The synthesis of ribosomes is one of the major cellular activities, and in eukaryotes, it takes place primarily, although not exclusively, in a specialized subnuclear compartment termed the nucleolus (125, 155). There, the rRNA genes are transcribed as precursors (pre-rRNAs), which undergo processing and covalent modification. Maturation of pre-rRNAs is intimately linked to their assembly with the ribosomal proteins (r-proteins). These processes depend on various cis-acting elements (6, 188), and they require a large number of nonribosomal protein trans-acting factors (97, 174, 193). Experimental evidence suggests that the basic outline of ribosome synthesis is conserved throughout eukaryotes. However, most of our knowledge comes from the combination of molecular genetic and biochemical approaches in the yeast Saccharomyces cerevisiae. This minireview is aimed at giving an insight into the functions of the many protein trans-acting factors involved in ribosome biogenesis in S. cerevisiae.
PRE-RRNA PROCESSING AND RIBOSOME ASSEMBLY PATHWAYS
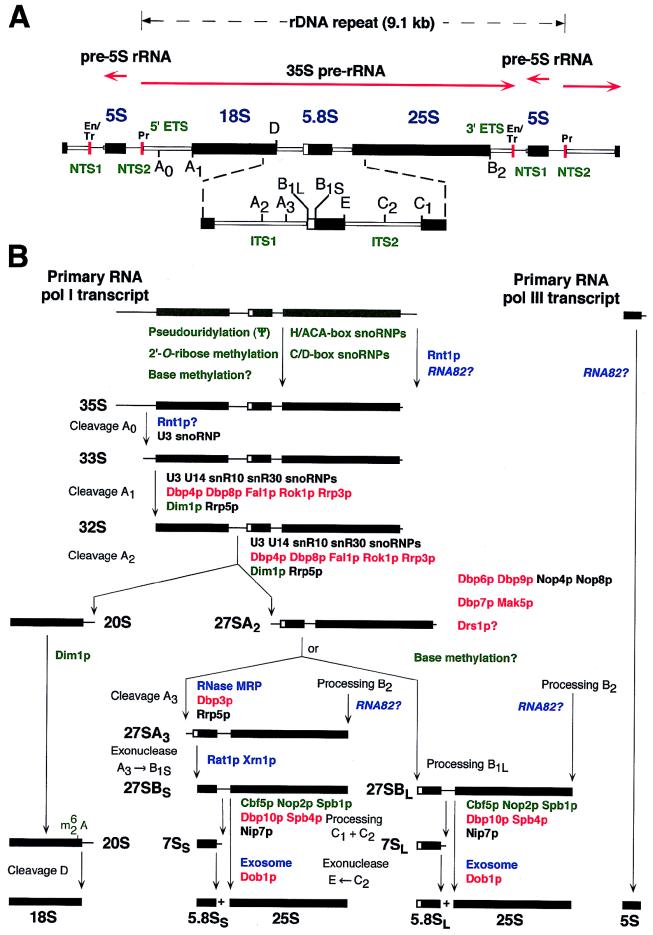
In S. cerevisiae, the large 60S ribosomal subunits are composed of 46 r-proteins and three rRNA species (5S, 5.8S, and 25S) while the small 40S ribosomal subunits contain 32 r-proteins and the 18S rRNA (142, 201). Three of the four rRNAs (18S, 5.8S, and 25S) are transcribed as a single large pre-rRNA by RNA polymerase I (RNA pol I), whereas the fourth rRNA (5S) is independently transcribed as a pre-rRNA by RNA pol III (201). All four rRNAs are encoded by a 9.1-kb rDNA unit, which is repeated 100 to 200 times on the long arm of chromosome XII (Fig. 1A). In the 35S pre-rRNA, which is the longest detectable precursor, the mature rRNA sequences are separated by two internal transcribed spacer (ITS) sequences, ITS1 and ITS2, and flanked by two external transcribed spacer (ETS) sequences, a 5′ ETS and a 3′ ETS (Fig. 1). The 35S pre-rRNA differs from the primary RNA pol I transcript at its 3′ end because transcription termination maps to nucleotide position +210 relative to the 3′ end of the mature 25S rRNA, while the 35S pre-rRNA is extended by 7 to 10 nucleotides (78, 187, 189). Maturation of the 35S pre-rRNA, which contains 10 known processing sites, is a multistep pathway that requires many different trans-acting factors (Fig. 1B and its legend) (97, 174, 193). Processing of the pre-5S rRNA is independent of 35S pre-rRNA maturation and kinetically faster than formation of mature 18S, 5.8S, and 25S rRNAs (146). The 5′ end of the mature 5S rRNA corresponds to that of the primary transcript, whereas the 3′ end is processed from a pre-5S rRNA that is extended by 7 to 13 nucleotides (141). Many specific nucleotides within the rRNA also undergo, mainly shortly after transcription, covalent modification. These modifications include isomerization of uridine to pseudouridine (Ψ) by base rotation (45 modified nucleotides), methylation of the 2′-hydroxyl group of sugar residues (2′-O-ribose methylation; 55 modified nucleotides), and base methylation (about 10 modified nucleotides) (9, 21, 84, 120, 135).
FIG. 1.
Pre-rRNA processing in S. cerevisiae. (A) Structure of an rDNA repeat unit. Each unit contains a large operon encoding the 18S, 5.8S, and 25S rRNAs, which is transcribed by RNA pol I (pr, promoter; tr, terminator; eh, enhancer), and an RNA pol III-transcribed 5S rRNA gene. Mature sequences of the RNA pol I transcript are separated by ITS1 and ITS2 and flanked by a 5′ ETS and a 3′ ETS. The 5S rRNA gene is located between two nontranscribed spacers (NTS1 and NTS2). Black boxes represent mature rRNAs, white thin bars represent the transcribed spacers, and lines represent the nontranscribed spacers. Processing sites are also indicated. (B) Pre-rRNA processing pathways. The primary RNA pol I transcript is processed at its 3′ end to yield the 35S pre-rRNA, which is the longest detectable pre-rRNA. The 35S pre-rRNA is first processed at sites A0, A1, and A2, which results in the separation of the pre-rRNAs destined for the small and large ribosomal subunits. The 20S pre-rRNA is matured by endonucleolytic cleavage at site D. The 27SA2 precursor is processed by two alternative pathways. In the major pathway, about 85% of 27SA2 pre-rRNA is cleaved at site A3 and then 5′→3′ exonucleolytically digested to site B1S. In the minor pathway, about 15% of the 27SA2 molecules are processed at site B1L. While processing at site B1 is completed, the 3′ end of mature 25S rRNA is generated by processing at site B2. The subsequent ITS2 processing of both 27SB species appears to be the same. Cleavage at sites C1 and C2 releases the mature 25S rRNA and the 7S pre-rRNA. The latter undergoes 3′→5′ exonucleolytic digestion to the 3′ end of the mature 5.8S rRNA. The pre-5S rRNA is processed at its 3′ end to generate the mature 5S rRNA. Pseudouridylation and 2′-O-ribose methylation take place on the primary transcript. Note that except for Dim1p, the timing of base methylation and the protein trans-acting factors involved therein have not yet been uncovered. Factors involved in pre-rRNA processing and modification are shown. Colors were assigned to each class of protein trans-acting factors as follows: blue, exo- and endonucleases; red, putative ATP-dependent RNA helicases; green, components involved in pre-rRNA modification; black, others. See the text for further details.
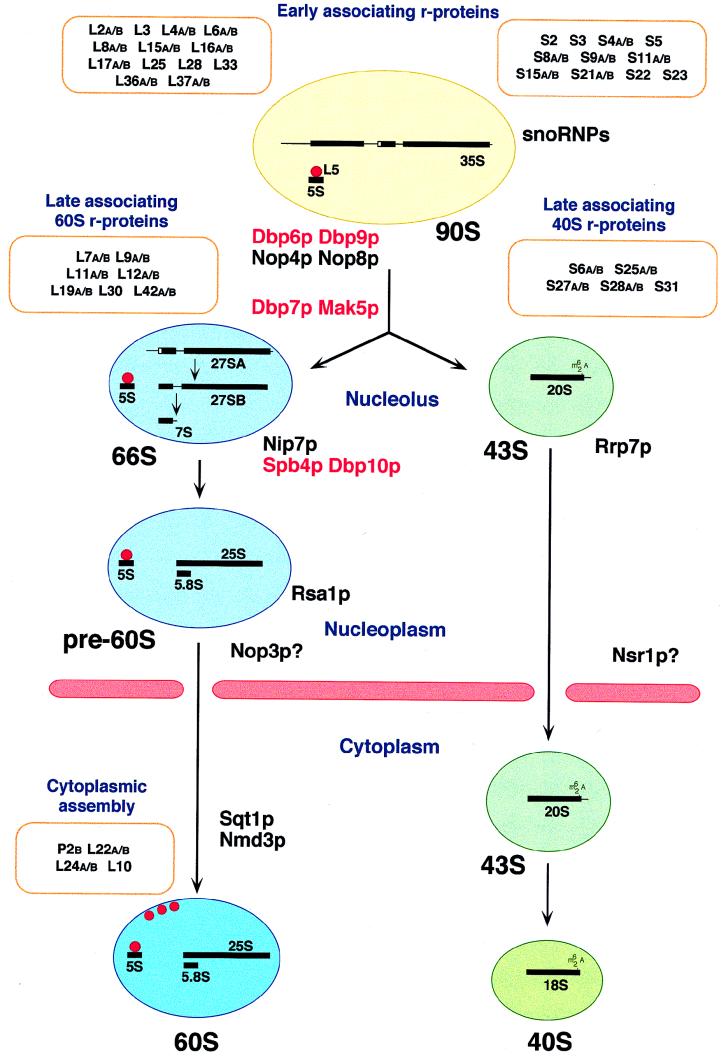
Pre-rRNA processing and modification do not take place on “naked” pre-rRNAs. Instead, pre-rRNAs associate with many of the r-proteins in the nucleolus to form preribosomal particles (182). In addition to r-proteins, the nucleolar preribosomes have long been known to contain non-r-proteins (182). The identity of these proteins has not been clearly established, but they presumably correspond to trans-acting factors required for pre-rRNA processing and modification or involved in the assembly of the pre-rRNAs with the r-proteins. Although the pre-rRNA processing pathway and its intermediates have been fairly well characterized, the assembly process of the rRNAs and the approximately 80 r-proteins into mature ribosomal subunits is still poorly understood (Fig. 2 and its legend) (93, 180–182, 184).
FIG. 2.
Ribosomal subunit assembly pathway in S. cerevisiae. In the nucleolus, the 35S pre-rRNA associates with many early-associating 40S and 60S r-proteins to form a 90S preribosome. The 5S rRNA forms a stable RNP with Rpl5p (L5, red dot). The 5S rRNA is probably already present in 90S preribosomes; therefore, Rpl5p may belong to the group of early-assembling 60S r-proteins. From the 90S particle, 66S and 43S preribosomes containing the 27S and 20S pre-rRNAs, respectively, are formed. The 66S and the 43S preribosomes contain the late-associating r-proteins. Preribosomes are most likely actively exported through nuclear pores. It has been demonstrated that export of 60S ribosomal subunits requires the nuclear or cytoplasmic Ran cycle and distinct nucleoporins (not shown; for details, see reference 70). It is believed that the 66S particle remains in the nucleus until the 7S pre-rRNA is processed to the mature 5.8S rRNA, while the 43S preribosome is rapidly exported to the cytoplasm, where the final maturation step in the synthesis of the 18S rRNA takes place. Assembly of 60S ribosomal subunits is also completed in the cytoplasm by the final incorporation of some r-proteins. At least three of them (also depicted as red dots), including the P2B and Rpl10p/Qsr1p (L10) r-proteins, can exchange on mature 60S ribosomal subunits in vivo. Preribosomes are drawn as balloons, and the nuclear envelope is depicted as pink rods. Protein trans-acting factors with likely involvement in the assembly of ribosomal subunits are colored as follows: red, putative ATP-dependent RNA helicases; black, others. Only those r-proteins for which an indication about the timing of their association with preribosomal particles is known are shown. See the text for further details.
PROTEIN TRANS-ACTING FACTORS INVOLVED IN RIBOSOME BIOGENESIS
To date, more than 60 protein trans-acting factors have been described to function in yeast ribosome biogenesis and most likely not all of them have been identified yet. Most of the protein trans-acting factors can be grouped into classes according to their experimental or predicted enzymatic functions and/or their physical or functional interactions with small nucleolar RNAs (snoRNAs) or other proteins. These classes include components of small nucleolar ribonucleoprotein particles (snoRNPs), rRNA-modifying enzymes, endo- and exonucleases, and putative RNA helicases. However, not all protein trans-acting factors fall into the above classes and many of these contain particular conserved motifs or domains that could mediate important functions such as RNA-protein or protein-protein interactions. A list of trans-acting factors involved in ribosome biogenesis and links to databases is accessible at reference 115a.
PROTEIN TRANS-ACTING FACTORS ASSOCIATED WITH SNORNAS
A large number of protein trans-acting factors that are required for ribosome biogenesis are quite stably associated with snoRNAs in the form of snoRNP complexes (Table 1). In yeast, there are about 100 different snoRNAs (116, 153), which can be structurally and functionally grouped into three families. Most snoRNAs belong to either the H/ACA-box or the C/D-box family, whereas the RNA component of RNase MRP constitutes one class by itself (see below) (11, 101, 176).
TABLE 1.
Protein components of snoRNPs
| Class and proteina | Essentialnessb | Associated snoRNA(s) | snoRNA stabilityc | Function | Particular feature(s) | Reference(s) |
|---|---|---|---|---|---|---|
| H/ACA-box snoRNP | ||||||
| Cbf5p | Y | H/ACA-box | − | pre-rRNA pseudouridylation and processing (A1, A2) | Putative rRNA Ψ-synthase, KKE/D motif repeats | 99 |
| Gar1p | Y | H/ACA-box | + | pre-rRNA pseudouridylation and processing (A1, A2) | Two GAR domains | 20, 61 |
| Nhp2p | Y | H/ACA-box | − | pre-rRNA pseudouridylation and processing (A1, A2) | Homology to Rpl30p | 63, 197 |
| Nop10p | Y | H/ACA-box | − | pre-rRNA pseudouridylation and processing (A1, A2) | 63, 197 | |
| Sbp1p | N | snR10, snR11 | ND | pre-rRNA processing (A1, A2) | RRM and GAR domain | 34 |
| Sen1p | Y | snR10, snR30, U3 | − | Maturation of diverse RNAs | Putative RNA helicase | 145, 185 |
| C/D-box snoRNP | ||||||
| Nop1p | Y | C/D-box | − | pre-rRNA 2′-O-methylation and processing (A0, A1, A2) | Orthology to human fibrillarin, GAR domain | 58, 177, 178 |
| Nop56p | Y | C/D-box | + | pre-rRNA processing (A0, A1, A2) | Homology to Nop58p, KKE/D motif repeats | 59, 96 |
| Nop58p/Nop5p | Y | C/D-box | − | pre-rRNA processing (A0, A1, A2) | Homology to Nop56p, KKE/D motif repeats | 59, 102, 202 |
| Sof1p | Y | U3 | + | pre-rRNA processing (A0, A1, A2) | WD repeats | 74 |
| Mpp10p | Y | U3 | + | pre-rRNA processing (A0, A1, A2) | 48, 108 | |
| Imp3p | Y | U3 | + | pre-rRNA processing (A0, A1, A2) | Similarity to Rps9Ap | 109 |
| Imp4p | Y | U3 | + | pre-rRNA processing (A0, A1, A2) | 109 | |
| Rrp9p | Y | U3 | ND | pre-rRNA processing | 190 | |
| Lcp5p | Y | U3 | + | pre-rRNA processing (A0, A1, A2) | Rich in charged residues | 200 |
| RNase MRP | ||||||
| Pop1p | Y | MRP, P | − | pre-tRNA and pre-rRNA (A3) processing | KS-rich regions | 27, 119 |
| Pop3p | Y | MRP, P | + | pre-tRNA and pre-rRNA (A3) processing | KS-rich regions | 27, 45 |
| Pop4p | Y | MRP, P | − | pre-tRNA and pre-rRNA (A3) processing | KS-rich regions | 27, 33 |
| Pop5p | Y | MRP, P | − | pre-tRNA and pre-rRNA (A3) processing | KS-rich regions | 27 |
| Pop6p | Y | MRP, P | − | pre-tRNA and pre-rRNA (A3) processing | KS-rich regions | 27 |
| Pop7p/Rpp2p | Y | MRP, P | − | pre-tRNA and pre-rRNA (A3) processing | Regions homologous to Rrp7p, KS-rich regions | 27, 169 |
| Pop8p | Y | MRP, P | − | pre-tRNA and pre-rRNA (A3) processing | KS-rich regions | 27 |
| Rpp1p | Y | MRP, P | − | pre-tRNA and pre-tRNA (A3) processing | 27, 168 | |
| Snm1p | Y | MRP | − | pre-rRNA processing (A3) | Potential zinc-binding domain, KS-rich regions | 157 |
The presence of a protein on this list is exclusively based on experimental evidence. Note that Sen1p is associated with both H/ACA- and C/D-box snoRNAs. A more complete list of trans-acting factors involved in ribosome biogenesis and links to databases is accessible at reference 115a.
Abbreviations: Y, essential (i.e., knockout is lethal); N, nonessential (i.e., knockout is not lethal).
Wild-type (+) or reduced (−) steady-state levels of the associated snoRNAs upon mutation or depletion of the respective protein trans-acting factor. ND, not determined.
H/ACA-box snoRNPs.
In yeast, most, if not all, of the 45 Ψ residues are present within the mature rRNA sequences (135). The site-specific pseudouridylation of rRNA requires H/ACA-box snoRNPs, in which the H/ACA-box snoRNAs function as guides that specify the uridines to be isomerized in the pre-rRNA. Site selection is mediated by short base pairings between the snoRNA and the pre-rRNA, and pseudouridylation occurs at a fixed distance of about 14 nucleotides from the conserved snoRNA sequence elements (box H and/or ACA triplet) (57, 131). One H/ACA-box snoRNA, snR30, seems to be exclusively required for the early pre-rRNA processing reactions at sites A1 and A2, while the H/ACA-box snoRNA snR10 participates in both early pre-rRNA processing reactions and a pseudouridylation event within 25S rRNA (130, 131, 153, 175).
Based on coimmunoprecipitation and purification experiments, the core protein components common to all H/ACA-box snoRNPs have been identified. This core particle probably contains two copies of each of four proteins (Cbf5p, Gar1p, Nhp2p, and Nop10p) and one H/ACA-box snoRNA (63, 197), and it is arranged in a symmetric two-domain structure (117, 197). However, the detailed architecture of H/ACA-box snoRNPs, as well as the order of their assembly, remains to be elucidated. The nucleolar protein Cbf5p might be the Ψ-synthase in each snoRNP, since it is homologous to a family of putative Ψ-synthases (87). Indeed, depletion of Cbf5p blocks Ψ formation in pre-rRNAs, which is most likely due to the loss of Cbf5p’s putative enzymatic function and the codepletion of H/ACA-box snoRNAs (99). Probably as a consequence of the concomitant codepletion of snR10 and snR30, Cbf5p depletion inhibits pre-rRNA processing at sites A1 and A2 and, to a lesser extent, at site A0 (99). Interestingly, depletion of Cbf5p also leads to slightly reduced levels of 25S rRNA and to accumulation of 27SB and 7S pre-rRNAs and of an aberrant extended 5.8S rRNA species (99). This latter phenotype is unique and not related to the loss of function of any known snoRNA. Furthermore, the depletion of neither Nhp2p nor Nop10p, which also results in the codepletion of H/ACA-box snoRNAs, shows such a particular phenotype (63). Therefore, it can be assumed that Cbf5p has other roles in ribosome biogenesis in addition to its function as an integral component of H/ACA-box snoRNPs, perhaps through interactions with other trans-acting factors during the assembly of preribosomes (25, 99). Indeed, the mammalian homolog of Cbf5p, termed NAP57, is associated in stoichiometric amounts with another nucleolar protein called Nopp140 (124), which is the homolog of the yeast nucleolar acidic and serine-rich protein Srp40p (123). A genetic interaction between Srp40p and H/ACA-box snoRNAs has been recently uncovered, thereby indirectly linking Srp40p and Cbf5p (122). Therefore, the Srp40p-Cbf5p combination seems to, indeed, mirror the Nopp140-NAP57 connection in mammalian cells. Although the function of Srp40p in ribosome biogenesis has not been analyzed so far, it has been suggested that Srp40p, in analogy to the proposed function of the Nopp140-NAP57 complex, might mediate traffic of diverse nuclear components, among them, snoRNAs and preribosomes (72, 122).
Gar1p is a nucleolar protein that contains two regions that are rich in glycine and arginine (GAR domain) (61). The GAR domain is also present in other nucleolar proteins involved in ribosome biogenesis (see below), and it is thought to mediate protein-RNA interactions (61). Neither one of the two GAR domains (one N terminal and one C terminal) of Gar1p is required for cell viability or for in vitro snoRNA binding (10, 61). Gar1p is required for pre-rRNA processing at sites A1 and A2 (61). More recently, it has been demonstrated that Gar1p is involved in global pre-rRNA pseudouridylation (20) and that it is specifically associated with all H/ACA-box snoRNAs (58). However, Gar1p is the only core component of H/ACA-box snoRNPs whose depletion does not alter the steady-state levels of snoRNAs (20, 61). Nevertheless, in the absence of Gar1p, the H/ACA-box snoRNPs are unable to efficiently associate with higher-order nucleolar particles. This suggests that Gar1p might work by establishing or maintaining interactions of snoRNPs with the pre-rRNA (cited in references 20 and 99). The putative RNA helicase Rok1p (see below) may assist Gar1p in this function, since ROK1 was identified in genetic screens for synthetic lethality with both a conditional gar1 allele and the snr10 null (ΔsnR10) mutation (191). Furthermore, Gar1p has been shown to interact with Cbf5p by the yeast two-hybrid system (cited in reference 63).
Nucleolar Nhp2p and Nop10p also specifically associate with all H/ACA-box snoRNAs (63, 197). Nhp2p shares homology with a 53-amino-acid region within the r-protein Rpl30p (formerly L32) that has been implicated in RNA binding (195). Therefore, it is assumed that Nhp2p directly binds to H/ACA-box snoRNAs (63). This binding (and/or the binding of the other H/ACA-box snoRNP components) probably limits further trimming and degradation of H/ACA-box snoRNAs by the exonucleases Xrn1p and Rat1p (see below) and other nucleases (18, 63, 140). Nop10p, on the other hand, contains no obvious protein motifs (63). Depletion of Nhp2p or Nop10p inhibits pre-rRNA processing at sites A1 and A2, and as in the case of Cbf5p depletion, this phenotype can be explained by the codepletion of snR10 and snR30 (63). In addition, since the stability of all H/ACA-box snoRNAs is affected, Nhp2p and Nop10p depletion also blocks Ψ formation (63). Moreover, as in cells lacking Cbf5p, Gar1p is rapidly degraded in the absence of either Nhp2p or Nop10p, indicating that the integrity of H/ACA-box snoRNPs is perturbed (63). However, Nhp2p or Nop10p depletion does not lead to the codepletion of Nop10p or Nhp2p, respectively (63).
Apart from the core components, two further proteins, Sen1p and Sbp1p, have been described to associate with certain H/ACA-box snoRNAs. The function of Sen1p, a putative RNA helicase, will be discussed below. The nonessential Sbp1p contains a central GAR domain followed by an RNA recognition motif (RRM) and acidic and serine-rich stretches; these motifs probably mediate the specific strong binding to snR10 and the weak binding to snR11 (34). In addition, the Δsbp1 strain has growth and pre-rRNA processing phenotypes that are similar to those of the Δsnr10 strain (cited in reference 34). However, whether Sbp1p is required for in vivo snR10 stability and/or snR10-associated pseudouridylation is not known.
As mentioned above, the snR30 H/ACA-box snoRNP seems to be solely required for the early pre-rRNA processing reactions at sites A1 and A2. Nevertheless, its core composition and structure are similar to those reported for the snR42 H/ACA-box snoRNP, which is exclusively involved in pseudouridylation (117, 197). In clear contrast to the snR10 and U3 snoRNPs (see below), it is unknown if additional proteins are required for the function of the snR30 snoRNP.
C/D-box snoRNPs.
Yeast rRNAs contain about 65 methyl groups; 55 of these are linked to the 2′-hydroxyl residue of ribose, while the rest of them are attached to the bases (9). Ribose methylation is, in analogy to pseudouridylation, carried out by specific snoRNPs that contain snoRNAs of the C/D-box family (82, 176). These snoRNAs have one or two sets of well-conserved short sequence elements, which are called box C and box D (or box C′ and box D′, for the second set), and about 10 to 20 nucleotides with sequence complementarity to the rRNA. The position of 2′-O-ribose methylation of rRNA is 5 nucleotides upstream of box D (or D′) (9, 83). Most of the 55 2′-O-ribose-methylated sites have been recently assigned to different guide C/D-box snoRNAs, all of them dispensable for cell viability (116, 153). In contrast to these, U3 and U14 are essential C/D-box snoRNAs (176). The U3 snoRNA seems to be exclusively required for pre-rRNA processing at sites A0 to A2, while the U14 snoRNA is involved in pre-rRNA processing reactions at sites A1 and A2 and in a 2′-O-ribose methylation event within the 18S rRNA (153, 176).
To date, three proteins (Nop1p, Nop56p, and Nop58p) have been identified that probably form the core of all C/D-box snoRNPs (Table 1) (58, 59, 102, 202). In clear contrast to the presence of the candidate Ψ-synthase Cbf5p in H/ACA-box snoRNPs, the putative methylase(s) has not yet been identified as a core component of C/D-box snoRNPs, although an enzymatic role for Nop1p cannot be excluded (132). Moreover, there are several proteins with predicted rRNA methyltransferase activity that are thought to be good candidates (Nop2p, Ncl1p, and Spb1p; see below) (68, 92, 203).
Nop1p, which contains an N-terminal GAR domain (156), is structurally and functionally equivalent to human fibrillarin (75). Although early work suggested that Nop1p efficiently precipitates both C/D-box and H/ACA-box snoRNAs (156, 177), it now seems clear that Nop1p is specifically associated with C/D-box snoRNAs (58). Depletion of Nop1p leads to codepletion of methylation guide snoRNAs (as demonstrated for U14 and snR190) but not of U3 snoRNA (177). Partial inactivation of the U3 snoRNP function, due to the loss of its Nop1p component, and the reduction in the levels of U14 might be responsible for the pre-rRNA processing defects at sites A0 to A2 that are detected upon depletion of Nop1p (177). Similarly, loss of methylation guide snoRNAs is consistent with impaired global methylation upon Nop1p depletion. Interestingly, it has been reported that in the nop1-3 mutant, global methylation is dramatically affected without having major effects on pre-rRNA processing or ribosome assembly (178). This indicates that 2′-O-ribose methylation, as well as Ψ synthesis, and pre-rRNA processing are independent events (20, 178). Other nop1 mutants affect the assembly of 60S ribosomal subunits without leading to clear pre-rRNA processing and methylation defects (178). Whether this phenotype is the consequence of the loss of particular snoRNA–pre-rRNA interactions or the impairment of another independent function of Nop1p remains to be elucidated. Indeed, Nop1p, but not any snoRNA, also binds in substoichiometric amounts to another trans-acting factor named Nop4p/Nop77p (15). Nop4p is a nucleolar protein, which has been identified both in a synthetic lethal screen with a nop1 allele and as a nucleolar antigen (15, 170). Nop4p is made up of a modular primary structure that consists of three N-terminal RRMs and a fourth incomplete RRM in the C-terminal part of the protein. The fourth RRM is followed by a region rich in lysine and arginine residues, while the first three RRMs are separated by two regions rich in acidic amino acids. This structure suggests that Nop4p is able to directly interact with RNA. Moreover, each RRM is required for optimal Nop4p function in vivo (171). Depletion of Nop4p delays 35S pre-rRNA processing and leads to a drastic reduction in the steady-state levels of the 27SA3 and 27SB pre-rRNAs (15). As a consequence, synthesis of mature 25S rRNA and production of 60S ribosomal subunits are reduced (170). It is, however, controversial if Nop4p is involved in rRNA methylation (15, 170). The above pre-rRNA processing phenotype is clearly different from that observed after depletion of Nop1p or the snoRNA U3 or U14. Most likely, Nop4p, in concert with Nop1p, plays a role in the early assembly of 60S ribosomal subunits.
Nucleolar Nop56p and Nop58p, which are homologous to each other, were also both identified in a synthetic lethal screen with nop1 alleles (59). Nop58p (also named Nop5p) has been simultaneously isolated in a screen for nucleolar antigens (202). The primary sequence of both proteins contains several repeats of the KKE/D motif, a feature shared with Cbf5p and the putative RNA helicase Dbp3p (198). In the case of Nop58p, these repeats are not required for cell growth, protein targeting to the nucleolus, or stable association with snoRNAs (59, 102). Depletion of Nop56p results in the inhibition of pre-rRNA processing at sites A0 to A2 and reduced steady-state levels of Nop1p; however, neither global 2′-O-ribose methylation nor the stability of C/D-box snoRNAs is affected (96). In contrast, the temperature-sensitive nop56-2 mutation leads to the accumulation of 27SB pre-rRNAs and to delayed conversion of these precursors to mature 25S rRNA, which most likely accounts for the observed deficiency in the formation of 60S ribosomal subunits (59). This pre-rRNA processing phenotype is similar to that observed upon Nop2p and Spb1p depletion (68, 92). The function of Nop58p has been studied in more detail. Nop58p specifically interacts with all C/D-box snoRNAs, including U3 and U14 (102, 202). Moreover, depletion of Nop58p results in the mislocalization of Nop1p (202) and in the instability of all C/D-box snoRNAs (102). Consistent with this, depletion of Nop58p leads to inhibition of the A0-to-A2 pre-rRNA processing reactions (102), therefore resulting in a reduction of 40S relative to 60S ribosomal subunits (202). Surprisingly, global methylation is not affected upon Nop58p depletion (102, 202), perhaps the reduced levels of C/D-box snoRNAs are sufficient to promote 2′-O-ribose methylation but can no longer support pre-rRNA processing (102).
Unfortunately, no information is available concerning the architecture of C/D-box snoRNPs. Due to the conserved nature of the C and D boxes and the adjacent terminal stem (C/D-box motif) (9), and because of the requirement of this C/D-box motif for snoRNA stability and function (83, 154), it is conceivable that either Nop1p, Nop56p, Nop58p, or the candidate 2′-O-ribose methylase(s) binds directly to this motif. However, there is no evidence yet for an interaction between any of the putative methyltransferases (apart from Nop1p [see below]) with C/D-box snoRNPs.
The U3 snoRNP is a particular C/D-box snoRNP. In addition to the core proteins, it contains unique proteins that are not present in other C/D-box snoRNPs (Table 1). Sof1p was the first described U3 snoRNP protein; it was isolated as an extragenic suppressor of the thermosensitive phenotype of a Δnop1 strain complemented with human fibrillarin (74). Sof1p is a nucleolar protein whose primary structure contains seven WD repeats, which could mediate protein-protein interactions, and a C-terminal part that is rich in charged amino acids (74). Apart from binding exclusively to the U3 snoRNA, Sof1p appears to associate with Nop1p in substoichiometric amounts, which is consistent with the fact that Nop1p is a core component of all C/D-box snoRNAs. Depletion of Sof1p leads to impaired pre-rRNA processing at sites A0 to A2 and inhibition of 18S rRNA production, but it does not result in instability of either U3 or Nop1p (74). Consistent with the specific association of Sof1p with U3, pre-rRNA methylation is not affected upon Sof1p depletion. Mpp10p and its interacting partners Imp3p and Imp4p (48, 108, 109), Lcp5p (200), and Rrp9p (190) are also U3 snoRNP components, and their depletion leads to inhibition of the early pre-rRNA cleavages. As reported for Sof1p, U3 stability is not affected upon depletion of Mpp10p, Imp3p, or Imp4p (48, 109). So far, there are at least nine proteins that are associated directly or indirectly with the U3 snoRNA; however, it remains to be determined whether all of these coexist in the same U3 snoRNP.
RRNA-MODIFYING ENZYMES
As mentioned above, posttranscriptional modification of mature rRNAs occurs mostly on the pre-rRNA. These modifications include Ψ formation, 2′-O-ribose methylation, and base methylation (9, 120, 135). In contrast to the putative rRNA Ψ-synthase Cbf5p (99, 197), which is a subunit of all H/ACA-box snoRNPs (see above), the candidate methylase(s) responsible for 2′-O-ribose methylation has not yet been identified. Proteins with predicted rRNA methyltransferase activity have been detected by sequence comparison to the human tumor-specific nucleolar protein p120 and to putative bacterial rRNA methyltransferases (86). In particular, Nop2p shows strong overall homology to p120 (39). Nucleolar Nop2p is required for the conversion of the 27SB pre-rRNA into mature 25S rRNA and, thus, for synthesis of 60S ribosomal subunits (68). However, although it seems that 2′-O-ribose methylation at a specific site in the 27S pre-rRNA is delayed upon Nop2p depletion, global methylation is not affected (68). Thus, Nop2p is unlikely to be the only candidate rRNA methyltransferase (68). Indeed, a similar pre-rRNA processing phenotype is observed when cells are depleted of the essential protein Spb1p, which also contains a putative S-adenosylmethionine (SAM)-binding motif (92). The closest S. cerevisiae homolog of Nop2p is Ncl1p, which contains, in addition to the SAM-binding motif, the evolutionarily conserved “NOL1/NOP2/fmu family signature” motif (203). The nonessential Ncl1p localizes to the nucleus, including the nucleolus, and is concentrated at the nuclear periphery. Disruption of NCL1 affects neither cell growth nor steady-state levels of ribosomal subunits; however, it leads to increased sensitivity to the aminoglycoside antibiotic paromomycin, which suggests that ribosome function is perturbed (31, 54, 138, 162). Niewmierzycka and Clarke have recently searched the complete S. cerevisiae genome with sequence motifs that are conserved in SAM-dependent methyltransferases. They identified 7 known methyltransferases, including Dim1p (see below), and 26 putative methyltransferases (132). Neither Nop2p, Ncl1p, nor Spb1p is among these putative methyltransferases, but interestingly, Nop1p (certain mutant forms of which affect global methylation [see above]) was found by this analysis. Therefore, it cannot be excluded, until otherwise demonstrated, that Nop1p itself is an rRNA 2′-O-ribose methyltransferase.
Little is known about base methylation in eukaryotes, and in contrast to 2′-O-ribose methylation and Ψ formation, it does not appear to involve guide snoRNA cofactors. The only base methylation studied in yeast is the evolutionarily conserved dimethylation of two adjacent adenosines (m6,2A1779 m6,2A1780) at the 3′ end of the 18S rRNA. The essential protein Dim1p is responsible for these modifications, and it is also required for pre-rRNA processing at sites A1 and A2 (95, 98), which generate the 20S pre-rRNA. Since dimethylation occurs on the 20S pre-rRNA (98), this observation is consistent with early reports that formation of m6,2A m6,2A is a late event in ribosome synthesis (21); it can be concluded that processing at sites A1 and A2 does not require prior dimethylation. In agreement with this model, the dim1-2 mutation strongly inhibits dimethylation while pre-rRNA processing is practically not affected at the permissive temperature. Thus, the enzymatic function of Dim1p can be separated from its involvement in pre-rRNA processing (100). The dim1-2 mutant strain displays wild-type growth at the permissive temperature, indicating that dimethylation is not essential for cell viability. On the other hand, cell extracts prepared from this strain are incompetent for translation in vitro, which suggests that dimethylation is necessary under suboptimal in vitro conditions but only fine tunes ribosomal function in vivo (100). Interestingly, pre-rRNA processing is insensitive to temperature-sensitive mutations in or depletion of Dim1p when transcription of the rDNA unit is driven by an RNA pol II PGK promoter, which suggests that Dim1p is not directly required for pre-rRNA processing. Therefore, it is very likely that an active repression system (quality control) blocks pre-rRNA processing in the absence of the binding of Dim1p to the pre-rRNA (100, 103). Moreover, all dim1 mutants are hypersensitive to the aminoglycoside antibiotics paromomycin and neomycin B, even under conditions in which neither dimethylation nor pre-rRNA processing is clearly affected (100). This suggests that Dim1p plays an additional role in ribosome assembly.
What is the role of these rRNA modifications? Ψ residues are functionally more versatile than uridine residues in that both N1 and N3 are available for hydrogen bonding. In contrast, 2′-O-ribose methylation has the opposite effect since it prevents the 2′ hydroxyl group of ribose from hydrogen bonding (120). However, 2′-O-methyl groups can stabilize RNA stems by conferring conformational rigidity on the nucleotide (101 and references therein). Strikingly, most pseudouridylated and methylated residues are clustered in the rRNA domains that carry out important functions during translation. This led to the proposition that rRNA modifications may have a direct role in the function of the ribosome as, for instance, in the peptidyl transfer reaction (120). Alternatively, these modifications may play roles in fine-tuning ribosome assembly (174). Deletion of single or multiple guide snoRNAs does not affect cell growth (116, 139, 153). However, rRNA folding and/or association with the r-proteins may still be less efficient in these strains, even though the growth disadvantage is not detectable under laboratory conditions. In addition, even when global 2′-O-ribose methylation is completely inhibited, as in the nop1-3 mutant at the permissive temperature, pre-rRNA processing, ribosome assembly, and export of ribosomal subunits to the cytoplasm continue (178). However, growth of the nop1-3 mutant strain is strongly affected at the nonpermissive temperature, which implies that ribosomes containing severely undermethylated rRNA are most likely deficient in some important translational function (120, 178). Intriguingly, in bacteria, partially functional 30S and 50S ribosomal subunits can be reconstituted with in vitro-transcribed 16S and 23S rRNAs, respectively (35, 62, 80). This suggests that, at least in prokaryotes, posttranscriptional modification of rRNAs is not essential for the assembly or function of ribosomal subunits.
ENDO- AND EXO-RNASES
Several endo- and exonucleases carry out the different processing reactions during maturation of pre-rRNAs. The largest detectable pre-rRNA is the 35S pre-rRNA, and it differs from the primary RNA pol I transcript at its 3′ end (Fig. 1). The 35S pre-rRNA is generated by endonucleolytic cleavage of the primary transcript between nucleotide positions +45 and +15 relative to the 3′ end of the mature 25S rRNA. This cleavage is then followed by shortening of the pre-rRNA up to position +7 (78). These first processing events are dependent on Rnt1p. The Δrnt1 strain, which displays a very strong slow-growth phenotype and temperature sensitivity (30), and the rnt1-1 mutant strain accumulate the primary transcript in vivo (2, 94). Rnt1p is a double-strand-specific endo-RNase that is homologous to bacterial RNase III (2, 8). In vivo, the Rnt1p cleavage sites map between nucleotide positions +14 and +49, which is in perfect agreement with the reported processing sites within the primary RNA pol I transcript (94). Moreover, recombinant Rnt1p is able to cleave a synthetic 3′ ETS substrate 21 nucleotides downstream of the 3′ end of the 25S rRNA (2), which is within the reported region where in vivo processing takes place. The so far unidentified product of the RNA82 gene is probably also required for 3′ ETS processing because the rna82-1 mutation affects 3′-end processing of transcripts derived from artificial rDNA minigene reporters. In the rna82-1 mutant strain, the majority of transcripts extend to nucleotide positions between +15 and +10, instead of being matured to the 3′ end of the 25S rRNA as observed for the wild-type strain (78).
The earliest processing event in the 35S pre-rRNA is the endonucleolytic cleavage at site A0, which is dependent on the U3 snoRNP (13, 69). In vitro, Rnt1p cleaves a model 5′ ETS substrate at site A0 in the absence of other factors and the rnt1-1 mutation leads to depletion of the 33S pre-rRNA (2). Therefore, it seems likely that Rnt1p is required for pre-rRNA processing at site A0. Recent data, however, seem to exclude a direct role for Rnt1p in cleavage at site A0 because, although kinetically delayed, cleavage at this site still occurs in the Δrnt1 mutant strain (94). Therefore, if Rnt1p is involved in cleavage at site A0, this function can be partially fulfilled by another endonuclease(s). This would then resemble very much the situation in Escherichia coli, where RNase III disruption is not lethal and other endonucleases carry out somewhat redundant functions (8). In addition, pre-rRNA processing at sites A1 and A2 is also delayed in the Δrnt1 strain (94). This could be the consequence of codepletion of the mature form of the U14 snoRNA, since Rnt1p is also involved in the correct processing of some snRNAs and snoRNAs from precursors, such as the dicistronic snR190-U14 precursor (1, 29, 30). However, the snR190-U14 precursor probably retains at least some functionality, since U14 is essential for cell viability and depletion of U14 does block pre-rRNA processing at sites A1 and A2 (113).
RNase MRP is an endo-RNase that cleaves pre-rRNA at site A3 in vivo and in vitro (32, 118, 158). The RNA component of RNase MRP, encoded by the NME1/RRP2 gene (115, 159, 160), is a unique snoRNA because it does not contain complementary sequences that allow base pairing with the pre-rRNA (see above). MRP RNA is structurally related to RNase P RNA; this suggests that RNase MRP arose in eukaryotes as a form of RNase P specialized for pre-rRNA processing (129). RNase MRP consists, in addition to MRP RNA, of nine integral protein subunits (Table 1) (27, 157), eight of these (Pop1p, Pop3p, Pop4p, Pop5p, Pop6p, Pop7p/Rpp2p, Pop8p, and Rpp1p) being shared between RNase MRP and RNase P (27). The only protein component unique to RNase MRP is Snm1p (157). Mutation in or depletion of the RNA or any of the protein components of RNase MRP, except Pop8p (not determined for Snm1p), leads to the inhibition of cleavage at site A3. Moreover, depletion of any protein component of RNase MRP, except Pop3p, leads to codepletion of MRP RNA (27, 33, 45, 119, 158, 168, 169). As a consequence, the synthesis of 5.8SS is impaired and the ratio of 5.8SS to 5.8SL, which is around 7:1 in wild-type cells (45), is strongly decreased. The same phenotype is seen in a mutant carrying a deletion of the A3 cleavage site (64). Intriguingly, cleavage at site A3 is nonessential for cell viability (64) whereas all known components of RNase MRP are essential. Therefore, it is likely that RNase MRP has additional, as yet unidentified, substrates (44, 45, 173). Early tRNA maturation reactions, including 5′-terminal processing by RNase P, occur in the nucleolus (16), which suggests the possibility of direct coordination between tRNA and ribosome biosynthesis. Interestingly, a mutation in the RNase P RNA subunit (rpr1) leads to the accumulation of an aberrant form of 5.8S rRNA that is 3′ extended for about 30 nucleotides (28). However, the precise role of RNase P in pre-rRNA processing remains obscure and it is possible that the rpr1 mutation affects pre-rRNA processing indirectly. Indeed, depletion of Rpr2p, an exclusive component of RNase P, decreases the stability of RNase P RNA and impairs 5′-end maturation of pre-tRNAs without affecting pre-rRNA processing (27).
It is noteworthy that the endo-RNases responsible for cleavage at processing sites A1, D, and A2 have not yet been identified. Moreover, it is only suspected that an endonucleolytic activity is required for processing at site B1L. In the case of ITS2 processing sites C1 and C2, it has to be assumed that at least one (if not both) of these sites is cleaved endonucleolytically. Unfortunately, no trans-acting factors that are involved in processing at sites B1L, C1, and C2 have been identified so far.
In contrast, the enzymes responsible for all exonucleolytic processing reactions have most likely been identified. The essential Rat1p and the nonessential Xrn1p are homologous and functionally equivalent proteins that have processive 5′→3′ exo-RNase activity in vitro (7, 76, 105, 165, 167). Xrn1p functions mainly in the cytoplasm, while Rat1p is a nuclear protein (66, 76). Xrn1p was also purified as a protein having DNA strand exchange and DNA G4 tetraplex-specific nuclease activity, and its gene was cloned by a number of different genetic selection techniques that are not obviously related (see reference 76 and references therein). Since the Δxrn1 strain is severely impaired in 5′→3′ degradation of mRNA (reviewed in reference 26), the myriad putative activities of Xrn1p can be rationalized by a model in which regulation of gene expression is globally deranged due to the inhibition of 5′→3′ mRNA decay pathways (76). Moreover, combination of the Δxrn1 mutation with mutations affecting 3′→5′ degradation of mRNAs leads to synthetic lethality, which confirms that the “essential” function of Xrn1p is its 5′→3′ exoribonucleolytic activity and further indicates that efficient mRNA turnover is required for cell viability (73). Rat1p has been isolated as required for the efficient nucleocytoplasmic trafficking of mRNAs (7), and it was also independently identified in two other, unrelated, genetic screens (47, 79). In analogy to the role of Xrn1p, it is suspected that the essential function of Rat1p is the 5′→3′ degradation of nuclear RNAs. The role of Xrn1p and Rat1p in pre-rRNA processing is the 5′→3′ exonucleolytic digestion of the 27SA3 pre-rRNA up to site B1S (7, 64). This reaction generates the 5′ end of the major form of 5.8S rRNA, the 5.8SS rRNA. Accordingly, 5′-extended forms of 5.8S rRNA accumulate in Δxrn1 single mutants and more drastically in Δxrn1 rat1-1 double mutants (64). In wild-type cells, 5.8SL rRNA, which is 7 to 8 nucleotides longer than 5.8SS, appears before 5.8SS rRNA. However, the synthesis of both 5.8S rRNA species is kinetically delayed in a Δxrn1 rat1-1 double mutant at the nonpermissive temperature (64); the reason for this is not clear, but it could reflect an involvement of Xrn1p and Rat1p in processing at site B1L. Another still obscure finding is that the putative RNA helicase Rok1p (see below) was isolated as a multicopy suppressor of the benomyl-hypersensitive phenotype of the Δxrn1 mutant strain (164). Xrn1p and Rat1p are also required for the degradation of several excised pre-rRNA spacer fragments, namely, the A0-A1, D-A2, and A2-A3 fragments (140, 166). The accumulation of these spacer fragments is in agreement with endonucleolytic processing at sites A0, A1, D, A2, and A3 (2, 32, 42, 64, 158, 192). Moreover, both Xrn1p and Rat1p, in conjunction with Rnt1p, are involved in the posttranscriptional processing of snoRNAs (such as snR190, U14, U18, U24, and several others) from larger precursors (30, 140, 144, 196).
Formation of the 3′ end of 5.8S rRNA from its 7S precursor occurs via a 3′→5′ exonucleolytic processing mechanism (127), which involves a protein complex called the exosome (126). This complex is composed of at least three 3′→5′ exonucleases (Rrp4p, Rrp41p/Ski6p, and Rrp44p) and eight putative 3′→5′ exonucleases (Csl4p, Mtr3p, Rrp40p, Rrp42p, Rrp43p, Rrp45p, Rrp46p, and Rrp6p) (5, 126). The modes of action of the three 3′→5′ exonucleases are distinct: Rrp4p acts by a hydrolytic mechanism and has distributive activity, whereas Rrp41p and Rrp44p have processive activities and act by a phosphorylytic or a hydrolytic mechanism, respectively (126). Mutation in or depletion of most of the exosome components impairs synthesis of 5.8S rRNA, and it leads to the accumulation of 3′-extended forms of the 5.8S rRNA, which may extend up to the 3′ end of the 7S pre-rRNA at site C2 (extension of around 140 nucleotides) (5, 126). Mutations in RRP6 (Rrp6p is homologous to the 3′→5′ exo-RNase RNase D from E. coli) result in the accumulation of a distinct 5.8S rRNA processing intermediate, termed 5.8S*, that has a normal 5′ end but retains about 30 nucleotides of ITS2 (22). Interestingly, this intermediate is quite similar to that detected upon mutation of the RNase P RNA (see above) (28). The exosome seems to be assisted by the putative RNA helicase Dob1p/Mtr4p (see below), since mutation in DOB1 or depletion of Dob1p and exosome components leads to a similar accumulation of the 7S pre-rRNA and depletion of the mature 5.8S rRNAs (42). Moreover, the slow-growth phenotype of the rrp4-1 mutant is synthetically enhanced by the dob1-1 mutation, which is in support of a functional interaction between the exosome and Dob1p (42). The exosome, together with Ski2p, which is the cytoplasmic homolog of Dob1p, is also required for the 3′→5′ degradation of cytoplasmic mRNAs (73). Therefore, the exosome may be involved in the degradation of nuclear RNA substrates other than the 7S pre-rRNA and even a role in nuclear poly(A)+ RNA turnover can be envisaged. Indeed, depletion of Dob1p or exosome components leads to the accumulation of the 5′-A0 spacer fragment (42), which is the only spacer fragment degraded in a 3′→5′ direction. However, the specific contribution of the different exosome components to the 3′→5′ exonucleolytic trimming of the 7S pre-rRNA, the 5′-A0 spacer fragment, and other possible RNA substrates remains to be determined. It is even likely that not all of the exosome’s exonucleases are simultaneously involved in the degradation of the same RNA substrate.
As mentioned in the Introduction, the pre-5S rRNA only needs to be processed at its 3′ end. Piper et al. proposed that the 3′ end of mature 5S rRNA is generated by a single endonucleolytic cleavage, which requires the product of the RNA82 gene (141). In a pulse-chase labeling experiment, rna82-1 mutant cells initially accumulate mainly the pre-5S rRNA, from which the additional nucleotides are then slowly removed by a 3′→5′ exonucleolytic mechanism to yield 5S rRNA species that are mostly only up to 3 nucleotides longer than in wild-type cells (141). A more recent study, however, disagrees with the above findings and rather supports a model in which 3′-end maturation of pre-5S rRNA relies exclusively on a 3′→5′ exonucleolytic activity, which simply trims the pre-5S rRNA to the mature form (112). In this model, the exonuclease activity is primarily blocked by a secondary structure (helix I) formed between the interacting 5′ and 3′ ends of the mature 5S rRNA. Association of the 5S rRNA with the 5S rRNA-binding protein Rpl5p, which results in the formation of the stable Rpl5p-5S RNP complex, appears not to directly influence the maturation process but rather seems to protect the 5S rRNA from degradation by other nucleases (43, 112).
PUTATIVE ATP-DEPENDENT RNA HELICASES
The largest class of trans-acting factors involved in ribosome biogenesis contains the RNA helicases of the DEAD-box and related families. These protein families are defined by several evolutionarily conserved motifs, and their members are involved in various RNA metabolic processes, including pre-mRNA splicing, translation initiation, RNA degradation, and ribosome biogenesis (40). Many of these proteins possess an RNA-dependent ATPase activity, which is sometimes only stimulated by specific RNA substrates (56, 134). Since an ATP-dependent RNA helicase activity could only be shown for a few of them (discussed in reference 71), they are collectively referred to as putative ATP-dependent RNA helicases. However, it is generally believed that putative RNA helicases act as ATP-dependent modulators of RNA structure (40).
Different functions can be envisaged for putative RNA helicases in ribosome biogenesis. (i) An RNA-unwinding activity could be required to establish and/or dissociate snoRNA–pre-rRNA base pairings, which are, in most cases, mutually exclusive with respect to the final folding of the rRNA in the mature ribosome. (ii) Putative RNA helicases may functionally assist endo- and exonucleases by rendering their substrates accessible for the processing reactions. (iii) Finally, they may recruit, rearrange, or dissociate trans-acting factors and r-proteins within preribosomal particles during the processing and assembly reactions by modulating specific intramolecular rRNA, rRNA-protein, or even protein-protein interactions. In the absence of such a putative RNA helicase, the lack or retardation of the required structural changes may lead to an abortive assembly, which can entail either disassembly of preribosomal particles and destabilization of pre-rRNA intermediates or accumulation of preribosomal particles and stabilization of pre-rRNA intermediates.
To date, 16 putative RNA helicases have been directly implicated in ribosome biogenesis in S. cerevisiae (40). Seven of them, Dbp4p (81), Dbp8p (37), Dhr1p (96), Dhr2p (96), Fal1p (89), Rok1p (191), and Rrp3p (133), are required for the early pre-rRNA cleavages at sites A0 to A2. Depletion of these putative RNA helicases leads to similar pre-RNA processing defects, even though these proteins carry out nonredundant functions. Therefore, it is likely that not all of them directly assist the cleavage reactions. The observed pre-rRNA processing phenotypes may also be the indirect consequence of defective assembly of an early preribosomal particle on the pathway to 40S ribosomal subunit formation. Unfortunately, data giving an indication of the functional environment of Dbp8p, Dhr1p, Dhr2p, Fal1p, or Rrp3p are still lacking. On the other hand, genetic observations suggest a functional interaction between Rok1p and both snR10 and Gar1p (191) and between Dbp4p and snoRNA U14 (114). However, there is no evidence for stable physical interactions between these putative RNA helicases and snoRNAs (81, 191) and they therefore probably associate only transiently. Recombinant Rrp3p (cited in reference 133) and Rok1p display ATPase activity in vitro (136, 143), and recombinant Dbp4p binds nonspecifically to RNA and has an intrinsic ATPase activity that is not stimulated by the addition of U14, 18S rRNA, or total RNA (81).
Nine putative RNA helicases act at different steps during the biogenesis of 60S ribosomal subunits. The nonessential Dbp3p is required for efficient pre-rRNA processing at site A3 (198). This site is located 5 nucleotides downstream of a proposed stable stem-loop structure (4, 205); thus, Dbp3p could be involved in the local unwinding of this structure, thereby facilitating the assembly of the RNase MRP complex at site A3 (198). Strains in which Dob1p/Mtr4p is defective have pre-rRNA processing defects that are similar to those of exosome mutants (see above) (42, 126), and the specific inhibition of the 3′→5′ exonucleolytic processing of the 7S pre-rRNA to mature 5.8S rRNA is most likely responsible for the underaccumulation of 60S ribosomal subunits in dob1 mutants. Given that Dob1p/Mtr4p does not tightly associate with the exosome, it may act as a cofactor that prevents stalling of the exosome by unwinding secondary structures (42, 172). The remaining seven putative RNA helicases are likely to be required for early, intermediate, or late structural rearrangement reactions within preribosomal particles that are mainly necessary for the formation of 60S ribosomal subunits. The pre-rRNA processing phenotypes observed upon depletion of Dbp6p and Dbp9p (instability of 35S pre-rRNA-derived processing intermediates and/or decreased formation and steady-state levels of 27SB precursors) suggest that these two proteins are essential for assembly reactions that occur within early preribosomal particles (36, 88). Moreover, dbp6 dbp9 double mutants are synthetically lethal and the slow-growth phenotype of some dbp6 mutants is exclusively suppressed by overexpression of Dbp9p (36). The nonessential Dbp7p (38), whose deletion confers a strong slow-growth phenotype, and Mak5p (206) most likely act downstream of Dbp6p and Dbp9p. Absence of Dbp7p or depletion of Mak5p results in a weaker decrease in 27SB pre-rRNA levels and in delayed conversion of the 27SA2 pre-rRNA to 27SB species. In contrast to the above phenotypes, depletion of Spb4p and Dbp10p leads to the accumulation of 27SB pre-rRNAs within 66S preribosomal particles (23, 41). The delayed conversion of 27SB species to mature 25S rRNA is more drastic in the absence of Spb4p; therefore, Spb4p may be responsible for structural rearrangements that facilitate cleavage at site C1 or C2. The role of Drs1p in the biogenesis of 60S ribosomal subunits is less clear, and it was only shown that formation of 25S rRNA is impaired in the cold-sensitive drs1-1 mutant (148). To characterize the functional environment of Drs1p, a genetic screen for synthetic lethality with drs1 alleles was carried out; this screen identified an essential nucleolar protein that is homologous to pescadillo, which is an essential gene for embryonic development in zebrafish (3, 149).
In contrast to the 16 putative RNA helicases described above, the involvement of the putative RNA helicase Sen1p in ribosome biogenesis is most likely indirect. Sen1p seems to be involved in pre-rRNA processing, since a mutation in SEN1 leads to delayed 35S pre-rRNA processing (186); however, the same mutation also affects the levels of intronic pre-tRNAs, snRNAs, and snoRNAs and furthermore leads to the mislocalization of several nucleolar proteins (185, 186). In addition, Sen1p has been reported to interact with both H/ACA- and C/D-box snoRNAs (186) and to be required for snoRNA maturation and stability (145).
FACTORS INVOLVED IN RIBOSOME ASSEMBLY?
The course of the assembly of the different r-proteins into preribosomal particles has been studied mainly by monitoring the kinetics of incorporation of individual r-proteins into cytoplasmic and nuclear ribosomal particles after pulse-labeling of yeast protoplasts with tritiated amino acids (93). These experiments indicate that a large number of r-proteins associate with the nucleolar preribosomes at early steps in ribosome maturation, whereas others assemble at later steps or are even only added in the cytoplasm (Fig. 2 and its legend) (93). It has also been shown that three large-subunit r-proteins on mature 60S subunits can exchange in vivo, and the exchangeability of Rpl10p/Qsr1p, in particular, which is required for 60S to 40S subunit joining, suggests the possibility of an additional translational regulatory mechanism (46, 209). Due to the lack of in vitro reconstitution assays for eukaryotic ribosomes, the above-mentioned studies could only be complemented by approaches that examined the abilities of the different r-proteins to either bind to pre-rRNA molecules in vitro (204) or dissociate from purified ribosomal particles by increasing concentration of ions (52, 106). The affinity of r-proteins for pre-rRNAs and the release of r-proteins by increasing concentration of ions are in general agreement with the order of their association with the preribosomal particles during in vivo ribosomal subunit assembly. However, the precise order of the assembly of the different r-proteins into preribosomal particles has not yet been unveiled. The roles of the protein trans-acting factors involved in ribosome assembly are even less clear. Moreover, it is often difficult to exclusively attribute an assembly function to a protein trans-acting factor, since ribosome assembly and pre-rRNA processing are intimately linked. Indeed, mutation in or depletion of r-proteins generally leads to a pre-rRNA processing phenotype (12, 128, 195). Therefore, we expect that some of the above-mentioned protein trans-acting factors also contribute in one way or another to ribosome assembly. In this section, we describe the protein trans-acting factors that could function mainly in promoting structural rearrangement reactions during the assembly of the preribosomal particles.
The function of only one protein trans-acting factor, Rrp7p, has so far been linked to the process of 40S ribosomal subunit assembly (Fig. 2). Depletion of Rrp7p leads to general inhibition of the early pre-rRNA cleavage reactions at A0 to A2, with the A2 site being the most affected. This results in a reduction of 18S rRNA synthesis and in a decreased ratio of 40S to 60S ribosomal subunits (12). Interestingly, the lethality of the Δrrp7 allele is suppressed by overexpression of the nearly identical proteins Rps27Ap and Rps27Bp (12), which belong to a small group of 40S subunit r-proteins that assemble late into pre-40S ribosomal particles (Fig. 2) (93). This suggests that Rrp7p is required for the correct assembly of Rps27A/Bp into pre-40S ribosomal particles (12).
Mutation in or depletion of protein trans-acting factors involved in the assembly of 60S ribosomal subunits often leads to an abortive assembly, which can either entail disassembly of pre-60S ribosomal particles and destabilization of pre-rRNA intermediates or accumulation of pre-60S ribosomal particles and stabilization of pre-rRNA intermediates. Disassembly of pre-60S ribosomal particles is thought to occur when the integrity of early-acting factors is affected, whereas accumulation of pre-rRNA intermediates can be attributed to the disfunctionality of late-acting factors (discussed in references 38, 41, and 88). In addition, it is conceivable that some late assembly events are also critical for the formation of stable pre-60S ribosomal particles or mature 60S ribosomal subunits. Depletion of Nop4p, Nop8p, and the putative RNA helicases Dbp6p and Dbp9p leads to decreased formation and steady-state levels of 27S pre-rRNAs (15, 36, 88, 170, 207). According to the above model, these protein trans-acting factors are supposed to act very early in the pathway of 60S ribosomal subunit assembly (Fig. 2). In agreement with a role of Dbp6p in early assembly events, two independent rpl3 mutants were identified in a synthetic lethal screen with conditional dbp6 alleles (91). RPL3 encodes the L3 r-protein of the large ribosomal subunit, which has been shown to belong to a group of r-proteins that associate early with preribosomal particles (Fig. 2) (93). Two further putative RNA helicases, Dbp7p and Mak5p, are most likely required for early assembly reactions downstream of Nop4p, Nop8p, Dbp6p, and Dbp9p (38, 206). In contrast, depletion of the putative RNA helicases Spb4p and Dbp10p leads to the accumulation of 27S pre-rRNAs within 66S preribosomal particles (23, 41). This is in agreement with their involvement in late assembly events. Delayed conversion of 27SB pre-rRNAs to mature 25S rRNAs and increased steady-state levels of 27SB pre-rRNAs are also seen upon depletion of Nip7p (208). Nip7p seems to be a multifunctional protein, since it is localized in the nucleus with nucleolar enrichment and in the cytoplasm, where it is associated with free 60S ribosomal subunits (208). Nip7p interacts with both the exclusively nucleolar protein Nop8p and the exosome component Rrp43p, and intriguingly, depletion of either of these proteins leads to pre-rRNA processing phenotypes that are clearly distinct from that observed upon depletion of Nip7p (126, 207). The ski6-2 mutation in another exosome component, Rrp41p/Ski6p, leads to the accumulation of a 38S particle containing 5′-truncated 25S rRNA but no 5.8S rRNA; otherwise, this mutant has normal amounts of 40S and 60S ribosomal subunits (14). This 38S particle corresponds either to an incompletely assembled or, more likely, to a partially degraded 60S ribosomal subunit. The latter origin of the 38S particle could be explained by deficient cytoplasmic turnover of “old” 60S ribosomal subunits due to the decreased 3′→5′ exonuclease activity of Ski6-2p.
The nonessential gene RSA1 was identified in a genetic screen for synthetic lethality with dbp6 alleles (90). In the Δrsa1 mutant, half-mer polysomes accumulate even though the pool of free 60S ribosomal subunits is only moderately decreased. This phenotype is similar to that observed in mutants defective in 60S to 40S subunit joining (50, 51). The presence of Rpl10p/Qsr1p in 60S ribosomal subunits is necessary for 60S to 40S subunit joining to occur (51). Accordingly, Rpl10p/Qsr1p is practically absent from free 60S ribosomal subunits in Δrsa1 mutant strains (90). Moreover, combination of the Δrsa1 and qsr1-1 mutations leads to a strong synthetic growth inhibition (90). Taken together, Rsa1p is likely to be involved in a late step of pre-60S ribosomal subunit assembly that is required for efficient recruitment of Rpl10p/Qsr1p. Sqt1p is a cytoplasmic protein containing multiple WD repeats that interacts strongly with Rpl10p/Qsr1p in a yeast two-hybrid system. Moreover, overexpression of Sqt1p suppresses the slow-growth phenotype of dominant negative qsr1 truncation mutants (50). Depletion of Sqt1p results in the formation of half-mer polysomes and decreased levels of Rpl10p/Qsr1p on free 60S ribosomal subunits (50). A likely explanation is that Sqt1p is involved in the assembly of Rpl10p/Qsr1p into 60S ribosomal subunits late in the assembly pathway. The function of Nmd3p is also genetically linked to Rpl10p/Qsr1p because dominant NMD3 mutants suppress the growth defect of a temperature-sensitive mutation in RPL10/QSR1 (77). Nmd3p is a cytoplasmic protein that sediments at the position of free 60S ribosomal subunits in sucrose gradients (67). Interestingly, the mature 25S rRNA is rapidly degraded in nmd3 mutant strains, as evidenced by pulse-chase analysis, which suggests that Nmd3p is required for the formation of stable 60S ribosomal subunits (67). The stability of 60S ribosomal subunits is also affected in strains depleted of Rpl5p, which assembles as the Rpl5p-5S RNP early into pre-60S ribosomal subunits (43).
UNCLASSIFIED PROTEIN TRANS-ACTING FACTORS
Rrp5p is perhaps one of the most interesting trans-acting factors belonging to this class. Rrp5p was identified in a screen for mutations showing synthetic lethality with the Δsnr10 allele. Rrp5p is the only factor that is simultaneously required for pre-rRNA processing at sites A0, A1, A2, and A3 (194) and therefore for the synthesis of both the mature 18S and 5.8SS rRNAs (194). Rrp5p is a large nucleolar protein composed of a modular structure of two different kinds of repeats. The N-terminal two-thirds of the protein contains 12 repeats of the so-called S1 RNA-binding motif (S1-RM), while the C-terminal part contains seven tetratricopeptide repeats (TPRs) (53, 179). S1-RM is most probably involved in the binding of single-stranded regions in the rRNA (24). The TPR motif is a degenerate sequence of 34 amino acids which can mediate inter- and intramolecular protein-protein interactions (104). Large portions of the S1-RM repeats and a short C-terminal fragment containing two TPRs can be individually deleted without dramatically affecting cell growth (53), suggesting some sort of functional redundancy among the repeats. Deletions in the S1-RM region specifically inhibit processing at site A3; therefore, this region of Rrp5p is required for the synthesis of 5.8SS rRNA (53). Mutations or deletions in the TPRs affect pre-rRNA processing at sites A0, A1, and A2, which leads to inhibition of 18S rRNA synthesis (53, 179). Most importantly, Rrp5p can be physically divided into two fragments that act in trans to functionally replace the integral protein (53, 179). One fragment contains most of the S1-RM repeats, and the other covers the rest of the protein. Thus, both mutational analysis and trans-complementation nicely demonstrate that the two domains of Rrp5p carry out two distinct functions. This supports a model in which Rrp5p acts as the “bridging factor” between the machineries required for A0 to A2 (snoRNP complexes) and A3 processing (RNase MRP complex) (173). So far, genetic analyses have shown a functional interaction between Rrp5p and some snoRNP components (snR10 and Rok1p) (53, 179, 194), but such an interaction with the RNase MRP complex still remains to be uncovered.
Two factors, Nop3p and Nsr1p, might mediate nucleocytoplasmic traffic of ribosomal components, although direct experimental evidence for such an involvement is still lacking. Nop3p (also Npl3p) is an essential nuclear (and nucleolar) member of the GAR domain protein family (150). In addition to its GAR domain, which is present in the C-terminal part of the protein, Nop3p contains two central RRMs (150). A more recent study indicates that Nop3p shuttles between the nucleus and the cytoplasm (107); however, the observed steady-state nuclear localization indicates that it must be rapidly reimported. In cells depleted of Nop3p, 27SB pre-rRNA processing is impaired and 27SB pre-rRNAs accumulate (150). In addition, nop3 mutants accumulate poly(A)+ RNA in the nucleus (107, 163). Therefore, it has been suggested that Nop3p acts as a carrier for poly(A)+ RNA export from the nucleus (107). Experimental evidence also suggests that Nop3p has an indirect role in protein uptake into the nucleus (19, 163). Taken together, the pre-rRNA processing defects observed upon Nop3p depletion can be explained in a model in which Nop3p is involved in nucleocytoplasmic transport of pre-60S ribosomes; however, the possibility cannot be excluded that Nop3p is required for the nuclear import of any r-protein or protein trans-acting factor. Nsr1p is a nucleolar protein that is structurally and functionally similar to mammalian nucleolin (110). Nucleolin is a major nucleolar protein that shuttles between the nucleus and the cytoplasm, recognizes nuclear localization signals, and participates in many processes involving RNA, including pre-rRNA processing (60, 110, 183). Like Nop3p, Nsr1p contains two central RRMs and a C-terminal GAR domain. The N-terminal part consists of a lysine-rich region followed by serine-rich segments and stretches enriched in acidic amino acids (85). Nsr1p is not essential for cell viability, but Δnsr1 cells are severely affected in growth (85, 110). In addition, due to delayed A0-to-A2 pre-rRNA processing, the Δnsr1 mutation results in reduced steady-state levels of 20S pre-rRNA and mature 18S rRNA and therefore in a deficiency of 40S relative to 60S ribosomal subunits (85, 111). As for Nop3p, the precise role of Nsr1p in ribosome biogenesis is unknown; Nsr1p either plays a direct role in pre-rRNA processing and/or ribosome assembly or participates in the import of an r-protein or another component required in the above-described processes (111).
Miscellaneous protein trans-acting factors such as Rrp1p (55) and Srd1p (65) have been identified genetically, whereas others were obtained by specific genetic screens that yielded classes of mutants which preferentially affect ribosome biogenesis; these classes include the drs, spb, and mak mutants (137, 147, 151). The drs (deficiency of ribosomal subunits) group was isolated by screening of a collection of cold-sensitive mutants for altered ratios of free 40S to 60S ribosomal subunits or qualitative changes in polysome profiles (147). The drs mutants were grouped into seven complementation groups, two affecting 60S ribosomal subunit production and five affecting 40S ribosomal subunit synthesis (147). Two DRS genes have been identified so far: DRS1 encodes a putative RNA helicase (see above) (148), and DRS2 encodes a plasma membrane ATPase whose function in ribosome biogenesis is unknown (147, 161). The spb mutants are extragenic suppressors of a mutation in PAB1, which encodes poly(A)+ binding protein 1 (151). Except for spb8 (17) and pbp1 (121), all described spb mutations interfere with synthesis of 60S ribosomal subunits (151). Spb2p is Rpl39p (152), Spb1p is a candidate rRNA methyltransferase (see above) (92), and Spb4p is a putative RNA helicase (see above) (41, 152). About 30 different mak complementation groups were identified as mutations that lead to loss of the M1 double-stranded RNA virus encoding the killer toxin (199). Like the spb mutants, most mak mutants are affected in the biogenesis of 60S ribosomal subunits and no mak mutants have been identified that are deficient in 40S ribosomal subunits (49, 137, 199). Some MAK genes encode r-proteins (i.e., Rpl3p, Rpl8Ap, and Rpl42Ap), while Mak5p, a putative RNA helicase (see above), and Mak21p are involved in the biogenesis of 60S ribosomal subunits (49, 206). Further studies are required to reveal how qualitative and quantitative alterations of 60S ribosomal subunits can specifically rescue a pab1 mutation or impede the propagation of M1 double-stranded RNA viruses.
CONCLUDING REMARKS AND PERSPECTIVES
Yeast molecular genetics has been a valuable tool for the identification and characterization of a large number of protein trans-acting factors involved in eukaryotic ribosome biogenesis. However, we still do not understand the precise functions of most of them and it is likely that many more protein trans-acting factors play a role in this complex process. Therefore, different biochemical and genetic methods, including the systematic functional analysis of yeast open reading frames, have to be exploited in order first to identify all of the protein trans-acting factors that are directly involved in ribosome biogenesis and second to reveal the network of physical interactions among the protein trans-acting factors themselves and their interactions with r-proteins and/or pre-rRNAs. This knowledge is a prerequisite for tackling the challenge of understanding the timing of the action and the substrate specificity of the different protein trans-acting factors during the pre-rRNA processing and ribosome assembly reactions. These efforts should allow us to eventually draw a more detailed picture of the assembly of the different r-proteins and the pre-rRNAs into preribosomes and their maturation to export- and translation-competent ribosomal subunits.
ACKNOWLEDGMENTS
We thank our many colleagues who have contributed unpublished results to this review, and we apologize to all those whose work could not be cited owing to space limitations. We are grateful to K. Tanner, D. Tollervey, and J. Venema for stimulating discussions and R. Boeck and K. Tanner for critical reading of the manuscript. J.D.L.C. thanks A. Vioque for encouragement.
The work in the authors’ laboratory was funded by grants from the Swiss National Science Foundation and the University of Geneva to P.L. We gratefully acknowledge C. Georgopoulos for support. J.D.L.C. thanks the MEC (Spain) for financial support.
REFERENCES
- 1.Abou Elela S, Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 3.Allende M L, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- 4.Allmang C, Henry Y, Wood H, Morrissey J P, Petfalski E, Tollervey D. Recognition of cleavage site A2 in the yeast pre-rRNA. RNA. 1996;2:51–62. [PMC free article] [PubMed] [Google Scholar]
- 5.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allmang C, Tollervey D. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J Mol Biol. 1998;278:67–78. doi: 10.1006/jmbi.1998.1693. [DOI] [PubMed] [Google Scholar]
- 7.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 8.Apirion D, Miczak A. RNA processing in prokaryotic cells. Bioessays. 1993;15:113–120. doi: 10.1002/bies.950150207. [DOI] [PubMed] [Google Scholar]
- 9.Bachellerie J-P, Cavaillé J. Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998. pp. 255–272. [Google Scholar]
- 10.Bagni C, Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J Biol Chem. 1998;273:10868–10873. doi: 10.1074/jbc.273.18.10868. [DOI] [PubMed] [Google Scholar]
- 11.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 12.Baudin-Baillieu A, Tollervey D, Cullin C, Lacroute F. Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol Cell Biol. 1997;17:5023–5032. doi: 10.1128/mcb.17.9.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benard L, Carroll K, Valle R C P, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergès T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertrand E, Houser-Scott F, Kendall A, Singer R H, Engelke D R. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeck R, Lapeyre B, Brown C E, Sachs A B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bortolin M-L, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bossie M A, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bousquet-Antonelli C, Henry Y, Gélugne J-P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brand R C, Klootwijk J, van Steenbergen T J M, de Kok A J, Planta R J. Secondary methylation of yeast ribosomal precursor RNA. Eur J Biochem. 1977;75:311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- 22.Briggs M W, Burkard K T D, Butler J S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 23.Burger, F., M.-C. Daugeron, and P. Linder. 1999. Unpublished data.
- 24.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 25.Cadwell C, Yoon H-J, Zebarjadian Y, Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol Cell Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chamberlain J R, Pagán-Ramos E, Kindelberger D W, Engelke D R. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chanfreau G, Abou Elela S, Ares M, Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chernoff Y O, Vincent A, Liebman S W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu S, Archer R H, Zengel J M, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu S, Zengel J M, Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- 34.Clark M W, Yip M L R, Campbell J, Abelson J. SSB-1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J Cell Biol. 1990;111:1741–1751. doi: 10.1083/jcb.111.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham P R, Richard R B, Weitzmann C J, Nurse K, Ofengand J. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S ribosomal subunit of Escherichia coli. Biochimie. 1991;73:789–796. doi: 10.1016/0300-9084(91)90058-9. [DOI] [PubMed] [Google Scholar]
- 36.Daugeron, M.-C., D. Kressler, and P. Linder. 1999. Unpublished data.
- 37.Daugeron, M.-C., and P. Linder. 1999. Unpublished data.
- 38.Daugeron M-C, Linder P. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA. 1998;4:566–581. doi: 10.1017/s1355838298980190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Beus E, Brockenbrough J S, Hong B, Aris J P. Yeast NOP2 encodes an essential nucleolar protein with homology to a human proliferation marker. J Cell Biol. 1994;127:1799–1813. doi: 10.1083/jcb.127.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 41.de la Cruz J, Kressler D, Rojo M, Tollervey D, Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deshmukh M, Tsay Y-F, Paulovich A G, Woolford J L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dichtl B, Stevens A, Tollervey D. Lithium toxicity in yeast is due the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dick F A, Eisinger D P, Trumpower B L. Exchangeability of Qsr1p, a large ribosomal subunit protein required for subunit joining, suggests a novel translational regulatory mechanism. FEBS Lett. 1997;419:1–3. doi: 10.1016/s0014-5793(97)01402-6. [DOI] [PubMed] [Google Scholar]
- 47.Di Segni G, McConaughy B L, Shapiro R A, Aldrich T L, Hall B D. TAP1, a yeast gene that activates the expression of a tRNA gene with a defective internal promoter. Mol Cell Biol. 1993;13:3424–3433. doi: 10.1128/mcb.13.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edskes H E, Ohtake Y, Wickner R B. Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60S ribosomal subunit biogenesis. J Biol Chem. 1998;273:28912–28920. doi: 10.1074/jbc.273.44.28912. [DOI] [PubMed] [Google Scholar]
- 50.Eisinger D P, Dick F A, Denke E, Trumpower B L. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1. Mol Cell Biol. 1997;17:5146–5155. doi: 10.1128/mcb.17.9.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisinger D P, Dick F A, Trumpower B L. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El-Baradi T T A L, Raué H A, de Regt V C H F, Planta R J. Stepwise dissociation of yeast 60S ribosomal subunits by LiCl and identification of L25 as a primary 26S rRNA binding protein. Eur J Biochem. 1984;144:393–400. doi: 10.1111/j.1432-1033.1984.tb08477.x. [DOI] [PubMed] [Google Scholar]
- 53.Eppens N A, Rensen S, Granneman S, Raué H A, Venema J. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA. 1999;5:779–793. doi: 10.1017/s1355838299990313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eustice D C, Wilhelm J M. Fidelity of the eukaryotic codon-anticodon interaction: interference by aminoglycoside antibiotics. Biochemistry. 1984;23:1462–1467. doi: 10.1021/bi00302a019. [DOI] [PubMed] [Google Scholar]
- 55.Fabian G R, Hopper A K. RRP1, a Saccharomyces cerevisiae gene affecting rRNA processing and production of mature ribosomal subunits. J Bacteriol. 1987;169:1571–1578. doi: 10.1128/jb.169.4.1571-1578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuller-Pace F V, Nicol S M, Reid A D, Lane D P. DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 58.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 59.Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Green R, Noller H F. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry. 1999;38:1772–1779. doi: 10.1021/bi982246a. [DOI] [PubMed] [Google Scholar]
- 63.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gélugne J-P, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hess S M, Stanford D R, Hopper A K. SRD1, S. cerevisiae gene affecting pre-rRNA processing contains a C2/C2 zinc finger motif. Nucleic Acids Res. 1994;22:1265–1271. doi: 10.1093/nar/22.7.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heyer W-D, Johnson A W, Reinhart U, Kolodner R D. Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol Cell Biol. 1995;15:2728–2736. doi: 10.1128/mcb.15.5.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ho J H-N, Johnson A W. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong B, Brockenbrough J S, Wu P, Aris J P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hughes J M X, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in Ran-cycle and nucleoporin mutants. J Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 72.Isaac C, Yang Y, Meier U T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacobs Anderson J S, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and Gβ domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jansen R P, Hurt E C, Kern H, Lehtonen H, Carmo-Fonseca M, Lapeyre B, Tollervey D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol. 1991;113:715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karl T, Önder K, Kodzius R, Pichová A, Wimmer H, Thür A, Hundsberger H, Löffler M, Klade T, Beyer A, Breitenbach M, Koller L. GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr Genet. 1999;34:419–429. doi: 10.1007/s002940050416. [DOI] [PubMed] [Google Scholar]
- 78.Kempers-Veenstra A E, Oliemans J, Offenberg H, Dekker A F, Piper P W, Planta R J, Klootwijk J. 3′-end formation of transcripts from the yeast rRNA operon. EMBO J. 1986;5:2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kenna M, Stevens A, McCammon M, Douglas M G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khaitovich P, Tenson T, Kloss P, Mankin A S. Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA. Biochemistry. 1999;38:1780–1788. doi: 10.1021/bi9822473. [DOI] [PubMed] [Google Scholar]
- 81.King, T. H., and M. J. Fournier. 1999. Personal communication.
- 82.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 83.Kiss-László Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klootwijk J, Planta R J. Analysis of the methylation sites in yeast ribosomal RNA. Eur J Biochem. 1973;39:325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- 85.Kondo K, Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992;267:16252–16258. [PubMed] [Google Scholar]
- 86.Koonin E V. Prediction of an rRNA methyltransferase domain in human tumor-specific nucleolar protein p120. Nucleic Acids Res. 1994;22:2476–2478. doi: 10.1093/nar/22.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koonin E V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kressler D, de la Cruz J, Rojo M, Linder P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1855–1865. doi: 10.1128/mcb.18.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kressler D, Doère M, Rojo M, Linder P. Synthetic lethality with conditional dbp6 alleles identifies the previously uncharacterized RSA1 gene, encoding a protein involved in a late nucleoplasmic step of 60S-ribosomal-subunit assembly. Mol Cell Biol. 1999;19:8633–8645. doi: 10.1128/mcb.19.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kressler, D., and P. Linder. 1999. Unpublished data.
- 92.Kressler, D., M. Rojo, P. Linder, and J. de la Cruz. Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 93.Kruiswijk T, Planta R J, Krop J M. The course of the assembly of ribosomal subunits in yeast. Biochim Biophys Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 94.Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lafontaine D, Delcour J, Glasser A-L, Desgrès J, Vandenhaute J. The DIM1 gene responsible for the conserved m6,2Am6,2A dimethylation in the 3′-terminal loop of 18S rRNA is essential in yeast. J Mol Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 96.Lafontaine, D., and D. Tollervey. 1999. Personal communication.
- 97.Lafontaine D, Tollervey D. trans-acting factors in yeast pre-rRNA and pre-snoRNA processing. Biochem Cell Biol. 1995;73:803–812. doi: 10.1139/o95-088. [DOI] [PubMed] [Google Scholar]
- 98.Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 99.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lafontaine D L J, Preiss T, Tollervey D. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol. 1998;18:2360–2370. doi: 10.1128/mcb.18.4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lafontaine D L J, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 102.Lafontaine D L J, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lafontaine D L J, Tollervey D. Regulatory aspects of rRNA modification and pre-rRNA processing. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998. pp. 281–288. [Google Scholar]
- 104.Lamb J R, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 105.Larimer F W, Stevens A. Disruption of the gene XRN1, coding for a 5′→3′ exoribonuclease, restricts yeast cell growth. Gene. 1990;95:85–90. doi: 10.1016/0378-1119(90)90417-p. [DOI] [PubMed] [Google Scholar]
- 106.Lee J C, Anderson R, Yeh Y C, Horowitz P M. Extraction of proteins from Saccharomyces cerevisiae ribosomes under nondenaturing conditions. Arch Biochem Biophys. 1985;237:292–299. doi: 10.1016/0003-9861(85)90280-2. [DOI] [PubMed] [Google Scholar]
- 107.Lee M S, Henry M, Silver P A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 108.Lee S J, Baserga S J. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc Natl Acad Sci USA. 1997;94:13536–13541. doi: 10.1073/pnas.94.25.13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee S J, Baserga S J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee W-C, Xue Z, Mélèse T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991;113:1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee W-C, Zabetakis D, Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee Y, Nazar R N. Ribosomal 5S rRNA maturation in Saccharomyces cerevisiae. J Biol Chem. 1997;272:15206–15212. doi: 10.1074/jbc.272.24.15206. [DOI] [PubMed] [Google Scholar]
- 113.Li H V, Zagorski J, Fournier M J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liang W-Q, Clark J A, Fournier M J. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol Cell Biol. 1997;17:4124–4132. doi: 10.1128/mcb.17.7.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lindahl L, Archer R H, Zengel J M. A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115a.Linder, P. 6 July 1999, posting date; 7 September 1999, revision date. [Online.] http://www.expasy.ch/linder/proteins.html. [1 October 1999, last date accessed.]
- 116.Lowe T M, Eddy S R. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 117.Lübben B, Fabrizio P, Kastner B, Lührmann R. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J Biol Chem. 1995;270:11549–11554. doi: 10.1074/jbc.270.19.11549. [DOI] [PubMed] [Google Scholar]
- 118.Lygerou Z, Allmang C, Tollervey D, Séraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 119.Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 120.Maden B E H, Hughes J M X. Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem. Chromosoma. 1997;105:391–400. doi: 10.1007/BF02510475. [DOI] [PubMed] [Google Scholar]
- 121.Mangus D A, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein regulates polyadenylation. Mol Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meier, U. T. 1999. Personal communication.
- 123.Meier U T. Comparison of the rat nucleolar protein Nopp140 with its yeast homolog SRP40. J Biol Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- 124.Meier U T, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mélèse T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 126.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 127.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 128.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Morrissey J P, Tollervey D. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- 130.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 132.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 133.O’Day C L, Chavanikamannil F, Abelson J. 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res. 1996;24:3201–3207. doi: 10.1093/nar/24.16.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.O’Day C L, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 135.Ofengand J, Fournier M J. The pseudouridine residues of rRNA: number, location, biosynthesis, and function. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998. pp. 229–253. [Google Scholar]
- 136.Oh J-Y, Kim J. ATP hydrolysis activity of the DEAD box protein Rok1p is required for in vivo ROK1 function. Nucleic Acids Res. 1999;27:2753–2759. doi: 10.1093/nar/27.13.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ohtake Y, Wickner R B. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Palmer E, Wilhelm J M, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 139.Parker R, Simmons T, Shuster E O, Siliciano P G, Guthrie C. Genetic analysis of small nuclear RNAs in Saccharomyces cerevisiae: viable sextuple mutant. Mol Cell Biol. 1988;8:3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Piper P W, Bellatin J A, Lockheart A. Altered maturation of sequences at the 3′ terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J. 1983;2:353–359. doi: 10.1002/j.1460-2075.1983.tb01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Planta R J, Mager W H. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 143.Puras-Lutzke, R. A., and J. Venema. 1999. Personal communication.
- 144.Qu L-H, Henras A, Lu Y-J, Zhou H, Zhou W-X, Zhu Y-Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J-P. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol Cell Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rasmussen T P, Culbertson M R. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Raué H A, Planta R J. Ribosome biogenesis in yeast. Prog Nucleic Acid Res Mol Biol. 1991;41:89–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- 147.Ripmaster T L, Vaughn G P, Woolford J L., Jr DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7901–7912. doi: 10.1128/mcb.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ripmaster T L, Vaughn G P, Woolford J L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Roman, J., C. Adams, P. Harnpicharnchai, and J. L. Woolford, Jr. 1999. Personal communication.
- 150.Russell I D, Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992;119:737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 152.Sachs A B, Davis R W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 153.Samarsky D A, Fournier M J. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:161–164. doi: 10.1093/nar/27.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 156.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schmitt M E, Clayton D A. Characterization of a unique protein component of yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain. Genes Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- 158.Schmitt M E, Clayton D A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schmitt M E, Clayton D A. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP and essential for cell viability. Genes Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- 160.Shuai K, Warner J R. A temperature sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res. 1991;19:5059–5064. doi: 10.1093/nar/19.18.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Siegmund A, Grant A, Angeletti C, Malone L, Nichols J W, Rudolph H K. Loss of Drs2p does not abolish transfer of fluorescence-labeled phospholipids across the plasma membrane of Saccharomyces cerevisiae. J Biol Chem. 1998;273:34399–34405. doi: 10.1074/jbc.273.51.34399. [DOI] [PubMed] [Google Scholar]
- 162.Singh A, Ursic D, Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 163.Singleton D R, Chen S, Hitomi M, Kumagai C, Tartakoff A M. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- 164.Song Y, Kim S, Kim J. ROK1, a high-copy-number plasmid suppressor of kem1, encodes a putative ATP-dependent RNA helicase in Saccharomyces cerevisiae. Gene. 1995;166:151–154. doi: 10.1016/0378-1119(96)80010-2. [DOI] [PubMed] [Google Scholar]
- 165.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′→3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 166.Stevens A, Hsu C L, Isham K R, Larimer F W. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J Bacteriol. 1991;173:7024–7028. doi: 10.1128/jb.173.21.7024-7028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stevens A, Poole T L. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J Biol Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 168.Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 1997;11:2926–2937. doi: 10.1101/gad.11.21.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stolc V, Katz A, Altman S. Rpp2, an essential protein subunit of nuclear RNase P, is required for processing of precursor tRNAs and 35S precursor rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:6716–6721. doi: 10.1073/pnas.95.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sun C, Woolford J L., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994;13:3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sun C, Woolford J L., Jr The yeast nucleolar protein Nop4p contains four RNA recognition motifs necessary for ribosome biogenesis. J Biol Chem. 1997;272:25345–25352. doi: 10.1074/jbc.272.40.25345. [DOI] [PubMed] [Google Scholar]
- 172.Tollervey, D. 1999. Personal communication.
- 173.Tollervey D. Genetic and biochemical analyses of yeast RNase MRP. Mol Biol Rep. 1996;22:75–79. doi: 10.1007/BF00988709. [DOI] [PubMed] [Google Scholar]
- 174.Tollervey D. trans-acting factors in ribosome synthesis. Exp Cell Res. 1996;229:226–232. doi: 10.1006/excr.1996.0364. [DOI] [PubMed] [Google Scholar]
- 175.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 177.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 179.Torchet C, Jacq C, Hermann-Le Denmat S. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA. 1998;4:1636–1652. doi: 10.1017/s1355838298981511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Trapman J, Planta R J. Maturation of ribosomes in yeast. I. Kinetic analysis by labelling of high molecular weight rRNA species. Biochim Biophys Acta. 1976;442:265–274. doi: 10.1016/0005-2787(76)90301-4. [DOI] [PubMed] [Google Scholar]
- 181.Trapman J, Planta R J, Raué H A. Maturation of ribosomes in yeast. II. Position of the low molecular weight rRNA species in the maturation process. Biochim Biophys Acta. 1976;442:275–284. doi: 10.1016/0005-2787(76)90302-6. [DOI] [PubMed] [Google Scholar]
- 182.Trapman J, Retèl J, Planta R J. Ribosomal precursor particles from yeast. Exp Cell Res. 1975;90:95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- 183.Tuteja R, Tuteja N. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- 184.Udem S A, Warner J R. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem. 1973;248:1412–1416. [PubMed] [Google Scholar]
- 185.Ursic D, DeMarini D J, Culbertson M R. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol Gen Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- 186.Ursic D, Himmel K L, Gurley K A, Webb F, Culbertson M R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.van der Sande C A F M, Kulkens T, Kramer A B, de Wijs I J, van Heerikhuizen H, Klootwijk J, Planta R J. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989;17:9127–9146. doi: 10.1093/nar/17.22.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.van Nues R W, Venema J, Rientjes J M J, Dirks-Mulder A, Raué H A. Processing of eukaryotic pre-rRNA: the role of the transcribed spacers. Biochem Cell Biol. 1995;73:789–801. doi: 10.1139/o95-087. [DOI] [PubMed] [Google Scholar]
- 189.Veldman G M, Klootwijk J, de Jonge P, Leer R J, Planta R J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980;8:5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Venema, J. 1999. Personal communication.
- 191.Venema J, Bousquet-Antonelli C, Gelugne J-P, Caizergues-Ferrer M, Tollervey D. Rok1p is a putative RNA helicase required for rRNA processing. Mol Cell Biol. 1997;17:3398–3407. doi: 10.1128/mcb.17.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Venema J, Henry Y, Tollervey D. Two distinct recognition signals define the site of endonucleolytic cleavage at the 5′-end of yeast 18S rRNA. EMBO J. 1995;14:4883–4892. doi: 10.1002/j.1460-2075.1995.tb00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 194.Venema J, Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 195.Vilardell J, Warner J R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Villa T, Ceradini F, Presutti C, Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Watkins N J, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weaver P L, Sun C, Chang T-H. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol Cell Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wiederkehr T, Prétôt R F, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Woolford J L, Jr, Warner J R. The ribosome and its synthesis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 202.Wu P, Brockenbrough J S, Metcalfe A C, Chen S, Aris J P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu P, Brockenbrough J S, Paddy M R, Aris J P. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene. 1998;220:109–117. doi: 10.1016/s0378-1119(98)00330-8. [DOI] [PubMed] [Google Scholar]
- 204.Yeh L-C C, Lee J C. Yeast ribosomal proteins L4, L17, L20 and L25 exhibit different binding characteristics for the yeast 35S precursor rRNA. Biochim Biophys Acta. 1998;1443:139–148. doi: 10.1016/s0167-4781(98)00202-4. [DOI] [PubMed] [Google Scholar]
- 205.Yeh L-C C, Thweatt R, Lee J C. Internal transcribed spacer 1 of the yeast precursor ribosomal RNA. Higher order structure and common structural motifs. Biochemistry. 1990;29:5911–5918. doi: 10.1021/bi00477a005. [DOI] [PubMed] [Google Scholar]
- 206.Zagulski, M., A.-M. Becam, D. Kressler, J. Rytka, and C. J. Herbert. 1999. Unpublished data.
- 207.Zanchin N I T, Goldfarb D S. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol Cell Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zanchin N I T, Roberts P, DeSilva A, Sherman F, Goldfarb D S. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zinker S, Warner J R. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J Biol Chem. 1976;251:1799–1807. [PubMed] [Google Scholar]