Abstract
Background
Ferroptosis is the latest-discovered, iron-dependent form of regulated cell death. It has been increasingly recognized that ferroptosis-related genes participate in oncogenesis and development of cancers, including breast cancer (BRCA). Thus, It is important to explore the biofunctions of ferroptosis-related genes in BRCA.
Methods
Transcriptome microarray datasets (GSE22358, GSE9014 and GSE8977, GSE2990 and GSE2034) and TCGA-BRCA were retrieved for analyses. And a variety of computational tools were used to identify the roles and associated biological functions in BRCA.
Results
Two ferroptosis-related genes (AKR1C1 and AKR1C3) were significantly expressed in GSE22358, GSE9014 and GSE8977. Higher expression of AKR1C1 was related with favorable prognosis. TCGA-BRCA further confirmed the expression of AKR1C1 and the prognostic value of AKR1C1. Co-expression analyses showed the most enriched GO term and KEGG pathways were termination of DNA-templated transcription and Fanconi anemia pathway. Subsequently, immunological analyses showed AKR1C1 was significantly associated with various immune infiltration cells; among these, dendritic cells, neutrophils, macrophages were the top three infiltrating cells. Furthermore, AKR1C1 was also associated with multiple immunostimulatory molecules and chemokines, including PD-1, PD-L1, CTLA4, B7-H3, VSIR, IL-6, BTLA, CXCL2, and CCR7. These results indicated the potential roles of AKR1C1 in the immune reaction during the pathogenesis of breast cancer.
Conclusion
This study firstly demonstrated that ferroptosis-related gene, AKR1C1, could be associated with immune microenvironment, thereby influencing the development and prognosis of patient with breast cancer.
Keywords: breast cancer, AKR1C1, ferroptosis, immune microenvironment, immune infiltration
Background
Breast cancer is the most common cancer in women, followed by nonmelanoma skin cancer,1 as well as the worldwide top three common malignancies with lung and colon cancer.2 About 1.7 million people were diagnosed with breast cancer per year with one in three of them dying from it.3 Despite significant developments of endocrine therapy and anti-HER2 targeting therapy, breast cancer remains one of the top three common causes of death in most countries, with raising mortality in some districts. Thus, the discovery of novel therapeutic targets in breast cancer is urgently needed for better patient prognosis.
Ferroptosis is the latest-discovered, iron-dependent form of regulated cell death, which is characterized by accumulating reactive oxygen species and lipid peroxidation products to lethal levels.4 Besides, ferroptosis plays a pivotal role in normal cells and tissues, it has been increasingly recognized that ferroptosis and related genes are also involved in oncogenesis and cancer development, including breast cancer.5,6 Studies have also shown that some aggressive malignancies, such as triple-negative breast cancer, are intrinsically sensitive to ferroptosis due to the nonapoptotic feature,7 which indicates ferroptosis-related therapies as a promising treatment for these malignancies. More recently, ferroptosis and related genes are proven to be a mediator in immune microenvironment and cancer immunotherapy,8 which provides a novel insight of pathophysiological function of ferroptosis. Aldo-keto reductases (AKRs) gene family (AKR1C1-3) has been introduced to potentially help to metabolize lipid peroxides by catalyzing the conversion of aldehydes and ketones to alcohols, therefore, inhibiting the ferroptosis.9 Meanwhile, two studies validated the role of AKR1C1 in ferroptosis in lung cancer and melanoma.10,11 However, no study reported the role of AKR1C1 in breast cancer with detailed mechanisms.
In order to reveal the importance of these ferroptosis-related genes in breast cancer, this study was conducted comprehensively, analyzing the clinical and immunological role of ferroptosis-related genes in breast cancer, which could provide a better understanding of the complex functions and mechanisms of ferroptosis-related genes in breast cancer.
Methods
Data Acquisition
Datasets of breast cancer were searched in the GEO database, which included all the ferroptosis-related genes and adequate clinical information. Three transcriptome microarray datasets (GSE22358, GSE9014, and GSE8977) were retrieved from gene expression omnibus (GEO) database for identification of ferroptosis-related genes. Meanwhile, a Venn diagram was used for illustrating co-differentially expressed genes (co-DEGs). Ferroptosis-related genes were retrieved from the reports from Liang et al’s group.12 Further validations were realized using transcriptome and clinical data of BRCA patients from TCGA database (http://cancergenome.nih.gov/), including 1097 tumor tissues and 114 normal tissues. RNA-seq data were downloaded with the TPM format and received log2 conversions. The expression of genes were compared using one-way ANOVA or Mann–Whitney U-test. GEPIA (http://gepia.cancer-pku.cn/),13 UALCAN (http://ualcan.path.uab.edu/analysis.html),14 and TNMplot (https://www.tnmplot.com/) were also used to access data from TCGA database.15
Correlation Analysis
Correlation analysis was realized by The LinkedOmics database (http://www.linkedomics.org/login.php).16 The LinkFinder module was used for screening the co-expressed genes with Pearson's correlation test, and the results were shown as volcano plot and heatmap. Subsequently, gene ontology biological process (GO_BP), gene ontology cellular component (GO_CC), gene ontology molecular function (GO_MF), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed by the gene set enrichment analysis (GSEA) in the LinkInterpreter module.
Immunological Analysis
Tumor IMmune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/)17 and the TISIDB database (http://cis.hku.hk/TISIDB)18 were used for validation of the abundance of tumor-infiltrating immune cells. In the meantime, the estimation of stromal and immune cells in malignant tumor tissues using expression data (ESTIMATE) algorithm was applied to generate immune score, representing the infiltration degree of immune cells in tumor tissue.
Statistical Analyses
R 3.3.0 and Statistical Package for Social Sciences 23.0 for Windows (BM Corporation, Armonk, NY, USA) were used to perform statistical analyses. One-way analysis of variance (ANOVA) was used, with homogeneous variance, to analyze differences in immune cell components between normal and breast cancer tissues, while Welch’s ANOVA was applied when variance was heterogeneous. Kaplan–Meier curve was used for survival analysis from Kaplan–Meier Plotter (http://kmplot.com/analysis/),19 where the log rank test was for comparisons between groups. The receiver operating characteristic (ROC) curve was used to investigate the value of genes in predicting patient survival. The area under the curve (AUC) was used for quantifying the prediction value. Meantime, univariate and multivariate Cox regression analyses were used to confirm the associations between patient characteristics and patient prognosis. Factors with P<0.05 were included in the multivariate Cox regression with the LR method. The main tools involved in this study were summarized in Table 1.
Table 1.
The Main Tools Involved in This Study
| Database | Samples | URL | References |
|---|---|---|---|
| GEPIA | Tissues | http://gepia.cancer-pku.cn/ | 13 |
| UALCAN | Tissues | http://ualcan.path.uab.edu/analysis.html | 14 |
| TNMplot | Tissues | https://www.tnmplot.com/ | 15 |
| LinkedOmics | Tissues | http://www.linkedomics.org/login.php | 16 |
| TIMER | Tissues | https://cistrome.shinyapps.io/timer/ | 17 |
| TISIDB | Tissues | http://cis.hku.hk/TISIDB | 18 |
| Kaplan–Meier Plotter | Tissues | http://kmplot.com/analysis | 19 |
Results
Identification of Candidate Ferroptosis-Related Genes
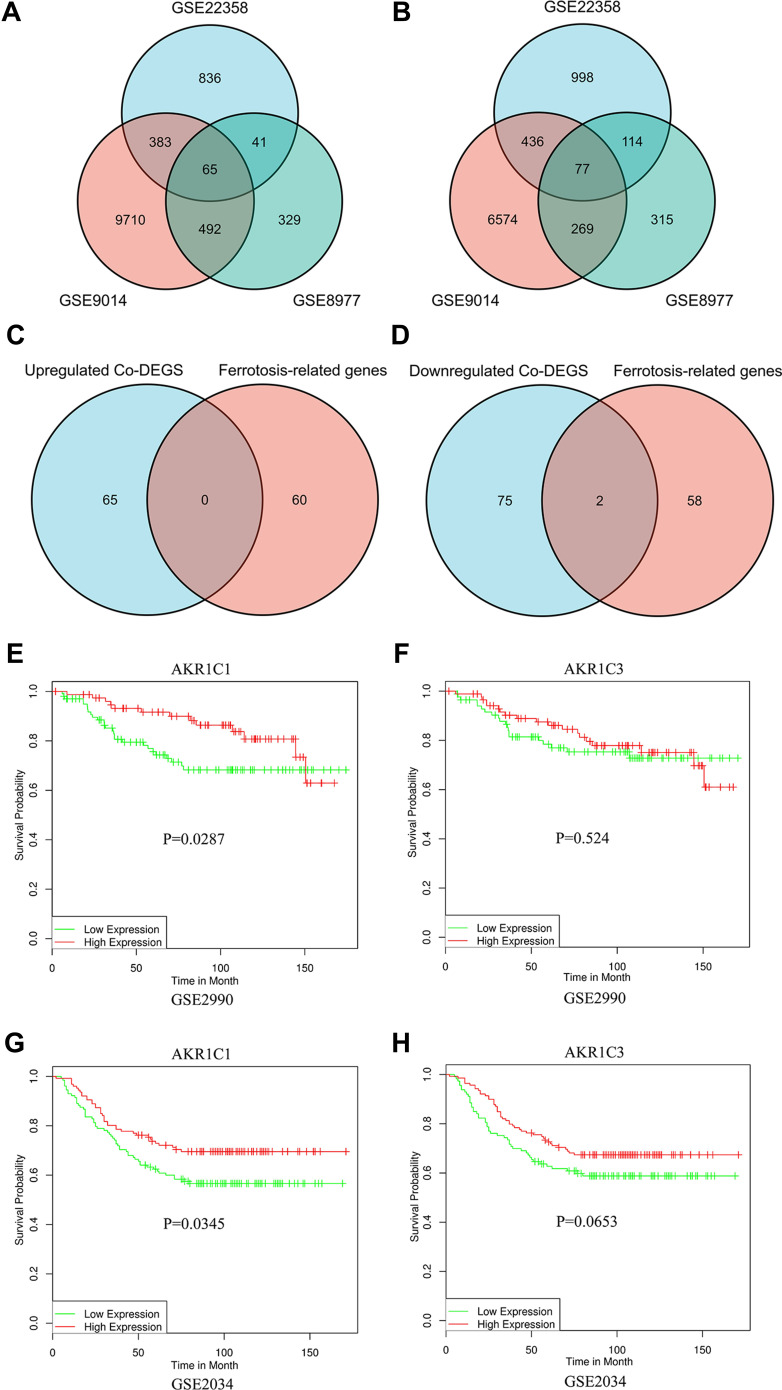
The data of GSE22358, GSE9014, and GSE8977 from GEO database were obtained to identify differentially expressed genes in breast cancer. One thousand, three hundred and twenty-five upregulated DEGS and 1625 downregulated DEGS were found in GSE22358, while 10,650 and 7356 in GSE9014, and 927 and 775 in GSE8977. As shown in Figure 1A and B, Venn plot showed 65 upregulated co-DEGS and 77 downregulated co-DEGS. Venn plots (Figure 1C and D) between co-DEGS and ferroptosis-related genes showed that two ferroptosis-related genes (AKR1C1 and AKR1C3) were downregulated. The prognostic value of AKR1C1 and AKR1C3 was subsequently investigated using overall survival (OS) data of breast cancer patients from GSE2990, GSE2034 (Figure 1E–H). Although both the higher AKR1C1 and AKR1C3 showed a better OS survival curve in breast cancer, the differences of AKR1C3 in both GSE2990 and GSE2034 were not significant. Therefore, further investigations were focused on AKR1C1.
Figure 1.
Identification of candidate ferroptosis-related genes. (A and B) Venn plots of to identify upregulated (A) and downregulated (B) co-DEGs. (C and D) Venn plots of to identify ferroptosis-related co-DEGs. (E–H). Kaplan–Meier curves of AKR1C1 and AKR1C3 on overall survival of GSE2990 and GSE2034.
Validation of the Expression Level and Clinical Significance of AKR1C1
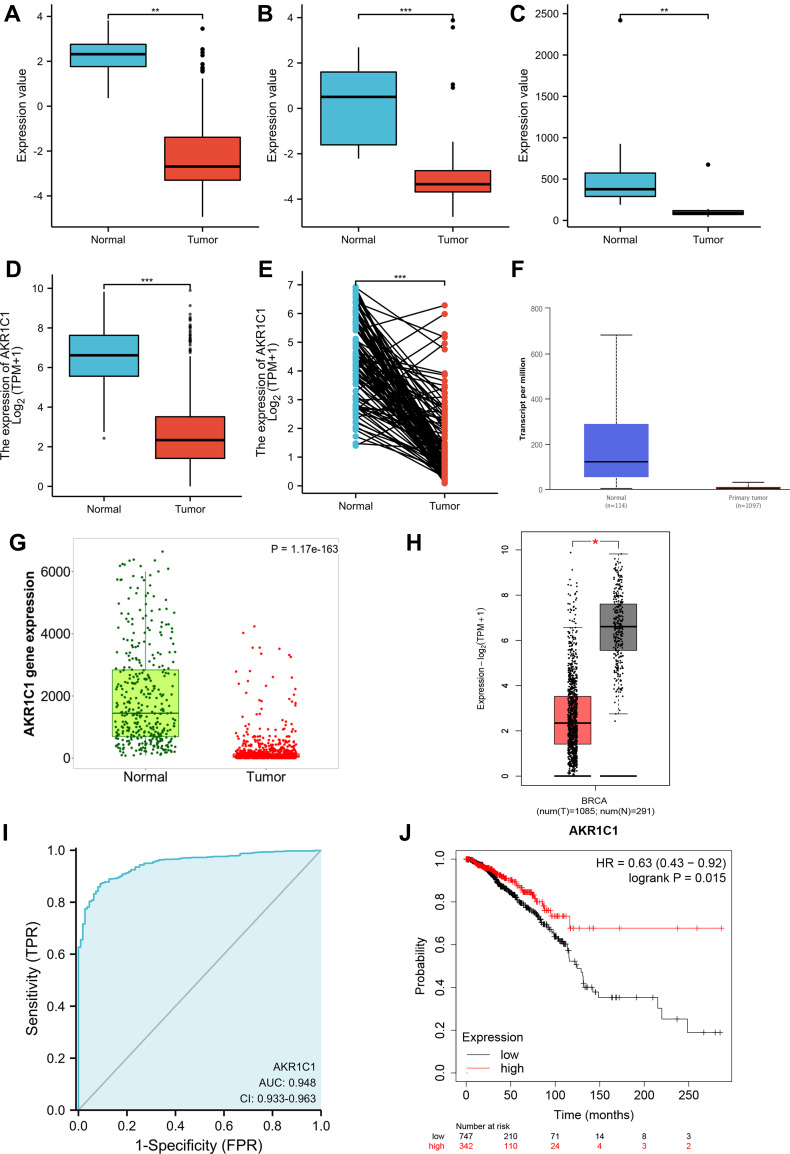
The expression level of AKR1C1 was significantly lower in breast cancer tissues compared with normal control in GSE22358 (Figure 2A), GSE9014 (Figure 2B), and GSE8977 (Figure 2C). To validate the significance and explore further information of AKR1C1, data from the BRCA cohort of TCGA database and normal breast tissues from GTEx were also used. The results showed a consistent decrease of AKR1C1 expression level in breast cancer, regardless of unpaired tissues (Figure 2D) or paired tissues (Figure 2E). In addition, several databases, such as UALCAN (Figure 2F), TNMplot (Figure 2G), and GEPIA (Figure 2H), were enrolled to validate the downregulated levels of AKR1C1 in BRCA patients. Meanwhile, the diagnostic and prognostic significance were also investigated. The baseline data of patient characteristics were shown in Table 2. Patients were divided into two groups according to the expression of AKR1C1. The median age of both low expression group and high expression group was 58 years old (P=0.778). There was no significant difference in TNM stage between the two groups. One hundred and thirty (24.6%) and 130 (24.4%) patients suffered from stage III or stage IV in the low expression group and high expression group (P=0.732). However, the low expression group had a higher positive rate of PR, ER, and HER2 compared with high expression group (P<0.001). The ROC curve showed a strong diagnostic value of AKR1C1 with AUC of 0.948 (95%CI: 0.933–0.963) (Figure 2I), while the survival curve of OS from Kaplan–Meier Plotter confirmed the high level AKR1C1 representing good patient’s prognosis (Figure 2J). Univariate and multivariate Cox regression analyses still showed independent prognostic value of AKR1C1 (Table 3).
Figure 2.
Validation of the expression level and clinical significance of AKR1C1. Expression comparisons of AKR1C1 between breast cancer and normal control in GSE22358 (A), GSE9014 (B), GSE8977 (C), unpaired TCGA-BRCA (D), and paired TCGA-BRCA (E). Expression of AKR1C1 in TCGA-BRCA validated by UALCAN (F), TNM plot (G), and GEPIA (H). The ROC curve of AKR1C1 on diagnosing breast cancer (I). The Kaplan–Meier curve of AKR1C1 on overall survival of TCGA-BRCA (J). *P<0.05; **P<0.01;***P<0.001.
Table 2.
The Characteristics of BRCA Patients from TCGA Database
| Characteristic | Low Expression of AKR1C1 (n=541) | High Expression of AKR1C1 (n=542) | P-value |
|---|---|---|---|
| Age, median (IQR) | 58 (48, 68) | 58 (49, 66) | 0.778 |
| Age, n (%) | 1.000 | ||
| ≤60 | 300 (55.5%) | 301 (55.5%) | |
| >60 | 241 (44.5%) | 241 (44.5%) | |
| T stage, n (%) | 0.083 | ||
| T1 | 129 (23.9%) | 148 (27.4%) | |
| T2 | 327 (60.7%) | 302 (55.8%) | |
| T3 | 61 (11.3%) | 78 (14.4%) | |
| T4 | 22 (4.1%) | 13 (2.4%) | |
| N stage, n (%) | 0.530 | ||
| N0 | 247 (46.9%) | 267 (49.7%) | |
| N1 | 186 (35.3%) | 172 (32%) | |
| N2 | 60 (11.4%) | 56 (10.4%) | |
| N3 | 34 (6.5%) | 42 (7.8%) | |
| M stage, n (%) | 0.914 | ||
| M0 | 462 (97.7%) | 440 (98%) | |
| M1 | 11 (2.3%) | 9 (2%) | |
| Pathologic stage, n (%) | 0.732 | ||
| Stage I | 84 (15.9%) | 97 (18.2%) | |
| Stage II | 314 (59.5%) | 305 (57.3%) | |
| Stage III | 120 (22.7%) | 122 (22.9%) | |
| Stage IV | 10 (1.9%) | 8 (1.5%) | |
| Histological type, n (%) | < 0.001 | ||
| Infiltrating ductal carcinoma | 413 (86.8%) | 359 (71.7%) | |
| Infiltrating lobular carcinoma | 63 (13.2%) | 142 (28.3%) | |
| PR status, n (%) | < 0.001 | ||
| Negative | 139 (27.3%) | 203 (38.7%) | |
| Indeterminate | 2 (0.4%) | 2 (0.4%) | |
| Positive | 368 (72.3%) | 320 (61%) | |
| ER status, n (%) | < 0.001 | ||
| Negative | 84 (16.5%) | 156 (29.7%) | |
| Indeterminate | 2 (0.4%) | 0 (0%) | |
| Positive | 424 (83.1%) | 369 (70.3%) | |
| HER2 status, n (%) | 0.012 | ||
| Negative | 253 (72.7%) | 305 (80.5%) | |
| Indeterminate | 4 (1.1%) | 8 (2.1%) | |
| Positive | 91 (26.1%) | 66 (17.4%) | |
| Anatomic neoplasm subdivisions, n (%) | 0.171 | ||
| Left | 293 (54.2%) | 270 (49.8%) | |
| Right | 248 (45.8%) | 272 (50.2%) | |
| Radiation therapy, n (%) | 0.184 | ||
| No | 225 (46.2%) | 209 (41.8%) | |
| Yes | 262 (53.8%) | 291 (58.2%) |
Table 3.
Univariate and Multivariate Analyses of Risk Factors with and OS in BRCA Patients
| Variables | HR (95%CI) | P-value |
|---|---|---|
| Univariate analyses | ||
| Age (years) (>60 vs ≤60) | 2.015 (1.460–2.781) | <0.001 |
| Gender (female vs male) | 1.228 (0.171–8.794) | 0.838 |
| T stage (T3 and T4 vs T1 and T2) | 1.760 (1.223–2.535) | 0.002 |
| N stage (N2 and N3 vs N0 and N1) | 2.299 (1.571–3.359) | <0.001 |
| M stage (M1 vs M0) | 4.859 (2.900–8.139) | <0.001 |
| AJCC tumor stage (III and IV vs I and II) | 1.624 (1.373–1.921) | <0.001 |
| AKR1C1 (high group vs low group) | 0.640 (0.440–0.931) | 0.020 |
| Multivariate analyses | ||
| Age (years) (>60 vs ≤60) | 2.276 (1.613–3.212) | <0.001 |
| T stage (T3 and T4 vs T1 and T2) | 0.985 (0.581–1.668) | 0.954 |
| N stage (N2 and N3 vs N0 and N1) | 1.075 (0.591–1.955) | 0.812 |
| AJCC tumor stage (III and IV vs I and II) | 1.654 (1.195–2.290) | 0.002 |
| AKR1C1 (high group vs low group) | 0.652 (0.440–0.964) | 0.032 |
Abbreviations: HR, hazard ratio; CI, confidence interval; OS, overall survival; AJCC, American Joint Committee on Cancer.
AKR1C1 Co-expression Analyses
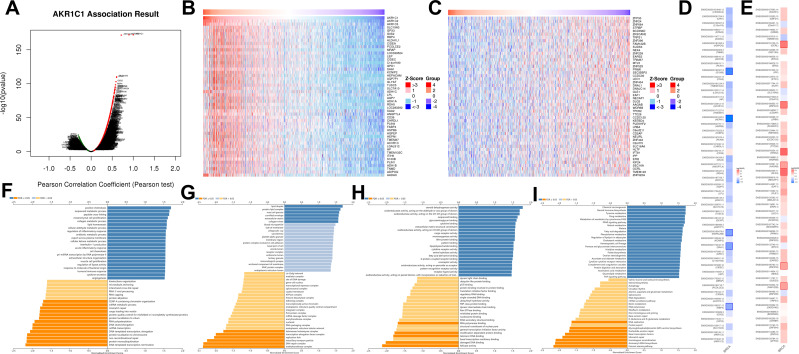
To explore the knowledge of biological function of AKR1C1 in breast cancer, the LinkedOmics tool was used to obtain the co-expression pattern of AKR1C1 in TCGA-BRCA. 7441 genes showed a positively correlation with AKR1C1, while 4424 genes a negatively correlation (Figure 3A). Figure 3B and C showed the top 50 genes positively and negatively co-expressed with AKR1C1, respectively. The prognostic values of these genes were subsequently investigated (Figure 3D and E). Most positively co-expressed genes (38 out of 50) had a protective hazard ratio, and most of the negatively co-expressed genes (39 of 50) had an unfavorable hazard ratio, which is consistent with the prognostic role of AKR1C1 in breast cancer.
Figure 3.
The co-expression genes with AKR1C1 from the LinkedOmics database. (A) Genes significantly associated with AKR1C1 by Pearson test. (B and C) Heat maps of top 50 genes positively and negatively related to AKR1C1A. Red represents positively linked genes and blue represents negatively linked genes. (D and E) Survival map of the top 50 genes positively (D) and negatively (E) associated with AKR1C1A in TCGA-BRCA. (F–I) GO terms for biological process, cellular component, molecular functions, and KEGG pathways of AKR1C1A by GSEA analyses.
Moreover, GSEA analysis was performed to explore the potential biological functions of AKR1C1. The enrichment results of GO terms and KEGG pathway were shown in Figure 3F–I. Termination of DNA-templated transcription, methyltransferase complex and helicase activity were the top enriched terms in biological process, cellular component, and molecular functions. Fanconi anemia pathway, aminoacyl-tRNA biosynthesis and homologous recombination were the top three enriched KEGG pathways.
The Roles of AKR1C1 in the Immune Microenvironment
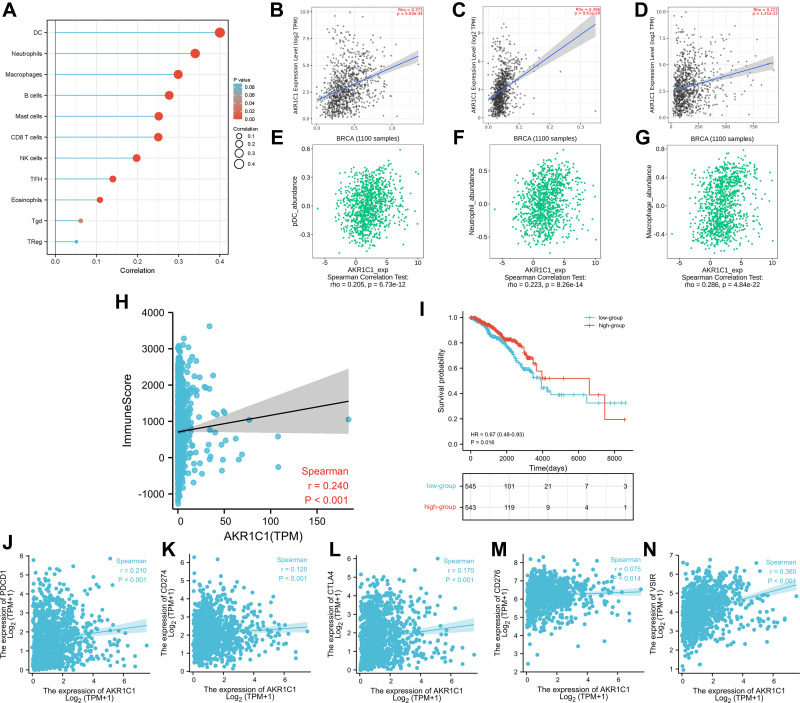
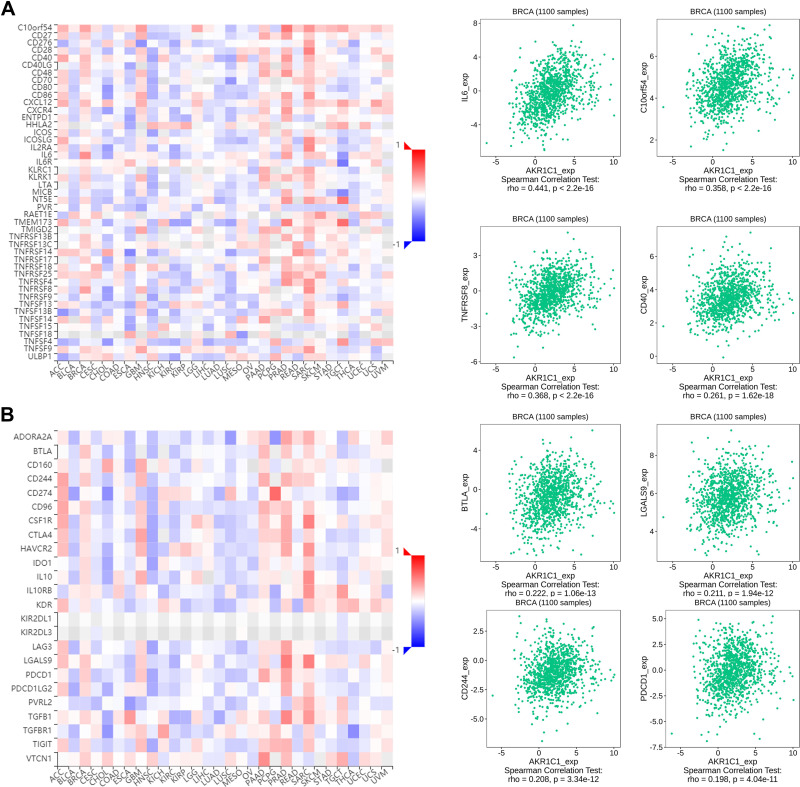
Due to the revealed association between ferroptosis and immune microenvironment,20 we subsequently attempted to explore the role of AKR1C1 in the immune microenvironment of breast cancer. We further investigated associations between AKR1C1 with immune infiltration cells using ssGSEA algorithm. Dendritic cells (DC), neutrophils, macrophages, B cells, mast cells, CD8 T cells, NK cells, TFH, and eosinophils were found positively correlated with AKR1C1 in breast cancer (Figure 4A). The top three cell type (DC, neutrophils, macrophages) were subsequently validated using the TIMER database and the TISIDB database, and the results were consistent (Figure 4B–G). Moreover, the ESTIMATE algorithm showed a positive correlation between AKR1C1 and the infiltration immune score in breast cancer (Figure 4H). We further investigated the prognostic value of the immune score, and the results showed patients with higher immune score had a better OS (Figure 4I), indicating AKR1C1 may improve the patient prognosis by influencing immune microenvironment in breast cancer. In addition, we also observed the positive associations between AKR1C1 levels and multiple immune checkpoint molecules, including PD-1 (Figure 4J), PD-L1 (Figure 4K), CTLA4 (Figure 4L), B7-H3 (Figure 4M), and VSIR (Figure 4N).
Figure 4.
The associations between immune infiltration with AKR1C1. (A) Immune infiltration cells associated with AKR1C1 using ssGESA algorithm. (B–D) Validations of top three cell (DC, neutrophils, macrophages) by the TIMER database. (E–G) Validations of top three cell (DC, neutrophils, macrophages) by the TISIDB database. (H) The association between AKR1C1 with immune score calculated by ESTIMATE algorithm. (I) The Kaplan–Meier curve of immune score on overall survival of TCGA-BRCA. (J–N) The association between AKR1C1 with PD-1, PD-L1, CTLA4, B7-H3, and VSIR.
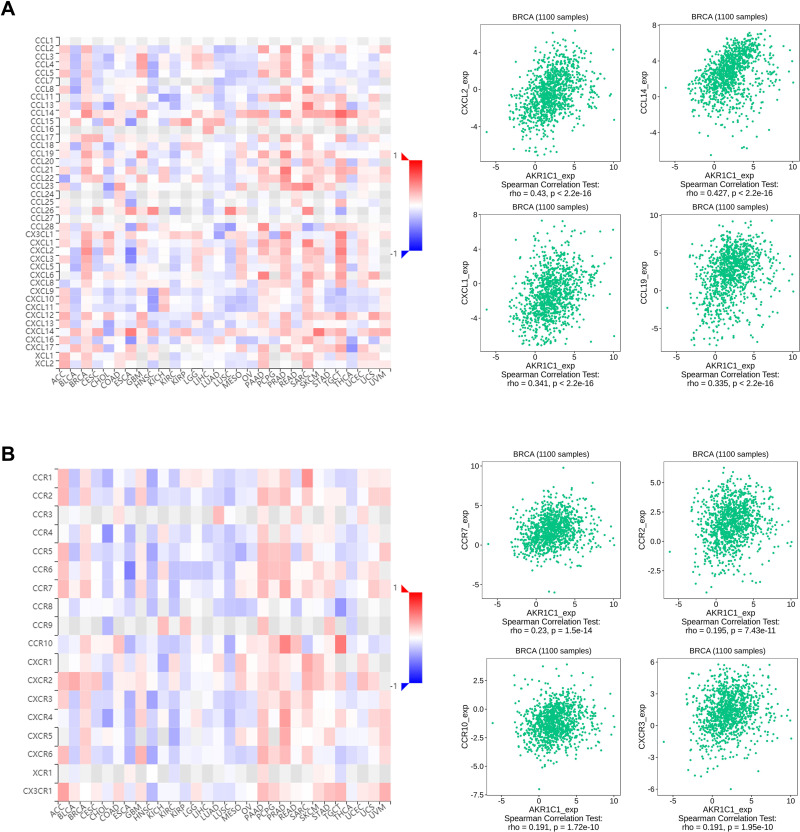
Further investigations were focused on broadening the recognition of immune role of AKR1C1 other than immune infiltration cells. TISIDB database was used to analyze the associations between AKR1C1 with immunostimulators, immunoinhibitors, chemokines, and receptors. As shown in Figure 5, multiple immunostimulators and immunoinhibitors showed associations with expression of AKR1C1, with IL-6 as the most significant immunostimulatory marker and BTLA as the most significant immunoinhibitory marker. Meanwhile, Figure 6 showed that CXCL2 was the most significant chemokine related to AKR1C1, and CCR7 was the most significant chemokine receptor. These results together suggested that AKR1C1 might play promising roles in the prognosis of patients with breast cancer through regulating the immune response.
Figure 5.
Associations of the AKR1C1 expression with immunomodulators from the TISIDB database. (A) Correlations between immunostimulators and AKR1C1 with the four most significant molecules. (B) Correlations between immunoinhibitors and AKR1C1 with the four most significant molecules.
Figure 6.
Associations of the AKR1C1 expression with chemokines and receptors from the TISIDB database. (A) Correlations between chemokines and AKR1C1 with the four most significant molecules. (B) Correlations between receptors and AKR1C1 with the four most significant molecules.
Discussion
In this study, we found a ferroptosis-related gene, AKR1C1, plays a significant role in the oncogenesis and development of breast cancer, functioning as a potential tumor suppressor. Further investigations showed that AKR1C1 was significantly associated with the immune microenvironment of breast cancer, indicating that AKR1C1 might make an impact on patient prognosis through altering immune microenvironment in breast cancer. These results suggested that AKR1C1 had potential value for use as a treatment target or a biomarker of patient prognosis in breast cancer.
There are several studies introducing the significance of ferroptosis and regulators in breast cancer. One of the features in ferroptosis is the inactivation of GPX4, which has also been proven in breast cancer.21 Surprisingly, the drug-tolerant breast cancer cells seem to be more vulnerable to inducing ferroptosis by GPX4 inhibition.22 Alternatively, lipid reactive oxygen species (ROS) is the main killer in ferroptosis. Among the regulators involved in lipid metabolism, ACSL4 is specifically associated with ferroptosis.23 Particularly, ACSL4 was reported to be a promoter of ferroptosis by enriching cellular membranes with long polyunsaturated n-6 fatty acids in breast cancer.24 In this study, we found the expression of AKR1C1 was downregulated in tumor tissues, which might indicate the treatment of inducing ferroptosis may be potential effective in breast cancer.
As an important part of tumor microenvironment (TME), immune microenvironment composed of various immune cells is widely recognized as a biomarker of tumor progression and predictors of clinical responses to therapeutic regimens, especially immunotherapy.25 DC cells are essential for antitumor immune responses due to their functions of cross-presenting tumor antigen to lymphocytes. In breast cancer, a high proportion of DC was reported in less aggressive tumors, which were explained by getting involved in T-cell dependent immune clearance, therefore influencing patient prognosis.26 TME neutrophils (TANs) may have antitumor functions and pro-tumorigenic functions at the same time.27 TANs can function against tumor cells by promoting inflammatory, while it also possesses the anti-inflammatory and immunosuppressive abilities, which may be regulated by TME.28 Similarly, tumor-associated macrophage (TAM) also displays multi-aspect functions ranging from antitumor responses and pro-tumorigenic responses.29 Only one previous study by Wenners et al referred the association between AKR1C1 expression and overall survival, however, it failed to investigate the further underlying mechanisms.30 In this study, we found that AKR1C1 was associated with the immune microenvironment of breast cancer, thus, it may make an impact on patient prognosis and tumor development. As far we know, this was the first study reporting the association between AKR1C1 and the immune microenvironment of breast cancer, suggesting that AKR1C1 could be developed as a therapeutic target or an indicator of immunotherapy.
There were several limitations in this study. This work revealed the significance of AKR1C1 in breast cancer and the association between AKR1C1 and the immune microenvironment by bioinformatic analyses. We strongly recommend further experimental and clinical research to comprehensively validate the biological role of AKR1C1 in influencing the immune microenvironment and patient prognosis of breast cancer.
Conclusion
In summary, this is the first study reporting a ferroptosis-related gene, AKR1C1, which may be associated with the immune microenvironment and therefore influence development of tumor and patient prognosis in breast cancer.
Funding Statement
This study is supported by grants from the China Postdoctoral Science Foundation (2021T140754, 2020M672521), the Natural Science Foundation of Hunan Province (2019JJ50932, 2020JJ5934), and the Postdoctoral Science Foundation of Central South University (248485).
Abbreviations
HCC, hepatocellular carcinoma; GEO, gene expression omnibus; co-DEGs, co-differentially expressed genes; GO_BP, gene ontology biological process; GO_CC, gene ontology cellular component; GO_MF, gene ontology molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; TIMER, Tumor IMmune Estimation Resource; ANOVA, one-way analysis of variance; DC, dendritic cells; ROS, reactive oxygen species; AKRs, aldo-keto reductases; TME, tumor microenvironment; TAN, TME neutrophils; TAM, tumor-associated macrophage.
Guarantor of the Article
The corresponding authors have control of the decision to publish and accept full responsibility for the conduct of the study. The data is available from the corresponding author Fada Xia.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
Extra informed consent is not essential as the data were all obtained from a public database. The authors did not have access to any identifying characteristics, which do not distort scientific meaning.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. doi: 10.1001/jama.2018.19323 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 4.Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19(7):405–414. doi: 10.1038/s41568-019-0149-1 [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Chen L, Chen C, et al. Targeting ferroptosis in breast cancer. Biomarker Res. 2020;8(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou Y, Palte MJ, Deik AA, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10(1):1617. doi: 10.1038/s41467-019-09277-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Green M, Choi JE, et al. CD8 T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wohlhieter CA, Richards AL, Uddin F, et al. Concurrent mutations in STK11 and KEAP1 promote ferroptosis protection and SCD1 dependence in lung cancer. Cell Rep. 2020;33(9):108444. doi: 10.1016/j.celrep.2020.108444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagliardi M, Cotella D, Santoro C, et al. Aldo-keto reductases protect metastatic melanoma from ER stress-independent ferroptosis. Cell Death Dis. 2019;10(12):902. doi: 10.1038/s41419-019-2143-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang J, Wang D, Lin H, et al. A novel ferroptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Int J Biol Sci. 2020;16(13):2430–2441. doi: 10.7150/ijbs.45050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49(W1):W242–W246. doi: 10.1093/nar/gkab418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartha Á, Győrffy B. TNMplot.com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int J Mol Sci. 2021;22(5):2622. doi: 10.3390/ijms22052622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi: 10.1093/nar/gkx1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210 [DOI] [PubMed] [Google Scholar]
- 19.Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11(1):6047. doi: 10.1038/s41598-021-84787-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi: 10.1038/cdd.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, Yang C, Jian L, et al. Sulfasalazine‑induced ferroptosis in breast cancer cells is reduced by the inhibitory effect of estrogen receptor on the transferrin receptor. Oncol Rep. 2019;42(2):826–838. [DOI] [PubMed] [Google Scholar]
- 22.Hangauer MJ, Viswanathan VS, Ryan MJ, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551(7679):247–250. doi: 10.1038/nature24297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478(3):1338–1343. doi: 10.1016/j.bbrc.2016.08.124 [DOI] [PubMed] [Google Scholar]
- 24.Doll S, Proneth B, Tyurina YY, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13(1):91–98. doi: 10.1038/nchembio.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019;20(4):840. doi: 10.3390/ijms20040840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broz ML, Binnewies M, Boldajipour B, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26(5):638–652. doi: 10.1016/j.ccell.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sionov RV, Fridlender ZG, Granot Z. The multifaceted roles neutrophils play in the tumor microenvironment. Cancer Microenviron. 2015;8(3):125–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vols S, Sionov RV, Granot Z. Always look on the bright side: anti-tumor functions of neutrophils. Curr Pharm Design. 2017;23(32):4862–4892. doi: 10.2174/1381612823666170704125420 [DOI] [PubMed] [Google Scholar]
- 29.Allen M, Louise Jones J. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. J Pathol. 2011;223(2):162–176. doi: 10.1002/path.2803 [DOI] [PubMed] [Google Scholar]
- 30.Wenners A, Hartmann F, Jochens A, et al. Stromal markers AKR1C1 and AKR1C2 are prognostic factors in primary human breast cancer. Int J Clin Oncol. 2016;21(3):548–556. doi: 10.1007/s10147-015-0924-2 [DOI] [PubMed] [Google Scholar]