Abstract
We present a compartmental extended SEIQRD metapopulation model for SARS-CoV-2 spread in Belgium. We demonstrate the robustness of the calibration procedure by calibrating the model using incrementally larger datasets and dissect the model results by computing the effective reproduction number at home, in workplaces, in schools, and during leisure activities. We find that schools and home contacts are important transmission pathways for SARS-CoV-2 under lockdown measures. School reopening has the potential to increase the effective reproduction number from (95 % CI) to (95 % CI) under lockdown measures. The model accounts for the main characteristics of SARS-CoV-2 transmission and COVID-19 disease and features a detailed representation of hospitals with parameters derived from a dataset consisting of 22 136 hospitalized patients. Social contact during the pandemic is modeled by scaling pre-pandemic contact matrices with Google Community Mobility data and with effectivity-of-contact parameters inferred from hospitalization data. The calibrated social contact model with its publically available mobility data, although coarse-grained, is a cheap and readily available alternative to social–epidemiological contact studies under lockdown measures, which were not available at the start of the pandemic.
Keywords: SARS-CoV-2, Compartmental SEIQRD model, Non-pharmaceutical interventions, Google Community Mobility data, COVID-19 hospital length-of-stay, Social contact effectivity, Schools closure
1. Introduction
After an initial outbreak in early 2020 in Wuhan, China, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread globally (Li et al., 2020a). SARS-CoV-2 is capable of sustained human-to-human transmission (Riou and Althaus, 2020) and may cause severe disease and death, especially in older individuals. The SARS-CoV-2 pandemic has, in general, shown a remarkably low incidence among children and young adults (Davies et al., 2020, Verity et al., 2020, Molenberghs et al., 2020). Furthermore, presymptomatic transmission is a major contributor to SARS-CoV-2 spread (Liu et al., 2020a, Wei et al., 2020). Both on March 15th, 2020, and on October 19th, 2020, the Belgian governments imposed social restrictions after testing & tracing methods had failed to prevent the large-scale spread of SARS-CoV-2. Recently, pharmaceutical interventions under the form of vaccinations have become available. If natural immunity wanes or if SARS-CoV-2 further mutates, it is expected that SARS-CoV-2 will become endemic (Shaman and Galanti, 2020). Hence, there is a need for well-informed models and knowledge build-up to assist policymakers in choosing the best cocktail of non-pharmaceutical and pharmaceutical interventions during future SARS-CoV-2 outbreaks.
Currently, four other models exist to inform policymakers in Belgium. The agent-based model (ABM) of Willem et al. (2020), the data-driven model by Barbe et al. (2020) and the compartmental models of Abrams et al. (2021) and Franco (2020). The models of Abrams et al. (2021) and Franco (2020) feature similar disease progression as our model but use different methods to model social contact. To account for structural uncertainty in the models, their outputs are currently combined into an ensemble to inform policymakers (Willem, 2021).
In this work, we present our compartmental, age-stratified, nation-level model which accounts for the main characteristics of SARS-CoV-2 disease. The model features a detailed representation of hospitals with residence times and mortalities derived from a large dataset of hospitalized patients in Belgium. We built a social contact model which scales pre-pandemic contact matrices from a study by Willem et al. (2012) with the Google Community Mobility data (Google LLC, 2020) and with effectivity-of-contact parameters derived from hospitalization data using an Markov Chain Monte Carlo (MCMC) method (Goodman and Weare, 2010). Tardiness in compliance with social restrictions is included using a delayed-ramp model and waning of humoral immunity is included by estimating the rate of seroreversion from two serological datasets. We find that the combination of the deterministic epidemiological model, which incorporates rigid a priori knowledge on disease dynamics, and the calibrated effectivity-of-contact parameters in the social contact model allows us to combine the ease of long-term extrapolation and scenario analysis of compartmental models with the flexibility of a data-driven model. The model does not require ad hoc tweaking and is computationally cheap, making it ideal to perform optimizations that require thousands of model evaluations. Further, due to the public nature of the Google Community Mobility data, the model provides a more rapidly deployable alternative to social epidemiological studies comparing mixing patterns during and after lockdown, such as Coletti et al. (2020) for Belgium, which were not available at the start of the pandemic.
Using a hospitalization dataset of 22 136 coronavirus disease 19 (COVID-19) patients in Belgian hospitals, we computed age-stratified hospital residence times and mortalities. Using the obtained parameters, we found the model was able to predict the total number of patients and the number of deceased patients in Belgian hospitals well. We calibrated the model to hospitalization data made publically available by the Belgian Scientific Institute of Public Health (Sciensano) and demonstrated the calibration procedure’s robustness. We computed the basic reproduction numbers () and the time to reach compliance to lockdown measures during both coronavirus disease 2019 (COVID-19) waves in Belgium. The average time to for anti-SARS-CoV-2 antibodies to wane (seroreversion), was estimated as 9.2 months (IQR: 7.2–12.1 months). Using the calibrated model, we computed the relative share of contacts and the effective reproduction numbers at home, at school, at work and during leisure activities to assess their effect on SARS-CoV-2 spread during both 2020 COVID-19 waves. We observed a strong correlation between school re-opening and increases in SARS-CoV-2 transmission. More precisely, schools have the potential to increase the effective reproduction number from (95% CI) to (95% CI) under lockdown measures.
Throughout the work, Belgium is used as a case but the scope of the work is extendable to other countries. Since February 2021, the effects of new SARS-CoV-2 strains and pharmaceutical interventions (vaccines) need to be accounted for. For this purpose, a model extension was developed and is currently used in the aforementioned model ensemble (Willem, 2021). However, due to the longevity of this work, we chose to limit the scope of this study to the effects of non-pharmaceutical interventions.
2. Materials and methods
2.1. The extended SEIQRD-model
2.1.1. Disease dynamics
The SEIR(D) model (Kermack et al., 1927) is a compartmental model that subdivides the human population into four groups: 1. susceptible individuals (S), 2. exposed individuals in the latent phase (E), 3. infectious individuals capable of transmitting the disease (I) and 4. individuals removed from the population either through immunization or death (R/D). Despite being a simple and idealized reality, the SEIR(D) dynamics are used extensively to predict the outbreak of infectious diseases and this was no different during the SARS-CoV-2 outbreak earlier this year (Wu et al., 2020b, Li et al., 2020a, Davies et al., 2020).
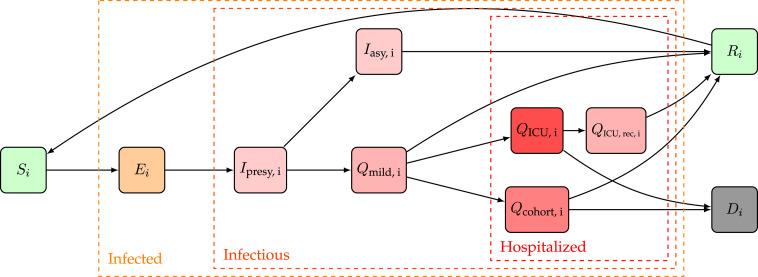
In this work, we extended the SEIRD model to incorporate more expert knowledge on SARS-CoV-2 disease dynamics. For that purpose, the infectious compartment was split into four parts. The first is a period of presymptomatic infectiousness because several studies have shown that presymptomatic transmission is a dominant transmission mechanism of SARS-CoV-2 (Wei et al., 2020, Liu et al., 2020a). After the presymptomatic period, three possible infectious outcomes are modeled: (1) Asymptomatic outcome, for individuals who show no symptoms at all, (2) Mild outcome, for individuals with mild symptoms who recover at home, and (3) Hospitalization, when mild symptoms worsen. Children and young adults have a high propensity to experience an asymptomatic or mild outcome, while older individuals have a higher propensity to be hospitalized (Liu et al., 2020a, Wei et al., 2020). Belgian hospitals generally have two wards for COVID-19 patients: (1) cohort, where patients are not monitored continuously and (2) Intensive care units (ICUs), for patients with the most severe symptoms. Intensive care includes permanent monitoring, the use of ventilators, or the use of extracorporeal membrane oxygenation (ECMO). Patients can perish in both hospital wards, but mortalities are generally lower in cohort. After a stay in an ICU, patients return to cohort for recovery in the hospital. During the recovery stay, mortality is limited. We assume that mildly infected individuals and hospitalized patients cannot infect susceptibles are thus quarantined. Because reinfections with SARS-CoV-2 have already been reported (Tillett et al., 2021, Prado-Vivar et al., 2020, Van Elslande et al., 2020, Gupta et al., 2020), and because it has already been estimated that anti-SARS-CoV-2 antibodies wane (Rosado et al., 2021, Wheatley et al., 2021), we incorporate waning antibody immunity by sending recovered individuals back to the susceptible population pool. The model dynamics are depicted in Fig. 1.
Fig. 1.
Extended SEIQRD dynamics used in this study. Here, stands for susceptible, for exposed, for presymptomatic and infectious, for asymptomatic and infectious, for mildly symptomatic and infectious, for cohort, for a recovery stay in cohort coming from IC, for Intensive Care Unit, for dead and for recovered. A subscript is used to denote the th age strata of the model, the model has a total of nine age strata. An overview of the model parameters can be found in Table 1 (supplementary materials).
2.1.2. Model structure and equations
In this work, we implemented the extended SEIQRD dynamics shown in Fig. 1 using ordinary differential equations (ODEs), without spatial stratification and with age-stratification. This was accomplished by defining a system of ordinary differential equations, one for every of the model compartments, each of which is further split into age-stratified metapopulations. The age groups have different contact rates with other age groups and the disease progresses differently for each age group, making the model behave realistically. Our model consists of 9 age classes, i.e., , , , , , , , , . Our choice for ODEs over network- or agent-based models is motivated mainly by the limited computational resources required to explore scenarios and perform optimizations requiring thousands of function evaluations. The model dynamics are translated into the following system of coupled ordinary differential equations,
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (7) |
| (8) |
| (9) |
| (10) |
| (12) |
| (13) |
for . Here, stands for total population (Table 1, supplementary materials), stands for susceptible, for exposed, for presymptomatic and infectious, for asymptomatic and infectious, for mildly symptomatic and infectious, for cohort, for a recovery stay in cohort coming from Intensive Care, for Intensive Care Unit, for dead and for recovered. A subscript to these variables is used to refer to one of the nine age strata in the model. Using the above notation, all model states are 9-dimensional vectors,
where denotes the number of susceptibles in age-class at time after the introduction of SARS-CoV-2 in the population. As initial condition, the whole population is assumed susceptible to SARS-CoV-2 and one exposed individual and one pre-symptomatic infectious individual in every age class is assumed, so for all . The time between the start of the simulation and the start of data collection is then estimated when calibrating the model. An overview of all model parameters, their values, and their meaning can be found in Table 1 (supplementary materials). In what follows, the most important model parameters and their chosen values are motivated.
2.1.3. Model parameters
Transmission rate and social contact data
The transmission rate of the disease depends on the product of four contributions (Eq. (1)). The first contribution, , is the fraction of contagious individuals in age group . We thus assume presymptomatic and asymptomatic individuals spread the disease, while mildly infected are assumed to self-quarantine and hospitalized individuals cannot infect health care workers. The second contribution, , is the average number of human-to-human interactions of an individual in age group , with an individual in age group per day. The sum of the first two contributions over all age groups , , is the number of contacts of an individual in age group that can result in SARS-CoV-2 transmission. This is multiplied with the number of susceptibles in age group (), and with , the probability of contracting COVID-19 when encountering a contagious individual, to compute the number of effective contacts at every timestep. We assume that the per-contact transmission probability is independent of age and we infer its value by calibrating our model to Belgian hospitalization data. In a model-based inference-based study by Davies et al. (2020), it was deduced that children were less susceptible to SARS-CoV-2 disease. Viner et al. (2021) analyzed 32 studies that reported on the susceptibility of children and found preliminary evidence that susceptibility to SARS-CoV-2 infection is lower in children. However, it assumed in our model that individuals of all ages to have an equal susceptibility to and transmissibility of SARS-CoV-2. The number of (pre-pandemic) human–human interactions, , are both place and age-dependent. These matrices assume the form of a 9x9 interaction matrix where an entry , denotes the number of social contacts age group has with age group per day. These matrices are available for homes (), schools (), workplaces (), in public transport (), during leisure activities () and during other activities (), from a study by Willem et al. (2012). The total number of prepandemic social interactions must be translated into an appropriately weighted sum of the contributions in different places, adequately describing pandemic social behavior (Section 2.3). The basic reproduction number , defined as the expected number of secondary cases directly generated by one case in a population where all individuals are susceptible to infection, is computed using the next-generation matrix (NGM) approach introduced by Diekmann et al., 1990, Diekmann et al., 2010. For our model, the basic reproduction number of age group is,
| (14) |
and the population basic reproduction number is calculated as the weighted average over all age groups using the demographic data in Table 1 (supplementary materials). The detailed algebra underlying the computation of Eq. (14) is presented in the supplementary materials (Section A.4).
Duration of infectiousness
The duration of infectiousness is determined by the number of days patients can spread viral particles. Several studies have reported patients have the highest viral load of the coronavirus at the time they are diagnosed and patient’s viral loads declining gradually over time (He et al., 2020, Liu et al., 2020b, Lescure et al., 2020, To et al., 2020, Zou et al., 2020). He et al. (2020) inferred the infectiousness profile of COVID-19 patients to be an approximately normal distribution, with the peak infectivity roughly at the time of symptom onset and infectiousness quickly declining within 7 days after symptom onset. A comparison of viral load between symptomatic and one asymptomatic case revealed similar viral loads, an indicator that asymptomatic individuals can be as infectious as symptomatic patients (Zou et al., 2020). He et al. (2020) further concluded that 44% of secondary cases were infected during the presymptomatic stage, a finding consistent with studies from other authors (Liu et al., 2020a, Wei et al., 2020). Wei et al. (2020) determined that presymptomatic transmission exposure occurred 1–3 days before the source patient developed symptoms . In Eq. (2), denotes the length of the latent, non-infectious period and in Eq. (3), is the length of the presymptomatic infectious period. In this work, we assume the incubation period, equal to , lasts 5.2 days (Liu et al., 2020a). The length of the presymptomatic period is fixed at 0.7 days, which corresponds to 44% of SARS-CoV-2 infections experiencing a presymptomatic infectious period of 2 days. The duration of infectiousness for mildly symptomatic cases () is assumed to be 7 days. The average duration of asymptomatic infectiousness, on which the basic reproduction number () depends (Eq. (14)), will be inferred from hospitalization data using an MCMC method (Section 2.4).
Disease severity and hospitalizations
The model parameter (Eq. (4)) is the probability of an individual in age group having a subclinical infection. Several authors have attempted to estimate the fraction of asymptomatic infections. Li et al. (2020b) estimated that 86% of coronavirus infections in China were undocumented in the weeks before their government instituted stringent quarantines. However, this figure includes an unknown number of mildly symptomatic cases and is thus an overestimation of the asymptomatic fraction. In Iceland, citizens were invited for testing regardless of symptoms. Of all people with positive test results, 43% were subclinical (Gudbjartsson et al., 2020). A systematic review and meta-analysis by Buitrago-Garcia et al. (2020) suggested a lower subclinical fraction of 31% (26%–37%, 95% CI). If the subclinical fractions per age group estimated by Davies et al. (2020) are applied to the Belgian population, an average subclinical fraction of 57% is obtained for Belgium. In this study, we applied the relative subclinical fraction per age group of Wu et al. (2020a) to obtain a population average subclinical fraction of 57% (Table 1, supplementary materials). In Eq. (5), is the fraction of mild cases that require hospitalization and in Eq. (8), is the fraction of the hospitalized which remain in cohort. In this study, the age-stratified hospitalization probabilities () were inferred from hospital mortality data (Table 1) and the age-stratified distributions between cohort and ICU () were computed using data from 22 136 patients treated in Belgian hospitals (Section 2.2). In Eq. (5), is the average time between first symptoms and hospitalization, which was previously estimated as 5–9 days by Linton et al. (2020) and as 4 days by To et al. (2020). In Eqs. (8), (9), (10), , , and are the age-stratified average lengths of a hospital stay in cohort and in an ICU. The subscript denotes the duration if the patient recovers, while subscript denotes the duration if the patient perishes. and are the age-stratified mortalities of patients in cohort and in ICU. In Eq. (10), denoted the age-stratified length of a recovery stay in cohort after a stay in ICU. The aforementioned hospitalization parameters are computed using data from 22 136 patients treated in Belgian hospitals. The methodology of the analysis is presented in Section 2.2, the results of the analysis are presented in Section 3.1
Testing, tracing and quarantine, waning antibody immunity
The effects of testing, tracing and quarantine are not explicitly implemented for this study. Reinfections with SARS-CoV-2 have been reported in single cases in the USA (Tillett et al., 2021), Ecuador (Prado-Vivar et al., 2020) and Belgium (Van Elslande et al., 2020). Further, two asymptomatic reinfections were also reported in Indian healthcare workers (Gupta et al., 2020). Rosado et al. (2021) estimated that antibodies could wane in 50% of recovered individuals after 1 year. Wheatley et al. (2021) found that both neutralizing and binding antibody responses decay after recovery from a mild COVID-19 infection. Although the long-term kinetics of the antibody response to SARS-CoV-2 will not be definitively quantified until infected individuals are followed years after a confirmed infection, and although the persistence of serum antibodies is unlikely to be the sole determinant of long-lasting immunity (memory T and B cells), it is clear that waning of antibodies best be included in our model. In Eqs. (1), (12), the rate of anti-SARS-CoV-2 antibody waning is denoted as , and its inverse is the average time for anti-SARS-CoV-2 antibodies to wane. Using serological data by Herzog et al. (2021) and the Belgian Scientific Institute of Public Health (Sciensano), the distribution of will be inferred using an MCMC method.
2.2. Analysis of hospital surveillance data
A subset of data from the Belgian COVID-19 clinical surveillance on hospitalizations by Van Goethem et al. (2020), which was anonymized and provided through a secured data transfer platform by the Belgian Scientific Institute of Public Health (Sciensano), is analyzed to compute age-stratified estimates of the following model parameters: the distribution of patients between the cohort and IC wards (), the residence times in the cohort and IC wards, in the case of recovery and in the case of death (, , , ), the residence time for a recovery stay in cohort after a stay in ICU (), the time between symptom onset and hospitalization () and the mortalities in the hospital, cohort and IC wards (, , ). The raw data consists of 52 327 patients hospitalized in Belgian hospitals between March 4th, 2020, and March 3rd, 2021. Data are reported for all hospitalized patients with a confirmed COVID-19 infection (diagnosed using reverse transcriptase–polymerase chain reaction, chest computed tomography, or rapid antigen test) and the reporting coverage on the period 15th of March - 27th of June was estimated to be rough 70% of all hospitalized COVID-19 cases (Van Goethem et al., 2020). The data gathered during the period March 14th, 2020 until June 12th, 2020 were previously analyzed by Faes et al. (2020). The added value of performing a similar analysis in this study is threefold: (1) To include the patient data gathered in the meantime. (2) To compute the age-stratified mortalities in the cohort and IC hospital wards (, ), as well as the age-stratified recovery time in cohort after a stay in ICU (), which were not included by Faes et al. (2020). (3) To obtain age-stratified estimates in nine ten-year age strata as compared to four age strata by Faes et al. (2020). For every patient the following data were provided: (1) age, (2) date of onset of symptoms (3) hospital admission date, (4) hospital discharge date, (5) date of ICU transfer, (6) the number of days spent in ICU, (7) outcome (recovered or deceased). Data from 30 191 patients were excluded from the analysis because one or more of the above entries were missing or because the computed residence times were negative. Patients that came from a nursing home were excluded from the analysis. Thus, in total, data from the remaining 22 136 patients were used (Figure 14). The confidence intervals of the mortalities (, and ) and the distribution between the cohort and IC ward () were computed using bootstrap resampling. For all hospital residence times, the shape and scale parameters of a Weibull distribution were fitted to the data. To determine if the duration of a cohort or ICU stay differed significantly and to determine if the mortalities in cohort and ICU differed significantly, a non-parametric Mann–Whitney U-test was used. Temporal changes in the estimated hospitalization parameters are not considered in this study. The results of the analysis are presented in Section 3.1.
2.3. Social contact model
The pandemic social behavior of the Belgian population must be translated into a linear combination of the aforementioned pre-pandemic interaction matrices of Willem et al. (2012). Mathematically, we must find tangible coefficients so that the linear combination of pre-pandemic interaction matrices, i.e.,
| (15) |
is a good representation of macroscopic social behavior during the pandemic. Ideally, pandemic contact matrices are used as these will better represent mixing behavior under lockdown measures. However, such matrices were not available at the start of the pandemic. Hence, our model builds upon pre-pandemic knowledge of social behavior to make a prediction on pandemic social behavior. In our model, the pre-pandemic matrices are scaled with mobility reductions from Google’s Community Mobility Reports (GCMRs) and with an additional parameter to account for the effectivity of the contact to contribute to SARS-CoV-2 spread.
Mobility reductions
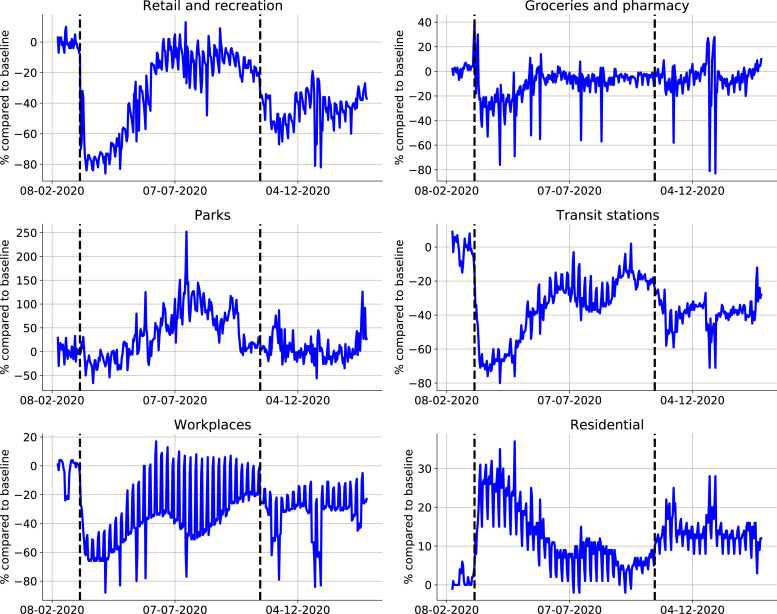
The GCMRs collates data from smartphone users accessing Google applications who allow recording of their location history (Aktay et al., 2020). The data are categorized into six discrete categories: (1) retail and recreation, (2) parks, (3) groceries and pharmacies, (4) workplaces, (5) transport and (6) residential areas. The GCMRs provide the percentage change in activity at each location category compared to that on baseline days before the start of the COVID-19 pandemic (a 5-week period running from 3 January 2020 to 6 February 2020) (Google LLC, 2020). The GCMRs are not age-stratified and do not correct for potential underrepresentation of older individuals in the data collection. In our model, the GCMRs for Workplaces, Transit stations, Retail & recreation and Groceries & pharmacy are used as proxies to scale the work (), transport (), leisure () and other () social contact matrices.
Two surges in COVID-19 cases were observed in Belgium, resulting in two lockdowns (Fig. 2). The first lockdown was imposed on March 15th, 2020, and lasted until May 4th, 2020, and involved the closure of schools, bars, clubs, restaurants, all non-essential shops, and a closure of the border to non-essential travel (Table 2, supplementary materials). From May 4th, 2020 until July 1st, 2020 the lockdown was gradually lifted. During the first lockdown, schools remained fully closed until May 18th, 2020, and were only re-opened to a very limited extent before the end of the school year on July 1st, 2020. The second lockdown was imposed on October 19th, 2020, and was gradually lifted in May 2021. Schools were closed on November 2nd, 2020, and re-opened on November 16th, 2020. Further, schools were closed during the Christmas holidays from December 18th, 2020 until January 4th, 2021. Universities remained fully closed since October 19th, 2020. Briefly summarized, the first 2020 COVID-19 wave consisted of (1) a rapid surge in cases, (2) a lockdown, and (3) a release of lockdown measures. The second 2020 COVID-19 wave consisted of (1) a rapid surge in cases, (2) a lockdown with schools closed, (3) a lockdown with varying school policies.
Fig. 2.
Mobility data extracted from the Google Community Mobility Reports. Dashed lines indicate the start of the first lockdown on Friday, March 13th, 2020, and the start of the second lockdown on Monday, October 19th, 2020. Increases in the categories residential and parks suggest increased activity around the home environment, while increases in the other categories are more indicative of increases in general mobility (Sulyok and Walker, 2020). The mobility reduction in workplaces is used to scale the work interaction matrix, the retail & recreation reduction is used to scale the leisure interaction matrix, the groceries & pharmacy reduction is used to scale the other interaction matrix, the transit stations reduction is used to scale the public transport mobility matrix.
During both lockdowns, mobility increases in the categories residential and parks were observed (Fig. 2). These are indicative of decreased community mobility, as these suggest increased activity around the home environment. The other four categories are more indicative of general mobility as they are related to activity around workplaces, retail outlets and use of public transportation (Sulyok and Walker, 2020). Thus, although the mobility figures indicate people spent more time at home, this does not mean people have more contacts at home (especially under stay-at-home orders). Amplifying the fraction of household contacts under lockdown measures would increase intergenerational mixing of the population under lockdown, which is unrealistic and will lead to overestimations of the hospitalizations. The inability to accurately capture the disease spread in home bubbles under lockdown measures is an inherent downside of compartmental epidemiological models. We have thus not scaled the home interaction matrix () with the residential mobility from the GCMRs.
Effectivity parameters
During the first lockdown, we estimated that the overall fraction of the social contacts that contributed to SARS-CoV-2 spread, from hereon referred to as the effectiveness of the contacts (), was approximately one third of what would be expected based on the GCMRs mobility reductions. During the first 2020 lockdown, work mobility decreased by 56%, the public transport mobility decreased by 65%, leisure mobility decreased by 72% and grocery (others) mobility decreased by 26% (Table 2, supplementary materials). Mathematically, the linear combination of pre-pandemic contact matrices able to adequately describe pandemic behavior was equal to,
| (16) |
Intuitively, the effectivity of a contacts in a given location may not scale linearly with the observed mobility reductions. The net effectivity of the contacts under lockdown measures depends on a combination of the pre-pandemic physical proximity and duration of the contact, the effectivity of preventive measures and on behavioral changes when lockdown measures are taken. As an example, the effects of alcohol gel and face masks might be large in the workplace and in grocery stores, but not at home or during leisure activities. To account for different effectivities of contacts in different places, we could introduce one additional parameter per contact matrix, bound between zero and one, and infer its distribution from the available hospitalization data. However, estimating six effectivity parameters is unfeasible because of identifiability issues. The effectivity parameters of public transport and other places could not be identified. This is most likely because very little contacts are made in those places (Mossong et al., 2008). Consequently, the effectivity parameters of public transport, other places and leisure contacts were aggregated to reduce the number of effectivity parameters from six to four. Finally, the linear combination of interaction matrices used to represent social contact under lockdown measures is,
| (17) |
Here, , , , , and denote the pre-pandemic contact matrices at home, in schools, in workplaces, on public transport, during leisure activities and during other activities (Willem et al., 2012). , , and denote the GCMRs mobility reductions in the respective categories and are updated at every timestep in the simulations. denotes the fraction of schools opened, as school opening cannot be deduced from the GCMRs. In spite of their limited re-opening on May 18th, 2020, schools are assumed to be closed during the first lockdown. , , , are the effectivity parameters at home, in schools, at work and during the combination of leisure, public transport and other activities.
Obedience to measures
In reality, compliance to social restrictions is gradual and cannot be modeled using a step-wise change of the social interaction matrix (Section 2.1.3). The added value of a social compliance model is to gradually introduce the effects of the effectivity parameters in the model when lockdown measures are taken. The added degrees of freedom aid in obtaining a better prediction of the peak in hospitalizations. In this study, we use a delayed ramp to model compliance, i.e.,
| (18) |
where,
where is the number of days before measures start having an effect and is the number of additional days after the time delay until full compliance is reached. Both parameters are calibrated to the daily number of hospitalizations in Belgium (Section 2.4). The difference denotes the number of days since a change in social policy.
2.4. Parameter identification
Aim of the calibration procedure
To demonstrate the robustness of the social contact model and calibration method, for each of the 2020 COVID-19 waves, we calibrate the model to a minimal dataset and then increase the amount of data used in the calibration procedure to assess if the model can adequately predict future hospitalizations and to assess if the posterior distributions of the effectivity parameters () converge. For the first 2020 COVID-19 wave, we calibrate the model using data until April 4th, 2020, and then extend the data range used in the calibration in two-week increments until July 1st, 2020. During the second 2020 COVID-19 wave, we calibrate the model until November 7th, 2020, and then extend the calibration to the date of schools re-opening until November 16th, 2020, the date of schools closing for Christmas holidays on December 18th, 2020 and we finally calibrate until February 1st, 2021. By February 1st, 2021, the full impact of school closure and decrease in work mobility during the holiday period is visible in the new hospitalizations. Extending the calibration beyond February 1st, 2021 is out of scope for this study, as the emergence of more contagious strains (B.1.1.7) and the national vaccination campaign need to be included from this point onward (Table 2, supplementary materials). As previously mentioned, the effectiveness of contacts in schools cannot be studied during the first 2020 COVID-19 wave because schools were only opened to a very limited extent before their final closure on July 1st, 2020.
Calibrated parameters
The model parameters , , , , , , are calibrated to the time-series of daily new hospitalizations (), which are available for download at https://epistat.sciensano.be/Data. The seroreversion rate, , is estimated using five serological measurements from Herzog et al. (2021) and eight serological measurements from Sciensano, spanning a period from March 30th, 2020 until July 7th, 2020. For the sake of computational efficacy, the model is first calibrated to the first 2020 COVID-19 wave in Belgium, then, the model states on September 1st, 2020 are used as the initial condition to initiate the calibration of the second 2020 COVID-19 wave. In this way, the calibration procedure is split between the first 2020 COVID-19 wave from March 15th, 2020 until July 1st, 2020, and the second 2020 COVID-19 wave from September 1st, 2020 until February 1st, 2021. In total, eight model parameters are calibrated to data.
Statistical model
We assume the data are independent and identically distributed (i.d.d.) sequences of Poisson variables. The resulting log-likelihood function is,
| (19) |
where the vector of parameters, , that maximizes the log-likelihood function must be found. In Eq. (19), denotes the model prediction, denotes the timeseries of data and represents the number of datapoints.
Calibration procedure
The fitting procedure is performed in two steps. Maximizing the result of Eq. (19) is computationally demanding and suffers from the presence of local maxima. We thus need an efficient way to scan through the nine-dimensional parameter space . A good technique to initially broadly identify the region where the global maximum is situated is Particle Swarm Optimization (PSO) (Kennedy and Eberhart, 1995). When a region of interest has been identified, we use the maximum-likelihood estimates as initial values for the ensemble sampler for Markov Chain Monte Carlo (MCMC) proposed by Goodman and Weare (2010). For all parameters, uniform prior distributions were used.
2.5. Effects of non-pharmaceutical interventions
To better compare the effects of mobility changes on the daily number of new hospitalizations, we compute the relative share of contacts and the effective reproduction number () at home, in schools, in workplaces and for the combination of leisure, public transport and other contacts. The number of effective contacts in the aforementioned places at time are equal to,
| (20) |
| (21) |
| (22) |
| (23) |
where , , , denote the number of effective contacts at home, in schools, at work or for the sum of leisure, public transport and other contacts. The relative share of contacts in location and for age group is computed as,
| (24) |
The effective reproduction number for age group , in place and at time is computed as,
| (25) |
Finally, the population average effective reproduction number in place , and the population average relative share of contacts in location , are computed as the weighted average over all age groups using the demographics listed in Table 1 (supplementary materials).
3. Results
3.1. Analysis of hospital surveillance data
The average time from symptom onset to hospitalization is 6.4 days (IQR 2.0–8.0 days). Of the 22 136 hospitalized patients, 3 624 patients (16.2%) required intensive care at some point during their stay and 18 512 (83.8%) remained in cohort. The overall mortality in the hospital is 21.4%, the mortality in cohort was significantly lower than the mortality in ICU (16.6% vs. 46.3%, ). One patient under 20 years old has died from COVID-19, mortality is generally low for young patients and increases with older age (Figure 16 and Table 4 of the supplementary materials). The average length of the stay in a cohort ward was 11.0 days (IQR: 4.0–13.0 days) and the average length of an ICU stay was 13.6 days (IQR: 4.0–19.0 days) (). The average cohort stay was 10.8 days (IQR: 4.0–12.0 days) if the patient had recovered and 11.8 days (IQR: 4.0–14.0 days) if the patient had died (). The average ICU stay was 12.0 days (IQR: 3.0–15.0 days) if the patient had recovered and 15.2 days (IQR: 5.0–21.0 days) if the patient had died (). Patients recovering from their ICU stay spend 11.2 additional days (IQR: 4.0–13.0 days) in cohort for a recovery and observation stay. Residence times in cohort are shorter than residence times in ICU. In both wards, deceased patients had longer stays than recovered patients (Figure 15 and Table 5 of the supplementary materials). Residence times in cohort and ICU increase with the patient’s age, the same goes for the length of a recovery stay after a stay in ICU. For example, a 20–30 year old patient is expected to spend 6.3 days (IQR: 2.0–7.0 days) in cohort while a 70–80 year old patient is expected to spend 12.6 days in cohort (IQR: 5.0–14.0 days) (Table 5, supplementary materials).
3.2. Model calibration
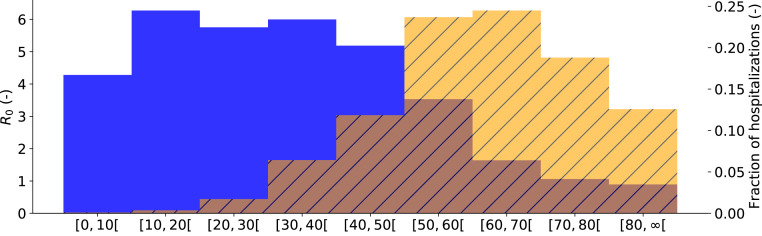
The population average basic reproduction number was computed as (IQR: 3.90–4.39) for the first 2020 COVID-19 wave and as (IQR: 3.64–3.75) for the second 2020 COVID-19 wave. Large differences in the basic reproduction number exist between the different age groups (Fig. 3). It is clear that the youths and working-aged population drive the SARS-CoV-2 pandemic while people of ages 70 or above can hardly sustain a SARS-CoV-2 pandemic amongst themselves, this is mainly because elderly individuals have limited social interactions (Fig. 3). Still, these individuals make up roughly 35% of all hospitalizations. The biggest risk group are the individuals aged 50 to 70, which make up roughly 50% of the expected hospitalizations. The high expected fraction of hospitalizations in this age group is due to a trade-off between social contact and hospitalization risk. These individuals have plenty of social contact and at the same time, have a high propensity to hospitalization.
Fig. 3.
Basic reproduction number per age group (), for Belgium (blue). Expected fraction of the total Belgian hospitalizations during the first COVID-19 wave, as predicted by the model, from March 15th, 2020 until July 1st, 2020 in age group (orange, striped). Youths and working-aged population drive the pandemic, while the senior population is mostly in need of hospital care. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Compliance to lockdown measures was similar for both 2020 COVID-19 waves, with an average delay of 0.22 (IQR: 0.07–0.31) and 0.39 (IQR: 0.20–0.52) days, and a time to reach full compliance to measures of 9.17 (IQR: 8.89–9.50) and 6.94 (IQR: 6.71–7.18) days respectively. Using the serological datasets by Herzog et al. (2021) and Sciensano, the average time to seroreversion () was estimated as 9.2 months (IQR: 7.2–12.1 months) (Figure 13, supplementary materials). The model was calibrated to the daily hospitalizations and serological data, however, to obtain estimates for the total number of patients in Belgian hospitals and the number of deceased patients in Belgian hospitals, the hospitalization parameters computed using the clinical surveillance dataset are propagated in the model using bootstrap sampling. In supplementary Figs. 9 and 10, the ability of the calibrated model to predict the number of daily hospitalizations, the total number of patients in Belgian hospitals, the total number of deaths in Belgian hospitals, and the seroprevalence in the Belgian population during both 2020 COVID-19 waves are demonstrated. The model’s ability to predict the number of hospital deaths in every age strata is demonstrated in Figures 11 and 12 of the supplementary materials.
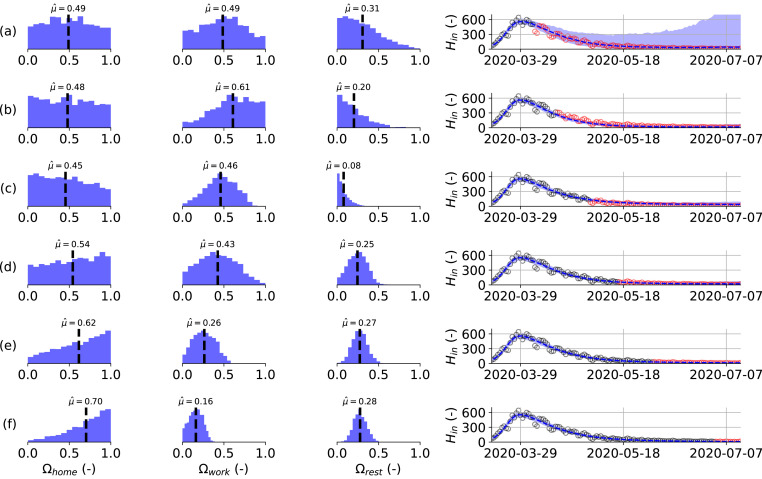
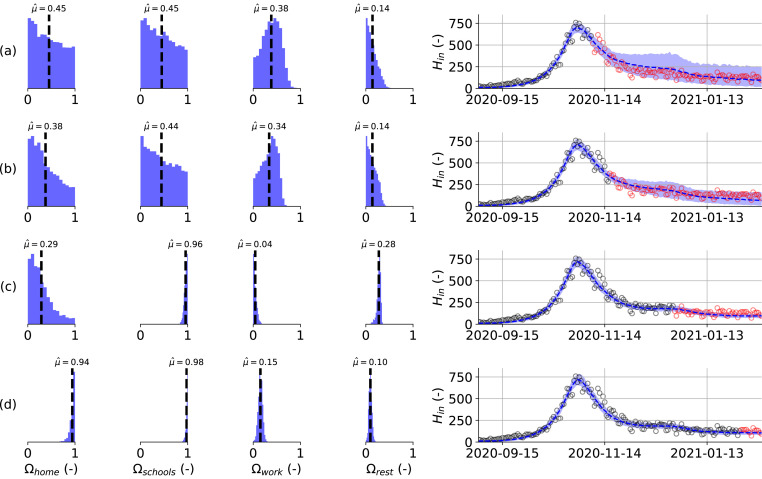
Fig. 5 summarizes the results of six model calibrations using hospitalization datasets of increasing length during the first 2020 COVID-19 wave. Here, Fig. 5(a) represents the minimal dataset, where the data range used for the calibration was equal to March 15th, 2020 until April 4th, 2020. Opposed is Fig. 5f, which uses the maximal dataset, using hospitalization data from March 15th, 2020 until July 1st, 2020. Using the minimal dataset (Fig. 5a), the posterior distributions are uninformative and model prediction uncertainty is large. Using additional data from April 15th, 2020 (Fig. 5b) onwards, the model captures the observed downward trend in the hospitalization data. Before the release of social restrictions on May 4th, 2020 (Fig. 5a–c), the posterior distributions seem to converge to distributions different from the ones found using the maximal dataset (Fig. 5f). However, during the gradual lifting of lockdown restrictions (Fig. 5d–f), the posterior distributions monotonically converge to their final distributions.
Fig. 5.
(left) Estimated posterior distributions for the effectivity of a contact at home (), in the workplace () and for the sum of leisure activities, other activities and public transport (), (right) together with the resulting model prediction for the daily hospitalizations from March 15th, 2020 until July 14th, 2020 (right). The effectivity of school contacts could not be deduced during the first 2020 COVID-19 wave because schools were only re-opened very limited before their final closure on July 1st, 2020. Calibration performed using the daily hospitalizations in Belgium until: (a) 2020-04-04, (b) 2020-04-15, (c) 2020-05-01, (d) 2020-05-01, (e) 2020-06-01 and (f) 2020-07-01. Calibration data in black, validation data in red. Model predictions are accurate in all but the minimal calibration dataset (a). Monotonic convergence of the effectivity parameter posteriors is reached quickly after lockdown release on May 4th, 2020 (d–f).(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Similarly, four calibrations on hospitalization datasets of increasing length during the second 2020 COVID-19 wave were performed and the results are summarized in Fig. 6. Once more, the minimal dataset (Fig. 6a), which uses data from September 1st, 2020 until November 7th, 2020 does not result in informative posterior distributions of the effectivity parameters. Uncertainty on the model prediction is large, but the mean model prediction is fairly accurate. As soon as schools are opened on November 16th, 2020, the daily hospitalizations evolve to a plateau. Despite large uncertainty on the model prediction, the emergence of the hospitalization plateau is captured in the uncertainty band, and the model thus provides a starting estimate using the minimal dataset. A similar conclusion can be drawn for the calibration using data until schools re-opening on November 16th, 2020 (Fig. 6b). When including data in the hospitalization dataset until schools closure for the Christmas holidays on December 18th, 2020 (Fig. 6c), the model correctly attributes the increased transmission to the opening of schools. In Fig. 6c, it can be seen that the effectivity parameter for schools has become almost equal to the maximum value of one. Although the posteriors of the effectivity parameters still differ significantly from their final distributions, the model provides an accurate prediction for the future evolution of the new hospitalizations during the Christmas holidays and until schools re-opening on January 4th, 2021. From the inference using the maximal dataset (Fig. 6d), it is clear that the model attributes high effectivities for contacts at home and in school and lower effectivities to the remaining leisure and workplace contacts.
Fig. 6.
Estimated posterior distributions for the effectivity of a contact at home (), at school (), in the workplace () and for the sum of leisure activities, other activities and public transport (left), together with the resulting model prediction for the daily hospitalizations from September 1st, 2020 until February 14th, 2021 (right). Calibration performed using the daily hospitalizations in Belgium until: (a) 2020-11-07, (b) 2020-11-16, (c) 2020-12-18, (d) 2021-02-01. Calibration data in black, validation data in red. Model predictions are accurate for all calibration datasets. Monotonic convergence of the schools effectivity parameter is reached a-posteriori schools re-opening (c).(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
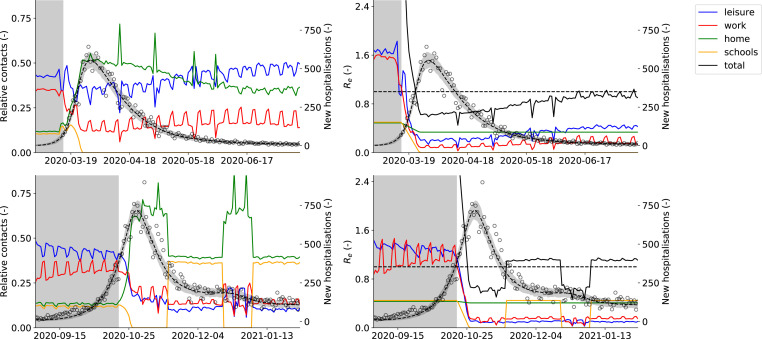
3.3. Effects of non-pharmaceutical interventions
To better compare the effects of non-pharmaceutical interventions between both 2020 COVID-19 waves, we computed the relative share of contacts and the effective reproduction number at home, in schools, in workplaces, and for the sum of leisure, public transport, and other contacts (Fig. 7). In this way, we can dissect the force of infection in our model, allowing us to assess the relative impact of contacts made at different locations on SARS-CoV-2 transmission. In pre-pandemic times, leisure and work contacts account for the bulk of total contacts, while under strict lockdown measures (March 15th, 2020 - May 4th, 2020 and October 19th, 2020 - November 16th, 2020), the contacts at home are the main driver of SARS-CoV-2 spread. The effective reproduction number under strict lockdown measures was estimated as (IQR: 0.48–0.76) for the first COVID-19 epidemic and was equal to (IQR: 0.61–0.69) for the second COVID-19 epidemic. Aside from the interactions at home, leisure contacts had the second most impact during the first COVID-19 wave, with roughly twice the impact of work contacts. When lifting social restrictions from May 4th, 2020 onwards, the relative contribution of home contacts gradually declines, while the contributions of work and leisure become more important. The effective reproduction number gradually increases and approaches the critical value of by the beginning of summer (average of June, 2020 , IQR: 0.77–1.00).
Fig. 7.
(First column) Relative share of contacts at home, in the workplace, in schools and for the sum of leisure activities, (Second column) effective reproduction number () at home, in the workplace, in schools and for the sum of leisure activities, other activities and public transport. The right axis denotes the predicted number of daily Belgian hospitalizations. The first row depicts the first COVID-19 wave in Belgium, from March 15th, 2020 until July 14th, 2020, while the second row depicts the second COVID-19 wave in Belgium, from September 1st, 2020 until February 1st, 2020. Mean and 95% confidence interval of 1000 model realizations. The background is shaded gray before lockdown measures were taken. During both lockdowns, home interactions have the largest share of effective contacts. During lockdown release, the relative importance of work and leisure contacts start increasing. Schools opening and closing has a large impact on the effective reproduction number, and can end a decreasing trend in hospitalizations.
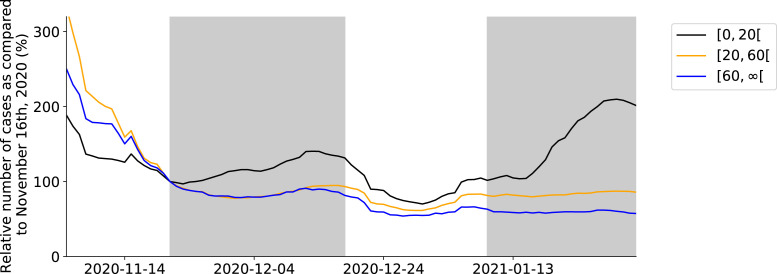
As soon as schools are re-opened on November 16th, 2020 during the second 2020 COVID-19 wave, a plateau in the daily number of hospitalizations emerges (Fig. 7). There were no other major policy changes around this time, except schools re-opening. Our model deduces this correlation by inferring posterior values of the effectivity of contacts in schools close to one, meaning school contacts were highly effective for SARS-CoV-2 transmission. Schools have an impact similar to the home interactions, with both contributing roughly 40% to the total number of effective contacts during the second COVID-19 wave. The opening of schools under lockdown can tip the scale, and push the effective reproduction number just above the critical value of . When schools are opened, the effective reproduction number increases from to , causing a stagnation of the daily hospitalizations. To further validate this result, we extracted the number of laboratory-confirmed cases in youths , the working population and the senior population from the Belgian Scientific Institute of Public Health (Sciensano). The time-series were normalized with the number of cases on November 21st, 20202 to allow a better comparison. The number of laboratory-confirmed cases amongst youths starts increasing as soon as schools are opened on November 16th, 2020 (Fig. 4). A similar pattern is observed during school closure and re-opening for the Christmas holidays, although it should be noted the relationship is more clouded. This is most likely the effect of Christmas and New Year celebrations and returning travelers. The use of a time-lagged cross-correlation revealed a significant lead-relationship between the number of cases in youths and the working population by 9 days, and a leading relationship between the number of cases amongst youths and the senior population by 13 days (Section A.5, supplementary materials). This indicates that as schools are reopened, SARS-CoV-2 percolates through social networks from younger to older individuals, eventually pushing the effective reproduction number above one.
Fig. 4.
Relative number of confirmed cases in youths, the working population and the senior population during the period November 2nd, 2020 until February 1st, 2021, as compared to the number of confirmed cases in each group on November 16th, 2020. The gray shade indicates schools were open.
4. Discussion
4.1. Analysis of hospital surveillance data
We computed hospitalization parameters using data from 22 136 patients in Belgian hospitals. The average time from symptom onset to hospitalization was estimated as 6.4 days. This estimate is in line with the previous estimate for Belgium of 5.7 days by Faes et al. (2020), and is in line with estimates for other regions such as 5–9 days for China (Linton et al., 2020), 4.4 days for Hong Kong, and 5.1 days for the UK (Pellis et al., 2021), especially when the interquartile range of 2.0–8.0 days is taken into account. Of the 22 136 hospitalized patients, 3 624 patients (16.2%) required intensive care at some point during their stay and 18 512 (83.8%) remained in cohort. The result is slightly lower than the estimate of Wu and McGoogan (2020) for China, who estimated that one-quarter of all hospitalized patients require intensive care. It should however be noted that the criteria for ICU admission and release might differ between countries. The ICU admission probabilities and mortalities in cohort and ICU indicate that COVID-19 has a much higher severity in older individuals, which is in line with estimates from other studies (CDC COVID-19 Response Team, 2020, Verity et al., 2020). In terms of hospital residence times, our estimates agree well with those made by Faes and colleagues (Faes et al., 2020). The average time spent in cohort was estimated as 11.0 days (3.4–15.6 days for the youngest versus oldest age groups), while the average time spent in ICU was estimated as 13.6 days (6.0–10.8 days for the youngest versus oldest age groups). The average time spent in ICU was lower in the 80 age group (10.8 days) than in the 70-80-year-olds (15.0 days). The residence time estimates are in line with Vekaria et al. (2021) who estimated a length of stay in England for COVID-19 patients not admitted to ICU of 8.4 days and for ICU length of stay of 12.4 days. It was previously reported by Faes et al. (2020) that the median residence time decreased after the first 2020 COVID-19 wave, however, we chose not to account for temporal changes in the hospital residence times and mortalities. The model predicted total number of patients and number of deaths in Belgian hospitals (Figures 9 and 10) would likely benefit from propagating time-dependent hospitalization parameters in the model. Vandromme et al. (2021) previously found that the average hospital residence times in Belgium have decreased between the first and second 2020 COVID-19 waves, which is mainly due to standardization of COVID-19 hospital treatment. In spite, the model predictions are sufficiently accurate to aid policymakers in the decision-making process.
4.2. Model calibration
We obtained an average basic reproduction number of (IQR: 3.90–4.39) for the first 2020 COVID-19 wave and of (IQR: 3.64–3.75) for the second 2020 COVID-19 wave, which is in line with the global consensus range of . The estimate for the second COVID-19 wave is slightly lower, and this is most likely because this estimate implicitly includes the effects of preventive measures and mentality changes that were gradually adopted during the first 2020 COVID-19 wave. The compliance to social measures was similar between both 2020 COVID-19 waves, little lag was observed (0.22 vs. 0.39 days) and the time to reach full compliance was of the same magnitude (9.17 vs 6.94 days). Thus, compliance to lockdown restrictions can be modeled using a ramp function without lag, eliminating one of the model’s parameters, namely (Eq. (18)). The seroreversion rate was estimated using two serological datasets. The data by Herzog et al. (2021) consists of residual blood samples sent to laboratories, while the dataset of Sciensano consists of blood samples from Red Cross blood donors. The dataset of Herzog et al. (2021) is likely biased towards sick individuals, while the dataset of Sciensano is biased towards healthy individuals. In the calibration procedure, both datasets were given equal weights to incorporate a truth in the middle heuristic. We estimated the average time to seroreversion as 9.2 months (IQR: 7.2–12.1 months), which is consistent with the finding that 50% of antibodies are most likely lost one year after the infection (Rosado et al., 2021). Using the same dataset, Abrams and colleagues (Abrams et al., 2021) have estimated the rate of antibody waning at 8 months using their SARS-CoV-2 model (informal communication). It should be noted that the incorporation of antibody waning completely ignores the effects of cellular immunity and that more research on the exact kinetics of the immune response is necessary. In spite, it is best to include waning immunity in SARS-CoV-2 models, especially when long time-horizons are considered in the simulations. In this study, a population average subclinical fraction of 57% was used, which was higher than estimated in a systematic review by Buitrago-Garcia et al. (2020) (31%) and higher than the estimate for the Icelandic population of Gudbjartsson et al. (2020) (43%). We expect that a higher subclinical fraction could be compensated by a higher per-case hospitalization risk () to obtain the same fit to the hospitalization data and thus, some unidentifiability between these model parameters exists. In spite, the calibrated combination of the per-case hospitalization risk and fraction of subclinical infections result in agreeance between the simulated and observed seroprevalence (Figures 9 and 10, supplementary materials).
We calibrated the model’s effectivity parameters (, , , ) on incrementally larger hospitalization datasets and found that the model provides accurate forecasts under the observed mobility changes, even when the posteriors still depend on the extent of the dataset. However, correct 3 effectivity parameters could only be deduced a posteriori events. This is because information on the effectiveness of contacts can only be obtained by observing the hospitalizations under changing policies. Examples are the effects of leisure and work relaxations during the first COVID-19 wave and the effect of schools re-opening during the second COVID-19 wave. From April 15th, 2020 onwards (Fig. 5, panel b) the ever decreasing trend in the daily hospitalizations is nicely captured even with posteriors seemingly converging to distributions different than those of the maximal dataset (panel f). Still, on May 1st 2020 (panel c), the model could have been used to accurately inform policymakers on the effects of lifting work and leisure restrictions just four days later. As soon as restrictions are lifted, the posteriors quickly converge to their final distributions. A similar observation is made with regard to the schools effectivity parameter. From November 7th, 2020 onwards (Fig. 6, panel a) the effect of schools re-opening is captured in the model uncertainty, in spite of deviant posterior distributions. From December 18th, 2020 onwards (panel c) the effect of schools re-opening is captured both in the model predictions and the effectivity parameters. Because accurate posteriors can only be inferred a posteriori, the modeler must assess if policy changes have been sufficient to deduce meaningful effectivity posteriors. This is important when performing scenario analysis, as incomplete knowledge of the effectivity posterior can significantly alter the results.
Scaling pre-pandemic contact matrices with public mobility data has proven to be a rapidly deployable and cheap alternative to the use of survey-based contact studies under lockdown measures, such as the one of Coletti et al. (2020) for Belgium. The social contact model is well-fit for the acute stages of the pandemic when these contact data are still being gathered. However, as the pandemic progresses, the survey-based contact studies are the preferred choice as the use of public mobility data is more coarse-grained. Because the GCMRs are not available for different age groups, they do not allow us to accurately capture how individuals of different ages have altered their behavior under lockdown measures. For example, the contact study by Coletti et al. (2020) shows that younger individuals tend to increase their contacts sooner than older individuals after the release of lockdown measures. These differential effects are still captured in our social contact model, albeit less accurate than the survey-based contact model, by the multiplication of the GCMRs with the pre-pandemic number of contacts. For example, the mobility reduction in workplaces is only applied to the matrix of work contacts, which only contains contacts for individuals between 20 and 60 years old. Further, because the GCMRs are collated smartphone data, one could expect the elderly population to be underrepresented due to lower smartphone usage. However, it is unlikely that this would drastically alter our study’s results because older individuals have fewer contacts than younger individuals and thus contribute less to overall SARS-CoV-2 spread.
4.3. Effects of non-pharmaceutical interventions
Finally, we would like to discuss the importance of schools in the SARS-CoV-2 pandemic. As previously mentioned in Section 3.3, there seems to be a strong correlation between school re-opening, the rise of laboratory-confirmed cases amongst youths, and the emergence of plateaus in the daily hospitalizations (Fig. 4, Fig. 7). Our model incorporates this correlation as high effectivities of school contacts. An increase in the effective reproduction number, from to , is observed when schools are re-opened. Several studies have found children to be less susceptible to a SARS-CoV-2 infection (Davies et al., 2020, Viner et al., 2021, Dattner et al., 2021). Because quantitative data was scarce at the time of writing, we incorporated no changes in susceptibility and infectiousness in children in this study. However, this will not alter the large impact schools seem to have on SARS-CoV-2 spread in our model. If the susceptibility and infectiousness in children is lowered, this will most likely be countered during the parameter inference, where we expect higher values for the effectivity of contacts of children in schools () to be inferred. Although the presented evidence is circumstantial, and correlation does not imply causation, schools seem to play a large role in SARS-CoV-2 spread. Thus, school closure seems an effective way of countering an epidemic SARS-CoV-2 trend.
5. Conclusions
We obtained an average basic reproduction number of (IQR: 3.90–4.39) and (IQR: 3.64–3.75) for both 2020 COVID-19 waves in Belgium. We found that SARS-CoV-2 strongly discriminates between individuals of different age groups, with youths and the working-aged population driving the pandemic, and the senior population needing hospital care. These results are in line with the established consensuses and highlight the model’s validity. Further, by propagating the hospitalization parameters computed using the clinical surveillance dataset, the model is able to accurately predict the number of daily hospitalizations, the total number of patients in Belgian hospitals, the total number of deaths in Belgian hospitals, and the seroprevalence in the Belgian population during both 2020 COVID-19 waves.
The combination of the deterministic epidemiological model, which incorporates a-priori knowledge on disease dynamics, and the social contact model whose infectivity parameters were inferred from the hospitalization data allow us to make the most out of the available pre-pandemic data and public mobility data. Our method is computationally cheap and does not require ad-hoc tweaking to obtain a good fit to the observed data. A disadvantage is that the effectivity parameter distributions only converge to their correct posterior distributions a posteriori policy changes. Still, even when using a very limited calibration dataset, the model is able to make fairly accurate predictions of the future number of hospitalizations, highlighting the robustness of the calibration method.
As soon as schools were re-opened on November 16th, 2020, the number of confirmed cases amongst youths starts increasing. A significant lead relationship between the number of cases amongst youths and the working population, and youths and the senior population was found. Our model incorporates this correlation as high effectivities of school contacts. When schools were re-opened under lockdown policies, the model indicates the effective reproduction number increased from to . Thus, school closure seems to be an effective measure to counter an epidemic SARS-CoV-2 trend.
CRediT authorship contribution statement
Tijs W. Alleman: Conceptualization, Software, Methodology, Investigation, Data curation, Writing – original draft. Jenna Vergeynst: Conceptualization, Software, Writing – review & editing, Project administration. Lander De Visscher: Methodology. Michiel Rollier: Methodology, Writing – review. Elena Torfs: Data collection, Data curation. Ingmar Nopens: Conceptualization, Funding acquisition, Project administration, Writing – review & editing. Jan M. Baetens: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research presented in this article would not have be possible without the selfless help of good colleagues. I want to thank Daniel Illana, Bram De Jaegher, Daan Van Hauwermeiren, Stijn Van Hoey and Joris Van den Bossche for their help in maintaining the GitHub repo, coding the visualizations and for teaching me the basics of object-oriented programming in Python. I would like to thank Mieke Descheppere, from the Ghent University hospital and Wim Verbeke, MD from AZ Delta Roeselare, for sharing their insights on hospital dynamics. We thank VZW 100 km Dodentocht Kadee for their financial support through the organization of the 2020 100 km COVID-Challenge.
Role of the funding source
This work was supported by the UGent Special Research Fund, Belgium, by the Research Foundation Flanders (FWO), Belgium, project number G0G2920N and by VZW 100 km Dodentocht Kadee, Belgium through the organization of the 2020 100 km COVID-Challenge. Further, the computational resources and services used in this work were also provided by the VSC (Flemish Supercomputer Center), funded by FWO, Belgium and the Flemish Government . The funding sources played no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Ethics statement
Permission for the clinical hospital surveillance was granted by the Committee on Medical Ethics of the Ghent University Hospital (BC-07507) and by the Belgian Federal Information Security Committee to the Belgian Collaborative Group on COVID-19 Hospital Surveillance. The need for informed consent was renounced, however, upon hospital discharge, patients were informed that their data would be curated by the Belgian Scientific Institute for Public Health (Sciensano) and used in the context of the COVID-19 public health crisis for policy-supporting research.
Footnotes
Date of school reopening 2021-11-16 plus one five-day incubation period.
Assuming the inferred posterior distributions of the maximal dataset are correct.
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.epidem.2021.100505.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
References
- Abrams Steven, Wambua James, Santermans Eva, Willem Lander, Kuylen Elise, Coletti Pietro, Libin Pieter, Faes Christel, Petrof Oana, Herzog Sereina A., Beutels Philippe, Hens Niel. Modelling the early phase of the belgian COVID-19 epidemic using a stochastic compartmental model and studying its implied future trajectories. Epidemics. 2021;35 doi: 10.1016/j.epidem.2021.100449. URL https://www.sciencedirect.com/science/article/pii/S1755436521000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktay Ahmet, Bavadekar Shailesh, Cossoul Gwen, Davis John, Desfontaines Damien, Fabrikant Alex, Gabrilovich Evgeniy, Gadepalli Krishna, Gipson Bryant, Guevara Miguel, Kamath Chaitanya, Kansal Mansi, Lange Ali, Mandayam Chinmoy, Oplinger Andrew, Pluntke Christopher, Roessler Thomas, Schlosberg Arran, Shekel Tomer, Vispute Swapnil, Vu Mia, Wellenius Gregory, Williams Brian, Wilson Royce J. 2020. Google COVID-19 community mobility reports: Anonymization process description (version 1.0) CoRR, abs/2004.04145 URL https://arxiv.org/abs/2004.04145. [Google Scholar]
- Barbe Kurt, Blotwijk Susanne, Cools Wilfried. Vrije Universiteit Brussel; 2020. Data-Driven Epidemiological Model to Monitor the Sustainability of Hospital Care: Technical Report ICDS300420. [Google Scholar]
- Buitrago-Garcia Diana, Egli-Gany Dianne, Counotte Michel J., Hossmann Stefanie, Imeri Hira, Ipekci Aziz Mert, Salanti Georgia, Low Nicola. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: A living systematic review and meta-analysis. Ford Nathan., editor. PLoS Med. 2020;17(9) doi: 10.1371/journal.pmed.1003346. URL https://dx.plos.org/10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC COVID-19 Response Team Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, february 12-march 16, 2020. Morb. Mortal. Wkly. Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletti Pietro, Wambua James, Gimma Amy, Willem Lander, Vercruysse Sarah, Vanhoutte Bieke, Jarvis Christopher I, Van Zandvoort Kevin, Edmunds John, Beutels Philippe, Hens Niel. CoMix: comparing mixing patterns in the Belgian population during and after lockdown. Sci. Rep. 2020;10(1):21885. doi: 10.1038/s41598-020-78540-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattner Itai, Goldberg Yair, Katriel Guy, Yaari Rami, Gal Nurit, Miron Yoav, Ziv Arnona, Sheffer Rivka, Hamo Yoram, Huppert Amit. The role of children in the spread of COVID-19: Using household data from bnei brak, Israel, to estimate the relative susceptibility and infectivity of children. PLoS Comput. Biol. 2021;17(2):1–19. doi: 10.1371/journal.pcbi.1008559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies Nicholas G, Klepac Petra, Liu Yang, Prem Kiesha, Jit Mark, Pearson Carl A B, Quilty Billy J, Kucharski Adam J, Gibbs Hamish, Clifford Samuel, Gimma Amy, van Zandvoort Kevin, Munday James D, Diamond Charlie, Edmunds W John, Houben Rein M G J, Hellewell Joel, Russell Timothy W, Abbott Sam, Funk Sebastian, Bosse Nikos I, Sun Yueqian Fiona, Flasche Stefan, Rosello Alicia, Jarvis Christopher I, Eggo Rosalind M, working Group CMMID COVID-19. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 2020 doi: 10.1038/s41591-020-0962-9. [DOI] [PubMed] [Google Scholar]
- Diekmann O, Heesterbeek J.A.P., J.A.J. Metz. On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. J. Math. Biol. 1990;28(4):365–382. doi: 10.1007/BF00178324. ISSN 0303-6812 (Print) [DOI] [PubMed] [Google Scholar]
- Diekmann O, Heesterbeek J.A.P., M.G. Roberts. The construction of next-generation matrices for compartmental epidemic models. J. R. Soc. Interface. 2010;7(47):873–885. doi: 10.1098/rsif.2009.0386. URL https://pubmed.ncbi.nlm.nih.gov/19892718 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2871801/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faes Christel, Abrams Steven, Van Beckhoven Dominique, Meyfroidt Geert, Vlieghe Erika, Hens Niel, on COVID-19 Hospital Surveillance Belgian Collaborative Group. Time between symptom onset, hospitalisation and recovery or death: Statistical analysis of Belgian COVID-19 patients. Int. J. Environ. Res. Public Health. 2020;17(20) doi: 10.3390/ijerph17207560. URL https://www.mdpi.com/1660-4601/17/20/7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco Nicolas. Covid-19 Belgium: Extended SEIR-QD model with nursing homes and long-term scenarios-based forecasts. MedRxiv. 2020 doi: 10.1101/2020.09.07.20190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman Jonathan, Weare Jonathan. Ensemble samplers with affine invariance. Commun. Appl. Math. Comput. Sci. 2010;5(1):65–80. doi: 10.2140/camcos.2010.5.65. [DOI] [Google Scholar]
- Google LLC . 2020. Google COVID-19 community mobility reports. URL https://www.google.com/covid19/mobility/ [Google Scholar]
- Gudbjartsson Daniel F., Helgason Agnar, Jonsson Hakon, Magnusson Olafur T., Melsted Pall, Norddahl Gudmundur L., Saemundsdottir Jona, Sigurdsson Asgeir, Sulem Patrick, Agustsdottir Arna B., Eiriksdottir Berglind, Fridriksdottir Run, Gardarsdottir Elisabet E., Georgsson Gudmundur, Gretarsdottir Olafia S., Gudmundsson Kjartan R., Gunnarsdottir Thora R., Gylfason Arnaldur, Holm Hilma, Jensson Brynjar O., Jonasdottir Aslaug, Jonsson Frosti, Josefsdottir Kamilla S., Kristjansson Thordur, Magnusdottir Droplaug N., le Roux Louise, Sigmundsdottir Gudrun, Sveinbjornsson Gardar, Sveinsdottir Kristin E., Sveinsdottir Maney, Thorarensen Emil A., Thorbjornsson Bjarni, Löve Arthur, Masson Gisli, Jonsdottir Ingileif, Möller Alma D., Gudnason Thorolfur, Kristinsson Karl G., Thorsteinsdottir Unnur, Stefansson Kari. Spread of SARS-CoV-2 in the icelandic population. N. Engl. J. Med. 2020;0(0):null. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Vivek, Bhoyar Rahul C, Jain Abhinav, Srivastava Saurabh, Upadhayay Rashmi, Imran Mohamed, Jolly Bani, Divakar Mohit Kumar, Sharma Disha, Sehgal Paras, Ranjan Gyan, Gupta Rakesh, Scaria Vinod, Sivasubbu Sridhar. Asymptomatic reinfection in 2 healthcare workers from India with genetically distinct severe acute respiratory syndrome coronavirus 2. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1451. ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Xi, Lau Eric H Y, Wu Peng, Deng Xilong, Wang Jian, Hao Xinxin, Lau Yiu Chung, Wong Jessica Y, Guan Yujuan, Tan Xinghua, Mo Xiaoneng, Chen Yanqing, Liao Baolin, Chen Weilie, Hu Fengyu, Zhang Qing, Zhong Mingqiu, Wu Yanrong, Zhao Lingzhai, Zhang Fuchun, Cowling Benjamin J, Li Fang, Leung Gabriel M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Herzog Sereina, De Bie Jessie, Abrams Steven, Wouters Ine, Ekinci Esra, Patteet Lisbeth, Coppens Astrid, De Spiegeleer Sandy, Beutels Philippe, Van Damme Pierre, Hens Niel, Theeten Heidi. Seroprevalence of IgG antibodies against SARS coronavirus 2 in Belgium – a serial prospective cross-sectional nationwide study of residual samples. MedRxiv. 2021 doi: 10.1101/2020.06.08.20125179. URL https://www.medrxiv.org/content/early/2021/04/07/2020.06.08.20125179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, J., Eberhart, R., 1995. Particle swarm optimization. In: Proceedings of ICNN’95 - International Conference on Neural Networks, Vol. 4, pp. 1942–1948 vol.4.
- Kermack William Ogilvy, McKendrick A.G., Walker Gilbert Thomas. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. A. 1927;115(772):700–721. doi: 10.1098/rspa.1927.0118. URL https://royalsocietypublishing.org/doi/abs/10.1098/rspa.1927.0118. [DOI] [Google Scholar]
- Lescure Francois-Xavier, Bouadma Lila, Nguyen Duc, Parisey Marion, Wicky Paul-Henri, Behillil Sylvie, Gaymard Alexandre, Bouscambert-Duchamp Maude, Donati Flora, Hingrat Quentin Le, Enouf Vincent, Houhou-Fidouh Nadhira, Valette Martine, Mailles Alexandra, Lucet Jean-Christophe, Mentre France, Duval Xavier, Descamps Diane, Malvy Denis, Timsit Jean-François, Lina Bruno, Van-der Werf Sylvie, Yazdanpanah Yazdan. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Qun, Guan Xuhua, Wu Peng, Wang Xiaoye, Zhou Lei, Tong Yeqing, Ren Ruiqi, Leung Kathy S.M., Lau Eric H.Y., Wong Jessica Y., Xing Xuesen, Xiang Nijuan, Wu Yang, Li Chao, Chen Qi, Li Dan, Liu Tian, Zhao Jing, Liu Man, Tu Wenxiao, Chen Chuding, Jin Lianmei, Yang Rui, Wang Qi, Zhou Suhua, Wang Rui, Liu Hui, Luo Yinbo, Liu Yuan, Shao Ge, Li Huan, Tao Zhongfa, Yang Yang, Deng Zhiqiang, Liu Boxi, Ma Zhitao, Zhang Yanping, Shi Guoqing, Lam Tommy T.Y., Wu Joseph T., Gao George F., Cowling Benjamin J., Yang Bo, Leung Gabriel M., Feng Zijian. Early transmission dynamics in Wuhan, China, of novel coronavirus–Infected pneumonia. N. Engl. J. Med. 2020;0(0):null. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Ruiyun, Pei Sen, Chen Bin, Song Yimeng, Zhang Tao, Yang Wan, Shaman Jeffrey. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020 doi: 10.1126/science.abb3221. URL https://science.sciencemag.org/content/early/2020/03/13/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton Natalie M, Kobayashi Tetsuro, Yang Yichi, Hayashi Katsuma, Akhmetzhanov Andrei R, Jung Sung-Mok, Yuan Baoyin, Kinoshita Ryo, Nishiura Hiroshi. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: A statistical analysis of publicly available case data. J. Clin. Med. 2020;9(2) doi: 10.3390/jcm9020538. ISSN 2077-0383 (Print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, null null, Funk S, Flasche S. The contribution of pre-symptomatic infection to the transmission dynamics of COVID-2019 [version 1; peer review: 1 approved] Wellcome Open Res. 2020;5(58) doi: 10.12688/wellcomeopenres.15788.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Yang, Yan Li-Meng, Wan Lagen, Xiang Tian-Xin, Le Aiping, Liu Jia-Ming, Peiris Malik, Poon Leo L M, Zhang Wei. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenberghs Geert, Faes Christel, Aerts Jan, Theeten Heidi, Devleesschauwer Brecht, Bustos Sierra Natalia, Braeye Toon, Renard Francoise, Herzog Sereina, Lusyne Patrick, Van der Heyden Johan, Van Oyen Herman, Van Damme Pierre, Hens Niel. Belgian Covid-19 mortality, excess deaths, number of deaths per million, and infection fatality rates (8 march - 9 may 2020) MedRxiv. 2020 doi: 10.1101/2020.06.20.20136234. URL https://www.medrxiv.org/content/early/2020/06/20/2020.06.20.20136234. [DOI] [Google Scholar]
- Mossong Jol, Hens Niel, Jit Mark, Beutels Philippe, Auranen Kari, Mikolajczyk Rafael, Massari Marco, Salmaso Stefania, Tomba Gianpaolo Scalia, Wallinga Jacco, Heijne Janneke, Sadkowska-Todys Malgorzata, Rosinska Magdalena, Edmunds W. John. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 2008;5(3):1. doi: 10.1371/journal.pmed.0050074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellis Lorenzo, Scarabel Francesca, Stage Helena B., Overton Christopher E., Chappell Lauren H.K., Fearon Elizabeth, Bennett Emma, Lythgoe Katrina A., House Thomas A., Hall Ian, null null. Challenges in control of COVID-19: short doubling time and long delay to effect of interventions. Philos. Trans. R. Soc. B. 2021;376(1829) doi: 10.1098/rstb.2020.0264. URL https://royalsocietypublishing.org/doi/abs/10.1098/rstb.2020.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Vivar Belen, Becerra-Wong Monica, Guadalupe Juan Jose, Marquez Sully, Gutierrez Bernardo, Rojas-Silva Patricio, Grunauer Michelle, Trueba Gabriel, Barragan Veronica, Cardenas Paul. COVID-19 re-infection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in south america. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3686174. URL https://www.ssrn.com/abstract=3686174. [DOI] [Google Scholar]
- Riou Julien, Althaus Christian L. Pattern of early human-to-human transmission of wuhan 2019 novel coronavirus (2019-nCoV), december 2019 to january 2020. Eurosurveillance. 2020;25(4) doi: 10.2807/1560-7917.ES.2020.25.4.2000058. URL https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado Jason, Pelleau Stéphane, Cockram Charlotte, Merkling Sarah Hélène, Nekkab Narimane, Demeret Caroline, Meola Annalisa, Kerneis Solen, Terrier Benjamin, Fafi-Kremer Samira, de Seze Jerome, Bruel Timothée, Dejardin François, Petres Stéphane, Longley Rhea, Fontanet Arnaud, Backovic Marija, Mueller Ivo, White Michael T. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: an antibody-based diagnostic and machine learning study. Lancet Microbe. 2021;2(2):e60–e69. doi: 10.1016/S2666-5247(20)30197-X. URL http://www.ncbi.nlm.nih.gov/pubmed/33521709 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC7837364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman Jeffrey, Galanti Marta. Will SARS-CoV-2 become endemic? Science. 2020;370(6516):527–529. doi: 10.1126/science.abe5960. URL https://science.sciencemag.org/content/370/6516/527. [DOI] [PubMed] [Google Scholar]
- Sulyok M., Walker M. Community movement and COVID-19: a global study using google’s community mobility reports. Epidemiol. Infect. 2020;148 doi: 10.1017/S0950268820002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett Richard L., Sevinsky Joel R., Hartley Paul D., Kerwin Heather, Crawford Natalie, Gorzalski Andrew, Laverdure Chris, Verma Subhash C., Rossetto Cyprian C., Jackson David, Farrell Megan J., Van Hooser Stephanie, Pandori Mark. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2021;21(1):52–58. doi: 10.1016/S1473-3099(20)30764-7. URL http://www.ncbi.nlm.nih.gov/pubmed/33058797 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC7550103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To Kelvin Kai-Wang, Tsang Owen Tak-Yin, Leung Wai-Shing, Tam Anthony Raymond, Wu Tak-Chiu, Lung David Christopher, Yip Cyril Chik-Yan, Cai Jian-Piao, Chan Jacky Man-Chun, Chik Thomas Shiu-Hong, Lau Daphne Pui-Ling, Choi Chris Yau-Chung, Chen Lin-Lei, Chan Wan-Mui, Chan Kwok-Hung, Ip Jonathan Daniel, Ng Anthony Chin-Ki, Poon Rosana Wing-Shan, Luo Cui-Ting, Cheng Vincent Chi-Chung, Chan Jasper Fuk-Woo, Hung Ivan Fan-Ngai, Chen Zhiwei, Chen Honglin, Yuen Kwok-Yung. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. URL https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(20)30196-1/fulltext#.XsE5ulFU1n8.mendeley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande Jan, Vermeersch Pieter, Vandervoort Kris, Wawina-Bokalanga Tony, Vanmechelen Bert, Wollants Elke, Laenen Lies, André Emmanuel, Van Ranst Marc, Lagrou Katrien, Maes Piet. Symptomatic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection by a phylogenetically distinct strain. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem Nina, Vilain Aline, Wyndham-Thomas Chloé, Deblonde Jessika, Bossuyt Nathalie, Lernout Tinne, Rebolledo Gonzalez Javiera, Quoilin Sophie, Melis Vincent, Van Beckhoven Dominique. Rapid establishment of a national surveillance of COVID-19 hospitalizations in Belgium. Arch. Public Health. 2020;78(1):121. doi: 10.1186/s13690-020-00505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandromme M, De Pauw R, Serrien B, Van Goethem N, Blot K. Belgian Federal Institute for Public Health; 2021. COVID-19 Clinical Hospital Surveillance Report. [Google Scholar]
- Vekaria Bindu, Overton Christopher, Wisniowski Arkadiusz, Ahmad Shazaad, Aparicio-Castro Andrea, Curran-Sebastian Jacob, Eddleston Jane, Hanley Neil, House Thomas, Kim Jihye, Olsen Wendy, Pampaka Maria, Pellis Lorenzo, Ruiz Diego Perez, Schofield John, Shryane Nick, Elliot Mark. Hospital length of stay for COVID-19 patients: Data-driven methods for forward planning. BMC Infect. Dis. 2021 doi: 10.21203/rs.3.rs-56855/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verity Robert, Okell Lucy C, Dorigatti Ilaria, Winskill Peter, Whittaker Charles, Imai Natsuko, Cuomo-Dannenburg Gina, Thompson Hayley, Walker Patrick G T, Fu Han, Dighe Amy, Griffin Jamie T, Baguelin Marc, Bhatia Sangeeta, Boonyasiri Adhiratha, Cori Anne, Cucunubá Zulma, FitzJohn Rich, Gaythorpe Katy, Green Will, Hamlet Arran, Hinsley Wes, Laydon Daniel, Nedjati-Gilani Gemma, Riley Steven, van Elsland Sabine, Volz Erik, Wang Haowei, Wang Yuanrong, Xi Xiaoyue, Donnelly Christl A, Ghani Azra C, Ferguson Neil M. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viner Russell M., Mytton Oliver T., Bonell Chris, Melendez-Torres G.J., Ward Joseph, Hudson Lee, Waddington Claire, Thomas James, Russell Simon, van der Klis Fiona, Koirala Archana, Ladhani Shamez, Panovska-Griffiths Jasmina, Davies Nicholas G., Booy Robert, Eggo Rosalind M. Susceptibility to SARS-CoV-2 infection among children and adolescents compared with adults: A systematic review and meta-analysis. JAMA Pediatr. 2021;175(2):143–156. doi: 10.1001/jamapediatrics.2020.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Wycliffe E., Li Zongbin, Chiew Calvin J., Yong Sarah E., Toh Matthias P., Lee Vernon J. Presymptomatic transmission of SARS-CoV-2 — Singapore, january 23–march 16, 2020. Morb. Mortal. Wkly. Rep. 2020;69:411–415. doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley Adam K, Juno Jennifer A, Wang Jing J, Selva Kevin J, Reynaldi Arnold, Tan Hyon-Xhi, Lee Wen Shi, Wragg Kathleen M, Kelly Hannah G, Esterbauer Robyn, Davis Samantha K, Kent Helen E, Mordant Francesca L, Schlub Timothy E, Gordon David L, Khoury David S, Subbarao Kanta, Cromer Deborah, Gordon Tom P, Chung Amy W, Davenport Miles P, Kent Stephen J. Evolution of immune responses to SARS-CoV-2 in mild-moderate COVID-19. Nature Commun. 2021;12(1):1162. doi: 10.1038/s41467-021-21444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem Lander. 2021. Restore. URL https://covid-en-wetenschap.github.io/restore.html. [Google Scholar]
- Willem Lander, Abrams Steven, Petrof Oana, Coletti Pietro, Kuylen Elise, Libin Pieter, Mogelmose Signe, Wambua James, Herzog Sereina A., Faes Christel, Beutels Philippe, Hens Niel. The impact of contact tracing and household bubbles on deconfinement strategies for COVID-19: an individual-based modelling study. MedRxiv. 2020 doi: 10.1038/s41467-021-21747-7. URL https://www.medrxiv.org/content/10.1101/2020.07.01.20144444v3.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem Lander, Van Kerckhove Kim, Chao Dennis L., Hens Niel, Beutels Philippe. A nice day for an infection? Weather conditions and social contact patterns relevant to influenza transmission. PLoS One. 2012;7(11):1–7. doi: 10.1371/journal.pone.0048695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Joseph T, Leung Kathy, Bushman Mary, Kishore Nishant, Niehus Rene, de Salazar Pablo M, Cowling Benjamin J, Lipsitch Marc, Leung Gabriel M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 2020;26(4):506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Joseph T, Leung Kathy, Leung Gabriel M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. URL http://www.sciencedirect.com/science/article/pii/S0140673620302609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Zunyou, McGoogan Jennifer M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Zou Lirong, Ruan Feng, Huang Mingxing, Liang Lijun, Huang Huitao, Hong Zhongsi, Yu Jianxiang, Kang Min, Song Yingchao, Xia Jinyu, Guo Qianfang, Song Tie, He Jianfeng, Yen Hui-Ling, Peiris Malik, Wu Jie. Sars-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. PMID: 32074444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.