Summary
This protocol describes the assembly and use of MitoPunch to deliver mitochondria containing mitochondrial DNA (mtDNA) into cells lacking mtDNA (ρ0 cells). MitoPunch generates stable isolated mitochondrial recipient clones with restored mtDNA and recovered respiration, enabling investigation of mtDNA mutations and mtDNA-nuclear DNA interactions in a range of cell types.
For complete details on the use and execution of this protocol, please refer to Sercel et al. (2021) and Patananan et al. (2020).
Subject areas: Cell Biology, Cell culture, Cell-based Assays, Metabolism, Biotechnology and bioengineering
Graphical abstract

Highlights
-
•
High-throughput mitochondrial transfer into transformed and primary recipient cells
-
•
Selection scheme to isolate stable mtDNA transplant recipient cells
-
•
MitoPunch device is easily assembled with minimal engineering expertise
-
•
MitoPunch enables new combinations of mitochondrial and nuclear genomes
This protocol describes the assembly and use of MitoPunch to deliver mitochondria containing mitochondrial DNA (mtDNA) into cells lacking mtDNA (ρ0 cells). MitoPunch generates stable isolated mitochondrial recipient clones with restored mtDNA and recovered respiration, enabling investigation of mtDNA mutations and mtDNA-nuclear DNA interactions in a range of cell types.
Before you begin
This protocol describes the process of using MitoPunch to deliver isolated HEK293T mitochondria into 143BTK− ρ0 recipient mammalian cells. We have used MitoPunch to generate stable isolated mitochondrial recipient (SIMR) cell lines with a wide range of both human and mouse cells, of both malignant and primary, non-immortalized origins. Few alterations are necessary to adapt the protocol to different cell lines and cell types, and guidance for doing so is included in the text below.
Construct MitoPunch apparatus
Timing: 30 min
-
1.Fabricate aluminum washer and 5 V solenoid housing.
-
a.Aluminum washer dimensions: outer diameter, 25 mm; inner diameter, 10 mm; thickness, 1.4 mm.
-
b.Aluminum housing dimensions: rectangular prism of 19.13 mm length, 12.80 mm width, and 30.50 mm height with an internal vertical cutout of 12.65 mm length, 10.20 mm depth, and 19.13 mm height, including two 2 mm diameter screw holes in the bottom of the housing and one 2 mm screw hole on either side of the cutout for securing the 5 V solenoid (Spark Fun, Cat#ROB-11015).
-
a.
Note: Ensure the 5 V solenoid fits securely in the machined housing. This may require close collaboration with a machinist.
-
2.
Connect mains power cord (Volex, Cat#1725010B1) to the power supply (MEAN WELL, Cat#RS-35-12) by removing the IEC 60320 C13 adapter, striping the wires, and attaching them appropriately to the load (L), neutral (N), and ground hookups.
-
3.
Connect the power supply to the power supply mini board (Futurelec, Cat#MINIPOWER) using a barrel style DC connector with wire leads (McMaster-Carr, Cat#8320N111).
-
4.
Insert hookup wires (Spark Fun, Cat#PRT-08867) into the positive and negative terminals of the mini board, then connect the mini board terminals to the breadboard (Spark Fun, Cat#12615).
-
5.
Install the switch (Spark Fun, Cat#COM-09190) across the DIP ravine on the breadboard and install the jumper wires (Spark Fun, Cat#PRT-00124) to complete the circuit.
-
6.
Connect the JST jumper 2 wire to the breadboard.
Note: See Figure 1 for circuit diagram and image of fully assembled device to assist with the wiring of the apparatus. See Figure 2 for step-by-step assembly of the device.
-
7.
Place the 5 V solenoid into the custom metal casing ensuring that the top of the solenoid is level with the top of the casing, and tighten screws (diameter, 2 mm) to hold it in place.
-
8.
Fit the 5 V solenoid in the casing onto a threaded plug with screws (Thor Labs, Cat#SM1PL) and screw plug into the bottom plate (Thor Labs, Cat#CP02T).
-
9.Insert 4 optomechanical assembly rods (Thor Labs, Cat#ER3) into the holes in the bottom plate.
-
a.With rods flush with the bottom cage plate, secure rods by tightening the screws found next to each rod slot.
-
a.
-
10.Remove internal ring from middle cage plate using a spanner wrench (Thor Labs, Cat#SPW602) and slide the middle cage plate (Thor Labs, Cat#CP02) down the assembly rods until the top surface of the middle plate is flush with the top surface of the solenoid.
-
a.Reinsert internal ring, tighten, and double check that the top of the middle cage plate is flush with the top of the solenoid.
-
a.
-
11.
Mount the custom washer atop the middle cage plate and check that the ring stably supports it.
-
12.
Insert the 2-pin JST connector from the breadboard into the MitoPunch 2-pin JST PH connector and test whether the piston actuates when the switch is depressed (see troubleshooting 1).
Figure 1.
MitoPunch circuit diagram
(A) Circuit diagram of the MitoPunch apparatus. Numbered nodes indicate where wires are inserted into the corresponding breadboard ports.
(B) Image of fully wired and completed MitoPunch apparatus.
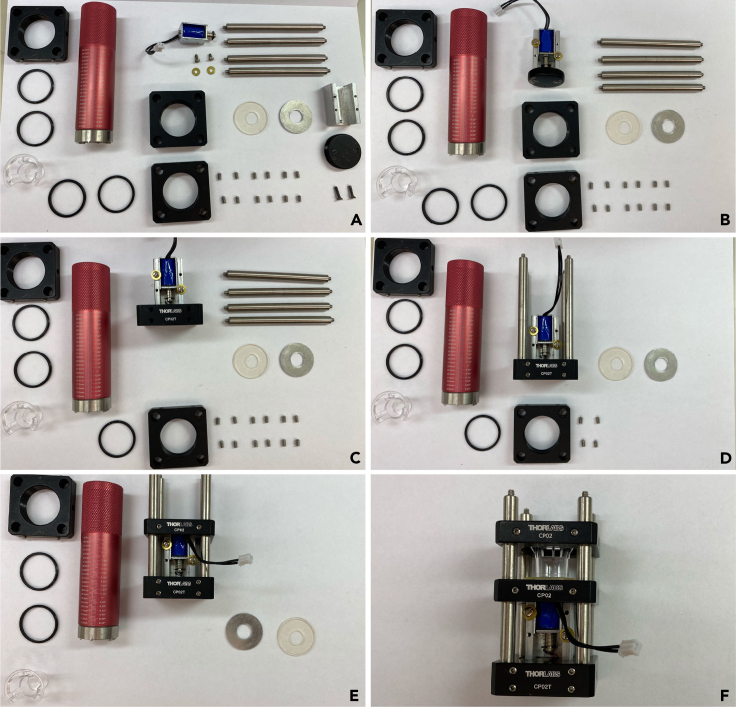
Figure 2.
Assembling the MitoPunch apparatus
(A) Layout of all MitoPunch components prior to assembly.
(B) MitoPunch solenoid mounted on the aluminum plug.
(C) Mounted MitoPunch solenoid screwed into base plate.
(D) MitoPunch base plate with optomechanical assembly rods inserted and fixed with screws.
(E) Middle plate mounted flush with the solenoid using screws on the optomechanical assembly rods.
(F) Fully assembled MitoPunch apparatus with top plate lowered onto PDMS reservoir ready for MitoPunch transfer.
Assemble polydimethylsiloxane (PDMS) reservoirs
Timing: 6 h
PDMS reservoirs are essential to hold the mitochondrial suspension for delivery into cells using MitoPunch and for delivering the pressure that enables a mitochondrial transfer. PDMS is a biocompatible polymer that is easily fabricated with minimal equipment and expertise (Berthier et al., 2012).
-
13.Weigh and mix Sylgard 184 PDMS base and curing agent (Dow Corning, Cat#1673921) at a 10:1 ratio.
-
a.Prepare a weigh boat on a scale. Tare the scale.
-
b.Pour 12 g PDMS base into the weigh boat.
-
c.Tare the scale.
-
d.Add 1.2 g curing agent into the weigh boat.
-
e.Mix thoroughly for about 1–2 min with a chemical spatula or similar implement.
-
a.
Note: PDMS should remain covered until cured to avoid the accumulation of dust and debris.
Note: For visual reference of PDMS fabrication, see Figure 3.
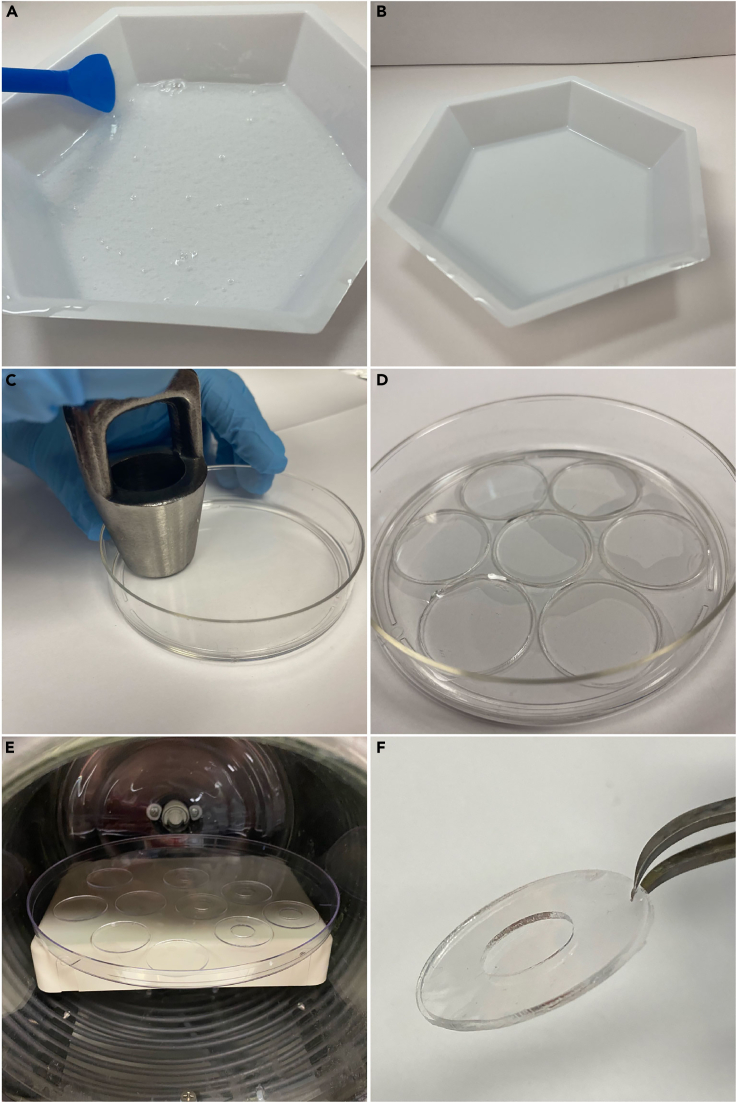
Figure 3.
Fabricating PDMS reservoirs for MitoPunch
(A) Uncured PDMS containing bubbles following thorough mixing of the PDMS base and curing agent.
(B) Uncured PDMS following degassing to remove all bubbles.
(C) Punching disks out of a cured layer of PDMS with a hole punch.
(D) Seven punched PDMS disks in a 10 cm dish.
(E) PDMS disks and rings on a 15 cm dish in an oxygen plasma cleaning chamber.
(F) Bonded PDMS reservoir after oxygen plasma treatment.
-
14.Degas PDMS.
-
a.Transfer the mixed PDMS into a vacuum desiccator.
-
b.Connect a vacuum pump to the desiccator and turn on the pump.
-
c.Degas PDMS until all visible air bubbles in the PDMS have disappeared, approximately 30 min.
-
a.
-
15.Pour and cure PDMS.
-
a.Prepare a clean 10 cm cell culture dish on a scale. Tare the scale.
-
b.Pour 6 g of PDMS into the dish. Be careful not to introduce bubbles. If bubbles are generated while pouring, use a 20 μL pipette tip to remove the bubbles or move the dish into the desiccator and repeat degas step.
-
i.Repeat with a second 10 cm dish using the remaining PDMS.
-
i.
-
c.Allow PDMS to uniformly coat the dishes (target thickness ∼1 mm).
-
d.Put the dishes with PDMS into an oven at 65°C for 2–4 h.
-
a.
CRITICAL: 10 cm dishes may melt if placed directly on a metal surface in the oven. Place dishes on autoclave safe plastic inside the oven to avoid melting.
-
16.Hole punch PDMS.
-
a.Take the cured PDMS out from the oven and allow to cool to 20°C–25°C.
-
b.For the bottom layer, cut out circular PDMS disks with a hole puncher of 25 mm diameter (McMaster-Carr, Cat#3418A25).
-
c.For the top layer, cut out circular PDMS disks with a hole puncher of 25 mm diameter. Then use another hole puncher of 10 mm diameter (McMaster-Carr, Cat#3418A1) to remove the center portion and create a PDMS ring.
-
a.
Note: Prepare an equal number of disks and rings for bonding.
-
17.Bond PDMS layers.
-
a.Remove dust and debris from the PDMS disks and PDMS rings with a piece of removable adhesive tape.
-
b.Place the PDMS disks and PDMS rings into an expanded plasma cleaner (Harrick Plasma, Cat#PDC-001) with the clean bonding side face up.
-
c.Close the chamber and set the plasma cleaner to high.
-
i.Maintain internal pressure between 300 mTorr and 500 mTorr with the RF level set on high for 1 min.
-
i.
-
d.Remove plasma treated PDMS disks and rings from the oxygen plasma chamber.
-
e.Carefully align and bond the disks and rings by applying firm pressure to ensure the two pieces are sealed together.Note: Join the two PDMS pieces by pressing the oxygen plasma treated faces together to ensure strong bonding.
-
f.Heat the bonded PDMS reservoir in the oven at 65°C for at least 2 h to complete bonding.
 CRITICAL: Excess exposure of the oxygen plasma treated faces of the PDMS pieces to air and debris before bonding will result in poor adhesion between the two components. Join the two parts as quickly as possible after oxygen plasma treatment for optimal bonding strength.
CRITICAL: Excess exposure of the oxygen plasma treated faces of the PDMS pieces to air and debris before bonding will result in poor adhesion between the two components. Join the two parts as quickly as possible after oxygen plasma treatment for optimal bonding strength.
-
a.
Sterilization of MitoPunch components
Timing: 2 h
-
18.Sterilize a number of PDMS reservoirs equal to the number of MitoPunch replicates to be performed.
-
a.Wash reservoirs three times with a 10% bleach solution.
-
b.Rinse three times with DI water.
-
c.Wash reservoirs three times with a 70% ethanol in water solution.
-
d.Rinse three times with DI water.
-
e.Package PDMS reservoirs into disposable sterilization pouches.
-
f.Autoclave packaged PDMS at 121°C for 30 min.
-
a.
Note: The wash steps can be completed in a 15 cm dish or a 50 mL conical tube for convenience prior to autoclaving in a disposable sterilization pouch.
-
19.Sterilize stainless steel forceps.
-
a.Wash forceps three times with a 10% bleach solution.
-
b.Rinse three times with DI water.
-
c.Wash forceps three times with a 70% ethanol in water solution.
-
d.Rinse three times with DI water.
-
e.Package forceps into a disposable sterilization pouch.
-
f.Autoclave packaged forceps at 121°C for 30 min.
-
a.
-
20.Sterilize high-vacuum grease.
-
a.Apply approximately 4 mL high-vacuum grease to a 5 cm glass petri dish.
-
b.Close the glass dish and seal with autoclave tape.
-
c.Autoclave high-vacuum grease at 121°C for 30 min.
-
a.
-
21.Sterilize glass cloning cylinders.
-
a.Place as many cloning cylinders as necessary into a 100 mL glass beaker.
-
b.Seal with aluminum foil and apply autoclave tape to the foil top.
-
c.Autoclave cloning cylinders at 121°C for 30 min.
-
a.
Thaw and expand mitochondrial donor and recipient cell lines
Timing: 1 week
-
22.Thaw mitochondrial donor and recipient cells according to standard protocols for the chosen lines.
-
a.Thaw at least 5 d prior to mitochondrial transfer to allow for expansion and mycoplasma testing of cell lines.
-
a.
-
23.Expand mitochondrial donor and recipient cell lines.
-
a.Expand recipient lines to have 1 × 105 cells per planned MitoPunch replicate.
-
b.Expand donor cells on T-225 plates until you have one T-225 plate for every two planned MitoPunch replicates, or ∼1.5 × 106 mitochondrial donor cells for each replicate transfer.
-
a.
CRITICAL: Donor cells harvested at high confluence are more likely to cause needle clogging during the mitochondrial isolation. Aim to harvest donor cells at ∼80%–90% confluence.
Test recipient cell sensitivity to different SIMR clone selection media conditions
Timing: 7 days
Isolation of SIMR clones following a MitoPunch transfer requires a restrictive medium selection that selectively permits only cells with functional mitochondria to grow. This selection scheme takes advantage of the dependence on electron transport chain activity for de novo nucleotide synthesis in mammalian cells (Gregoire et al., 1984). Two selection medium formulations, SIMR selection medium and galactose SIMR selection medium, are used depending on the sensitivity of mitochondrial recipient cells to this selection. SIMR selection medium contains 4.5 g/L glucose, whereas the galactose SIMR selection medium is glucose-free and supplemented with galactose so that glycolysis generates no net ATP (Aguer et al., 2011). Below is a protocol to measure the sensitivity of a chosen ρ0 recipient cell line in these two restrictive mediums in preparation for post-MitoPunch selection.
-
24.Seed ∼2 × 104 cells in a 12-well plate with the recipient cell line in uridine-supplemented complete medium.
-
a.Incubate ∼24 h to allow cells to adhere.
-
a.
Note: This seeding density depends upon the growth rate of the chosen recipient cell line. Alter the seeding density such that the cells would reach confluence ∼5 d post seeding under normal conditions.
-
25.Exchange the uridine-supplemented complete medium for 2 mL SIMR selection medium.
-
a.Exchange medium with fresh SIMR selection medium daily.
-
a.
-
26.Monitor the wells twice daily for 7 d.
-
a.Record the time point when cells begin to die.
-
a.
-
27.
Continue to feed wells and monitor cell viability until all cells have died or 7 d have passed.
Alternatives: In the event that living cells remain after 7 d, re-plate the experiment as outlined in step 23, and begin with the following alternative protocol.
-
28.Exchange the uridine-supplemented complete medium for 2 mL galactose SIMR selection medium.
-
a.Exchange medium with fresh galactose SIMR selection medium daily.
-
a.
-
29.Monitor the wells five times daily for 7 d.
-
a.Record the time point when cells begin to die.
-
a.
-
30.
Continue to feed wells and monitor cell viability until all cells have died or 7 d have passed.
Note: Galactose SIMR selection medium is far more restrictive than SIMR selection medium and cells can die within hours of first treatment. Careful monitoring of this process is key to timing the selection of MitoPunch experiments to successfully isolate SIMR clones. If cells do not die under either medium, see troubleshooting 2.
Seed mitochondrial recipient cells on PET filters
Timing: 1 days
-
31.Collect and count mitochondrial recipient cells.
-
a.Remove culture medium from recipient cell flasks.
-
b.Wash plate with 1X Dulbecco’s Phosphate-Buffered Salt Solution (DPBS) (Corning, Cat#21031CV).
-
c.Aspirate DPBS and apply StemPro Accutase Cell Dissociation Reagent (Accutase) (Thermo Fisher Scientific, Cat#A1110501).Note: Use preferred dissociation reagent for the chosen recipient cell type.
-
d.Incubate in 37°C humidified incubator for 5 min.
-
e.Resuspend cells in 8.5 mL medium. Transfer cell suspension to 15 mL conical tube.
-
f.Centrifuge at 500 × g for 5 min, aspirate supernatant, and resuspend in culture medium.
-
g.Count cells using a hemocytometer.
-
h.Calculate volume of cell suspension needed to seed 1 × 105 recipient cells.
-
a.
-
32.Seed recipient cells.
-
a.Insert 3.0 μm PET filter inserts (Corning, Cat# 353181) into wells of a 12-well dish and dispense 1.5 mL medium to the well outside of the filter insert.
-
b.Pipette volume of cell suspension containing 1 × 105 cells into filter insert. Add volume of medium to filter insert to bring volume inside filter insert to 500 μL.
-
a.
CRITICAL: Ensure even plating of recipient cells on the filter insert for consistent MitoPunch results. See Figure 4A for an example of an evenly seeded well 24 h post-seeding.
Figure 4.
Filter insert seeding and harvesting
(A) Filter insert without cells in DPBS.
(B) Filter insert seeded with 1 × 105 143BTK− ρ0 cells imaged after washing with DPBS.
(C) Filter insert seeded with 1 × 105 143BTK− ρ0 cells imaged after washing with DPBS and ∼7 min incubation with Accutase.
Scale bars represent 200 μm.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| DMEM w/ 4.5 g/L glucose, sodium pyruvate, and L-glutamine | Corning | Cat#10013CM |
| DMEM, no glucose | Thermo Fisher Scientific | Cat#11966025 |
| Fetal Bovine Serum | Omega Scientific | Cat#FB-11 |
| Fetal Bovine Serum, dialyzed, US origin | Thermo Fisher Scientific | Cat#26400-044 |
| Penicillin-Streptomycin Solution, 100× | Corning | Cat#30-002-CI |
| GlutaMAX Supplement | Thermo Fisher Scientific | Cat#35050061 |
| MEM Non-Essential Amino Acids Solution (100X) | Thermo Fisher Scientific | Cat#11140050 |
| Uridine | Acros Organics | Cat#140770250 |
| D-(+)-Galactose | Millipore Sigma | Cat#G5388-100G |
| Dulbecco’s Phosphate-Buffered Salt Solution 1X | Corning | Cat#21031CV |
| Sodium Pyruvate (100 mM) | Corning | Cat#25000Cl |
| Dulbecco's Phosphate-Buffered Salt Solution, w/ Calcium and Magnesium | Thermo Fisher Scientific | Cat#14040133 |
| StemPro Accutase Cell Dissociation Reagent | Thermo Fisher Scientific | Cat#A1110501 |
| 16% Formaldehyde | Thermo Fisher Scientific | Cat#28906 |
| Crystal Violet | Fisher Chemical | Cat#C581-25 |
| Methanol | Fisher Chemical | Cat#A412-4 |
| Critical commercial assays | ||
| Qproteome Mitochondria Isolation Kit | QIAGEN | Cat#37612 |
| Experimental models: Cell lines | ||
| 143 BTK– osteosarcoma ρ0; human, female | (Patananan et al., 2020) | N/A |
| Primary dermal fibroblast normal ρ0; human, male, neonatal | (Patananan et al., 2020) | N/A |
| HEK293T dsRed; human, female | Miyata et al., 2014 | N/A |
| Other | ||
| Breadboard - Full-Size (Bare) | SparkFun | Cat#12615 |
| Solenoid - 5V | SparkFun | Cat#ROB-11015 |
| Momentary Pushbutton Switch - 12mm Square | SparkFun | Cat#COM-09190 |
| Hook-up Stranded Wire - Black (22 AWG) | SparkFun | Cat#PRT-08867 |
| JST Jumper 2 Wire Assembly | SparkFun | Cat#PRT-09914 |
| Jumper Wire Kit - 140pcs | SparkFun | Cat#PRT-00124 |
| Power Supply Mini Board | Futurelec | Cat#MINIPOWER |
| 35W Single Output Switching Power Supply | MEAN WELL | Cat#RS-35-12 |
| Mains Power Cord, NEMA 5-15P to IEC 60320 C13, 2.3 m, 10 A, 125 VAC, Black | Volex | Cat#1725010B1 |
| Push-In Plugs with Wire Leads, 2.1 mm End ID, 12V DC | McMaster-Carr | Cat#8320N111 |
| Spanner Wrench for SM1-Threaded Retaining Rings | Thorlabs | Cat# SPW602 |
| Externally SM1-Threaded Plug | Thorlabs | Cat#SM1PL |
| SM1-Threaded 30 mm Cage Plate, 0.50" Thick, 2 Retaining Rings, 8–32 Tap | Thorlabs | Cat#CP02T |
| Cage Assembly Rod, 3" Long, Ø6 mm | Thorlabs | Cat#ER3 |
| SM1-Threaded 30 mm Cage Plate, 0.35" Thick, 2 Retaining Rings, 8–32 Tap | Thorlabs | Cat#CP02 |
| Custom Aluminum Washer | University of California, Los Angeles, Machine Shop | N/A |
| Custom Aluminum Case | University of California, Los Angeles, Machine Shop | N/A |
| SYLGARD™ 184 Silicone Elastomer Kit 1.1 KG Kit | Dow Corning | Cat#1673921 |
| Polypropylene Desiccator with Stopcock | Thermo Fisher Scientific | Cat#53100250 |
| 230 mm Ceramic Metal Desiccator Plate | Thermo Fisher Scientific | Cat#53120230 |
| 10 mm Hole punch | McMaster-Carr | Cat#3418A1 |
| 25 mm Hole punch | McMaster-Carr | Cat#3418A25 |
| Expanded Plasma Cleaner | Harrick Plasma | Cat#PDC-001 |
| 12–Well 3.0 μm Transparent PET Membrane Cell Culture Inserts | Corning | Cat#353181 |
| 26 G Blunt-ended needle | VWR | Cat#89134-164 |
| 3 mL Luer-Lok Syringe | BD Biosciences | Cat#309657 |
| High-vacuum grease | Dow Corning | Cat#1597418 |
| Cloning cylinders | Fisher Scientific | Cat# 09-552-21 |
Materials and equipment
Preparation of complete medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM w/ 4.5 g/L glucose, sodium pyruvate, and L-glutamine (Corning, C#10013CM) | - | 870 mL |
| Penicillin Streptomycin solution 100× (Corning, Cat#30-002-CI) | 1× | 10 mL (from 100× stock) |
| GlutaMAX Supplement (Thermo Fisher Scientific, Cat#35050061) | 1× | 10 mL (from 100× stock) |
| MEM Non-essential Amino Acids 100× (Thermo Fisher Scientific, Cat#11140050) | 1× | 10 mL (from 100× stock) |
| Fetal Bovine Serum (FBS) (Omega Scientific, Cat#FB-11) | 10% | 100 mL |
| Total | n/a | 1000 mL |
Medium can be stored at 4°C for up to 1 month.
Preparation of uridine-supplemented complete medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM w/ 4.5 g/L glucose, sodium pyruvate, and L-glutamine | - | 869 mL |
| Uridine (Acros Organics, Cat#140770250) | 50 mg/L | 1 mL (from 50 mg/mL stock) |
| Penicillin Streptomycin solution | 1× | 10 mL (from 100× stock) |
| GlutaMAX Supplement | 1× | 10 mL (from 100× stock) |
| MEM Non-essential Amino Acids | 1× | 10 mL (from 100× stock) |
| Fetal Bovine Serum (FBS) | 10% | 100 mL |
| Total | n/a | 1000 mL |
Medium can be stored at 4°C for up to 1 month.
Note: Uridine stocks are prepared and filtered through a 0.22 μm filter in advance then stored at –20°C for up to 1 year.
Preparation of SIMR selection medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM w/ 4.5 g/L glucose, sodium pyruvate, and L-glutamine | - | 870 mL |
| Penicillin Streptomycin solution | 1× | 10 mL (from 100× stock) |
| GlutaMAX Supplement | 1× | 10 mL (from 100× stock) |
| MEM Non-essential Amino Acids | 1× | 10 mL (from 100× stock) |
| Dialyzed Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, Cat#26400-044) | 10% | 100 mL |
| Total | n/a | 1000 mL |
Medium can be stored at 4°C for up to 1 month.
Preparation of galactose SIMR selection medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM, no glucose (Thermo Fisher Scientific, Cat#11966025) | - | 430 mL |
| Galactose (Millipore Sigma, Cat#G5388-100G) | 4.5 g/L | 2.25 g |
| Penicillin Streptomycin solution | 1× | 5 mL (from 100× stock) |
| GlutaMAX Supplement | 1× | 5 mL (from 100× stock) |
| MEM Non-essential Amino Acids | 1× | 5 mL (from 100× stock) |
| Sodium Pyruvate (Corning, Cat#25000CI) | 1× | 5 mL (from 100× stock) |
| Fetal Bovine Serum (FBS) | 10% | 50 mL |
| Total | n/a | 500 mL |
Medium can be stored at 4°C for up to 1 month.
Note: The galactose should be added by dissolving in 40 mL of DMEM and then adding back to the medium before filtering through a 0.22 μm PES filter.
Preparation of paraformaldehyde
| Reagent | Final concentration | Amount |
|---|---|---|
| 16% Paraformaldehyde (Thermo Fisher Scientific, Cat#28906) | 4% | 1 mL |
| Dulbecco's Phosphate-Buffered Salt Solution 1X | - | 3 mL |
| Total | 4% | 4 mL |
Paraformaldehyde solution should be made freshly before use.
CRITICAL: Paraformaldehyde is a toxin and carcinogen that should be handled only in a chemical fume hood for proper ventilation with appropriate PPE including safety glasses, a lab coat, closed-toed shoes, and nitrile gloves. All waste containing paraformaldehyde should be labeled and disposed of through proper channels. Paraformaldehyde should be stored in a flammable storage cabinet.
Preparation of crystal violet staining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Methanol | 20% | 20 mL |
| Deionized water | 80% | 80 mL |
| Crystal Violet (Fisher Chemical, Cat#C581-25) | 0.5% w/v | 5 g |
| Total | n/a | 100 mL |
Crystal violet solution can be kept in the flammable chemical cabinet for up to 6 months.
CRITICAL: Crystal violet staining solution contains methanol which is a flammable toxin and should be stored in a flammable storage cabinet. Appropriate PPE including safety glasses, a lab coat, closed-toed shoes, and nitrile gloves should be used when preparing and working with this solution. All waste containing crystal violet staining solution should be labeled and disposed of through proper channels.
Step-by-step method details
Mitochondrial preparation
Timing: 2–3 h
MitoPunch requires a freshly isolated stock of cell-free mitochondria for transfer into recipient cells. This protocol uses the Qiagen Qproteome Mitochondrial Isolation Kit (Qiagen, Cat#37612) with minor optimizations for efficiency and purity in this application; however, other mitochondrial isolation methods are also compatible with MitoPunch. Here we define purity as the absence of intact mitochondrial donor cells that escape through the disruption and mitochondrial isolation process. We have measured the purity of our mitochondrial isolates in Figure 3-figure supplement 1 of (Sercel et al., 2021).
-
1.Harvest mitochondrial donor cells.
-
a.Aspirate culture medium from T-225 flasks.
-
b.Pipette 20 mL DPBS into each flask.
-
c.Release cells from the flasks using a cell scraper.
-
d.Suspend scraped cells in DPBS and collect into 50 mL conical tubes.
-
e.Centrifuge tubes at 500 × g for 10 min at 4°C. Aspirate supernatant.
-
f.Wash cell pellets in 10 mL DPBS and centrifuge again at 500 × g for 10 min at 4°C. Aspirate supernatant.
-
a.
CRITICAL: Keep samples on ice when outside the centrifuge for the remainder of the mitochondrial isolation process.
-
2.Disrupt mitochondrial donor cells using the Qiagen Qproteome Mitochondrial Isolation Kit.Note: The buffers used in the following steps are from the Qiagen Qproteome Mitochondrial Isolation Kit and must be handled according to manufacturer protocol.
-
a.Resuspend cell pellets in ice-cold Lysis Buffer at a concentration of ∼1 × 107 cells per 1 mL and transfer to 2 mL tubes.
-
i.Prepare Lysis Buffer with 1:100 Protease Inhibitor Solution prior to resuspension.
-
i.
-
b.Incubate on end-over-end shaker for 10 min at 4°C.Note: Ensure microcentrifuge is chilled to 4°C during this incubation in preparation for the next step.
-
c.Centrifuge the samples at 1000 × g for 10 min at 4°C and aspirate supernatant without disrupting the pellet.
-
d.Resuspend each cell pellet in 1.5 mL ice cold Disruption Buffer with a 1 mL pipette tip.
-
i.Prepare Disruption Buffer with 1:100 Protease Inhibitor Solution prior to resuspension.
-
i.
-
e.Disrupt cells using a blunt-ended 26G (VWR, Cat#89134-164) needle attached to a 3 mL Luer-Lok syringe.
-
i.Slowly pull the entire lysate into the syringe and expel the volume 10 times, avoiding the formation of bubbles in the suspension.Note: If clogging of the disruption needle occurs, see troubleshooting 3.
-
i.
-
a.
-
3.Mitochondrial purification.
-
a.Centrifuge lysate at 1000 × g for 10 min at 4°C and transfer supernatant to a new sterile 1.5 mL tube.Optional: Repeat this step to reduce the probability of intact cells contaminating the mitochondrial preparation.Note: Pellet will be diffuse and easily disrupted. Take care in removing supernatant to minimize the mass of pellet that is carried over to the next step.Note: Supernatants should be combined at this point.
-
b.Centrifuge the supernatant at 6000 × g for 10 min at 4°C. Carefully aspirate supernatant.Note: The pellet now contains isolated mitochondria.
-
c.Resuspend the pellet with 1 mL Mitochondrial Storage Buffer using a 1 mL pipette tip and centrifuge at 6000 × g for 20 min at 4°C.
 CRITICAL: Prepare the MitoPunch apparatus during this centrifugation step to minimize time between resuspension of mitochondrial isolate and MitoPunch transfer as MitoPunch efficiency is negatively impacted by older mitochondrial preparations.
CRITICAL: Prepare the MitoPunch apparatus during this centrifugation step to minimize time between resuspension of mitochondrial isolate and MitoPunch transfer as MitoPunch efficiency is negatively impacted by older mitochondrial preparations. -
d.Remove supernatant. The mitochondrial mass should appear as a compact, pin-head-sized pellet at the bottom of the 1.5 mL tube.
-
e.Resuspend the mitochondrial pellet in a volume of DPBS with calcium and magnesium (Thermo Fisher Scientific, Cat#14040133) equal to 120 μL times the number of MitoPunch replicates and keep on ice.
-
a.
MitoPunch mitochondrial transfer
Timing: 1 h
The following instructions describe the process of delivering freshly isolated mitochondria to recipient cells using the MitoPunch apparatus. Briefly, the mechanism by which MitoPunch delivers isolated mitochondria to recipient cells requires pressure generated by a solenoid-driven mechanical plunger. Isolated mitochondria are loaded into a PDMS reservoir to which a cell culture filter insert seeded with recipient cells is sealed. The solenoid is activated which drives the mechanical plunger into the underside of the PDMS reservoir, causing the internal pressure of the reservoir to increase. This increase in pressure forces mitochondrial suspension directly through the pores in the filter insert and into the recipient cell cytoplasm. We have previously quantified the transfer efficiency of MitoPunch by delivering fluorescently labeled isolated mitochondria to 143 BTK− ρ0 and BJ ρ0 recipient cells and measuring the fraction of the recipients positive for the fluorescent marker by imaging flow cytometry (Sercel et al., 2021). We observe ∼50% of 143 BTK− ρ0 recipient cells and ∼15% of BJ ρ0 recipients as positive for the fluorescent mitochondria. Additionally, we measured the mean and median number of fluorescent mitochondrial puncta present in recipient cells by imaging flow cytometry. We observed a mean of ∼5 and a median of ∼3 puncta for 143 BTK− ρ0 recipients and a mean of ∼2 and a median of ∼1 puncta for BJ ρ0 recipients. Finally, it is important to keep the isolated mitochondrial sample on ice while performing MitoPunch transfers and to minimize the time that isolated mitochondria are kept on ice prior to transfer.
-
4.
Spray 70% ethanol into a tissue and wipe down the MitoPunch apparatus before entering it into a biosafety cabinet.
-
5.
Connect MitoPunch apparatus to breadboard and connect breadboard to power supply (Figure 1B). Connect power supply to the 120 V wall outlet and ensure device has power by testing piston actuation.
-
6.
Remove a sterile PDMS reservoir from an autoclave bag and place on MitoPunch apparatus.
-
7.
Wash PDMS reservoir 3 times with 120 μL DPBS with calcium and magnesium, pipetting up and down 3 times with each wash, being sure to cover the entire surface area of the reservoir.
Note: This step is important to remove any traces of bleach or ethanol from pre-autoclave sterilization and to help the hydrophobic PDMS more readily hold the mitochondrial suspension evenly without pooling into a large droplet.
-
8.
Pipette 120 μL mitochondrial suspension or DPBS control into the PDMS reservoir.
-
9.Remove 12-well dish of recipient-cell-seeded filter inserts from the incubator and enter it into the biosafety cabinet. Using sterile forceps, remove one filter insert and secure it into the top plate of the MitoPunch apparatus.
-
a.To secure, remove the top internal ring from the upper plate of the MitoPunch and place the plastic wings of the filter insert on the remaining ring such that the filter insert is supported within the hole of the top plate. Carefully screw the top internal ring until tight against the filter wings.
-
a.
-
10.
Lower the top plate along the optomechanical assembly rods such that the filter insert contacts the PDMS reservoir and ensure it is firmly pressed against the PDMS, creating a seal.
-
11.
Actuate the MitoPunch piston using the switch on the breadboard. Leave the piston in the protracted position for 3 s, release the piston switch, remove the filter insert from the apparatus, and replace it in the 12-well dish in its original medium.
CRITICAL: Apply gentle pressure to the top plate with a hand while actuating the solenoid to maintain a seal between the filter insert and the PDMS.
-
12.
After completing all MitoPunch transfers, place the 12-well dish in a humidified 37°C incubator for 2 h prior to collecting the recipient cells.
Recipient cell collection
Timing: 1 h
Mitochondrial recipient cells need to be harvested and plated on 10 cm dishes after the 2 h incubation that follows mitochondrial transfer for expansion and selection.
-
13.Aspirate medium from the wells containing filter inserts.
-
a.Carefully aspirate medium from within the filter insert, being mindful to not contact the cell layer on the PET membrane.
-
b.Aspirate medium from outside the filter insert, making sure to collect the medium retained at the contact point between the bottom of the filter insert and the well.
-
a.
Note: The PET filter can retain medium and reduce the efficacy of dissociation reagents if not handled properly.
CRITICAL: Change aspirator tip between samples to avoid contamination of intact cells from the mitochondrial preparation that may be present on the outside of the filter insert.
-
14.Wash filter inserts with DPBS without calcium and magnesium.
-
a.Apply 0.5 mL DPBS gently to the filter insert and 1.5 mL DPBS to the well outside the filter insert.
-
b.Carefully aspirate DPBS from within the filter insert and then from outside the insert, being sure to remove any residual liquid retained between the bottom of the filter and the well.
-
a.
CRITICAL: Ensure that DPBS reaches both sides of the filter insert.
CRITICAL: Change aspirator tip between samples to avoid contamination of intact cells from the mitochondrial preparation that may be present on the outside of the filter insert.
-
15.Apply Accutase dissociation reagent to the filters and incubate for 5 min in a 37°C humidified incubator.
-
a.Pipette 0.5 mL Accutase to the filter insert and 1.5 mL to the well outside the insert.
-
a.
CRITICAL: Ensure that Accutase reaches both sides of the filter insert.
Note: Visually inspect the plate under an inverted microscope to verify that the cells have released from the filter. If dissociation is incomplete (Figure 4B), return plate to the incubator for 3 min and repeat until cells have released from the membrane (Figure 4C).
-
16.Once the cells are released from the membrane, apply 0.5 mL uridine-supplemented medium to the filter insert and gently disrupt the cell layer with a 1 mL pipette tip.
-
a.Do not draw liquid from outside the filter to reduce the risk of intact mitochondrial donor cell contamination.
-
a.
-
17.
Pipette the full volume from the filter insert to a 10 cm dish containing 10 mL warm uridine-supplemented complete medium. If few cells are observed in the dish, see troubleshooting 4.
-
18.
Place 10 cm dishes in a 37°C humidified incubator.
SIMR clone selection
Timing: 1 week
Recipient cells need to be cultured in a dialyzed, nucleotide-free selection medium to isolate stable mitochondrial recipient cells. The selection medium is not permissive for ρ0 cells as they are incapable of generating nucleotides de novo. A negative ρ0 control is essential for determining the endpoint of the following selection protocol.
-
19.
Incubate the 10 cm dishes of MitoPunch recipient cells in a 37°C humidified incubator with uridine-supplemented complete medium for 3 d following transfer.
CRITICAL: Monitor confluence daily and do not allow cultures to exceed 80% confluence. If cultures reach >80% confluence, proceed with step 20.
-
20.Exchange the complete medium for SIMR selection medium.
-
a.Exchange the medium daily with 10 mL fresh SIMR selection medium and record the confluence of the ρ0 negative control.
-
a.
Note: Small colonies will form on the MitoPunch recipient plates and be visible by microscopy typically 3–5 d into SIMR selection medium treatment.
-
21.
Continue daily feeding with SIMR selection medium until the ρ0 control plate is no longer viable, typically 5–7 d under SIMR selection medium treatment.
Note: Proceed to either clone isolation or crystal violet staining as soon as the control plate is no longer viable.
Alternatives: When the recipient cells used were determined to require galactose SIMR selection medium in Test recipient cell sensitivity to different SIMR clone selection media conditions, proceed to step 22.
-
22.At 5 d post-delivery, feed cells with galactose SIMR selection medium.Note: Follow selection scheme developed in “Test recipient cell sensitivity to different SIMR clone selection media” for the specific recipient cells used.
-
a.Selection medium should be exchanged daily and condition of the cells noted.
-
a.
-
23.Continue galactose selection until the DPBS control transfer has completely died, up to 7 d post-transfer.
-
a.After death of the control, exchange the selection medium for SIMR selection medium to allow clones to grow large enough to harvest.
-
a.
Note: When selection fails to slow cell growth and induce cell death, see troubleshooting 5. In the event no clones appear after selection, see troubleshooting 6.
SIMR clone harvesting
Timing: 1 h
After the selection process, isolate SIMR clones from 10 cm dishes and passage them onto 6-well dishes for expansion. This must be done before clones overgrow as large clones can merge with neighboring clones or become internally over confluent and die.
-
24.
Select clones for isolation that are not adjacent to or in contact with other clones.
Note: When selecting multiple clones, mark the location of desired clones with a permanent marker or a microscope mounted object marker.
-
25.
Aspirate the medium from the dish and wash with 5 mL of DPBS. Aspirate the DPBS.
-
26.
For each clone to be isolated, coat one side of a cylinder in sterilized high-vacuum grease. Gently adhere each cylinder to the dish and fully encompass one clone within the cylinder.
-
27.
Slowly add 200 μL of Accutase to each cylinder and incubate for 5 min in a 37°C humidified incubator.
CRITICAL: Watch for any leaks from the bottom of the cylinder while adding dissociation reagent and immediately stop when one appears. Any leaked dissociation reagent can prematurely disrupt other clones adhered to the plate.
-
28.After incubation, verify that the cells have fully dissociated from the dish under an inverted microscope.
-
a.When the cells have not fully dissociated, incubate for another 3 min.
-
a.
-
29.
Once the cells have fully dissociated, use a micropipettor to move the cell suspension into a well of a 6-well dish containing 2 mL of warm complete medium.
-
30.
Incubate the newly plated clones in a 37°C humidified incubator and continue to expand until ready for use in downstream analysis.
Crystal violet staining
Timing: 2 h
This protocol enables visualization of clone morphology, number, and distribution. Plates treated with this protocol can be stored indefinitely away from light. Additionally, when stain does fade with time, plates can be re-stained with additional crystal violet to increase color saturation for future imaging.
-
31.
Aspirate medium from the plate, taking care to not disturb clones.
-
32.
Apply 1 mL of freshly diluted 4% paraformaldehyde to the dish and tilt the dish to fully coat the surface.
-
33.
Incubate at RT in a chemical fume hood for 15 min.
-
34.
Aspirate the 4% paraformaldehyde from the dish and dispose of in the appropriate chemical waste container.
-
35.
Apply 2 mL of crystal violet solution to each plate and tilt to fully coat. Incubate at RT in the chemical fume hood for 30 min.
-
36.
Remove excess crystal violet using a serological pipette and dispose of it in the appropriate chemical waste container.
-
37.
Rinse each plate twice with DI water to remove excess crystal violet solution.
-
38.
Dry plates upside-down on an absorbent surface to allow any residual crystal violet solution to drip off the plate.
-
39.
Store plates away from light until ready to analyze.
Expected outcomes
Mitochondrial preparations performed as written in this protocol typically yield 60–100 μg of isolated protein per T-225 plate of donor cells. MitoPunch transfer of HEK293T mitochondria into 143BTK− ρ0 cells can generate between dozens to hundreds of independent SIMR clones (Figure 5). Other cell lines used as donor or recipient cells, particularly transferring mutant mtDNA-bearing mitochondria, can reduce the SIMR clone yield (Patananan et al., 2020; Sercel et al., 2021).
Figure 5.
Crystal violet staining of SIMR clones
Images of Crystal Violet stained SIMR clones generated by MitoPunch transfer of HEK293T mitochondria or DPBS with calcium and magnesium vehicle into 143BTK− ρ0 and NDF ρ0 recipient cells on 10 cm dishes after SIMR clone selection.
Limitations
Using this protocol, MitoPunch can only deliver isolated mitochondria into adherent cells that can attach to the PET filter membrane. Additionally, MitoPunch is unable to generate SIMR clones in recipient cells that already contain mtDNA unless a secondary antibiotic selection is applied simultaneously with the SIMR restrictive medium scheme (Dawson et al., 2020). The pressure generated by the MitoPunch apparatus as described in this protocol is fixed, and some cell types may require different levels of pressure to generate SIMR clones efficiently (Sercel et al., 2021). MitoPunch delivery into ρ0 primary fibroblasts yields fewer SIMR clones than transfers into transformed ρ0 cells, and SIMR clones isolated from MitoPunch transfer into ρ0 primary fibroblasts can be difficult to expand before the cells reach the Hayflick limit and senesce. Furthermore, mtDNA and nDNA of different haplotypic backgrounds may suffer from biochemical or genetic incompatibilities and reduce the SIMR generation efficiency of MitoPunch transfers using material from such sources. Combinations of mitochondria and recipient cells from different species face incompatibility that prohibits SIMR clone generation. Lastly, isolated donor mitochondria must be freshly isolated prior to every delivery, as extended time in suspension or flash-freezing prior to MitoPunch delivery have a negative effect on SIMR generation efficiency (data not shown).
Troubleshooting
Problem 1
Construct MitoPunch apparatus: MitoPunch piston is not actuating.
Potential solution
Ensure the electrical wiring of the MitoPunch device is properly connected and remains connected between samples. Always test fire the MitoPunch device before placing the PDMS reservoir on the platform. Additionally, verify that the piston is installed in the correct orientation such that the piston is driven up toward the PDMS reservoir when actuated.
Problem 2
Test recipient cell sensitivity to different SIMR clone selection media conditions: Restrictive medium selection does not reduce cell viability of recipient cells.
Potential solution
When the recipient cells are not completely mtDNA deficient the restrictive media will not effectively induce cell cycle arrest and death. Verify that the recipient cells are ρ0 prior to beginning MitoPunch using qPCR to measure mtDNA copy number and respirometry to quantify mitochondrial electron transport chain activity.
Problem 3
Mitochondrial preparation: Needle clogs persistently during mechanical isolation step of mitochondrial isolation.
Potential solution
Mitochondrial donor cell cultures that reach 100% confluency cause more clogging than cells from plates at lower confluency. Only grow donor cell cultures to 80%–90% confluence before harvesting for mitochondrial isolation.
Problem 4
Recipient cell collection: Few cells are recovered after MitoPunch and collection from filter inserts.
Potential solution
Carefully remove all cell growth medium from inside, outside, and below the PET filter insert. Medium is easily trapped between the bottom surface of the insert and the 12-well culture dish, and this excess medium can impact the efficacy of the dissociation reagent used to collect cells. Additionally, ensure dissociation reagent is applied to both sides of the filter insert as MitoPunch recipient cells grown on the filters extend projections through the pores in the membrane and must be exposed to reagent from both sides for complete dissociation.
Problem 5
SIMR clone selection: Restrictive medium selection does not reduce cell viability of recovered MitoPunch recipient cells.
Potential solution
This problem can be caused by three main factors. The first is allowing the recipient cell cultures to reach 100% confluence before introducing restrictive medium. Begin medium selection before cultures reach 80% confluence. The second is not feeding recipient cell cultures with restrictive medium with sufficient frequency. Apoptotic cells on the dishes will release nucleotides into the medium which can be scavenged by nascent SIMR cells and ρ0 cells alike, leading to constant cell viability throughout the selection process. When daily feeding is insufficient, increase the frequency of media exchange to ensure selection occurs. Third, not all lines are sufficiently sensitive to SIMR selection medium and require galactose SIMR selection medium to induce selection. Follow the steps in Test recipient cell sensitivity to different SIMR clone selection media conditions to determine the sensitivity of the chosen recipient cell line to the selection scheme.
Problem 6
SIMR clone selection: No SIMR clones are observed following MitoPunch and restrictive medium selection.
Potential solution
When no SIMR clones are observed, verify that the mitochondrial isolation yielded coupled mitochondria. Perform a mitochondrial isolation and measure the respiration rate of the mitochondrial isolate using a Seahorse Extracellular Flux Analyzer. When the isolation does not yield coupled mitochondria, verify that the buffers used in the isolation protocol are stored at the appropriate temperatures and are supplemented with protease inhibitor.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael A. Teitell (mteitell@mednet.ucla.edu).
Materials availability
This study did not generate new unique reagents. Dimensions of custom fabricated parts necessary to replicate this work are included in the text of this protocol.
Acknowledgments
A.J.S. is supported by the NIH (T32GM007185 and T32CA009120). A.N.P. is supported by the NIH (T32CA009120) and American Heart Association (18POST34080342). P.-Y.C. is supported by the NSF (CBET 1404080), the NIH (RO1GM114188), and the Air Force Office of Scientific Research (FA9550-15-1-0406). M.A.T. is supported by the Air Force Office of Scientific Research (FA9550-15-1-0406), the Department of Defense (W81XWH2110139), the NIH (R01GM114188, R01GM073981, R01CA185189, R21CA227480, R01GM127985, and P30CA016042), and CIRM (RT3-07678). We thank Yen-Ju Lin, Xiang Zhang, and Marvin Tan for their assistance with polymer fabrication.
Author contributions
A.J.S. and A.J.N. drafted and edited the manuscript and figures. T.-H.W., P.-Y.C., and M.A.T. developed the core MitoPunch technology. A.J.S., A.N.P., and T.-H.W. optimized and implemented the protocol. P.-Y.C. provided funds and infrastructure to conduct the work. M.A.T. provided funds and infrastructure to conduct the work, edited the manuscript, and advised throughout the optimization of the protocol.
Declaration of interests
P.-Y.C. and M.A.T. are co-founders, board members, shareholders, and consultants for NanoCav, LLC. Patents related to this technology include (1) Efficient delivery of large cargos into cells on a porous substrate, issued Nov. 12, 2019, US 10,472,651 (UCLA); and (2) Mechanical transfection devices and methods, issued Sep. 1, 2020, US 10,760,040 (NanoCav). The other authors declare no competing interests.
Data and code availability
This study did not generate or analyze new code or datasets.
References
- Aguer C., Gambarotta D., Mailloux R.J., Moffat C., Dent R., McPherson R., Harper M.E. Galactose enhances oxidative metabolism and reveals mitochondrial dysfunction in human primary muscle cells. PLoS One. 2011;6:e28536. doi: 10.1371/journal.pone.0028536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier E., Young E.W., Beebe D. Engineers are from PDMS-land, biologists are from polystyrenia. Lab. Chip. 2012;12:1224–1237. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- Dawson E.R., Patananan A.N., Sercel A.J., Teitell M.A. Stable retention of chloramphenicol-resistant mtDNA to rescue metabolically impaired cells. Sci. Rep. 2020;10:14328. doi: 10.1038/s41598-020-71199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire M., Morais R., Quilliam M.A., Gravel D. On auxotrophy for pyrimidines of respiration-deficient chick embryo cells. Eur. J. Biochem. 1984;142:49–55. doi: 10.1111/j.1432-1033.1984.tb08249.x. [DOI] [PubMed] [Google Scholar]
- Miyata N., Steffen J., Johnson M.E., Fargue S., Danpure C.J., Koehler C.M. Pharmacologic rescue of an enzyme-trafficking defect in primary hyperoxaluria 1. Proceedings of the National Academy of Sciences. 2014;111:14406–14411. doi: 10.1073/pnas.1408401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patananan A.N., Sercel A.J., Wu T.H., Ahsan F.M., Torres A., Jr., Kennedy S.A.L., Vandiver A., Collier A.J., Mehrabi A., Van Lew J. Pressure-driven mitochondrial transfer pipeline generates mammalian cells of desired genetic combinations and fates. Cell Rep. 2020;33:108562. doi: 10.1016/j.celrep.2020.108562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercel A.J., Patananan A.N., Man T., Wu T.H., Yu A.K., Guyot G.W., Rabizadeh S., Niazi K.R., Chiou P.Y., Teitell M.A. Stable transplantation of human mitochondrial DNA by high-throughput, pressurized isolated mitochondrial delivery. Elife. 2021;10:e63102. doi: 10.7554/eLife.63102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze new code or datasets.