM MG132, 100 X Halt protease inhibitor cocktail (ThermoFisher, Waltham, MA, USA) on ice for 15 min. Cell lysis was completed by adding 0.1 volume of 10% NP40 to the cell suspension followed by 10 s of vigorous vortexing and centrifugation at 12,000×g for 10 min at 4 °C. The supernatant (cytosol) was removed to a fresh microfuge tube and protein concentration was determined based on the bicinchoninic acid assay. Spontaneous IRP1 and IRP2 RNA binding activities was assessed by incubating 5 μg total protein with saturating levels (1 nM) of 32P-labeled RNA from the L-ferritin IRE as previously described [12]. Total IRP1 mRNA binding activity was measured by incubating 1 μg of cytosolic protein to saturating levels of 32P-labeled RNA in the presence of 4% β-mercaptoethanol. Absolute quantitation of mRNA binding activity was determined using Optiquant Acquisition and Analysis software (Packard Bioscience, Meriden, CT, USA).
2.7. Protein expression analysis
Following their respective treatments, total protein was collected from SW13 cells by lysing cells in radioimmunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 2 mM EDTA, 150 mM NaCl) supplemented with 1X Halt Protease inhibitor cocktail (ThermoFisher Waltham, MA, USA), 1 mM DTT, 1 mM Citrate, 1 mM phenylmethylsulphonyl fluoride (PMSF), 10 μM Mg132. Samples were vortexed every 5 min for 20 min followed by centrifugation at 14,000×g for 15 min at 4 °C. The protein containing supernatant was collected and total protein concentration was determined based on the bicinchoninic acid assay.
To assay for differences in IRP target protein expression, 30 μg total protein was solubilized in 2 X Laemmli sample buffer (0.01% Bromophenol blue, 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.125 M Tris-HCl pH 6.8) and heated at 95 °C for 5 min before being separated by electrophoresis at 150 V on a Mini-PROTEAN TGX Stain-Free precast 4–20% polyacrylamide gel (Bio-Rad, Hercules, CA, USA). Separated proteins were then transferred to a PVDF membrane at 300 mA for 75 min. Following confirmation of equal transfer by Ponceau-S staining, membranes were blocked for 1 h in 5% nonfat milk in 1X TBS-0.01% Tween-20 at room temperature. Blots were then incubated in primary CD71/TFRC (Cell signaling, #D7S5Z), SLC40A1 (ferroportin) (Invitrogen, #PA5-22993), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz, #0411) antibodies overnight at 4 °C at 1:1000 dilution in 5% non-fat dry milk in 1X Tris-buffered saline with 0.01% Tween-20 before incubation with appropriate HRP-linked secondary antibodies, either anti-rabbit IgG #7074P2 or anti-mouse IgG #7076P2 (Cell Signaling Technology, Danvers, MA, USA) at room temperature for 1 h at 1:10,000 dilution. Membranes were then immersed in SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher, Waltham, MA, USA) and visualized using a FluorChem R ProteinSimple fluorescence imaging system (R&D Systems, Minneapolis, MN, USA).
2.8. Oxidative stress detection assay
To assess the influence of HDAC inhibitor induced EMT on ROS production, SW13 cells were seeded into pre-collagen coated 8-well chamber slides (Ibidi, Fitchburg, WI, USA) at 4000 cells/well and treated with 2 nM FK228 for 48 h. Cells were washed once with Hank's Balanced Salt Solution (HBSS) (Cellgro, Lincoln, NE, USA) prior to incubation with 5 μM CellRox™ Deep Red Reagent (ThermoFisher, Waltham, MA, USA) for 30 min at 37 ᵒC. Cells were then washed twice with HBSS, fixed with 4% paraformaldehyde, and slides were mounted ProLong™ Gold Antifade Mountant with DAPI (Invitrogen, Waltham, MA, USA). Oxidized probe was visualized at 40X using a BZ-X700 Fluorescent Microscope (Keyence, Osaka, Japan) with fluorescence 640 nmexcitation/665 nmemission. Low photobleach settings and exposure were held consistent through imaging process.
2.9. Lipid peroxidation assay
To determine how HDAC inhibitor treatment influences lipid peroxidation with and without co-treatment with erastin, a potent ferroptosis inducer, SW13 cells were plated at 4000 cells/well into a black-walled 96-well plate and treated with 2 nM FK228, 5 μM erastin, or a combination of 2 nM FK228 and 5 μM erastin for 24 and 48 h. Following a single wash with HBSS, levels of peroxidized lipids were assayed by incubating treated cells with 5 μM BODIPY 581/591 C11 for 20 min at 37 °C before washing once again with HBSS and measuring differences in fluorescence intensity at 488 nmexcitation/510 nmemission and 581 nmexcitation/591 nmemission wavelengths using Biotek Synergy H1 (Biotek, Winooski, VT, USA) plate reader. The fluorescence ratio, 591 nm[reduction]/510 nm[oxidation], was calculated and normalized to cell viability, which was assessed in parallel plates using PrestoBlue reagent (ThermoFisher, Waltham, MA, USA) as described above.
2.10. Statistical analyses
Student's t-test was used to analyzed differences in total iron, IRP mRNA binding activity, and relative mRNA expression between control and HDAC inhibitor-converted mesenchymal-like SW13 cells. Differences in cell growth, proliferation, viability, and lipid peroxidation following HDAC inhibitor treatment and iron treatments were analyzed using one-way ANOVA. When statistically significant effects were identified by ANOVA, post hoc analyses were performed to make pairwise comparisons using the Tukey HSD method. All tests were performed using SPSS v23.0 software (IBM-SPSS; Chicago, IL, USA). Descriptive statistics were calculated for all variables and include mean ± SEM. Differences will be considered statistically significant at the 95% confidence level (alpha = 0.05). All experiments were repeated 3 times, with n = 3 per group.
3. Results
3.1. HDAC inhibitor treatment induces EMT and promotes iron accumulation in SW13 cells
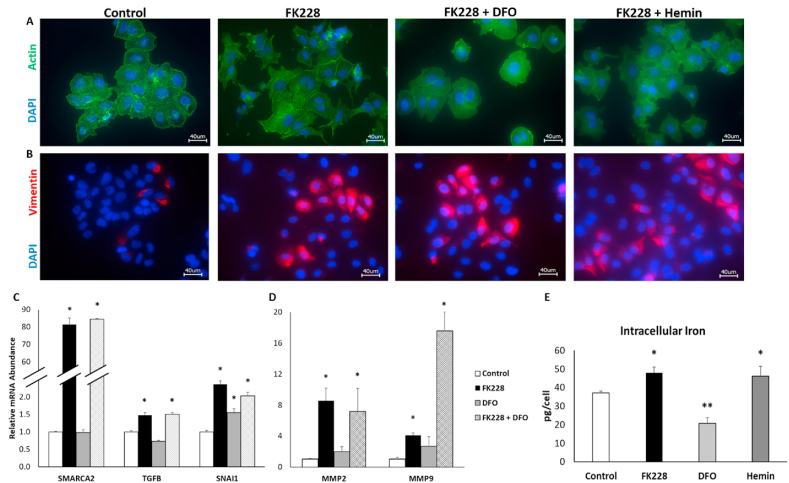
To confirm that the HDAC inhibitor induced SW13 phenotype conversion models canonical EMT signaling pathways, cellular morphology and EMT markers were assessed after treatment with the potent HDAC inhibitor, FK228, for 48 h. Under control conditions, the majority of SW13 cells display an epithelial-like cortical actin cytoskeleton (Fig. 1A, left panel). FK228 treatment results in cytoskeleton reorganization consistent with EMT, including fiber-like actin organization, and loss of cell-cell tight junctions (Fig. 1A), as well as increased expression of vimentin (Fig. 1B). A small amount of vimentin was detectable in the control cells (Fig. 1B left panel). However, this is consistent with the epigenetically plastic nature of SW13 cells and previous observation that ∼2–3% of the SW13 cell population will spontaneously transition to a mesenchymal phenotype under standard culture conditions [8]. Notably, not all cells treated with FK228 for 48 h expressed vimentin, but we have previously shown that it takes 72 h to achieve 100% conversion of the entire SW13 cell population to the SW13+ phenotype [9].
Fig. 1.
HDAC inhibitor mediated EMT in SW13 cells is not influenced by changes in cellular iron availability. (A) EMT-like changes in actin (green) morphology induced by treatment with 2 nM FK228 are not altered by iron chelation (FK228 + DFO) or iron supplementation (FK228 + Hemin). (B) Increased expression of the mesenchymal marker, vimentin by FK228 treatment is not influenced by co-treatment with DFO or hemin. (C) Increased mRNA expression of the mesenchymal markers SMARCA2, TGFβ1, and SNAI1 following HDAC inhibitor treatment were not influence by iron chelation. (D) The mRNA expression of matrix metalloproteinase enzymes MMP2 and MMP9 was also increased by HDAC inhibitor treatment and unaffected by iron chelation. (E) Increased intracellular iron accumulation following HDAC inhibitor mediated EMT (FK228) were consistent with levels observed following iron supplementation (Hemin). Images were taken using a 40X objective lens. Data are presented as mean ± SEM. * Denotes significant difference compared to control, p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
SW13+ phenotype conversion was further confirmed by the strong induction of SMARCA2 mRNA expression upon FK228 treatment (Fig. 1C), whereas SMARCA2 mRNA expression in untreated SW13 cells is nearly undetectable. To support our hypothesis that the SW13 mesenchymal phenotype shift was occurring via an EMT-like process, we analyzed EMT gene expression markers by qPCR. Indeed, the relative mRNA expression of both transforming growth factor-β 1 (TGF-β1), which is critical for EMT induction, and the EMT related transcription factor, snail family transcriptional repressor 1 (SNAI1), were significantly increased 1.5-fold and 2.4-fold, respectively (Fig. 1C). Moreover, the expression of matrix metallopeptidase 2 (MMP2) and MMP9 were also significantly increased by FK228 treatment, which supports our previous findings that the more mesenchymal-like SW13+ cell have increased invasive capacity (Fig. 1D) [9].
Recent studies in the cancer biology field have identified iron as an important component of EMT by demonstrating that mesenchymal-like cancer stem cells have an increased dependence on intracellular iron [1,4]. Therefore, we investigated the influence of SW13 phenotype conversion on total intracellular iron. HDAC inhibitor treatment significantly increased total intracellular iron levels by 29.5% in SW13 cells (Fig. 1E). Intriguingly, the increase in intracellular iron following 48 h of HDAC inhibitor treatment was equivalent to the increase in intracellular iron observed following 24 h of treatment with 40 μM of the iron supplement, hemin, indicating that SW13+ cells are indeed accumulating excess iron (Fig. 1E). These findings also indicate that SW13 cells can serve as a model for exploring the link between iron and EMT.
3.2. Altering iron availability does not interfere with HDAC inhibitor mediated SW13+ cell type conversion
Iron chelation has previously been shown to attenuate TGFβ1-induced EMT [13]. Thus, we examined how alterations in iron availability would influence HDAC inhibitor induced EMT in SW13 cells. To do so, SW13 cells were treated with 2 nM FK228 for 24 h before the addition of either an iron chelator (DFO) or supplemental iron (hemin). Then co-treatment continued for another 24 h before assessing changes in morphology and the expression of EMT markers. Following co-treatment with FK228 and DFO, while some more fiber-like cortical actin structures consistent with an SW13+ phenotype were observed, more cell-cell tight junctions appeared to remain intact (Fig. 1A). Conversely, the heme iron supplemented FK228 treated cells appeared indistinguishable from cells treated with FK228 alone. Similarly, induction of vimentin expression by FK228 was not significantly impacted by DFO or hemin treatment (Fig. 1B).
As iron chelation appeared to have a more significant impact on HDAC inhibitor mediated EMT morphology in SW13 cells, we investigated the influence of DFO treatment on the expression of EMT markers. Cells were co-treated with FK228 and DFO as described above and the expression of SMARCA2, TGFβ1, and SNAIL1 mRNA was measured by qPCR. Iron chelation did not impact the HDAC inhibitor mediated induction of any of the EMT markers examined (Fig. 1C). In fact, DFO treatment alone was sufficient to significantly increase the mRNA expression of SNAI1 above that of control levels (Fig. 1C). Similarly, DFO co-treatment did not influence the HDAC inhibitor mediated induction of the invasive marker MMP2, and actually amplified the increased expression of MMP9 (Fig. 1D). These findings suggest that in SW13 cells, once EMT is initiated, limiting cellular iron availability is not sufficient to block the EMT process.
3.3. Iron regulatory proteins contribute to iron accumulation during HDAC inhibitor mediated SW13 cell EMT
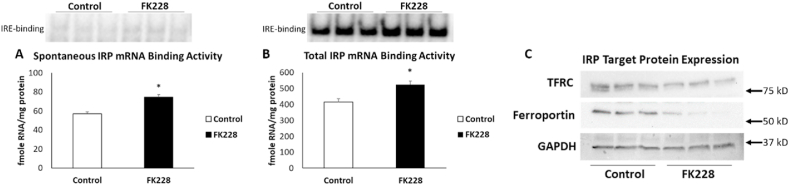
Intracellular iron homeostasis is regulated by iron regulatory proteins (IRP1 and IRP2). These proteins bind mRNA in an iron-dependent fashion, thereby “sensing” intracellular iron status and, accordingly, coordinating the uptake, storage, and utilization of iron [14]. Thus, to investigate the mechanisms contributing to increased intracellular iron following SW13 cell EMT, we analyzed the activity of IRPs following HDAC inhibitor treatment. In SW13 cells, spontaneous IRP mRNA binding activity significantly increased by 14%, from 57 ± 2 to 74 ± 3 fmol RNA/mg protein following 2 nM FK228 treatment for 24 h (Fig. 2A).
Fig. 2.
IRP mRNA binding activity is increased and ferroportin expression is decreased following HDAC inhibitor mediated EMT in SW13 cells. Spontaneous (A) and total (B) mRNA binding activity and expression of the IRP target proteins TFRC and ferroportin (C) in SW13 cells that were treated with DMSO (Control) or with 2 nM FK228 for 24 h were measured by gel-shift assay and Western blot, respectively. GAPDH was used as a loading control. Data are presented as mean ± SEM. * Denotes significant difference compared to control, p < 0.05.
The addition of the reductant β-mercaptoethanol (β-ME) results in disassembly of the Fe–S cluster in IRP1 and slight increase in the binding of the latent IRP2 thus allowing for the measurement of the total IRP1/2 protein present [15]. Following the addition of β-ME to the same cytoplasmic extracts used above, we observed a significant increase in total IRP mRNA binding capacity ∼14%, from 415 ± 20 to 525 ± 22 fmol/mg (Fig. 2B). These findings suggest that the observed increase is likely due to an increase in the total abundance of IRPs rather than a spontaneous increase in mRNA binding. Regardless of mechanism, increased IRP mRNA binding is expected to increase the expression of the iron uptake protein, TFRC, and decrease the expression of the iron export protein, ferroportin. While we did not observe the expected corresponding increase in the expression of TFRC, ferroportin protein expression was significantly decreased in SW13 cells following HDAC inhibitor treatment (Fig. 2C). These finding suggests that the accumulation of iron in SW13 cells during EMT is not due to increased iron uptake, but rather a reduced ability to export intracellular iron.
3.4. Both iron chelation and iron supplementation reduce proliferation and promote cell death in HDAC inhibitor treated SW13 cells
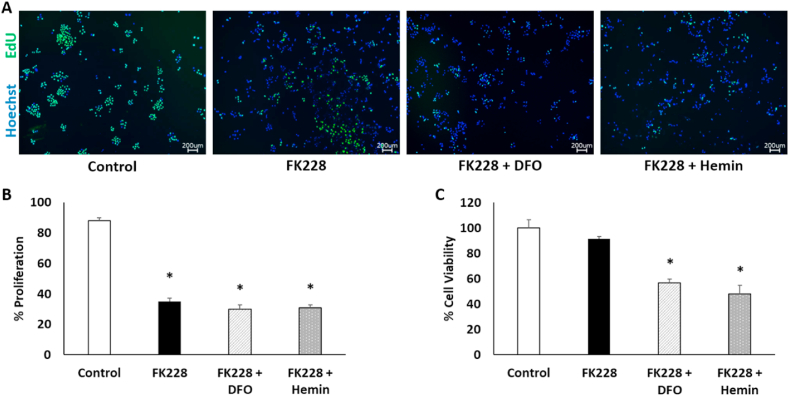
HDAC inhibitors have been extensively studied for their anti-cancer properties (reviewed in Ref. [16]. However, while HDAC inhibitors induce toxicity and slow cell growth in some cell types, they can also increase metastatic properties and chemotherapy resistance in others [17,18]. As iron is also critical for tumor cell growth, we examined whether manipulation of intracellular iron availability could improve HDAC inhibitor treatment outcomes. SW13 cell proliferation and viability were assessed following treatment with 2 nM FK228 for 24 h followed by co-treatment with 50 μM DFO or 40 μM hemin as described above. As expected, proliferation was significantly reduced following FK228 treatment alone (Fig. 3A–B). However, co-treatment with either DFO or hemin did not further decrease proliferation rates (Fig. 3A–B).
Fig. 3.
Both iron chelation and iron supplementation reduce cell viability in HDAC inhibitor treated SW13 cells. (A) Images and (B) quantitation of EdU-positive cells in SW13 cells following treatment with DMSO (Control) or 2 nM FK228 treatment for 24 h and then co-treatment with 50 μM DFO (FK228 + DFO) or 40 μM hemin (FK228 + Hemin) for another 24 h. (C) Percent cell viability was measured by fluorometric assay. Images were taken through an 4X objective lens. Data are presented as mean ± SEM. *Denotes statistical significance, p < 0.05.
To understand whether the observed decrease in proliferation was due to decreased cell division, or instead the result of increased cell death because of iron chelation or iron toxicity, we next assessed cell viability. Viability was unaffected by FK228 treatment alone, but notably both iron chelation and iron supplementation in combination with FK228 treatment significantly decreased cell viability as indicated by a reduced number of metabolically active cells (Fig. 3C). Collectively, these results indicate that HDAC inhibitor-induced mesenchymal-like SW13+ cells are more sensitive to alterations in iron availability and suggest that manipulation of iron-dependent pathways could be used to enhance the therapeutic efficacy of HDAC inhibitors.
3.5. SW13 cells exhibit increased oxidative stress during HDAC inhibitor mediated EMT
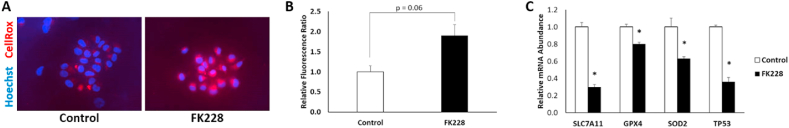
Iron is an essential but potentially toxic nutrient due to its propensity to form free radicals. To investigate mechanisms that could contribute to increased sensitivity to alterations in iron availability following HDAC inhibitor induced EMT in SW13 cells we used a fluorescent probe to measure cellular oxidative stress and found that SW13 cells have significantly higher levels of reactive oxygen species (ROS) following 48 h of FK228 treatment (Fig. 4A). HDAC inhibition has previously been shown to reduce the expression of solute carrier family 7 member 11 (SLC7A11), a crucial component of the cystine import system, system xc−, which is essential for glutathione biosynthesis, and subsequently the production of essential antioxidants such as glutathione peroxidase 4 (GPX4) [19,20]. Consistent with these findings, we found that HDAC inhibitor converted SW13+ cells had significantly reduced mRNA expression of SLC7A11 and GPX4 (Fig. 4B), suggesting that SW13+ cells may have a reduced capacity for lipid peroxide repair [21]. Intriguingly, HDAC inhibitor converted SW13+ cells also had reduced mRNA expression of the mitochondrial ROS detoxifier superoxide dismutase 2 (SOD2), and the tumor suppressor p53 (TP53) (Fig. 4B), which can further contribute to higher cellular oxidative stress levels [22]. Altogether, the weakened antioxidant defenses and increased iron accumulation suggests that mesenchymal-like SW13+ cells may be more susceptible to ferroptosis, a form of cell death driven by the iron-dependent accumulation of lipid ROS.
Fig. 4.
HDAC inhibitor-converted mesenchymal-like SW13 cells have increased levels of ROS and reduced expression of antioxidant defense genes. (A) ROS levels in SW13 following treatment with DMSO (Control) or 2 nM FK228 treatment were measured by incubating cells with 5 μM CellRox deep red reagent for 30 min and (B) quantitated relative to Hoechst stained nuclei with ImageJ. Images were taken through an 40X objective lens. (C) The relative mRNA abundance of SOD2, SLC7A11, TP53, and GPX4 were significantly decreased by FK228 treatment. Data are presented as mean ± SEM. *Denotes significance compared to control, p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. HDAC inhibition enhances erastin-induced cell death in mesenchymal-like SW13 cells
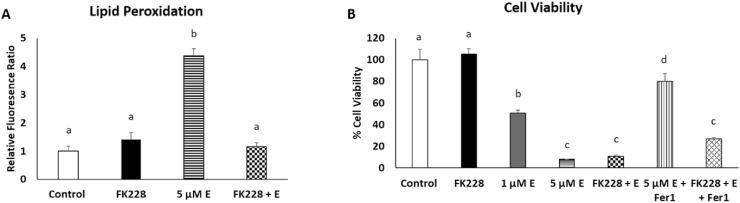
To test the hypothesis that SW13+ cells would have increased susceptibility to ferroptotic cell death, differences in lipid peroxidation and cell viability were measured in SW13 cells were treated with either FK228, 1 μM or 5 μM erastin, or a combination of FK228 and 1 μM erastin. Erastin functions by inhibiting system xc−-mediated cystine import, thereby abolishing glutathione biosynthesis, and crippling cellular antioxidant defense systems, which ultimately leads to an accumulation of peroxidized lipids and ferroptotic cell death [23]. Intriguingly, after 48 h, lipid peroxidation was increased only in those cells treated with 5 μM erastin (Fig. 5A) despite the fact that cell viability was dramatically reduced in the cells co-treated with FK228 and the low dose of erastin (Fig. 5B). Indeed, there was significantly more cell death in SW13 cells that were co-treated with FK228 and the low dose of erastin than when they were treated with erastin alone at an equivalent dose (Fig. 5B). To demonstrate that these cells were succumbing to ferroptosis, we performed rescue experiments by co-treating the high dose erastin treated cells and FK228 + low dose erastin treated cells with ferrostatin-1 (Fer1), a potent inhibitor of ferroptosis [23]. The increase in cell viability to near control levels in the cells treated with erastin and Fer1 indicates that indeed, the majority of cell death in the erastin treated cells can be attributed to ferroptosis alone (Fig. 5B). However, the addition of Fer1 to the FK228 + erastin treated cells elicited only a modest effect on cell viability.
Fig. 5.
HDAC inhibitor treatment synergistically promotes cell death following ferroptosis induction. (A) Lipid peroxidation levels were assessed by measuring relative changes in fluorescence of unoxidized versus oxidized C11-BODIPY probe following treatment with DMSO (control), 2 nM FK228, 1 μM erastin 5 μM erastin (5 μM E), or a combination of 2 nM FK228 and 1 μM erastin (FK228 + E) for 48 h. Changes in relative amounts of oxidized probe were normalized relative to cell viability. (B) Cell viability was measured following treatment with DMSO (control), 2 nM FK228, 1 μM erastin (1 μM E) or 5 μM erastin (5 μM E), or a combination of 2 nM FK228 and 1 μM erastin (FK228 + E). Rescue experiments were performed by adding 10 μM Fer1 to the 5 μM erastin treated cells (5 μM E + Fer1) and the combination of 2 nM FK228 and 1 μM erastin treated cells (FK228 + E + Fer1). Superscripts denote statistical difference, p < 0.05. Treatments with a shared superscript are not statistically significant. Data are presented as mean ± SEM.
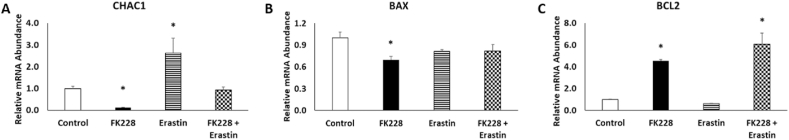
These findings led us to question whether cells co-treated with FK228 and erastin were succumbing to ferroptotic cell death at all. Thus, we sought to further characterize the type of cell death which is occurring by investigating the relative roles of ferroptosis and apoptosis. Gene expression analysis revealed that mRNA expression of ChaC glutathione specific gamma-glutamylcyclotransferase 1 (CHAC1), a marker of ferroptosis, was elevated in SW13 cells treated with 5 μM erastin, but not in SW13 cells treated with FK228 alone, or co-treated with FK228 + 1 μM erastin (Fig. 6A). These findings suggest that ferroptosis may not be the only cause of reduced cell viability in the cells treated with both FK228 and erastin. As HDAC inhibitor treatment has also been shown to promote apoptosis in cancer cell lines, we examine the expression of the pro-apoptotic and anti-apoptotic genes, BCL2 associated X, apoptosis regulator (BAX) and BCL2 apoptosis inhibitor (BCL2), respectively. Interestingly, BAX mRNA expression was modestly, but significantly decreased, in cells treated with FK228 alone (Fig. 6B), while BCL2 mRNA expression was increased nearly 5-fold in cells treated with FK228 alone and the FK228 + erastin combination (Fig. 6C). These findings are consistent with previous research demonstrating that BCL2 is overexpressed during EMT [24,25], and provides insight into previously observed chemotherapeutic resistance in SW13+ cells [18].
Fig. 6.
HDAC inhibitor treatment promotes anti-apoptotic mRNA expression in SW13 cells. The relative abundance of (A) ferroptotic, (B) pro-apoptotic, and (C) anti-apoptotic mRNAs were measured by qPCR following treatment with DMSO (Control), 2 nM FK228, 5 μM erastin, or a combination of 2 nM FK228 and 1 μM erastin for 48 h. Data are presented as mean ± SEM. *Denotes significance compared to control, p < 0.05.
4. Discussion
In this study, we demonstrate that HDAC inhibition induces EMT in SW13 cell through the activation of canonical EMT-driving transcription factors, TGFβ1 and SNAI1. In agreement with previous findings [6], we show that the more mesenchymal-like SW13+ cells have higher levels of intracellular iron than their epithelial SW13- counterparts. Thus, SW13 cells can serve as a model system for exploring the relationship between iron and EMT.
Using this model, we show that increased intracellular iron levels following EMT may be attributed to increased IRP mRNA binding activity and the subsequent downregulation of the iron export protein, ferroportin. Interestingly, both spontaneous and total IRP mRNA binding increased about 14% each following HDAC inhibitor treatment. As human IRP1 and IRP2 do not separate during standard gel shift analyses we cannot determine the individual contributions to the increase in spontaneous and total IRP mRNA binding. However, as IRP1 is regulated by insertion and removal of an Fe–S cluster, whereas IRP2 is regulated at the level of protein stability [14], it is tempting to speculate that the equivalent increase in spontaneous IRP binding and total IRP protein levels are due to increased IRP2 stability alone. Future research should interrogate mechanisms that could promote IRP2 stability during HDAC inhibitor induced EMT.
Increased levels of intracellular iron, such as those observed in the mesenchymal-like SW13+ cells, can contribute to the formation of ROS that can both drive EMT [13], but also damage the DNA of cells, ultimately leading to cell death [26]. Herein, we show that reduced antioxidant gene expression in SW13+ cells also limits their capacity to repair ROS-mediated damage, which increases their susceptibility to death following ferroptosis induction. These results are in agreement with previous work demonstrating that mesenchymal cells have increased ferroptosis sensitivity [7] and that HDAC inhibitor treatment could be used synergistically to augment ferroptotic cell death [27]. However, the question remains as to whether the increased intracellular iron levels proceed ROS accumulation during EMT, or if high levels of ROS are disrupting Fe–S cluster synthesis to increase IRP mRNA binding to ultimately promote mesenchymal cell iron accumulation. Moreover, because both HDAC inhibitor treatment and alterations in cellular iron availability can promote apoptosis, which in turn can elicit ROS generation in mitochondria [26,28,29], future studies should seek to mechanistically determine how HDAC inhibitor treatment augments cell death following ferroptosis induction.
In this regard, consideration of the TP53 status of the SW13 cell line is essential to place these results into a wider context. Activation of wild-type P53 is a well characterized inducer of ferroptosis (reviewed in Ref. [30]). A key feature of this ferroptosis is the downregulation of SLC7A11 independently from P53-mediated apoptosis and senescence [19]. Intriguingly, our data show a decrease in TP53 and SLC7A11 mRNA levels suggesting that the P53 level is not a key determinant of ferroptosis in this system. However, it should be noted that SW13 cells are homozygous for an H193Y gain-of-function (GOF) TP53 mutation and accumulate large amounts of mutant P53 protein [31]. So, it is possible that the reduced level of GOF p53 in SW13 cells after treatment may be sufficient to mediate ferroptosis.
The expression of a GOF p53 mutation in SW13 is relevant to this work in two ways. First, EMT is a complex process which is altered by many inputs but mutant p53 proteins clearly promote EMT [32,33]. Second, it should be noted that GOF p53 hotspot mutations show increased sensitivity to ferroptosis and some do not appear to act by canonical iron regulatory protein pathways [34]. Thus, the expression of the unusual H193Y GOF p53 mutation in the SW13 cell line may have an alteration in the normal p53 mediated ferroptosis pathway. In this context, it is important to note that the ability of p53 to mediate ferroptosis appears to be in the N-terminal trans-activation domain and not associated with the DNA binding domain where mutations endowing p53 with GOF occur [35].
Lastly, the complex role of HDAC inhibitor treatment in cancer biology also needs to be considered. As epigenetic regulators which alter the expression of a substantial portion (5–20%) of the genome [36], the majority of reports suggest that they promote EMT [17,[37], [38], [39], [40]] although some suggest the opposite [41,42]. This complexity may, in part, be due to the fact that HDAC inhibitor treatment will increase the levels of lysine acetylation in signaling proteins regulated by this post-translational modification. Thus, there are likely to be many factors which are involved in the ability of HDAC inhibitor treatment to promote EMT and influence iron metabolism.
5. Conclusions
The importance of these studies is underscored by the fact that iron is an essential, yet potentially toxic nutrient. Thus, the capacity to manipulate iron availability in specific cell types, without compromising systemic iron homeostasis is an essential component to the development of iron-targeted therapeutic strategies. Herein, we show that HDAC inhibitor treatment can augment ferroptosis sensitivity in SW13 cells by inducing EMT and altering intracellular iron levels. The implications of these findings are significant as many other cell types have also been shown to adopt a mesenchymal phenotype following HDAC inhibitor treatment [17,38,43]. Moreover, as several HDAC inhibitors are already used clinically to treat a wide variety of diseases, the findings from these studies could be used to develop iron-target therapeutic strategies that could improve clinical outcomes for multiple disorders.
Authors’ contributions
T.O. and E.H. performed the main experiments were involved in data analysis, preparation of figures and photos, and original draft preparation. D.L. and W.C. helped with data analysis and interpretation and manuscript review and editing. M.M designed the research and was responsible for project administration, funding acquisition and manuscript preparation.
Funding information
This research was supported by start-up funding from the Vice President of Research's office at Oklahoma State University and by the National Institutes of Health, National Cancer Institute grant R03 CA259595.
Declaration of competing interest
None of the authors have any conflicts of interests to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102149.
Contributor Information
Thais Oliveira, Email: thais.g.oliveira@okstate.edu.
Evan Hermann, Email: evanhermann93@gmail.com.
Daniel Lin, Email: dingbo.lin@okstate.edu.
Winyoo Chowanadisai, Email: winyoo.chowanadisai@okstate.edu.
Elizabeth Hull, Email: ehullx@midwestern.edu.
McKale Montgomery, Email: mckale.montgomery@okstate.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.El Hout M., Dos Santos L., Hamai A., Mehrpour M. A promising new approach to cancer therapy: Targeting iron metabolism in cancer stem cells. Semin. Canc. Biol. 2018;53:125–138. doi: 10.1016/j.semcancer.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Nieto M.A. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 3.Skrypek N., Goossens S., De Smedt E., Vandamme N., Berx G. Epithelial-to-Mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33(12):943–959. doi: 10.1016/j.tig.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Mai T.T., Hamai A., Hienzsch A., Caneque T., Muller S., Wicinski J., Cabaud O., Leroy C., David A., Acevedo V. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 2017;9(10):1025–1033. doi: 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rychtarcikova Z., Lettlova S., Tomkova V., Korenkova V., Langerova L., Simonova E., Zjablovskaja P., Alberich-Jorda M., Neuzil J., Truksa J. Tumor-initiating cells of breast and prostate origin show alterations in the expression of genes related to iron metabolism. Oncotarget. 2017;8(4):6376–6398. doi: 10.18632/oncotarget.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schonberg D.L., Miller T.E., Wu Q., Flavahan W.A., Das N.K., Hale J.S., Hubert C.G., Mack S.C., Jarrar A.M., Karl R.T. Preferential iron Trafficking characterizes glioblastoma stem-like cells. Canc. Cell. 2015;28(4):441–455. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viswanathan V.S., Ryan M.J., Dhruv H.D., Gill S., Eichhoff O.M., Seashore-Ludlow B., Kaffenberger S.D., Eaton J.K., Shimada K., Aguirre A.J. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547(7664):453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamichi-Nishina M., Ito T., Mizutani T., Yamamichi N., Watanabe H., Iba H. SW13 cells can transition between two distinct subtypes by switching expression of BRG1 and Brm genes at the post-transcriptional level. J. Biol. Chem. 2003;278(9):7422–7430. doi: 10.1074/jbc.M208458200. [DOI] [PubMed] [Google Scholar]
- 9.Davis M.R., Daggett J.J., Pascual A.S., Lam J.M., Leyva K.J., Cooper K.E., Hull E.E. Epigenetically maintained SW13+ and SW13- subtypes have different oncogenic potential and convert with HDAC1 inhibition. BMC Canc. 2016;16:316. doi: 10.1186/s12885-016-2353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 11.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis M.R., Shawron K.M., Rendina E., Peterson S.K., Lucas E.A., Smith B.J., Clarke S.L. Hypoxia inducible factor-2 alpha is translationally repressed in response to dietary iron deficiency in Sprague-Dawley rats. J. Nutr. 2011;141(9):1590–1596. doi: 10.3945/jn.111.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z., Zhang D., Yue F., Zheng M., Kovacevic Z., Richardson D.R. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1) J. Biol. Chem. 2012;287(21):17016–17028. doi: 10.1074/jbc.M112.350470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson C.P., Shen M., Eisenstein R.S., Leibold E.A. Mammalian iron metabolism and its control by iron regulatory proteins. Biochim. Biophys. Acta. 2012;1823(9):1468–1483. doi: 10.1016/j.bbamcr.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hentze M.W., Kuhn L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 1996;93(16):8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckschlager T., Plch J., Stiborova M., Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 2017;18(7) doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin K.T., Wang Y.W., Chen C.T., Ho C.M., Su W.H., Jou Y.S. HDAC inhibitors augmented cell migration and metastasis through induction of PKCs leading to identification of low toxicity modalities for combination cancer therapy. Clin. Canc. Res. : Official J. Am. Assoc. Canc. Res. 2012;18(17):4691–4701. doi: 10.1158/1078-0432.CCR-12-0633. [DOI] [PubMed] [Google Scholar]
- 18.Montgomery M.R., Hull E.E. Alterations in the glycome after HDAC inhibition impact oncogenic potential in epigenetically plastic SW13 cells. BMC Canc. 2019;19(1):79. doi: 10.1186/s12885-018-5129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22(4):266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., Leite de Oliveira R., Huijberts S., Bosdriesz E., Pencheva N., Brunen D., Bosma A., Song J.Y., Zevenhoven J., Los-de Vries G.T. An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell. 2018;173(6):1413–1425. doi: 10.1016/j.cell.2018.04.012. e1414. [DOI] [PubMed] [Google Scholar]
- 21.Dixon S.J., Stockwell B.R. The hallmarks of ferroptosis. Annu. Rev. Cell Biol. 2019;3:35–54. [Google Scholar]
- 22.Tang D., Chen X., Kang R., Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31(2):107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K., Sun B., Zhao X., Wang X., Li Y., Qiu Z., Gu Q., Dong X., Zhang Y., Wang Y. Hypoxia induced epithelial-mesenchymal transition and vasculogenic mimicry formation by promoting Bcl-2/Twist1 cooperation. Exp. Mol. Pathol. 2015;99(2):383–391. doi: 10.1016/j.yexmp.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Zuo J., Ishikawa T., Boutros S., Xiao Z., Humtsoe J.O., Kramer R.H. Bcl-2 overexpression induces a partial epithelial to mesenchymal transition and promotes squamous carcinoma cell invasion and metastasis. Mol. Canc. Res. : MCR. 2010;8(2):170–182. doi: 10.1158/1541-7786.MCR-09-0354. [DOI] [PubMed] [Google Scholar]
- 26.Walter P.B., Knutson M.D., Paler-Martinez A., Lee S., Xu Y., Viteri F.E., Ames B.N. Iron deficiency and iron excess damage mitochondria and mitochondrial DNA in rats. Proc. Natl. Acad. Sci. U.S.A. 2002;99(4):2264–2269. doi: 10.1073/pnas.261708798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zille M., Kumar A., Kundu N., Bourassa M.W., Wong V.S.C., Willis D., Karuppagounder S.S., Ratan R.R. Ferroptosis in neurons and cancer cells is similar but differentially regulated by histone deacetylase inhibitors. eNeuro. 2019;6(1) doi: 10.1523/ENEURO.0263-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim B.M., Choi J.Y., Kim Y.J., Woo H.D., Chung H.W. Desferrioxamine (DFX) has genotoxic effects on cultured human lymphocytes and induces the p53-mediated damage response. Toxicology. 2007;229(3):226–235. doi: 10.1016/j.tox.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 29.Richa S., Dey P., Park C., Yang J., Son J.Y., Park J.H., Lee S.H., Ahn M.Y., Kim I.S., Moon H.R. A new histone deacetylase inhibitor, MHY4381, induces apoptosis via generation of reactive oxygen species in human prostate cancer cells. Biomol Ther (Seoul) 2020;28(2):184–194. doi: 10.4062/biomolther.2019.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gnanapradeepan K., Basu S., Barnoud T., Budina-Kolomets A., Kung C.P., Murphy M.E. The p53 tumor suppressor in the control of metabolism and ferroptosis. Front. Endocrinol. 2018;9:124. doi: 10.3389/fendo.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reincke M., Karl M., Travis W.H., Mastorakos G., Allolio B., Linehan H.M., Chrousos G.P. p53 mutations in human adrenocortical neoplasms: immunohistochemical and molecular studies. J. Clin. Endocrinol. Metab. 1994;78(3):790–794. doi: 10.1210/jcem.78.3.8126158. [DOI] [PubMed] [Google Scholar]
- 32.Pecoraro A., Carotenuto P., Russo G., Russo A. Ribosomal protein uL3 targets E2F1 and Cyclin D1 in cancer cell response to nucleolar stress. Sci. Rep. 2019;9(1):15431. doi: 10.1038/s41598-019-51723-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Q., Su Z., Gu W., Rustgi A.K. Mutant p53 on the path to metastasis. Trends Cancer. 2020;6(1):62–73. doi: 10.1016/j.trecan.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson L.R., Oliveira T.G., Hermann E.R., Chowanadisai W., Clarke S.L., Montgomery M.R. Distinct TP53 mutation types exhibit increased sensitivity to ferroptosis independently of changes in iron regulatory protein activity. Int. J. Mol. Sci. 2020;21(18) doi: 10.3390/ijms21186751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jennis M., Kung C.P., Basu S., Budina-Kolomets A., Leu J.I., Khaku S., Scott J.P., Cai K.Q., Campbell M.R., Porter D.K. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30(8):918–930. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith K.T., Workman J.L. Histone deacetylase inhibitors: anticancer compounds. Int. J. Biochem. Cell Biol. 2009;41(1):21–25. doi: 10.1016/j.biocel.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Jiang G.M., Wang H.S., Zhang F., Zhang K.S., Liu Z.C., Fang R., Wang H., Cai S.H., Du J. Histone deacetylase inhibitor induction of epithelial-mesenchymal transitions via up-regulation of Snail facilitates cancer progression. Biochim. Biophys. Acta. 2013;1833(3):663–671. doi: 10.1016/j.bbamcr.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Kong D., Ahmad A., Bao B., Li Y., Banerjee S., Sarkar F.H. Histone deacetylase inhibitors induce epithelial-to-mesenchymal transition in prostate cancer cells. PloS One. 2012;7(9) doi: 10.1371/journal.pone.0045045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Z., Liao X., Shao Z., Feng M., Yuan J., Wang S., Gan S., Ha Y., He Z., Jie W. Short-term stimulation with histone deacetylase inhibitor trichostatin a induces epithelial-mesenchymal transition in nasopharyngeal carcinoma cells without increasing cell invasion ability. BMC Canc. 2019;19(1):262. doi: 10.1186/s12885-019-5482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Xu M.Q., Jiang X.L., Mei X.Y., Liu X.G. Histone deacetylase inhibitor SAHA-induced epithelial-mesenchymal transition by upregulating Slug in lung cancer cells. Anti Canc. Drugs. 2018;29(1):80–88. doi: 10.1097/CAD.0000000000000573. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava R.K., Kurzrock R., Shankar S. MS-275 sensitizes TRAIL-resistant breast cancer cells, inhibits angiogenesis and metastasis, and reverses epithelial-mesenchymal transition in vivo. Mol. Canc. Therapeut. 2010;9(12):3254–3266. doi: 10.1158/1535-7163.MCT-10-0582. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Xu J., Wang H., Wu L., Yuan W., Du J., Cai S., Trichostatin A. A histone deacetylase inhibitor, reverses epithelial-mesenchymal transition in colorectal cancer SW480 and prostate cancer PC3 cells. Biochem. Biophys. Res. Commun. 2015;456(1):320–326. doi: 10.1016/j.bbrc.2014.11.079. [DOI] [PubMed] [Google Scholar]
- 43.Ji M., Lee E.J., Kim K.B., Kim Y., Sung R., Lee S.J., Kim D.S., Park S.M. HDAC inhibitors induce epithelial-mesenchymal transition in colon carcinoma cells. Oncol. Rep. 2015;33(5):2299–2308. doi: 10.3892/or.2015.3879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.