Abstract
The mechanisms that promote brain dysfunction after acute liver failure (ALF) are not clearly understood. The small noncoding RNAs known as microRNAs (miRNAs) significantly control mRNA translation and thus normal and pathological functions in the mammalian body. To understand their significance in ALF, we currently profiled the expression of miRNAs in the cerebral cortex of mice sacrificed at coma stage following treatment with azoxymethane. Of the 470 miRNAs profiled using microarrays, 37 were significantly altered (20 up- and 17 down-regulated) in their expression in the ALF group compared to sham group. In silico analysis showed that the ALF-responsive miRNAs target on average 231 mRNAs/miRNA (range: 3 to 840 targets). Pathways analysis showed that many miRNAs altered after ALF target multiple mRNAs that are part of various biological and molecular pathways. Glutamatergic synapse, Wnt signaling, MAP-kinase signaling, axon guidance, PI3-kinase-AKT signaling, T-cell receptor signaling and ubiquitin-mediated proteolysis are the top pathways targeted by the ALF-sensitive miRNAs. At least 28 ALF-responsive miRNAs target each of the above pathways. We hypothesize that alterations in miRNAs and their down-stream mRNAs of signaling pathways might play a role in the induction and progression of neurological dysfunction observed during ALF.
Keywords: Liver disease, miRNA, Non-coding RNA, Hepatic encephalopathy, Neuroprotection, Molecular signaling
INTRODUCTION
Acute liver failure (ALF; also known as fulminant hepatic failure) resulting from liver toxicity or viral infection is associated with a high mortality rate. A major cause of death after ALF is a rapid progressing brain edema leading to increased intracranial hypertension and brainstem herniation (Jalan, 2003; Jayakumar et al., 2014). The edema and the ensuing hepatic encephalopathy (HE) are thus the serious neurological complications of ALF (Hazell and Butterworth, 1999; Scott et al., 2013) requiring emergency medical intervention, typically liver transplantation (Cordoba and Blei, 1996).
Despite significant progress over the years in understanding the functional pathways that are affected in ALF, the precise cause of HE and brain edema in ALF remain largely unknown. Studies indicate that the pathophysiology of HE involves a primary gliopathy in which astrocytes and microglia are targeted as a result of the associated hyperammonemia (Jiang et al., 2009; Rao et al., 2013). Recent studies also suggest that extracerebral mechanisms including a systemic response involving pro-inflammatory cytokines are a driving force in the evolving maladies (Bernal and Wendon, 2013; Tranah et al., 2013). Nevertheless, it is also evident from neuropathological studies that astrocytic swelling is the major hallmark of ALF.
MicroRNAs (miRNAs) are a group of small noncoding, evolutionarily conserved RNAs that are known to contribute in a major way to the control of thousands of downstream genes (Goodall et al., 2013; Vemuganti, 2013). Although their mechanism of action is not completely understood, miRNAs are thought to modulate translation by targeting the 3’UTRs of mRNAs and transcription by binding to consensus seed sequences in the promoter regions of DNA (Truesdell et al., 2012; Yan and Ma, 2012; Vemuganti, 2013). Many studies also showed that alterations in the levels of miRNAs are linked to brain damage in chronic and acute conditions including Alzheimer disease, schizophrenia, stroke, spinal cord injury and traumatic brain injury (please see the recent reviews (Goodall et al., 2013; Vemuganti, 2013)).
In order to better understand the cerebral regulatory mechanisms associated with alterations in gene expression in HE, we have profiled the expression levels of 470 miRNAs in the cerebral cortex of the azoxymethane-treated mouse, a well-established model of ALF (Matkowskyj et al., 1999; Belanger et al., 2006). Using microT-CDS and miRPath web tools of the DNA Intelligence Analysis (DIANA) algorithm (Papadopoulos et al., 2009), we identified the down-stream targets and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways common to several miRNAs altered after ALF that might play a role in mediating the associated neurological dysfunction.
MATERIALS AND METHODS
ALF induction
Adult male C57/BL6 mice (25–30g) were bred at Centro Multidisciplinar para Investigação Biológica na Área da Ciência de Animais de Laboratório (CEMIB), State University of Campinas (Universidade Estadual de Campinas, UNICAMP), and were handled in accordance with Brazilian animal welfare guidelines using protocols approved by CEMIB. Mice were treated with 100 μg/g azoxymethane (Sigma-Aldrich, Oakville, ON, Canada) dissolved in saline as previously described (Matkowskyj et al., 1999). Control animals received an equivalent volume of saline. All mice were carefully monitored throughout involving administration of glucose where necessary to maintain normoglycemia and the use of heating pads to control body temperature at 37°C. Mice were euthanized at the onset of coma stage in ALF mice (defined as loss of consciousness accompanied by a loss of corneal reflex), and the frontal cortex dissected, flash-frozen in 2-methylbutane maintained at −30°C over dry ice and stored at −80°C prior to study.
MicroRNA profiling
MicroRNA profiling was conducted as described previously (Dharap et al., 2009). In brief, total RNA was extracted from the frontal cortex using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) as per the manufacturer’s protocol. From each sample, 5 μg total RNA was size fractionated on a centrifugal filter (YM-100 Microcon; Millipore USA). To the small RNAs (<300 nt), poly-A tails were added at the 3’end mediated by poly(A) polymerase, and the nucleotide tags were ligated to the poly-A tails. Each sample was hybridized to a microarray (LC Sciences, Houston, TX) that contained 470 known miRNA probes (12 repeats of each probe) from the Sanger miRBase version 20 (http://microrna.sanger.ac.uk/sequences/). The miRNA hybridization data was corrected by subtracting the background (calculated from the median of 5% to 25% of the lowest-intensity cells) and normalized to the statistical median of all detectable transcripts using the locally-weighted regression (LOWESS) method which balances the intensities of Cy5 labeled transcripts so that the differential expression ratios can be properly calculated (Bolstad et al., 2003). For subtracting, the background was defined on each array as the average signal of the BKG0 spots (chemical linkers without the probes). The hybridization intensities above exp(5) (~150) were considered as significant as described earlier (Vagin et al., 2006) and intensities below 30,000 were considered as non-saturated, as established with titration of several synthetic 20-nt RNA oligos (external controls) spiked into each sample. In addition, on each array there were 16 sets of spatially distributed internal control probes. These include PUC2PM-20B and PUC2MM-20B which are the perfect match and the single-base mismatch sequences, respectively. The stringency of the intensity ratio of the PUC2PM-20B and PUC2MM-20B is expected to be larger than 30 indicating proper hybridization in each case. For proper analysis of signal intensities on each chip, both the internal controls and the test miRNA probes were repeated 12 times. On a microarray, the hybridization signal was linearly obtained from 1 to ~66,000 units. A miRNA transcript was considered detectable if it met the following criteria. (a) Signal intensity higher than 3 times the maximal background signal, (b) spot CV <0.5 (CV was calculated as (standard deviation)/(signal intensity)) and (c) the signals from at least 50% of the 12 redundant repeating probes above detection level. To avoid false positives, any spot that deviated >50% from the average value of the 12 repeating spots and/or spots with CV >0.5 were eliminated. The data from different groups was normalized and analyzed statistically using ANOVA. To increase the validity of the data, we generated cross-comparison matrices. For example, the data from the 3 vehicle treated mice was cross-compared with the 3 azoxymethane treated mice to generate a matrix of 9 comparisons.
Bioinformatics
The number of mRNA targets for each miRNA were predicted with the algorithm microT-CDS from DIANA tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index) (Betel et al., 2008) by computing the optimal sequence complementarity between mature miRNAs and mRNAs using a weighted dynamic programming algorithm. For searching the targets of the ALF-sensitive miRNAs, we used very high stringency parameters of p<0.001 and a microT-CDS threshold of 0.8. The miRNA sequences and the 3’UTR sequences of mRNAs used were from Sanger mirBASE database and from the NCBI database. For predicting the KEGG pathways of mRNAs that are targeted by ALF-altered miRNAs, we used DIANA miRPath algorithm combined with DIANA microT 4.0 (Lewis et al., 2005; Papadopoulos et al., 2009).
RESULTS
Physiological changes
Treatment with AOM led to all animals developing hypoglycemia and mild hypothermia that was controlled with careful monitoring. Development of the coma stage in these animals occurred 17.6 ± 0.5 h (mean ± SEM) following drug administration, involving a progression from onset of ataxia to loss of consciousness and then absence of corneal reflex.
Effect of ALF on miRNA expression
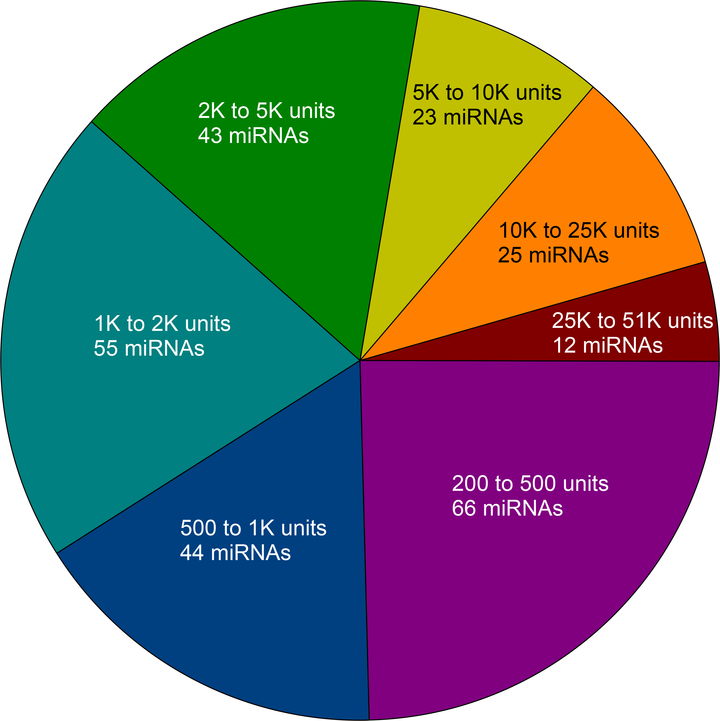
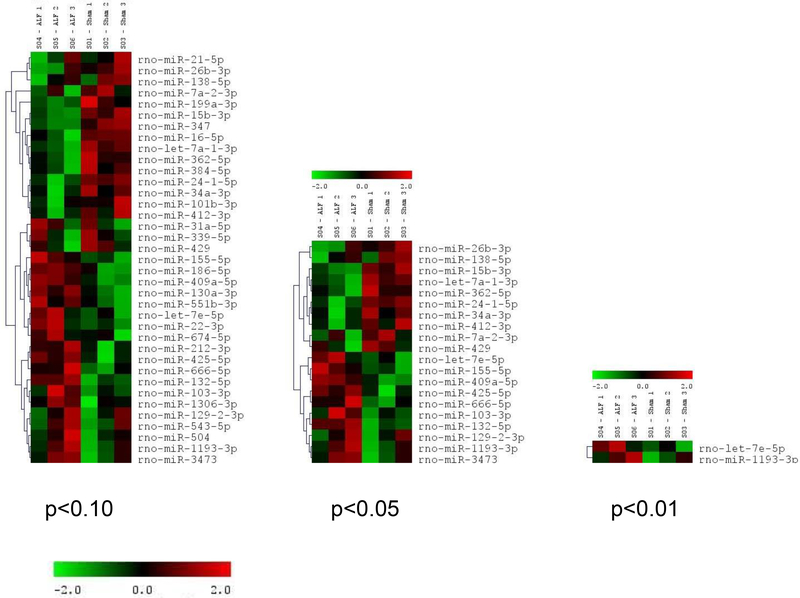
Of the 470 miRNAs analyzed, 268 (57%) were expressed (>200 units on a scale of 1 to 51,000) in the cerebral cortex of sham-operated mice (n=3). Of these, 12 were expressed at a very high level (>25,000 units), 25 at a high level (10,000 to 25,000 units), 23 at a medium to a high level (5,000 to 10,000 units), 43 at a medium level (2,000 to 5,000 units), 55 at a low to a medium level (1,000 to 2,000), 44 at a low level (500 to 1,000 units) and 66 at a very low level (200 to 500 units) (Fig. 1). To evaluate the effect of ALF, the miRNA expression profiles were analyzed when the mice were in coma at 18h after the administration of azoxymethane. In the cerebral cortex of the mice in ALF, 20 miRNAs showed a statistically significant increased expression and 17 miRNAs showed a statistically significant decreased expression compared to sham (Fig. 2; n =3/group). By a very high-stringency in silico analysis using microT-CDS web tool of the DIANA algorithm, the ALF-responsive miRNAs were observed to target 231 mRNAs/miRNA on average with a range 3 targets (miR-132–5p) to 840 targets (miR-186–5p) (Table 1).
Fig. 1:
The pie-chart shows the relative expression levels of miRNAs in the normal rat cerebral cortex (each value is a mean of n = 3). Of the 470 miRNAs analyzed, 268 (57%) were observed to be expressed at >200 units on a scale of 1 to 51,000 units.
Fig. 2:
Hierarchical clustering of the miRNAs altered after ALF in comparison to sham controls (n=3 microarrays/group). The figure shows the miRNAs altered in the ALF group at 3 different statistically significant levels (p<0.01, p<0.05 and p<0.10). In the ALF group 37 miRNAs were observed to be significantly altered compared to sham (20 miRNAs were up-regulated and 17 miRNAs were down-regulated). The color code in the heat maps is linear with green as the lowest and red as the highest. The miRNAs that were increased in expression were shown in green to red while the miRNAs that were decreased in expression were shown from red to green. The individual expression signal of each miRNA in each array was clustered using Euclidean distance function. The dendrograms (tree diagrams) show the grouping of miRNAs according to the order in which they were joined during the clustering. The miRNAs with most similar expression patters were placed next to each other.
Table 1:
The miRNAs altered after ALF target several mRNAs that participate in biologically critical functional pathways
| miRNA | # of targets | # of associated pathwaysa |
|---|---|---|
|
| ||
| Up-regulated | ||
| miR-186-5p | 840 | 14 |
| miR-155-5p | 291 | 10 |
| let-7e-5p | 481 | 9 |
| miR-31a-5p | 180 | 8 |
| miR-1193-3p | 116 | 3 |
| miR-3473 | 270 | 7 |
| miR-130a-3p | 358 | 5 |
| miR-425-5p | 153 | 4 |
| miR-666-5p | 6 | 3 |
| miR-543-5p | 63 | 3 |
| miR-504 | 79 | 2 |
| miR-103-3p | 258 | 1 |
| miR-551b-3p | 5 | 1 |
| miR-674-5p | 257 | 1 |
| miR-212-3p | 389 | 1 |
| miR-22-3p | 253 | 0 |
| miR-129-2-3p | 126 | 0 |
| miR-409a-5p | 33 | 0 |
| miR-1306-3p | 18 | 0 |
| miR-132-5p | 3 | 0 |
| Down-regulated | ||
| let-7a-1-3p | 468 | 18 |
| miR-16-5p | 533 | 14 |
| miR-384-5p | 881 | 13 |
| miR-138-5p | 240 | 7 |
| miR-101b-3p | 598 | 6 |
| miR-339-5p | 190 | 5 |
| miR-199a-3p | 206 | 5 |
| miR-7a-2-3p | 267 | 4 |
| miR-21-5p | 153 | 2 |
| miR-429 | 68 | 2 |
| miR-362-5p | 84 | 2 |
| miR-15b-3p | 50 | 2 |
| miR-34a-3p | 145 | 1 |
| miR-412-3p | 144 | 1 |
| miR-24-1-5p | 50 | 0 |
| miR-26b-3p | 83 | 0 |
| miR-347b | ** | ** |
The # of targets of miRNAs and the associated pathways were identified by in silico analysis using the algorithms microT-CDS and mirPath from DIANA tools. The parameters for the target identification were set at a very high-stringency of p<0.001 and a microT-CDS threshold of 0.8.
We considered only the top pathways that showed highest number of associated genes targeted by the miRNAs altered after ALF.
The algorithm didn’t predict any targets or pathways for this miRNA.
Convergent functional pathways downstream to the miRNAs altered after ALF
The molecular and functional pathways rather than the individual proteins are the more reliable controllers of the physiological and pathological outcomes. Furthermore, the effects of miRNAs can be combinatorial as the 3’UTRs of most mRNAs contain binding sites for multiple miRNAs and many miRNA can target several mRNAs; thus proteins of a pathway can be influenced at various steps by miRNAs altered after ALF. Hence, we combined the targets predicted by the microT-CDS tool with the mirPath tool of the DIANA algorithm to identify the biological pathways downstream to miRNAs altered after ALF. The targets of 29 of the ALF-responsive miRNAs are observed to be members of multiple biological pathways (Table 1). Thirteen of these miRNAs targeted mRNAs in 5 or more biological pathways each (Table 1). The major biological functional categories targeted by miRNAs altered after ALF are glutamatergic synapse, Wnt signaling, MAP-kinase signaling, axon guidance, PI3-kinase-AKT signaling, T-cell receptor signaling and ubiquitin-mediated proteolysis. The mRNAs of each of these pathways are targeted by at least 28 of the miRNAs altered after ALF (Table 2). Surprisingly, 12 of the 36 pathways targeted by ALF-sensitive miRNAs are associated with molecular signaling (Table 2). We observed that many mRNAs that are members of various biological pathways are targeted by multiple ALF-responsive miRNAs (Table 3). For example, glycogen synthase kinase-beta (GSK3β) that participates in at least 10 signaling pathways was observed to be targeted by 5 miRNAs (Table 3). Each of the growth factors brain derived neurotrophic factor (BDNF) and insulin-like growth factor (IGF) were observed to be targeted by 7 miRNAs altered after ALF (Table 3). Furthermore, nuclear factor of activated T-cells (Nfat5) that is a major player of T cell receptor signaling, and monocyte enhancer factor 2c (Mef2c) that participates in MAPK signaling are observed to be targeted by 9 and 8 ALF-responsive miRNAs, respectively (Table 3).
Table 2:
Pathways targeted by miRNAs altered after ALF
| KEGG pathway | p-value | #genes | #miRNAs |
|---|---|---|---|
|
| |||
| Glutamatergic synapse | 2.2E-05 | 39 | 31 |
| Wnt signaling pathway | 1.3E-23 | 69 | 30 |
| MAPK signaling pathway | 3.0E-14 | 90 | 30 |
| Axon guidance | 9.0E-63 | 76 | 29 |
| PI3K-Akt signaling pathway | 1.6E-24 | 124 | 28 |
| T cell receptor signaling pathway | 1.5E-20 | 49 | 28 |
| Ubiquitin mediated proteolysis | 5.5E-13 | 55 | 28 |
| Tight junction | 5.2E-06 | 48 | 28 |
| Regulation of actin cytoskeleton | 5.3E-22 | 86 | 27 |
| Protein processing in endoplasmic reticulum | 3.5E-04 | 55 | 27 |
| VEGF signaling pathway | 5.4E-24 | 34 | 26 |
| Chemokine signaling pathway | 1.4E-07 | 62 | 26 |
| ErbB signaling pathway | 2.0E-36 | 49 | 25 |
| Neurotrophin signaling pathway | 6.9E-17 | 52 | 25 |
| Cholinergic synapse | 4.4E-08 | 44 | 25 |
| Dopaminergic synapse | 8.4E-08 | 47 | 25 |
| Jak-STAT signaling pathway | 6.4E-05 | 47 | 25 |
| Focal adhesion | 1.6E-25 | 82 | 25 |
| mTOR signaling pathway | 3.2E-22 | 33 | 24 |
| Fcγ receptor-mediated phagocytosis | 2.3E-22 | 43 | 23 |
| Long-term depression | 2.9E-05 | 25 | 22 |
| TGF-beta signaling pathway | 6.3E-22 | 43 | 20 |
| Hedgehog signaling pathway | 2.8E-19 | 27 | 20 |
| Apoptosis | 1.0E-04 | 30 | 20 |
| Circadian rhythm | 5.0E-12 | 17 | 18 |
| Synaptic vesicle cycle | 9.9E-04 | 22 | 16 |
The table shows only pathways that contain at least 15 mRNAs targeted by the miRNA altered after ALF. The pathways were determined by bioinformatics analysis using the algorithm mirPath from DIANA tools. The algorithm combines the gene targets of the selected miRNAs into a superset (union). The resulting set is then incorporated in the enrichment analysis and combined the previously calculated significance levels to provide a merged P-value for each pathway, by applying Fisher’s combined probability method. KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, Mitogen activated protein kinase; PI3-K, phosphoinositide 3-kinase; AKT, protein kinase-B; VEGF, vascular endothelial growth factor; Jak-STAT, Janus kinase-Signal transducer and activator of transcription; mTOR, mammalian target of rapamycin; TGF, transforming growth factor.
Table 3:
Many mRNAs associated in the pathways are targeted by multiple miRNAs altered after ALF
| Gene | # of miRNAs | miRNAs | Pathway(s) | |
|---|---|---|---|---|
| Up-regulated | Down-regulated | |||
| Gsk3β | 5 | miR-3473; miR-155-5p; miR-186-5p | let-7a-1-3p; miR-101b-3p | Wnt signaling pathway Axon guidance PI3K-Akt signaling pathway T cell receptor signaling pathway Chemokine signaling pathway ErbB signaling pathway Neurotrophin signaling pathway Dopaminergic synapse Focal adhesion Hedgehog signaling pathway |
| Sos1 | 5 | miR-155-5p; miR-130a-3p; miR-212-3p | miR-384-5p; miR-339-5p | MAPK signaling pathway PI3K-Akt signaling pathway T cell receptor signaling pathway Regulation of actin cytoskeleton Chemokine signaling pathway ErbB signaling pathway Neurotrophin signaling pathway Jak-STAT signaling pathway Focal adhesion |
| Acvr1c | 6 | let-7e-5p; miR-425-5p; miR-674-5p; | miR-7a-2-3p; miR-101b-3p; miR-199a-3p | TGF-beta signaling pathway |
| Bdnf | 7 | miR-425-5p; miR-155-5p; miR-543-5p; | miR-7a-2-3p; let-7a-1-3p; miR-384-5p; miR-16-5p | MAPK signaling pathway Neurotrophin signaling pathway |
| Cask | 6 | miR-1193-3p; miR-425-5p; miR-674-5p; miR-186-5p; miR-543-5p | miR-16-5p | Tight junction |
| Cul2 | 6 | miR-103-1-5p; miR-31-5p | miR-412-3p; miR-384-5p; miR-101b-3p; miR-16-5p | Ubiquitin mediated proteolysis ErbB signaling pathway |
| Igf1 | 7 | let-7e-5p; miR-425-5p; miR-3473; miR-155-5p; miR-186-5p | miR-16-5p; miR-199a-3p | mTOR signaling pathway |
| Mapk8 | 6 | let-7e-5p | miR-34a-3p; miR-384-5p; miR-101b-3p; miR-16-5p; miR-199a-3p | Wnt signaling pathway MAPK signaling pathway Protein processing in endoplasmic reticulum ErbB signaling pathway Neurotrophin signaling pathway Dopaminergic synapse Focal adhesion |
| Mef2c | 8 | let-7e-5p; miR-155-5p; miR-674-5p; miR-31-5p; miR-186-5p; miR-212-3p | let-7a-1-3p; miR-21-5p | MAPK signaling pathway |
| Nfat5 | 9 | let-7e-5p; miR-22-3p; miR-674-5p; miR-186-5p; miR-212-3p; miR-543-5p | miR-7a-2-3p; let-7a-1-3p; miR-384-5p | Axon guidance T cell receptor signaling pathway VEGF signaling pathway |
| Wasl | 6 | let-7e-5p; miR-130a-3p; miR-186-5p | let-7a-1-3p; miR-412-3p; miR-16-5p | Regulation of actin cytoskeleton Chemokine signaling pathway Fc gamma R-mediated phagocytosis |
For this analysis, we used a stringency p-value of 0.001 and the microT threshold of 0.8 in microT-CDS and Diana mirPath algorithms. We presented only the genes that are targeted by at least 5 ALF-sensitive miRNAs. Acvr1c, activin A receptor, type IC; Bdnf, brain derived neurotrophic factor; Cask, calcium/calmodulin-dependent serine protein kinase (MAGUK family); Cul2, cullin 2; Gsk3β, glycogen synthase kinase 3 beta; Igf1, insulin-like growth factor 1; Mapk8, mitogen-activated protein kinase 8; Mef2c, myocyte enhancer factor 2C; Nfat5, nuclear factor of activated T cells 5; Sos1, son of sevenless homolog 1 (Drosophila); Wasl, Wiskott-Aldrich syndrome-like (human).
DISCUSSION
This is the first study showing that expression of several miRNAs change in brain during the development of ALF and many of the altered miRNAs target mRNAs that constitute pathways known to control cellular and molecular mechanisms that are essential for homeostatic brain function. While the pathological and psychiatric consequences of ALF/HE are well-documented (Hazell and Butterworth, 1999), the molecular mechanisms that underlie the brain dysfunction are unclear.
The major function attributed to miRNAs is to control the translation of mRNAs by binding to the seed sequences in their 3’UTRs (Bartel, 2009). As the seed sequences are only 5 to 8 nucleotides long, miRNAs function in a redundant manner either sequentially or simultaneously. Most miRNAs have consensus binding sites in the 3’UTRs of dozens of mRNA. Furthermore, many mRNA 3’UTRs contain binding sites for multiple miRNAs. Hence, to understand the magnitude of the effects of sets of miRNAs altered in a pathological condition, it is important to identify the pathways they target. In the present study, glutamatergic synapse is the KEGG pathway that was observed to be targeted by 31 miRNAs altered after ALF. This indicates that miRNAs might be responsible for the previously reported glutamatergic synaptic dysfunction in HE in which extracellular glutamate levels are increased concomitant with a loss of GLT-1 (Knecht et al., 1997; Michalak et al., 1997), which is the predominant glutamate transporter in cerebral cortex. Although the precise functional significance of the glutamatergic system in ALF is unclear, impaired tricarboxylic acid (TCA) cycle function may be linked to this. Early studies on brain mitochondria indicated that ammonia at high levels inhibits the TCA cycle enzymes including the rate-limiting α-ketoglutarate dehydrogenase (α-KGDH) (McKhann and Tower, 1961; Lai and Cooper, 1986). This has potentially important implications since impaired energy metabolism due to α-KGDH inhibition can compromise glutamate transport, which is dependent on maintenance of an intact Na+-K+ electrochemical gradient across the astrocyte cell membrane. Thus, reduced energy status due to hyperammonemia after ALF may be a contributing factor to the changes in miRNAs associated with the glutamatergic synapse as well as other biological pathways observed in the present study. Several ALF-responsive miRNAs also target cholinergic synapse, dopaminergic synapse and synaptic vesicle release KEGG pathways indicating that the pathological changes associated with HE might also be due to failure and/or dysfunction of other neurotransmitter systems in addition to glutamatergic system. Interestingly, tight junctions also provide a major target of a number of ALF-responsive miRNAs. While the exact mechanism remains to be evaluated, the hyperammonemia associated with ALF is known to alter blood-brain barrier (BBB) permeability (Chen et al., 2013; Cui et al., 2013; Scott et al., 2013). Thus, it is conceivable that miRNAs might influence the mRNAs/proteins that control tight junctional integrity and as a result contribute to altered BBB permeability and potentially astrocytic swelling, a major feature of ALF.
Humans and animals developing HE are also known to show altered circadian rhythm (Campbell et al., 1979; Cordoba et al., 1998; Montagnese et al., 2014) and this is one of the top KEGG pathways observed to be targeted by 18 of the miRNAs altered after ALF. A striking observation of the present study is that miRNAs altered in ALF target in different permutations and combinations multiple signaling pathways including MAP-kinase signaling, PI3-kinase signaling, VEGF signaling, ErbB signaling, JAK-STAT signaling, TGF-β signaling, Hedgehog signaling and mTOR signaling. This disrupted second messenger and growth factor signaling mediated by miRNAs might be in part responsible for the neuropsychiatric squeal observed in HE patients (Hazell and Butterworth, 1999; Patel et al., 2012). Both glial cells and neurons express Wnt receptors and Wnt signaling is known to minimize apoptosis by reducing cytochrome C release from mitochondria and by preventing β-catenin/Tcf transcriptional pathway (Chen et al., 2001). Hence, our observation that 30 of the ALF-sensitive miRNAs target 69 mRNAs of Wnt signaling pathway and apoptosis pathway might be adaptations to increase CNS cell survival during the acute stage of liver failure. Indeed, a significant feature of ALF is the absence of neuronal cell death, despite the presence of profound astrocytic swelling. Of importance, MAP-kinase signaling is a major pathway and many of its members regulate second messenger signal transduction, metabolism and gene expression (Johnson and Lapadat, 2002). Altered levels of 30 miRNAs that target 90 mRNAs of MAP-kinase pathway observed presently might be an adaptation to minimize the cellular stress that potentially disrupts the physiological kinase activity leading to deregulated cell signaling. Interestingly, recent studies have demonstrated that ammonia-induced oxidative stress, which is a major detrimental process occuring in ALF (Detry et al., 2005; Sathyasaikumar et al., 2007; Skowronska and Albrecht, 2013), activate MAP-kinases and inhibition of MAP-kinase phosphorylation reduces astrocyte swelling. Thus, altered MAP-kinase activation by these miRNAs may have an important role to play in determining cell volume regulation after ALF.
The 2 mRNAs targeted maximally by miRNAs altered after ALF are Nfat5 and Mef2c, and these are important modulators of inflammation. Furthermore, inflammation is known to be a major pathological consequence of HE (Jiang et al., 2009; Bemeur and Butterworth, 2013; Tranah et al., 2013), and we observed that ALF-responsive miRNAs target many inflammation-related pathways including T-cell receptor signaling, Fcγ receptor-mediated phagocytosis, focal adhesion and chemokine signaling. As mentioned above, ALF is known to promote energy stress and our pathways analysis shows that the ALF-responsive multiple miRNAs target the highly energy-intensive mTOR pathway as well as the ubiquitin-mediated proteolysis pathway that plays a major role in clearing the unfolded/misfolded proteins (Vadysirisack and Ellisen, 2012; Caldeira et al., 2014). Hence, the miRNAs altered after ALF might be involved in activating energy-conservation cascades and to prevent endoplasmic reticulum oxidative stress.
One interesting consequence of exposure to AOM is significant disruption of the BBB (Jayakumar et al., 2013). Although previous studies have not reported major structural disruption of the BBB in cases of human ALF (Kato et al., 1992) or in various models of the disorder (Traber et al., 1987; Gove et al., 1997), several groups have reported evidence of mild or subtle opening of the barrier in which increased endothelial vesicular profiles consistent with transendothelial transport are apparent (Potvin et al., 1984;.Kato et al., 1992). Thus, while increased BBB permeability is a feature of AOM treatment, a similar effect is also present in ALF in general although on a smaller scale. In addition, to our knowledge there is no reported greater effect of AOM in terms of brain edema, a major pathological hallmark of ALF than in other well-established models of the disorder. Together, this suggests that AOM-induced BBB disruption is not a likely significant contributor to the pathophysiology of ALF-induced HE. Rather, it may be perhaps more of a passive consequence of other major changes occurring in brain. Further studies are required to examine this possibility.
In conclusion, the present study shows that HE-mediated brain dysfunction in ALF has a molecular basis and might be controlled by miRNAs. However, further studies are needed to decipher the precise functional significance of the above pathways and their members in the cerebral pathophysiology associated with this disorder.
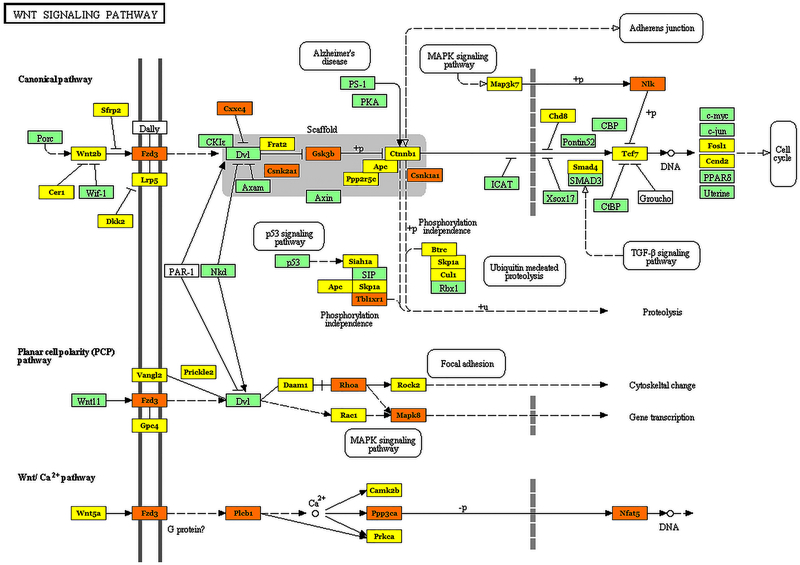
Fig. 3:
Effect of ALF-responsive miRNAs on members of the Wnt signaling pathway. The genes shown in orange are targeted by multiple miRNAs, while the genes shown in yellow are targeted by at least one miRNA and the genes shown in green are not targeted by miRNAs altered after ALF.
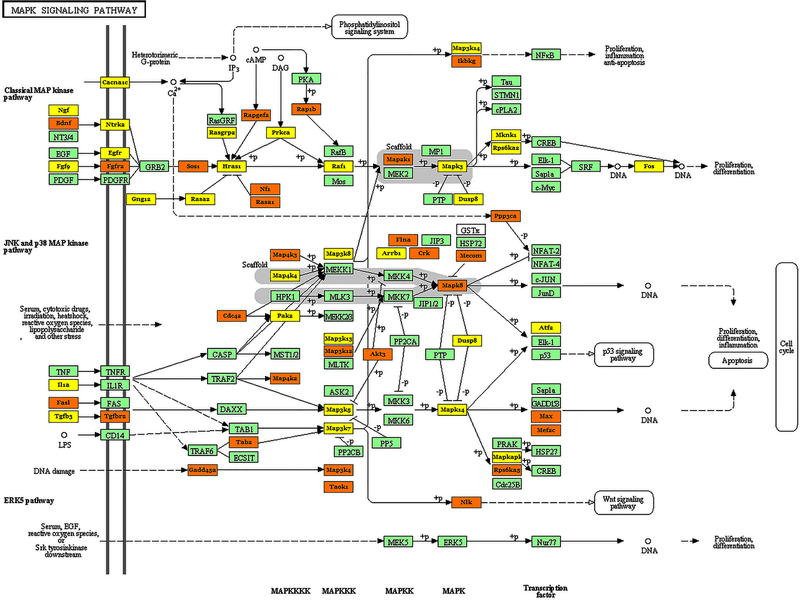
Fig. 4:
Effect of ALF-responsive miRNAs on members of the MAP-kinase signaling pathway. The genes shown in orange are targeted by multiple miRNAs, while the genes shown in yellow are targeted by at least one miRNA and the genes shown in green are not targeted by miRNAs altered after ALF.
Acknowledgements
This study was partially supported by a grant (2011/02263-3) to ASH from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). RV is partially supported by NIH grants NS074444, NS079585, NS 082957 and NS083007 and the Department of Neurological Surgery, University of Wisconsin Medical School. VRS is the recipient of a doctoral scholarship from FAPESP (2012/15405-3). ASH is a Visiting Professor in the Department of Neurology at Universidade Estadual de Campinas (UNICAMP), São Paulo, Brazil. His laboratory at the University of Montreal is supported by the Canadian Institutes of Health Research.
REFERENCES
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger M, Cote J, Butterworth RF (2006) Neurobiological characterization of an azoxymethane mouse model of acute liver failure. Neurochem Int 48:434–440. [DOI] [PubMed] [Google Scholar]
- Bemeur C, Butterworth RF (2013) Liver-brain proinflammatory signalling in acute liver failure: role in the pathogenesis of hepatic encephalopathy and brain edema. Metab Brain Dis 28:145–150. [DOI] [PubMed] [Google Scholar]
- Bernal W, Wendon J (2013) Acute liver failure. New Eng J Med 369:2525–2534. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucl Acid Res 36:D149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. [DOI] [PubMed] [Google Scholar]
- Caldeira MV, Salazar IL, Curcio M, Canzoniero LM, Duarte CB (2014) Role of the ubiquitin-proteasome system in brain ischemia: friend or foe? Prog Neurobiol 112:50–69. [DOI] [PubMed] [Google Scholar]
- Campbell A, Ziparo V, James JH, Fischer JE (1979) Loss of day-night rhythm in rats after portacaval shunt. Surg Forum 30:388–390. [PubMed] [Google Scholar]
- Chen HJ, Jiao Y, Zhu XQ, Zhang HY, Liu JC, Wen S, Teng GJ (2013) Brain dysfunction primarily related to previous overt hepatic encephalopathy compared with minimal hepatic encephalopathy: resting-state functional MR imaging demonstration. Radiology 266:261–270. [DOI] [PubMed] [Google Scholar]
- Chen S, Guttridge DC, You Z, Zhang Z, Fribley A, Mayo MW, Kitajewski J, Wang CY (2001) Wnt-1 signaling inhibits apoptosis by activating beta-catenin/T cell factor-mediated transcription. J Cell Biol 152:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba J, Blei AT (1996) Brain edema and hepatic encephalopathy. Semin Liver Dis 16:271–280. [DOI] [PubMed] [Google Scholar]
- Cordoba J, Cabrera J, Lataif L, Penev P, Zee P, Blei AT (1998) High prevalence of sleep disturbance in cirrhosis. Hepatology 27:339–345. [DOI] [PubMed] [Google Scholar]
- Cui W, Sun CM, Liu P (2013) Alterations of blood-brain barrier and associated factors in acute liver failure. Gastroenterol Res Pract 2013:841707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detry O, Gaspar Y, Cheramy-Bien JP, De Roover A, Honore P, Meurisse M, Defraigne JO, Pincemail J (2005) Oxidative stress in the liver and the brain of rats in fulminant hepatic failure. Transpl Proc 37:2883–2885. [DOI] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R (2009) Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab 29:675–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ (2013) Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front Cell Neurosci 7:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gove CD, Hughes RD, Ede RJ, Williams R (1997) Regional cerebral edema and chloride space in galactosamine-induced liver failure in rats. Hepatology 25:295–301. [DOI] [PubMed] [Google Scholar]
- Hazell AS, Butterworth RF (1999) Hepatic encephalopathy: An update of pathophysiologic mechanisms. Proc Soc Exp Biol Med 222:99–112. [DOI] [PubMed] [Google Scholar]
- Jalan R (2003) Intracranial hypertension in acute liver failure: pathophysiological basis of rational management. Semin Liver Dis 23:271–282. [DOI] [PubMed] [Google Scholar]
- Jayakumar AR, Valdes V, Tong XY, Shamaladevi N, Gonzalez W, Norenberg MD (2014) Sulfonylurea receptor 1 contributes to the astrocyte swelling and brain edema in acute liver failure. Transl Stroke Res 5:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Desjardins P, Butterworth RF (2009) Direct evidence for central proinflammatory mechanisms in rats with experimental acute liver failure: protective effect of hypothermia. J Cereb Blood Flow Metab 29:944–952. [DOI] [PubMed] [Google Scholar]
- Jayakumar AR, Ruiz-Cordero R, Tong XY, Norenberg MD (2013) Brain edema in acute liver failure: role of neurosteroids. Arch Biochem Biophys 536, 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911–1912. [DOI] [PubMed] [Google Scholar]
- Kato M, Hughes RD, Keays RT, Williams (1992) Electron microscopic study of brain capillaries in cerebral edema from fulminant hepatic failure. Hepatology. 15:1060–1066. [DOI] [PubMed] [Google Scholar]
- Knecht K, Michalak A, Rose C, Rothstein JD, Butterworth RF (1997) Decreased glutamate transporter (GLT-1) expression in frontal cortex of rats with acute liver failure. Neurosci Lett 229:201–203. [DOI] [PubMed] [Google Scholar]
- Lai JC, Cooper AJ (1986) Brain alpha-ketoglutarate dehydrogenase complex: kinetic properties, regional distribution, and effects of inhibitors. J Neurochem 47:1376–1386. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. [DOI] [PubMed] [Google Scholar]
- Matkowskyj KA, Marrero JA, Carroll RE, Danilkovich AV, Green RM, Benya RV (1999) Azoxymethane-induced fulminant hepatic failure in C57BL/6J mice: characterization of a new animal model. Amer J Physiol 277:G455–462. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Tower DB (1961) Ammonia toxicity and cerebral oxidative metabolism. Amer J Physiol 200:420–424. [DOI] [PubMed] [Google Scholar]
- Michalak A, Knecht K, Butterworth RF (1997) Hepatic encephalopathy in acute liver failure: role of the glutamate system. Adv Exp Med Biol 420:35–43. [DOI] [PubMed] [Google Scholar]
- Montagnese S, De Pitta C, De Rui M, Corrias M, Turco M, Merkel C, Amodio P, Costa R, Skene DJ, Gatta A (2014) Sleep-wake abnormalities in patients with cirrhosis. Hepatology 59:705–712. [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG (2009) DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics 25:1991–1993. [DOI] [PubMed] [Google Scholar]
- Patel D, McPhail MJ, Cobbold JF, Taylor-Robinson SD (2012) Hepatic encephalopathy. Br J Hosp Med (Lond) 73:79–85. [DOI] [PubMed] [Google Scholar]
- Potvin M, Finlayson MH, Hinchey EJ, Lough JO, Goresky CA (1984) Cerebral abnormalities in hepatectomized rats with acute hepatic coma. Lab Invest 50:560–564. [PubMed] [Google Scholar]
- Rao KV, Brahmbhatt M, Norenberg MD (2013) Microglia contribute to ammonia-induced astrocyte swelling in culture. Metab Brain Dis 28:139–143. [DOI] [PubMed] [Google Scholar]
- Sathyasaikumar KV, Swapna I, Reddy PV, Murthy Ch R, Dutta Gupta A, Senthilkumaran B, Reddanna P (2007) Fulminant hepatic failure in rats induces oxidative stress differentially in cerebral cortex, cerebellum and pons medulla. Neurochem Res 32:517–524. [DOI] [PubMed] [Google Scholar]
- Scott TR, Kronsten VT, Hughes RD, Shawcross DL (2013) Pathophysiology of cerebral oedema in acute liver failure. World J. Gastroenterol 19:9240–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronska M, Albrecht J (2013) Oxidative and nitrosative stress in ammonia neurotoxicity. Neurochem Int 62:731–737. [DOI] [PubMed] [Google Scholar]
- Traber PG, Dal Canto M, Ganger DR, Blei AT (1987) Electron microscopic evaluation of brain edema in rabbits with galactosamine-induced fulminant hepatic failure: ultrastructure and integrity of the blood-brain barrier. Hepatology 7:1272–1277. [DOI] [PubMed] [Google Scholar]
- Tranah TH, Vijay GK, Ryan JM, Shawcross DL (2013) Systemic inflammation and ammonia in hepatic encephalopathy. Metab Brain Dis 28:1–5. [DOI] [PubMed] [Google Scholar]
- Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S (2012) MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep 2:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadysirisack DD, Ellisen LW (2012) mTOR activity under hypoxia. Meth Molec Biol 821:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313:320–324. [DOI] [PubMed] [Google Scholar]
- Vemuganti R (2013) All’s well that transcribes well: non-coding RNAs and post-stroke brain damage. Neurochem Int 63:438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan BX, Ma JX (2012) Promoter-associated RNAs and promoter-targeted RNAs. Cell Molec Life Sci 69:2833–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]