Abstract
The DNA polymerase α-primase complex is the only enzyme that provides RNA-DNA primers for chromosomal DNA replication in eukaryotes. Mouse DNA polymerase α has been shown to consist of four subunits, p180, p68, p54, and p46. To characterize the domain structures and subunit requirements for the assembly of the complex, we constructed eukaryotic polycistronic cDNA expression plasmids expressing pairwise the four subunits of DNA polymerase α. In addition, the constructs contained an internal ribosome entry site derived from poliovirus. The constructs were transfected in different combinations with vectors expressing single subunits to allow the simultaneous expression of three or four of the subunits in cultured mammalian cells. We demonstrate that the carboxyl-terminal region of p180 (residues 1235 to 1465) is essential for its interaction with both p68 and p54-p46 by immunohistochemical analysis and coprecipitation studies with antibodies. Mutations in the putative zinc fingers present in the carboxyl terminus of p180 abolished the interaction with p68 completely, although the mutants were still capable of interacting with p54-p46. Furthermore, the amino-terminal region (residues 1 to 329) and the carboxyl-terminal region (residues 1280 to 1465) were revealed to be dispensable for DNA polymerase activity. Thus, we can divide the p180 subunit into three domains. The first is the amino-terminal domain (residues 1 to 329), which is dispensable for both polymerase activity and subunit assembly. The second is the minimal core domain (residues 330 to 1279), required for polymerase activity. The third is the carboxyl-terminal domain (residues 1280 to 1465), which is dispensable for polymerase activity but required for the interaction with the other three subunits. Taken together, these results allow us to propose the first structural model for the DNA polymerase α-primase complex in terms of subunit assembly, domain structure, and stepwise formation at the cellular level.
In mammalian cells, six distinct DNA polymerases, α, β, γ, δ, ɛ, and ζ, have been cloned so far (3, 13, 42). Among these, DNA polymerases α, δ, and ɛ are considered to be involved in chromosomal DNA replication. DNA polymerase α is the only enzyme that is tightly coupled to DNA primase. Therefore, DNA polymerase α has been considered to provide RNA-DNA primers for the initiation of leading-strand synthesis as well as Okazaki fragment synthesis on the lagging strand (12, 34, 42). By use of the simian virus 40 (SV40) in vitro DNA replication system, it was shown that DNA polymerase α plays a role in the initiation of DNA synthesis by providing RNA-DNA primers for both leading-strand synthesis and lagging-strand synthesis and that DNA polymerase δ extensively elongates these primers through a polymerase switch mechanism (40). However, even though the precise roles of DNA polymerases α and δ have been established for the SV40 DNA replication system, the way in which these enzymes function during replication of the chromosome is still not clear. Namely, we are ignorant about the architecture of the subunit assemblies in the replication complexes, the way in which the activities of these complexes are regulated, the coordination that must exist between these polymerases at the replication fork, and which DNA polymerase, δ or ɛ, participates in the elongation of the leading strand and lagging strand (3, 4, 34).
DNA polymerases α, δ, ɛ, and ζ contain amino acid sequences that are conserved among a wide range of DNA polymerases, indicating that these polymerases belong to the class B DNA polymerase family (32, 42, 44). During this decade, molecular cloning analysis has shown that the large subunits of all these DNA polymerases comprise the catalytic activity, whereas the functions of the smaller subunits, with the exception of the primase subunit, still remain uncertain (12, 34, 42). However, the second-largest subunits of DNA polymerases α, δ, and ɛ display significant homology, suggesting that these subunits may have pivotal functions that were conserved during evolution (2, 20). Characterization of the domain structures and subunit requirements for complex assembly should help us to determine the common properties and distinctive features of members of the class B DNA polymerase family.
To understand the molecular mechanism of eukaryotic DNA replication, we focused our attention on the DNA polymerase α-primase complex. Mouse DNA polymerase α is made up of four subunits (22, 36, 37). The largest subunit, p180, and the smallest subunit, p46, comprise the DNA polymerase and DNA primase activities, respectively (8, 9, 29). The other subunits, p68 and p54, have no known enzymatic activity. Recently, it was suggested that the replication activity of the DNA polymerase α-primase in human cells was regulated by cyclin-dependent kinase phosphorylation of p68, although the regulatory mechanism was not elucidated (39). To identify the precise functions of these subunits in cells, we exploited a cDNA expression system using mammalian cultured cells and found that p68 facilitates not only p180 protein synthesis through cotranslational interaction but also translocation of p180 into the nucleus as a p180-p68 heterodimer (23). Moreover, we found that p54 can carry p46 into the nucleus through the so-called piggyback binding transport mechanism (24). Thus, using the cDNA expression system in mammalian cultured cells, we showed that interactions involving specific combinations of p46 and p54 and of p68 and p180 are essential for the nuclear translocation of DNA polymerase α. However, study of the interactions among three or all of the subunits was hampered by the difficulty in obtaining continuous expression of more than two subunits in mammalian cells. Moreover, reconstitution of the tetrameric complex from individually purified subunits has not been possible to date. To characterize the subunit-subunit interactions more directly, we designed a cDNA expression system with eukaryotic polycistronic plasmids containing an internal ribosomal entry site (IRES) from poliovirus and examined the interactions between these subunits in vivo. Using triple and quadruple transfections, we were able to determine the subcellular distribution of the four subunits in detail, investigate the interactions among the four subunits, and resolve the domain organization of the p180 molecule. Taken together, these results have allowed us to postulate a structural model for the DNA polymerase α-primase complex, which succeeds in explaining its subunit assembly, domain structure, and stepwise formation at the cellular level.
MATERIALS AND METHODS
Materials.
All restriction enzymes and Klenow fragment were purchased from Takara (Ohtsu, Japan); phenylmethylsulfonyl fluoride was from Sigma; Expand high-fidelity DNA polymerase and 12CA5 anti-hemagglutinin (HA) antibody were from Boehringer Mannheim; anti-T7 tag antibody was from Novagen; horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse immunoglobulin G (IgG) antibodies were from MBL (Nagoya, Japan); fluorescein isothiocyanate (FITC)- or Texas red-conjugated goat anti-rabbit or anti-mouse IgG antibodies were from Vector Inc.; fetal bovine serum was from Intergen; calf serum was from HyClone; anti-six-His monoclonal antibody and cobalt-chelating Sepharose (TALON) were from Clontech. The expression vectors pcDEBΔ, pSRHisABC, and IRES were gifts from Y. Nakabeppu (26), S. Ohno (1), and A. Nomoto (30), respectively. DNA polymerase α-specific hybridoma SJK132-20 was purchased from the American Type Culture Collection (6). NIH 3T3 cells were from T. Akiyama. Unless otherwise stated, all other chemicals and reagents were obtained from Wako Chemicals (Osaka, Japan).
Construction of the expression vector.
cDNAs for the four subunits of mouse DNA polymerase α-primase complex were introduced into pcDEBΔ, which contains the SRα promoter (38), to generate plasmids pSRα46, pSRα54, pSRα68, and pSRα180 as described previously (16, 24).
IRES-derived polycistronic expression plasmids were constructed with poliovirus IRES (30). Restriction enzyme sites were introduced by PCR with two primers: 5′-CCGAGCTCTAGAGGCCCACGTGGCGGCTA-3′ and 5′-GGAGCGCTAGCAAACAGATAGATAATGA-3′. The 650-bp PCR product was digested with XbaI and NheI and subcloned into XbaI-digested pSRα54 (pSRαIRES-54). The 1.6-kb BglII-EcoRV-digested pSRα46 was filled in with Klenow enzyme. Then, pSRαIRES-54 was digested with XbaI, filled in with Klenow enzyme, and ligated with the blunt-ended fragment containing the cDNA for p46. The resultant plasmid containing the cDNAs of both p54 and p46 was designated pI-pri. To coexpress HA-tagged p46 with p54, an HA tag was introduced into the amino terminus of p46 by PCR with the primers 5′-GAGA TCTAGATGTACCCATACGACGTTCCTGACTACGCGGAGCCATTTGA TCCTGCGGA-3′ and 5′-GCTGTTTTCTCTCGAGATCTTTTTG-3′. The PCR products were digested with XbaI and HindIII and were used to replace the original fragment in pSRα46. The resulting construct was designated pSRα46-HA. Then, the 2.2-kb XhoI-KpnI-digested pSRα54 containing the cDNA for p54 and the 650-bp KpnI-NheI-digested PCR product containing the IRES were subcloned into XhoI-XbaI-digested pSRα46-HA, and the resulting plasmid was designated pI-pri(HA). To express the nuclear-translocation-deficient p54-p46, the amino-terminal nuclear localization signal (NLS) of p54 was disrupted by site-directed mutagenesis as described previously (24). The resulting plasmid was designated pI-pri(-NLS).
The ScaI-BalI-digested PCR fragment containing the IRES and a 2.0-kb SmaI-digested pSRα68 containing the cDNA for p68 were subcloned into the EcoRV site of pSRα180. The plasmid encoding the two cDNAs for p180 and p68 was designated pI-pol.α.
For the six-His-tagged mutants, PCR fragments were subcloned into pSRHisABC expression plasmids containing triple tags (six-His tag, T7 tag, and Express tag) under the control of the SRα promoter (1). For construction of six-His-tagged full-length p180 (H-p180), a 4.2-kb EcoRI-EcoRV fragment of pSRα180 was blunt ended with Klenow enzyme. pSRHisC was digested with BglII, blunt ended with Klenow enzyme, and ligated with the blunt-ended fragment containing the cDNA for p180. For the construction of the amino-terminal and the carboxyl-terminal truncation mutants, H-core, H-ΔN400, and H-ΔN600, the initiation methionine and restriction enzyme sites were introduced by PCR with the same 3′ primer, 5′-GCATTTGAATGGATCCTAATCCTTGTATT-3′, and different 5′ primers, 5′-AGTTCTAGATATCATGAGTAATCTCCCATTG-3′, 5′-GGAGATCTATGAAATTTGACCTAAA-3′, and 5′-GGAGATCTAAAGAA-3′. To construct H-ΔC200, the 2.5-kb fragment of BamHI-digested H-core fragment was subcloned into BglII-digested pSRHisC. H-ΔC was constructed by PCR with the primers 5′-GCATTTGAATGGATCCTAATCCTTGTATT-3′ and 5′-AAGCCGGGACACCATTG-3′. The PCR products were digested with BamHI and subcloned into BamHI-digested pSRα180. Then, Aor51HI-MluI-digested fragment containing a carboxyl-terminal deletion mutant of p180 was filled in with Klenow enzyme and subcloned into PvuII-digested pSRHisC. To construct the amino-terminal deletion mutant H-ΔN, a 1.4-kb BalI-KpnI-digested H-p180 fragment was subcloned into the BalI-KpnI-digested H-core.
To express the carboxyl-terminal fragment (residues 1235 to 1465), PCR-amplified products obtained with primers 5′-GATGGATCCGATGCTGTACTCATT-3′ and 5′-GGCCTCCTTGGATCCTTCCCG-3′ were digested with BamHI and subcloned into BglII-digested pSRHisB.
Mutants with cysteine residues replaced with alanine residues in the putative zinc fingers were constructed by PCR by the overlap extension technique (15). The following primer sets were used to introduce mutations: H-AZ, 5′-CTTTGTCCTTCATGTGGAACTGAAAATATTTAT-3′ and 5′-GGCCTCCTTGGATCCTTCCCG-3′; and H-ZA, 5′-CTGTGTCCAGTCTGCATGAAAGCTGTGCTTAGA-3′ and 5′-GGCCTCCTTGGATCCTTCCCG-3′. The double mutant H-AA was constructed by PCR with H-AZ as template DNA and primers for H-ZA.
The identity of each of these constructs was confirmed by DNA sequencing on an Applied Biosystems 377A automatic DNA sequencer.
Cell culture and transfection.
COS-1 cells, which are derived from the African green monkey kidney cell line CV-1 by transformation with an origin-defective SV40 virus, were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum in a 5% CO2 incubator. NIH 3T3 cells were cultured in medium supplemented with 10% calf serum instead of fetal bovine serum. Transfection was performed by electroporation as described elsewhere (31). The level of protein expression was analyzed 48 h after transfection unless otherwise indicated.
DNA polymerase assay.
The DNA polymerase assay was carried out as described previously (36). Briefly, 5 μg of protein was incubated for 1 h at 37°C with 0.5 mg of DNase I-activated calf thymus DNA per ml in a buffer containing 20 mM Tris-HCl (pH 8.0); 10% glycerol; 60 mM KCl; 5 mM MgCl2; 3.3 mM 2-mercaptoethanol; 0.2 mg of bovine serum albumin per ml; 100 μM (each) dATP, dCTP, and dGTP; and 50 μM [3H]dTTP (0.1 Ci/mmol). The incorporation of [3H]dTMP was measured with a Whatman DE81 paper disc as described elsewhere (35).
Glycerol density gradient sedimentation.
Proteins were extracted from NIH 3T3 cells with 0.3 M KCl in extraction buffer as described previously (37). Aliquots containing 200 μg of protein in 50 μl were layered onto 2 ml of a linear 15 to 35% glycerol gradient in a buffer containing 25 mM potassium phosphate (pH 7.5), 300 mM KCl, 1 mM MgCl2, and 0.5% Triton X-100. Centrifugation was at 55,000 rpm for 16 h at 4°C (Beckman TLS-55), and 30 fractions were collected from the top of the gradient. Western blot analysis of each fraction was done with antibodies specific for each subunit (23, 24).
Indirect immunofluorescence staining.
Cells were grown on chamber slides (Nunc) coated with poly-l-lysine, washed with phosphate-buffered saline (PBS), and fixed with 3.7% formaldehyde in PBS for 10 min on ice. Cells were then washed with PBS and permeabilized sequentially with 50, 75, and 95% ethanol on ice for 5 min each. The slides were then blocked with PBS containing 5% normal goat serum (blocking buffer) for 30 min at room temperature; incubated with anti-p46 (0.4 μg/ml), anti-p54 (0.3 μg/ml), anti-p68 (1.3 μg/ml), or anti-p180 (diluted 1:3,000) antibody in blocking buffer for 1 h at room temperature; and washed three times with PBS for 5 min each time. Cells were then incubated with FITC-conjugated secondary antibody for 1 h at room temperature, washed three times with PBS, and preserved in Vectorshield (Vector Inc.). DNA staining was performed by adding 1 μg of bis-benzimide (Hoechst 33258) per ml into the final PBS wash. The samples were examined with an Olympus PROVIS AX70 fluorescence microscope. For double staining studies, SJK132-20 monoclonal antibody (ascitic fluid), anti-HA antibody, and Texas red-conjugated secondary antibody were used at a 1:400 dilution, 5 μg/ml, and 1.5 μg/ml, respectively.
Preparation of cell extracts and Western blot analysis.
After transfection, COS-1 cells were washed with PBS, scraped from the plates in PBS, centrifuged for 5 min, and resuspended in extraction buffer as described previously (37). The insoluble materials were separated by centrifugation. Precipitates were resuspended in Laemmli sample buffer (17). The samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically onto 0.45-μm-pore-size polyvinylidene difluoride membranes (Millipore). After incubation of the membranes with either anti-p46 (0.13 μg/ml), anti-p54 (0.3 μg/ml), anti-p68 (0.3 μg/ml), anti-p180 (1:3,000), anti-HA (1:1,200), anti-six-His tag (1:3,000), or anti-T7 tag (1:3,000) antibodies in TBS (Tris-buffered saline; 50 mM Tris-HCl [pH 7.5] and 150 mM NaCl) containing 5% (wt/vol) dried milk for 1 h at room temperature, the membranes were washed three times with TBS containing 0.05% Tween 20. The membranes were then incubated for 1 h at room temperature with horseradish peroxidase-conjugated goat secondary antibody in TBS containing 5% dried milk and washed again. Detection of the protein bands was performed with the enhanced chemiluminescent reagent SuperSignal (Pierce) according to the manufacturer’s instructions. Kaleidoscope prestained standards (Bio-Rad) were used as molecular weight standards.
Pull-down assay with six-His tags.
Fifty micrograms of COS-1 cell extracts was mixed with cobalt-chelating Sepharose (TALON; Clontech) for 4 h at 4°C in 200 μl of PK buffer (20 mM potassium phosphate [pH 7.5], 100 mM KCl, 0.1% NP-40, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.2 μg of aprotinin per ml, 0.2 μg of leupeptin per ml, 0.1 μg of antipain per ml, and 0.1 μg of pepstatin A per ml). After washing with PK buffer containing 10 mM imidazole, six-His-tagged proteins were eluted with 30 μl of PK buffer containing 200 mM imidazole and then subjected to SDS-PAGE and Western blot analysis with anti-six-His tag or anti-T7 tag monoclonal antibody.
Coimmunoprecipitation analysis.
Fifty micrograms of COS-1 cell extracts was immunoprecipitated with 1 μl of anti-p46 antibody (0.13 μg) which had been preadsorbed to protein G-Sepharose (Pharmacia) for 4 h at 4°C in 200 μl of PK buffer. After washing with PK buffer, precipitates were dissolved with 30 μl of 2× Laemmli sample buffer and then subjected to SDS-PAGE and Western blot analysis with anti-six-His tag or anti-T7 tag monoclonal antibody.
RESULTS
The eukaryotic polycistronic cDNA expression system.
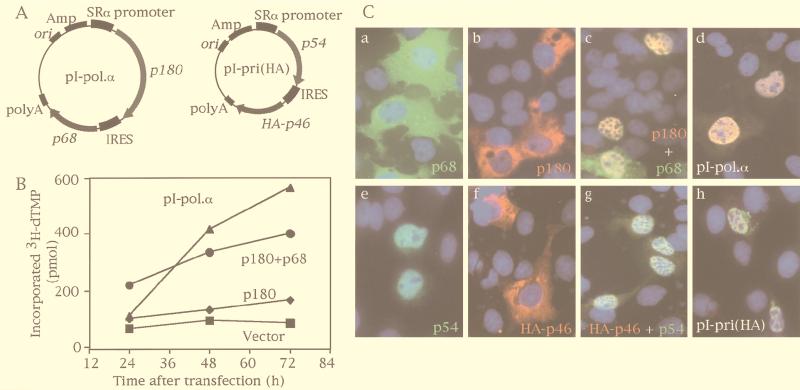
Although transient transfection by electroporation of COS-1 cells with one plasmid was very efficient and reproducible, simultaneous transfections with three or four plasmids were very difficult, because the amount of transfected plasmid varied from one cell to another. To overcome the problem of multisubunit transfection, we constructed a eukaryotic polycistronic cDNA expression plasmid containing an IRES. An IRES was discovered at first in the picornavirus genome, where it allows the initiation of translation in a cap-independent manner (7, 25, 28, 30). Using the IRES derived from poliovirus, we constructed pI-pol.α, which was designed to coexpress p180 and p68 as a heterodimer, and pI-pri, which was designed to express p54 and p46 as a heterodimer (Fig. 1A). These plasmids are suitable for the transfection of three or four subunits simultaneously because all of the cells expressing p180 will contain p68 and all of the cells expressing p54 will contain p46 at constant ratios. To verify that coexpressed p180 and p68 were assembled into a functional DNA polymerase, we measured DNA polymerase activity. Transiently transfected COS-1 cells were harvested at the indicated times, and the DNA polymerase activity of the extracts was determined with activated calf thymus DNA as a substrate. While singly expressed p180 showed a slight increase of polymerase activity, p180 coexpressed with p68 exhibited a marked increase of activity in a time-dependent manner, reflecting the enhanced protein synthesis of p180 in the presence of p68 as found previously (23). When pI-pol.α expression vector was transfected, the extracts exhibited the same level of polymerase activity as the extracts containing coexpressed p180 and p68 (Fig. 1B). Therefore, we confirmed that p180-p68 was expressed efficiently as a functional heterodimer complex by using pI-pol.α.
FIG. 1.
The cDNA expression system with IRES-derived expression plasmids. (A) Constructs of IRES-derived plasmids used for the expression of two subunits simultaneously. (B) Time course of DNA polymerase activity in the cell extracts transfected with various constructs. COS-1 cells were transfected with pcDEBΔ (■), pSRα180 alone (⧫), pSRα180 and pSRα68 (●), or pI-pol.α (▴); incubated for 24, 48, and 72 h; and then lysed with a solution containing 20 mM potassium phosphate (pH 7.5), 300 mM KCl, 10% glycerol, 0.05% Triton X-100, and 0.1 mM EDTA. After centrifugation, the supernatant was assayed to estimate DNA polymerase activity. Five micrograms of protein of COS-1 extracts was incubated with [3H]dTTP and DNase I-activated calf thymus DNA for 1 h at 37°C, and incorporation of radioactive material was measured. (C) Subcellular distribution of transiently overexpressed subunits of DNA polymerase α-primase complex. pSRα68 (a), pSRα180 (b), pSRα180 and pSRα68 (c), pI-pol.α (d), pSRα54 (e), pSRα46-HA (f), pSRα54 and pSRα46-HA (g), or pI-pri(HA) (h) was transfected into COS-1 cells, and the proteins expressed were detected by immunofluorescence analysis with anti-p68 (a, c, and d) or anti-p54 (e, g, and h) polyclonal antibodies and FITC-conjugated anti-rabbit IgG antibody or SJK132-20 anti-p180 monoclonal antibody (b, c, and d) or 12CA5 anti-HA tag antibody (f, g, and h) and Texas red-conjugated anti-mouse IgG antibody. DNA was stained blue by Hoechst 33258, and pictures were merged.
To determine the subcellular distribution of the subunits transiently coexpressed by pI-pri and pI-pol.α, immunohistochemical experiments were carried out. As we reported previously, p180, p68, and p46 were predominantly localized in the cytoplasm when expressed individually (Fig. 1C, a, b, and f) (23, 24). Only p54 was localized in the nucleus (Fig. 1C, e) (24). In contrast, when p46 and p54 or p68 and p180 were coexpressed, each coexpressed subunit was exclusively colocalized in the nucleus (Fig. 1C, c and g). Thus, specific interactions between the four subunits of DNA polymerase α, namely, between p46 and p54 and between p68 and p180, result in translocation of all of these subunits into the nucleus. When pI-pol.α or pI-pri(HA) was transfected, the subunits expressed by each vector were predominantly colocalized in the nucleus (Fig. 1C, d and h). Thus, the IRES-derived expression plasmids produced heterodimers in the nucleus efficiently.
Simultaneous expression of three or four of the subunits of DNA polymerase α in COS-1 cells.
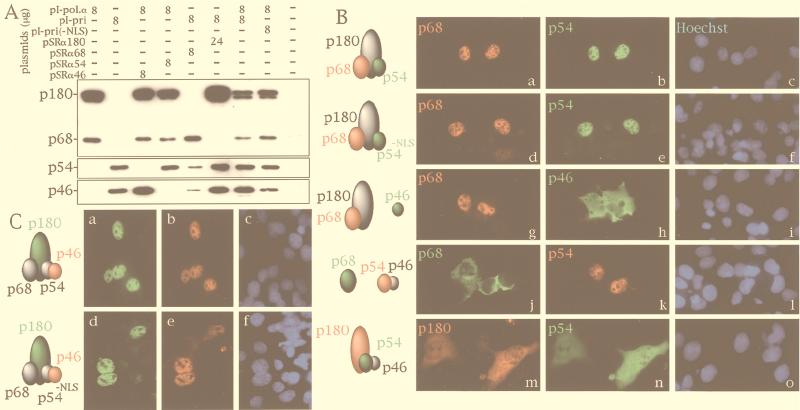
Using one of the IRES-derived plasmids (pI-pol.α and pI-pri) along with a vector expressing only a single subunit (pSRα46, pSRα54, pSRα68, or pSRα180), we were able to study the expression of three subunits at the same time. The expressed proteins were analyzed by Western blotting and by immunohistochemical techniques as described for Fig. 2A and B. Three or four subunits were coexpressed simultaneously by using specific combinations of the IRES-derived plasmids and single-subunit expression vectors (Fig. 2A). When p180, p68, and p54 were expressed together, all of the subunits were colocalized in the nucleus (Fig. 2B, a to c). When the amino-terminal NLS of p54 was disrupted by site-directed mutagenesis (24), the nuclear localization of these three subunits did not change (Fig. 2B, d to f). These results suggested that these three subunits were assembled into a trimeric complex in the cytoplasm and that the complex was then translocated into the nucleus. On the other hand, when p180, p68, and p46 were expressed together, p180-p68 was localized in the nucleus but p46 remained in the cytoplasm (Fig. 2B, g to i). When p68, p54, and p46 were expressed together, p54-p46 was localized in the nucleus but p68 remained in the cytoplasm (Fig. 2B, j to l). These results indicated that simultaneous expression of either p180, p68, and p46 or p68, p54, and p46 did not result in the formation of trimeric complexes in the cytoplasm. Only the dimeric complexes p180-p68 and p54-p46 containing nuclear-translocation-proficient subunits were translocated into the nucleus, while the nonassembled subunits p46 and p68 remained in the cytoplasm. These observations indicate that DNA polymerase α is assembled by the binding of p46 to p180-p68 via p54 and by the binding of p68 to p54-p46 via p180.
FIG. 2.
Coexpression of three or four of the subunits of DNA polymerase α in COS-1 cells. (A) Western blot analysis of transfected COS-1 extracts. Two, three, or all of the subunits of DNA polymerase α were coexpressed in COS-1 cells. Forty-eight hours after transfection, the cells were lysed with a solution containing 20 mM potassium phosphate (pH 7.5), 300 mM KCl, 10% glycerol, 0.05% Triton X-100, and 0.1 mM EDTA. Ten micrograms of protein from the extract was subjected to SDS-PAGE followed by Western blot analysis with anti-p46, anti-p54, anti-p68, and anti-p180 antibodies. To express p180-p54-p46, an excess amount of pSRα180 was used because the protein level of p180 decreased considerably in the absence of p68 (23). (B) Subcellular distribution of three subunits of DNA polymerase α coexpressed in COS-1 cells. The subunits shown by green letters were detected by FITC-conjugated anti-mouse IgG antibodies. The other subunits, shown by red letters, were visualized by Texas red-conjugated anti-rabbit IgG antibody. The rightmost panels show nuclear staining by Hoechst 33258. (C) Coexpression of the four subunits. pI-pol.α was cotransfected with pI-pri or pI-pri(-NLS), which encoded the nuclear-translocation-deficient primase. Then, the subcellular distribution of ectopically expressed proteins was determined. p180 was stained by anti-p180 monoclonal antibody and FITC-conjugated anti-mouse IgG antibody. p46 was detected by anti-p46 antibody and Texas red-conjugated anti-rabbit IgG antibody. Nuclear staining with Hoechst 33258 is shown in panels c and f.
When p180, p54, and p46 were expressed together, unexpected results were obtained. Although p54 and p46 can translocate as a heterodimer into the nucleus by virtue of their NLS (Fig. 1C, h), coexpression of p54-p46 with p180 resulted in localization of p54-p46 in the cytoplasm (Fig. 2B, m to o). This result suggests that the NLS of primase was occluded by the presence of p180.
We next undertook coexpression of the four subunits. When pI-pol.α and pI-pri were cotransfected into COS-1 cells, the four subunits were colocalized in the nucleus (Fig. 2C, a to c). Similarly, when the NLS of p54 was disrupted by site-directed mutagenesis, the four subunits were still translocated into the nucleus (Fig. 2C, d to f). Therefore, in the tetrameric complex, the NLS of p54 would appear to be dispensable for translocation into the nucleus. These results suggest that in the cytoplasm the heterodimeric complexes p54-p46 and p180-p68 could form a tetrameric complex, which was then translocated into the nucleus.
Identification of the binding domain of p180 required for the interaction with p54-p46 by immunohistochemical analysis.
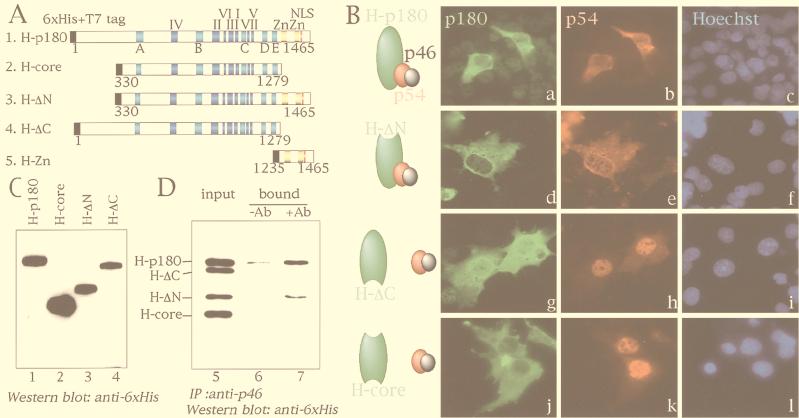
To verify the effect of p180 on the subcellular distribution of p54-p46, a series of deletion mutants of p180 containing six-His and T7 tags were designed as depicted in Fig. 3A and coexpressed with p54-p46 in COS-1 cells. An alignment of amino acid sequences of DNA polymerase α from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Trypanosoma brucei, humans, mice, and Oryza sativa allowed us to choose domain boundaries for construction of mutants with deletions (22, 44). The subcellular distribution of the deletion mutants was determined by immunodetection with an anti-six-His monoclonal antibody. The expression of the transiently overexpressed proteins was confirmed by Western analysis as shown in Fig. 3C. In the presence of six-His-tagged p180, p54-p46 was predominantly localized in the cytoplasm (Fig. 3B, a to c). However, when the carboxyl-terminal region of p180 was deleted (H-ΔC and H-core), p54-p46 was localized in the nucleus while p180 remained in the cytoplasm (Fig. 3B, g to l). In the case of the amino-terminal deletion mutants (H-ΔN), colocalization of p54-p46 and p180 in the cytoplasm was observed (Fig. 3B, d to f). Thus, when the carboxyl-terminal region of p180 was truncated, p54-p46 could enter the nucleus independently of p180, indicating that the carboxyl-terminal region of p180 is crucial for interaction with p54-p46.
FIG. 3.
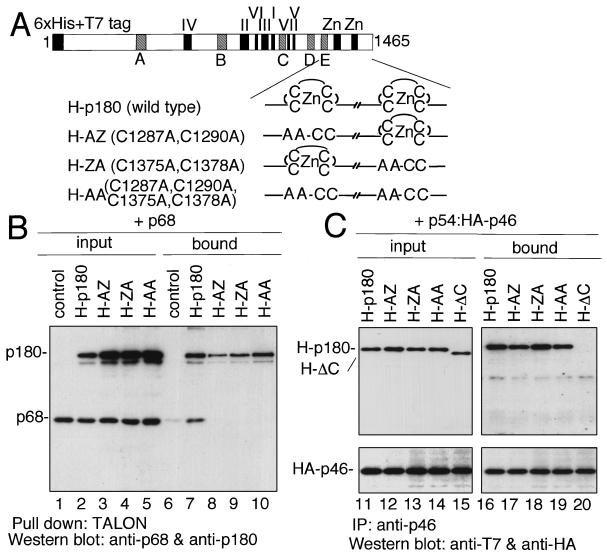
Identification of the p180 domain required for the interaction with p54-p46. (A) Schematic representation of p180 mutant constructs. H-p180, H-ΔN, H-ΔC, and H-core indicate full-length p180 with six-His tag and T7 tag at the amino terminus, amino-terminally deleted p180 with six-His tag and T7 tag at the amino terminus, carboxyl-terminally deleted p180 with six-His tag and T7 tag at the amino terminus, and both amino-terminally and carboxyl-terminally deleted p180 with six-His tag and T7 tag at the amino terminus, respectively. H-Zn, which contains only carboxyl-terminal putative zinc finger regions, is also shown for the experiments shown in Fig. 4. The seven highly conserved regions of class B DNA polymerases are indicated by blue boxes with roman numerals (I to VII) (32, 43). The five conserved regions in eukaryotic DNA polymerase α are indicated by light blue boxes with letters (A to E) (22). Putative zinc finger motifs and a putative NLS (23) are depicted by yellow boxes and a red line, respectively. Six-His and T7 tags are shown by solid boxes. Numbers indicate amino acid positions of p180. (B) Subcellular distribution of ectopically expressed p180 mutants and p54-p46. H-p180 (a to c), H-ΔN (d to f), H-ΔC (g to i), and H-core (j to l) were cotransfected with pI-pri into COS-1 cells, and the expressed proteins were detected simultaneously by indirect immunofluorescence analysis with anti-p54 polyclonal antibody and Texas red-conjugated anti-rabbit IgG antibody or anti-six-His monoclonal antibody and FITC-conjugated anti-mouse IgG antibody. Nuclear staining with Hoechst is shown in the rightmost panels. (C) Western blot analysis of transiently expressed p180 mutants. Extracts (10 μg of protein) were subjected to SDS-PAGE followed by Western blot analysis with anti-six-His monoclonal antibody. Lane numbers correspond to p180 mutant constructs shown in panel A. (D) Coimmunoprecipitation assay. Extracts (50 μg of protein) containing p54-p46 were mixed with a total of 250 μg of the extracts described for panel C (75 μg of the extracts containing H-p180, 75 μg of the extracts containing H-ΔN, 75 μg of the extracts containing H-ΔC, and 25 μg of the extracts containing H-core), incubated on ice for 2 h, and immunoprecipitated with (lane 7) or without (lane 6) anti-p46 antibody and protein G-Sepharose. One-third of the precipitates were subjected to Western blot analysis with anti-six-His monoclonal antibody. Lane 5 contains 5 μg of the input proteins. Ab, antibody; IP, immunoprecipitation.
To verify the interaction between the carboxyl-terminal region of p180 and p54-p46, immunoprecipitation analysis was carried out. Whole-cell extracts of cells transfected with the p180 mutant constructs were mixed, incubated with the extract containing p54-p46 heterodimer, and immunoprecipitated with anti-p46 antibody, and coprecipitated proteins were detected by Western blot analysis with the anti-six-His tag antibody. As shown in Fig. 3C, it is clear that full-length p180 and H-ΔN were coprecipitated with p46, while H-ΔC and H-core were not. These results are consistent with the immunohistochemical characterization which indicated that the carboxyl-terminal region of p180 (residues 1235 to 1485) is essential for its interaction with p54-p46.
Identification of the interaction domain between p180 and p68.
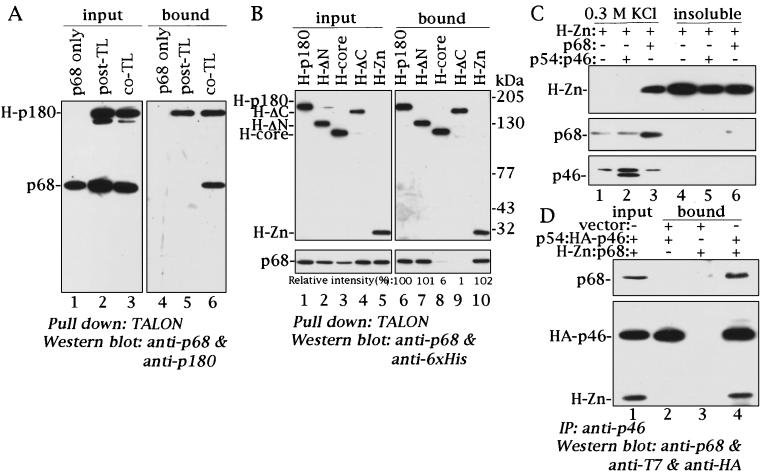
To investigate the domain of p180 required for interaction with p68, coprecipitation analysis was performed. In contrast to the p180 and p54-p46 interaction, singly expressed p180 in COS-1 cells did not form a complex with singly expressed p68 after mixing of the two extracts. However, when the two subunits were coexpressed in COS-1 cells, we found that p180 and p68 were tightly associated with each other and formed a complex, as shown in Fig. 4A. To determine the domain of p180 necessary for the interaction with p68, deletion mutants of p180 were constructed. Since all of the constructs described in the previous report (23) were capable of interacting with p68, further deletion mutants were designed, and their interaction with p68 was studied by pull-down analysis. Cell extracts from cells coexpressing the deletion mutants and p68 were precipitated with cobalt-chelating Sepharose (TALON), and coprecipitation of p68 was detected by Western blot analysis. When the carboxyl-terminal region of p180 was deleted (H-ΔC and H-core), p68 did not coprecipitate, as shown in Fig. 4B. In contrast, when H-ΔN was cotransfected with p68, p68 precipitated with H-ΔN as well as with full-length p180. Moreover, the carboxyl-terminal region alone (H-Zn, residues 1235 to 1465; shown in Fig. 3A, row 5) could bind tightly to p68. Thus, the carboxyl-terminal region of p180 is both necessary and sufficient for binding to p68.
FIG. 4.
Identification of the p180 domain required for the interaction with p68. (A) Extracts containing singly expressed p68 (50 μg of protein) were mixed in the presence (lanes 1 and 4) or absence (lanes 2 and 5) of singly expressed six-His-tagged p180 (50 μg of protein), incubated on ice for 1 h, pulled down with cobalt-chelating Sepharose (TALON), and then eluted with a solution containing 200 mM imidazole. Extracts containing coexpressed H-p180 and p68 (50 μg of protein) were also pulled down in parallel (lanes 3 and 6). One-third of the eluates were subjected to Western blot analysis with anti-p180 and anti-p68 antibodies (right panel). The left panel shows results with 5 μg of the input proteins. TL, translation. (B) Coprecipitation assay with p68 and six-His- and T7-tagged p180 mutants. Extracts containing coexpressed p68 and various mutants of the six-His- and T7-tagged p180 (50 μg of protein) were coprecipitated with cobalt-chelating Sepharose and eluted with 200 mM imidazole. One-third of the eluates were subjected to SDS-PAGE followed by Western blot analysis with anti-T7 tag monoclonal antibody (upper panels) or anti-p68 polyclonal antibody (lower panels). Precipitated protein levels of p68 were quantitated by densitometric scanning of the blot and are presented at the bottom as relative values compared to that of p68 coprecipitated with H-p180. (C) H-Zn can be expressed as a soluble form in the presence of p68. The carboxyl-terminal domain containing two zinc finger motifs was expressed alone (lanes 1 and 4) or in the presence of p54-p46 (lanes 2 and 5) or p68 (lanes 3 and 6). Transfected cells were lysed with a solution containing 20 mM potassium phosphate (pH 7.5), 300 mM KCl, 10% glycerol, 0.05% Triton X-100, and 0.1 mM EDTA, and insoluble materials were separated by centrifugation. Precipitates were resuspended in Laemmli sample buffer (17). Ten micrograms of protein of the soluble fraction and 10% of the corresponding insoluble fractions were subjected to SDS-PAGE followed by Western blotting with anti-six-His, anti-p68, and anti-p46 antibodies. (D) Coimmunoprecipitation assay with H-Zn–p68 and HA-tagged p46-p54. Fifty micrograms of the extracts containing coexpressed H-Zn–p68, coexpressed p54-HA-tagged p46, and vector control was mixed as shown on the top of the figure and immunoprecipitated (IP) with anti-p46 antibody. One-third of the precipitates were subjected to Western blot analysis with anti-T7, anti-HA, and anti-p68 antibodies. Lane 1 contains 6 μg of the extracts containing H-Zn–p68 and HA-tagged p46-p54 as input proteins.
In the course of our cDNA expression study, we noticed that the carboxyl-terminal fragment of p180 (H-Zn) could be expressed in a soluble form only in the presence of p68. Singly expressed H-Zn or H-Zn coexpressed with p54-p46 was scarcely detectable in the soluble fraction (Fig. 4C). These results suggested that the interaction between p68 and the carboxyl-terminal region of p180 changes the conformation of p180, so that both are expressed as a stable complex. Since singly expressed p180 cannot form a complex with p68, the conformational change of the carboxyl-terminal region most likely occurs in the presence of p68. An alternative hypothesis is that the carboxyl-terminal region of p180 either is very hydrophobic or contains very hydrophobic regions that reduce the solubility of this region. Upon binding to p68, these hydrophobic residues will become part of the core.
Since we obtained soluble H-Zn–p68 subcomplex by cotransfection into COS-1 cells, we undertook coimmunoprecipitation analysis with H-Zn–p68 and p54-p46 to verify that the carboxyl-terminal region of p180 alone is responsible for interactions with p68 and p54-p46. Coexpressed H-Zn–p68 and coexpressed p54 and HA-tagged p46 were mixed for 1 h and immunoprecipitated with anti-p46 antibody, and coprecipitated proteins were detected by Western blot analysis with the anti-T7, anti-HA, and anti-p68 antibodies. As shown in Fig. 4D, H-Zn–p68 was precipitated with anti-p46 antibody in the presence of p54-p46. Therefore, we conclude that H-Zn alone can mediate the coprecipitation of p68 and p54-p46.
Putative zinc finger motifs in the carboxyl-terminal region of p180 are required for the interaction with p68 but not with p54-p46.
The carboxyl-terminal region of p180 contains two zinc finger motifs which are highly conserved among eukaryotic DNA polymerases including α, δ, ɛ, and ζ (32, 42). To assess the role of these motifs in the formation of the DNA polymerase α complex, we performed mutational analysis of the putative zinc finger motifs by replacing highly conserved cysteine residues in the zinc finger motifs with alanine residues as shown in Fig. 5A. The interaction between the substitution mutants and p68 or p54-p46 was assayed by coprecipitation experiments. We demonstrated that substitution of either one of the two zinc fingers completely abolished the interaction with p68 (Fig. 5B). In contrast, the mutant proteins were able to associate with p54-p46 (Fig. 5C). The H-ΔC construct in lanes 15 and 20 was used as a negative control for p180 interaction with p54-p46. Therefore, we conclude that the interaction between p68 and p180 is absolutely dependent upon both of the putative zinc finger motifs, while the p54-p46 interaction occurs independently of the zinc finger motifs. We confirmed the results of the coprecipitation studies by an immunohistochemical analysis. When the substitution mutants were coexpressed with p68, all of the mutants were localized in the cytoplasm whereas wild-type p180 and p68 were colocalized in the nucleus. In contrast, p54-p46 was localized in the cytoplasm in the presence of the substitution mutants or wild-type p180 (data not shown).
FIG. 5.
The putative zinc finger motifs in the carboxyl-terminal domain of p180 are required for the interaction with p68 but not with p54-p46. (A) Schematic representation of p180 mutant constructs. Highly conserved motifs are depicted as described in the legend to Fig. 3A. (B) Coprecipitation assay with p68 and six-His- and T7-tagged p180 mutants. Fifty micrograms of the extracts containing p68 alone (lanes 1 and 6), coexpressed p68 and six-His-tagged p180 (lanes 2 and 7), H-AZ (lanes 3 and 8), H-ZA (lanes 4 and 9), and H-AA (lanes 5 and 10) was coprecipitated with cobalt-chelating Sepharose and eluted with 200 mM imidazole. One-third of the eluates were subjected to SDS-PAGE followed by Western blot analysis with anti-p180 and anti-p68 polyclonal antibodies (lanes 6 to 10). The left panel contains 10 μg of the input proteins (lanes 1 to 5). (C) Coimmunoprecipitation assay with p54-HA-tagged p46 and six-His- and T7-tagged p180 mutants. Fifty micrograms of the extracts containing coexpressed p54-HA-tagged p46 and six-His- and T7-tagged p180 (lanes 11 and 16), H-AZ (lanes 12 and 17), H-ZA (lanes 13 and 18), H-AA (lanes 14 and 19), and H-ΔC (lanes 15 and 20) was immunoprecipitated (IP) with anti-p46 antibody. One-tenth of the precipitates were subjected to Western blot analysis with anti-T7 (upper panel of lanes 16 to 20) and anti-HA (lower panel of lanes 16 to 20) monoclonal antibodies. The left panels contain 5 μg of the input proteins (lanes 11 to 15).
Domain of p180 required for DNA polymerase activity.
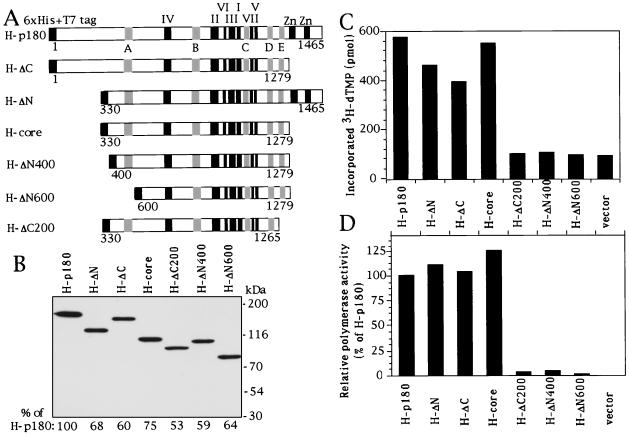
To determine the minimal domain required for DNA polymerase activity, COS-1 cells were transfected with constructs expressing the truncated p180 listed in Fig. 6A, and extracts were prepared at 48 h posttransfection. Five micrograms of each extract was subjected to SDS-PAGE followed by Western blot analysis with anti-T7 monoclonal antibody. Expression levels of the exogenous proteins were quantitated by densitometric scanning of the blot and are presented at the bottom of Fig. 6B as relative values compared to that of the full-length p180 (H-p180). For DNA polymerase activity, 5 micrograms of the extracts from the transfected COS-1 cells was assayed (Fig. 6C). The activity of the extract from cells transfected with the vector only indicates the level of endogenous DNA polymerase activity derived from host cells. DNA polymerase activities due to the exogenously expressed mutant p180 were obtained from the results of Fig. 6C by subtracting the endogenous DNA polymerase activity. The values were further normalized by the relative expression levels of respective proteins (obtained in Fig. 6B) and indicated as percentages of the H-p180 activity as shown in Fig. 6D. Interestingly, we were able to detect polymerase activity for the deletion mutants H-ΔN and H-ΔC. Moreover, polymerase activity could still be detected even when both regions were deleted (H-core). When the sizes of the deleted regions were further increased (H-ΔN400, H-ΔN600, and H-ΔC200), DNA polymerase activity was abolished completely. According to these results, we conclude that the minimal core domain for polymerase activity spans residues 330 to 1279 and that the amino-terminal region (1 to 329) and carboxyl-terminal region (1280 to 1465), including the zinc finger motifs, are dispensable for intrinsic DNA polymerase activity.
FIG. 6.
Identification of the minimal core domain required for DNA polymerase activity. (A) Schematic representation of p180 mutant constructs. Highly conserved motifs are depicted as described in the legend to Fig. 3A. (B) Western blotting. COS-1 cells were transfected with constructs expressing the truncated p180 listed in panel A, and extracts were prepared at 48 h posttransfection. Five micrograms of each extract was subjected to SDS-PAGE followed by Western blot analysis with anti-T7 monoclonal antibody. Expression levels of the exogenous proteins were quantitated by densitometric scanning of the blot and are presented at the bottom as relative values compared to that of the full-length p180 (H-p180). (C) DNA polymerase activity of transfected COS-1 extracts. Five micrograms of the extracts from the transfected COS-1 cells was incubated with [3H]dTTP and DNase I-activated calf thymus DNA for 1 h at 37°C, and the incorporated radioactivity was measured. The activity of the extract from cells transfected with the vector only indicates the level of endogenous DNA polymerase activity derived from host cells. (D) DNA polymerase activities due to the exogenously expressed mutant p180 were obtained from the results shown in panel C by subtracting the endogenous DNA polymerase activity. The values were further normalized by the relative expression levels of respective proteins (obtained for panel B) and indicated as percentages of the H-p180 activity.
Detection of free p68 and free p54-p46 in NIH 3T3 extracts.
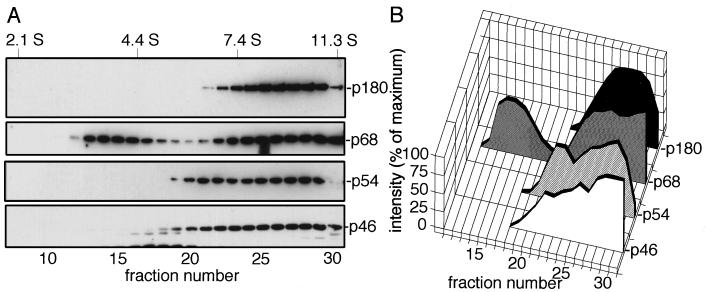
To gain insights into the endogenous profile of the mouse DNA polymerase α-primase complex, Western blot analysis was performed with antibodies specific for each subunit. The availability of antibodies against each subunit of DNA polymerase α enabled us to determine the endogenous level of each subunit in the cell. Whole-cell extracts of NIH 3T3 cells extracted with 0.3 M KCl were fractionated by 15 to 35% glycerol gradient centrifugation and subjected to SDS-PAGE followed by Western blot analysis. In the logarithmically growing cultures of NIH 3T3 cells, less than 5% of p180 was associated with the insoluble fraction after extraction with 0.3 M KCl as determined by Western blot analysis with anti-p180 antibody (data not shown). As shown in Fig. 7A, four subunits cosedimented together around fractions 25 to 28, indicating the presence of the tetrameric DNA polymerase α complex. Interestingly, we also observed an additional signal for p68 in fractions 12 to 16. Singly expressed p68 in COS-1 cells sedimented to a similar position on the gradient (23). Thus, p68 is present in two forms in NIH 3T3 cells: as a monomer and as part of the tetrameric complex. A proportion of the p54 and p46 subunits also sedimented as a broad peak in fractions 20 to 29, which were distinct from the fractions (25 to 28) containing the tetrameric complex. The Western blot signals were quantified by densitometry, and the results are presented in Fig. 7B. p54 and p46 subunits sedimented as a peak (fractions 20 to 23) in addition to the tetrameric peak (fractions 25 to 28), strongly suggesting that a subcomplex of primase exists in NIH 3T3 cells. We observed similar patterns in other cell lines, including FM3A, CV-1, and COS-1 cells (data not shown).
FIG. 7.
Fractionation of the endogenous four subunits of DNA polymerase α in NIH 3T3 cells by glycerol density gradient centrifugation. Lysates of NIH 3T3 cells (200 μg of protein) were fractionated by 15 to 35% glycerol gradient sedimentation. The fractions were subjected to Western blotting. Protein markers run in a parallel gradient were chicken lysozyme (2.1 S), bovine serum albumin (4.4 S), yeast alcohol dehydrogenase (7.4 S), and bovine catalase (11.3 S). (A) Western blotting with antibodies specific for each subunit. (B) Densitometric analyses. Western blots were examined by scanning densitometry, and the results are presented as percentages of the maximum intensity of each signal.
DISCUSSION
In this report, we designed a eukaryotic polycistronic cDNA expression system to investigate the subunit-subunit interactions and domain structures of DNA polymerase α. We transfected three or all of the subunits simultaneously in COS-1 cells, determined their subcellular distribution in detail, and uncovered how the subunits are assembled into the tetrameric complex. Moreover, we identified the minimal domains of p180 required for DNA polymerase activity and for interaction with p68 and p54-p46 by deletion and substitution mutation analyses. Taken together, these results allow us to present for the first time a model of not only DNA polymerase α subunit assembly but also the steps leading up to complex formation. Certain elements of the complex formation of DNA polymerase α may also be shared by other eukaryotic replicative DNA polymerases.
Although multisubunit complexes can be coexpressed in cultured cells by simply transfecting constructs with mixtures of plasmids expressing a single subunit, the simultaneous expression of two or three subunits at a constant level in one cell has been hard to obtain. We overcame this problem by using IRES-derived expression plasmids, which allowed the expression of two open reading frames under the control of a single promoter and a single poly(A) signal in a polycistronic manner. We used the IRES derived from the poliovirus type 1 Mahoney strain. This IRES has been shown to possess strong activity for cap-independent translation and to be less dependent on the distance between the IRES and the initiating methionine for translation (7). This expression system enabled us to determine the subcellular distribution of the four subunits of DNA polymerase α in detail.
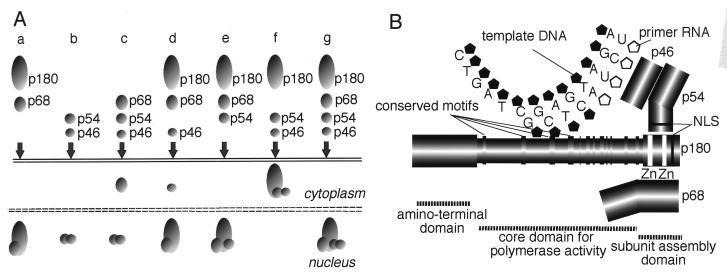
The subcellular distribution of various combinations of two, three, or four of the subunits of DNA polymerase α is summarized in Fig. 8A. We previously found that specific associations between p180 and p68 and between p54 and p46 were essential for their translocation into the nucleus (Fig. 8A, a and b) (23, 24). Here, we have extended these findings by coexpressing three or even four of the subunits simultaneously by using polycistronic expression plasmids (Fig. 8A, c to g). According to these results, we found that p46 interacts with p180-p68 via p54 and that p68 interacts with p54-p46 via p180. As shown in Fig. 8A, f, p54-p46 was localized in the cytoplasm in the presence of p180, although p54-p46 alone can enter the nucleus by virtue of the NLS. In the absence of the carboxyl-terminal region of p180, p54-p46 was translocated into the nucleus independently of the p180 mutants, indicating that three subunits form a trimer through the carboxyl-terminal region of p180. Although our coexpression system allowed us to characterize the interaction between p54 and p180 successfully, the reason why the p180-p54-p46 heterotrimer is localized in the cytoplasm remains unclear. One possibility is that the NLS of p54 is located at the interface of the binding site between p54 and p180, which could result in inactivation of the NLS of p54 in the p180-p54-p46 heterotrimer. Alternatively, inactivation of the NLS of p54 might result from conformational changes in p54 induced by p180. Since the NLS of p54 has been shown to possess strong activity as an authentic NLS and to be resistant to conformational changes such as those resulting from deletions and gene fusions (24), we prefer the former possibility. However, further experiments will be needed to clarify these points.
FIG. 8.
Schematic representations of the results of the present study. (A) Model for the nuclear transport pathway of mouse DNA polymerase α-primase complex which shows the subcellular distribution of two, three, or four of the coexpressed subunits of DNA polymerase α. (B) Organization of the subunit assembly of DNA polymerase α. The p180 molecule can be divided into three domains. The first is the amino-terminal domain (residues 1 to 329), which is dispensable for polymerase activity and subunit assembly. The second is the core domain (residues 330 to 1279), which is sufficient for DNA polymerase activities such as template recognition, substrate binding, and the phosphoryl transfer reaction. The third is the carboxyl-terminal domain (residues 1235 to 1465), which is dispensable for polymerase activity but required for assembly of the complex. p68 binds directly to the putative zinc finger motifs in the carboxyl-terminal domain, whereas p54-p46 associates with the carboxyl-terminal domain independently of the zinc finger motifs. The interaction between p46 and p180 is mediated by p54.
In NIH 3T3 cells, we found significant amounts of free p68 and free p54-p46 in addition to the tetrameric complex (Fig. 7). The presence of free p68 and free p54-p46 suggests the following series of events in the assembly of the subunit structure of the tetrameric complex. In the absence of p180, free p68 is localized in the cytoplasm, where it awaits p180 protein synthesis (Fig. 8A, c). Once p180 is synthesized, p68 rapidly associates with p180 and is translocated into the nucleus, where it binds to the p54-p46 heterodimer (Fig. 8A, e). Alternatively, all of the binding steps could take place in the cytoplasm before translocation of the tetrameric complex into the nucleus. Although there exists no evidence in favor of either of these hypotheses, we prefer the latter one, as the NLS-disrupted p54-p46 heterodimer was found to form a complex with p180-p68 in the cytoplasm before being translocated into the nucleus (Fig. 2C). To determine the limiting step in tetramer assembly, it will be necessary to characterize the transcriptional regulation of each subunit, since the levels of newly synthesized p180 and p54-p46 appear to be critical for complex formation. Recently, it was reported that DNA polymerase α recruitment to chromatin during G1 is independent of cdc6-dependent prereplicative complex formation (10). In addition, it was found that loading of DNA polymerase α onto chromatin is dependent on cdc45 (21). Thus, it is becoming clear that loading of DNA polymerase α onto chromatin is a crucial step for the initiation of DNA replication. However, for the moment, the relationship between chromatin loading and nuclear translocation of DNA polymerase α is still unclear. Therefore, further characterization of complex formation and nuclear translocation of DNA polymerase α should help us to understand the precise role of the tetrameric complex of DNA polymerase α in the nucleus during G1 phase.
To extract endogenous DNA polymerase α from NIH 3T3 cells, we prepared whole-cell extracts with 0.3 M KCl. Although 0.3 M KCl extraction has no effect on the assembly of the tetramer of DNA polymerase α under in vitro conditions, we cannot rule out the possibility that 0.3 M KCl extraction changes in the intracellular environment. To determine physiological functions of an endogenous single subunit or a subcomplex from four subunits of DNA polymerase α, further experiments will be needed.
Taking advantage of cDNA expression systems in mammalian cultured cells, we coexpressed several mutant forms of p180 with p68 and found that the carboxyl-terminal region of p180 is both essential and sufficient for interaction with p68. Moreover, the putative zinc finger motifs were shown to be essential for this interaction. Thus, these results, as well as the immunohistochemical analysis described above, allow us to present a model for the organization of the DNA polymerase α complex in Fig. 8B. According to this model, p46 associates with the p54 bound to the carboxyl-terminal region of p180, and p68 interacts with the putative zinc finger motifs of p180 independently of p54-p46. All these interactions depend on the carboxyl-terminal region of p180. Since enzymatic activity was independent of the other subunits and involved only the core domain, this model is consistent with the spatial arrangement between the core domain and the nonenzymatic subunits. Hence, we can divide the p180 molecule into three domains: (i) the amino-terminal domain (residues 1 to 329), which is dispensable for both polymerase activity and assembly with the other subunits; (ii) the core domain (residues 330 to 1279), which is capable of all the reactions involved in polymerase activity, such as template recognition, substrate binding, and phosphoryl transfer reaction; and (iii) the carboxyl-terminal domain (residues 1235 to 1465), which is dispensable for polymerase activity but essential for assembly of the complex. Recently, crystallographic studies of DNA polymerase gp43 of bacteriophage RB96, which is a member of the class B DNA polymerase family, showed that the central region of this protein containing the highly conserved motifs had a U or hand-like shape (41). Thus, it is tempting to speculate that the core domain of mouse DNA polymerase α also folds into a structure like that of gp43. Identification of the minimal core domain in this study should contribute to the structural characterization of class B DNA polymerase in higher eukaryotes.
Recently, Dua et al. reported that the carboxyl-terminal region containing the zinc finger motifs of DNA polymerase II in S. cerevisiae is essential for the interaction with DPB2 by two-hybrid analysis (11). In addition, it has been reported that the second-largest subunits of DNA polymerases α, δ, and ɛ share conserved motifs, implying that these subunits belong to a superfamily (2, 20). Taken together, these observations suggest that the putative zinc finger conserved among eukaryotic class B DNA polymerases is the binding site of the second-largest subunit, which is also conserved among eukaryotic class B DNA polymerases.
Characterization of the subunit assembly of DNA polymerase α was reported previously by Copeland and Wang (9) and Longhese et al. (19). Using the baculovirus expression system, Copeland and Wang showed that in the absence of p54, p46 cannot be copurified with p180, which suggested that p54 is important for the interaction between p46 and p180. Using temperature-sensitive mutants of DNA primase in S. cerevisiae, Longhese et al. showed that the protein level of purified p46 decreases in proportion to that of p54, suggesting that p54 is essential for the interaction of p46 with p180. All of these results are consistent with our above results showing that p46 binds to p180 through p54. Our observations extend their findings by showing that the carboxyl-terminal region of p180 is important for the physical interaction with p54 and confirm the notion that p54 functions as the crucial tethering point of the complex. In addition, we found that p68 is not capable of interacting with primase in vivo. The interaction between p68 and primase has not been studied to date.
Although the interactions within the multisubunit complex are central to our understanding of the molecular mechanism of a wide range of enzymes, reconstitution of such complexes from individually purified proteins has been difficult to accomplish in many cases. For example, the Ku antigen heterodimer can be assembled into a complex only when the two subunits are coexpressed simultaneously in insect cells (27). Trimeric RP-A was assembled efficiently only when the three subunits were coexpressed in bacteria or insect cells (14, 33). Coexpression of the subunits of the yeast origin recognition complex was essential for the formation of a stable complex (5, 18). Thus, expression of subunits of protein complexes through transcription-coupled translation has been considered to be crucial for their stability. Therefore, in this respect our cDNA expression system is ideal, as it allows the coordinated transcription-coupled translation in vivo of the subunits of multisubunit complexes. Thus, our system should be a useful tool to define the assembly of multisubunit complexes such as the origin recognition complex, the minichromosomal maintenance complex, RF-C, and other multisubunit DNA polymerases in vivo.
ACKNOWLEDGMENTS
We thank Yusaku Nakabeppu for providing the pcDEBΔ expression vector, Shigeo Ohno for the pSRHisABC expression plasmids, Akio Nomoto for IRES, and Tetsu Akiyama for NIH 3T3 cells. We also thank Kaoru Sugasawa for helpful discussions and Yasue Ichikawa at the Biodesign DNA sequencing facility in the Institute of Physical and Chemical Research (RIKEN) for DNA sequencing.
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan and a special grant for promotion of research from RIKEN and the Biodesign Research Program of RIKEN. T.M. was a special postdoctoral researcher of RIKEN.
REFERENCES
- 1.Akimoto K, Takahashi R, Moriya S, Nishioka N, Takayanagi J, Kimura K, Fukui Y, Osada S, Mizuno K, Hirai S, Kazlauskas A, Ohno S. EGF or PDGF receptors activate atypical PKCλ through phosphatidylinositol 3-kinase. EMBO J. 1996;15:788–798. [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Koonin E V. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker T A, Bell S P. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 4.Bambara R A, Murante R S, Henricksen L A. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 5.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 6.Bensch K G, Tanaka S, Hu S-Z, Wang T S-F, Korn D. Intracellular localization of human DNA polymerase α with monoclonal antibodies. J Biol Chem. 1982;257:8391–8396. [PubMed] [Google Scholar]
- 7.Borman A M, Bailly J-L, Girard M, Kean K M. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland W C, Wang T S-F. Catalytic subunit of human DNA polymerase α overproduced from baculovirus-infected insect cells. Structural and enzymological characterization. J Biol Chem. 1991;266:22739–22748. [PubMed] [Google Scholar]
- 9.Copeland W C, Wang T S-F. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem. 1993;268:26179–26189. [PubMed] [Google Scholar]
- 10.Desdouets C, Santocanale C, Drury L S, Perkins G, Foiani M, Plevani P, Diffley J F X. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase α. EMBO J. 1998;17:4139–4146. doi: 10.1093/emboj/17.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dua R, Levy D L, Campbell J L. Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase ɛ in DNA replication and the S/M checkpoint pathway. J Biol Chem. 1998;273:30046–30055. doi: 10.1074/jbc.273.45.30046. [DOI] [PubMed] [Google Scholar]
- 12.Foiani M, Lucchini G, Plevani P. The DNA polymerase α-primase complex couples DNA replication, cell-cycle progression and DNA-damage response. Trends Biochem Sci. 1997;22:424–427. doi: 10.1016/s0968-0004(97)01109-2. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs P E, McGregor W G, Maher V M, Nisson P, Lawrence C W. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 15.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 16.Izumi M, Miyazawa H, Harakawa S, Yatagai F, Hanaoka F. Identification of a point mutation in the cDNA of the catalytic subunit of DNA polymerase α from a temperature-sensitive mouse FM3A cell line. J Biol Chem. 1994;269:7639–7644. [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee D G, Bell S P. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longhese M P, Jovine L, Plevani P, Lucchini G. Conditional mutations in the yeast DNA primase genes affect different aspects of DNA metabolism and interactions in the DNA polymerase α-primase complex. Genetics. 1993;133:183–191. doi: 10.1093/genetics/133.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mäkiniemi M, Pospjech H, Kilpeläinen S, Jokela M, Vihinen M, Syväoja J E. A novel family of DNA-polymerase-associated B subunits. Trends Biochem Sci. 1999;24:14–16. doi: 10.1016/s0968-0004(98)01327-9. [DOI] [PubMed] [Google Scholar]
- 21.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazawa H, Izumi M, Tada S, Takada R, Masutani M, Ui M, Hanaoka F. Molecular cloning of the cDNAs for the four subunits of mouse DNA polymerase α-primase complex and their gene expression during cell proliferation and the cell cycle. J Biol Chem. 1993;268:8111–8122. [PubMed] [Google Scholar]
- 23.Mizuno T, Ito N, Yokoi M, Kobayashi A, Tamai K, Miyazawa H, Hanaoka F. The second-largest subunit of the mouse DNA polymerase α-primase complex facilitates both production and nuclear translocation of the catalytic subunit of DNA polymerase α. Mol Cell Biol. 1998;18:3552–3562. doi: 10.1128/mcb.18.6.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuno T, Okamoto T, Yokoi M, Izumi M, Kobayashi A, Hachiya T, Tamai K, Inoue T, Hanaoka F. Identification of the nuclear localization signal of mouse DNA primase: nuclear transport of p46 subunit is facilitated by interaction with p54 subunit. J Cell Sci. 1996;109:2627–2636. doi: 10.1242/jcs.109.11.2627. [DOI] [PubMed] [Google Scholar]
- 25.Molla A, Jang S K, Paul A V, Reuer Q, Wimmer E. Cardioviral internal ribosomal entry site is functional in a genetically engineered dicistronic poliovirus. Nature. 1992;356:255–257. doi: 10.1038/356255a0. [DOI] [PubMed] [Google Scholar]
- 26.Nakabeppu Y, Oda S, Sekiguchi M. Proliferative activation of quiescent Rat-1A cells by ΔFosB. Mol Cell Biol. 1993;13:4157–4166. doi: 10.1128/mcb.13.7.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono M, Tucker P W, Capra J D. Production and characterization of recombinant human Ku antigen. Nucleic Acids Res. 1994;22:3918–3924. doi: 10.1093/nar/22.19.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 29.Schneider A, Smith R W P, Kautz A R, Weisshart K, Grosse F, Nasheuer H-P. Primase activity of human DNA polymerase α-primase. Divalent cations stabilize the enzyme activity of the p48 subunit. J Biol Chem. 1998;273:21608–21615. doi: 10.1074/jbc.273.34.21608. [DOI] [PubMed] [Google Scholar]
- 30.Shiroki K, Ishii T, Aoki T, Ota Y, Yang W X, Komatsu T, Ami Y, Arita M, Abe S, Hashizume S, Nomoto A. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J Virol. 1997;71:1–8. doi: 10.1128/jvi.71.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorimachi H, Toyama-Sorimachi N, Saido T C, Kawasaki H, Sugita H, Miyasaka M, Arahata K, Ishiura S, Suzuki K. Muscle-specific calpain, p94, is degraded by autolysis immediately after translation, resulting in disappearance from muscle. J Biol Chem. 1993;268:10593–10605. [PubMed] [Google Scholar]
- 32.Spicer E K, Rush J, Fung C, Reha-Krantz L J, Karam J D, Konigsberg W H. Primary structure of T4 DNA polymerase. Evolutionary relatedness to eucaryotic and other procaryotic DNA polymerases. J Biol Chem. 1988;263:7478–7486. [PubMed] [Google Scholar]
- 33.Stigger E, Dean F B, Hurwitz J, Lee S H. Reconstitution of functional human single-stranded DNA-binding protein from individual subunits expressed by recombinant baculoviruses. Proc Natl Acad Sci USA. 1994;91:579–583. doi: 10.1073/pnas.91.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugino A. Yeast DNA polymerases and their role at the replication fork. Trends Biochem Sci. 1995;20:319–323. doi: 10.1016/s0968-0004(00)89059-3. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki M, Enomoto T, Hanaoka F, Yamada M. Dissociation and reconstitution of a DNA polymerase α-primase complex. J Biochem (Tokyo) 1985;98:581–584. doi: 10.1093/oxfordjournals.jbchem.a135314. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki M, Enomoto T, Masutani C, Hanaoka F, Yamada M, Ui M. DNA primase-DNA polymerase α assembly from mouse FM3A cells. Purification of constituting enzymes, reconstitution, and analysis of RNA priming as coupled to DNA synthesis. J Biol Chem. 1989;264:10065–10071. [PubMed] [Google Scholar]
- 37.Takada-Takayama R, Tada S, Hanaoka F, Ui M. Peptide mapping of the four subunits of the mouse DNA polymerase α-primase complex. Biochem Biophys Res Commun. 1990;170:589–595. doi: 10.1016/0006-291x(90)92132-j. [DOI] [PubMed] [Google Scholar]
- 38.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voitenleitner C, Rehfuess C, Hilmes M, O’Rear L, Liao P-C, Gage D A, Ott R, Nasheuer H-P, Fanning E. Cell cycle-dependent regulation of human DNA polymerase α-primase activity by phosphorylation. Mol Cell Biol. 1999;19:646–656. doi: 10.1128/mcb.19.1.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Sattar A K, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 42.Wang T S-F. Eukaryotic DNA polymerases. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- 43.Wong S W, Wahl A F, Yuan P-M, Arai N, Pearson B E, Arai K, Korn D, Hunkapiller M W, Wang T S-F. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokoi M, Ito M, Izumi M, Miyazawa H, Nakai H, Hanaoka F. Molecular cloning of the cDNA for the catalytic subunit of plant DNA polymerase α and its cell-cycle dependent expression. Genes Cells. 1997;2:695–709. doi: 10.1046/j.1365-2443.1997.1560354.x. [DOI] [PubMed] [Google Scholar]