Abstract
Light signal perceived by the red/far-red absorbing phytochrome (phy) family of photoreceptors regulates plant growth and development throughout the life cycle. Phytochromes regulate the light-triggered physiological responses by controlling gene expression both at the transcriptional and post-transcriptional levels. Recent large-scale RNA-seq studies have demonstrated the roles of phys in altering the global transcript diversity by modulating the pre-mRNA splicing in response to light. Moreover, several phy-interacting splicing factors/regulators from different species have been identified using forward genetics and protein-protein interaction studies, which modulate the light-regulated pre-mRNA splicing. In this review, we summarize our current understanding of the role of phys in the light-mediated pre-mRNA splicing and how that contributes to the regulation of gene expression to promote photomorphogenesis.
Keywords: pre-mRNA splicing, photomorphogenesis, Phytochrome signaling, Splicing factor, Arabidopsis
Introduction
One of the most important environmental factors that has a profound effect on plant growth and development is light. At young seedling stage, the perception of light enables plants to switch from skotomorphogenesis (a dark-adapted developmental program characterized by long hypocotyl, small and unopened cotyledons) to photomorphogenesis (a light-adapted developmental program characterized by short hypocotyl, open expanded and green cotyledons suitable for photosynthetic growth). At later stages of growth, light plays a crucial role in regulating shade avoidance, flowering time, and eventually senescence. Plants have several classes of photoreceptors including, the phototropins (PHOTs) and cryptochromes (CRYs) for UV-A and blue-light, UV resistant locus 8 (UVR8) for UV-B, and phytochromes (phys) for red/far-red-light [1]. Plants utilize these photoreceptors to perceive minute changes in light quality, quantity, direction and overall duration, and integrate the surrounding information to modulate adaptative growth and development for reproductive success.
Phytochromes are ubiquitous across the plant kingdom and many bacterial lineages [2]. They consist of a small multigene family (designated PHYA to PHYE in Arabidopsis thaliana) encoding ~125 kDa soluble proteins that can form selective homo- and hetero-dimers [3–5]. They are synthesized in their inactive red light absorbing Pr form and in response to red-light they are photoconverted to the biologically active far-red-light absorbing Pfr form. The pfr form translocates into nucleus, forms nuclear photobodies (PB) and induces large scale gene expression changes to promote photomorphogenesis. As a pivotal light sensor, phys employ multiple layers of regulations including transcriptional, post-transcriptional, translational, post-translational modifications and ultimately protein degradation/stabilization to control the transcriptome and proteome that drives the growth and developmental re-programming [6,7]. While much of the emphasis of the past decades has been on the transcriptional regulation, recent studies indicate a broader impact of the phys on the modulation of post-transcriptional pre-mRNA splicing [8–10].
Alternative splicing (AS) can lead to an intron removal or retention, or the use of alternative 5’- and 3’-splice site (SS) of an exon and fine-tune the global gene expression in response to a range of internal and external cues. AS uses variable SS selection to generate two or more spliced mRNA isoforms from one pre-mRNA and enables organisms to generate more complex transcriptome and proteome without increasing the gene number and the genome size [11,12]. These include include exon skipping, intron retention, mutually exclusive exons, alternative 5’ and 3’ SS (Figure 1A, B). Pre-mRNA splicing is a tightly regulated process carried-out by highly conserved spliceosome machinery, a dynamic multi-megadalton ribonuclear protein complex consisting of ~200 proteins and five small nuclear ribonucleoproteins (snRNPs; U1, U2, U4, U5 and U6) [13]. In addition, auxiliary splicing regulatory proteins such as heterogenous nuclear ribonucleoproteins (hnRNPs) and serine/arginine-rich (SR) proteins variably complex with the core spliceosome machinery for target identification and modulation of appropriate splicing event [11,12]. Every intron contains core splicing signals consisting of the conserved 5’-SS, 3’-SS, a branched point (BP) adenine (A), and a poly-pyrimidine tract (PPT) (Figure 1A), which collectively participate in the splicing reaction [13]. In addition, majority of the pre-mRNAs destined for AS contains cis-acting splicing regulatory elements (SREs) that confer gene-specific regulation of splicing. These include exonic or intronic splicing enhancers (ESE or ISE) and exonic or intronic splicing silencers (ESS or ISS) (Figure 1A) [11,12]. The activities of the cis-acting SREs are dependent upon the interaction with the trans-acting auxiliary splicing regulatory proteins, and therefore, these auxiliary splicing regulators through upstream protein-protein and downstream protein-RNA interactions modulate the final outcome of AS. Thus, SREs and auxiliary splicing regulators play a critical role in a tissue or cell-type specific pre-mRNA AS [11,14]. A comprehensive analysis of the Arabidopsis genome revealed that around 20% of whole genome is intronic region and significantly large number of genes contain at least one intron [15]. Moreover, a recent study on the prevalence of AS in Arabidopsis genome identified at least 61% of all multi-intronic genes undergo AS, revealing a more prevalent significance of pre-mRNA AS regulation in shaping the overall physiology and morphology of plants [14,16].
Figure 1. Schematics of pre-mRNA splicing and major types of alternative splicing (AS).
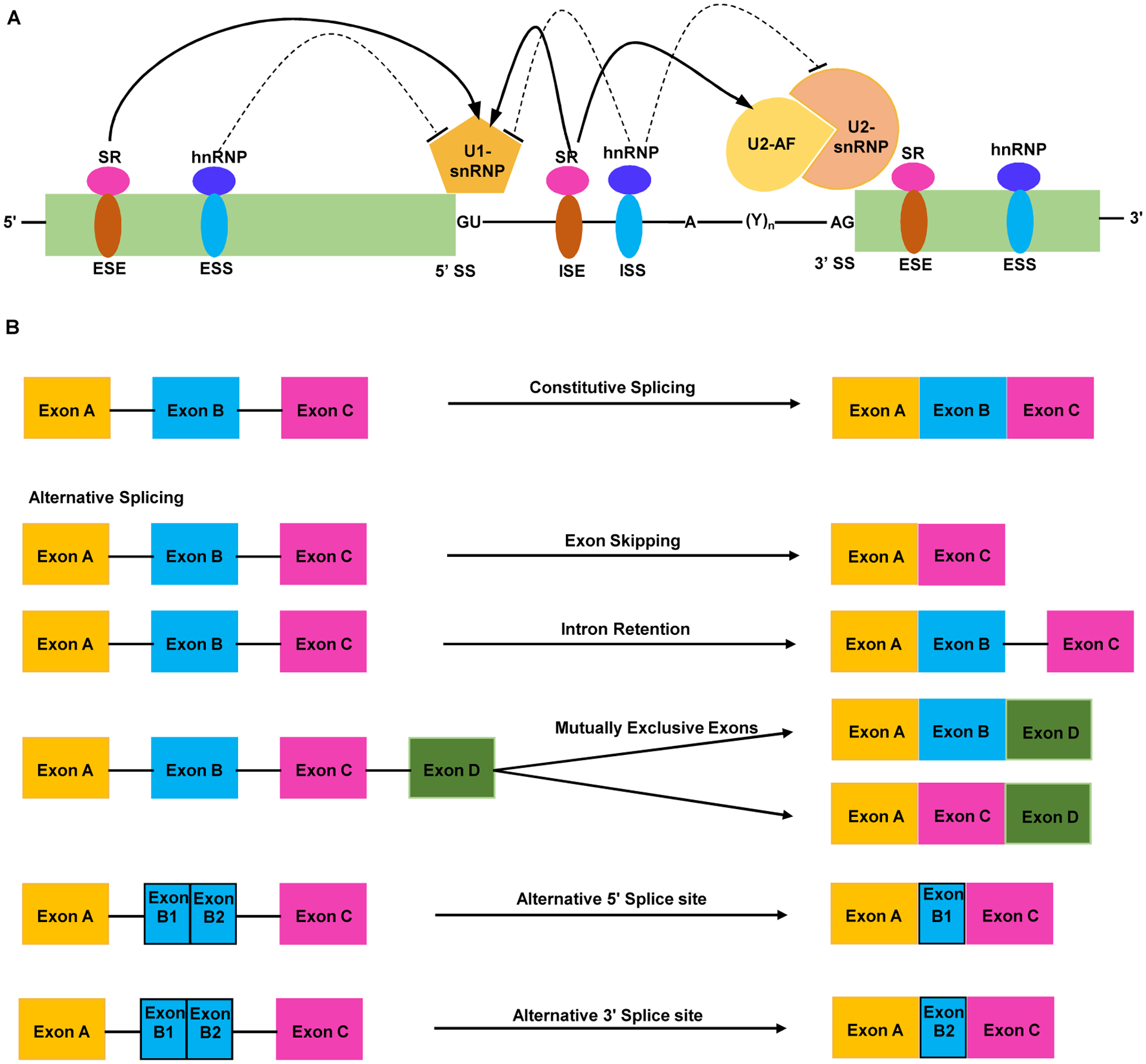
(A) Majority of the pre-mRNAs contain one or more introns flanking the exons on either side and consensus sequences defining the 5’-splice site (5’-SS) with a conserved GU, 3’-SS with a conserved AG, a branched point (BP) adenine (A) 18 to 40 nucleotide upstream of the 3’-SS, and a poly-pyrimidine tract (PPT) following the BP. Majority of the exons and introns contain regulatory cis-acting elements including, exonic/intronic splicing enhancers (ESE/ISE), exonic/intronic splicing silencers (ESS/ISS). Trans-acting regulators such as serine/arginine-rich (SR) proteins bind to the ESE/ISEs to promote splicing, while heterogeneous nuclear ribonucleoproteins (hnRNPs) bind to the ESS/ISS and repress the splicing. U1-small nuclear ribonucleoproteins (U1-snRNPs) target the 5’-SS, while the U2-snRNPs and U2- associated factors target the 3’-SS. Appropriate exon-intron junction and the splicing is determined by the core spliceosome assembly and the corresponding cis-acting elements bound by the trans-acting regulators interacting with the U1 and/or U2-snRNPs.
(B) Drawings show constitutive splicing and different forms of alternative splicing common in plants such as exon skipping, intron retention, mutually exclusive exons, alternative 5’-SS and 3’-SS selection. Black lines denote introns and different colored rectangles indicate exons.
Light-mediated pre-mRNA splicing
Light can affect pre-mRNA splicing both as an environmental signal as well as an energy source. A retrograde signal emanating from chloroplast has been shown to control splicing of light- and circadian clock-regulated genes [17]. By using various photosynthetic electron transport inhibitors, these authors showed that a reduced pool of plastoquinones intiates the chloroplast retrograde signaling. Recently, it was shown that several hundred genes undergo AS during early photomorphogenesis, and energy availability plays an important role in this regulation by controlling the rate of transcriptional elongation by RNA polymerase II [18,19]. Rate of transcription determines the binding opportunity of splicing and auxiliary regulatory factors to the target pre-mRNA sequences, and thereby determines the fate of AS [19,20]. Depending on the specific pre-mRNAs, slower rate of elongation stimulates either higher exon skipping or higher exon inclusion. It was shown that light promotes the RNA Polymerase II elongation on the target genes, while in darkness the rate of transcription of those target genes is substantially lower. Thus, by altering the transcription kinetics, light controls AS events to modulate appropriate transcript isoforms and the optimal physiological responses to the environmental cues [19]. While these are more long-term effects of light on splicing, an immediate response of light acting as a signal has been demonstrated using pulses or continuous light [21,22].
Molecular details on the photoreceptor-mediated control of pre-mRNA splicing are still in its early stage. Among all known major photoreceptors, regulatory role of phys in pre-mRNA splicing is best studied till date [8,10]. Genome-wide analyses of red-light dependent and phy-regulated AS uncovered significant changes in AS pattern in hundreds of genes within 1 hr of continuous red-light (cRL) exposure, including a group of RNA splicing-related genes such as several SR proteins, U1 and U2 auxiliary factors at the seedling stage [22]. Interestingly, the cRL-dependent differential AS pattern observed in one of the SR protein genes, RS31, could also be replicated under 2-mins pulse of red-light (pRL) and more importantly altered AS pattern could be suppressed by 2-min pulse of far-red-light (pFRL) immediately following pRL. In addition, pulses of red and far-red light have been shown to regulate AS patterns of 226 genes during seed germination, many of which are associated with mRNA processing [23]. Among these, the red light-mediated AS changes of AtSR30, AtSR31, AtSR31a and AtU2AF65A were phyB-independent, while the AS changes of the light signaling component PIF6 and the DORMANCY ASSOCIATED PROTEIN 1 (DRM1) were phyB-dependent, supporting previous conclusion that the AS changes of PIF6 contributes to seed dormancy [24]. These data strongly suggest that phys in response to red-light might target regulatory auxiliary splicing factors under early light exposure and through which it modulates the genome-wide AS pattern under prolonged light conditions [22]. Several studies have also identified some of the critical components of light signaling pathways as targets of phy-modulated AS (Table 1) [8,10]. Shikata et al. identified that SUPPRESOR OF phyA-105 3 (SPA3) is one of the targets of phys-regulated AS [22]. It was shown that phys promote the retention of intron 4 of SPA3 and also selection of alternative 5’-SS within intron 4, both of which introduces pre-mature stop codons resulting in truncated non-functional SPA3 proteins [22]. Recently, a distinctive role of phyB in the regulation of PIF3 level through AS and its corresponding effect on the translational inhibition has been reported [25]. Overaccumulation of the active phyB stimulates a specific AS of PIF3 mRNA resulting in intron retention (IR) in the 5’ untranslated region (5’ UTR). Retained intron contain multiple upstream open reading frame (uORF), which in-turn inhibits downstream expression of PIF3 protein and PIF3 activity under prolonged red-light conditions. Thus, several phy signaling components including many transcription factors are early targets of AS that drives altered transcriptional reprogramming to promote photomorphogenesis.
Table 1:
Pre-mRNA alternative splicing of light-related genes from Arabidopsis and Physcomitrella patens
| Gene | Alternative splicing (AS) type | Regulator | References |
|---|---|---|---|
| PIF3 | IR | phy-regulated | Dong et al., 2020 |
| PIF6 | IR | Unknown | Penfield et al., 2010 |
| PHYA | IR | phy-regulated | Shikata et al., 2014 |
| COP1 | AltA | phy-regulated | Shikata et al., 2014; Zhou et al., 1998 |
| SPA3 | IR | phy-regulated | Shikata et al., 2014 |
| CRY2 | AltA | Phy-regulated | Shikata et al., 2014 |
| HY5 | AltD; IR | Light-regulated | Mancini et al., 2016 |
| HYH | IR | Phy-regulated | Shikata et al., 2014 |
| RRC1 | ES | Light-regulated | Hartmann et al., 2016; Xin et al., 2019 |
| ELF3 | IR | Light-regulated | Xin et al., 2017; Xin et al., 2019 |
| FUS6 | IR | Light-regulated | Xin et al., 2019 |
| SPT1 | IR | Light-regulated | Xin et al., 2019 |
| BBX25 | IR | Light-regulated | Xin et al., 2019 |
| FRY1 | IR | Light-regulated | Xin et al., 2019 |
| DET1 | IR; AltA;AltD | Light-regulated | Wu et al., 2014 |
| HY5/HYH | IR; ES; AltA | Light-regulated | Wu et al., 2014 |
| PIFs | IR | Light-regulated | Wu et al., 2014 |
| DDB1 | IR | Light-regulated | Wu et al., 2014 |
| CSN1–8 | IR; AltA | Light-regulated | Wu et al., 2014 |
| NPH3 | IR; AltA | Light-regulated | Wu et al., 2014 |
| BBX22 | IR | Light-regulated | Wu et al., 2014 |
| COP1 | IR; AltA | Light-regulated | Wu et al., 2014 |
| ELF3 | IR | Light-regulated | Wu et al., 2014 |
IR: Intron Retention; AltA: Alternative acceptor site; AltD: Alternative donor site; ES: Exon skipping
In the last few years at least a few of the splicing factors/regulators involved in the phy-mediated modulation of pre-mRNA splicing have been identified based on different genetic and proteomic approaches from plants and moss [26–30]. SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS), a potential ortholog of Drosophila and human splicing factor 45, was the first bona fide phy-interacting splicing factor identified from Arabidopsis [27]. A subsequent study involving an affinity purification followed by mass spectrometry analyses to identify SFPS-interacting proteins has identified another splicing factor REDUCED RED-LIGHT RESPONSES IN CRY1 CRY2 BACKGROUND 1 (RRC1) [28], which was previously described from a genetic screen [26]. Both SFPS and RRC1 forms discrete nuclear speckles, which in part co-localize with the red-light-induced phyB photobodies, and also interact physically in response to red-light. Interestingly, SFPS and RRC1 also co-localize and interact in vivo with multiple 3’-SS determining U2-associated factors, suggesting that these two splicing factors might play a role in 3’-SS determination (Figure 2). Phenotypically, sfps and rrc1 mutant alleles display light hyposensitive hypocotyl growth and early flowering, and interestingly, sfps/rrc1 double mutant phenocopy parental single mutants, implying that these two proteins function coordinately in part to modulate optimal light signaling (Figure 2). RRC1 itself is the target of light-regulated AS, generating RRC1.1 and RRC1.2 isoforms [18]. RRC1.1 isoform translates to produce functional protein, while RRC1.2 contain a premature stop codon and might encode a nonfunctional protein. Light typically favors RRC1.1 over RRC1.2 and therefore, light-irradiation results in higher accumulation of a functional RRC1.1 in wild type plants. Strikingly, SFPS was shown to regulate light-dependent AS of RRC1. In sfps-2 mutant background, RRC1.1 isoform was predominant, while RRC1.2 was non-detectable under both dark and light conditions [28], implying the presence of a self-reinforcing circuitry.
Figure 2. Model shows phytochrome-modulated pre-mRNA splicing in Arabidopsis.
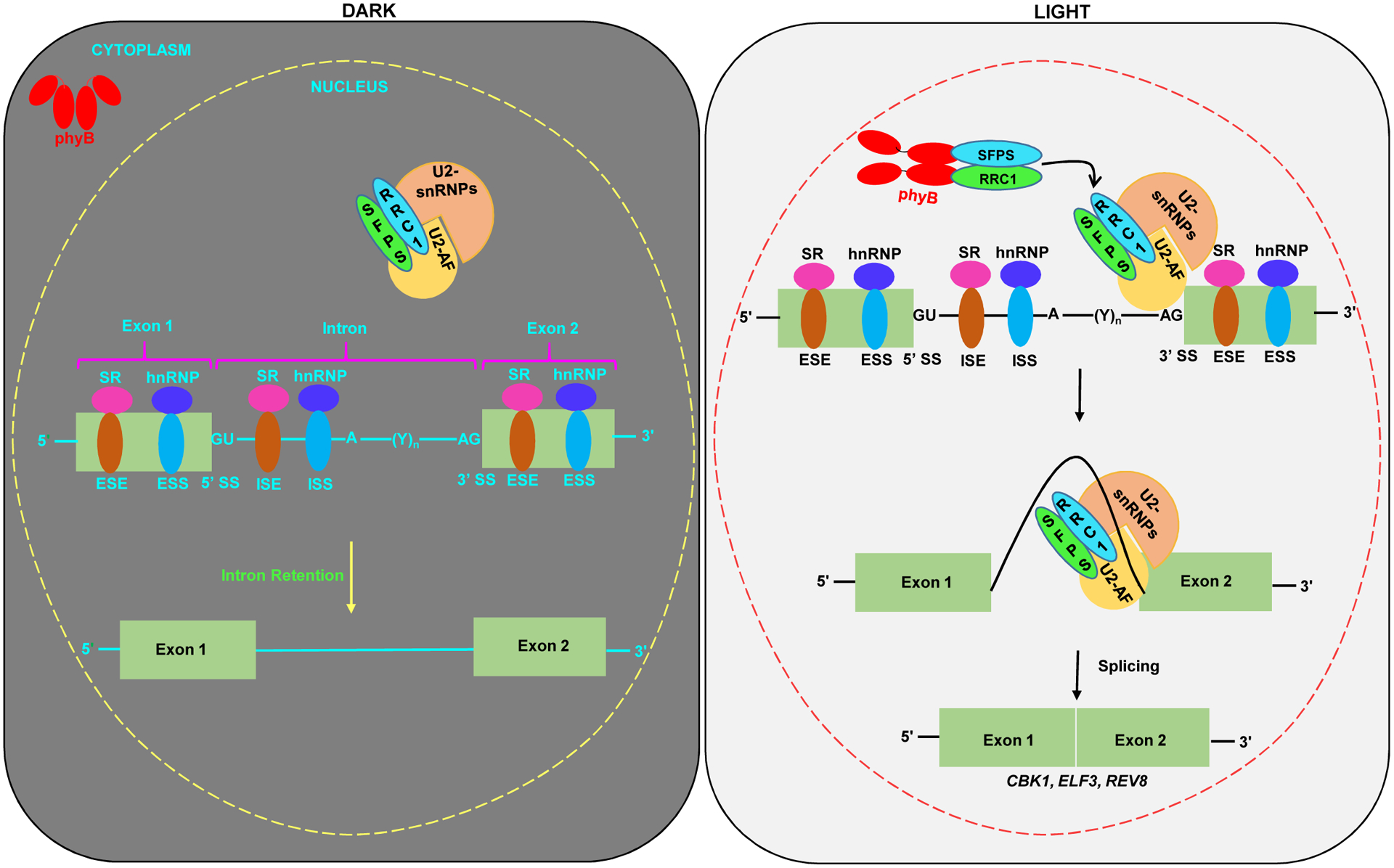
SFPS and RRC1 are two known splicing factors that directly interact with phytochrome B (phyB) and regulate phytochrome-modulated pre-mRNA splicing in Arabidopsis. In the dark (left panel), SFPS and RRC1 interact with each other and also form complexes with U2-snRNP/U2-AF and target hundreds of co-regulated pre-mRNAs for splicing. Moreover, SFPS and RRC1 independently target a large number of distinct pre-mRNAs for splicing in both dark and light conditions. When plants are exposed to light (right panel), activated phyB interacts with SFPS/RRC1 complex in nucleoplasm and photobodies, which might lead to the targeting of different sets of pre-mRNAs for splicing and/or prevention of splicing by unknown mechanism. Because SFPS/RRC1 can associate with U2AFs, and the U2AFs associate with U2 snRNP, it is still unclear whether phyB remains in the nucleoplasm and photobodies or phyB also directly participates on the spliceosome complex throughout the process of target identification and splicing. It is also possible that upon interaction with the SFPS/RRC1 complex, phyB induces biochemical changes to the SFPS and RRC1 proteins, and releases the complex for appropriate target selection and subsequent splicing.
SFPS and RRC1 control the gene expression and pre-mRNA splicing of a large number of genes both under dark and light conditions [28]. A comparison of SFPS and RRC1 regulated splicing events identified hundreds of co-regulated splicing events, both under the dark and light conditions, reiterating the fact that these two proteins function in the same complex collaboratively to regulate pre-mRNA splicing of a subset of genes to regulate photomorphogenesis. Therefore, it is possible that the red-light dependent interaction with phyB might serve as a regulatory switch to guide the SFPS and RRC1 to specific targets. Analyses of the splicing defects uncovered a significant enrichment in IR events in sfps and rrc1 mutants, while all other forms of AS defects were recorded to a lesser extent. A comprehensive gene ontology (GO) analyses established a significant enrichment of multiple light signaling related GO categories including circadian clock, transcription activity, light stimulus and photosynthesis in both sfps and rrc1, pointing to a critical regulatory role of SFPS and RRC1 in light signaling by direct interaction with phyB [27,28].
In addition to Arabidopsis, light-responsive changes in pre-mRNA AS events are also observed in the moss Physcomitrella patens [29–31]. Light instantaneously induces AS in P. patens and predominantly favors IR. Interestingly, light modulates AS with transcript selectivity in genes with a function related to splicing and light signaling to regulate photomorphogenesis. An RNA-seq survey of light-dependent alternatively spliced light signaling genes identified a total of 36 genes including HY5/HYH, PIFs, DET1, DDB1, CSNs and COP1 covering the breadth of photomorphogenic gene regulation from chromatin-remodeling to regulated protein degradation (Table 1) [31]. P. patens contains seven phys (PpPHY1 to PpPHY7), of which PpPHY1 and PpPHY3 are clustered as phyA-type and the remaining five as phyB-type phys [32]. Analyses of mutants defective in all seven Ppphy revealed a primary role of this group of red-light photoreceptors in pre-mRNA AS, and a further analysis of individual PpPHYs suggest a more prominent role of phyB-type PpPHYs in red-light mediated AS. A large-scale protein-protein interaction studies have recently identified two splicing regulators, PphnRNPs (heterogeneous nuclear ribonucleoproteins), which interact with light-activated PpPHY4 to regulate light-mediated AS (Figure 3) [29,30]. Red-light activated PpPHY4 interacts with a splicing regulator PphnRNP-H1 and moreover, PphnRNP-H1 interacts with PpPRP39–1 (pre-mRNA-processing factor 39–1), a component of the U1-snRNP spliceosome complex, with higher affinity in the presence of activated PpPHYs (Figure 3). It is proposed that such interaction induces the dissociation of PpPRP39–1 from spliceosome complex, possibly altering the downstream molecular events [29]. In the subsequent studies, the same group identified PphnRNP-F1 as another splicing regulator that interacts with the activated PpPHY4 to regulate light-mediated AS in P. patens. RNA-seq analyses revealed that PpPRP39–1, PphnRNP-H1 and PphnRNP-F1 modulate light-dependent AS largely in an overlapping manner to that of PpPHY4 [29,30], thus, confirming a more coordinated role of PpPHYs and its interacting splicing regulators in light-mediated AS to promote developmental plasticity in P. Patens (Figure 3).
Figure 3. Model shows phytochrome-modulated pre-mRNA splicing in Physcomitrella patens.

PphnRNP-H1 and PphnRNP-F1 are the two splicing regulators known to modulate phytochrome-dependent pre-mRNA splicing in P. patens. In the dark (left panel), U1-snRNP/U1C/PRP39 associated spliceosome complex promotes the AS in hundreds of target pre-mRNAs in a phytochrome-independent manner. In response to light irradiation (right panel), activated phytochromes interact with PphnRNP-H1/PphnRNP-F1. Phytochromes also promote the high-affinity interaction between PphnRNP-H1 and PRP39, due to which PRP39 dissociates from U1snRNP/U1C complex. This might lead to the reduced activity of U1-snRNP/U1C complex and intron retention in target pre-mRNAs. Moreover, a purine-rich GAA motif is one of the bona fide exonic splicing silencer (ESS), which recruits hnRNP-F1 to promote intron retention in affected transcripts.
Future perspectives
As the phy-mediated modulation of AS in plants is still in its nascent stage, further studies are necessary to uncover additional phy-interacting splicing factors/regulators and their combined impact on light-mediated global AS. Since, presently known Arabidopsis and P. patens phy-interacting splicing factors/regulators form complexes with 3’-SS targeting U2-snRNPs and 5’-SS targeting U1-snRNPs, respectively [27–30], it is possible that either light-mediated AS might have evolved in parallel in these two species or the corresponding homologous genes might be present in each species, which need to be identified and characterized.
One of the most fundamental questions is how phys control splicing. Light-activated phys largely modulate the abundance and/or activity of its target proteins upon interaction by inducing post-translation modifications, such as phosphorylation [7]. The abundance of any of the phy-interacting splicing factors/regulators (e.g., SFPS/RRC1) is not reported to be altered in response to either dark or light treatment [27–30]. However, several large-scale proteomic studies have identified multiple phosphorylation sites within SFPS and RRC1 [33,34]. Therefore, it is possible that the interaction between activated phy and SFPS/RRC1 might lead to phosphorylation within SFPS/RRC1 and subsequent modulation of their splicing activity. However, this hypothesis needs to be examined in future.
To thoroughly understand the role of phy-interacting splicing factors/regulators, it is pertinent to identify their target pre-mRNAs by ascertaining splicing factors/regulators-RNA interactions under in vivo conditions. CLIP (Cross-Linking and ImmunoPrecipitation) and its variants has long-established to be the method of choice for such studies. Although highly competent, these methods have a list of drawbacks and are less efficient in detecting rare or low-abundant RNA targets [35]. Recently, a highly versatile TRIBE (Target of RNA- binding proteins Identified By Editing) and its variant HyperTRIBE has been developed to study in vivo targets of RNA binding proteins in mammalian system [36,37]. Although this method has been optimized and applied in mammalian system, it could potentially be used in plant system with slight modifications to identify the possible target transcripts of plant splicing factors/regulators.
Acknowledgements
We thank members of the Huq laboratory for critical reading of the manuscript. This work was supported by grants from the National Science Foundation (MCB-2014408) and National Institute of Health (NIH) (GM-114297) to E.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest Statement
Praveen Kathare: Conceptualization, Writing- Original draft preparation, Figure preparation.
Enamul Huq: Conceptualization, Writing- Reviewing and Editing.
References:
- 1.Paik I, Huq E: Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Seminars in Cell & Developmental Biology 2019, 92:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rockwell N, Lagarias J: Phytochrome evolution in 3D: deletion, duplication, and diversification. New Phytologist 2019, doi: 10.1111/nph.16240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathews S, Sharrock RA: Phytochrome gene diversity. Plant Cell Environment 1997, 20:666–671. [Google Scholar]
- 4.Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA: Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 2009, 21:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharrock RA, Clack T: Heterodimerization of type II phytochromes in Arabidopsis. Proc Natl Acad Sci U S A 2004, 101:11500–11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu S-H: Gene Expression Regulation in Photomorphogenesis from the Perspective of the Central Dogma. Annual Review of Plant Biology 2014, 65:311–333. [DOI] [PubMed] [Google Scholar]
- 7.Cheng MC, Kathare PK, Paik I, Huq E: Phytochrome signaling networks. Annual Review of Plant Biology 2021, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Y-L, Tu S-L: Alternative Splicing and Cross-Talk with Light Signaling. Plant and Cell Physiology 2018, 59:1104–1110. [DOI] [PubMed] [Google Scholar]
- 9.Matsushita T: Regulation of Alternative Splicing by Phytochrome. In Phytochromes: Methods and Protocols. Edited by Hiltbrunner A: Springer New York; 2019:143–148. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Lin C, Gu L: Light Regulation of Alternative Pre-mRNA Splicing in Plants. Photochemistry and Photobiology 2017, 93:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Rio DC: Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem 2015, 84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhary S, Khokhar W, Jabre I, Reddy ASN, Byrne LJ, Wilson CM, Syed NH: Alternative Splicing and Protein Diversity: Plants Versus Animals. Frontiers in Plant Science 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson ME, Charenton C, Nagai K: RNA Splicing by the Spliceosome. Annual Review of Biochemistry 2020, 89:359–388. [DOI] [PubMed] [Google Scholar]
- 14.Syed NH, Kalyna M, Marquez Y, Barta A, Brown JWS: Alternative splicing in plants--coming of age. Trends in plant science 2012, 17:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas BJ, Wortman JR, Ronning CM, Hannick LI, Smith RK Jr., Maiti R, Chan AP, Yu C, Farzad M, Wu D, et al. : Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC biology 2005, 3:7–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M: Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome research 2012, 22:1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JWS, Barta A, Kalyna M, et al. : A Chloroplast Retrograde Signal Regulates Nuclear Alternative Splicing. Science 2014, 344:427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmann L, Drewe-Boß P, Wießner T, Wagner G, Geue S, Lee H-C, Obermüller DM, Kahles A, Behr J, Sinz FH, et al. : Alternative Splicing Substantially Diversifies the Transcriptome during Early Photomorphogenesis and Correlates with the Energy Availability in Arabidopsis. The Plant Cell 2016, 28:2715–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study established that light-induced skoto-to-photomorphogenesis switch promotes massive transcriptomic reprogramming by modulating alternative splicing (AS) events. Moreover, they showed that in vast majority of photomorphogenesis-related AS events, light favors protein-coding transcript variant over variant containing Non-sense Mediated mRNA Decay (NMD) features. The authors also concluded that energy availability plays a cruicla role in AS.
- 19.Godoy Herz MA, Kubaczka MG, Brzyżek G, Servi L, Krzyszton M, Simpson C, Brown J, Swiezewski S, Petrillo E, Kornblihtt AR: Light Regulates Plant Alternative Splicing through the Control of Transcriptional Elongation. Molecular Cell 2019, 73:1066–1074.e1063. [DOI] [PubMed] [Google Scholar]; This study highlighted the fact that light modulates alternative splicing in affected genes in part by controlling their rate of transcription elongation, thus, establishing that the kinetic coupling mechanism identified in mammalian system is also functional in plant system.
- 20.Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu X-D, Bentley DL: Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes & Development 2014, 28:2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancini E, Sanchez SE, Romanowski A, Schlaen RG, Sanchez-Lamas M, Cerdán PD, Yanovsky MJ: Acute Effects of Light on Alternative Splicing in Light-Grown Plants. Photochemistry and Photobiology 2016, 92:126–133. [DOI] [PubMed] [Google Scholar]
- 22.Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T: Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proceedings of the National Academy of Sciences 2014, 111:18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study performed in-depth RNA-seq analyses of wild type vs phyAphyB double mutant and showed that light-activated phytochromes not only regulate transcription but also post-transcriptional alternative splicing events.
- 23.Tognacca RS, Servi L, Hernando CE, Saura-Sanchez M, Yanovsky MJ, Petrillo E, Botto JF: Alternative Splicing Regulation During Light-Induced Germination of Arabidopsis thaliana Seeds. Frontiers in Plant Science 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penfield S, Josse E-M, Halliday KJ: A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Molecular Biology 2010, 73:89–95. [DOI] [PubMed] [Google Scholar]
- 25.Dong J, Chen H, Deng XW, Irish VF, Wei N: Phytochrome B Induces Intron Retention and Translational Inhibition of PHYTOCHROME-INTERACTING FACTOR3. Plant Physiology 2020, 182:159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study revelead a unique regulatory mechanism of phytochrome B (phyB) to inhibit PIF3 function under continuous red-light conditions. It was shown that the overaccumulated phyB induces an intron retention within the 5’-untranslated region (UTR), which in-turn inhibits PIF3 protein synthesis and PIF3 function.
- 26.Shikata H, Shibata M, Ushijima T, Nakashima M, Kong S-G, Matsuoka K, Lin C, Matsushita T: The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. The Plant Journal 2012, 70:727–738. [DOI] [PubMed] [Google Scholar]
- 27.Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, Huq E: SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proceedings of the National Academy of Sciences 2017, 114:E7018–E7027. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified the first phytochrome B-interacting splicing factor SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS) from Arabidopsis. SFPS modulates light-mediated and phytochrome-dependent alternative splicing in target transctiptome to fine-tune photomorphogenic responses in plants
- 28.Xin R, Kathare PK, Huq E: Coordinated Regulation of Pre-mRNA Splicing by the SFPS-RRC1 Complex to Promote Photomorphogenesis. The Plant Cell 2019, 31:2052–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that a RRC1 (REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND1), a splicing factor, interacts with phytochrome B and also forms a functional complex with SFPS (SPLICING FACTOR FOR PHYTOCHROME SIGNALING). It was established that a functional SFPS-RRC1 complex co-regulate pre-mRNA splicing of subset of genes to regulate photomorphogenesis.
- 29.Shih C-J, Chen H-W, Hsieh H-Y, Lai Y-H, Chiu F-Y, Chen Y-R, Tu S-L: Heterogeneous Nuclear Ribonucleoprotein H1 Coordinates with Phytochrome and the U1 snRNP Complex to Regulate Alternative Splicing in Physcomitrella patens. The Plant Cell 2019, 31:2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified the first phytochrome 4 (PHY4)-interacting splicing regulator hnRNP-H1 (heterogeneous nuclear ribonucleoprotein H1) from Physcomitrella patens. PHY4 associated hnRNP-H1 interacts with a positive splicing factor PRP39–1 (pre-mRNA-processing factor 39–1) and induces its dissociation from the core spliceosome; thus, repressing splicing and promoting intron retention in target transctiptome. They establsihed that phytochrome through cascade of protein-protein interactions targets spliceosome assembly to modulate pre-mRNA splicing and thereby photomorphogenesis.
- 30.Lin B-Y, Shih C-J, Hsieh H-Y, Chen H-C, Tu S-L: Phytochrome Coordinates with a hnRNP to Regulate Alternative Splicing via an Exonic Splicing Silencer. Plant Physiology 2020, 182:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study performed large-scale protein-protein interaction study and identified phytochrome 4 (PHY4)-interacting splicing regulator hnRNP-F1 (heterogeneous nuclear ribonucleoprotein F1) from Physcomitrella patens. They showed that PHY4 and hnRNP-F1 co-regulate ~70% of intron retention (IR) events under red-light treated conditions. They also identified purine-rich exonic splicing silencer GAA motif in majority of transctipts with IR and showed that hnRNP-F1 forms RNA-protein complex to repress splicing in affected transcriptome.
- 31.Wu H-P, Su Y-s, Chen H-C, Chen Y-R, Wu C-C, Lin W-D, Tu S-L: Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome biology 2014, 15:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the first systematic in-depth RNA-seq analyses to study light-mediated changes in alternative splicing (AS) in the Physcomitrella patens. They established that light instantly switches AS and preferably favors intron retention. They identified large number of regulatory genes involved in light signaling as prefered targets of light-mediated changes in AS.
- 32.Xu T, Yuan J, Hiltbrunner A: PHYTOCHROME INTERACTING FACTORs in the moss Physcomitrella patens regulate light-controlled gene expression. Physiologia Plantarum 2020, 169:467–479. [DOI] [PubMed] [Google Scholar]
- 33.Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y: Large scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Molecular Systems Biology 2008, 4:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K: Large-Scale Comparative Phosphoproteomics Identifies Conserved Phosphorylation Sites in Plants. Plant Physiology 2010, 153:1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler EC, Van Nostrand EL, Yeo GW: Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. WIREs RNA 2018, 9:e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman R, Xu W, Jin H, Rosbash M: Identification of RNA-binding protein targets with HyperTRIBE. Nature Protocols 2018, 13:1829–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu W, Rahman R, Rosbash M: Mechanistic implications of enhanced editing by a HyperTRIBE RNA-binding protein. RNA 2018, 24:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]


