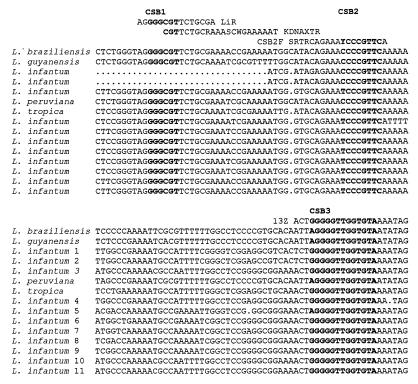
In a recent paper (5), Noyes et al. discussed a nested-PCR-based approach for identification and analysis of Leishmania kinetoplast DNA (kDNA) minicircle classes using the analysis of minicircle variable-region (VR) restriction profiles.
The kDNA of trypanosomatids consists of catenated minicircle (approximately 5 × 103 to 5 × 104 per associate) and maxicircle (20 to 50 per associate) molecules (8). It was shown that the minicircles encode guide RNA (gRNA) molecules involved in the RNA editing of maxicircle cryptogenes (for a review, see reference 9). To date, only this one important cellular function has been demonstrated for minicircles but the enigma of their redundancy remains topical. The prognosticated number of minicircle classes in Leishmania is more than 60 (4). This number comes from the hypothetical number of maxicircle cryptogenes requiring editing of pre-mRNA transcripts. Recently the nonrandom distribution of minicircle classes has been demonstrated for the following set of Leishmania tropica isolates: MHOM/SU/74/SAF/K-27, MHOM/TU/85/TAT3, MHOM/KE/84/NLB299, MRAT/IQ/72/MRCB-JBF, and MHOM/NA/76/ROSSI-II (3). Moreover, it has been proposed that some minicircle classes might be preferentially amplified for expression of required gRNA. The results obtained by Noyes et al. lead to the same conclusion. We think that analysis of the sequence (5) (GenBank accession no. AF032997) (which is in fact a minicircle VR) for the presence of specific gRNA genes (2a) or regulatory sequences (2) may shed more light on the functional role of these molecules.
We have analyzed the presented sequence (5) (GenBank accession no. AF032997) using the Parasite Genome Blast Server (5a) computational tool. The results of our search have revealed a rather high level of homology of the L. infantum MHOM/TN/80/IPT1 minicircle (5) with L. donovani MHOM/IQ/88/RTC6 (clone 11; GenBank accession no. AJ010074), L. donovani MHOM/SD/85/FORSTER (clone 7; GenBank accession no. AJ010080), and L. infantum (clone 10543; GenBank accession no. Z32846) minicircle DNAs (up to 90, 68, and 68%, respectively). We speculate that these minicircles might present one interspecies class for expression of the one certain gRNA type.
All Trypanosomatidae kDNA minicircles are organized by a uniform scheme and contain one or several highly conserved regions (CR). There are three highly conserved blocks (CSB) within CRs: CSB1 (GGGCGT), CSB2 (CCCCGTTC), and CSB3 (GGGGTTGGTGTA) (with interspecies homology of ca. 90 to 100%). CSB1 and CSB3 are almost identical in all species investigated to date, whereas CSB2 is less universal (6, 7). Such an organization model provides us with at least two different strategies for PCR analysis of minicircles with the outward-oriented PCR primers specific to CSB1 and CSB3 regions (for amplification of VRs adjacent to the neighboring CRs) (1) or to the CSB3 region (for amplification of CRs and VRs simultaneously) (3).
Noyes et al. (5) used another two-step-PCR approach. The first step was amplification of the minicircle template using the nested CSB2 primers. The second round was performed by using CSB1-CSB3 primers. We think it would be of interest to discuss the possibility of nonrandom amplification of a single minicircle class during the nested (CSB2) PCR. Our experience shows that using outward-oriented CSB1 and CSB3 primers leads to the detectable amplification of all minicircular templates.
REFERENCES
- 1.de Bruijn M H, Barker D C. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensiscomplex by amplification of kinetoplast DNA. Acta Trop. 1992;52:45–58. doi: 10.1016/0001-706x(92)90006-j. [DOI] [PubMed] [Google Scholar]
- 2.Fu G, Kolesnikov A A. Leishmania gymnodactyli and Leishmania infantum minicircles contain the same guide RNA genes as do minicircles of Leishmania tarentolae. Mol Biochem Parasitol. 1994;67:171–174. doi: 10.1016/0166-6851(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 2a.Guide RNA Database. 19 October 1998, revision date. [Online.] http://www.biochem.mpg.de/∼goeringe/. [6 March 1999, last date accessed.]
- 3.Kolesnikov A A, Yurchenko V Y, Martinkina L P, Schoenian G. A new class of mitochondrial minicircular DNA in Leishmania tropica. Mol Biol (Moscow) 1997;31:458–462. [PubMed] [Google Scholar]
- 4.Maslov D A, Sturm N R, Niner B M, Gruszynski E S, Peris M, Simpson L. An intergenic G-rich region in Leishmania tarentolaekinetoplast maxicircle DNA is a pan-edited cryptogene encoding ribosomal protein S12. Mol Cell Biol. 1992;12:56–57. doi: 10.1128/mcb.12.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noyes H A, Reyburn H, Bailey J W, Smith D. A nested-PCR-based schizodeme method for identifying leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropicain Pakistan. J Clin Microbiol. 1998;36:2877–2881. doi: 10.1128/jcm.36.10.2877-2881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Parasite Genome Blast Server. 3 February 1999, revision date. [Online.] Parasite Genome Blast Server computational tool. http://www.ebi.ac.uk/parasites/parasite_blast_server.html. [4 March 1999, last date accessed.]
- 6.Ray D S. Conserved sequence blocks in kinetoplast minicircles from diverse species of trypanosomes. Mol Cell Biol. 1989;9:1365–1367. doi: 10.1128/mcb.9.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheline C, Ray D S. Specific discontinuities in Leishmania tarentolaeminicircles map within universally conserved sequence blocks. Mol Biochem Parasitol. 1989;37:151–157. doi: 10.1016/0166-6851(89)90147-3. [DOI] [PubMed] [Google Scholar]
- 8.Simpson L. Kinetoplast DNA in trypanosomid flagellates. Int Rev Cytol. 1986;99:119–179. doi: 10.1016/s0074-7696(08)61426-6. [DOI] [PubMed] [Google Scholar]
- 9.Simpson L. The genomic organization of guide RNA genes in kinetoplastid protozoa: several conundrums and their solutions. Mol Biochem Parasitol. 1997;86:133–141. doi: 10.1016/s0166-6851(97)00037-6. [DOI] [PubMed] [Google Scholar]