Abstract
Since its first discovery in December 2019, the global coronavirus disease 2019 (COVID-19) pandemic caused by the novel coronavirus (SARS-CoV-2) has been posing a serious threat to human life and health. Diagnostic testing is critical for the control and management of the COVID-19 pandemic. In particular, diagnostic testing at the point of care (POC) has been widely accepted as part of the post restriction COVID-19 control strategy. Lateral flow assay (LFA) is a popular POC diagnostic platform that plays an important role in controlling the COVID-19 pandemic in industrialized countries and resource-limited settings. Numerous pioneering studies on the design and development of diverse LFA-based diagnostic technologies for the rapid diagnosis of COVID-19 have been done and reported by researchers. Hundreds of LFA-based diagnostic prototypes have sprung up, some of which have been developed into commercial test kits for the rapid diagnosis of COVID-19. In this review, we summarize the crucial role of rapid diagnostic tests using LFA in targeting SARS-CoV-2-specific RNA, antibodies, antigens, and whole virus. Then, we discuss the design principle and working mechanisms of these available LFA methods, emphasizing their clinical diagnostic efficiency. Ultimately, we elaborate the challenges of current LFA diagnostics for COVID-19 and highlight the need for continuous improvement in rapid diagnostic tests.
Keywords: COVID-19, SARS-CoV-2, Lateral flow assay, Immunoassay, Rapid diagnostic test
Graphical abstract
1. Introduction
Since it was first reported in December 2019, the coronavirus disease 2019 (COVID-19) caused by the novel coronavirus (SARS-CoV-2) has rapidly evolved into a pandemic because of its high person-to-person transmission [1,2]. As of April 28, 2021, over 148 million COVID-19 cases and over 3.1 million deaths were confirmed in 223 countries and territories. Despite various emergency measures taken by many countries, the spread of the SARS-CoV-2 virus remains uncontrolled in many countries and regions due to the emergence of SARS-CoV-2 mutants [3]. Currently, although some specific drugs or vaccines can treat or prevent SARS-CoV-2 infection, developing technologies, such as rapid diagnostic tests, vaccine, specific medicine for infections, and intelligent tracking strategies, for managing the spread of the virus and reducing community transmission has drawn increasing interest in the scientific community [4,5]. In particular, large-scale diagnostic testing is critical in containing the COVID-19 epidemic [6]. In this content, simple, convenient, rapid, sensitive, and accurate detection techniques that can target SARS-CoV-2 must be developed urgently [7]. In most countries, real-time reverse transcriptase quantitative polymerase chain reaction (rRT-qPCR), which detects SARS-CoV-2 RNA, is used as the gold standard for COVID-19 diagnosis [8,9]. However, obtaining the test results of rRT-qPCR is time consuming (>24 h) [10,11]. In addition, rRT-qPCR requires specialized equipment, certified molecular testing laboratories with biosafety level 3, and skilled technicians and has a high false-negative rate of up to 30%. Therefore, a suitable detection system that can be used for SARS-CoV-2 detection at the point of care (POC) is important for the prevention and control of the COVID-19 pandemic.
Despite the strong demand for the POC diagnostic testing in managing COVID-19, developers must consider and evaluate multiple parameters to provide an easy-to-use device with optimal performance or market readiness [12,13]. Inspired by the idea of “old drug in new use,” a number of well-developed and widely used techniques have been employed for SARS-CoV-2 detection. Typical representatives of these analytical technologies include enzyme-linked immunosorbent assay (ELISA) [14], lateral flow assay (LFA) [15], biosensors, and isothermal amplification-assisted nucleic acid test [16,17]. LFA has obtained a great success in COVID-19 diagnosis because it well matches the ASSURED criteria of the WHO for POC testing [18]. However, the success of LFA lies in its general design that has remained almost unchanged since their first use as pregnancy test in the 1970s [19,20]. Although almost all LFA strips depend on the capillary forces to move the sample along a test strip to generate a measurable signal, its fabrication is far from being straightforward. As an “old” and well-known technology, many online resources and guidelines have been provided by public and private entities [21].
As a complement to the current rRT-qPCR assay, LFA-based rapid diagnostic tests have recently attracted extensive attention in curbing the spread and resurgence of COVID-19 since they can quickly identify new COVID-19 infections by analyzing specific biomarkers of SARS-CoV-2, such as nucleic acids, antibodies and antigens [22]. Currently, numerous rapid diagnostic technologies based on LFA have been developed, part of which have been developed as rapid test kits for detecting SARS-CoV-2 and have played a big role in managing the COVID-19, especially in resource-limited settings with the low accessibility of rRT-qPCR assay [23,24]. Given the crucial role of LFA in assisting COVID-19 diagnostics, it is of importance to be knowledgeable regarding the status and further prospect of the LFA platform for POC diagnostics of COVID-19. Although there have been a large number of publications about the rapid diagnostics tests of COVID-19, they only address some of the key concerns regarding molecular [11,[25], [26], [27]] and serological diagnostic techniques [[28], [29], [30]] and biosensing strategies [[31], [32], [33], [34]] that have arisen in responding to the recent COVID-19 pandemic [35]. To our best knowledge, a review paper focusing on the critical role of LFA-assisted rapid diagnostic tests in managing COVID-19 remains missing. Herein, we provide the first systematic and comprehensive review of emerging rapid diagnostic tests involving LFA techniques, and their application potential to improve COVID-19 diagnosis by targeting SARS-CoV-2-specific viral RNA, antibodies, antigens, and whole viruses (Scheme 1 ). The key insights of the reported LFA strategies on the design principle, working mechanism, methodological performance, and clinical diagnostic efficiency are detailedly discussed. Additionally, the status of commercially available LFA test kits for SARS-CoV-2 detection is further summarized. The ending of this review will be organized to elaborate the challenges and future improvements in COVID-19 diagnosis.
Scheme 1.
Schematic representation for the POC diagnostics of COVID-19 powered by LFA.
2. LFA for viral nucleic acid detection
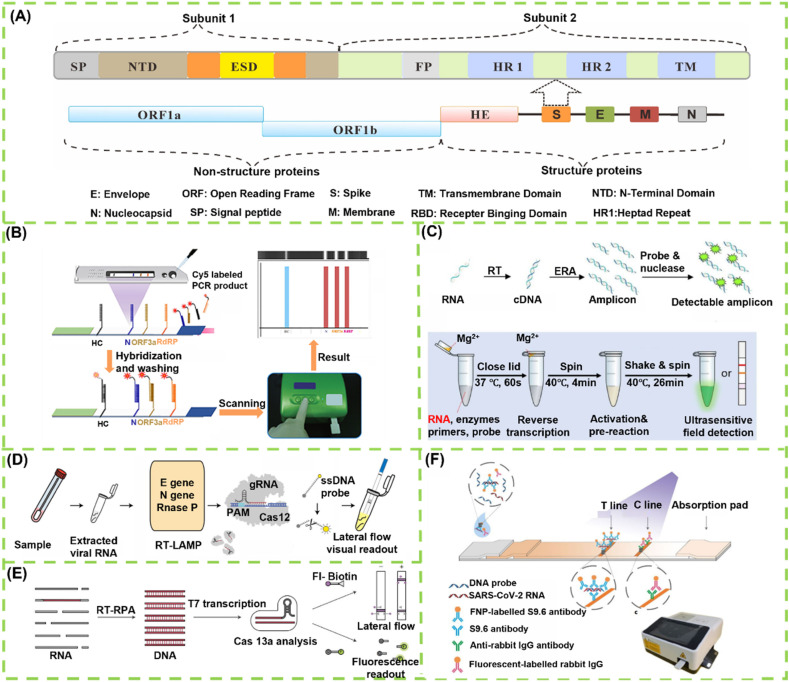
SARS-CoV-2 is a single-stranded positive-sense RNA (ssRNA) virus, and its genetic makeup contains ∼29.9 nucleotides with the gene order of ORF1a/b, spike (S), envelope (E), membrane (M), and nucleocapsid (N) in the 5ʹ to 3ʹ direction (Fig. 1 A) [36,37]. The availability of SARS-CoV-2 genetic materials and the deep understanding of their biological functions have provided potential opportunities for researchers to develop various nucleic acid-based detection techniques [38,39]. At present, real-time RT-qPCR, which involves the reverse transcription of RNA and the amplification of specific complementary DNA (cDNA) fragments, is the most widely used strategy for COVID-19 diagnosis. This method detects SARS-CoV-2 RNA and has become the gold standard of clinical COVID-19 diagnosis because it can provide quantitative information on viral loads in a highly sensitive and selective manner [40]. However, the accessibility of rRT-qPCR in less-developed or remote regions largely limits its broad appeal. In addition, this approach requires long testing time to give result feedback, specialized laboratories, skilled technicians and expensive equipment [41,42]. Therefore, developing a series of simple and rapid POC nucleic acid testing techniques is highly desired. The ability of LFA to sense nucleic acids in a rapid and portable format has important applications ranging from field detection to clinical diagnostics [43]. However, the SARS-CoV-2 concentration in specimen is very low and hard to be directly detected using LFA [44]. Therefore, the integration of nucleic acid amplification technologies, such as RT-PCR [45], isothermal amplification [46], and clustered regularly interspaced short palindromic repeats (CRISPR) [47], with LFA is a promising strategy to simplify the traditional molecular test, wherein LFA can allow the direct visual interpretation of nucleic acid amplification results [48]. Given their considerable advantages of portability, rapidity, cost effectiveness, and user friendliness, advanced nucleic acid amplification-assisted LFA techniques for detecting viral RNA against SARS-CoV-2 have been developed recently [49]. Except for nucleic acid amplification, introducing signal amplification strategies is another common approach to improve the nucleic acid detection ability of LFA [50]. When these technologies are integrated into a system, the COVID-19 diagnosis of an unknown sample can be achieved with high sensitivity and selectivity within a short time. In this section, we briefly discuss several specific examples wherein nucleic acid/signal amplification strategies were exploited to enhance LFA test, facilitating the diagnosis of SARS-CoV-2 infection (Table 1 ).
Fig. 1.
LFA for detection of viral RNA: (A) The genome information of SARS-CoV-2 and its corresponding protein structure [36]; (B) Working principle and flow of RT-PCR-enhanced LFA sensor for multiple-detection of N, ORF3a, and RdRP gene to diagnose SARS-CoV-2 [53]; (C) The work principle of RT-RPA combined LFA in detecting SARS-CoV-2 [38]; (D) Schematic of SARS-CoV-2 DETECTR workflow, including conventional RNA extraction, RT-LAMP preamplification, CRISPR-Cas12-based enhancement for E gene, N gene and RNase P, and LFA-based visual detection [48]; (E) RT-RPA and CRISPR co-enhanced SHERLOCK detector in detecting SARS-CoV-2 [79]; (F) The principle of S9.6 antibody-based signal amplification for amplifying LFA [50].
Table 1.
Representative LFAs for the detection of viral genes.
| Target gene | Sample | Probe | Enhancement strategy | LOD | Sensitivity | Specificity | Ref. |
|---|---|---|---|---|---|---|---|
| RdRp, ORF3a, and N genes | Nasopharyngeal swabs and sputum | Cy5 | PCR | 10 copies/test | 100% | 99% | [53] |
| ORF1ab and N genes | Oropharynx swab | FITC | LAMP | 12 copies/test | 100% | 100% | [62] |
| N and S genes | Nasopharyngeal swabs | FAM | RPA | 1 ag | / | / | [170] |
| RdRp, ORF1b and ORF1ab genes | Nasopharyngeal swabs and saliva | FAM | RPA-CRISPR | 10 copies/test | / | / | [171] |
| N gene | Nasopharyngeal and throat swab | AuNP | RPA-CRISPR | 100 copies/test | 90% | 95% | [79] |
| N gene | Nasopharyngeal swabs | / | LAMP-CRISPR | / | 94.9% | / | [172] |
| E and N gene | Oropharyngeal swabs | / | LAMP-CRISPR | 10 copies μL−1 | / | / | [48] |
| S, N and Orf1ab genes | Nasopharyngeal swab | AuNP | RPA-CRISPR | 42 copies/test | 97.1% | 100% | [49] |
| N gene | Nasopharyngeal swab | / | RPA-CRISPR | 10 copies mL−1 | [173] | ||
| N and E gene | Nasopharyngeal swabs | / | LAMP-CRISPR | 10 copies/test | 86% | 100% | [39] |
| E, N and ORF1ab gene | Throat swabs | FNP | Signal amplification | / | 100% | 99% | [50] |
2.1. LFA combined with RT-PCR amplification
In a typical RT–qPCR assay for SARS-CoV-2, the reverse transcription (RT) reaction can convert viral RNA to complementary DNA (cDNA) in the presence of reverse transcriptase [51]. The cDNA is then exponentially amplified by PCR, and the obtained DNA product is analyzed in real time using fluorescent probes. Although RT–qPCR provides adequate sensitivity to enable the early diagnosis of infection, the requirement of expensive instruments, skilled technicians, thermal cycler, and the high test cost poses challenges in POC applications [52]. As an alternative to traditional RT-qPCR, Yu et al. developed a multiplex LFA for the simultaneous detection of three genomic regions of SARS-CoV-2, namely RdRp, ORF3a, and N genes within 30 min using the PCR product obtained by the single-tube RT-PCR. Fig. 1B shows the working principle and analytical procedure of the developed LFA, wherein three complementary sequences to the RdRp, ORF3a, and N genes and one hybridization control (HC) sequence are immobilized on the 9G membrane as test lines and control line, respectively [53]. The prepared LFA could directly detect the Cy5-labeled RT-PCR products. COVID-19 negative is confirmed if LFA shows the Cy5 fluorescence signals only for the HC probe, whereas COVID-19 positive is identified if any one of the probes, two probes, or all three probes show Cy5 fluorescence signals. Through combining RT-PCR amplification, the developed LFA allows a highly sensitive detection of SARS-CoV-2 with an LOD of 10 copies per test for each gene. The clinical diagnostic efficacy of this LFA method for the detection of SARS-CoV-2 was further evaluated using 162 clinical samples, and the detection results were compared with those obtained by the commercial assay. Results show that the percent positive agreement, percent negative agreement, and overall percent agreement of this LFA can reach 100% (94.2–100%), 99.0% (94.6–100%), and 99.4% (96.6–100%), respectively, indicating high consistency with the commercial method for the test of SARS-CoV-2 in clinical specimens. Although LFA can achieve the rapid detection of SARS-CoV-2 within 12 min after a PCR, the total time for completing the viral detection still takes approximately 2 h (100 min PCR process and 12 min LFA test) after extraction. In addition, the simultaneous detection by targeting more than one gene can effectively avoid the false-negative results encountered by conventional RT-qPCR, but the presented approach in this work still requires a PCR machine to amplify the genomic copies, greatly constraining its potential for POC use [54].
2.2. LFA integrated with isothermal amplification
Although RT-PCR is the gold standard for detecting viral RNA, its requirement of thermal cycling, long turnaround time, and complex operation limits its applications for the POC diagnostics of COVID-19 patients [46,55]. Isothermal nucleic acid amplification is a simple process that can rapidly amplify small amounts of nucleic acid at a constant temperature [56,57]. In addition, it has been developed as an alternative strategy to PCR with the characteristics of high specificity, high sensitivity, convenience, and low cost. Currently available isothermal amplification techniques mainly include loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), rolling circle amplification, and strand displacement amplification [56,[58], [59], [60]]. The application potentials of these techniques for improving the detection of various targets, such as DNA, RNA, proteins, small molecules, metal ions, and cells, have been well demonstrated [61]. Without the need of a thermal cycler, these isothermal nucleic acid amplification strategies are amenable for POC applications. The incorporation of isothermal amplification methods into a portable LFA contributes to achieving on-site test of viral RNA with high sensitivity. Recently, several enhanced LFA approaches by isothermal amplification have been developed to manage COVID-19 [62,63]. Xia et al. reported an ultrasensitive field-deployable method (RT-RPA-LFA) to detect SARS-CoV-2 gene by employing RT–enzymatic recombinase amplification (RT–ERA), a modified RPA introduced by GenDx Biotech (Fig. 1C). In their study, a multienzyme RT–ERA reaction system comprising ribonuclease (RNase) inhibitor, reverse transcriptase, recombinase, polymerase, single-stranded DNA-binding protein, creatine kinase, and nuclease was designed and employed to amplify and detect the target RNA [38]. To achieve the field or even household-deployable detection, the authors further designed a nfo-affinity probe system for viral RNA test through coupling with LFA, wherein a pair of nfo forward and reward primers was applied to amplify an amplicon within the N gene and the amplified product was detected by a nfo-affinity probe. The visual LOD of this approach is as low as 1 ag of the N-gene RNA. Similarly, several improved RT-RPA-amplified LFA sensors have been reported for the rapid, sensitive, specific, and direct visual detection of the SARS-CoV-2 N and S genes in recent years [64].
As the most popular isothermal amplification technique, LAMP has been widely used to address the challenges of conventional PCR assay in resource-limited settings [65]. In general, LAMP exponentially amplifies nucleic acid targets at a relatively constant temperature of 60°C-65°C. Thus, it is suitable for the development of POC settings [66]. Given its high efficiency, excellent specificity, and speediness, LAMP combined with RT (RT-LAMP) has been proposed for measuring diverse RNA viruses [67,68]. Inspired by these studies, RT-LAMP-based methods have been developed to enable the rapid detection of SARS-CoV-2. In specific, several RT-LAMP-enhanced fluorescent/colorimetric assays target different SARS-CoV-2 gene regions [69,70]. However, these reported RT-LAMP assays suffer the risk of false positive results because of the use of traditional monitoring techniques, such as agarose gel electrophoresis, SYBR Green, calcein, or pH indicators that are not specific for SARS-CoV-2 LAMP products [55,71]. Thus, a new LAMP-based approach that can provide a rapid and more objective result is desired to accelerate COVID-19 diagnostics. In response, an RT-LAMP method combined with nanoparticle-based LFA (RT-LAMP-LFA) was fabricated for the highly sensitive detection of SARS-CoV-2. Compared with traditional monitoring techniques, the application of LFA can effectively avoid the detection of non-specific amplification by using only specific reactants against target amplicons in the detected complexes. Zhu et al. developed a multiplex RT-LAMP-LFA biosensor for COVID-19 diagnosis. In this case, the target genes of ORF1ab and N against SARS-CoV-2 were simultaneously amplified using two LAMP primer sets in an isothermal reaction and detected by the designed multiplex LFA strips with two test lines of anti-FITC and anti-digoxigenin (DIG) antibodies and one control line of Biotin-BSA. In the presence of target genes, ORF1ab-RT-LAMP products were labeled with FITC and biotin, and N-RT-LAMP products were labeled with DIG and biotin. The obtained FITC- and DIG-labeled products were then captured by immobilized antibodies. With the nanoparticle accumulation on the test line under the assistance of a streptavidin–biotin system, a characteristic crimson band appeared, thus enabling the multiplex detection of the ORF1ab and N genes. The LOD of this biosensor was 12 copies (for each target) per reaction with no cross-reaction to non-SARS-CoV-2 templates [62]. The application potential of this method for clinical COVID-19 diagnosis was further demonstrated by analyzing129 RT-qPCR-confirmed respiratory samples, including 33 COVID-19-positive samples and 96 COVID-19-negative samples. Results show that the sensitivity and specificity of this RT-LAMP-LFA are 100%, indicating that this method is a promising diagnostic tool for determining SARS-CoV-2 infection.
2.3. LFA incorporated with isothermal amplification and CRISPR
Although isothermal amplification approaches have emerged as effective and appealing alternatives to PCR, the analytical performances of these methods are often compromised by the nonspecific amplification from nontarget sequences or primer dimers because they measure all nucleic acids and are not specific for the amplification products of target genes. Thus, an efficient solution to avoid the detectable signals caused by the nonspecific amplification is to achieve the specific determination of the amplified products [11]. Since its discovery in the 1980s, CRISPR has become the most popular and powerful genome editing system and is highly dependent on the RNA-guided activity of CRISPR-associated (Cas) proteins [72,73]. The CRISPR/Cas9 system is widely regarded as an adaptive immune defense system in most prokaryotes that can resist foreign genetic materials and provides the basis for a genome editing to modify genes in a sequence-specific, targeted way. Owing to its unique sequence-specific characteristics, the CRISPR/Cas technology has recently obtained increasing interest in biodetection and biosensing since its first use in nucleic acid testing [74,75]. To date, miscellaneous variants of CRISPR/Cas-based detection systems have been designed and explored to target various analytes from nucleic acids to non-nucleic acids and exhibit a great potential to transform traditional analytical methods [49,76]. A successful strategy is exemplified by the integration of isothermal amplification approaches with CRISPR, wherein the presence of CRISPR can significantly decrease the risk of nonspecific detection encountered by conventional isothermal amplification, thus facilitating the diagnostic sensitivity and specificity without compromising the analytical sensitivity. Based on this design concept, many CRISPR-coupled isothermal amplification techniques amplify target genes and enhance SARS-CoV-2 detection [77,78]. For example, Broughton et al. reported a CRISPR-Cas12-based LFA for the detection of SARS-CoV-2 from respiratory swab RNA extracts (Fig. 1D). This newly developed assay, named as “SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR),” was implemented by conducting the simultaneous RT–LAMP for SARS-CoV-2 RNA, followed by the Cas12 detection of predefined nucleotide sequences targeting the E and N genes of SARS-CoV-2 [48]. After the cleavage of a single-stranded DNA reporter, the detection of the virus can be visualized by the LFA test strip with an LOD of 10 copies per μL input. The proposed DETECTR assay was further validated to show high consistency with RT-qPCR assay by blindly analyzing 60 nasopharyngeal swab samples. Another common CRISPR-Cas-based diagnostic system, the so-called “Specific High-sensitivity Enzymatic Reporter unLOCKing (SHERLOCK),” was also exploited to target SARS-CoV-2 by using extracted nucleic acids as input and Cas13 as detection [79]. This design for SARS-CoV-2 detection was performed by introducing RT–RPA to amplify target gene segments, followed by the T7 transcription and Cas13-mediated cleavage of a single-stranded RNA reporter to measure the amplified product with fluorescent and LFA readouts (Fig. 1E). Within the characterized LOD of 42 RNA copies per reaction, the developed SHERLOCK can provide 100% and 97% sensitivities for fluorescent and LFA readout and 100% specificity for both. To reduce the risk of RNase contamination, a multiplex SHERLOCK LFA test strip was further designed for the simultaneous assay of SARS-CoV-2 RNA and RNase presence by incorporating an internal control for RNase contamination. The multiplexed design can promote the potential application of SHERLOCK for SARS-CoV-2 detection in resource-limited settings.
2.4. Enhanced LFA by signal amplification strategies
As mentioned earlier, the limitation of using LFA to directly detect SARS-CoV-2 RNA is its low concentration of target genes in specimens. The above strategies use target amplification to improve LFA, and the detection sensitivity of these methods for SARS-CoV-2 is dramatically enhanced. Unlike nucleic acid amplification by accumulating abundant target genes, signal amplification by using high-performance probes is another commonly used strategy to improve LFA detection. Compared with target amplification, signal amplification provides substantial benefits in terms of workflow and minimizing cross-contamination [80,81]. The S9.6 monoclonal antibody features high binding affinity and selectivity to DNA–RNA hybrids, suggesting that these hybrids can be captured by the S9.6 monoclonal antibody [82,83]. Using this design principle, Wang et al. conducted an amplification-free hybrid capture fluorescence immunoassay (HC-FIA) on an LFA test strip for the improved detection of SARS-CoV-2 RNA [50], wherein S9.6 antibody-labeled europium-chelate-based fluorescent nanoparticles were designed as signal-amplification probes to capture and detect the formed DNA–RNA hybrids between the DNA probe and the SARS-CoV-2 RNA with a fluorescent signal readout (Fig. 1F). The LOD of this HC-FIA for the N, E or ORF1ab regions of SARS-CoV-2 is 500 copies mL−1 for clinical throat swab and 1000 TU mL−1 for pseudoviruses. In a multi-center randomized double-blind trial, this approach achieved 100% sensitivity and 99% specificity in analyzing 734 clinical samples. Notably, the developed HC-FIA has been proposed and approved by the National Medical Products Administration and the European Conformity as a commercial test kit for SARS-CoV-2 diagnosis. Although this method provides many obvious advantages, such as no nucleic acid extraction, RT, amplification, and labeling, the signal amplification potential of the S9.6 antibody-assisted strategy is largely compromised by the limited binding sites and steric hindrance.
Viral nucleic acid test usually depends on three operating steps: sample treatment, nucleic acid amplification, and result readout [65]. The usage of LFA readout to replace conventional readouts of fluorescence signals or gel electrophoresis has largely simplified the requirement for specialized equipment and extra operating steps. However, both the need for sample treatment for preparing high-purity templates and the selection of appropriate amplification reactions for ensuring enough accumulation of target amplicons impede the development of POC nucleic acid detection. Hence, developing a one-pot molecular diagnostic workflow that integrates all steps is very important, thus decreasing the operating procedures and shortening the detection time for POC applications. Besides, the possible risk of RNA degradation and amplification inhibition could cause the loss of LFA sensitivity and thus yield false-negative results, which is a common problem encountered by RNA-based assays. Therefore, alternative methods to bypass the requirement for RNA extraction should be considered in further studies.
3. LFA for antibody test
Confirmed diagnosis of SARS-CoV-2 infections by molecular tests has been challenged because of its poor positive rates. In addition to testing the viral nucleic acid, detecting specific antibodies in blood has been proven to be a complement to confirm SARS-CoV-2 infection [84,85]. Antibodies are produced as a defense mechanism by the immune system against SARS-CoV-2 infection [86]. Maturation of the immune response usually takes a long time, followed by the dynamic variations of antibody response [87]. During this process, various specific immunoglobulins that are produced in response to SARS-CoV-2, including IgA, IgM, and IgG, appear and last for different times [88,89]. Thus, by theory, one or all immunoglobulins can be targeted to diagnose COVID-19 [90,91]. Compared with nucleic acid and antigen tests, antibody tests exhibit many outstanding advantages, including wider detection window, easier and safer for operators to collect blood samples than respiratory samples, higher stability and less susceptibility to degradation, and more uniform distribution of antibodies in blood [92]. In addition, antibody testing can detect ongoing or past infections, leading to an improved understanding of transmission dynamics and increased efficacy and accuracy of COVID-19 prevention [93]. Besides, different from antigen and RNA tests, targeting the antibody response can serve as a public health tool to characterize the host immune response to SARS-CoV-2 infection and to confirm the acquired immunity to COVID-19 in the population. Over the past few decades, various serological tests have been developed to monitor antibody responses to pathogens in body fluids, especially serum or plasma [94,95]. These tests mainly involve the use of different platforms, including ELISA [96], LFA [97], microfluidic [98], and biosensors [99]. Among them, serological test for antibodies against pathogens using LFA has received great attention due to its unique superiorities, including timely feedback of test results and field tests. Recently, lateral flow immunoassay (LFIA)-based serological test has become one of the most mainstream options for the specific determination of antibodies against SARS-CoV-2 [100]. Numerous commercial LFIA kits for targeting SARS-CoV-2 antibody have been developed and approved for clinical use [97]. In this section, we review the recent advances of LFIA-based serological assay for the rapid and specific detection of SARS-CoV-2 antibodies and enumerate the clinical value of detecting IgM, IgG, IgA, and total antibodies to provide a guideline for LFIA development. Table 2 summaries several representative LFIA methods that were reported for the detection of SARS-CoV-2 antibodies.
Table 2.
Representative LFAs for antibody test.
| Target | Sample | Probe | Assay time (min) | Sensitivity | Specificity | Ref. |
|---|---|---|---|---|---|---|
| IgM | Whole blood | AuNP | 15 | 100% | 93.3% | [97] |
| IgG | Serum | Lanthanide-doped polysterene nanoparticles | 10 | 100% | 93.6% | [105] |
| IgG | Serum | AuNP | 15–20 | 69.1% | 100% | [104] |
| IgM and IgG | Serum | SiO2@Au@QD nanobeads | 15 | 100% | 100% | [116] |
| IgM and IgG | Serum and plasma | Eu(III) fluorescent microsphere | 10 | IgM: 98.68% IgG: 98.72% |
IgM: 93.10% IgG: 100% |
[117] |
| IgM and IgG | Whole blood | AuNP | 15 | 85.29% | 100.00% | [114] |
| IgM and IgG | Serum | AuNP | 15 | 95.85% | 97.47% | [111] |
| IgM and IgG | Whole blood and serum | Selenium nanoparticle | 5 | 93.33% | 97.34% | [112] |
| IgA | Serum and saliva | AuNP and HRP | 15 | / | / | [110] |
| Total antibody | Serum | AuNP | 20 | 94.6% | 100% | [130] |
| Total antibody | Serum | AuNP with a CISG enhancement | 17 | 89.9% | 100% | [132] |
3.1. IgM, IgG, and their combination
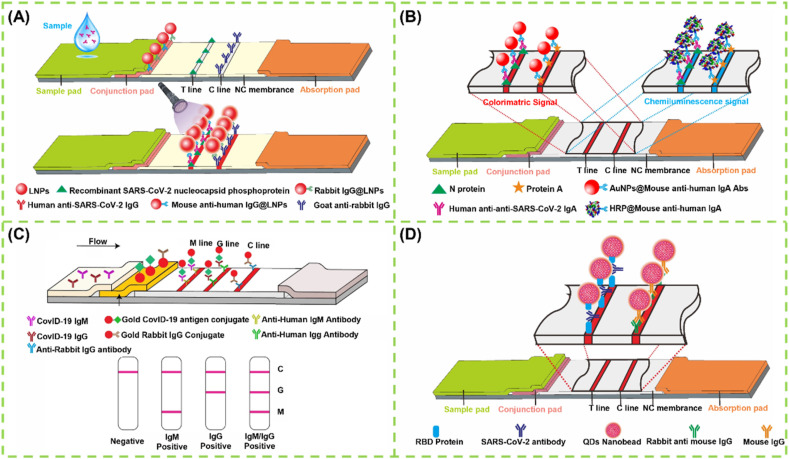
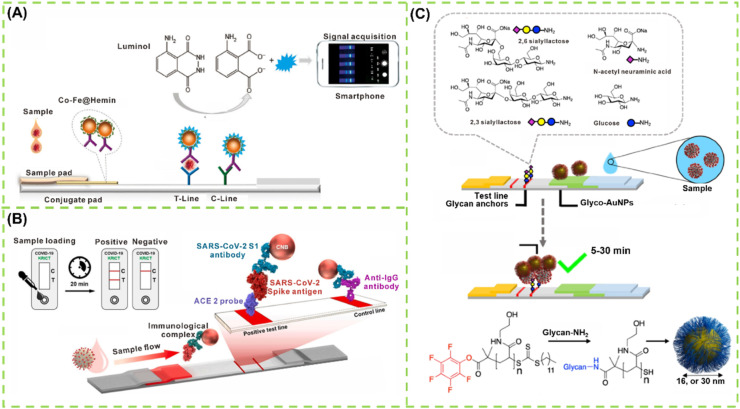
IgM first appears in blood after SARS-CoV-2 infection and increases rapidly. It begins to be detectable about 5–10 days after onset of symptoms [101]. Thus, IgM detection is regarded as an indicator of early-stage infection. Developing a rapid detection method for IgM against SARS-CoV-2 will provide a valuable supplementary in the diagnosis of COVID-19. Under this consideration, LFIA for IgM detection has aroused the attention of scholars. Huang et al. established an AuNP-based LFIA for the rapid and on-site detection of the IgM antibody against SARS-CoV-2 by using an indirect immunochromatographic method [97]. In this case, the SARS-CoV-2 N protein was sprayed onto the test line to capture the IgM antibody, and then the captured IgM was detected by anti-human IgM-conjugated AuNPs. The analytical performance of this AuNP-based LFIA was estimated using RT-qPCR-confirmed positive or negative serum samples. All five SARS-CoV-2-positive samples tested positive, whereas 13 out of 14 SARS-CoV-2-negative samples tested negative, indicating 100% sensitivity and 93.3% specificity. Although this method has an acceptable performance for detecting the SARS-CoV-2 IgM antibody in actual serum, the sample number used in this work for verifying its clinical diagnostic efficiency is relatively limited. In general, IgG antibody production follows the IgM response closely and lasts much longer to act as the body's immune defense system for avoiding reinfection of the same pathogen [102]. Thus, targeting the specific IgG response against SARS-CoV-2 in serum has been extensively used as complementary to the imperfect nucleic acid diagnosis strategy [103]. In addition, monitoring the variations of IgG concentration in serum is beneficial to evaluating the therapeutic response and prognosis and the measurements of protective antibodies upon vaccination. Altogether, development of rapid, low-cost, portable, and user-friendly methods to monitor the IgG antibody against SARS-CoV-2 is an urgent requirement for managing COVID-19. Under the circumstances, LFA for detecting IgG antibody has attracted increasing research interests because of its unique superiority. For example, Wen et al. developed a rapid and sensitive LFIA for the measurement of IgG antibody against SARS-CoV-2 virus by spraying recombinant SARS-CoV-2 N protein onto the test line of the strip, with anti-human IgG-coupled AuNPs as signal probes [104]. Using a similar design strategy, an improved LFIA has been recently established for the sensitive fluorescent sensing of SARS-CoV-2 IgG [105], with lanthanide-doped polysterene nanoparticles as alternative labels of AuNPs readout (Fig. 2 A).
Fig. 2.
LFA for detection of antibodies: (A) Design and fabrication of a LNP-based LFIA for detection of IgG [105]; (B) AuNP-based LFIA for co-detection of SARS-CoV-2 IgM and IgG for diagnosis of CVID-19 [110]; (C) Schematic of chemiluminescence LFIA for detection of IgA [113]; (D) QD nanobeads as the fluorescence probes to fabricate LFIA for detection of SARS-CoV-2 total antibody [131].
Given that the concentrations of IgM and IgG significantly increase during the acute and convalescent phases of COVID-19, the detectable rates of IgM and IgG in patients increase to 94.1% and 100% within 19 days after symptom onset, respectively [106]. Moreover, the IgM level decreases rapidly in recovered patients by 3 weeks, whereas IgG titer levels remain high even after 2 months [107]. Therefore, the simultaneous detection of SARS-CoV-2-specific IgM and IgG can not only realize the accurate identification of early SARS-CoV-2 infection but also allow monitoring the disease progression. Encouraged by this, several multiplexed LFIA systems have been reported for the simultaneous detection of IgM and IgG in patient serum [108,109]. Li et al. presented the first multiplexed AuNP-based LFIA for the rapid and simultaneous detection of IgM and IgG antibodies against SARS-CoV-2 in blood samples (Fig. 2B). In this design, anti-human IgM and anti-human IgG antibodies are simultaneously immobilized onto the membrane as M and G test lines to capture the IgM and IgG antibodies, respectively, and then the captured antibodies are visually detected with SARS-CoV-2 recombinant S protein-conjugated AuNP probes [110]. The clinical efficacy uses of this method were validated by testing 525 clinical blood samples, covering 397 clinically confirmed SARS-CoV-2-infected patients and 128 non-SARS-CoV-2-infected patients, collected from eight medical centers. When only testing IgM or IgG, the sensitivities of this method are 82.6% and 70.5%, respectively, whereas the overall sensitivities obviously increase to 88.66% by using the combined IgM and IgG detection, implying the higher accuracy and sensitivity of the simultaneous determination of IgM and IgG antibodies SARS-CoV-2 in comparison with single target detection. Inspired by this pioneering work, increasing efforts have been devoted to improve the multiple sensing capability of LFIA for the sensitive and simultaneous detection of SARS-CoV-2-specific IgM and IgG [111,112]. Among these studies, except for the common AuNPs [113,114], other nanomaterials including selenium nanoparticle, quantum dots (QDs) [115] or QD nanobeads [116], lanthanide fluorescent microsphere [117], aggregation-induced emission based polymeric nanoparticles [118], core–shell SiO2@AgNPs [119], and magnetic nanoparticles [120], have also been used in LFIA for enhancing the detection of SARS-CoV-2 antibodies with the help of sensitive signal outputs of colorimetry, fluorescence, surface enhanced Raman scattering, and giant magnetoresistance. In addition, the serologic test performance of these reported multiple LFIA for COVID-19 can be further improved by integrating with a machine-learning classifier to control the false positive rate at a targeted level [121].
3.2. IgA
IgA plays a key role in respiratory mucosal epithelial protection and homeostasis regulation, separating the external environment from the inside of the body [122]. The primary function of IgA is known as immune exclusion, a process that can restrict the entry of microorganisms and antigens into the fragile mucosal barrier [123,124]. In comparison with IgM, the IgA antibody kinetics was deepened from the COVID-19 onset. IgA response appeared and increased earlier, peaked at the third week, and remained strong and persistent. A previous work indicated that IgA antibodies offer higher sensitivity than IgM because serum IgA production is time dependent and in larger amounts than IgM production [125]. Thus, the benefits of IgA antibodies for diagnosing SARS-CoV-2 are its early detection and high sensitivity. Moreover, IgA levels are closely related to the severity of disease [110]. As a complement to antibody detection, targeting IgA in serum or saliva is drawing increasing attention. In this regard, Roda et al. fabricated a dual optical/chemiluminescent LFIA for the detection of salivary and serum IgA, wherein a recombinant N protein was used to capture the SARS-CoV-2 antibodies in specimens and a labeled anti-human IgA was applied to sense the captured IgA (Fig. 2C). For optical detection, the AuNPs were adopted as colorimetric signal reporters. For chemiluminescent assay, horseradish peroxidase (HRP) label was designed to catalyze the luminol/H2O2/enhancer substrate to enable chemiluminescent signal outputs [113]. The analysis performance of this method was well demonstrated by measuring 25 serum and 9 saliva samples collected from infected and/or recovered individuals. Compared with colorimetric detection, the chemiluminescent detection showed higher testing ability, uncovering the existence of salivary IgA in infected patients. These results revealed that the reported IgA-LFIA can serve as a noninvasive screening tool for early monitoring immune response to COVID-19, which may help narrow the COVID-19 serological gap.
3.3. Total antibodies
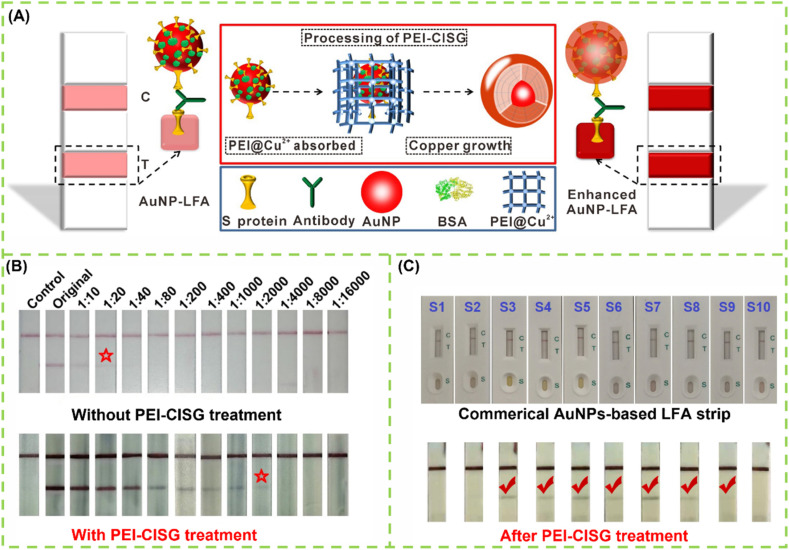
As with the reported SARS and MERS viruses, the sequential production of IgM and IgG is questionable when the immune system encounters SARS-CoV-2 [126]. IgM responses are prior to IgG, together with IgG, later than IgG, or were absent [127]. In addition, IgA response is relatively limited [128]. Thus, the separate detection of IgA, IgM, and IgG is hard to distinguish among early, middle, and past infections. A previous work on the serological response variability to SARS-CoV-2 infection found that the individual immunoglobulin test is useless in defining the stages of infection but can markedly decrease the detection sensitivity because the amount of each immunoglobulin produces a lower signal than that caused by the sum of all the contributions [129]. This finding suggests that the diagnostic sensitivity of COVID-19 can be improved by targeting total antibody response stimulated by the SARS-CoV-2 infection. In addition, the detection of total antibodies against SARS-CoV-2 by a double-antigen sandwich immunoassay can effectively avoid the potential serological cross-reaction with autoantibodies in patients with autoimmune disease, which is often encountered by conventional IgG or IgM immunoassay using anti-human IgG or IgM antibodies as recognition elements. Given that, the serological test of SARS-CoV-2 gradually trends to detect total antibody, instead of each immunoglobulin separately [130]. The double antigen sandwich immunoassay format was proposed to detect total antibody. In this proposed method, antigen pre-immobilized strip and antigen-labeled signal reporter were used to capture and measure antibodies against SARS-CoV-2 in samples, followed by the formation of a sandwich immunocomplex. Li et al. constructed a double-antigen sandwich fluorescent LFIA for the detection of total antibodies against SARS-CoV-2 by using a recombinant spike receptor-binding domain (RBD) protein pre-immobilized as the test line and RBD protein-coated QD nanobeads as fluorescent signal reporters (Fig. 2D). Compared with conventional AuNP-based LFIA, QD nanobead-based fluorescent approach exhibits about 32-fold improvement in detection sensitivity, thus contributing to reducing the false-negative rate confronted by AuNP-based LFIA [131]. In addition, our group designed a controllable copper in situ growth strategy (namely PEI-CISG) to enhance traditional AuNP-based LFIA and improve COVID-19 diagnosis (Fig. 3 A). In our design, after executing the conventional double-antigen sandwich LFIA with AuNPs as labels, an additional copper in situ growth step was conducted by using polyethyleneimine as a structure-directing agent to regulate the thermodynamics of anisotropic Cu nanoshell growth on the AuNP surface [132]. With the in-situ deposition of Cu nanoshell, the optical signal intensity of the captured AuNPs at the test line is significantly amplified, thus resulting in an enhanced sensitivity. Fig. 3B shows that the PEI-CISG technology provides about two orders of magnitude improvement in sensitivity relative to the unamplified LFIA. Importantly, seven pseudo-negative samples detected by traditional AuNP-based LFIA have been successfully identified as positive after PEI-CISG amplification without the presence of any false positives (Fig. 3C). Nonetheless, increasing the sample number and simplifying the signal amplification procedure should be considered in further work.
Fig. 3.
(A) Schematic illustration of the principle and process of PEI-CISG-enhanced LFIA for detection of SARS-CoV-2 total antibody; (B) LFA and PEI-CISG-enhanced LFA in detecting diluted SARS-CoV-2 positive serum samples; (C) The performance of PEI-CISG-enhanced LFA in eliminating false negative results in diagnosis of SARS-CoV-2 [132].
In the early stages of the COVID-19 epidemic, LFIA has played critical roles in rapid screening and discriminating SARS-CoV-2-infected people by measuring the specific antibodies. However, the performance of these developed serological assays varies widely among different test groups, manufacturers, assay timing, and target antibodies, which need further evaluation. Antibody testing for SARS-CoV-2 may be compromised by the cross-reaction with other human coronaviruses or underlying conditions and thus leads to false-positive results. More importantly, with the rapid development of global mass immunization in the crowd, targeting antibody response to the SARS-CoV-2 infection cannot act as an effective way for improving COVID-19 diagnosis because of its invalidity to identify SARS-CoV-2-vaccinated or SARS-CoV-2-infected people. Even so, serosurveillance studies can still be used to support the investigations of tracking the incidence and prevalence change, understanding the dynamic decay of antibodies after recovery, monitoring the titer of neutralizing antibodies during convalescent plasma therapy, and characterizing the seroprevalence and vaccine coverage [133].
4. LFA for antigen detection
Unlike nucleic acid and antibody detection that has demonstrated explosive growth within a short time, the development of antigen test is relatively backward. Until September 11, 2020, the WHO provides an interim guidance on the potential role of antigen detection in the diagnosis of SARS-CoV-2 infection [134]. Compared with RT-qPCR, the detection of SARS-CoV-2 antigens is less expensive and faster, which is ideally suitable for developing countries that experience challenges in conducting RT-qPCR [135]. Antigen tests are designed to directly detect SARS-CoV-2 virus and its related proteins produced by replicating virus in respiratory secretions. By theory, the whole SARS-CoV-2 virus and its structural proteins can be used as antigens for COVID-19 diagnosis [136,137]. These antigens can be detected directly by using various biological samples, such as tissue swabbed from the anterior nasal cavity, oropharynx, or even directly from saliva. Currently, S and N proteins are the most valuable antigen biomarkers for COVID-19 diagnosis, and researchers are working on developing specific antigen-based tests for detecting the presence of SARS-CoV-2 by using S and N proteins [138,139]. Some commercial rapid test kits, including the BIOCREDIT COVID-19 Ag test by RapiGEN (Gyeonggi-do, Korea), the PanbioTM COVID-19 Ag by Abbott (Cologne, Germany), and the SARS-CoV-2 rapid antigen test by Roche, have been recently approved. In this section, we summarize the recent advances on LFA for the direct detection of SARS-CoV-2 specific antigen (Table 3 ).
Table 3.
Representative LFAs for the detection of viral antigens.
| Target | Sample | Probe | Assay time (min) | LOD | Sensitivity | Specificity | Ref. |
|---|---|---|---|---|---|---|---|
| N protein | Nasopharyngeal swab | / | 10 | / | 75.6% | 100% | [151] |
| N protein | Nasal swab | Red latex beads | 20 | 0.65 ng mL−1 | / | / | [149] |
| N protein | Nasopharyngeal swabs | AgNP | 15 | 250 pg mL−1 | 57.6% | 99.5% | [150] |
| N protein | Nasopharyngeal swab | AuNP | 30 | 56 copies μL−1 | 82.2% | 100% | [153] |
| N protein | Oropharyngeal swabs | Europium (III) chelate microparticles | 10 | / | 100% | 100% | [152] |
| S protein | Nasopharyngeal swab | AuNP | 30 | 5 μg mL−1 | / | / | [144] |
| S protein | Saliva and nasal swab | Co–Fe@hemin-peroxidase nanozyme | 16 | 0.1 ng mL−1 | / | / | [174] |
| S protein | Nasal swabs | Red cellulose nanobeads | 20 | 1.86 × 105 copies mL−1 | / | / | [143] |
4.1. S protein
Recent studies have indicated that a large number of glycosylated S proteins cover the surface of SARS-CoV-2 and bind to the host cell receptor angiotensin-converting enzyme II (ACE2), mediating viral entry [140]. S protein has become the main target for developing vaccine [141]. Thus, diagnosing COVID-19 by using S protein as the antigen is specific and sensitive. Using a double-antibody sandwich immunoassay strategy, Liu et al. established a nanozyme-based chemiluminescent LFIA for the rapid and sensitive detection of SARS-CoV-2 S protein (Fig. 4 A). In this case, a Co–Fe@hemin-peroxidase nanozyme possessing peroxidase-mimicking activity was synthesized by the hydrothermal method and then applied as a nanozyme label to catalyze the luminol/H2O2/enhancer substrate to produce chemiluminescent signals [142]. After conjugating with anti-S protein antibody, this nanozyme specifically senses the SARS-CoV-2 S protein captured on a prepared strip with the formation of a sandwich immunocomplex. The LODs for recombinant S antigen and pseudovirus are 0.1 ng mL−1 and 360 TCID50 mL−1, respectively. However, the preparation of perfectly matched paired antibodies to S protein is time consuming, costly, and random. Similar to SARS-CoV, SARS-CoV-2 is also proven to co-localize in animal cells with angiotensin-converting enzyme II (ACE2) as a cellular entry receptor, but the S protein of SARS-CoV-2 displays higher binding affinity with ACE2 than that of SARS-CoV, implying that ACE2 can serve as an affinity element to recognize the RBD of S protein. Combined with the specific antibodies of SARS-CoV-2 S1 to form the matched pairs, Lee et al. developed a sandwich LFIA for the rapid detection for SARS-CoV-2 S1 protein. As indicated in Fig. 4B, ACE2 is immobilized on the nitrocellulose membrane as the test line and the antibodies are labeled with red cellulose nanobeads (CNBs) as detection probes. ACE2 and anti-SARS-CoV-2 S1 antibodies were paired to capture and detect S1 protein in LFIA without cross-reaction to other coronavirus S1 proteins [143]. The LOD of the fabricated LFIA with the matched pair consisting of ACE2 and antibody is 1.86 × 105 copies mL−1 in specimens. Coronaviruses usually present homotrimeres of spike glycoproteins on their surface, and the crucial step for SARS-CoV-2 to engage the host cell is the sialic acid binding by the S1 spike protein subunits. For example, human coronavirus (strain OC43) can bind 9-O-acetylated sialic acid, and MERS S1 can bind α2,3′-linked sialic acids. These findings imply that the glycan anchoring of coronaviruses by sialic acid recognition may provide the possibility for its detection. Baker et al. have recently discovered that N-acetyl neuraminic acid has high affinity toward the SARS-CoV-2 S glycoprotein and exploited this interaction as the capture and detection unit in a LFA device, namely, glyco-LFA. Fig. 4C depicts the design principle of this glyco-LFA, wherein BSA-glycoconjugate was immobilized on the test strip and sialic acid derivative-modified AuNPs (named glyconanoparticles) were used to interrogate SARS-CoV-2 S1 antigen [144]. After optimizing the glycan position on sialic acid, the designed sialic acid derivatives could specifically capture SARS-CoV-2 S protein rather than SARS-CoV S protein. Encouragingly, the proposed glyco-LFA can successfully detect a virus mimic particle containing SARS-CoV-2 S1 antigen with an LOD of the S protein around 5 μg mL−1 and a SARS-CoV-2 S protein-presenting pseudotyped lentivirus at 1.5 × 104 transduction units per mL within 30 min.
Fig. 4.
LFA-based sensors for detection of SARS-CoV-2 S protein: (A) Working principle of nanozyme-based LFA sensor for detection of SARS-CoV-2 S protein [142]; (B) Schematic of ACE2 receptor and antibody co-fabricated LFA for detection of SARS-CoV-2 S protein [143]; (C) Molecular structure of glycan capture units, the formation of glyconanoparticles, and the design concept of glyconanoparticles-based LFA [144].
4.2. N protein
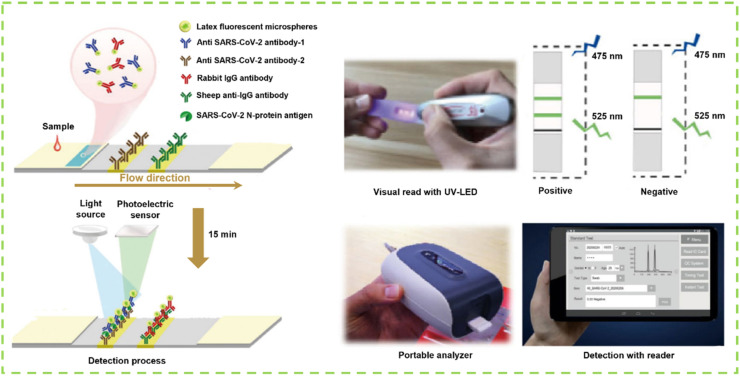
N protein is the predominant structural protein of SARS-CoV-2 virus [145]. During virus assembly, N protein can be exposed and released in large amounts into the blood, nasopharyngeal aspirate, throat wash samples, feces, and urine, making it act as one of the targets for the early detection of SARS-CoV-2 infection [146,147]. Thus, detecting N antigen is an effective strategy for the early screening of suspected COVID-19 patients [148]. Recently, several antigen rapid diagnostic immunoassays have been reported for targeting SARS-CoV-2 N protein, and most of them are based on the double-antibody sandwich principle on a simple LFIA platform [149]. The developed LFIA test strip involves the use of AuNPs [150], latex beads [149], and fluorescent microparticles [151] as signal reporters. For instance, Zhang et al. developed a fluorescent LFIA test strip for SARS-CoV-2 N protein through coupling fluorescent microsphere labeling technology with immunochromatographic technology (Fig. 5 ). In their work, anti-SARS-CoV-2 N protein monoclonal antibody-1 was sprayed onto the strip as the test line, and anti-SARS-CoV-2 N protein monoclonal antibody-2 was conjugated with fluorescent microspheres as the detection probe. With the design principle of double antibody sandwich, the LOD of this strip is 100 ng mL−1 for recombinant N protein and 1 × 103 TCID50 mL−1 for activated SARS-CoV-2 virus [152]. Combined with silver amplification technology, Miyakawa et al. designed an enhanced SARS-CoV-2 antigen rapid diagnostic test for N antigen based on the AuNP-based LFIA. The silver-enhanced LFIA strip provides higher analytical sensitivity in testing N protein in nasopharyngeal swab samples than the unamplified AuNP-based LFIA [153].
Fig. 5.
The whole detection process of fluorescent microsphere-based double-antibody sandwiched LFIA for the diagnosis of SARS-CoV-2 including oropharyngeal swab sampling, visual read with the UV-LED, and quantitative detection by the portable detector [152].
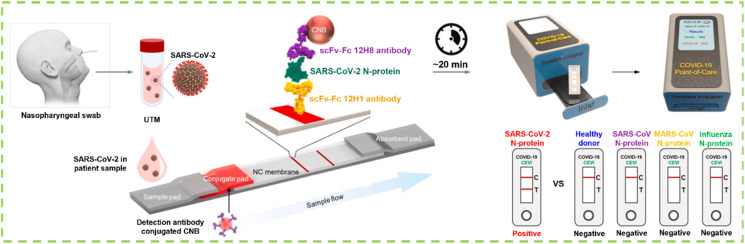
As mentioned earlier, the matched antibody pairs are required for the specific detection of protein antigen. However, the screening of high-affinity antibody via in vivo affinity maturation in the mammalian immune system is hard and random, especially a pair of perfectly matched antibodies. By contrast, the preparation of high-affinity antibodies via in vitro display technologies is simpler and more controllable [154]. Kim et al. used phage display technology to prepare single-chain variable fragment (scFv)-crystallizable fragment (Fc) fusion proteins (scFv-Fc) as antibodies for the specific detection of SARS-CoV-2 N protein combined with the portable LFIA platform (Fig. 6 ). The obtained scFv-Fc antibodies can selectively bind to the SARS-CoV-2 N antigen [155]. After the careful screening of a scFv-Fc antibody pair, the antibody pair of 12H8 as the captured antibody and 12H1 as the detected antibody was obtained. Using the best pair of scFv-Fc antibodies, the LOD of the prepared LFIA using CNBs as signal probes for SARS-CoV-2 virus is as low as 2.5 × 104 pfu with no cross-reaction to the N protein of other coronaviruses. These results demonstrate the feasibility of this LFIA-based biosensor using scFv-Fc fusion proteins for rapid and specific COVID-19 diagnosis.
Fig. 6.
Schematic diagram of the fabrication of phage display technology-assisted LFIA using scFv-Fc fusion proteins as capture antibody (12H1) and detection antibody (12H8) with CNBs as signal reporters for the diagnosis of SARS-CoV-2 [155].
Generally, antigen testing confirms the SARS-CoV-2 infection by analyzing its viral proteins in different types of specimens. As the COVID-19 pandemic continues, many SARS-CoV-2 variants have been discovered and characterized with higher infectiousness than the original virus. The presence of these variants is challenging the accuracy and reliability of antigen rapid diagnostic tests in various settings. In addition, the available data on antigen performance in the clinic remains limited. Therefore, antigen validations and performance evaluations of these reported LFA-based antigen diagnostic tests in clinical studies should be encouraged to support rapid identification and management of COVID-19 by antigen testing, thus helping alleviate some of the bottlenecks encountered by molecular testing.
5. Commercial LFA kits for the diagnosis of SARS-CoV-2 infection
The on-going global COVID-19 pandemic caused by SARS-CoV-2 has been underway for over one year [156]. The rapid, sensitive, specific, and accurate detection of SARS-CoV-2 is still urgently needed worldwide to control the pandemic and resurgence of COVID-19 [157]. In response, as a popular rapid diagnostic test, LFA has played an important role in the diagnosis of SARS-CoV-2 infection. To date, many available LFA-based commercial rapid diagnostic kits have been approved for diagnosing COVID-19. Table 4 provides a detailed list of commercial kits from different countries and regions that cover three types of tests, namely, nucleic acids, antibodies, and antigens. This information, including target analytes, detection time, clinical specimens, sensitivity, specificity, manufacturers, and referenced websites is summarized. Nonetheless, with the rapid development of LFA technology toward SARS-CoV-2 detection, this list is incomplete and requires constant updating. On the other hand, although many commercial LFA strips are provided for SARS-CoV-2 detection, the careful comparison and test selection of different commercial LFA kits are still needed to screen the most suitable test. In addition, newly developed LFA strip prototypes with excellent commercial potential should be encouraged to conduct further in-depth studies and large-scale clinical validations. More importantly, the advanced sensitization technologies are urgently needed to integrate with LFA-based COVID-19 diagnosis kits for improving their detection accuracy and negative rate. Thus, these methods are expected to lay the foundation for controlling the COVID-19 pandemic.
Table 4.
Representative commercial LFA test kits from different countries and regions for the diagnosis of COVID-19.
6. Conclusions and future perspectives
The diagnosis of SARS-CoV-2 infection remains the main driving force to alleviate the COVID-19 pandemic. Rapid testing needs should be provided at the early stage to control the spread and resurgence of COVID-19, thus contributing to the rapid identification of new COVID-19-infected persons and the implementation of their isolation and quarantine measures. In this regard, LFA-assisted rapid diagnostic tests play an important role in targeting SARS-CoV-2 specific biomarkers, such as viral RNA, antibodies, antigens, and whole virus. Various rapid diagnostic technologies using the portable LFA platform have sprung up, some of which have been developed into test kits for rapidly diagnosing COVID-19. As a helpful complement to the current rRT-qPCR assay, LFA-based test products are becoming popular and widely available in low-resource settings. This review is the first to provide an overview of rapid diagnostic tests that involve LFA techniques and their application potential in improving COVID-19 diagnosis. The key insights of the developed LFA methods on the design principle, working mechanism, methodological performance, and clinical diagnostic efficiency have been discussed in detail.
Although the LFA rapid detection technology holds great promise for COVID-19 diagnostics, several challenges remain to be addressed to improve the performance of LFA in controlling the COVID-19 epidemic. Continuous improvement of diagnostic test preparation based on LFA is essential for the rapid detection of SARS-CoV-2. First, further optimization of these LFA tests and extensive clinical validation are still required prior to their clinical use due to the relatively limited sample numbers for methodological evaluation and the use of non-fresh blood samples, which may cause the statistical bias of detected results. Second, COVID-19 LFA may no longer be limited to detecting a single type of target. For example, the simultaneous detection of antigens and antibodies against SARS-CoV-2 by a single LFA test strip is beneficial for pathology studies and vaccine development of COVID-19. Third, the needs of developing non-laboratory, community-type, house-hold, and POC diagnostic methods are accelerating the integration of the LFA platform with advanced technologies, such as artificial intelligence, machine learning, smartphone, and big data [158]. Fourth, rational resource allocation can further enhance the community management of COVID-19. Understanding the diagnostic performance of each LFA test strip when different targets are used as test objects is important to achieve this goal. A growing number of studies have indicated the emergence of SARS-CoV-2 mutations, like SARS-CoV-2 D614G, which appears to the chief culprit for the second-wave pandemic. Developing LFA-based biosensing technologies for targeting the SARS-CoV-2 mutants is thus crucial for preventing further spread.
The current COVID-19 pandemic worldwide has exposed our weaknesses in coupling with emerging infectious diseases, especially in rapid test technologies. As the global healthcare system continues to function, the urgent need for innovative technologies for accurate and rapid diagnosis of infectious diseases, particularly emerging ones, remains critical. Except for as a complementary to molecular tests to confirm diagnosis, LFA can provide a timely and convenient way to understand and respond to new infectious diseases. Recent benefits and advancements in addressing various infectious diseases have pushed the LFA technology to compete with molecular diagnostics. Nevertheless, many questions and challenges regarding LFA technology remain to be resolved, comprising their variable sensitivity and specificity and sample pretreatment. Thus, continuous efforts should be devoted to facilitating new LFA-based rapid diagnostic assay development by introducing innovative signal transducer strategies, such as spin-enhanced nanodiamond [159], background-free long-lasting phosphorescence [160] and near-infrared Ⅰ or Ⅱ emission [161], and new signal amplification technologies, like nanoparticle accumulation and enzymatic amplification, thereby transcending the current nucleic acid diagnostics and achieving rapid field diagnosis [162]. In recent years, many research groups have authorized several key review papers that focus on the improvements of LFA from different perspectives, including the recognition molecule [163,164], the nanoparticle design [162,165,166], the signal amplification strategy [81,167], the strip device engineering [168], and the multiplexing [169]. In addition to assay improvement, the selection of appropriate targets against pathogens, such as nucleic sequences, antigens, and antibody types, requires careful consideration for enhanced sensitivity and specificity. Future's goal to respond to new infectious diseases using LFA is to construct a highly integrated POC device with sample-to-answer portability and higher clinical sensitivity and specificity. However, it should be noted that healthcare professionals need to balance the benefits of obtaining rapid diagnostic results using LFA for immediate clinical intervention and management and public health action against the risk of false positive or negative results. With the joint efforts of the world, the battle against various infectious diseases, like COVID-19, will be won.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful for financial support from the National Natural Science Foundation of China (32001788, 31760485, 31901780), the Scientific Research Foundation of Education Department of Jiangxi Province (GJJ200221), the Interdisciplinary Innovation Fund of Natural Science, Nanchang University (9166-27060003-ZD01), and Jiangxi Provincial Natural Science Foundation (20202ACB215004).
References
- 1.Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y. Science. 2020;370:4250. [Google Scholar]
- 2.Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S. Nature. 2020;583:437. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 3.Schett G., Manger B., Simon D., Caporali R. Nat. Rev. Rheumatol. 2020;160:465. doi: 10.1038/s41584-020-0451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le T.T., Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M., Mayhew S. Nat. Rev. Drug Discov. 2020;19:305. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 5.Cao X. Nat. Rev. Immunol. 2020;20:269. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tromberg B.J., Schwetz T.A., Pérez-Stable E.J., Hodes R.J., Woychik R.P., Bright R.A., Fleurence R.L., Collins F.S. N. Engl. J. Med. 2020;383:1071. doi: 10.1056/NEJMsr2022263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakhar D., Kaur I., Kaul S. J. Eur. Acad. Dermatol. Venereol. 2020;34:242. doi: 10.1111/jdv.16412. [DOI] [PubMed] [Google Scholar]
- 8.Smyrlaki I., Ekman M., Lentini A., de Sousa N.R., Papanicolaou N., Vondracek M., Aarum J., Safari H., Muradrasoli S., Rothfuchs A.G. Nat. Commun. 2020;11:4812. doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. J. Am. Med. Assoc. 2020;323:1502. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindson J. Nat. Rev. Gastroenterol. Hepatol. 2020;17:259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y.C., Lee Y.T., Yang T., Sun J.R., Shen C.F., Cheng C.M. Bioeng. Transl. Med. 2020;5:10177. doi: 10.1002/btm2.10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H., Zhang H., Ni S., Korabečná M., Yobas L., Neuzil P. Trac. Trends Anal. Chem. 2020;130:115984. doi: 10.1016/j.trac.2020.115984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y., Zhou Y., Xiong S., Zeng L., Huang X., Leng Y., Xiong Y. Sensor. Actuator. B Chem. 2020;305:127439. [Google Scholar]
- 15.Wu Y., Zhou Y., Huang H., Chen X., Leng Y., Lai W., Huang X., Xiong Y. Sensor. Actuator. B Chem. 2020;316:128107. [Google Scholar]
- 16.Xiong Y., Zhang J., Yang Z., Mou Q., Ma Y., Xiong Y., Lu Y. J. Am. Chem. Soc. 2019;142:207. doi: 10.1021/jacs.9b09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan S., Hu J., Li Y., Huang X., Xiong Y. Food Chem. 2020;342:128327. doi: 10.1016/j.foodchem.2020.128327. [DOI] [PubMed] [Google Scholar]
- 18.Peeling R.W., Mabey D., Herring A., Hook E.W. Nat. Rev. Microbiol. 2006;4:909. doi: 10.1038/nrmicro1555. [DOI] [PubMed] [Google Scholar]
- 19.Parolo C., de la Escosura-Muñiz A., Merkoçi A. Biosens. Bioelectron. 2013;40:412. doi: 10.1016/j.bios.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Jiménez-de-la-Cuesta D., Mauritsen T. Nat. Geosci. 2019;12:902. [Google Scholar]
- 21.Parolo C., Sena-Torralba A., Bergua J.F., Calucho E., Fuentes-Chust C., Hu L., Rivas L., Álvarez-Diduk R., Nguyen E.P., Cinti S. Nat. Protoc. 2020;15:3788. doi: 10.1038/s41596-020-0357-x. [DOI] [PubMed] [Google Scholar]
- 22.Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M. Nat. Mater. 2021;20:593. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montesinos I., Gruson D., Kabamba B., Dahma H., Van den Wijngaert S., Reza S., Carbone V., Vandenberg O., Gulbis B., Wolff F. J. Clin. Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silveira M.F., Barros A.J., Horta B.L., Pellanda L.C., Victora G.D., Dellagostin O.A., Struchiner C.J., Burattini M.N., Valim A.R., Berlezi E.M. Nat. Med. 2020;26:1196. doi: 10.1038/s41591-020-0992-3. [DOI] [PubMed] [Google Scholar]
- 25.Nouri R., Tang Z., Dong M., Liu T., Kshirsagar A., Guan W. Biosens. Bioelectron. 2021;190:113012. doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Nat. Rev. Microbiol. 2020;19:171. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poortahmasebi V., Zandi M., Soltani S., Jazayeri S.M. Front. Emerg. Med. 2020;4:57. [Google Scholar]
- 28.Taleghani N., Taghipour F. Biosens. Bioelectron. 2020;174:112830. doi: 10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C.Y.-P., Lin R.T., Renia L., Ng L.F. Front. Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastos M.L., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.-P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merkoçi A., Li C.-z., Lechuga L.M., Ozcan A. Biosens. Bioelectron. 2021;178:113046. doi: 10.1016/j.bios.2021.113046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahshid S.S., Flynn S.E., Mahshid S. Biosens. Bioelectron. 2021;176:112905. doi: 10.1016/j.bios.2020.112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji T., Liu Z., Wang G., Guo X., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu L., Li D., Ramadan S., Li Y., Klein N. Biosens. Bioelectron. 2020;170:112673. doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. Sci. Total Environ. 2020;743:140444. doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suleman S., Shukla S.K., Malhotra N., Bukkitgar S.D., Shetti N.P., Pilloton R., Narang J., Tan Y.N., Aminabhavi T.M. Chem. Eng. J. 2021;414:128759. doi: 10.1016/j.cej.2021.128759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexandersen S., Chamings A., Bhatta T.R. Nat. Commun. 2020;11:6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia S., Chen X. Cell Discov. 2020;6:34. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M., Hamdan S.M. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xi H., Juhas M., Zhang Y. Biosens. Bioelectron. 2020;167:112494. doi: 10.1016/j.bios.2020.112494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamiolkowski D., Mühleisen B., Müller S., Navarini A.A., Tzankov A., Roider E. Lancet. 2020;396:598. doi: 10.1016/S0140-6736(20)31754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell K.H., Tornatore J.M., Lawrence K.E., Illuzzi J.L., Sussman L.S., Lipkind H.S., Pettker C.M. J. Am. Med. Assoc. 2020;323:2520. doi: 10.1001/jama.2020.8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong L., Tian J., He S., Zhu C., Wang J., Liu C., Yang J. J. Am. Med. Assoc. 2020;323:1846. doi: 10.1001/jama.2020.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M. Nat. Biotechnol. 2020;38:1164. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han M.S., Byun J.-H., Cho Y., Rim J.H. Lancet Infect. Dis. 2021;21:165. doi: 10.1016/S1473-3099(20)30424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., de Ramirez S.A.S., Valera E., Cunningham B.T., King W.P., Bashir R. Proc. Natl. Acad. Sci. U.S.A. 2020;117:22727. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R. Cell. 2020;181:865. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. Nat. Biotechnol. 2020;38:870. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong E., Jiang L., Tian T., Hu M., Yue H., Huang M., Lin W., Jiang Y., Zhu D., Zhou X. Angew. Chem. Int. Ed. 2020;10:5367. doi: 10.1002/anie.202014506. [DOI] [PubMed] [Google Scholar]
- 50.Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L. Nat. Biomed. Eng. 2020;4:1150. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- 51.Jung Y., Park G.-S., Moon J.H., Ku K., Beak S.-H., Lee C.-S., Kim S., Park E.C., Park D., Lee J.-H. ACS Infect. Dis. 2020;6:2513. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- 52.Choi J.R. Front. Chem. 2020;8:517. doi: 10.3389/fchem.2020.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu S., Nimse S.B., Kim J., Song K.-S., Kim T. Anal. Chem. 2020;92:14139. doi: 10.1021/acs.analchem.0c03202. [DOI] [PubMed] [Google Scholar]
- 54.de Kock R., Baselmans M., Scharnhorst V., Deiman B. Eur. J. Clin. Microbiol. Infect. Dis. 2020;40:807. doi: 10.1007/s10096-020-04076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baek Y.H., Um J., Antigua K.J.C., Park J.-H., Kim Y., Oh S., Kim Y.-I., Choi W.-S., Kim S.G., Jeong J.H. Emerg. Microb. Infect. 2020;9:998. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomita N., Mori Y., Kanda H., Notomi T. Nat. Protoc. 2008;3:877. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 57.Jia H., Li Z., Liu C., Cheng Y. Angew. Chem. Int. Ed. 2010;49:5498. doi: 10.1002/anie.201001375. [DOI] [PubMed] [Google Scholar]
- 58.Lobato I.M., O'Sullivan C.K. Trac. Trends Anal. Chem. 2018;98:19. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schweitzer B., Roberts S., Grimwade B., Shao W., Wang M., Fu Q., Shu Q., Laroche I., Zhou Z., Tchernev V.T. Nat. Biotechnol. 2002;20:359. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou W., Hu L., Ying L., Zhao Z., Chu P.K., Yu X.-F. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spits C., Le Caignec C., De Rycke M., Van Haute L., Van Steirteghem A., Liebaers I., Sermon K. Nat. Protoc. 2006;1:1965. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 62.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varona M., Eitzmann D.R., Anderson J.L. Anal. Chem. 2021;93:4149. doi: 10.1021/acs.analchem.0c05355. [DOI] [PubMed] [Google Scholar]
- 64.Qian J., Boswell S.A., Chidley C., Lu Z.-x., Pettit M.E., Gaudio B.L., Fajnzylber J.M., Ingram R.T., Ward R.H., Li J.Z. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-19258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang C., Zheng T., Wang H., Chen W., Huang X., Liang J., Qiu L., Han D., Tan W. Anal. Chem. 2021;93:3325. doi: 10.1021/acs.analchem.0c05059. [DOI] [PubMed] [Google Scholar]
- 66.Hsieh K., Patterson A.S., Ferguson B.S., Plaxco K.W., Soh H.T. Angew. Chem. Int. Ed. 2012;51:4896. doi: 10.1002/anie.201109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott A.T., Layne T.R., O'Connell K.C., Tanner N.A., Landers J.P. Anal. Chem. 2020;92:13343. doi: 10.1021/acs.analchem.0c02666. [DOI] [PubMed] [Google Scholar]
- 68.Fang X., Chen H., Yu S., Jiang X., Kong J. Anal. Chem. 2011;83:690. doi: 10.1021/ac102858j. [DOI] [PubMed] [Google Scholar]
- 69.Mattioli I.A., Hassan A., Oliveira O.N., Jr., Crespilho F.N. ACS Sens. 2020;5:3655. doi: 10.1021/acssensors.0c01382. [DOI] [PubMed] [Google Scholar]
- 70.El Jaddaoui I., Allali M., Raoui S., Sehli S., Habib N., Chaouni B., Al Idrissi N., Benslima N., Maher W., Benrahma H. Expert Rev. Mol. Diagn. 2021;21:141. doi: 10.1080/14737159.2021.1886927. [DOI] [PubMed] [Google Scholar]
- 71.van Dongen J.E., Berendsen J.T., Steenbergen R.D., Wolthuis R.M., Eijkel J.C., Segerink L.I. Biosens. Bioelectron. 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rahimi H., Salehiabar M., Barsbay M., Ghaffarlou M., Kavetskyy T., Sharafi A., Davaran S., Chauhan S.C., Danafar H., Kaboli S. ACS Sens. 2021;6:1430. doi: 10.1021/acssensors.0c02312. [DOI] [PubMed] [Google Scholar]
- 73.Zhu X., Wang X., Li S., Luo W., Zhang X., Wang C., Chen Q., Yu S., Tai J., Wang Y. ACS Sens. 2021;6:881. doi: 10.1021/acssensors.0c01984. [DOI] [PubMed] [Google Scholar]
- 74.Ramachandran A., Huyke D.A., Sharma E., Sahoo M.K., Huang C., Banaei N., Pinsky B.A., Santiago J.G. Proc. Natl. Acad. Sci. U.S.A. 2020;117:29518. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ning B., Yu T., Zhang S., Huang Z., Tian D., Lin Z., Niu A., Golden N., Hensley K., Threeton B. Sci. Adv. 2021;7:3703. doi: 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L., Zhou J., Wang Q., Wang Y., Kang C. Theranostics. 2021;11:649. doi: 10.7150/thno.51479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garcia-Venzor A., Rueda-Zarazua B., Marquez-Garcia E., Maldonado V., Moncada-Morales A., Olivera H., Lopez I., Zuñiga J., Melendez-Zajgla J. Front. Med. 2021;8:125. doi: 10.3389/fmed.2021.627679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., Li Y., Wu Y., Zeng B., Zhang K. Anal. Chem. 2021;93:3393. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- 79.Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T. Nat. Biomed. Eng. 2020;4:1140. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 80.Fu J., Zhou Y., Huang X., Zhang W., Wu Y., Fang H., Zhang C., Xiong Y. J. Agric. Food Chem. 2020;68:1118. doi: 10.1021/acs.jafc.9b07076. [DOI] [PubMed] [Google Scholar]
- 81.Zhou Y., Ding L., Wu Y., Huang X., Lai W., Xiong Y. Trac. Trends Anal. Chem. 2019;112:147. [Google Scholar]
- 82.Sanz L.A., Chédin F. Nat. Protoc. 2019;14:1734. doi: 10.1038/s41596-019-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhatia V., Barroso S.I., García-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A. Nature. 2014;511:362. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 84.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., May B., De Freitas J.K., Glicksberg B.S., Mason C.E. Nat. Med. 2020;26:1609. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilk C.M. Nat. Rev. Immunol. 2020;20:350. doi: 10.1038/s41577-020-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang C., Li W., Drabek D., Okba N.M., van Haperen R., Osterhaus A.D., van Kuppeveld F.J., Haagmans B.L., Grosveld F., Bosch B.-J. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. Nature. 2020;588:682. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gaebler C., Wang Z., Lorenzi J.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y. Nature. 2021;591:639. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K. Science. 2020;369:1010. [Google Scholar]
- 90.Watson J., Richter A., Deeks J. BMJ. 2020;370:3325. doi: 10.1136/bmj.m3325. [DOI] [PubMed] [Google Scholar]
- 91.Ravi N., Cortade D.L., Ng E., Wang S.X. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jalandra R., Yadav A.K., Verma D., Dalal N., Sharma M., Singh R., Kumar A., Solanki P.R. Biomed. Pharmacother. 2020;129:110446. doi: 10.1016/j.biopha.2020.110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petherick A. Lancet. 2020;395:1101. doi: 10.1016/S0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou Y., Huang X., Zhang W., Ji Y., Chen R., Xiong Y. Biosens. Bioelectron. 2018;102:9. doi: 10.1016/j.bios.2017.10.046. [DOI] [PubMed] [Google Scholar]
- 95.Huang X., Zhou Y., Ding L., Yu G., Leng Y., Lai W., Xiong Y., Chen X. Small. 2019;15:1903861. doi: 10.1002/smll.201903861. [DOI] [PubMed] [Google Scholar]
- 96.Leirs K., Tewari Kumar P., Decrop D., Pérez-Ruiz E., Leblebici P., Van Kelst B., Compernolle G., Meeuws H., Van Wesenbeeck L., Lagatie O. Anal. Chem. 2016;88:8450. doi: 10.1021/acs.analchem.6b00502. [DOI] [PubMed] [Google Scholar]
- 97.Huang C., Wen T., Shi F.-J., Zeng X.-Y., Jiao Y.-J. ACS Omega. 2020;5:12550. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ng A.H., Fobel R., Fobel C., Lamanna J., Rackus D.G., Summers A., Dixon C., Dryden M.D., Lam C., Ho M. Sci. Transl. Med. 2018;10:6076. doi: 10.1126/scitranslmed.aar6076. [DOI] [PubMed] [Google Scholar]
- 99.Patris S., Vandeput M., Kauffmann J.-M. Trac. Trends Anal. Chem. 2016;79:239. [Google Scholar]
- 100.Krammer F., Simon V. Science. 2020;368:1060. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 101.Lee Y.-L., Liao C.-H., Liu P.-Y., Cheng C.-Y., Chung M.-Y., Liu C.-E., Chang S.-Y., Hsueh P.-R. J. Infect. 2020;81:55. doi: 10.1016/j.jinf.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. J. Med. Virol. 2020;92:2243. doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uyoga S., Adetifa I.M., Karanja H.K., Nyagwange J., Tuju J., Wanjiku P., Aman R., Mwangangi M., Amoth P., Kasera K. Science. 2021;371:79. doi: 10.1126/science.abe1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wen T., Huang C., Shi F.-J., Zeng X.-Y., Lu T., Ding S.-N., Jiao Y.-J. Analyst. 2020;145:5345. doi: 10.1039/d0an00629g. [DOI] [PubMed] [Google Scholar]
- 105.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y. Anal. Chem. 2020;92:7226. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 106.Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., Kong J. Anal. Chem. 2020;92:9454. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- 107.Hou H., Wang T., Zhang B., Luo Y., Mao L., Wang F., Wu S., Sun Z. Clin. Transl. Immunol. 2020;9:1136. doi: 10.1002/cti2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spicuzza L., Montineri A., Manuele R., Crimi C., Pistorio M.P., Campisi R., Vancheri C., Crimi N. J. Infect. 2020;81:53. doi: 10.1016/j.jinf.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Abreu M.C., Choquet C., Petit H., Bouzid D., Damond F., Marot S., Ferre V.M., Burrel S., Boutolleau D., Houdou-Fidouh N. J. Infect. 2020;81:816. doi: 10.1016/j.jinf.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Biosens. Bioelectron. 2020;172:112765. doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu C., Mao B., Martinez V., Chen X., Li Y., He L., Chen S., Guo X., Shen X., Bao X. RSC Adv. 2020;10:28041. doi: 10.1039/d0ra04107f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Z., Zheng Z., Hu H., Zhou Q., Liu W., Li X., Liu Z., Wang Y., Ma Y. Lab Chip. 2020;20:4255. doi: 10.1039/d0lc00828a. [DOI] [PubMed] [Google Scholar]
- 113.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W. J. Med. Virol. 2020;92:1518. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeng L., Li Y., Liu J., Guo L., Wang Z., Xu X., Song S., Hao C., Liu L., Xin M. Mater. Chem. Front. 2020;4:2000. [Google Scholar]
- 115.Liu B., Li J., Tang X., Wu Z., Lu J., Liang C., Hou S., Zhang L., Li T., Zhao W. MedRxiv. 2020 doi: 10.1101/2020.07.21.20159392. [DOI] [Google Scholar]
- 116.Wang C., Yang X., Gu B., Liu H., Zhou Z., Shi L., Cheng X., Wang S. Anal. Chem. 2020;92:15542. doi: 10.1021/acs.analchem.0c03484. [DOI] [PubMed] [Google Scholar]
- 117.Feng M., Chen J., Xun J., Dai R., Zhao W., Lu H., Xu J., Chen L., Sui G., Cheng X. ACS Sens. 2020;5:2331. doi: 10.1021/acssensors.0c00927. [DOI] [PubMed] [Google Scholar]
- 118.Wang G., Yang L., Li C., Yu H., He Z., Yang C., Sun J., Zhang P., Gu X., Tang B.Z. Mater. Chem. Front. 2021;5:2452. [Google Scholar]
- 119.Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C. Sensor. Actuator. B Chem. 2020;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bayin Q., Huang L., Ren C., Fu Y., Ma X., Guo J. Talanta. 2021;227:122207. doi: 10.1016/j.talanta.2021.122207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rosado J., Pelleau S., Cockram C., Merkling S.H., Nekkab N., Demeret C., Meola A., Kerneis S., Terrier B., Fafi-Kremer S. Lancet Microbe. 2020;5:2452. doi: 10.1016/S2666-5247(20)30197-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fagarasan S., Honjo T. Nat. Rev. Immunol. 2003;3:63. doi: 10.1038/nri982. [DOI] [PubMed] [Google Scholar]
- 123.Bidgood S.R., Tam J.C., McEwan W.A., Mallery D.L., James L.C. Proc. Natl. Acad. Sci. U.S.A. 2014;111:13463. doi: 10.1073/pnas.1410980111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moor K., Diard M., Sellin M.E., Felmy B., Wotzka S.Y., Toska A., Bakkeren E., Arnoldini M., Bansept F., Dal Co A. Nature. 2017;544:498. doi: 10.1038/nature22058. [DOI] [PubMed] [Google Scholar]
- 125.Infantino M., Manfredi M., Grossi V., Lari B., Fabbri S., Benucci M., Fortini A., Damiani A., Mobilia E.M., Panciroli M. J. Med. Virol. 2021;93:1436. doi: 10.1002/jmv.26422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yan Y., Chang L., Wang L. Rev. Med. Virol. 2020;30:2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Cell. Mol. Immunol. 2020;17:773. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu H.-q., Sun B.-q., Fang Z.-f., Zhao J.-c., Liu X.-y., Li Y.-m., Sun X.-z., Liang H.-f., Zhong B., Huang Z.-f. Eur. Respir. J. 2020;56:2000799. doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu J.-L., Tseng W.-P., Lin C.-H., Lee T.-F., Chung M.-Y., Huang C.-H., Chen S.-Y., Hsueh P.-R., Chen S.-C. J. Infect. 2020;81:435. doi: 10.1016/j.jinf.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cavalera S., Colitti B., Rosati S., Ferrara G., Bertolotti L., Nogarol C., Guiotto C., Cagnazzo C., Denina M., Fagioli F. Talanta. 2020;223:121737. doi: 10.1016/j.talanta.2020.121737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li C., Zou Z., Liu H., Jin Y., Li G., Yuan C., Xiao Z., Jin M. Talanta. 2020;223:122064. doi: 10.1016/j.talanta.2020.122064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou Y., Chen Y., Liu Y., Fang H., Huang X., Leng Y., Liu Z., Hou L., Zhang W., Lai W. Biosens. Bioelectron. 2020;171:112753. doi: 10.1016/j.bios.2020.112753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou Y., Chen Y., Liu W., Fang H., Li X., Hou L., Liu Y., Lai W., Huang X., Xiong Y. Sensor. Actuator. B Chem. 2021;343:130139. doi: 10.1016/j.snb.2021.130139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mak G.C., Lau S.S., Wong K.K., Chow N.L., Lau C., Lam E.T., Chan R.C., Tsang D.N. J. Clin. Virol. 2021;134:104712. doi: 10.1016/j.jcv.2020.104712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weitzel T., Legarraga P., Iruretagoyena M., Pizarro G., Vollrath V., Araos R., Munita J.M., Porte L. Trav. Med. Infect. Dis. 2021;39:101942. doi: 10.1016/j.tmaid.2020.101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pekosz A., Parvu V., Li M., Andrews J.C., Manabe Y.C., Kodsi S., Gary D.S., Roger-Dalbert C., Leitch J., Cooper C.K. Clin. Infect. Dis. 2021:ciaa1706. doi: 10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schildgen V., Demuth S., Lüsebrink J., Schildgen O. Pathogens. 2021;10:38. doi: 10.3390/pathogens10010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2020;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S. J. Clin. Microbiol. 2020;58:461. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G. ACS Nano. 2020;14:5135. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 141.Tan H.-X., Juno J.A., Lee W.S., Barber-Axthelm I., Kelly H.G., Wragg K.M., Esterbauer R., Amarasena T., Mordant F.L., Subbarao K. Nat. Commun. 2021;12:1403. doi: 10.1038/s41467-021-21665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan J., Yan X. Biosens. Bioelectron. 2021;173:112817. doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. Biosens. Bioelectron. 2020;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baker A.N., Richards S.-J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gallo A., Lewandowski J.R., Stansfeld P.J., Straube A. ACS Cent. Sci. 2020;6:2046. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. Nature. 2020;584:457. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 146.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C. Cell. 2020;183:739. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dagotto G., Yu J., Barouch D.H. Cell Host Microbe. 2020;28:364. doi: 10.1016/j.chom.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rikhtegaran Tehrani Z., Saadat S., Saleh E., Ouyang X., Constantine N., DeVico A.L., Harris A.D., Lewis G.K., Kottilil S., Sajadi M.M. PLoS One. 2020;15 doi: 10.1371/journal.pone.0237828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. Anal. Chem. 2020;92:11305. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 150.Mertens P., De Vos N., Martiny D., Jassoy C., Mirazimi A., Cuypers L., Van den Wijngaert S., Monteil V., Melin P., Stoffels K. Front. Med. 2020;7:225. doi: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Diao B., Wen K., Zhang J., Chen J., Han C., Chen Y., Wang S., Deng G., Zhou H., Wu Y. Clin. Microbiol. Infect. 2020;27:289. doi: 10.1016/j.cmi.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang C., Zhou L., Du K., Zhang Y., Wang J., Chen L., Lyu Y., Li J., Liu H., Huo J. Front. Cell. Infect. Microbiol. 2020;10:553837. doi: 10.3389/fcimb.2020.553837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miyakawa K., Funabashi R., Yamaoka Y., Jeremiah S.S., Katada J., Wada A., Takei T., Shimizu K., Ozawa H., Kawakami C. MedRxiv. 2021 doi: 10.1101/2021.01.27.21250659. [DOI] [Google Scholar]
- 154.Lippow S.M., Wittrup K.D., Tidor B. Nat. Biotechnol. 2007;25:1171. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim H.-Y., Lee J.-H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Park E.C., Kim H.G. Biosens. Bioelectron. 2020;175:112868. doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Parker E.P., Shrotri M., Kampmann B. Nat. Rev. Immunol. 2020;20:650. doi: 10.1038/s41577-020-00455-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H., Nasrallah G.K. Viruses. 2020;12:582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fozouni P., Son S., de León Derby M.D., Knott G.J., Gray C.N., D'Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I. Cell. 2020;184:323. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Miller B.S., Bezinge L., Gliddon H.D., Huang D., Dold G., Gray E.R., Heaney J., Dobson P.J., Nastouli E., Morton J.J. Nature. 2020;587:588. doi: 10.1038/s41586-020-2917-1. [DOI] [PubMed] [Google Scholar]
- 160.Gu L., Shi H., Bian L., Gu M., Ling K., Wang X., Ma H., Cai S., Ning W., Fu L. Nat. Photonics. 2019;13:406. [Google Scholar]
- 161.Li D., Jing P., Sun L., An Y., Shan X., Lu X., Zhou D., Han D., Shen D., Zhai Y. Adv. Mater. 2018;30:1705913. doi: 10.1002/adma.201705913. [DOI] [PubMed] [Google Scholar]
- 162.Huang X., Aguilar Z.P., Xu H., Lai W., Xiong Y. Biosens. Bioelectron. 2016;75:166. doi: 10.1016/j.bios.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 163.Liu Y., Zhan L., Qin Z., Sackrison J., Bischof J.C. ACS Nano. 2021;15:3593. doi: 10.1021/acsnano.0c10035. [DOI] [PubMed] [Google Scholar]
- 164.Chen A., Yang S. Biosens. Bioelectron. 2015;71:230. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 165.Nguyen V.-T., Song S., Park S., Joo C. Biosens. Bioelectron. 2020;152:112015. doi: 10.1016/j.bios.2020.112015. [DOI] [PubMed] [Google Scholar]
- 166.Bishop J.D., Hsieh H.V., Gasperino D.J., Weigl B.H. Lab Chip. 2019;19:2486. doi: 10.1039/c9lc00104b. [DOI] [PubMed] [Google Scholar]
- 167.Ye H., Liu Y., Zhan L., Liu Y., Qin Z. Theranostics. 2020;10:4359. doi: 10.7150/thno.44298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shirshahi V., Liu G. Trac. Trends Anal. Chem. 2021;136:116200. [Google Scholar]
- 169.Wu Y., Zhou Y., Leng Y., Lai W., Huang X., Xiong Y. Biosens. Bioelectron. 2020;157:112168. doi: 10.1016/j.bios.2020.112168. [DOI] [PubMed] [Google Scholar]
- 170.Shelite T., Uscanga-Palomeque A., Castellanos A., Melby P., Travi B. J. Virol. Methods. 2020;296:114227. doi: 10.1016/j.jviromet.2021.114227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.-S.F., Kemball M.E. Nat. Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brandsma E., Verhagen H.J., van de Laar T.J., Claas E.C., Cornelissen M., van den Akker E. J. Infect. Dis. 2021;223:206. doi: 10.1093/infdis/jiaa641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Javalkote V.S., Kancharla N., Bhadra B., Shukla M., Soni B., Goodin M., Bandyopadhyay A., Dasgupta S. Methods. 2020 doi: 10.1016/j.ymeth.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan J., Yan X. Biosens. Bioelectron. 2020;173:112817. doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]