Abstract
The multicopy rDNA array gives origin to the nucleolus, a large non-membrane bound organelle that occupies a substantial space of the cell nucleus. The rDNA/nucleolus have emerged as a coordinating hub in which seemingly disparate cellular functions converge and from which a variety of cellular and organismal phenotypes emerge. However, the role of the nucleolus as a determinant and organizer of nuclear architecture and other epigenetic states of the genome has not been well understood. Here, we discuss the role of the rDNA and the nucleolus in nuclear organization and function; from nucleolus associated domains, to the regulation of imprinted loci and X chromosome inactivation, and rDNA contact maps that anchor and position the rDNA relative to the rest of the genome. Nucleolus influence on nuclear organization is bound to modulate diverse biological processes; from metabolism to cell proliferation, genome-wide gene expression, the maintenance of epigenetic states, and aging.
A Multifunctional Powerhouse at the Core of the Cell Nucleus
The rDNA is the most abundantly transcribed segment of the genome, producing over 70% of all cellular RNAs in an eukaryotic cell [1]. While each transcribed mRNA of the highly expressed ribosomal protein genes are translated into a larger number of ribosomal proteins, each transcribed rRNA molecule is used only once as it is assembled with ribosomal proteins to produce >2,000–10,000 ribosomes per minute [1, 2]. This imposes a disproportionate demand for rRNAs that must be dutifully supplied during ribosome biogenesis to ensure cell survival and proliferation. Maintenance of remarkably high rRNA transcription and ribosomal assembly rates is dependent upon the nucleolus, a large non-membrane bound nuclear organelle that is dedicated to the task of rRNA transcription and ribosome assembly and occupies a substantial and disproportionate space of the cell nucleus.
Despite essential and well-document roles of the rDNA/nucleolus in ribosome biogenesis, little is known about the rDNA/nucleolus influence on nuclear organization and function of the genome. For instance, nucleolar associated domains (NADs) were identified about 10 years ago [3, 4], but their evolutionary conservation and stability through development and cell stress have remained uncertain. Recent studies have used proximity ligation approaches to identify rDNA contacts within the nucleus, which also need to be reconciled with NAD localization and behavior [5–7]. Furthermore, copy number of the 45S rDNA, an attribute that is relevant to nucleolus function, impacts cellular senescence and genome stability [8, 9], apart from being associated with the induction and silencing of hundreds to thousands of genes dispersed throughout the genome [10–12]. Similarly, rDNA methylation and nucleolar size vary across individuals and are associated with age and longevity [13–15] but NADs themselves appear stable through cell senescence [16]. Here we review how nucleolar elements are organized, how rDNA arrays are epigenetically regulated, and how both interact and influence the architecture of the nucleus, with implications for cellular function and organismal health.
The rDNA Encodes the Nucleolus
The nucleolus originates from the nucleolus organizer regions (NOR) that are comprised of rDNA repeat clusters typically dispersed across several chromosomes (Figure 1). In humans, NORs are present in the short arm of five acrocentric chromosomes (13, 14, 15, 21 and 22), whereas in laboratory mice they are found on six chromosomes (11, 12, 15, 16, 18, 19). The fundamental repeat unit consists of a ~13 KB rDNA core that encodes the Pol I transcribed 45S rRNA genes and ~30 KB intergenic spacers (IGS). The IGS may contain regulatory elements, replication fork barriers, and other RNAs transcribed by Pol II [17]. The core 45S rDNA produces nascent 45S transcripts that are processed into 18S, 5.8S and 28S rRNA components which, together with the 5S RNA, are loaded into the newly formed ribosomes that are assembled within the nucleolus [18]. The 5S RNA genes, typically localized somewhere else in the genome, are transcribed by RNA polymerase III in the euchromatic compartment of the nucleus or at the periphery of the nucleolus [6]. In some organisms the 5S rDNA is located inside the 45S rDNA array [19, 20]. The evolutionary pressures and functional constraints that contribute to a linked rDNA array with intermixed 45S-5S units (as seen in some plants and fungi) or the separate 45S and 5S arrays (as seen in mammals and Drosophila) have remained poorly known [5, 6].
Figure 1. Organization of the nucleolus organizer region (NOR) and the rDNA array.
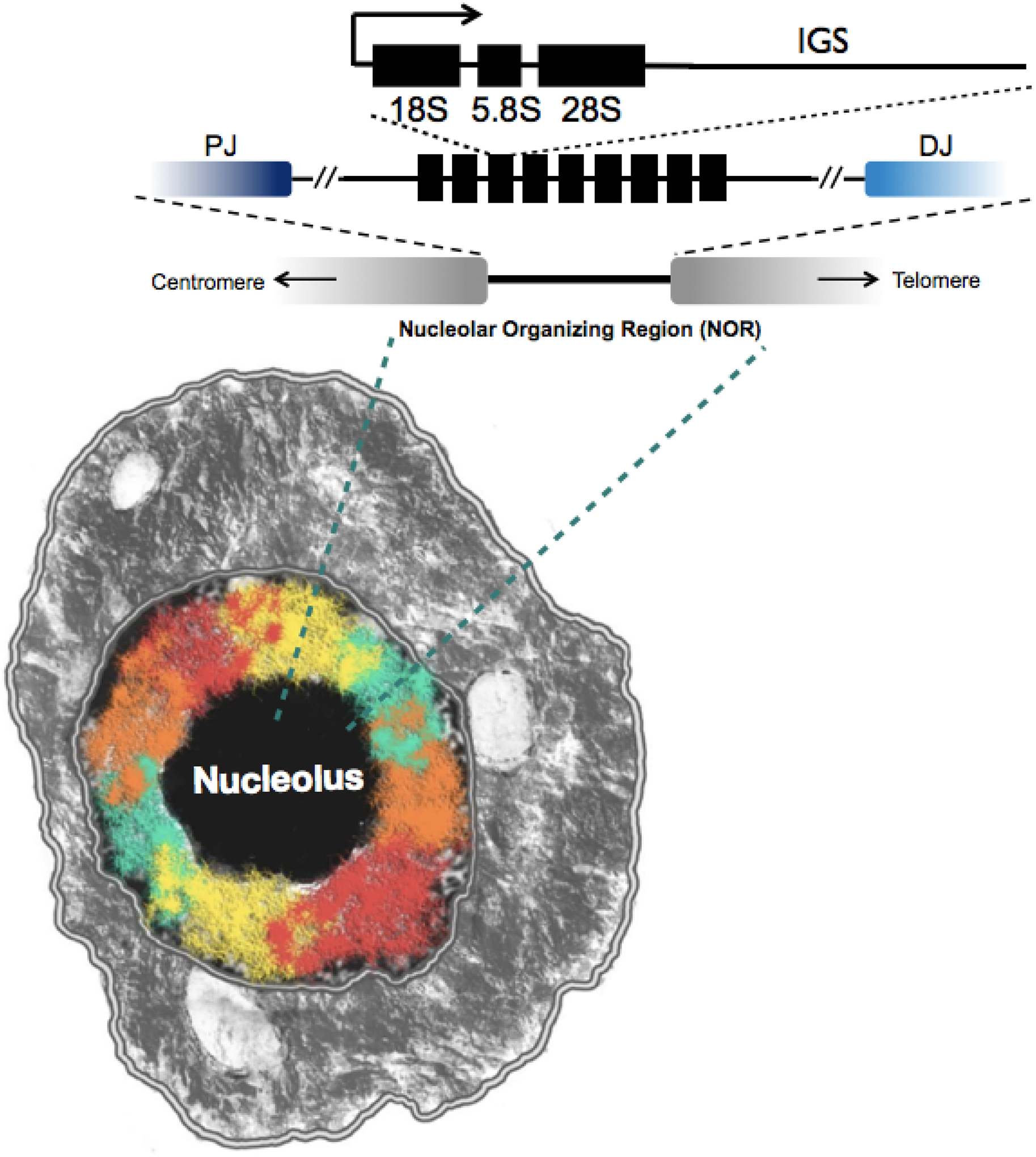
Structure of the rDNA array; IGS = non-transcribed intergenic spacer. Proximal (PJ) and distal (DJ) junction link the rDNA array to the neighboring genomic region. NORs reside in 5 human chromosomes and are structurally complex rather than neatly organized as shown above: repeat units can be truncated, inverted, and contain small and large indels. NOR gives origin the nucleolus (black circle), a relatively large non-membrane bound organelle inside the cell nucleus. The colors represent chromatin from other chromosomes. A single nucleolus or multiple nucleoli can exist per cell.
Dynamic Structural Organization of the Nucleolus
The nucleolus consists of a large granular component (GC), with one or a few dense fibrillar components (DFCs), each of which has a fibrillar center (FC) (Figure 2). In addition, a layer of heterochromatin surrounds the nucleolus, forming the peri-nucleolar chromatin [21]. From this shell around the nucleolus, strands of chromatin enter the organelle, forming the intranucleolar chromatin (Figure 2). The organization of nucleolus elements into substructures has not been completely understood [18]. There is consensus, however, that the majority of the rDNA is located in either FC or DFC [22, 23], as well as in peri-nucleolar heterochromatin and in loops of rDNA from peri-nucleolar heterochromatin to the interior of nucleolus (also called intranucleolar stretches) [18]. Overall, the rDNA outside the nucleolus is viewed as transcriptionally inactive [24–26], with intranucleolar stretches containing both active and silenced genes [25]. One hypothesis is that nascent rRNA transcripts are found in one of the fibrillary parts, but mainly in DFC [27] (Figure 2), with the nascent rRNA generating the DFC [28]. Another hypothesis is that the rDNA and its transcription localize to the FC. This is supported by early data showing Pol I localization to the FC [29]. However, some have argued that the rDNA and its transcription are located in DFC [30] where fibrillarin (FBL) is located and contributes to keeping chromatin in an opened state [31–33].
Figure 2. Organization and diversity of the nucleolus.
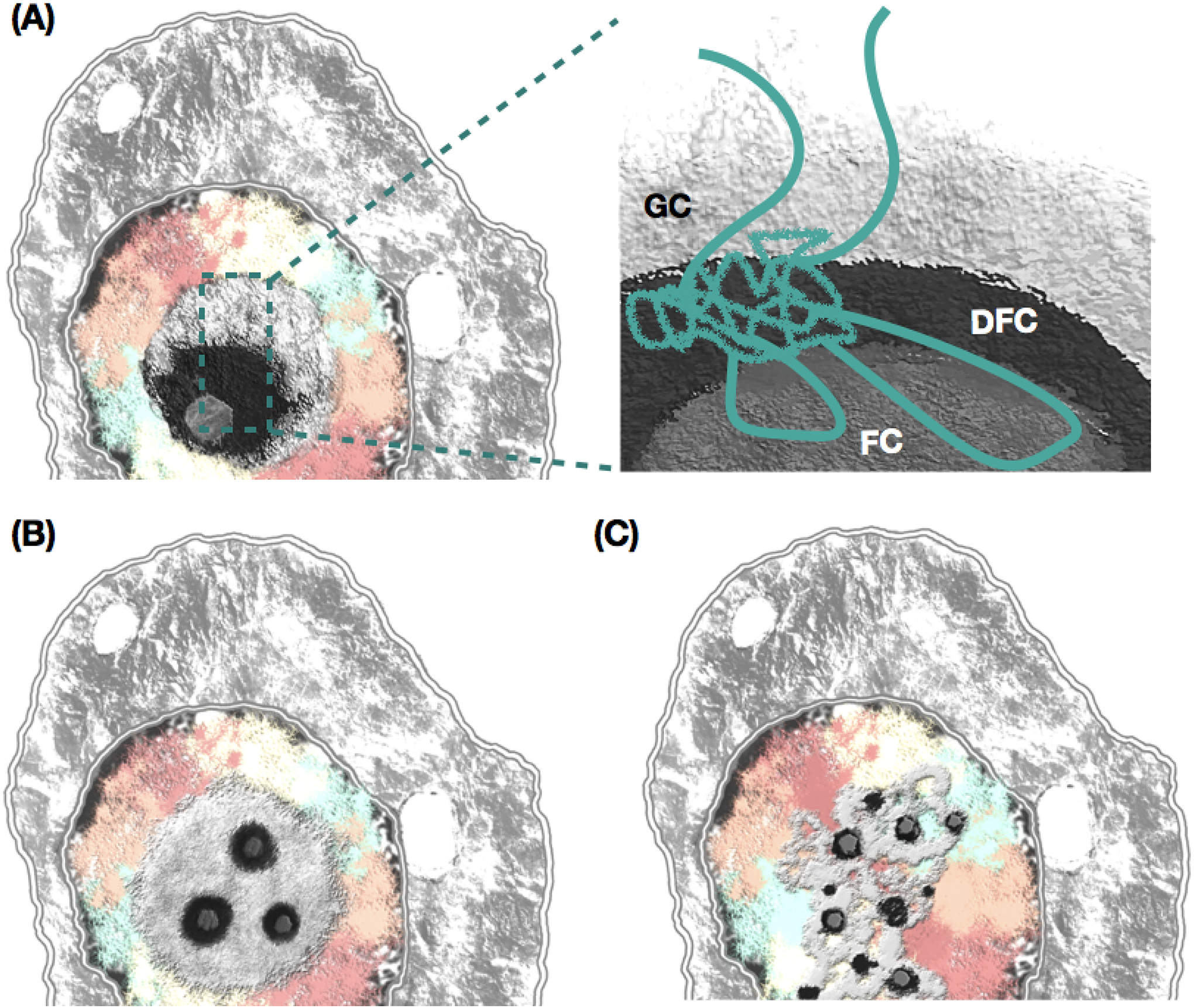
The nucleolus is structurally organized with at least 3 three sub-regions: fibrillar centers (FCs), dense fibrillar components (DFCs), and granular components (GCs). A single nucleolus or multiple nucleoli can exist per cell. (a) In one hypothesis, the IGS is localized at the Dense Fibrillar Components (DFCs), with transcribed 45S units extending into the Fibrilar Center (FC) and possibly into granular components (GCs). (a-c) The nucleolus displays morphological diversity across cells and conditions. Note that a single or multiple FC and DFC foci can exist per nucleolus and can occur at the center or near the periphery of the organelle; reticulate nucleoli have also been documented.
The nucleolus displays substantial structural diversity during development, across tissues, as well as a dynamic organization with assembly and disassembly of components during the cell cycle, DNA repair, and response to stress (Figure 2, Figure 3). A variety of nucleolar proteins, chromatin modulators and transcription regulators such as FBL, nucleophosmin (NPM1), Pol I, and UBF contribute to determining nucleolus structures [34]. Proteomic analysis revealed over 500 proteins that localize to the nucleolus [35–37]. UBF plays an essential role in the aggregation of nucleolar proteins [38–40], with ectopically located UBF resulting in nucleolus-like structures, with pseudo NOR and Pol I machinery [41]. The distal flanking region of the rDNA might also be relevant in nucleolar aggregation and assembly/disassembly of rDNA from different chromosomes. Experimental data showed, for instance, that the distal junction associates with the nucleolus even when ectopically positioned [42]; it localizes closer to the periphery of the organelle and is transcriptionally active. However, rDNA association can occur even without the flanking regions [43, 44]. Thus, flanking regions might be involved in NOR coalescence but not in its association with other nucleolar components and nucleolus formation [18]. Furthermore, inactive NORs localize outside the nucleolus [25]; the higher the proportion of active NOR in a NOR bearing chromosome, the closer it is to the nucleolus, suggesting that transcription activity is a factor in nucleolus association [24, 45].
Figure 3. Nucleolar-organization and re-organization upon DNA damage.
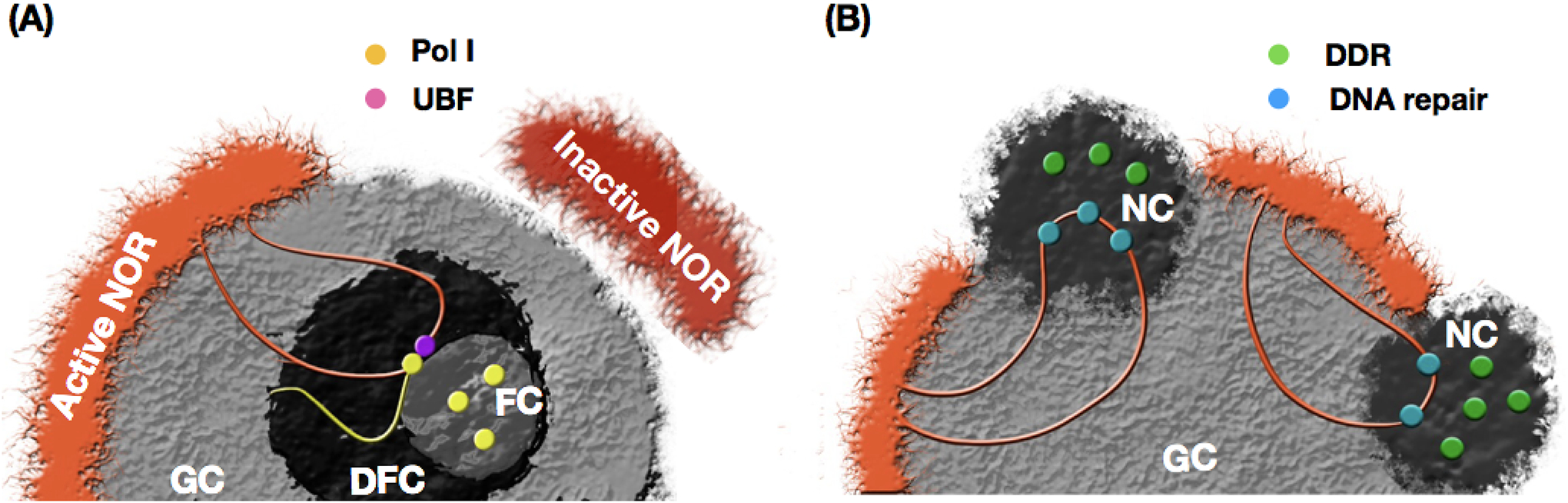
(a) Genomic regions that surround the nucleolus (e.g., NADs) preferentially acquire a silenced condensed state. The rDNA in NOR may invaginate into the internal layers of the nucleolus, forming the intranucleolar stretches that contain both inactive and translated genes. In normal conditions, active NORs form the fibrillar center (FC) and dense fibrillar center (DFC), inside the granular component (GC). (b) Nucleolar-reorganization upon DNA damage. The nucleolar cap (NC) is formed, replacing FC and DFC, and the rDNA is dislocated to the periphery of the nucleolus, where the DNA repair machinery can access and repair the rDNA.
During cell cycle, the nucleolus disassembles as the cell reaches metaphase and subsequently reassembles [43, 48]. The assembly process begins in early telophase with pre-rRNA processing factors associating with NORs [49]. At the end of mitosis, nucleolar proteins aggregate, forming prenucleolar bodies (PNB) [34], which will then associate with NORs to form the nucleolus. While the association of PNB depends on Pol I, the beginning of the process with the pre-rRNA processing factors does not [49]. The nucleolus is also responsive to DNA damage [50, 51], with Pol I transcription inhibition [52] in response to double strand breaks (DSBs) both in rDNA [52] and outside the nucleolus [53]. This disruption of the transcription process leads to substantial internal dynamics and modification of nucleolar structures, with FC and DFC going to GC periphery, where it matches with DNA repair machineries and numerous DNA damage response (DDR) proteins (Figure 3), forming the nucleolar cap [50, 52, 54, 55]. Changes in nucleolar organization upon DNA repair have also been observed, with rDNA moving to the periphery and nucleolar components (nucleolin and nucleophosmin) translocating to and out of the nucleolus in an ATM dependent manner [46]. Nucleolar/rDNA changes through the cell cycle and during DNA repair are likely to substantial alter the nuclear landscape. All in all, dynamic changes to the nucleolus [18, 46, 47] during the cell cycle, DNA repair, or tissue differentiation are expected to influence the architecture of the entire nucleus and play a role modulating epigenetic, transcriptional, metabolic, and other functional states of the cell.
Epigenetic States of the rDNA
The rDNA is found in three different transcriptional states: active, poised and inactive. The poised state has open chromatin and unmethylated promoter but inactive nucleosome conformation – it has silenced units that are more easily activated [56, 57]. These epigenetic states of the rDNA are controlled by multiple elements. One of these elements is UBF, without which NORs become inactive and unbound to nucleoli [58]. Another important element is the transcription termination factor 1 (TTF1), which binds to terminator elements upstream and downstream of the rDNA. There are two factors that may bind to TTF1: Cockayne syndrome B protein (CSB) and the nucleolar remodeling complex (NoRC). When TTF1 binds to CSB, it promotes histone methylation, leading to activation of chromatin, whereas TTF1 binding to NoRC promotes both DNA methylation and histone deacetylation, leading to rDNA silencing [59].
CpG methylation is presumed to be an important regulator of rDNA transcription in humans and mice. Methylated DNA in the nucleolus preferentially localizes in the periphery of the organelle [60] and possibly on loops of rDNA (intranucleolar stretches) that extend from the perinucleolar heterochromatin into the interior of nucleolus. rDNA methylation is maintained by the DNA methyltransferase 1 (Dnmt1); cells lacking Dnmt1, but not Dnmt3b, show DNA methylation loss at rDNA genes as well as a disrupted and disorganized nucleolus [61]. The data show the relevance of Dnmt1 function on epigenetic states of rDNA and the structure of nucleolus. On the other hand, DNMT3a and DNMT3b were not initially localized to nucleoli in NIH 3T3 cells [62]; however more recent data indicated that both DNMT3a and DNMT3b interact with the Nucleolar Remodeling Complex (NoRC) and are recruited to nucleoli [63].
The insulator factor CTCF is a zinc finger protein that mediates folding and looping of the genome. CTCF is key to both genome-wide nuclear organization and to the epigenetic control of the rDNA [64, 65]. The protein binds to thousands of sites along the genome as well as to specific sites in the rDNA [66]; it contributes to rDNA activation through changes in nuclear and nucleolar structure and also by facilitating UBF loading onto the rDNA [65–68]. CTCF binds both UBF and a site immediately upstream from the spacer promoter [65], leading to increased Pol I activity; One hypothesis is that CTCF enhances UBF binding to this region, promoting rDNA transcription. Further research has provided additional evidence to support the hypothesis that CTCF promotes rDNA expression, with overexpression of CTCF causing increased rRNA transcription in a Pol I-dependent way [68]. CTCF might recruit cohesin, a protein that is primarily known for its role in sister chromatid cohesion during chromosome segregation and DNA repair, but that also plays a key role in epigenetic regulation of the genome [69]. When together, the CTCF-cohesin complex promotes the formation of loops in the rDNA [70], leading to promoter-enhancer interactions [71, 72] as well as chromatin insulation [64]. Intriguingly, variation in CTCF mRNA abundance across individuals in human populations is negatively associated with rDNA copy number [11]. Regulatory feedbacks that directly link CTCF mRNA/protein abundance to rDNA copy number have remained an intriguing possibility that could significantly and globally impact nuclear architecture.
Nucleolus-Associated Domains
Non-rDNA parts of the genome have long been hypothesized to interact with the nucleolus, such as the centromere of human chromosomes 1 and 9 [73] (Figure 4a). Seminal work with yeast also showed that tRNA genes preferentially localize at the periphery of the nucleolus [74–76]. However, sequencing approaches enabled more recent agnostic analyses of nucleolus associated chromatin. Accordingly, genome-wide identification of genomic segments associated with the nucleolus was reported in two seminal studies that conducted deep-sequencing of DNA retrieved from isolated human nucleoli [3, 4]. The studies identified nucleolus-associated domains (NADs) that preferentially localized around the nucleolus. Both studies found that NADs are enriched in olfactory receptor genes, zinc finger genes, and immunoglobulin gene families. Another study identified nucleolus associated regions in Arabdopsis and found that they are enriched in pseudogenes and tRNA genes [77]. Additionally, studies in human cells [3] [4] also noted an overlap between NADs and lamina-associated domains (LAD), a finding that is concordant with data showing that genes in LADs may temporarily move to the nucleolar periphery [78]. LAD and NAD territories have a disproportionate number of silenced genes [79, 80]. Thus, it has been hypothesized that LAD and NAD regulate tissue and development-specific gene expression as genes move to and from heterochromatic and euchromatic states. LAD territory in the nuclear periphery is organized by lamin B [79]. Lamin B has in turn also been linked with an organizational role in the nucleolus, according to evidence of its binding to nucleolin and nucleophosmin and of nucleolar disruption in its absence [81]. However, recent studies of genes specific to NAD showed that one third of them do not overlap with LAD [16]. This is expected because some LADs are constitutively maintained in the nuclear periphery [82]. Further detailed analyses of NADs and their dynamic during development, across tissues, and during the cell cycle, DNA repair, and response to stress are bound to provide novel insights on nuclear organization.
Figure 4. The nucleolus as an organizer of the nuclear space and coordinator of disparate nuclear functions and regulatory programs.
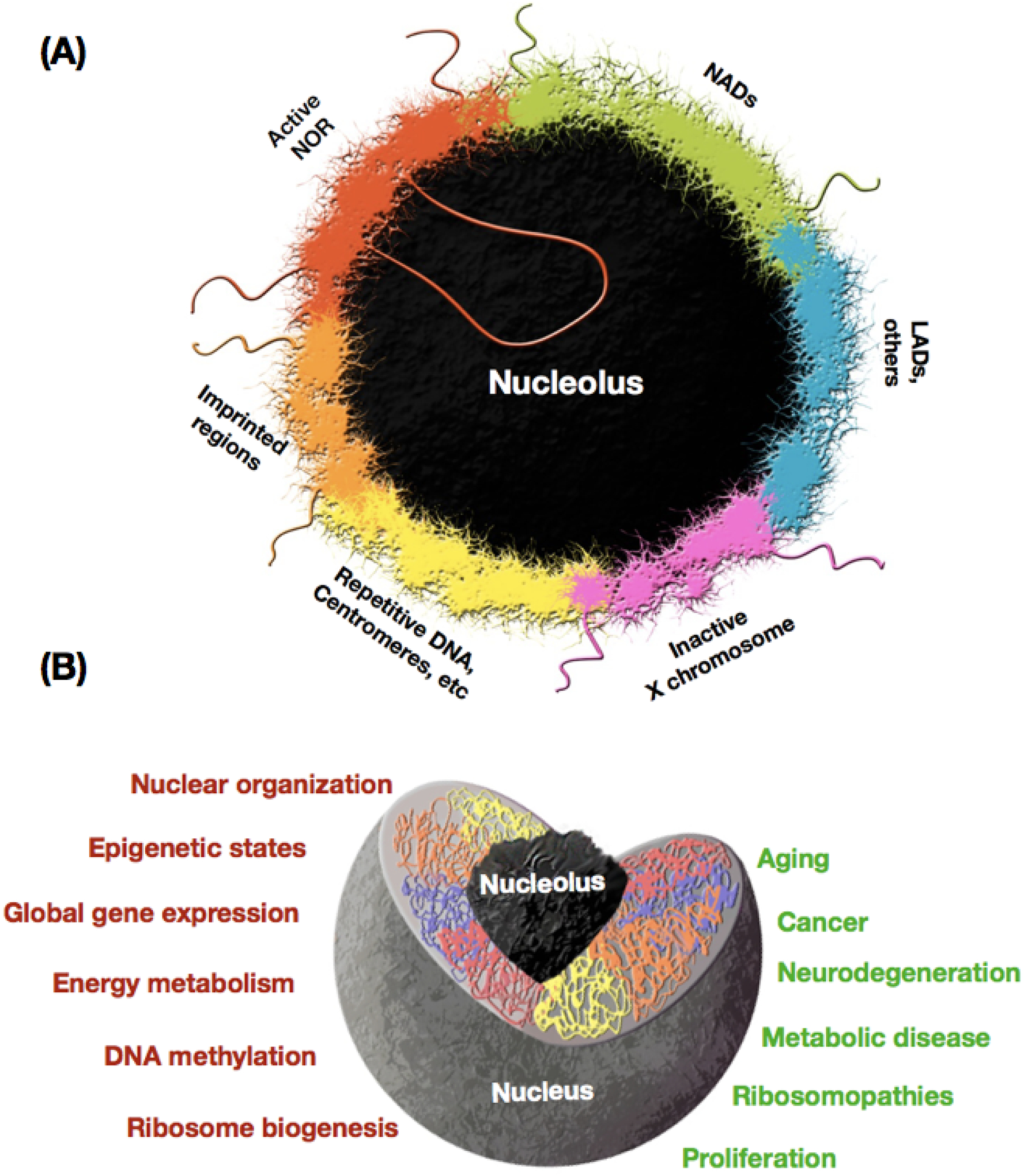
(a) Nucleolus-associated domains (NADs) contain >1,000 Pol II transcribed genes. NADs have a significant overlap with lamina-associated domains (LADs). The inactive X chromosome and some imprinted regions are also localized in the periphery of the nucleolus. Repetitive segments including telomeric and centromeric sequences preferentially associate with the nucleolus and form a layer of condensed heterochromatin around the organelle. Other segments include those harboring developmentally regulated genes and multigenic families (e.g., olfactory receptors) that might use nucleoli proximity to maintain the inactive state of specific members. (b) The nucleolus modulates a variety of biological processes and functional states of the cell, and has been linked to a diverse of organismal and cellular phenotypes.
A recent study analyzed NADs during cell senescence in a fibroblast model [16]. Surprisingly, almost no change in NAD identity was detected [16]. However, satellite repeat cluster in centromeric and pericentromeric regions, which generally localize in perinucleolar heterochromatin, showed impaired interactions with nucleolus. Furthermore, H3K9me3-marked heterochromatin, typical of centromeric and pericentromeric satellite repeat clusters in nucleolus-associated chromatin, was shown to be lost and rearranged in the nucleus. These changes in nucleolar localization of satellite repeats and histone marks are likely responsible for senescence-associated distension of satellites (SADS), which associates with increased transcription of these regions during aging.
The clustering of NAD around the nucleolus emerges from many factors. Research in yeast, for example, have found that repeat elements, which are enriched in the NAD regions [3, 4], interact with ETS2 in the nucleolus, locking the chromosomes in a specific conformation around the organelle [83]. The insulator factor CTCF has also a role in tethering genomic regions to the nucleolus periphery in a nucleophosmin 1 (NLP1)-dependent way [84]. This is further supported by the fact that NLP1 binds centromere regions [85], is enriched in NADs, and that NLP1 depletion triggers nucleolus disruption [86]. Another mechanism suggested is the binding of satellite RNA to centromere-specific proteins, CENPC1 and INCENP, directing them to the nucleolus during interphase and to kinetochore assemblies during mitosis [87]. Some histone modifications, such as H4K20me3, H3K9me3 and H3K27me3 are enriched in NADs and may be a binding site to scaffolding proteins, such as heterochromatin protein 1 (HP1) and Polycomb group (PcG) proteins [88]. It remains unclear, however, if these histone modifications drive NAD association to the nucleolus or emerge as a byproduct of NAD localization in the periphery of the nucleolus.
The typical assumption is that perinucleolar localization induces a certain chromatin state and gene silencing, at least around large segments of heterochromatic NADs [89]. This hypothesis is further corroborated by a recent study in which euchromatin is associated with nuclear speckles, whereas repressive heterochromatin is associated with the nucleolar periphery [90]. Repressive domains are present on rDNA bearing chromosomes of A. thaliana and partially correspond to genomic regions flanking the rDNA arrays [90, 91]. Accordingly, NOR-bearing chromosomes 2 and 4 (NOR2 and NOR4) of A. thaliana are enriched in NADs. In leaf tissues that exclusively transcribe rDNA repeats from chromosome 4, NOR4 is associated with the nucleolus, whereas chromosome 2 rDNA repeats are transcriptionally repressed and excluded from the nucleolus. Induced expression of NOR2-derived rDNA, which is normally inactive, leads to its nucleolar association and global reorganization of the short arm of chromosome 2 inside the nucleus [90]. This is accompanied by lowered expression of chromosome 2 genes [91]. Nevertheless, tRNA localization in the nucleolar periphery is suggested to facilitate tRNA expression [74–76]. This needs to be reconciled with observations that proximity to tRNAs and nucleolar localization inhibits pol II and pol III mediated transcription of certain genes [76]. Similarly, it has been suggested that ribosomal protein gene (RPG) localization proximal to the nucleolus could facilitate coordinated expression of RPGs and rRNAs [6], although nucleolar proximity could also instead be used to inactivate certain RPGs. Thus, genes and NORs with tissue and development specific expression might be regulated through changes in their position relative to the nucleolus. These ideas are in accordance with findings that targeted modification of rDNA copy number impacts gene expression across the whole genome [10].
rDNA Contact Maps and Nuclear Architecture
Proximity-ligation technology such as Hi-C has enabled the analysis of chromatin interactions, providing a clearer understanding of chromosomal organization in the interphase nucleus and its relation to functional states of the cell [92, 93]. Multiple Hi-C datasets were recently assembled to uncover rDNA array interactions with the genome [5, 6]. Accordingly, rDNA contacts preferentially occur at repressed genes, in repetitive or insulator segments, and encompass CTCF binding regions. These observations have also been replicated in 4C data [7] and are concordant with evidence suggesting that the nucleolus and its periphery have a mostly repressive chromatin environment.
Furthermore, interaction between 5S and 45S rDNA arrays were absent in both Hi-C [5, 6] and 4C [7] data despite concerted copy number variation between these two regions [94]. However, there was a considerable overlap between the contact sites of the 5S and 45S rDNA through the genome, probably reflecting some amount of spatial proximity between the arrays. Sites of joint 5S and 45S contact might point to potential mechanism for concerted copy number variation and, possibly, 5S-45S co-regulation [6]. Nevertheless, the observation that direct 5S and 45S rDNA contacts were lacking [5–7] needs to be reconciled with reports of 5S rDNA localization within NADs. On the other hand, Hi-C studies observed a great number of contacts between the rDNA and genes encoding proteins that localize to the mitochondria [6]. Differential expression of mitochondrial protein genes had already been linked with rDNA in fruit-flies [10] and humans [11]. These intriguing observations suggest an evolutionary conserved and physiologically relevant relationship between the rDNA arrays and the mitochondria. The 5S rDNA was also enriched in contact with mitochondrial protein genes and might have an even closer relationship with the mitochondria, with the import of 5S RNAs into the mitochondria [95].
Ribosomal protein genes (RPG) comprised another class with closer than average rDNA proximity, although the magnitude of the shift is rather small [6]. RPG-nucleolus proximity could partially reflect coordination between RPG and rRNA expression in order to produce ribosomal components in a stoichiometric manner. On the other hand, the proximity of some RPGs to the nucleolus could reflect a regulatory mechanism to inactivate specific ribosomal proteins variants. Indeed, localization to the nucleolar periphery appears to be used for the selective inactivation of specific alleles in certain imprinted loci. For instance, the Kcnq1ot long ncRNA is exclusively transcribed from the paternal chromosome and drives the targeting of the paternal locus to the nucleolar periphery, where a 1-Mbp region containing ten protein-coding genes is silenced [96, 97]. Similarly, expression of the Xist long ncRNA drives X chromosome association to the nucleolar periphery and is required to maintain the inactivated X chromosome in a silenced state [98]
Changes in rDNA interactions with the genome during progression to malignancy in a Myc-driven B-cell lymphoma model were documented with 4C-seq [7]. The model is particularly interesting because oncogenic Myc localizes to the nucleolus, physically associates with the rDNA, remodels rDNA chromatin looping structures, induces TTF-1, and activates RNA Pol I transcription as quiescent cells re-enter the cell cycle [99, 100]. The transition from quiescence to cell cycle entry and proliferation is mediated by Myc-dependent attachment of the rDNA to the nuclear matrix via the rDNA non-transcribed intergenic spacer (IGS) region [100]. Matrix-attached rDNA repeats are hypomethylated and presumably active or poised for transcription whereas hypermethylated silenced rDNA repeats are not recruited to the matrix [100]. Interestingly, genes encoding protein components of the ribosomes displayed decreased contact density with the rDNA and increased mRNA expression upon Myc activation and malignancy progression [7].
In summary, while most studies with deep sequencing of nucleoli and 4C/Hi-C proximity ligation are in fairly good agreement regarding key observations, some discrepancies have remained. These differences likely reflect the different methods used (NADs as identified with deep sequencing of nucleoli vs proximity ligation) but might also emerge from differences in the cell type studied, their stage in the cell cycle, or the DNA repair activity of the cell. Continued studies with common protocols and a greater variety of cell types and conditions are bound to help elucidate these discrepancies and link rDNA/nucleolus to specific metabolic states of the cell. Overall, the findings shed light into a crucial component of nuclear organization, reaffirming the role of the nucleolus in modifying and maybe coordinating genomic expression, cellular function, and pathological alterations in human diseases.
rDNA Contact Maps and Human Diseases
Nucleolar dysfunction has been implicated in a variety of human diseases (Figure 4b). In cancer, structural alterations of the nucleolus have long been documented [101]. In addition, many oncogenes and tumor-suppressor genes have been shown to physically interact or directly influence the nucleolus and the rDNA arrays [102–104]. Furthermore, cancer lineages have expanded 5S rDNA and contracted 45S rDNA arrays relative to normal adjacent tissue from the same individual [105, 106]. A smaller rDNA array might in turn enable the release of rDNA bound proteins such as Myc [107], CTCF [67] and histones [67] which could then be more abundantly supplied to other parts of the genome, possibly altering global nuclear organization and cellular function. Thus, genome-wide transcriptional consequences of the rDNA in cancers could be partially exerted by changes in nuclear organization that are mediated by rDNA copy number, as has been suggested in fruit-flies [10] and humans [11]. Indeed, recent research with a Myc-driven B-cell lymphoma model has shown an UBF-mediated increase in the open chromatin state of the rDNA and altered nucleolar interaction with NADs [7]. Genes associated with B-cell differentiation displayed increased interaction with NADs and decreased expression, while genes involved with cell growth and metabolism displayed increased expression. These alterations are compatible with a causal role of rDNA mediated changes in nuclear architecture during carcinogenesis. Because accelerated ribosome biogenesis is presumably necessary to the higher demand for protein synthesis in rapidly proliferating cells, the decrease of rDNA copy number is compensated by upregulation of genes responsible to increased nucleolar activity [106, 108, 109]. Accordingly, the Myc gene, which was shown to be overexpressed in several cancers [110–116], promotes upregulation of rRNAs by mediating the binding of DNA consensus elements to selectivity factor 1 (SL1). This, in turn, recruits Pol I and increases rRNA transcription, leading to cell cycle reentry [107, 117]. Recent evidence further supports the hypothesis of rDNA-mediated changes in nuclear organization [7] as well as increased nucleolar activity in cancers [7] [106], despite rDNA copy number loss [105, 106]. The data suggests that carcinogenesis is accompanied by the conversion of rDNA chromatin to an open state [7], with activation of poised rDNA genes and concomitant changes in nuclear organization and global gene expression.
The nucleolus has also been implicated in neurodegenerative diseases. Nucleolar/rDNA activity is important, for instance, to keep with the demand of high protein synthesis during cell regeneration from previous axon damage and maintenance of synapse plasticity [118]. The inactivation of RNA polymerase I enzyme, responsible for rDNA transcription, has been linked to neurodegeneration [119]. Defects in rDNA transcription due to dysfunctional UBF has been linked to Huntington Disease pathology [120]. Also, neurons from brains affected by either Alzheimer’s disease (AD) or mild cognitive impairment show smaller nucleoli and reduced or defective ribosomes [121]. Likewise, alterations in AIDA-1, a synaptonuclear factor that increases nucleoli number and protein synthesis in an activity-dependent way, results in synaptic defects in mice. Genome-wide studies have linked AIDA-1 variants to neuropsychiatric disorders, such as schizophrenia, bipolar disorders, and AD [122, 123]. Dementia with Lewy bodies (DLB) neurons had increased rDNA CN but stable rDNA CpG methylation in parietal cortex [118]. AD showed increased CN in frontal and parietal cortex, but this time associated with hypermethylation and, therefore, silencing [124]. There is an apparent hypertrophy of nucleoli in old individuals with pathological signatures of AD but asymptomatic for the disease [125]. One hypothesis is that increased nucleolus activity protects from cognitive impairment despite AD pathology [125], possibly explaining why pathological affected areas in AD and Parkinson disease might also display higher rDNA CN, as those would be the remaining positively selected cells [118].
Ribosomopathies might also be influenced by epigenetic states of the nucleolus and the organelle’s impact across the whole genome. Diamond Blackfan anemia is characterized by haploinsufficiency of ribosome proteins, leading to nucleolar stress and activation of the p53 pathway [126]. Treacher-Collins Syndrome is another ribosomopathy. It results from mutation in TOC1, a gene responsible for rRNA processing [127]. Mutations in TOC1 also lead to apoptosis by nucleolar stress via a p53 mechanism [128, 129]. Finally, recent hypotheses link nucleolar dysfunction and autoimmune diseases [130]. Accordingly, stressors during development might lead to nucleolar expansion, altering nuclear architecture and engulfing the inactive X chromosome of females. This process purportedly increases the formation of autoantigens, which generally pass through the nucleolus [131, 132]. Polyamines, typically increased in cell stress, might help stabilize the autoantigens and further contribute to the initiation of an autoimmune response.
Concluding Remarks
The rDNA and the nucleolus have long been viewed as an organelle “merely” responsible for the production of ribosomes and the expression of the vast majority of all RNAs in the cell. While these processes are essential and represent in and by themselves the single major source of energy expenditure by the cell, the rDNA/nucleolus is also increasingly recognized as an organizer hub that coordinates and controls seemingly disparate cellular functions and biological processes (Figure 4). These include processes such as maintenance of genomic integrity, DNA repair and recombination, telomere maintenance, heterochromatin stability, control of repeat element expression, maintenance of X chromosome inactivation, allelic exclusion in imprinting, genome wide transcriptional regulation, modulation of epigenetic states. The nucleolus is also a keystone in nuclear organization, not only providing a port for the anchoring of certain components of heterochromatin and gene silencing but also impacting epigenetic states and gene expression throughout the whole nucleus. The rDNA/nucleolus have also been implicated in cancer, metabolic diseases, progeria, neurodegeneration and aging. All in all, much work and many surprises are bound to lie ahead as investigators connect specific disease etiologies and cellular states to changes in nuclear architecture and epigenetic states of the genome that are driven by the rDNA/nucleolus (see Outstanding Questions).
Outstanding Questions.
What determines the localization of specific nucleolar associated domains to the nuclear periphery? How dynamic is NAD localization and how does it change during development, stress response, aging, across tissues, and in specific diseases? Are NADs evolutionarily conserved?
What determines rDNA-genome and rDNA-gene contacts? How dynamic are these contacts? Do they change during development, stress response, aging, across tissues, and in specific diseases? Are rDNA contacts evolutionarily conserved?
How do specific proteins (e.g., CTCF) organize NAD structure and localization as well as rDNA contacts with the rest of the genome.
How does proximity with the nucleolus affect transcriptional and epigenetic states of specific genes and segments of the genome? What are the mechanisms through which the rDNA/nucleolus impact global chromosomal organization and chromatin states in the nucleus?
Highlights.
The rDNA is the basis for nucleolus biogenesis, organization, and function; the rDNA is evolutionarily ultra-conserved yet displays extensive copy number variation across individuals.
The nucleolus is a dynamic non-membrane bound nuclear organelle that displays structural diversity during the cell cycle, DNA repair, and under stress.
The rDNA/nucleolus are coordinating centers for cellular functions (e.g., heterochromatin maintenance, metabolism) that go well beyond those of ribosome biogenesis.
The rDNA/nucleolus as organizers of chromatin territories, exert influence over X-chromosome inactivation, imprinting, genome-wide gene expression, and epigenetic states of the genome.
The rDNA/nucleolus are re-emerging as crucial determinants underlying or modulating cancer, neurodegeneration, and aging.
Acknowledgements
We thank the many investigators who have contributed to our current understanding of rDNA/nucleolar biology. We apologize that we could not ever do justice and acknowledge all relevant work due to space constraints. We also thank Amina Bedrat for critical comments and feedback on an earlier version of the manuscript. Work in the Lemos lab has been partially supported by a NIGMS grant R01GM122088; and NIEHS grant R01ES027981. Amanda V. Cerqueira is partially supported by the Leman program.
References
- 1.Warner JR, The economics of ribosome biosynthesis in yeast. Trends Biochem Sci, 1999. 24(11): p. 437–40. [DOI] [PubMed] [Google Scholar]
- 2.Lewis JD and Tollervey D, Like attracts like: getting RNA processing together in the nucleus. Science, 2000. 288(5470): p. 1385–9. [DOI] [PubMed] [Google Scholar]
- 3.van Koningsbruggen S, et al. , High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell, 2010. 21(21): p. 3735–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Németh A, et al. , Initial genomics of the human nucleolus. PLoS Genet, 2010. 6(3): p. e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu S and Lemos B, A Portrait of Ribosomal DNA Contacts with Hi-C Reveals 5S and 45S rDNA Anchoring Points in the Folded Human Genome. Genome Biol Evol, 2016. 8(11): p. 3545–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu S and Lemos B, The long-range interaction map of ribosomal DNA arrays. PLoS Genet, 2018. 14(3): p. e1007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diesch J, et al. , Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation. Commun Biol, 2019. 2: p. 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi T, A new role of the rDNA and nucleolus in the nucleus--rDNA instability maintains genome integrity. Bioessays, 2008. 30(3): p. 267–72. [DOI] [PubMed] [Google Scholar]
- 9.Ide S, et al. , Abundance of ribosomal RNA gene copies maintains genome integrity. Science, 2010. 327: p. 693–696. [DOI] [PubMed] [Google Scholar]
- 10.Paredes S, et al. , Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet, 2011. 7(4): p. e1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons JG, et al. , Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat Commun, 2014. 5: p. 4850. [DOI] [PubMed] [Google Scholar]
- 12.Michel AH, et al. , Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev, 2005. 19(10): p. 1199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M and Lemos B, Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res, 2019. 29(3): p. 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiku V, et al. , Small nucleoli are a cellular hallmark of longevity. Nat Commun, 2017. 8: p. 16083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchwalter A and Hetzer MW, Nucleolar expansion and elevated protein translation in premature aging. Nat Commun, 2017. 8(1): p. 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillinger S, Straub T, and Németh A, Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. PLoS One, 2017. 12(6): p. e0178821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindström MS, et al. , Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene, 2018. 37(18): p. 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schöfer C and Weipoltshammer K, Nucleolus and chromatin. Histochem Cell Biol, 2018. 150(3): p. 209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley R, et al. , Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci U S A, 2016. 113(35): p. 9882–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia S and Kovařík A, Dancing together and separate again: gymnosperms exhibit frequent changes of fundamental 5S and 35S rRNA gene (rDNA) organisation. Heredity (Edinb), 2013. 111(1): p. 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manuelidis L and Borden J, Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridization and three-dimensional reconstruction. Chromosoma, 1988. 96(6): p. 397–410. [DOI] [PubMed] [Google Scholar]
- 22.Thiry M and Thiry-Blaise L, In situ hybridization at the electron microscope level: an improved method for precise localization of ribosomal DNA and RNA. Eur J Cell Biol, 1989. 50(1): p. 235–43. [PubMed] [Google Scholar]
- 23.Wachtler F, et al. , Human ribosomal RNA gene repeats are localized in the dense fibrillar component of nucleoli: light and electron microscopic in situ hybridization in human Sertoli cells. Exp Cell Res, 1992. 198(1): p. 135–43. [DOI] [PubMed] [Google Scholar]
- 24.Wachtler F, et al. , On the position of nucleolus organizer regions (NORs) in interphase nuclei. Studies with a new, non-autoradiographic in situ hybridization method. Exp Cell Res, 1986. 167(1): p. 227–40. [DOI] [PubMed] [Google Scholar]
- 25.Leitch AR, et al. , Different patterns of rDNA organization at interphase in nuclei of wheat and rye. J Cell Sci, 1992. 101 (Pt 4): p. 751–7. [DOI] [PubMed] [Google Scholar]
- 26.Weipoltshammer K, et al. , Intranuclear anchoring of repetitive DNA sequences: centromeres, telomeres, and ribosomal DNA. J Cell Biol, 1999. 147(7): p. 1409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koberna K, et al. , Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J Cell Biol, 2002. 157(5): p. 743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raska I, et al. , Does the synthesis of ribosomal RNA take place within nucleolar fibrillar centers or dense fibrillar components? A critical appraisal. J Struct Biol, 1995. 114(1): p. 1–22. [DOI] [PubMed] [Google Scholar]
- 29.Scheer U and Rose KM, Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc Natl Acad Sci U S A, 1984. 81(5): p. 1431–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hozák P, et al. , Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J Cell Sci, 1994. 107 (Pt 2): p. 639–48. [DOI] [PubMed] [Google Scholar]
- 31.Ochs RL, et al. , Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell, 1985. 54(2): p. 123–33. [DOI] [PubMed] [Google Scholar]
- 32.Jansen RP, et al. , Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol, 1991. 113(4): p. 715–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobol M, et al. , UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus, 2013. 4(6): p. 478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Németh A and Grummt I, Dynamic regulation of nucleolar architecture. Curr Opin Cell Biol, 2018. 52: p. 105–111. [DOI] [PubMed] [Google Scholar]
- 35.Pendle AF, et al. , Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell, 2005. 16(1): p. 260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen JS, et al. , Nucleolar proteome dynamics. Nature, 2005. 433(7021): p. 77–83. [DOI] [PubMed] [Google Scholar]
- 37.Andersen JS, et al. , Directed proteomic analysis of the human nucleolus. Curr Biol, 2002. 12(1): p. 1–11. [DOI] [PubMed] [Google Scholar]
- 38.Ueshima S, Nagata K, and Okuwaki M, Internal associations of the acidic region of upstream binding factor control its nucleolar localization. Molecular and Cellular Biology, 2017. 37(22): p. e00218–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen D and Huang S, Nucleolar Components Involved in Ribosome Biogenesis Cycle between the Nucleolus and Nucleoplasm in Interphase Cells. The Journal of Cell Biology, 2001. 153(1): p. 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dousset T, et al. , Initiation of Nucleolar Assembly Is Independent of RNA Polymerase I Transcription. Molecular Biology of the Cell, 2017. 11(8): p. 2513–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mais C, et al. , UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev, 2005. 19(1): p. 50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Floutsakou I, et al. , The shared genomic architecture of human nucleolar organizer regions. Genome Res, 2013. 23(12): p. 2003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grob A, Colleran C, and McStay B, Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev, 2014. 28(3): p. 220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karpen GH, Schaefer JE, and Laird CD, A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev, 1988. 2(12B): p. 1745–63. [DOI] [PubMed] [Google Scholar]
- 45.Kalmárová M, et al. , Positioning of NORs and NOR-bearing chromosomes in relation to nucleoli. J Struct Biol, 2007. 160(1): p. 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warmerdam DO, van den Berg J, and Medema RH, Breaks in the 45S rDNA Lead to Recombination-Mediated Loss of Repeats. Cell Rep, 2016. 14(11): p. 2519–27. [DOI] [PubMed] [Google Scholar]
- 47.Tchelidze P, et al. , Nucleolar sub-compartments in motion during rRNA synthesis inhibition: Contraction of nucleolar condensed chromatin and gathering of fibrillar centers are concomitant. PLoS ONE, 2017. 2(11): p. e0187977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smirnov E, et al. , Reproduction of the FC/DFC units in nucleoli. Nucleus, 2016. 7(2): p. 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dousset T, et al. , Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell, 2000. 11(8): p. 2705–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hutten S, et al. , An intranucleolar body associated with rDNA. Chromosoma, 2011. 120(5): p. 481–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wild T, et al. , A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol, 2010. 8(10): p. e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kruhlak M, et al. , The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature, 2007. 447(7145): p. 730–4. [DOI] [PubMed] [Google Scholar]
- 53.Harding SM, Boiarsky JA, and Greenberg RA, ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition. Cell Rep, 2015. 13(2): p. 251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shav-Tal Y, et al. , Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell, 2005. 16(5): p. 2395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Sluis M and McStay B, Nucleolar reorganization in response to rDNA damage. Curr Opin Cell Biol, 2017. 46: p. 81–86. [DOI] [PubMed] [Google Scholar]
- 56.Xie W, et al. , The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci U S A, 2012. 109(21): p. 8161–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salifou K, et al. , The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nature Communications, 2016. 7:10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McStay B, Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev, 2016. 30(14): p. 1598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santoro R, Li J, and Grummt I, The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet, 2002. 32(3): p. 393–6. [DOI] [PubMed] [Google Scholar]
- 60.Masiello I and Biggiogera M, Ultrastructural localization of 5-methylcytosine on DNA and RNA. Cell Mol Life Sci, 2017. 74(16): p. 3057–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Espada J, et al. , Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res, 2007. 35(7): p. 2191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen T, Tsujimoto N, and Li E, The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol, 2004. 24(20): p. 9048–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitz KM, et al. , Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev, 2010. 24(20): p. 2264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips JE and Corces VG, CTCF: master weaver of the genome. Cell, 2009. 137(7): p. 1194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van de Nobelen S, et al. , CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin, 2010. 3(1): p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herdman C, et al. , A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genetics, 2017. 13(7):e1006899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zentner GE, et al. , Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res, 2011. 39(12): p. 4949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang K, et al. , Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J Biol Chem, 2013. 288(36): p. 26067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parelho V, et al. , Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell, 2008. 132(3): p. 422–33. [DOI] [PubMed] [Google Scholar]
- 70.de Wit E, et al. , CTCF Binding Polarity Determines Chromatin Looping. Mol Cell, 2015. 60(4): p. 676–84. [DOI] [PubMed] [Google Scholar]
- 71.Guo Y, et al. , CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci U S A, 2012. 109(51): p. 21081–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kagey MH, et al. , Mediator and cohesin connect gene expression and chromatin architecture. Nature, 2010. 467(7314): p. 430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stahl A, et al. , Chromosomal constitution of nucleolus-associated chromatin in man. Hum Genet, 1976. 35(1): p. 27–34. [DOI] [PubMed] [Google Scholar]
- 74.Bertrand E, et al. , Nucleolar localization of early tRNA processing. Genes Dev, 1998. 12(16): p. 2463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson M, et al. , Nucleolar clustering of dispersed tRNA genes. Science, 2003. 302(5649): p. 1399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L, et al. , Silencing near tRNA genes requires nucleolar localization. J Biol Chem, 2005. 280(10): p. 8637–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pontvianne F, et al. , Identification of Nucleolus-Associated Chromatin Domains Reveals a Role for the Nucleolus in 3D Organization of the A. thaliana Genome. Cell Rep, 2016. 16(6): p. 1574–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ragoczy T, et al. , Functional redundancy in the nuclear compartmentalization of the late-replicating genome. Nucleus, 2014. 5(6): p. 626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shimi T, et al. , The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev, 2008. 22(24): p. 3409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guelen L, et al. , Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature, 2008. 453(7197): p. 948–51. [DOI] [PubMed] [Google Scholar]
- 81.Sen Gupta A and Sengupta K, Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function. Mol Cell Biol, 2017. 37(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Steensel B and Belmont AS, Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell, 2017. 169(5): p. 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Sullivan JM, et al. , Repeated elements coordinate the spatial organization of the yeast genome. Yeast, 2009. 26(2): p. 125–38. [DOI] [PubMed] [Google Scholar]
- 84.Yusufzai TM, et al. , CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell, 2004. 13(2): p. 291–8. [DOI] [PubMed] [Google Scholar]
- 85.Foltz DR, et al. , The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol, 2006. 8(5): p. 458–69. [DOI] [PubMed] [Google Scholar]
- 86.Holmberg Olausson K, Nistér M, and Lindström MS, Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription. J Biol Chem, 2014. 289(50): p. 34601–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wong LH, et al. , Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res, 2007. 17(8): p. 1146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Németh A and Längst G, Genome organization in and around the nucleolus. Trends Genet, 2011. 27(4): p. 149–56. [DOI] [PubMed] [Google Scholar]
- 89.Nemeth A and Langst G, Genome organization in and around the nucleolus. Trends Genetics, 2011. 27: p. 149–156. [DOI] [PubMed] [Google Scholar]
- 90.Quinodoz SA, et al. , Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell, 2018. 174(3): p. 744–757.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Picart-Picolo A, Picault N, and Pontvianne F, Ribosomal RNA genes shape chromatin domains associating with the nucleolus. Nucleus, 2019. 10(1): p. 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lieberman-Aiden E, et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 2009. 326(5950): p. 289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Berkum NL, et al. , Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp, 2010(39). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gibbons JG, et al. , Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci U S A, 2015. 112(8): p. 2485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smirnov A, et al. , Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev, 2011. 25(12): p. 1289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mohammad F, et al. , Kcnq1ot1 / Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Molecular and Cellular Biology, 2008. 28: p. 3713–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fedoriw A, et al. , Differentiation-Driven Nucleolar Association of the Mouse Imprinted Kcnq1 Locus. G3: GENES, GENOMES, GENETICS, 2012. 2(12): p. 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang L, Huynh K, and Lee J, Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell, 2007. 129(4): p. 693–706. [DOI] [PubMed] [Google Scholar]
- 99.Shiue C, Berkson R, and Wright A, c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene, 2009. 28(16): p. 1833–1842. [DOI] [PubMed] [Google Scholar]
- 100.Shiue C, Nematollahi-Mahani A, and Wright A, Myc-induced anchorage of the rDNA IGS region to nucleolar matrix modulates growth-stimulated changes in higher-order rDNA architecture. Nucleic Acids Research, 2014. 42(9): p. 5505–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pianese G, Beitrag zur histologie und aetiologie des carcinoms. Vol. 1. 1896: G. Fischer. [Google Scholar]
- 102.Hannan KM, et al. , Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene, 2000. 19(43): p. 4988–99. [DOI] [PubMed] [Google Scholar]
- 103.Zhai W and Comai L, Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol, 2000. 20(16): p. 5930–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boon K, et al. , N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J, 2001. 20(6): p. 1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu B, et al. , Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet, 2017. 13(6): p. e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang M and Lemos B, Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet, 2017. 13(9): p. e1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grandori C, et al. , c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol, 2005. 7(3): p. 311–8. [DOI] [PubMed] [Google Scholar]
- 108.Pogue-Geile K, et al. , Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol, 1991. 11(8): p. 3842–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miluzio A, et al. , Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell, 2011. 19(6): p. 765–75. [DOI] [PubMed] [Google Scholar]
- 110.Brodeur GM, et al. , Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science, 1984. 224(4653): p. 1121–4. [DOI] [PubMed] [Google Scholar]
- 111.Baker VV, et al. , c-myc amplification in ovarian cancer. Gynecol Oncol, 1990. 38(3): p. 340–2. [DOI] [PubMed] [Google Scholar]
- 112.Kozma L, et al. , Investigation of c-myc and K-ras amplification in renal clear cell adenocarcinoma. Cancer Lett, 1997. 111(1–2): p. 127–31. [DOI] [PubMed] [Google Scholar]
- 113.Herms JW, et al. , c-myc oncogene family expression in glioblastoma and survival. Surg Neurol, 1999. 51(5): p. 536–42. [DOI] [PubMed] [Google Scholar]
- 114.Parrella P, et al. , Detection of c-myc amplification in uveal melanoma by fluorescent in situ hybridization. Invest Ophthalmol Vis Sci, 2001. 42(8): p. 1679–84. [PubMed] [Google Scholar]
- 115.Bitzer M, et al. , C-myc gene amplification in different stages of oesophageal squamous cell carcinoma: prognostic value in relation to treatment modality. Anticancer Res, 2003. 23(2B): p. 1489–93. [PubMed] [Google Scholar]
- 116.Abba MC, et al. , The c-myc activation in cervical carcinomas and HPV 16 infections. Mutat Res, 2004. 557(2): p. 151–8. [DOI] [PubMed] [Google Scholar]
- 117.Arabi A, et al. , c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol, 2005. 7(3): p. 303–10. [DOI] [PubMed] [Google Scholar]
- 118.Hallgren J, et al. , Neurodegeneration-associated instability of ribosomal DNA. Biochim Biophys Acta, 2014. 1842(6): p. 860–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Parlato R, et al. , Activation of an endogenous suicide response after perturbation of rRNA synthesis leads to neurodegeneration in mice. J Neurosci, 2008. 28(48): p. 12759–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee J, et al. , Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington’s disease. Cell Death Differ, 2011. 18(11): p. 1726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hetman M and Pietrzak M, Emerging roles of the neuronal nucleolus. Trends Neurosci, 2012. 35(5): p. 305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jordan BA, et al. , Activity-dependent AIDA-1 nuclear signaling regulates nucleolar numbers and protein synthesis in neurons. Nat Neurosci, 2007. 10(4): p. 427–35. [DOI] [PubMed] [Google Scholar]
- 123.Parra-Damas A and Saura CA, Synapse-to-Nucleus Signaling in Neurodegenerative and Neuropsychiatric Disorders. Biol Psychiatry, 2019. [DOI] [PubMed] [Google Scholar]
- 124.Pietrzak M, et al. , Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS One, 2011. 6(7): p. e22585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Iacono D, et al. , The Nun study: clinically silent AD, neuronal hypertrophy, and linguistic skills in early life. Neurology, 2009. 73(9): p. 665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellis SR, Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim Biophys Acta, 2014. 1842(6): p. 765–8. [DOI] [PubMed] [Google Scholar]
- 127.Gonzales B, et al. , The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum Mol Genet, 2005. 14(14): p. 2035–43. [DOI] [PubMed] [Google Scholar]
- 128.Dixon J, et al. , Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci U S A, 2006. 103(36): p. 13403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jones NC, et al. , Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med, 2008. 14(2): p. 125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brooks WH and Renaudineau Y, Epigenetics and autoimmune diseases: the X chromosome-nucleolus nexus. Front Genet, 2015. 6: p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ochs R and Press R, Centromere autoantigens are associated with the nucleolus. Exp Cell Res, 1992. 200(2): p. 339–350. [DOI] [PubMed] [Google Scholar]
- 132.Cai Y, et al. , Broad Susceptibility of Nucleolar Proteins and Autoantigens to Complement C1 Protease Degradation. Journal of Immunology, 2017. 199(12): p. 3981–3990. [DOI] [PubMed] [Google Scholar]


