Abstract
During the course of the SARS-CoV-2 pandemic reports of mutations with effects on spreading and vaccine effectiveness emerged. Large scale mutation analysis using rapid SARS-CoV-2 Whole Genome Sequencing (WGS) is often unavailable but could support public health organizations and hospitals in monitoring transmission and rising levels of mutant strains. Here we report a novel WGS technique for SARS-CoV-2, the EasySeq™ RC-PCR SARS-CoV-2 WGS kit. By applying a reverse complement polymerase chain reaction (RC-PCR), an Illumina library preparation is obtained in a single PCR, thereby saving time, resources and facilitating high-throughput screening. Using this WGS technique, we evaluated SARS-CoV-2 diversity and possible transmission within a group of 173 patients and healthcare workers (HCW) of the Radboud university medical center during 2020. Due to the emergence of variants of concern, we screened SARS-CoV-2 positive samples in 2021 for identification of mutations and lineages. With use of EasySeq™ RC-PCR SARS-CoV-2 WGS kit we were able to obtain reliable results to confirm outbreak clusters and additionally identify new previously unassociated links in a considerably easier workaround compared to current methods. Furthermore, various SARS-CoV-2 variants of interest were detected among samples and validated against an Oxford Nanopore sequencing amplicon strategy which illustrates this technique is suitable for surveillance and monitoring current circulating variants.
Keywords: COVID-19, SARS-CoV-2, WGS, RC-PCR, Mutation, Lineage
1. Introduction
In December 2019 China reported a group of patients with a severe respiratory illness caused by a thus far unknown coronavirus. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as the causative agent [1]. Since then the outbreak has evolved into a pandemic with more than a hundred million infections and almost 4 million deaths worldwide by June 2021 [2]. Healthcare systems, governments and society as a whole are under pressure, working to reduce the spread of SARS-CoV-2 by large scale testing and vaccination. The start of vaccination coincided with reports of new variants of SARS-CoV-2, variants with specific mutations in the spike protein reported to be associated with either an increase in infectiousness or a possible reduction of vaccine effectiveness [3], [4], [5], [6], [7]. Defined variants of concern or interest are already being detected in multiple countries and the proportion of mutants in the population is increasing. This poses a new challenge besides the already large-scale testing of the community. Current testing is based on RT-PCR detection of SARS-CoV-2 in naso- or oropharyngeal swabs. If tested SARS-CoV-2 positive, instructions are to self-isolate at home and source finding and contact tracing is performed. In a hospital setting, the same procedures are in place to identify patients and personnel at risk of infection. Contact tracing is time consuming both in- and outside a hospital setting and when numbers of infections are high, the public health capacity reaches the limits of feasibility of thorough source and contact tracing investigations [8]. Routine sequencing of the SARS-CoV-2 genome from positive samples provides crucial insights into viral evolution and can support outbreak analysis [9, 10]. Current Whole Genome Sequencing (WGS) workflows often require cumbersome preparation, are laborious to implement for high throughput screening complicating widespread implementation. Using the Reverse Complement Polymerase Chain Reaction (RC-PCR) EasySeq™ RC-PCR SARS-CoV-2 WGS kit (NimaGen, Nijmegen, The Netherlands), which integrates tiled target amplification with Illumina library preparation, has a simple workflow with minimal hands-on time. The current study evaluates the technique and shows the application in the detection of variants of interest. Additionally, a set of epidemiologically linked cases was used to illustrate its added value in detecting potential transmission events in public health and hospital settings.
2. Material and methods
In this study we assessed the performance of the novel RC-PCR sequencing technology applied to SARS-CoV-2, EasySeq™ RC-PCR SARS-CoV-2 WGS kit (NimaGen, Nijmegen, The Netherlands).
3. Samples
3.1. March – September 2020
Naso- and oropharyngeal swabs collected in UTM or GLY medium of 173 SARS-CoV-2 positive and 15 negative samples were collected from healthcare workers and patients at the Radboud university medical center. Among these samples, 6 outbreak clusters defined by our hospital infection prevention and control (IPC) team were included. 64 samples were collected and tested on behalf of the local public health service. These were samples of people living in the defined public health region surrounding our hospital.
3.2. January – May 2021
Naso- and oropharyngeal swabs collected in UTM or GLY medium from patients and healthcare workers who tested SARS-CoV-2 positive at the Radboud university medical center during from January to May 2021 were included (n = 171) to determine lineage and the presence of variants of interest within our hospital population.
3.3. Variant panel
Seven cultivated SARS-CoV-2 samples of various lineages previously sequenced by the national public health authority of the Netherlands (RIVM) using Oxford Nanopore technologies amplicon strategy [10].
All personal data of patients, HCW and public health service samples was anonymized. Cluster information was provided anonymously by the IPC team and the regional public health service.
Detailed descriptions on the included samples can be found in Supplementary Data.
4. Real-time polymerase chain reaction (RT-PCR)
SARS-CoV-2 RT-PCR was performed on all samples during routine diagnostics. RNA was isolated using Roche COBAS 4800 (Roche Diagnostics Corporation) with a CT/NG extraction kit according to the manufacturers protocol. RT-PCR with primers targeting the envelope (E-gene) was used as described by Corman et al., and performed on a LightCycler 480 (Roche Diagnostics Corporation) using Roche Multiplex RNA Virus Mastermix [11].
4.1. Reverse complement polymerase chain reaction (RC-PCR)
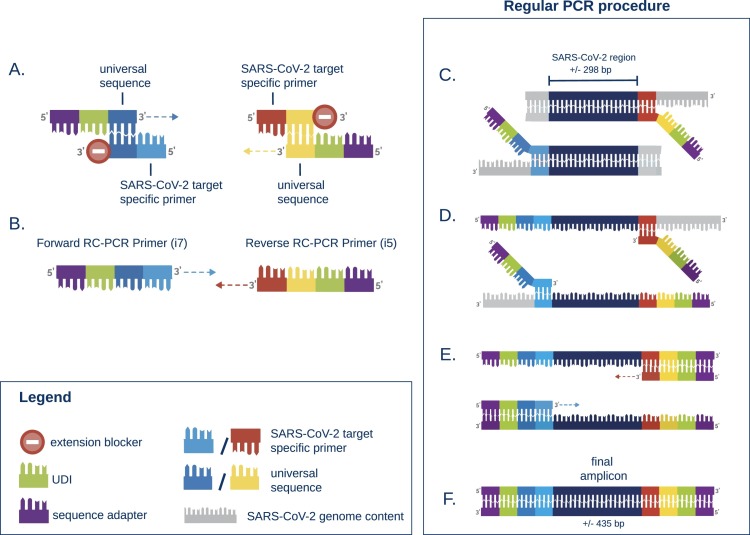
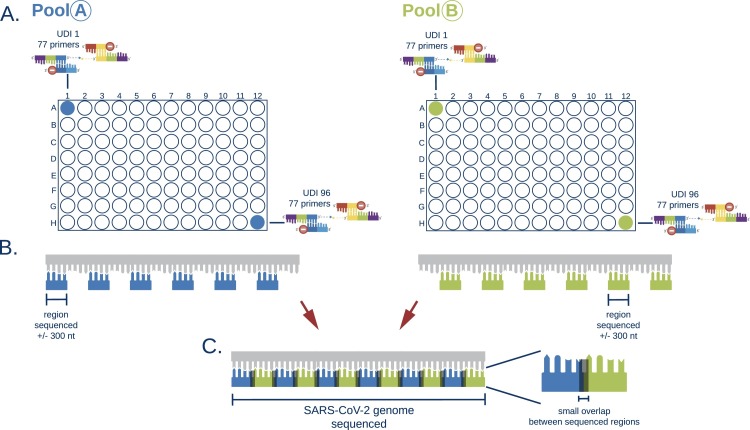
For all samples, RNA isolation was repeated on the MagnaPure 96 (Roche Diagnostics Corporation) using Small Volume isolate protocol with 200 µl of sample and eluting isolated RNA in 50 µl. cDNA-synthesis was performed using either Multiscribe RT (Applied Biosystems, USA) or LunaScript® RT SuperMix Kit (New England Biolabs, USA) with respectively 5 or 6 µl of RNA input. Whole genome sequencing (WGS) was performed in 6 independent runs using the EasySeq™ RC-PCR SARS-CoV-2 WGS kit (NimaGen, Nijmegen, The Netherlands). Fig. 1, Fig. 2 show a detailed description of the technology in which two types of oligo's are used to start the targeted amplification. The universal sequence hybridizes with the SARS-CoV-2 target specific primer creating the RC-PCR primer which includes the specific SARS-CoV-2 primers with Unique Dual Index (UDI) and adapter sequences. This in contrast to other techniques where multiple steps are needed to add sequence adapters and UDI's. Thus, a regular PCR-system can be used to produce SARS-CoV-2 specific amplicons ready for sequencing. The kit uses 155 (v1) and 154 (v2–v3) newly designed primer pairs with a tiling strategy as previously implemented in the ARTIC protocols [12]. The primer pairs are divided in two pools, A and B. Pool A contains 78 or 77 primers (v1 and v2-v3 respectively) and Pool B contains 77 primers. This strategy requires two separate RC-PCR reactions but ensures there is minimal chance of forming chimeric sequences or other PCR artifacts (Fig. 2). After the PCR, samples of each plate are pooled into an Eppendorf tube, resulting in two tubes, for pool A and B, respectively. These are individually cleaned using AmpliClean™ Magnetic Bead PCR Clean-up Kit (NimaGen, Nijmegen, The Netherlands). Afterwards, quantification using the Qubit double strand DNA (dsDNA) High Sensitivity assay kit on a Qubit 4.0 instrument (Life Technologies) is performed and pool A and B are combined. The amplicon fragment size in the final library will be around 435 bp with a 298 bp SARS-CoV-2 genomic insert. Next Generation Sequencing (NGS) was performed on an Illumina MiniSeq® using a Mid Output Kit (2 × 149 or 2 × 151-cycles) (Illumina, San Diego, CA, USA) by loading 0.8 pM on the flowcell. The first two sequence runs were conducted using version 1 of the EasySeq™ RC-PCR SARS-CoV-2 WGS kit on a large variety of Ct-values (Ct 16 – 41) using the standard protocol provided by NimaGen. The additional sequence runs were conducted using version 2 or version 3 of the EasySeq™ SARS-CoV-2 WGS kit using a balanced library pooling strategy based on estimated cDNA input according to the manufacturer's protocol.
Fig. 1.
Schematic representation of the RC-PCR technology used in EasySeq™ SARS-CoV-2 WGS kit (NimaGen, Nijmegen, The Netherlands). The protocol consists of one single PCR-like reaction consisting of 2 steps. The schematic is adapted from Kieser et al., (Kieser et al., 2020) A. Two types of oligo's are present, 1) the universal tail which includes a Unique Dual Index (UDI), sequence adapter, and universal sequence. 2) The RC primer which contains an extension blocker, universal sequence, and the reverse complement of the SARS-CoV-2 genomic target sequence. B. The universal sequences anneal and form a SARS-CoV-2 specific PCR primer. C – E. A regular PCR in which the SARS-CoV-2 specific amplicons are created. F. The final amplicons are ready to sequence on an illumina sequencer.
Fig. 2.
Schematic overview of the EasySeq™ RC-PCR SARS-CoV-2 WGS kit. A. For 96 samples the kit provides a pool A plate and a pool B plate. These plates contain the RC-probe and the universal oligo's with an UDI per well. Identical UDI's between pool A and B are used. B. For covering the SARS-CoV-2 genome a tiling method similar to the ARTIC protocol is used. (DNA Pipelines R&D, 2020) The kit provides two distinct plates to separate into amplicon pools that do not overlap. This greatly enhances accurate sequencing output. C. The same wells of pool A and B share the same index and allow the combination of corresponding sequencing results to cover the SARS-CoV-2 genome.
5. SARS-CoV-2 WGS 2020
A custom designed variant pipeline (version 0.3.3) was developed and used to process the EasySeq™ Illumina paired-end reads (https://github.com/JordyCoolen/easyseq_covid19). In short, sequence reads were cleaned using fastp (version 0.20.1) [13]. Cleaned reads were mapped to SARS-CoV-2 reference NC_045512.2 using bwa mem (version 0.7.17) [14]. Bamclipper (version 1.0.0) [15] is used to clip the bam file from EasySeqtm RC-PCR SARS-CoV-2 specific primer pairs. Bcftools (version 1.9) [16] and KMA (version 1.3.9) [17] are used to perform the variant calling, using a mutation frequency ≥ 75%, QUAL score of ≥ 20 and depth of ≥ 5. Variant calls are annotated using snpEff (version 5.0) [18] and reference NC_045512.2. Consensus output was generated using bcftools consensus (version 1.9). Sequence statistics were calculated using faCount (version 377). Lineage determination was performed using pangolin (version 2.3) with pangoLEARN (2021‐02‐18) (github.com/cov-lineages/pangolin). Consensus sequences were aligned using mafft (version 7.474) [19]. Phylogeny was inferred using IQTREE (version 2.0.3) [20] with setting –ufboot 1000 -m GTR + F + I + G4. Phylogenetic tree visualization and annotation was performed using iTOL (version 5.6.3) [21].
6. SARS-CoV-2 WGS 2021
Adjustments to the variant pipeline were made to create the consensus sequence and call variants resulting in EasySeq variant pipeline (version 0.8.1) (https://github.com/JordyCoolen/easyseq_covid19). Adjustments made in short, for variant calling lofreq (version 2.1.5) was used with settings mutation frequency ≥ 50%, QUAL score of ≥ 20 and depth of ≥ 10. Lineage determination was performed using pangolin (version 3.0.5) with pangoLEARN (2021‐06‐05) (github.com/cov-lineages/pangolin). Genome read coverage and spike S read coverage were calculated using command multiBamSummary bins of deeptools (version 3.5.0) [22]. Samples with ≥90% genome coverage are used for analysis and clinical and public health reporting.
7. Results
In this study we performed Illumina sequencing of up to 96 samples using various versions of the EasySeq™ RC-PCR SARS-CoV-2 WGS kit. The total turnaround time is about 26 h, consisting of 1 h hands-on time for preparing 96 samples, 6.5 h for performing the RC-PCR, and 1 h of hands-on time for pooling and sample clean-up, approximately 17 h of DNA sequencing and up to 1 h of analysis.
7.1. Technical evaluation – mean sequence depth plots
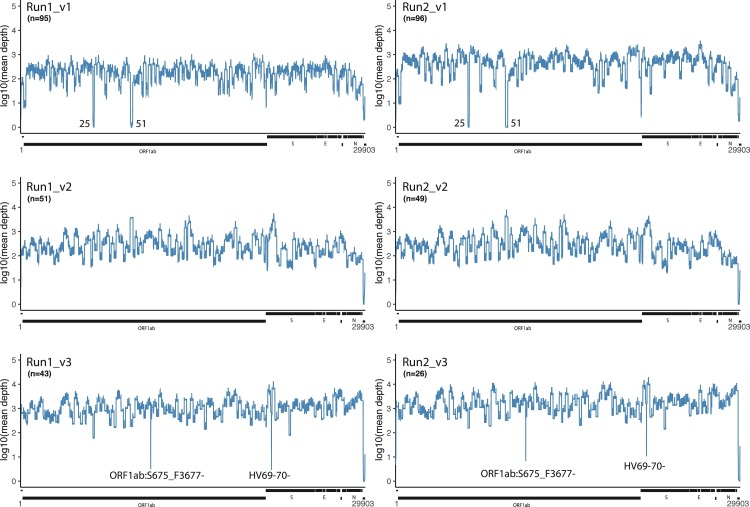
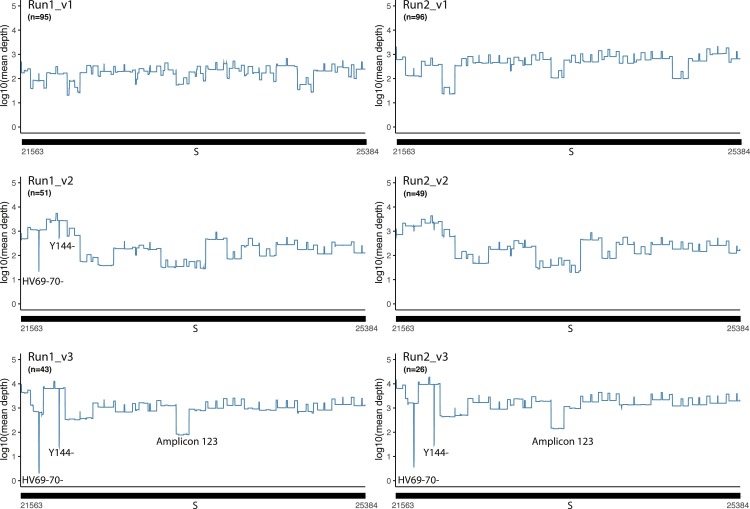
The mean sequence depth of the SARS-CoV-2 genome is plotted for 3 versions of the EasySeq™ RC-PCR WGS kit, each 2 runs (Fig. 3 ). Mean depths are centered on a depth of 2–3 log10. Version 1 (v1) of the EasySeq™ RC-PCR kit was not able to amplify all coding regions, 6280–6407 (amplicon 35) and 9525–9737 (amplicon 51) both located on ORF1ab were missed (Fig. 3). For version 2 (v2), new amplicons were designed and added resulting in coverage of all coding regions as illustrated in Fig. 3. As for version 3 (v3) another low covered region can be observed which is the dominant ORF1ab:S3675_F3677- in the B.1.1.7 (Alpha) variant (Run1_v3). In v3, a new amplicon design of the primers covering the Spike HV69–70 deletion has been designed to more accurately detect this region. HV69–70- is clearly visible in Fig. 3(v3). To give a better representation of the coverage of the Spike gene, mean sequence depth plots specifically on the Spike gene were generated (Fig. 4 ).
Fig. 3.
Graphical representation of mean sequence depth obtained using EasySeq™ RC-PCR SARS-CoV-2 WGS kit over multiple illumina sequence runs and versions. The line graph represents the mean sequence depth in log10 scale of the SARS-CoV-2 genome. The top two graphs are based on data produced using v1 kit of EasySeq™, middle two graphs are based on data using v2 and the last two of v3. Below each plot a representation of the SARS-CoV-2 ORFs are given. 25 and 51 indicate the amplicons dropouts in v1. ORF1ab:S675_F3677- and S:HV69–70- low coverage is due to a known deletions in B.1.1.7 (alpha).
Fig. 4.
Graphical representation of mean sequence depth on the spike gene. The line graph represents the mean sequence depth in log10 scale of the spike gene. Top two graphs using v1, middle two graphs v2, and the last two using v3 of EasySeq™ RC-PCR SARS-CoV-2 WGS kit. HV69–70- and Y144- indicate the region of known alpha variant deletions. Region indicated the amplicon 123 region which resulted in a lower coverage compared to average in v3.
Results of the mean coverage plots of the Spike gene show a mean sequence depth of around 2–3 log10 for v1 and 2, and in v3 on average one log10 higher. The coverage plot of v3 three regions can be observed with lower coverages. The first two regions are S:HV69–70- and S:Y144-, which are known deletions of the Alpha (B.1.1.7) variant and dominant during time of screening [23]. The third region is a larger region (23,289–23,431), this region has lower coverage due to less efficient amplification of amplicon 123. This Alpha variant was not present during using v1 and only limited present during v2 (Fig. 4).
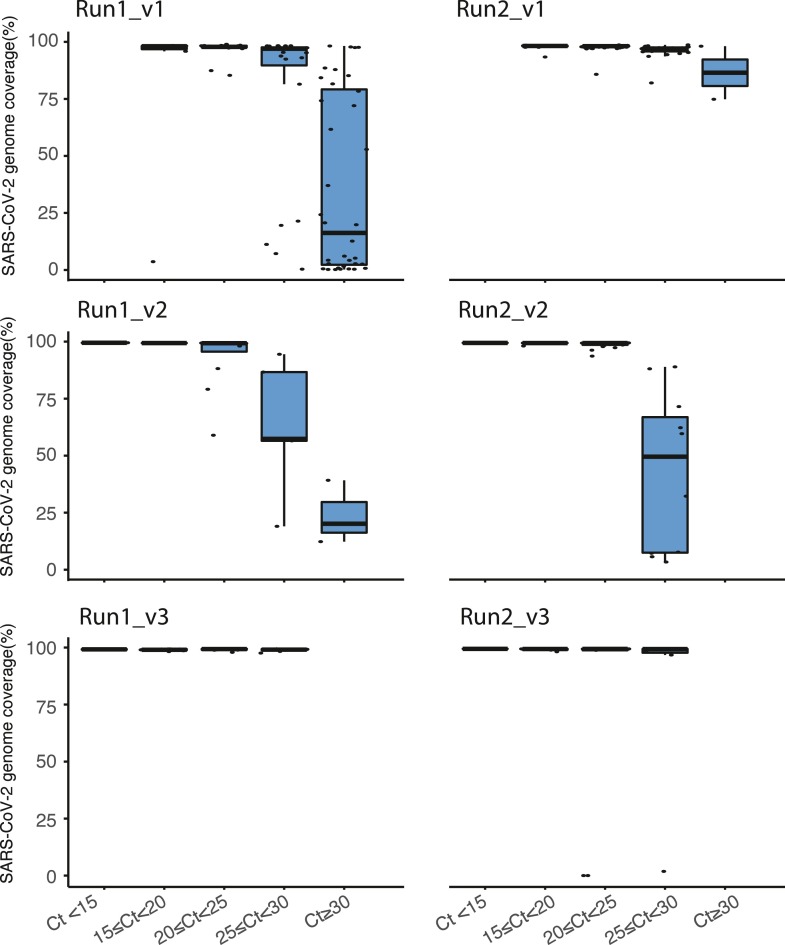
Additionally, the effect of viral load on the SARS-CoV-2 genome coverage was examined (Fig. 5 ). For this, samples were divided in five Ct groups (Ct < 15, 15 ≤ Ct ≤ 20, 20 ≤ Ct ≤ 25, 25 ≤ Ct ≤ 30, and Ct ≥ 30). For the runs with v1, all samples were included regardless Ct value. The runs performed with v2 and 3 of the kit, only sample with Ct values up to 30 were included. Variation is seen between runs and versions, for instance, the runs using v2 mis more regions with higher Ct values ≥ 25, whereas version 3 in which Lunascript was used for the RT reaction shows much better coverage on higher Ct values tested up to Ct 30.
Fig. 5.
. Boxplots of the Ct value as determined by EasySeq™ to illustrate the effect of viral load on the SARS-CoV-2 genome coverage. For the top two runs (v1 of EasySeq™), all samples regardless of Ct value are included. For the other four runs (v2 and v3 of EasySeq™) samples up to Ct 30 were included. For reporting to clinic and public health services samples with genome coverage <90% were excluded.
8. SARS-CoV-2 phylogenetic analysis (samples March – September 2020)
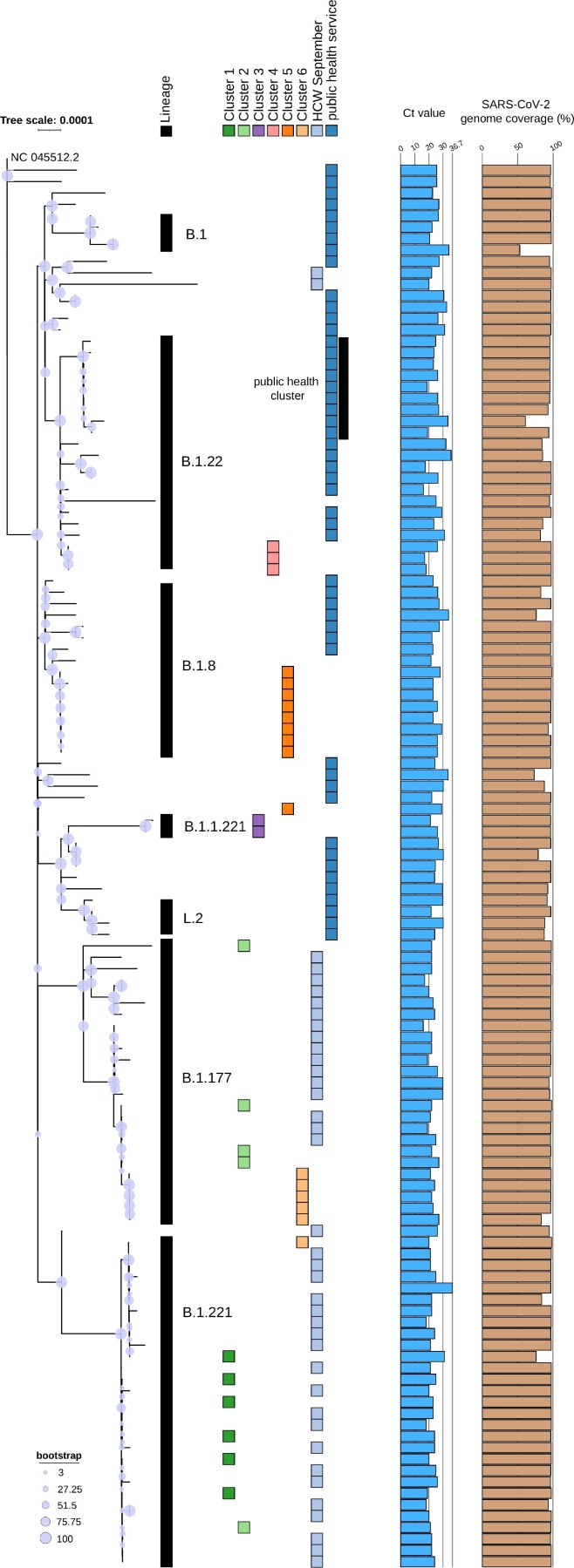
173 samples were tested SARS-CoV-2 positive of which 123 were samples of patients and HCW in our hospital and 64 samples from the public health region. Those samples were sequenced in Run1_v1 and Run2_v2 and are depicted in the phylogenetic analysis of Fig. 6. This clearly illustrates the relationship between samples and clusters sequenced in 2020. Seven distinct lineages have been detected, B.1, B.1.22, B.1.8, B.1.1.221, L.2, B.1.177 and B.1.221. Those samples collected early in the pandemic during March and April 2020 (community samples from public health service) are separated from the others, especially compared to the samples from September 2020 (Cluster 1,2,3,6, and the HCW). In four out of six outbreak clusters (Cluster 3,4,5,6) defined by the infection prevention team, sequencing results support previously identified epidemiological information. However, some samples within these epidemiologically defined clusters were excluded based on phylogenetic placement, for instance, one sample of Cluster 5 is not part of lineage B.1.8 (Fig. 6).
Fig. 6.
Phylogenetic tree of 123 SARS-CoV-2 genomes rooted at the NC045512.2 reference genome. Sample groups of six epidemiologically identified clusters in the hospital are indicated by the colored blocks. Additional Ct values and genome coverage are plotted in barplots to illustrate the diversity in viral load between the samples and the high genome coverage that can be achieved by v1 of EasySeq™ RC-PCR SARS-CoV-2 WGS kit, respectively.
Within the 64 community samples, samples from nine people clustered together (Fig. 6, part of lineage B.1.22). The public health service confirmed a cluster seen within this group of samples.
9. SARS-CoV-2 lineage and variant screening (January – May 2021)
Lineage and variant screening results of samples obtained using two individual sequence runs (Run1_v3 and Run1_v3) performed in 2021 are given to illustrate the method is able to detect circulating variants. Results show circulation of various lineages; P.1 (Gamma), C.36.3, B.1.525 (Eta), B.1.351 (Beta), and B.1.1.7 (Alpha) (Fig. 7A). The most dominant lineage circulating during that period was B.1.1.7 (Alpha) being 78% (53/68) of all screened samples. Fig. 7A demonstrates the corresponding Spike gene mutation detected during the variant screening. Results show that for B.1.1.7 (Alpha) all associated Spike mutation were detected in tested samples (S:HV69–70-, S:Y144-, S:N501Y, S:A570Y, S:P681H, S:T716I, S:S982A, and S:D1118H) [23]. Furthermore, in 96.4% of all samples of the Alpha variant Spike mutation S:D614G was observed (Fig. 7A). Additionally, mutation S:D614G was found in all circulating lineages.
Fig. 7.
Variant and mutation detection of the SARS-CoV-2 located on the spike gene. A). Samples collected in 2021 using v3. B). Seven samples from a variant panel were sequenced using v3 to validate and compare the results obtained with deposited sequences generated using Oxford nanopore technologies amplicon sequencing and performed by the national public health authority of the Netherlands (RIVM). ONT: Oxford nanopore technologies.
10. SARS-CoV-2 lineage and variant detection verification
We tested seven samples to validate if lineage determination and Spike variant detection matches between another broadly used sequencing method. The SARS-CoV-2 samples were sequenced by the national public health authority of the Netherlands (RIVM) using Oxford Nanopore Technologies (ONT) sequencing using an amplicon strategy as described by Oude Munnink et al., [10]. Results show 100% consensus on lineage outcome and 100% identical detection for all Spike gene mutations specific to each lineage (Fig. 7B).
11. Discussion
This study describes the first application of Reverse Complement-PCR implemented in the EasySeq™ RC-PCR SARS-CoV-2 WGS kit to sequence the SARS-CoV-2 genome. This novel method combines target amplification and indexing in a single procedure, directly creating a sequencing ready Illumina library. Using this method, epidemiological clusters from the hospital and the community were supported by phylogenetic outbreak analysis. Additionally, circulating SARS-CoV-2 lineages and defined variants of concern could be identified and monitored. Using RC-PCR, samples with Ct values up to 30 as determined by RT-PCR could be sequenced with a high SARS-CoV-2 genome coverage. With optimization of the protocols, bioinformatic analysis, and the kit itself, it is expected that performance can be increased. As was already seen with the switch from Multiscribe RT to Lunascript RT for the reverse transcriptase, changes to primers between kit versions and optimization of the bioinformatic analysis resulting in higher genome coverage at higher Ct values and a more accurate mutation detection.
Previous studies showed the benefit of using WGS of SARS-CoV-2 for outbreak investigation purposes and to study transmission routes [10, [24], [25], [26], [27], [28]]. Several methods have been optimized for this purpose. The ARTIC Illumina method, a tiling multiplex PCR approach, was the first that enabled WGS of SARS-CoV-2 using Illumina sequencers [29]. The technique has subsequently been optimized and analysis, albeit in small sample numbers, concluded that it delivers sufficient quality to perform phylogenetic analysis [30], [31], [32]. It had been used as targeted and random RT-PCR screening with subsequent sequencing of the population in order to study the spread of SARS-CoV-2 through the community [24]. Sikkema et al., showed the use of SARS-CoV-2 sequencing in healthcare associated infections and identified multiple introductions into Dutch hospitals through community-acquired infections [9].
During this pandemic many advances have been made in WGS of SARS-CoV-2 [33]. It should be noted that for the EasySeq™ RC PCR SARS-CoV-2 WGS kit two of the primer pairs using v1 of EasySeq™ were suboptimal. Improvements were seen in version 2 and 3 of the kit, which resulted in an increase in genome coverage to a maximum 98.9% and 99.5% respectively between versions 2 and 3. Currently, EasySeq™ is able to retrieve nearly 100% genome coverage, our study shows that the technology is very useful for phylogenetic analysis and mutation and variant detection of SARS-CoV-2. The EasySeq™ RC-PCR SARS-CoV-2 WGS kit uses 155 amplicons which makes it susceptible to amplicon dropouts (regions of no sequence coverage) as a result of accumulation of mutation on primer binding locations, this was observed in this study but also has been reported using other amplicon designs [34,35,36]. The introduction of the SARS-CoV-2 B1.1.7 (Alpha) variant caused problems to properly detect S:HV69–70- using v1 and v2. The limitation of detecting S:HV69–70- has been solved by the new design in v3. In line with the mutation rate of SARS-CoV-2 probably more adjustments to the RC-PCR primers have to be made in the future to ensure retrieval of full SARS-CoV-2 genome sequences which is apparent to amplicon-based assays [34]. Vice versa, the high number of amplicons also limits the size of the dropout making it less vulnerable for losing a large portion of the SARS-CoV-2 genome.
Regardless of high or low infection rates, real-time sequencing of SARS-CoV-2 positive samples could be used to target infection prevention measures nationwide and locally [37]. Its application can range from incidental cluster analysis to support or reject epidemiological related cases to real-time surveillance in the community or health care institutes. The latter surveillance strategies have already been implemented in the Netherlands recently due to the emergence of new variants of interest especially related to infectiousness, clinical outcome and vaccine effectiveness [38, 39].
With this study we evaluated the performance of the EasySeq™ RC-PCR SARS-CoV-2 WGS kit, although this gives a good impression of how well this novel RC-PCR technology works we want to emphasize that there are limitations to this evaluation. It is difficult to compare sensitivity of the assay to other available SARS-CoV-2 WGS strategies. Other studies also demonstrate its performance by comparing the genome completeness to Ct values however, one-on-one comparison of these values is limited [33]. Preferably DNA copies or viral copies as viral load would be more valuable but are difficult to obtain.
In conclusion, this study shows the first application of RC-PCR in the field of medical microbiology and infectious diseases. Results confirm the robustness of the method which requires less hands-on time compared to current sequencing methods and can be used for high throughput sequencing of SARS-CoV-2. WGS of SARS-CoV-2 accompanied with bioinformatic analysis support the identification of chains of transmission of SARS-CoV-2 and the spread of different lineages including mutation profiles and variant detection. This enables a rapid, targeted and adaptive response to an ongoing outbreak that has great impact on public health and society.
12. Author contributions
J.P.M.C. and F.W. conducted the research, performed analysis, wrote manuscript and created the figures. L.F.J.vG., C.P.B-R., E.C.T.H.T., N.vdG-B., J.L.A.H. proofreading and provided clinical information and samples of patients and HCWs. A.T.and J.H. conducted the contact tracing and proofreading of the manuscript. H.F.L.W., J.C.R-L., M.S., and W.J.G.M supervised the study and drafted the manuscript.
13. Data availability
Tailored variant analysis pipeline can be found on https://github.com/JordyCoolen/easyseq_covid19 SARS-CoV-2 metadata and GISAID is available in the Supplementary Data.
14. Ethical approval
N/A
15. Funding/support
The EasySeq™ RC-PCR SARS-CoV-2 WGS version 1 kit was supplied by NimaGen and sequencing of the first two Illumina libraries was performed by NimaGen. Further sequencing was performed by the Department of Medical Microbiology at the Radboud university medical center for the purpose of using the technology in routine diagnostics and support the national surveillance program. Therefore, no other funding was applied for.
16. Role of funder/sponsor
NimaGen had no role in the design and conduct of the study; collection, management, data analysis; preparation or approval of the manuscript.
Declaration of Competing Interest
The authors have no conflict of interest to disclose.
Acknowledgements
We would like to thank all the support and work done by our coworkers at Medical Microbiology Radboudumc, Bart van den Bosch, Ellen Koenraad. Coworkers at Human Genetics Radboudumc: Ronny Derks, Amber den Ouden, Duaa Elmelik, Michiel Oorsprong and Marcel Nelen. Coworkers at Bioinformatics Human Genetics, Radboudumc: Steven Castelein en Christian Gilissen. Technical support from NimaGen: Rick Lammerts, Walter van der Vliet, Simon van Reijmersdal and Joop Theelen. John Sluimer and Adam Meijer for providing the variant panel samples.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.104993.
Appendix. Supplementary materials
References
- 1.Zhu N., et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019:2020. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toovey O.T., et al. Introduction of Brazilian SARS-CoV-2 484 K. V2 related variants into the UK. J. Infect. 2021 doi: 10.1016/j.jinf.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.COVID, T., An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe, 2020. [DOI] [PMC free article] [PubMed]
- 5.Rambaut A., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson E.C., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021 doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemmermann N.A., et al. SARS-CoV-2 genome surveillance in Mainz, Germany, reveals convergent origin of the N501Y spike mutation in a hospital setting. medRxiv. 2021 [Google Scholar]
- 8.McLachlan, S., et al., The fundamental limitations of COVID-19 contact tracing methods and how to resolve them with a Bayesian network approach. 2020.
- 9.Sikkema R.S., et al. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect. Dis. 2020;20(11):1273–1280. doi: 10.1016/S1473-3099(20)30527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munnink B.B.O., et al. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat. Med. 2020;26(9):1405–1410. doi: 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- 11.Corman V.M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D.N.A. Pipelines R&D, B.F., Diana Rajan, Emma Betteridge, Lesley Shirley, Michael Quail, Naomi Park, Nicholas Redshaw, Iraad F Bronner, Louise Aigrain, Scott Goodwin, Scott Thurston, Stefanie Lensing, Charlotte Beaver, Ian Johnston. COVID-19 ARTIC v3 illumina library construction and sequencing protocol V.1. 2020 10th september 2020]; Available from: https://www.protocols.io/view/covid-19-artic-v3-illumina-library-construction-an-beuzjex6?version_warning=no.
- 13.Chen S., et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997, 2013.
- 15.Au C.H., et al. BAMClipper: removing primers from alignments to minimize false-negative mutations in amplicon next-generation sequencing. Sci. Rep. 2017;7(1):1–7. doi: 10.1038/s41598-017-01703-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dona M.S., et al. Powerful differential expression analysis incorporating network topology for next-generation sequencing data. Bioinformatics. 2017;33(10):1505–1513. doi: 10.1093/bioinformatics/btw833. [DOI] [PubMed] [Google Scholar]
- 17.Clausen P.T., Aarestrup F.M., Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. 2018;19(1):1–8. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cingolani P., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: sNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K., Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26(15):1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minh B.Q., et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilar-Gamboa F.R., et al. Genomic sequences and analysis of five SARS-CoV-2 variants obtained from patients in Lambayeque. Peru. Microbiol. Resour. Announc. 2021;10(1) doi: 10.1128/MRA.01267-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramírez F., et al. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rambaut A., et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Genom. Epidemiol. 2020:1–5. [Google Scholar]
- 24.Gudbjartsson D.F., et al. Spread of SARS-CoV-2 in the Icelandic population. N. Engl. J. Med. 2020;382(24):2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meredith L.W., et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis. 2020;20(11):1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quick J., et al. Multiplex PCR method for MinION and illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12(6):1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richard M., et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. bioRxiv. 2020 doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefanelli P., et al. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020;25(13) doi: 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quick, J., nCoV-2019 sequencing protocol 2020. Publisher Full Text, 2020.
- 30.Batty E.M., et al. Comparing library preparation methods for SARS-CoV-2 multiplex amplicon sequencing on the illumina MiSeq platform. BioRxiv. 2020 [Google Scholar]
- 31.Pillay, S. Illumina nextera DNA flex library construction and sequencing for SARS-CoV-2: adapting COVID-19 ARTIC protocol. 2020 September 30th 2020]; Available from: https://www.protocols.io/view/illumina-nextera-dna-flex-library-construction-and-bhjgj4jw.
- 32.Pillay S., et al. Whole genome sequencing of SARS-CoV-2: adapting illumina protocols for quick and accurate outbreak investigation during a pandemic. Genes (Basel) 2020;11(8) doi: 10.3390/genes11080949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker D.J., et al. CoronaHiT: high-throughput sequencing of SARS-CoV-2 genomes. Genome Med. 2021;13(1):1–11. doi: 10.1186/s13073-021-00839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itokawa K., et al. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS ONE. 2020;15(9) doi: 10.1371/journal.pone.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed N.E., et al. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford nanopore rapid barcoding. Biol. Methods Protoc. 2020;5(1) doi: 10.1093/biomethods/bpaa014. p. bpaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nemudryi A., et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.RIVM. Current information about COVID-19 (novel coronavirus). 2020 October 13th 2020 October 15th 202]; Available from: https://www.rivm.nl/en/novel-coronavirus-covid-19/current-information.
- 38.Gong Y.N., et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes Infect. 2020;9(1):1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C., et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92(6):667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tailored variant analysis pipeline can be found on https://github.com/JordyCoolen/easyseq_covid19 SARS-CoV-2 metadata and GISAID is available in the Supplementary Data.