Abstract
Ribosomes that stall inappropriately during protein synthesis harbor proteotoxic components linked to cellular stress and neurodegenerative diseases. Molecular mechanisms that rescue stalled ribosomes must selectively detect rare aberrant translational complexes and process the heterogeneous components. Ribosome-associated quality control (RQC) pathways eliminate problematic mRNAs and nascent proteins on stalled translational complexes. In addition, recent studies have uncovered general principles of stall recognition upstream of quality control pathways and fail-safe mechanisms that ensure nascent proteome integrity. Here, we discuss developments in our mechanistic understanding of the detection and rescue of stalled ribosomal complexes in eukaryotes.
Keywords: ribosome stalling, ribosome collisions, ribosome-associated quality control (RQC)
Rescuing stalled ribosomes
Protein synthesis must be tightly regulated to maintain cellular and organismal health. Rescuing ribosomes that stall aberrantly during translation is required to prevent proteotoxicity (see Glossary), mitochondrial dysfunction, and neurodegeneration [1–10]. Ribosome rescue involves several considerations. First, selectively detecting aberrant translational complexes is a non-trivial task. Ribosomes may stall for diverse reasons, and stalled ribosomes are rare among much more abundant functional counterparts. Second, rescue mechanisms must account for all components of stalled translational complexes, including messenger RNA (mRNA), an incompletely-synthesized nascent protein attached to transfer RNA (tRNA), the ribosomal subunits, and associated translation factors. Because any factor may contribute to stalling, cells would ideally triage each component for recycling or degradation. Finally, in addition to rescuing individual ribosomes, excessive ribosome stalling should activate cellular responses to minimize proteotoxic burden.
Dedicated eukaryotic ribosome rescue mechanisms were revealed by observations that problematic mRNAs (Box 1) and the nascent proteins they produce are degraded specifically in a translation-dependent manner (reviewed in [10–13]). Work over the last 5 years has rapidly advanced our understanding of what happens upstream and in parallel to these degradative pathways, such as how specific mRNA sequences cause ribosomes to stall, and how cells selectively detect stalled ribosomes, scrutinize translational factors, and ensure aberrant nascent protein degradation (Figure 1). This review focuses on recent developments in our mechanistic understanding of ribosome stalling and rescue in yeast and mammalian cells.
Box 1 – Ribosome stalling and mRNA degradation.
Ribosomal subunit dissociation triggers mRNA surveillance
Defective mRNAs that stall ribosomes are cleared to eliminate risk of future stalls [10–13]. Although mRNA surveillance was the first quality control mechanism linked to aberrant translation, mechanistic understanding of mRNA surveillance now lags behind other aspects of ribosome-associated quality control (RQC). Stall-inducing mRNA features and surveillance mechanisms also may differ between yeast, where the most work has been done, and metazoans. In yeast, degradation of 3’-truncated mRNAs depends on Dom34 and the Ski2/3/8 complex, an adaptor for the RNA exosome [34,88], suggesting that ribosomal subunit dissociation mediated by Dom34 is a prerequisite for mRNA degradation. Loss of the C-terminal domain of Ski7, an eEF1A-like GTPase homolog and exosome cofactor, further stabilizes 3’-truncated mRNAs in a dom34Δ background [34], indicating a possible parallel degradation mechanism. The HBS1L3 isoform may be the mammalian homolog of Ski7 and similarly interacts with the exosome [35].
Degradation of non-truncated stall-inducing mRNAs, such as on poly(A) mRNA lacking stop codons [14,88], requires Hel2-mediated ubiquitination of uS10 [37,50]. mRNA fragments generated by Hel2-dependent endonucleolytic cleavages [48] are not protected by a poly(A) tail or a 5’-cap and are readily degraded by the exosome or the 5’–3’ exonuclease Xrn1 [16,88]. Recent genetic screens identified yeast Cue2 (NONU-1 in Caenorhabditis elegans) as this endonuclease [49,89]. Cue2 contains a SMR hydrolase domain and ubiquitin-binding CUE domains that may recognize Hel2-ubiquitinated ribosomes, although this remains to be conclusively demonstrated. While endonucleolytic cleavage was originally considered an obligate step for mRNA degradation, deleting Cue2 abrogates the generation of low-abundance mRNA fragments but does not stabilize stalling reporter mRNA levels [49]. Thus, endonucleolytic cleavage is not required and may not constitute the major pathway for mRNA degradation. Instead, ribosome clearance by the RQT complex leading to exonucleolytic mRNA degradation appears more prevalent [49]. In mammalian cells, mRNA degradation of poly(A) stalling reporters is less obvious [41], and further work is needed to understand analogous pathways in other eukaryotes.
Slowed translation triggers degradation of nonoptimal mRNAs
mRNAs containing nonoptimal codons slow translation and are preferentially degraded. This sensing mechanism involves Not5, a subunit of the Ccr4-Not deadenylase complex, which binds the E-site of ribosomes in the post-translocated state that occurs after E-site tRNA exit but before A-site tRNA binding [90]. The lack of cognate aminoacylated-tRNA may stabilize this normally transient conformation, providing time for Not5 binding to selectively sense ribosomes that slow translation on nonoptimal codons.
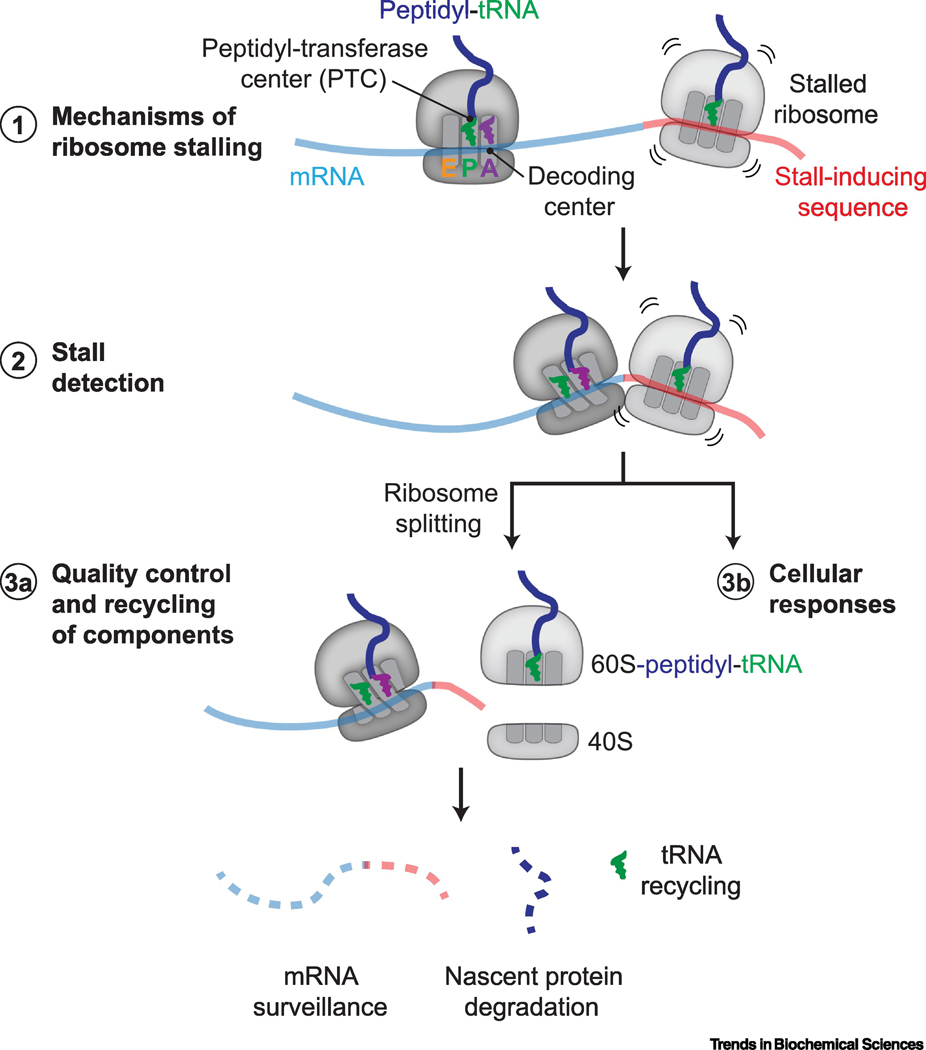
Figure 1 |. Progress on ribosome rescue in eukaryotes.
Recent work has revealed (1) insights into how ribosomes stall while translating specific mRNA sequences (red). The aminoacyl (A), peptidyl (P), and exit (E) tRNA binding sites, decoding center, and peptidyl-transferase center (PTC) on an 80S ribosome are indicated. (2) Ribosome rescue requires factors that specifically detect molecular signatures of ribosome stalling, such as an empty decoding center or ribosome collisions. Ribosome rescue factors activate (3a) local quality control and recycling mechanisms to process individual components of stalled ribosomal complexes and (3b) cellular stress responses to reduce proteotoxic burden.
How ribosomes stall
mRNA defects are the best understood causes of ribosome stalling. Stall-inducing mRNA features include truncations within coding sequences and specific mRNA sequences such as poly(A) tracts [10–14]. Indeed, premature polyadenylation of up to ~1–5% of mRNAs may be the most common cause of ribosome stalling in eukaryotes [14,15]. Stable secondary structures and repeats of CGA arginine codons are also routinely used as reporters of ribosome stalling in yeast, although these specific features may not stall metazoan ribosomes (Box 1) [10–13,16].
Recent studies revealed how eukaryotic ribosomes stall on poly(A) and certain endogenous mRNA sequences [17–19]. Poly(A) mRNA was originally thought to stall ribosomes through interactions between the basic lysines encoded by AAA codons and the negatively charged ribosomal exit tunnel [20]. However, neither poly-lysine encoded by AAG codons nor basic poly-arginine tracts are sufficient to stall mammalian ribosomes [21]. Analysis of different combinations of AAG and AAA codons in vitro revealed that ribosome stalling requires the simultaneous presence of lysines in the exit tunnel and poly(A) mRNA in the decoding center [19,21] (Figure 2A). In yeast, a stalling sequence in the SDD1 mRNA also requires “coincidence detection” of both nascent polypeptide and mRNA features [17].
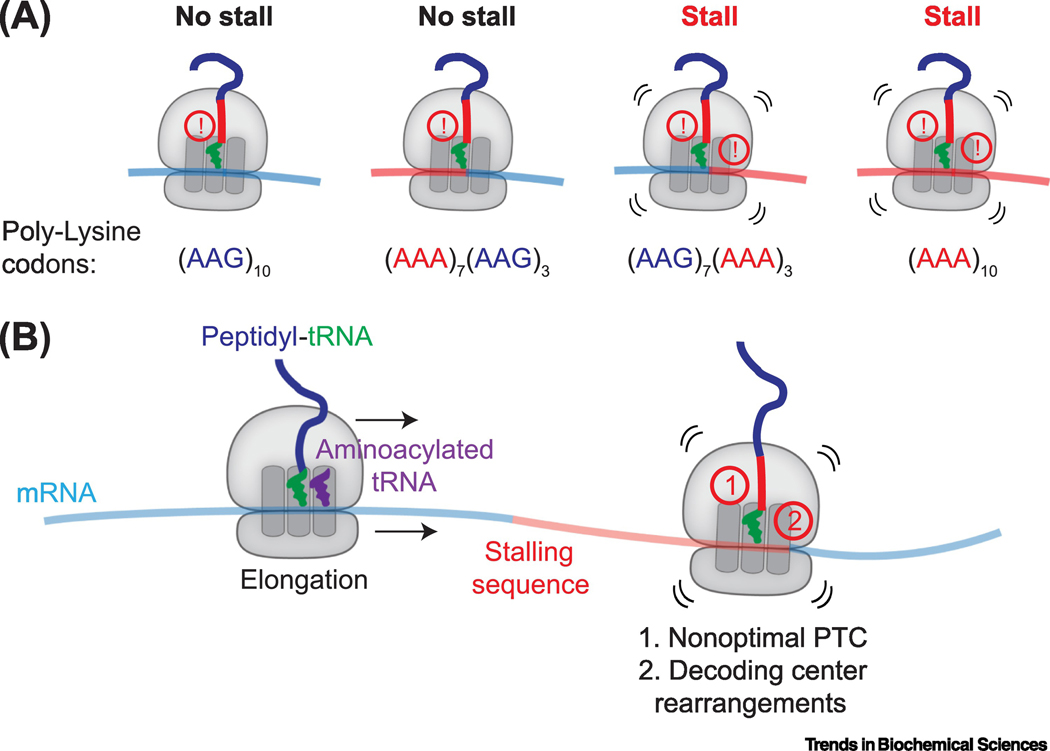
Figure 2 |. Coincidence detection leads to ribosome stalling.
A,In vitro translation of reporters containing 10 lysine residues encoded by different combinations of AAA and AAG codons demonstrate that the simultaneous presence of lysines in the ribosomal exit tunnel and poly(A) mRNA in the decoding center is required to stall mammalian ribosomes [19]. Specifically, ribosomes slow but do not terminally stall when lysines are present in the exit tunnel and AAG codons are in the decoding center (far left; indicated by single !), but strong stalling is detected when lysines occupy the exit tunnel and AAA codons are in the decoding center (far right; indicated by two !). (AAG)10 and (AAA)10 refer to 10 AAG or AAA codons, respectively; (AAA)7(AAG)3 refers to 7 AAA codons followed by 3 AAG codons; (AAG)7(AAA)3 refers to 7 AAG codons followed by 3 AAA codons. B, The coincidence detection model for ribosome stalling suggests that 1) nascent peptide interactions with the ribosomal exit tunnel lead to nonoptimal conformations at the peptidyl transferase center (PTC) that slow translation and allow 2) specific mRNA sequences to adopt intrinsic structures that remodel the decoding center at the A-site of the ribosome, resulting in translational arrest.
The coincidence detection model is supported by cryo-electron microscopy (cryoEM) structures showing perturbations in both the peptidyl-transferase center (PTC) and mRNA decoding center of ribosomes stalled on poly(A) or SDD1 mRNA [17–19] (Figure 2B). In the PTC of poly(A)-stalled ribosomes, the side chain of the lysine attached to P-site tRNA points towards the A-site [18,19], opposite to the orientation seen in elongating ribosomes [22]. This orientation may disfavor peptidyl-transfer, although further work is needed to understand how A-site tRNA binding influences these conformations. In the SDD1-stalled ribosome, interactions between the nascent protein and the ribosomal exit tunnel may displace key 28S ribosomal RNA (rRNA) bases in the PTC into a suboptimal conformation for peptide bond formation [17]. Notably, a parasite with a disproportionately high number of poly(A)-containing open reading frames (ORFs) that apparently do not cause ribosome stalling also show differences in the ribosomal exit tunnel that reduce nascent peptide interactions [23].
Nascent protein-induced PTC deformations likely slow translation, prolonging the amount of time the ribosome resides at one location on the mRNA. This lengthened dwell period may allow certain mRNA sequences to adopt intrinsic structures that rearrange the decoding center and terminally stall the ribosome. For example, in the poly(A)-stalled ribosome, four adenosines form a single-stranded helix that stacks with A1825 and C1698 (Saccharomyces cerevisiae A1756 and C1634) of 18S rRNA, which are ‘flipped out’ from their positions during decoding [18,19]. Helix formation is favored for poly(A) RNA [24], explaining why AAA but not AAG codons in the decoding center lead to ribosome stalling. Further, the structure of the SDD1-stalled ribosome revealed a similar helical mRNA conformation of CGAA in the A-site [17]. A more severe defect at the PTC of SDD1-stalled ribosomes may compensate for the time needed for CGAA to adopt this conformation. Thus, an interplay of nascent protein and mRNA interactions contributes to ribosome stalling propensity.
Coincidence detection does not apply to all instances of ribosome stalling. For example, ribosomes stalled on the XBP1 stalling sequence show nascent protein-ribosome interactions but no remodeling of the decoding center [25], while yeast ribosomes stalled on CGA codons do not show obvious PTC defects [18]. In addition to mRNA features, ribosomes may stall for reasons such as deficiencies in aminoacylated tRNAs, mutated or damaged translation factors, ribosome or translation factor inhibitors, and during cellular stresses such as oxidation or UV exposure [4,6,10–13,22,26–28]. Additional insights into how specific rescue mechanisms respond to ribosomes that stall for different reasons may come from analyzing the functional and structural effects of translation inhibitors that inhibit distinct steps of translation elongation [22,26,27].
Detecting stalled ribosomes
Ribosome rescue factors must selectively recognize rare stalled ribosomes among abundant functional counterparts. At least two molecular features, an empty mRNA channel in the ribosomal A site and unique composite interfaces formed by ribosome collisions, distinguish stalled ribosomes from translation-competent ribosomes and recruit specific rescue factors (Figure 3A).
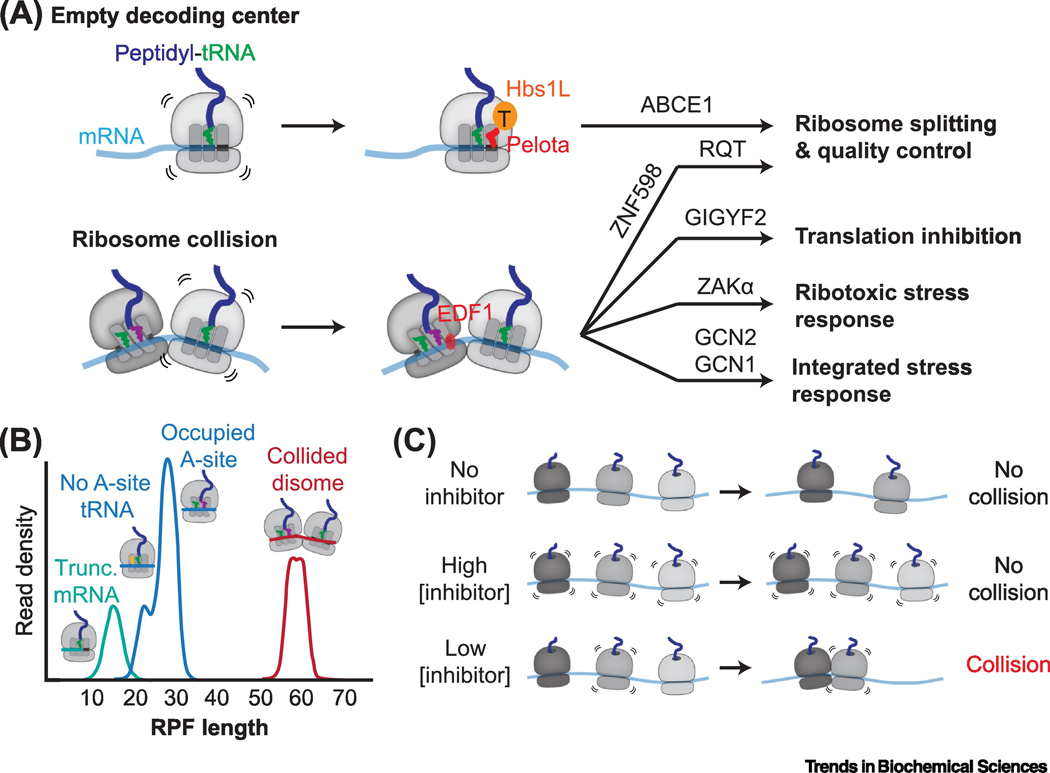
Figure 3 |. Detecting stalled ribosomes.
A, Two unique molecular signatures of stalled ribosomes are an empty decoding center (top) and ribosome collision interfaces (bottom). Each are recognized by specific ribosome rescue factors that activate molecular pathways leading to dissociation of the stalled ribosome into 60S and 40S ribosomal subunits and activation of ribosome-associated quality control (RQC) pathways. Ribosome collisions also activate cellular pathways that inhibit translation in cis and stress response pathways that downregulate translation (integrated stress response) and regulate cellular survival (ribotoxic stress response). (top) The Hbs1L-Pelota complex engages ribosomes with an empty decoding center, leading to ribosomal subunit dissociation via the ribosome recycling factor ABCE1. (bottom) EDF1 selectively binds collided ribosomes, possibly stabilizing the collision interface to recruit downstream factors. Collision-dependent ribosome rescue factors include the E3 ubiquitin ligase ZNF598, which ubiquitylates small ribosomal subunit proteins leading to ribosomal subunit dissociation by the RQT complex, the translation initiation repressor GIGYF2, the MAP3 kinase ZAKα, and the GCN2 co-activator GCN1. B, Ribosome-protected fragments (RPFs) after nuclease treatment distinguish different ribosomal populations. RPF peaks at 21 and 28 nucleotides correspond to translating ribosomes that lack or contain A-site tRNA respectively (blue) [27]. Ribosomes lacking A-site mRNA protect ~16 nucleotides (teal) [32], and collided ribosomes protect ~60 nucleotides (red) [41–43]. C, Effects of translation elongation inhibitor concentrations on ribosome collisions. Treating cells with a high concentration of an elongation inhibitor ‘freezes’ translating ribosomes, preventing collisions. A low concentration of an elongation inhibitor stalls only a subset of ribosomes, resulting in collisions with uninhibited trailing ribosomes [37–40].
Ribosome splitting factors detect ribosomes with an empty decoding center
The Hbs1L-Pelota (yeast Hbs1-Dom34) complex was among the first eukaryotic factors identified to recognize stalled ribosomes [10–13,16,29]. Hbs1L and Pelota are homologous, or structurally related, to the eEF1A-like family of translational GTPases and their partner decoding factors, respectively. This group includes the translation elongation factor eEF1A-aminoacylated tRNA (aa-tRNA) complexes and the termination factors eRF3-eRF1. Each GTPase-decoding factor complex engages the GTPase center and A-site of ribosomes based on mRNA features in the decoding center. While aa-tRNAs pair with sense codons and eRF1 recognizes stop codons, Pelota/Dom34 extends a β-loop into the mRNA entry channel [22,30,31], which requires either the absence or displacement of mRNA. Supporting the model that Pelota/Dom34 rescues ribosomes on truncated mRNAs, yeast lacking Dom34 show an increase in 16-nucleotide ribosome-protected fragments following nuclease treatment, a signature of ribosomes lacking mRNA in the A-site [32] (Figure 3B). In vitro, Hbs1L-Pelota preferentially acts on stalled ribosomes with short downstream mRNA sequences [33]. Thus, Hbs1L-Pelota recognizes ribosomes with empty decoding centers, conceptually similar to the activity of certain bacterial ribosome rescue factors (reviewed in [11]).
After recognition, Hbs1L hydrolyzes GTP and dissociates from stalled ribosomes, which allows Pelota to recruit the recycling ATPase ABCE1 (yeast Rli1) to dissociate stalled ribosomes into 40S and 60S subunits [29,33,34]. Ribosomal subunit dissociation generates specific substrates for mRNA and protein quality control (see below and Box 1). Pelota also interacts with GTPBP2, a mammalian Hbs1L homolog required to rescue ribosomes that stall due to mutation of a neuron-specific arginyl-tRNAUCU [6]. Hbs1L cannot compensate for loss of GTPBP2, suggesting that these GTPases have nonoverlapping functions in resolving different types of stalled ribosomes. Other members of this family, such as GTPBP1 and Hbs1L3, a splicing isoform of Hbs1L, also are implicated in distinct aspects of ribosome rescue, although the details of their functions remain to be clarified [35,36].
Ribosome collisions are a universal consequence of ribosome stalling
Most stalled ribosomes contain mRNA downstream of the A-site and are not optimal Hbs1L-Pelota substrates. Accumulating evidence shows that the collision of a translating ribosome into a stalled ribosome generates unique composite interfaces that are selectively recognized by ribosome rescue factors. Ribosome collisions are a general signature of ribosome stalling irrespective of the original cause of translational arrest, and two experimental strategies have proved instrumental in studying how cells detect and respond to ribosome collisions. First, the frequency of ribosome collisions can be manipulated with different concentrations of translation elongation inhibitors. For example, while a high concentration of an irreversible inhibitor stalls all translating ribosomes, low concentrations stall only a subset, causing collisions with uninhibited ribosomes (Figure 3C) [37–40]. Second, unlike multiple ribosomes independently translating the same mRNA, collided ribosome interactions are resistant to nuclease treatment, permitting biochemical isolation of collided ribosomes and “disome” profiling to identify collision sites (Figure 3B) [38,40–43].
Ribosome collisions trigger ribosomal protein ubiquitination and ribosomal subunit dissociation
The first ribosome collision sensor identified was the E3 ubiquitin ligase ZNF598 (yeast Hel2) [37,38]. A link between Hel2 and ribosome rescue emerged from a genetic screen for factors that stabilize a ribosome stalling reporter [44]. Cells lacking ZNF598/Hel2 show increased readthrough of stalling sequences [44,45], presumably because ribosomes that are not rescued slowly resume translation. ZNF598/Hel2 selectively ubiquitinates small ribosomal subunit proteins near the mRNA entry channel: yeast Hel2 ubiquitinates ribosomal proteins uS3 and uS10, while in human cells, ZNF598 preferentially ubiquitinates uS10 and eS10 [45–48]. ZNF598/Hel2-mediated ubiquitination leads to ribosomal subunit dissociation and, in yeast, endonucleolytic cleavage of stall-inducing mRNAs by Cue2 (Box 1) [37,45,48,49]. Mapping 3’ mRNA fragments revealed that Hel2-dependent cleavages occur at 30-nucleotide periodicities from the stall site, consistent with the spacing between different numbers of collided ribosomes [37]. In addition, only low levels of translation inhibitors resulted in ZNF598/Hel2-dependent ribosomal protein ubiquitination [37,38]. Critically, in vitro reconstitutions showed that ZNF598 recruitment and ubiquitination occurs only on nuclease-resistant collided ribosomes [38]. Further, cryo-EM structures of collided ribosomes showed extensive 40S-40S interactions involving the ribosomal protein RACK1 (yeast Asc1) and ribosomal proteins ubiquitinated by ZNF598/Hel2 [10–12,38,50]. Although density for ZNF598/Hel2 was not visualized in these structures, this composite interface likely facilitates specific recognition of collided ribosomes.
Hel2-bound ribosomes co-purify the ribosome-associated quality control (RQC)-triggering (RQT) complex comprised of Slh1, Cue3, and YWR023W (human ASCC3, ASCC2, and TRIP4/ASC-1) [48,51–53]. The RQT complex dissociates ZNF598/Hel2-ubiquitinated ribosomes into 60S and 40S subunits [17,53], generating the same substrates for quality control as Hbs1L-Pelota. Slh1 contains a helicase domain required for ribosomal subunit dissociation, and Cue3 contains a ubiquitin-binding CUE domain which may bind Hel2-ubiquitinated ribosomes. In vitro reconstitutions showed that the leading stalled ribosome is preferentially ubiquitinated by ZNF598/Hel2 and selectively dissociated by the RQT complex [17,53]. After clearance of the leading ribosome, trailing ribosomes may resume elongation, which may distinguish problematic mRNAs that would cause trailing ribosomes to also stall from defective translational components that would stall only one ribosome.
Ribosome collisions lead to local translation inhibition
Recognition that ribosome collisions are a universal signal of ribosome stalling prompted the search for other factors that preferentially associate with collided ribosomes; ultimately, this led to two independent identifications of EDF1 (yeast Mbf1) and GIGYF2 [40,54]. Knocking down either GIGYF1/2 or EDF1 resulted in increased translation of stalling reporters [40,54,55], supporting a model that ribosome collisions selectively inhibit translation in cis to minimize additional collisions and aberrant protein production from that mRNA. Cryo-EM structures revealed EDF1/Mbf1 bound near the A-site of the mRNA channel specifically of rotated trailing collided ribosomes, changing the path of mRNA through interactions with 18S rRNA, uS3, and mRNA [40,56]. Further, EDF1 facilitates recruitment of GIGYF2 [40,54], which represses translation initiation by interacting with 4EHP, a competitor of the cap-binding initiation factor eIF4E [57]. GIGYF2 also may directly or indirectly to influence mRNA turnover in yeast, which lack 4EHP, and in mammals [55,58].
Ribosome collisions activate cellular stress responses
Translation inhibition by ribosome-inactivating proteins, small molecule translation inhibitors, and UV irradiation also activate the ribotoxic stress response (RSR) mediated by the mitogen-activated protein kinases (MAPKs) p38 and JNK that regulate inflammation and cell death [39,59]. Indicative of a link to ribosome collisions, only intermediate levels of translation elongation inhibitors activate the RSR in a mechanism dependent on autophosphorylation of the MAP-3-kinase (MAP3K) ZAKα in human cells [39]. ZAKα generally associates with polysomes through an isoform-specific C-terminal domain, but is specifically and acutely autophosphorylated in conditions that induce ribosome collisions via a mechanism that may involve EDF1 [39,40,60].
Collided ribosomes also may play a role in activating GCN2, which phosphorylates eIF2α to repress cap-dependent translation as part of the integrated stress response [39,61,62]. GCN2 responds to amino acid starvation through incompletely understood mechanisms that may include sensing deacylated tRNAs and ribosome stalling [61]. For example, in a neurodegenerative mouse model, the combined loss of function of a neuron-specific arginine tRNA and GTPBP2 lead to increased ribosome stalling and GCN2 activation [6,63]. Intermediate concentrations of translation elongation inhibitors also selectively activate GCN2 independently of amino acid deprivation via a mechanism that, in human cells, may involve a structural role for ZAKα [39,62]. In vitro, purified ribosomes or the C-terminal acidic tails of ribosomal P-stalk proteins are sufficient to activate mammalian GCN2 to a greater extent than deacylated tRNAs [64,65]. Disrupting P-stalk proteins in cells also selectively impairs activation of the integrated stress response by GCN2 [64]. Notably, a recent cryo-EM study showed GCN1, a GCN2 activator, directly bound to collided yeast disomes [56]. GCN1 spans both the leading and collided ribosomes: the C-terminus approaches the P-stalk of the leading ribosome, while the N-terminus contacts the P-stalk of the trailing ribosome. Because the P-stalk is part of the ribosomal GTPase center that is blocked by elongation factor binding, translational arrests that cause ribosome collisions and disfavor translation factor binding may preferentially activate GCN2.
Coordinating different responses to ribosome collisions
Collisions are a universal consequence of ribosome stalling, yet different causes of ribosome stalling trigger distinct consequences [39,41,56,62]. The principles that determine the choice and order of pathways remain poorly understood, but may involve contributions from the extent of ribosome collisions throughout the cell, competition between factors for collided ribosomal interfaces, and stabilization of distinct transient ribosomal conformations due to different causes of ribosome stalling. For instance, EDF1 is 50-fold more abundant than GIGYF2 or ZNF598, providing an advantage to engage collided ribosomes first [40,54]. Early engagement by EDF1 and GIGYF2 recruitment would acutely inhibit translation to minimize further collisions, while ZNF598 recruitment and ribosomal protein ubiquitination probably requires more time to effect dissociation of leading stalled ribosomes for mRNA and protein quality control. Although GIGYF2 also independently interacts with ZNF598 [54,55,57], deleting either ZNF598 or GIGYF2 enhances the other response [40,54,55], hinting at nuanced or redundant regulation beyond simple competition or recruitment.
These local events appear to be prioritized over stress responses [39,41,62], where ribosome collisions may acutely activate ZAKα and GCN2 to minimize translational load and promote cellular survival, while prolonged widespread collisions eventually activate downstream RSR kinases and cell death [39]. Different translational aberrancies also may stabilize distinct ribosomal conformations recognized by specific rescue factors, such as the role of P-stalk protein access in GCN2 activation [64,65]. Thus, assigning the appropriate response to ribosome stalling stems from a combination of structural and kinetic considerations. At the level of an individual ribosome, stall recognition may occur based on an empty decoding site that arises from translating a truncated mRNA or prolonged residence times of typically short-lived ribosomal conformations. Another layer of stall recognition leverages the unique interfaces formed by ribosome collisions. Future work will determine more precisely how these responses are regulated based on how often collisions occur, how long collision interfaces persist, specific ribosomal conformations that mediate accessibility to structural components such as the P-stalk, and the relative abundance and activity of ribosome rescue factors.
Quality control of stalled ribosomal complex components
Dissociation of stalled ribosomes into 40S and 60S subunits downstream of recognition by Hbs1L-Pelota or ZNF598 trigger mRNA surveillance (Box 1) and ribosome-associated nascent protein quality control (RQC) mechanisms to degrade the substrates and products of aberrant translation [10–13]. Other components, such as tRNAs and ribosomal subunits, are assumed to be recycled by mechanisms that also may scrutinize and eliminate faulty translational factors.
RQC degrades nascent proteins
Nascent proteins on stalled ribosomes are either truncated or contain non-native sequences. After ribosomal subunit dissociation, the tRNA and folded polypeptide domains at either end of the ribosome exit tunnel trap aberrant nascent peptidyl-tRNAs on the 60S ribosomal subunit [10–12]. NEMF (yeast Rqc2) selectively binds the 60S-tRNA interface that distinguishes these complexes from 80S ribosomes and empty 60S subunits (Figure 4A) [10–12,66,67]. NEMF also prevents 40S rejoining and helps recruit the E3 ubiquitin ligase Listerin (yeast Ltn1). The Listerin RING domain resides directly outside the ribosomal exit tunnel to catalyze K48-linked polyubiquitiylation on the nascent protein for proteasomal degradation [10–12,44,66–68]. Genetic screens for RQC factors also identified a functional role for TCF25 (yeast Rqc1), possibly involving suppressing non-K48 ubiquitin linkages [7,44,69].
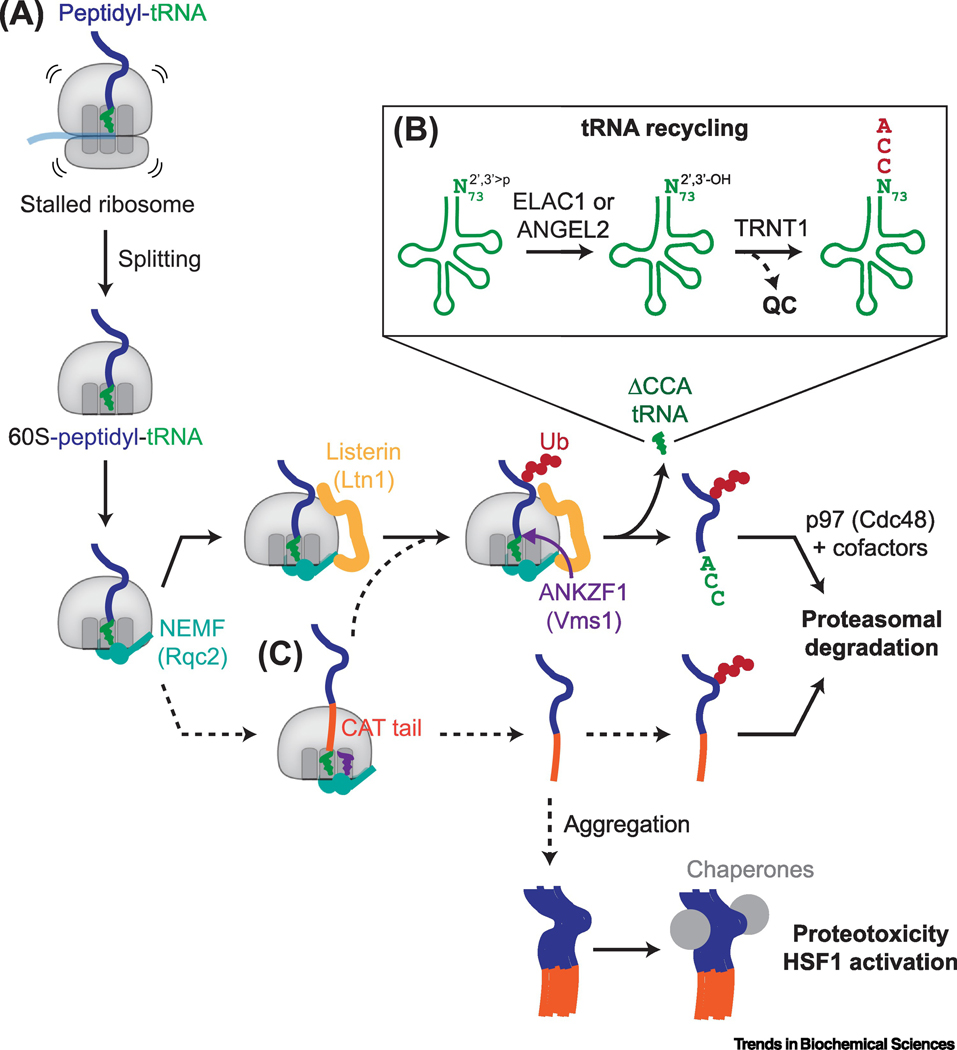
Figure 4 |. Quality control and recycling of peptidyl-tRNAs on stalled ribosomes.
A, Ribosomal subunit dissociation downstream of stall recognition by Hbs1L-Pelota or ZNF598 traps the peptidyl-tRNA on the 60S ribosomal subunit. NEMF (yeast Rqc2) binds the exposed 60S-tRNA interface, which prevents 40S rejoining and facilitates recruitment of the E3 ubiquitin ligase Listerin (yeast Ltn1) to polyubiquitinate the nascent chain. The polyubiquitinated nascent chain may be released by the endonuclease ANKZF1 (yeast Vms1) for proteasomal degradation via a process involving the AAA+ ATPase p97 (yeast Cdc48) and its ubiquitin binding cofactors. B, ANKZF1 cleaves the peptidyl-tRNA just before the invariant 3’CCA nucleotides, generating a CCA-less tRNA intermediate with a terminal 2’,3’-cyclic phosphate (2’,3’>p) that is recycled in a two-step pathway involving the removal of the 2’,3’>p by the phosphatases ELAC1 or ANGEL2, followed by re-addition of the 3’-CCA by TRNT1. TRNT1 also possesses proofreading capabilities that can target defective tRNAs for degradation (QC). C, Rqc2 can catalyze the non-templated addition of C-terminal alanine and possibly threonine residues (CAT tails). CAT tails can push out lysines within the ribosome exit tunnel for ubiquitylation by Listerin. Alternatively, CAT-tailed nascent chains may be released from 60S ribosomal subunits. Off the ribosome, CAT tails may serve as degrons leading to proteasomal degradation, or are otherwise prone to aggregation, which may titrate away chaperones and, in yeast, activate the heat shock response via HSF1.
Once ubiquitinated, nascent proteins must be released from the 60S subunit for proteasomal degradation. These latter steps require the AAA+ ATPase p97 (yeast Cdc48) and its ubiquitin-binding cofactors [44,70] (Figure 4A). While it seemed possible that p97 could ‘pull’ the nascent protein from the ribosome, recent studies showed that ANKZF1 (yeast Vms1), a homolog of eRF1, separates the nascent protein from tRNA on 60S subunits [71,72]. Indeed, in a yeast cryo-EM structure [73], Vms1 resides in the A-site with its catalytic domain reaching towards the PTC, resembling eRF1. In addition, the ribosome biogenesis factor Arb1 (mammalian ABCF2) occupies the E-site and extends a ‘leg domain’ towards the peptidyl-tRNA that may help position it for release. Deleting Vms1 leads to nascent protein aggregation and mitochondrial dysfunction [2,74]. However, the proportion of RQC substrates released by ANKZF1, and how ANKZF1/Vms1, which independently interacts with Cdc48 [74], coordinates with other RQC factors is not known. In vitro, ANKZF1/Vms1 activity is slow relative to Listerin-mediated ubiquitination [69,75], possibly avoiding premature release of non-ubiquitinated nascent proteins. Vms1-released nascent proteins freely ‘fall off’ 60S subunits [75], suggesting that p97 is not required for extraction and may be most relevant in unfolding substrates immediately upstream of proteasomal degradation.
Scrutiny and recycling of tRNAs
Biochemical reconstitutions revealed ANKZF1/Vms1 to be an endonuclease [69,75] that selectively cleaves off the 3’-CCA nucleotides of the peptidyl-tRNA on 60S RQC complexes [75]. Because the 3’-CCA is invariant on all tRNAs, ANKZF1 can act on ribosomes stalled at any codon and generate a CCA-less tRNA intermediate incompatible with aminoacylation that is recycled in a two-step pathway (Figure 4B) [75]. In the first step, a 2’,3’-cyclic phosphate on the 3’ nucleotide ribose of the cleaved tRNA is converted to 2’-OH and 3’-OH. Two enzymes may perform this function. One is ELAC1, a mammalian homolog of RNase Z that appears to have specialized activity to remove 2’,3’-cyclic phosphate from CCA-less tRNAs [76]. ELAC2, the other RNase Z homolog, cleaves tRNA precursors during tRNA biogenesis but has poor 2’,3’-cyclic phosphate removal function, while Trz1, the only RNase Z in yeast, can perform both functions. Consistent with this assignment, cells lacking ELAC1 accumulate ANKZF1-dependent CCA-less tRNA intermediates when treated with translation inhibitors. 2’,3’-cyclic phosphate removal also may be carried out by ANGEL2, which has promiscuous 2’,3’-cyclic phosphatase activity against multiple RNA species in vitro [77]. After removal of the 2’,3’-cyclic phosphate, the CCA-adding enzyme TRNT1 adds back the 3’-CCA nucleotides [75]. TRNT1 also has a proofreading function which adds tandem CCA repeats to tRNAs with a destabilized acceptor stem, targeting them for rapid removal [78]. Thus, the integrity of tRNAs cleaved on stalled ribosomes is specifically checked during recycling.
How cells process other components of stalled ribosomal complexes, such as the ribosomal subunits and translation factors, remain open questions. In yeast, mutating certain key rRNA bases in the PTC or decoding center is compatible with ribosome assembly but results in rapid rRNA degradation [79]. In particular, nonfunctional rRNA decay (NRD) of mutated 18S rRNA requires translation, Asc1, and Dom34, factors linked to ribosome rescue [80,81], as well as ubiquitination of ribosomal proteins [82]. Thus, similar concepts as those discussed above may apply to studying how lesser understood forms of ribosome damage are selectively recognized for quality control.
mRNA-independent polypeptide elongation ensures nascent protein degradation
The unexpected observation of two tRNAs in the cryo-EM structure of the yeast RQC complex led to the discovery that Rqc2 mediates the non-templated addition of C-terminal alanine and threonine residues (“CAT” tails) to nascent proteins that are not efficiently released from 60S subunits [66] (Figure 4C). Remarkably, this mRNA-independent peptide elongation function is conserved in gram-positive bacteria via the function of RqcH, the only known bacterial RQC factor, which adds C-terminal poly-alanine tails that serve as degrons for proteolysis [83] (Box 2). In eukaryotes, CAT tails may facilitate Ltn1-mediated ubiquitination by ‘pushing out’ lysines buried in the ribosomal exit tunnel [84,85], or by increasing the accessibility of lysines in structured domains for ubiquitylation [86]. Recent work suggests that physiological CAT tails in mammals also may primarily contain alanines [9] and serve as degrons off the ribosome recognized by independent quality control factors [86]. The details and regulation of these quality control mechanisms remain subjects of investigation. If not degraded, CAT-tailed proteins are prone to aggregation, which are proteotoxic and titrates away chaperones to activate the heat shock response [1,2,4,7,87]. Thus, Rqc2 homologs and C-terminal degrons may comprise a ‘core’ ribosome-associated protein quality control mechanism that was elaborated on by the ubiquitin-proteasome system in eukaryotes.
Box 2 – RQC at organelles and in bacteria.
In eukaryotes, cytosolic ribosomes may target to organelles that can influence ribosome-associated quality control (RQC). Principles of RQC also may apply to stalled bacterial and mitochondrial ribosomes.
RQC at the ER
Approximately 1/3 of the proteome cotranslationally target to the endoplasmic reticulum (ER), where ribosomes dock at the Sec61 translocon. RQC is intact on non-stop ER-targeted model substrates [91–93], and Listerin can ubiquitinate translocating nascent proteins on Sec61-bound ribosomes in vitro [94]. While Sec61 does not sterically occlude Listerin binding, successful ER-RQC may have additional considerations such as the capacity of the nascent protein to backslide across the ER membrane to increase accessibility of lysines for ubiquitination, or additional factors to extract translocated portions of the nascent protein for degradation. ER-RQC may also be influenced by mechanisms that regulate translation to help insert, chaperone and fold membrane proteins [95,96].
Independent of RQC, inducing ribosome stalling also specifically increases UFMylation of uL24 on ER-bound ribosomes [97,98]. UFMylation is a metazoan-specific post-translational modification in which the ubiquitin-like protein UFM1 is added onto substrate lysines. Ribosome UFMylation may facilitate lysosome-dependent nascent protein degradation [97], although the prevalence and mechanisms of this pathway remains to be determined.
RQC at the outer mitochondrial membrane
Mitochondrial function is especially sensitive to defects in RQC, particularly in conditions that promote accumulation of CAT-tailed nascent proteins [2,4,73]. Deleting Vms1 and Ltn1 in yeast causes growth defects under respiration-dependent conditions, in which nascent mitochondrial proteins form detergent-insoluble aggregates that sequester mitochondrial chaperones and disrupt mitochondrial quality control [2]. Interestingly, this defect is rescued by deleting or mutating Rqc2 to abolish CAT-tailing [2]. Similarly, in fly models, ribosome stalling and accumulation of CAT-tailed respiratory complex I-30 kDa protein (C-I30) disrupts ATP production [4].
RQC-like mechanisms on bacterial and mitochondrial ribosomes
Although bacteria do not have the ubiquitin-proteasome system, the Rqc2 homolog RqcH specifically adds C-terminal alanines to nascent proteins on 50S ribosomal subunits that signal for proteolysis [83]. Two recent cryo-EM studies clarified the mechanism of Ala-tailing by RqcH [99,100]. These studies revealed that RqcH-bound 50S ribosomal subunits containing a canonical P-site tRNA, but not A/P-tRNA, specifically engage the Hsp15 family protein RqcP, suggesting that RqcP may facilitate non-canonical tRNA translocation. Whether similar factors mediate CAT-tailing in eukaryotes remains to be determined. A RQC-like pathway also may rescue stalled mitochondrial ribosomes [101], in which the MTRES1 protein selectively recognizes large ribosomal subunit-peptidyl-tRNA complexes and engages mtRF-R to release nascent proteins.
Concluding Remarks
Over the last decade, RQC has been an exciting field full of fast-paced, elegant, and unexpected discoveries. Since the report of a ribosome-associated ubiquitin ligase [68], the field has gained clear molecular insights into how rescue factors specifically recognize and act on aberrantly stalled ribosomes. Findings in recent years particularly highlight the complexity and intricate interplay of rescue mechanisms, from the conserved role of mRNA-independent polypeptide elongation in nascent protein quality control, a mechanism that specifically scrutinizes tRNAs as they are recycled, and how multiple factors recognize ribosome collisions as a universal proxy of ribosome stalling to activate different responses. Future progress (Outstanding Questions) promises to reveal how ribosome rescue pathways deviate and collaborate with each other, how ribosome rescue mechanisms operate in different cellular contexts (Box 2) and cell types, and how these functions relate to neurodegenerative diseases.
Outstanding questions.
How prevalent are ribosome stalling sequences in the transcriptome of different cell types? What are the functions of endogenous ribosome stalling sequences such as SDD1 and how are these genes regulated?
How do defects in translational components such as tRNAs, ribosomal proteins, rRNA, and elongation factors affect translation? Are there translation-dependent quality control mechanisms that sense and degrade these components?
Beyond EDF1 and GCN1, how do other ribosome collision sensors selectively detect collided interfaces? What is the molecular basis for ZAKα and GCN2 activation?
What principles determine how different pathways are prioritized at collided ribosomal interfaces to activate the appropriate cellular responses?
Do additional factors aid Rqc2 in mRNA-independent polypeptide elongation on large ribosomal subunits in eukaryotes? Is the composition of Rqc2-generated tails regulated, and what factors recognize Rqc2-generated tails off the ribosome in different organelles and eukaryotes?
How does subcellular localization influence ribosome stalling and ribosome-associated quality control mechanisms?
How do defects in ribosome rescue lead to neurodegeneration?
Highlights.
Errors that occur during protein synthesis must be detected rapidly and either corrected or eliminated to maintain protein homeostasis.
Ribosomes may stall during protein synthesis due to local reasons such as a damaged mRNA, or due to general cellular stresses such as amino acid depletion. Understanding how differently-stalled ribosomes activate the appropriate cellular responses is an area of active investigation.
Ribosome collisions are a universal signature of ribosome stalling. Unique composite interfaces formed by collided ribosomes specifically recruit ribosome rescue factors that enact local quality control mechanisms, inhibit translation, and trigger cellular responses to mitigate proteotoxicity.
Individual components of stalled ribosome are scrutinized and triaged for degradation or recycling to ensure clearance of defective components.
Acknowledgements
We thank V. Chu and S. Sedor for critical reading and Shao lab members for discussions.
Glossary
- A-site
aminoacyl-tRNA binding site on the ribosome
- Coincidence detection
a process in which an output is achieved only by simultaneous detection of two signals that may be spatially separated. In the context of ribosome stalling on certain mRNA sequences such as poly(A) tracts, coincidence detection of perturbations in both the peptidyl-transferase center and the decoding center of a ribosome leads to stable stalling
- Decoding center
the site on the small ribosomal subunit where codon:anticodon pairing of mRNA and A-site tRNAs occur. 18S rRNA bases and ribosomal proteins at the decoding center make interactions to facilitate accurate decoding during translation
- Disome
a unit of two ribosomes, often used to refer to two collided ribosomes
- Integrated stress response (ISR)
a eukaryotic stress response characterized by the phosphorylation of the translation initiation factor eIF2α, resulting in the downregulation of general translation and upregulation of specific stress response genes. In yeast, the ISR is activated by GCN2. In mammals, four kinases including GCN2 may activate the ISR in response to a variety of cellular or environmental stresses, including nutrient depletion, ER stress, and viral infection
- Nonfunctional rRNA decay (NRD)
rapid degradation of rRNA on mature but defective ribosomes
- P-site
peptidyl-tRNA binding site on the ribosome
- P-stalk proteins
ribosomal proteins uL10, P1, and P2 comprising a pentameric structure on the large ribosomal subunit that interacts with translational GTPases that bind the A-site
- Peptidyl-transferase center (PTC)
site of peptide bond formation on the large ribosomal subunit
- Proteotoxicity
adverse cellular effects due to the accumulation of misfolded, damaged, or aggregated proteins
- Ribosome collision
when a translating ribosome runs into a stalled ribosome
- Ribosome-associated quality control (RQC)
a eukaryotic cellular pathway responsible for proteasomal degradation of incompletely-synthesized nascent proteins on stalled ribosomes. RQC is triggered by ribosomal subunit dissociation and selective recognition of large ribosomal subunit-peptidyl-tRNA complexes
- Ribosome-protected fragment (RPF)
mRNA fragments protected by ribosomes after nuclease digestion and whose sequences are identified in the process of ribosome profiling
- Ribotoxic stress response (RSR)
a cellular response in which ribosomal damage or inhibition activates mitogen- and stress-activated protein kinases that regulate inflammation and cell death
- RNA exosome
a conserved multisubunit complex that degrades RNA in the 3’–5’ direction
- RQC-triggering (RQT) complex
a protein complex comprising Slh1, Cue3, and YWR023W in yeast (ASCC3, ASCC2, and TRIP4/ASC-1 in humans) that selectively dissociates stalled ribosomes ubiquitinated by ZNF598/Hel2 into ribosomal subunits
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choe Y-J et al. (2016) Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 [DOI] [PubMed] [Google Scholar]
- 2.Izawa T. et al. (2017) Cytosolic Protein Vms1 Links Ribosome Quality Control to Mitochondrial and Cellular Homeostasis. Cell 171, 890–903.e18 [DOI] [PubMed] [Google Scholar]
- 3.Sitron CS et al. (2020) Aggregation of CAT tails blocks their degradation and causes proteotoxicity in S. cerevisiae. PLOS ONE 15, e0227841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z. et al. (2019) MISTERMINATE Mechanistically Links Mitochondrial Dysfunction with Proteostasis Failure. Mol. Cell 75, 835–848.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu J. et al. (2009) A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. U. S. A 106, 2097–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishimura R. et al. (2014) Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Defenouillère Q. et al. (2016) Rqc1 and Ltn1 Prevent C-terminal Alanine-Threonine Tail (CAT-tail)-induced Protein Aggregation by Efficient Recruitment of Cdc48 on Stalled 60S Subunits. J. Biol. Chem 291, 12245–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin PB et al. (2020) NEMF mutations that impair ribosome-associated quality control are associated with neuromuscular disease. Nat. Commun 11, 4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Udagawa T. et al. (2021) Failure to Degrade CAT-Tailed Proteins Disrupts Neuronal Morphogenesis and Cell Survival. Cell Rep. 34, 108599. [DOI] [PubMed] [Google Scholar]
- 10.Joazeiro CAP (2019) Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol 20, 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inada T. (2020) Quality controls induced by aberrant translation. Nucleic Acids Res. 48, 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandman O. and Hegde RS (2016) Ribosome-associated protein quality control. Nat. Struct. Mol. Biol 23, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoemaker CJ and Green R. (2012) Translation drives mRNA quality control. Nat. Struct. Mol. Biol 19, 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frischmeyer PA et al. (2002) An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 [DOI] [PubMed] [Google Scholar]
- 15.Ozsolak F. et al. (2010) Comprehensive Polyadenylation Site Maps in Yeast and Human Reveal Pervasive Alternative Polyadenylation. Cell 143, 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doma MK and Parker R. (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuo Y. et al. (2020) RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1. Nat. Struct. Mol. Biol 27, 323–332 [DOI] [PubMed] [Google Scholar]
- 18.Tesina P. et al. (2020) Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J. 39, e103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrasekaran V. et al. (2019) Mechanism of ribosome stalling during translation of a poly(A) tail. Nat. Struct. Mol. Biol 26, 1132–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J. and Deutsch C. (2008) Electrostatics in the Ribosomal Tunnel Modulate Chain Elongation Rates. J. Mol. Biol 384, 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arthur LL et al. (2015) Translational control by lysine-encoding A-rich sequences. Sci. Adv 1, e1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao S. et al. (2016) Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Cell 167, 1229–1240.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavlovic Djuranovic S. et al. (2020) Plasmodium falciparum translational machinery condones polyadenosine repeats. eLife 9, e57799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang TTL et al. (2019) The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases. Nat. Struct. Mol. Biol 26, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanmuganathan V. et al. (2019) Structural and mutational analysis of the ribosome-arresting human XBP1u. eLife 8, e46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Q. and Shao S. (2018) In vitro reconstitution of translational arrest pathways. Methods 137, 20–36 [DOI] [PubMed] [Google Scholar]
- 27.Wu CC-C et al. (2019) High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol. Cell 73, 959–970.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan LL et al. (2019) Oxidation and alkylation stresses activate ribosome-quality control. Nat. Commun 10, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoemaker CJ et al. (2010) Dom34:Hbs1 Promotes Subunit Dissociation and Peptidyl-tRNA Drop-Off to Initiate No-Go Decay. Science 330, 369–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker T. et al. (2011) Structure of the no-go mRNA decay complex Dom34–Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol 18, 715–720 [DOI] [PubMed] [Google Scholar]
- 31.Hilal T. et al. (2016) Structural insights into ribosomal rescue by Dom34 and Hbs1 at nearatomic resolution. Nat. Commun 7, 13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guydosh NR and Green R. (2014) Dom34 Rescues Ribosomes in 3′ Untranslated Regions. Cell 156, 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisareva VP et al. (2011) Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30, 1804–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuboi T. et al. (2012) Dom34:Hbs1 Plays a General Role in Quality-Control Systems by Dissociation of a Stalled Ribosome at the 3′ End of Aberrant mRNA. Mol. Cell 46, 518–529 [DOI] [PubMed] [Google Scholar]
- 35.Kowalinski E. et al. (2016) Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex. Mol. Cell 63, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terrey M. et al. (2020) GTPBP1 resolves paused ribosomes to maintain neuronal homeostasis. eLife 9, e62731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simms CL et al. (2017) Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell 68, 361–373.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juszkiewicz S. et al. (2018) ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol. Cell 72, 469–481.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu CC-C et al. (2020) Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 182, 404–416.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinha NK et al. (2020) EDF1 coordinates cellular responses to ribosome collisions. eLife 9, e58828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meydan S. and Guydosh NR (2020) Disome and Trisome Profiling Reveal Genome-wide Targets of Ribosome Quality Control. Mol. Cell 79, 588–602.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han P. et al. (2020) Genome-wide Survey of Ribosome Collision. Cell Rep. 31, 107610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao T. et al. (2021) Disome-seq reveals widespread ribosome collisions that promote cotranslational protein folding. Genome Biol. 22, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandman O. et al. (2012) A Ribosome-Bound Quality Control Complex Triggers Degradation of Nascent Peptides and Signals Translation Stress. Cell 151, 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juszkiewicz S. and Hegde RS (2017) Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol. Cell 65, 743–750.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundaramoorthy E. et al. (2017) ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol. Cell 65, 751–760.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garzia A. et al. (2017) The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun 8, 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuo Y. et al. (2017) Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D’Orazio KN et al. (2019) The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. eLife 8, e49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikeuchi K. et al. (2019) Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38, e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sitron CS et al. (2017) Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA 23, 798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto S. et al. (2020) Identification of a novel trigger complex that facilitates ribosome-associated quality control in mammalian cells. Sci. Rep 10, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juszkiewicz S. et al. (2020) The ASC-1 Complex Disassembles Collided Ribosomes. Mol. Cell 79, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Juszkiewicz S. et al. (2020) Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. eLife 9, e60038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hickey KL et al. (2020) GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control. Mol. Cell 79, 950–962.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pochopien AA et al. (2020) Structure of Gcn1 bound to stalled and colliding 80S ribosomes. bioRxiv DOI: 10.1101/2020.10.31.363135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morita M. et al. (2012) A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell. Biol 32, 3585–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber R. et al. (2020) 4EHP and GIGYF1/2 Mediate Translation-Coupled Messenger RNA Decay. Cell Rep. 33, 108262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouyang D-Y et al. (2005) Activation of c-Jun N-terminal Kinases by Ribotoxic Stresses. Mol. Immunol 2, 419–425 [PubMed] [Google Scholar]
- 60.Vind AC et al. (2020) ZAKα Recognizes Stalled Ribosomes through Partially Redundant Sensor Domains. Mol. Cell 78, 700–713.e7 [DOI] [PubMed] [Google Scholar]
- 61.Masson GR (2019) Towards a model of GCN2 activation. Biochem. Soc. Trans 47, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan LL and Zaher HS (2020) Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol. Cell 81, 614–628.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishimura R. et al. (2016) Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. eLife 5, e14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harding HP et al. (2019) The ribosomal P-stalk couples amino acid starvation to GCN2 activation in mammalian cells. eLife 8, e50149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inglis AJ et al. (2019) Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci 116, 4946–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen PS et al. (2015) Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shao S. et al. (2015) Structure and assembly pathway of the ribosome quality control complex. Mol. Cell 57, 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bengtson MH and Joazeiro CAP (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuroha K. et al. (2018) Release of Ubiquitinated and Non-ubiquitinated Nascent Chains from Stalled Mammalian Ribosomal Complexes by ANKZF1 and Ptrh1. Mol. Cell 72, 286–302.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verma R. et al. (2013) Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife 2, e00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verma R. et al. (2018) Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature 557, 446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rendón OZ et al. (2018) Vms1p is a release factor for the ribosome-associated quality control complex. Nat. Commun 9, 2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su T. et al. (2019) Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis. Nature 570, 538–542 [DOI] [PubMed] [Google Scholar]
- 74.Heo J-M et al. (2010) A Stress-Responsive System for Mitochondrial Protein Degradation. Mol. Cell 40, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yip MCJ et al. (2019) Mechanism for recycling tRNAs on stalled ribosomes. Nat. Struct. Mol. Biol 26, 343–349 [DOI] [PubMed] [Google Scholar]
- 76.Yip MCJ et al. (2020) ELAC1 Repairs tRNAs Cleaved during Ribosome-Associated Quality Control. Cell Rep. 30, 2106–2114.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pinto PH et al. (2020) ANGEL2 is a member of the CCR4 family of deadenylases with 2′,3′-cyclic phosphatase activity. Science 369, 524–530 [DOI] [PubMed] [Google Scholar]
- 78.Wilusz JE et al. (2011) tRNAs Marked with CCACCA Are Targeted for Degradation. Science 334, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.LaRiviere FJ et al. (2006) A Late-Acting Quality Control Process for Mature Eukaryotic rRNAs. Mol. Cell 24, 619–626 [DOI] [PubMed] [Google Scholar]
- 80.Cole SE et al. (2009) A Convergence of rRNA and mRNA Quality Control Pathways Revealed by Mechanistic Analysis of Nonfunctional rRNA Decay. Mol. Cell 34, 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Limoncelli KA et al. (2017) ASC1 and RPS3: New actors in 18S non-functional rRNA decay. RNA DOI: 10.1261/rna.061671.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sugiyama T. et al. (2019) Sequential Ubiquitination of Ribosomal Protein uS3 Triggers the Degradation of Non-functional 18S rRNA. Cell Rep. 26, 3400–3415.e7 [DOI] [PubMed] [Google Scholar]
- 83.Lytvynenko I. et al. (2019) Alanine Tails Signal Proteolysis in Bacterial Ribosome-Associated Quality Control. Cell 178, 76–90.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kostova KK et al. (2017) CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357, 414–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Osuna BA et al. (2017) In vitro analysis of RQC activities provides insights into the mechanism and function of CAT tailing. eLife 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sitron CS and Brandman O. (2019) CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat. Struct. Mol. Biol 26, 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yonashiro R. et al. (2016) The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife 5, e11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Hoof A. et al. (2002) Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 [DOI] [PubMed] [Google Scholar]
- 89.Glover ML et al. (2020) NONU-1 Encodes a Conserved Endonuclease Required for mRNA Translation Surveillance. Cell Rep. 30, 4321–4331.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buschauer R. et al. (2020) The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 368, eaay6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Izawa T. et al. (2012) Roles of dom34:hbs1 in nonstop protein clearance from translocators for normal organelle protein influx. Cell Rep. 2, 447–453 [DOI] [PubMed] [Google Scholar]
- 92.Crowder JJ et al. (2015) Rkr1/Ltn1 Ubiquitin Ligase-mediated Degradation of Translationally Stalled Endoplasmic Reticulum Proteins. J. Biol. Chem 290, 18454–18466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arakawa S. et al. (2016) Quality control of nonstop membrane proteins at the ER membrane and in the cytosol. Sci. Rep 6, 30795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.von der Malsburg K. et al. (2015) The ribosome quality control pathway can access nascent polypeptides stalled at the Sec61 translocon. Mol. Biol. Cell 26, 2168–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lakshminarayan R. et al. (2020) Pre-emptive Quality Control of a Misfolded Membrane Protein by Ribosome-Driven Effects. Curr. Biol 30, 854–864.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trentini DB et al. (2020) Role for ribosome-associated quality control in sampling proteins for MHC class I-mediated antigen presentation. Proc. Natl. Acad. Sci 117, 4099–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang L. et al. (2020) UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 30, 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walczak CP et al. (2019) Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl. Acad. Sci 116, 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crowe-McAuliffe C. et al. (2020) Structural Basis for Bacterial Ribosome-Associated Quality Control by RqcH and RqcP. Mol. Cell 81, 115–126.e7 [DOI] [PubMed] [Google Scholar]
- 100.Filbeck S. et al. (2020) Mimicry of Canonical Translation Elongation Underlies Alanine Tail Synthesis in RQC. Mol. Cell 81, 104–114.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Desai N. et al. (2020) Elongational stalling activates mitoribosome-associated quality control. Science 370, 1105–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]