Abstract
A perilous increase in the number of bacterial infections has led to developing throngs of antibiotics for increasing the quality and expectancy of life. Pseudomonas aeruginosa is becoming resistant to all known conventional antimicrobial agents thereby posing a deadly threat to the human population. Nowadays, targeting virulence traits of infectious agents is an alternative approach to antimicrobials that is gaining much popularity to fight antimicrobial resistance. Quorum sensing (QS) involves interspecies communication via a chemical signaling pathway. Under this mechanism, cells work in a concerted manner, communicate with each other with the help of signaling molecules called auto-inducers (AI). The virulence of these strains is driven by genes, whose expression is regulated by AI, which in turn acts as transcriptional activators. Moreover, the problem of antibiotic-resistance in case of infections caused by P. aeruginosa becomes more alarming among immune-compromised patients, where the infectious agents easily take over the cellular machinery of the host while hidden in the QS mediated biofilms. Inhibition of the QS circuit of P. aeruginosa by targeting various signaling pathways such as LasR, RhlR, Pqs, and QScR transcriptional proteins will help in blocking downstream signal transducers which could result in reducing the bacterial virulence. The anti-virulence agent does not pose an immediate selective pressure on growing bacterium and thus reduces the pathogenicity without harming the target species. Here, we review exclusively, the growing emergence of multi-drug resistant (MDR) P. aeruginosa and the critical literature survey of QS inhibitors with their potential application of blocking P. aeruginosa infections.
Key Words: Anti-virulence drugs, Biofilm inhibitors, Multidrug resistance, Pseudomonas aeruginosa, Quorum sensing inhibitors
Introduction
In the last few decades, there has been an alarming increase of reports documented for microbial infections. The mortality caused by pathogenic microorganisms that are currently targeted through known antimicrobials is also a matter of great concern as the microbial populations (bacteria, fungi, viruses, and parasites) have developed strategies to combat antimicrobial drugs worldwide. This has led to an ineffective treatment regime and resulted in the development of resistant strains of microorganisms causing deadly infections. Especially, these resistant microbes have shown fragile access in immune-compromised patients. In this category, Pseudomonas aeruginosa is more frequently seen to be associated with healthcare infections (1-3). The versatility of this pathogen to cause several infections is accepted worldwide as it majorly affects aged/immune-compromised patients (elderly and infant patients), HIV patients, individuals undergoing organ transplantation, and people with severe burns and wounds (4, 5). Unlike other bacteria, MDR opportunistic pathogen, P. aeruginosa, can grow in niches with high antibiotic pressure as well as may disturb the host-microbiota that may lead to an increase in bacterial virulence or pathogenicity. This causes the bacterium to survive in adverse conditions thus causing high morbidity and mortality due to antibiotic resistance (6, 7). Being a multidrug-resistant strain, P. aeruginosa is becoming more difficult to eradicate. The resistance acquired by this pathogen towards several antibiotics is majorly mediated through two types of mechanisms (8). The first mechanism involves a transfer of plasmid among bacteria carrying genes that express β-lactamases or aminoglycosides modifying enzymes (9). The second mechanism involves the mutation in the bacterial genome that causes a targeted mutation in the pathogen. For instance, the gyrase gene present in bacterial membranes is responsible for quinolone resistance due to variable expression of transport protein (regulation of efflux pumps and porins) (10). In both cases, the pathogen develops resistance against the antibiotics that affect bacterial growth; subsequently, bacteria develop their community for enhanced survival which serves as a shield reducing the antimicrobial compound’s activity. For this reason, a different approach is needed to be developed for blocking P. aeruginosa infections without interfering with the growth cycle of the pathogen. In recent times, the scientific community identified a novel and efficient strategy called “anti-virulence strategy” that focuses on the inhibition of expression of virulence factors that causes acute and chronic infections, without killing a pathogen (11-13). This non-killing approach renders a low rate of resistance as the survival of pathogens will not be affected by the active drug itself. Thus, in turn, the bacterial community would not be increased to inactivate the active drugs (14, 15). There are several review articles in the literature which are based on QS in P. aeruginosa and its associated virulence. Recently, a study (2019) highlighted the importance of P. aeruginosa biofilm and its relationship with QS (16). Similarly, another study (2017) described the importance of bacterial QS that can be targeted to modulate virulence among pathogens (17). Along similar lines, a study (2017) also supports the role played by QS in P. aeruginosa virulence (18). On the other hand, recently a review (2019) highlighted the importance of naturally derived quorum sensing inhibitors (QSIs) in blocking different signaling pathways in P. aeruginosa (19). A systematic review of the various signaling pathways and QS regulators in P. aeruginosa was published by Venturi (2006) (20).
We have performed an in-depth literature survey on the QS process in P. aeruginosa and its regulation. In this review, we have compiled the data based on various statistical reports published on the growing emergence of resistance in P. aeruginosa among clinical samples in varied timelines. This study will help in understanding how resistance develops in this organism for different categories of antibiotics. In addition to this, the current scenario of resistance patterns is alarming and reflects the dire need to develop anti-pseudomonal drugs. We also tried to review and gather the literature on the investigated QSI compounds (chemical and natural origin) targeting LasR, RhL, Pqs, and biofilms to mitigate P. aeruginosa infections in an alternative way. This review will help the researchers working from the biological or chemical point of view to understand the growing prevalence of antibiotic-resistant P. aeruginosa and the ways to curb these resistant strains by a process of QS inhibition. It will indeed help clinicians and public health professionals to improve their knowledge of the sensitivity or susceptibility of different antibiotics against resistant strains of P. aeruginosa.
Prevalence of drug-resistant of P. aeruginosa
P. aeruginosa is an opportunistic microorganism that causes infection among ill patients, immune-compromised patients, patients compromised by age (infant and elderly patients). Data from the National Nosocomial Infections Surveillance System from 1986-2003 reported P. aeruginosa as the second most common cause of pneumonia (18.1%), the third most common cause of urinary tract infection (16.3%), and the eighth-most frequently isolated pathogen from the bloodstream (3.4%) (21). While the overall proportion of infections caused by P. aeruginosa remained stable during the 1986-2003 period, however, the proportion of resistant isolates had shown an alarming increase in the consequent years. P. aeruginosa resistance to imipenem, quinolones, and third-generation cephalosporins increased by 15.0, 9.0, and 20.0%, respectively. Similarly, a national surveillance study of intensive care unit (ICU) patients from 1993 to 2002, reported a significant increase in multidrug-resistance towards at least three to four agents like imipenem, ceftazidime, ciprofloxacin, and tobramycin. These infections are often problematic, life-threatening, and cause a large number of deaths because of their inherent ability to resist all classes of antimicrobial agents (22, 23). A study was conducted at National Taiwan University Hospital (NTUH) in 2006 where the PDR (pan drug-resistant) strain of P. aeruginosa was isolated, this strain has shown resistance to all effective antimicrobial agents including cefepime, ceftazidime, imipenem, meropenem, piperacillin-tazobactam, ciprofloxacin, and levofloxacin leading to resistance of P. aeruginosa to all commercially available antimicrobial agents in Taiwan (24). Five years after this report, a national survey on infectious-diseases was conducted by the Infectious Disease Society of America (IDSA), Emerging Infections Network, in 2011, where it was found that more than 60.0% of participants are reported to have infections with a pan-resistant infectious agent, which is untreatable. Many public health organizations have already declared that the human population will face the “catastrophic consequences” of the antibiotic resistance era which will cause havoc for the human civilization (25, 26). Various global organizations like the Centers for Disease Control and Prevention (CDC), IDSA, World Economic Forum, and the World Health Organization (WHO) have announced antibiotic resistance to be a global public health concern (6).
Researchers (2012) observed that the prevalence of P. aeruginosa resistant isolates to antimicrobials has increased considerably and the resistance rate of P. aeruginosa to antimicrobials such as amikacin, ceftazidime, cefepime, imipenem, and ciprofloxacin was found to be 53.3%, 43.3%, 40.0%, 40.0%, and 33.3%, respectively (27). A study reported in 2014, describes the following resistance rates to cefepime 64.8%, piperacillin 45%, ciprofloxacin 38.9%, levofloxacin 36.1%, gentamicin 37.3%, and amikacin 30% (28). In 2014, EARSS reports showed a high percentage of resistance in P. aeruginosa in eastern and southern parts of Europe especially in Germany, Hungary, and Slovakia (29). In 2015, another study conducted for three years from 2013 to 2015, examined P. aeruginosa isolates against various antimicrobial agents and reported increasing resistance to a variety of antibiotics, including third and fourth generation cephalosporins such as ceftazidime and cefepime, respectively. A high level of resistance has been reported to β-lactam antibiotics in the United States, Europe, and South America. In the research period, resistance developed by cefepime was significantly increased each year, i.e. 31.6% in 2013, 44.2% in 2014, and 64.5% in 2015, whereas observed resistance to ceftazidime was 59.8% in 2013, 37.3% in 2014, and 42.0% in 2015. The difference in resistance rate towards antimicrobials usually relates to the frequency of use and prescribing practices of hospitals (30). As reported in a study in 2015 on the prevalence of antibiotic resistance among the P. aeruginosa population, statistics have shown that the highest resistance is produced against quinolones including ofloxacin (61.3%), ciprofloxacin (60.0%), and levofloxacin (56.4%). Secondly, the aminoglycosides class of compounds (e.g., amikacin and gentamicin) have shown higher rates of resistance to P. aeruginosa (31). In 2016, a study was conducted on patients admitted to the ICU of the Tertiary Care Hospital in eastern India for one year (2012-13). The prevalence found among patients to develop nosocomial infections was 24.3% where UTIs were predominant in patients followed by respiratory tract and skin infections (32). A study (2016) reported low to moderate rates of drug resistance to commonly used anti-pseudomonal drugs in P. aeruginosa isolates ranging from 4.9% to 30.6%. P. aeruginosa showed resistance towards piperacillin-tazobactam, ticarcillin, imipenem, cefepime, amikacin, and meropenem with a prevalence of 4.9%, 22.3%, 19%, 8.3%, 7.4%, and 30.6%, respectively, irrespective of the site of infection. The prevalence of multidrug resistance was 10.7% (33). A study (2017) reflected an increased percentage of drug-resistance in P. aeruginosa in patients with community-acquired pneumonia (CAP) (34).
A study by Lila et al. (2017) showed an increase of P. aeruginosa carbapenem resistance from 2013 to 2015 for imipenem (25.2% in 2013, 26.5% in 2014, and 37.7% in 2015) and meropenem (20.1% in 2013, 23.4% in 2014, and 38.3% in 2015) (35). Similarly, increased rates of imipenem resistance among P. aeruginosa (10.2% in 2013, 31.6% in 2014, and 22.1% in 2015) were reported in Croatia, studied by Barsic et al. (2004) (36). Benie et al. (2017) evaluated P. aeruginosa multidrug-resistant (PAMDR) contaminating animal products. All strains of P.aeruginosa isolated from bovine meat, fresh and smoked fish expressed resistance to almost all antibiotics. The prevalence of P.aeruginosa multidrug-resistant was 47.8%, 33.1%, and 20.0%, respectively, in bovine meat, fresh fish, and smoked fish. The percentage of resistance showed by P. aeruginosa strains was 98.4% for aztreonam, 51.4% ticarcillin-clavulanic acid, 50.4% ticarcillin, 31.4% piperacillin, 33.6% ciprofloxacin, 17.0% cefepime, 6.9% ceftazidim 7.2% imipenem, 4.5% colistin and 0.0% fosfomycin (37). In 2017, the Government of India declared P. aeruginosa as one of the most important pathogens in National Programme for the Containment of Antimicrobial Resistance (5 Year plan, 2012-2017) under National Centre for Disease control. In 2017, WHO published a list of pathogens in which carbapenem-resistant P. aeruginosa stands at the second position as critical pathogens. Among different anti-pseudomonas drugs tested, interquartile range showed that almost all are highly susceptible to colistin (96.25-100) whereas less susceptible to gentamicin (24-46.5), ceftazidime (31-55), and cefepime (26-58.75). Under Carbapenems such as imipenem (43-72.5) and meropenem (33-69) interquartile range was observed which were found moderately susceptible (38-43). In 2018, another investigation was conducted by Lila et al. (2018) on P. aeruginosa isolates at the University Clinical Center of Kosovo (UCCK) using pulse-field gel electrophoresis (PEGE) for identification of anti-microbial susceptibility. The level of resistance was found to be lowest for carbapenems and highest for aminoglycosides. The results exhibited a high sensitivity of amikacin (52.7%), gentamicin (56.6%), and tobramycin (54.5%) towards P. aeruginosa. In the same study, piperacillin-tazobactam showed resistance ranges from 26.6% to 44.1% (44). Andrea et al. (2019) observed the prevalence and antibiotic resistance profiles of P. aeruginosa. The samples were isolated from healthy captive ophidians and also correlated the statistical associations with farming conditions. From this study, the prevalence of multidrug-resistant P. aeruginosa strains, as well as strains isolated from young samples and adult samples, were found to be 35.5% and 59.9% respectively where widespread resistance has been observed for cephalosporins, polymyxins, and sulfonamides (45).
Pathogenicity and virulence of P. aeruginosa
Microbiology
P. aeruginosa is a Gram-negative, non-fermentative, rod-shaped bacterium, a member of the γ-subdivision of the Proteobacteria (26). P. aeruginosa cells measure 0.5 to 1.0 μM by 3 to 4 μM. They are motile due to the presence of one or two polar flagella, grow on a wide variety of culture media over a wide range of temperatures ranging from 0–42 °C. The optimal temperature required for growth is 37 °C, which is also the normal human body temperature. It is a strict aerobe but can grow anaerobically in a nitrate-rich medium. It forms colonies that appear colored according to the pigment overproduced like the production of pigments a) pyocyanin, responsible for bluish-green, b) fluorescein, responsible for greenish-yellow color, and c) phenazine, a yellow color water-soluble pigment (46). It has been recognized as a ubiquitous organism because of its extremely ordinary survival and adaptation abilities in a wide array of environmental conditions. As an opportunistic human pathogen, P. aeruginosa has a remarkable capacity to cause diseases in susceptible hosts. It is the major colonizing microbial pathogen for cystic fibrosis (CF) patients and a common infectious agent in nosocomial infections, in infections of patients with severe burns, cancer, transplantation, AIDS, and other immuno-compromising conditions. P. aeruginosa is also noted for its conversion from non-mucoid (environmental) to mucoid (clinical) phenotype and its resistance to various antibiotics. P. aeruginosa has been found to cause a variety of infections in clinical practice besides chronic CF lung infection, including common acute septicemia from a burn or surgical wound infection, urinary tract infection, corneal ulceration (from wearing contact lenses), endocarditis (caused by intravenous drug use, etc.), and pneumonia (from use of a ventilator and endotracheal tube) (47-49). The morphology of P. aeruginosa is diagrammatically represented in Figure 1.
Figure 1.
Morphology of Pseudomonas aeruginosa representing cell-associated and extracellular virulence factors
Epidemiology: nosocomial infections caused by P. aeruginosa
P. aeruginosa is a common cause of hospitalized infections in immune-compromised patients. The major source of infection is medical equipment and cross colonization from other patients. This bacterium contaminates the medical devices and forms biofilm which poses a serious problem to the patients. For instance, patients may develop catheter-associated urinary tract infections (CAUTIs) as P. aeruginosa forms a biofilm on the facets of indwelling catheters. The colonial pathogen causes direct damage to the host tissue and increases the bacterium’s competitiveness (50, 51). P. aeruginosa can infect frequently the respiratory tract, blood cells, urinary tract, ear, and soft skin tissues. However, eyes, heart, CNS, bones, and joints are among the sites where the chances of infection are rare. The main cause of infection at these sites is due to trauma following surgery or by the over usage of a drug or any other thing that makes the tissue vulnerable at an immune-compromised state (52, 53).
Hospital-acquired pneumonia
Hospital-acquired pneumonia is the most common life-threatening infection majorly associated with mechanical ventilation and secondly with ICU in hospitals. Ventilator-associated pneumonia (VAP) usually occurs in patients who stay on ventilators for more than 48 hr causing a significant increase in the duration of stay in hospitals cost and death rate. VAP caused by P. aeruginosa is associated with trachea-bronchial colonization which is very difficult to eradicate with conventional antibiotics due to the involvement of complex genes in drug resistance, which leads to higher case fatality rates (54-58). P. aeruginosa is also considered to be a major cause of permanent blocking of the airways of CF patients, which results in recurrence of lung infections and also decrease in lung function, increasing morbidity and mortality rates (59-61). The infection is mainly associated with a genetic mutation in a protein namely cystic fibrosis transmembrane conductance regulator (CFTR). CFTR is a chloride channel that maintains homeostasis in epithelial cells. The disruption in the regulation of chloride ion transport across membrane results in impaired mucociliary clearance due to an increase in sodium absorption, causing obstruction and mucus hypoxia hence supports colonization of P. aeruginosa. Patients with chronic obstructive pulmonary disease (COPD) are also susceptible to respiratory tract infection by P. aeruginosa and show similar symptoms to CF patients (62-65). In addition, various studies reported that P. aeruginosa produces Pyoverdine (a siderophore, ion-chelating molecule) (66, 67) which functions as a signal molecule since it persuades the expression of virulence and biofilm formation causing chronic lung infections in patients with CF (68, 69).
Blood infections
Although very few studies reported different sources of infection for bloodstream infections (BSI) with P. aeruginosa, it is considered to be a serious life-threatening condition and a major cause of the increased rate of morbidity and mortality, as the incidence of BSI caused by P. aeruginosa is increasing. One of the studies reported that respiratory tract and central venous catheters were found to be the most frequent sources of BSI. Other risk factors include immuno-compromised patients in ICU, lung cancer, septic shock, pneumonia, having severe disease, delayed antimicrobial therapy, and multidrug resistance (70-72).
Urinary tract infections
Urinary Tract Infections (UTIs) are also another common type of acute and chronic infections caused by P. aeruginosa, they generally occur after catheterization, instrumentation, or surgery. Urinary tract catheterization is known to be a major cause of nosocomial acquired-UTI by P. aeruginosa as the pathogen utilizes catheters as a medium of bacteria entry resulting in attaching to catheter surface and biofilm formations (73-77).
Skin and soft tissue infections
Multidrug resistant P. aeruginosa is the most common cause of severe wound and burn infections and is associated with high morbidity and mortality rates worldwide. Various studies reported nosocomial outbreaks of the pathogen in surgical wounds resulting in post-operative wound infections (78-80). Some severe soft tissue infections have also been investigated which are associated with P. aeruginosa such as follicular dermatitis or folliculitis (a condition described as an itchy rash with a red base and white pustules), nail disease (onychosis) also known as green nail syndrome, paronychial infection (associated with prolonged exposure to moisture), onycholysis and onychomycosis in post-surgical patients, burn wound sepsis, pyoderma, dermatitis, and ecthyma gangrenosum. Mild skin infections have been reported in some previously healthy persons caused by P. aeruginosa adulteration in swimming pools, hot tubs, and other water sources (81-86).
Eye infections
P. aeruginosa is the main root of bacterial keratitis and it occurs in patients with several medical conditions such as pre-existing ocular diseases, post-ocular surgery, and patients using contact lenses. After adhesion, P. aeruginosa damages corneal epithelial cells and internalize rapidly. The contact lens may damage the epithelial surface of the cornea resulting in corneal keratitis in case of prolonged use or contamination of contact lens and improper handling or care by patients. Some studies reported that there is a rare occurrence of infections like endophthalmitis and neonatal ophthalmia in some patients caused by P. aeruginosa (87-91).
Ear infections
It is well known that P. aeruginosa is the most common cause of ear infections namely otitis externa (swimmer’s ear), which involves inflammation of external auditory occurring on prolonged exposure to moisture or associated with swimming in contaminated recreation pools and/or the insertion of foreign objects such as cotton buds, etc. Other types of infections caused by the pathogen are canal chronic supportive otitis media and malignant external otitis (92-94).
Miscellaneous
Other than above mentioned common infections, P. aeruginosa also contributes to some rare infections. The infection of the blood caused by any bacteria is called bacteremia and septicemia and the common symptoms observed in Pseudomonas infection of lungs and blood are fever, chilling, fatigue, muscle and joint pain, and cough with or without sputum accompanied by difficulty in breathing. P. aeruginosa also causes meningitis and brain abscess, infections related to the central nervous system, which are rare and secondary to neurosurgery or trauma. The pathogen also causes infections affecting bones and joints resulting in the development of several rare disease conditions such as steno-articular pyoarthrosis, vertebral osteomyelitis, symphysis pubis infection, osteochondritis of the foot, and chronic contiguous osteomyelitis. Rarely seen in drug addict patients, P. aeruginosa affects the heart leading to endocarditis (95-100).
Virulence factors in P. aeruginosa
The bacteria adhere to the host tissue with the help of pili, flagella, exo-enzymes, and exopolysaccharides. Colonization of this bacterial species is promoted by glycoprotein consisting of N-acetyl glucosamine (GlcNAc), N-acetylgalactosamine (GalNAc), D-mannose, L-fucose, and N-acetylneuraminic acid (NeuAc) sugar motifs (46). The various virulent factors which are responsible for the pathogenicity of P. aeruginosa are:
Protease, which causes ulceration and infections.
Exotoxin spread infections in the wounds of burn patients.
Phospholipase, which is associated with chronic pulmonary colonization.
Exotoxin A has also been shown to induce host cell death by apoptosis; it is an immunotoxin that targets tumor cells for anticancer therapy.
Lipases and phospholipases break down surfactant lipids and the phospholipids of host cell membranes.
The blue-green pigment pyocyanin gives P. aeruginosa colonies their distinct color and causes oxidative stress to the host, disrupting host catalase, and mitochondrial electron transport.
Purified pyocyanin has been shown in vitro to induce apoptosis in neutrophils (101,102).
P. aeruginosa causes acute infections mainly in three steps, i.e., adhesion, invasion, and systemic spreading. It utilizes cell-associated and extracellular virulent factors to attack the host cell which causes damage to the host skin and reduces the efficiency of the immune system. In immunocompromised patients, the pathogen adheres to epithelial cells and utilizes sugar-binding proteins such as fimbriae (Polar, Type IV pili), flagella, and lectins (LecA and LecB) for the production of elastases, LasA, and LasB which exert cytotoxic effects on respiratory cells and promote bacterial adhesion to airway mucosa. These produced enzymes, hydrolase elastin, an essential protein of connective tissue that is considered to be an important factor of lung innate immunity (103-108). Also, P. aeruginosa facilitates the production of rhamnolipids and hemolytic phospholipases C responsible for the dissolution of phospholipids (phosphatidylcholine) present in the eukaryotic cell membrane and lungs. Moreover, the pathogen synthesizes the redox toxin pyocyanin which obstructs multiple mammalian cell functions such as cell respiration, metal-ion uptake, etc (105,109,110). After colonization at the site of infection, the same can spread the infection in the whole body through systemic circulation using the same virulence factors involved in adhesion and invasion steps leading to the development of biofilms (a heterogeneous structure consists of exopolysaccharide, rhamnolipids, extracellular DNA and proteins) at the colonized sites of host tissues with improved adhesion and stabilization, which causes the establishment of chronic infection and creates a physical barrier to several biocides, the immune system, UV light and antimicrobial agents (105). Moreover, the overall bacterial community formed in biofilm is not homogeneous. The cells present in the middle of the heterogeneous matrix are dormant and comparatively less metabolically active than the cells located on the surface due to low access to oxygen. Taking this fact into consideration, the effect of antibiotics becomes less effective as these agents can only kill pathogens with an active metabolism, for instance, cells on the surface of the biofilm. The bacterial cells attached to the inner layer of the biofilm remaining unaffected by antibiotics are then called persisters. As the concentration of antibiotics reaches sub-inhibitory levels, the persisters tend to switch their metabolic pathway on to repopulate the tissue causing the unmanageable infections which are very difficult to eradicate (111-113). Besides, patients with severe underlying diseases reducing physical (burn patients, mechanically ventilated patients) and/or immune defense mechanisms (neutropenia, AIDS patients) are at serious risk for the evolution of localized infections toward systemic disease, which is associated with dramatically elevated mortality. Just as varied as the clinical diseases caused by P aeruginosa, this typical nosocomial pathogen possesses and produces a large variety of both cell-associated and extracellular virulence factors. It is important to realize that the pathogenesis of P. aeruginosa is not related to a single virulence factor, but the precise and delicate interplay between different factors, leading from efficient colonization and biofilm formation to tissue necrosis, invasion, and dissemination through the vascular system, as well as activation of both local and systemic inflammatory responses. Extensive studies have shown that P. aeruginosa is armed with a large arsenal of virulence factors (described in the following paragraphs), enabling it to breach the human innate immune system, to intoxicate host cells, and to modulate human adaptive immune mechanisms, serving the goal of establishing a systemic infection or more localized chronic colonization (114, 115). In this review, we will discuss the various virulence determinants that have been suggested to play a role during the pathogenesis of P. aeruginosa infections. The various virulence factors produced by P. aeruginosa have been diagrammatically represented in Figure 2.
Figure 2.
Schematic representation of key virulence factors of Pseudomonas aeruginosa
Mechanism of antibiotic resistance development by P. aeruginosa
Generally, the three major mechanisms of antibiotics resistance in P. aeruginosa can be classified into intrinsic resistance, extrinsic (acquired) resistance, and adaptive resistance.
Intrinsic resistance
The intrinsic resistance mechanism in P. aeruginosa involves restricted outer cell membrane permeability and expression of efflux pumps that expel antimicrobial agents out of the cell and also promote the production of antibiotics-inactivating enzymes (116). The four major mechanisms responsible for intrinsic resistance of P. aeruginosa towards antibiotics are 1) target mutation, 2) restrict cell wall uptake, 3) efflux pump, and 4) drug inactivation.
Extrinsic or acquired resistance
In this mechanism, bacteria attain resistance by mutational changes at the genetic level via horizontal gene transfer. The extrinsic mechanism significantly contributes to the development of multi-drug resistant pathogens leading to extreme difficulty in the eradication of microorganisms, which results in boosting cases of persistent infections (117, 118). A study reported that there are two ampG homologs in P. aeruginosa namely ampG (PA4393) and ampG1 (PA4218). ampG is only a functional protein and its inactivation by mutational change leads to a non-inducible and low-level β-lactamase expression (119).
Adaptive antibiotic resistance
This type of resistance mechanism is associated with increased ability of the pathogen for survival against antibiotics attack due to transient alterations in gene expressions in response to environmental stimuli and the mechanism gets reversed when the stimuli are removed. In P. aeruginosa to represent the adaptive resistance, the best-mentioned mechanisms are biofilm formation and development of persisters leading to persistent infections in CF patients (120, 121).
• Biofilm formation
Biofilms are specific and organized communities of cells under the control of signaling molecules, rather than random accumulations of cells resulting from cell division. These biological communities can be embedded in an extracellular matrix that is self-produced. Biofilms may help maintain the role of bacteria as pathogenic by evading host immune mechanisms, resisting antimicrobial treatment, and withstanding competitive pressure from other organisms. Consequently, biofilm-related infections are difficult to treat as they are less sensitive to anti-microbial agents. Biofilm production is also associated with a high level of antimicrobial resistance of the associated organisms (122-125).
• Persistent cell-induced resistance
This involves the formation of bacterial persister cells in the presence of high concentrations of antibiotics. Though, these persister cells (phenotypic variant) are not genetically resistant to antibiotics but are formed as a result of the heterogeneous response to the environment among the bacterial community which is genetically identical (115, 126-129).
Biofilms of P. aeruginosa
Biofilms are communities of microorganisms protected by a self-synthesized layer of complex polysaccharides, proteins, lipids, and extracellular DNA, collectively called the extracellular polymeric substance (130). Being in a biofilm, microbes are covered by a lot of advantages, including, but not limited to physical protection from the host immune system and antimicrobials/antibiotics, retention of water and tolerance to desiccation, nutrient sorption and storage, high extracellular enzymatic activity, adhesion to the infection site, and cell aggregation leading to coordination of virulence factor expression via QS (131-133). Particularly troubling to the medical field, it has been estimated that as much as 80.0% of all human bacterial infections are biofilm-associated, including more than 90.0% of all chronic wound infections (134, 135). Additionally, the biofilm mode of microbial life is responsible for up to a 1000-fold increase in antibiotic tolerance due to the physical impedance and enzymatic inactivation of the drugs, coupled with lowered metabolic rates in many biofilm-associated cells. Thus, biofilm infections are highly recalcitrant and are associated with chronic, non-healing infections (136, 137). Biofilms cause clinical problems of concern because they increase resistance to antifungal therapy; one hypothesis of the mechanism of biofilm resistance is the presence of the matrix that restricts the penetration of drugs through the formation of a diffusion barrier and only the most superficial layers are in contact with lethal doses of antibiotics (138). P. aeruginosa can form a biofilm in various environments. Biofilms have been known to have a rather complex structure with (to a certain level) differentiated bacterial populations and increased resistance against hostile environmental factors, including host immune mechanisms and treatments such as antibiotics. Evidence indicates that P. aeruginosa forms a biofilm in CF lungs where the bacterium lives in an anaerobic environment, as opposed to the aerobic biofilm formed in laboratory conditions. The biofilm mode of growth is recognized as an important bacterial trait that is relevant to infections (122,139). The biofilm formed by P. aeruginosa is shown in Figure 3.
Figure 3.
Pseudomonas aeruginosa biofilms confocal image (surface material: coverslip, taken by Dr Shaminder Singh using a Nikon A1 Confocal Laser Microscope System)
Many infections involve the formation of bacterial biofilms, which are bacterial communities that settle and proliferate on surfaces and are covered by exopolymers. Once established, biofilms are difficult to eradicate and become a source of secondary infection. The dose of antibiotics needed in this situation will often exceed the highest deliverable dose, which makes efficient treatment impossible (140).
Role of quorum sensing in P. aeruginosa virulence
Several new approaches are being actively developed for curbing P. aeruginosa infections over conventional antibiotic chemotherapy in clinical practice. Some of them are based on QS and biofilm inhibition, which is characterized under anti-virulence strategies.
Quorum sensing mechanism
QS phenomenon involves microbial behaviors or responses that are governed by microbial cell density. This mechanism occurs in both Gram-positive as well as Gram-negative bacteria. Such community behaviors are usually determined by secreting signaling molecules, so-called auto-inducers (AIs), accumulation of which is a measure of cell density and nutrient concentration such as iron and phosphate. QS has a pivotal role in biofilms of all kinds (141). Bacteria produce and release small diffusible molecules, usually termed signals, which have two main consequences. First, the uptake of these molecules into cells regulates (auto-induction) a whole variety of behaviors, including the production of a range of exofactors that are released from the cells to aid growth, motility, and/or biofilm formation. Second, the uptake of these molecules also leads to an increase in the production of the signal molecule itself (auto-regulation). The production of these signals or autoinducer molecules, therefore, leads to positive feedback at high cell densities, which results in a considerable increase in the production of signal and QS controlled factors (Figure 2). The hypothesis here is that producing certain extracellular factors is most beneficial at high cell densities and that QS provides a mechanism that allows cells to increase the production of extracellular factors at high cell density (142-144).
In many cases, autoinducers and other molecules are not only responsible for same-species communication but also for the more complex interspecies cross-talk. The diversity of inter-kingdom signaling occurring in a myriad of environments has been classified into four categories:
(1) One-way sensing: one organism senses and responds to a diffusible signal produced by a second organism;
(2) Co-opting for a signal: one organism uses the signal produced by another to regulate its gene expression;
(3) Modulation of a signal: one organism alters the production or stability of a signal from another organism; and
(4) Two-way communication: multiple signals are exchanged between organisms (145) as shown in Figure 4.
Figure 4.
Examples of uni and bi-directional signaling interactions
Quorum sensing in P. aeruginosa
The behavior of P. aeruginosa is monitored by a complex regulatory mechanism called Quorum sensing (QS) in acute and chronic infections (12). The co-ordination of specific gene expression in the community involves the interaction of diffused molecular signals. The quorum-sensing system depends on 3 basic principles in the bacterium. First, the production of AIs also called signaling molecules by the bacterial population. At low cell density, these signaling molecules diffuse away and therefore are present at concentrations below the threshold required for detection. At high cell density, the cumulative production of signaling molecules results in high concentration locally facilitating detection and response. Second, these AIs are detected by the receptors present in the cell (cytoplasm or membrane). Third, the detection of AIs facilitates AI production to potentiate the expression of genes. This feed-forward auto-induction loop promotes the development of the population (146-148). In P. aeruginosa, the quorum-sensing circuit is controlled by the expression of gene systems viz. four different QS channels interlinked to each other for disseminating virulence, biofilm production, and synthesis of signal molecules. The channels are las, Rhl, Iqs, and Pqs where these systems employ transcriptional regulators such as LasR, RhlR, IqsR, and PqsR, respectively (also known as Multiple virulence factor regulator, MvfR). This MvfR binds to specific AIs to aggravate the expression of selected genes to cause virulence. The expression of different QS systems took place in response to the varying levels of cell density (Figure 4). Furthermore, the las, rhl and Pqs based systems coordinate biofilm formation. The las system utilizes N-(3-oxododecanoyl)-L homoserine lactone (3-oxo-C12-HSL) as a signal molecule that induces the expression of LasA and LasB elastases, alkaline protease, MvfR, RhlR, IqsR, and the cognate synthetase LasI. The Rhl system uses a molecule of N-butanoyl-L-homoserine lactone (C4-HSL) as an auto-inducer (belonging to an acyl-homoserine lactone (AHL)) which facilitates the synthesis of rhamnolipids, LasB elastase, pyocyanin, hydrogen cyanide RhlI (related signal molecule biosynthetic protein), and down-regulation of mvfR. This chemical triggers the production of inflammatory mediators. However, in the case of chronic infections, the rhl system is expressed and maintained for a longer duration (149-151). The recently discovered Iqs system employs 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde (IQS), which is supposed to regulate the Pqs system (152). The Pqs system makes use of quinolone signals (QS) molecule which helps in the synthesis of pyocyanin, hydrogen cyanide, as well as LecA lectin, the enzyme required for Pqs biosynthesis and the expression of RhlR and LasR. Along with the production of acyl-homoserine lactone as QS signals in P. aeruginosa, the other class of autoinducers is 4-hydroxy 2-alkyl quinolones (HAQs) and derivatives of 4-hydroxy-2-heptylquinoline (HHQ), including di-hydroxy derivatives like 2-heptyl-3,4-dihydroxyquinoline (152-155). All the different types of QS mechanisms such as lasR, rhls, and Pqs have been depicted in Figure 5, 6, and 7, respectively.
Figure 5.
Different quorum sensing systems in Pseudomonas aeruginosa
Figure 6.
Diagrammatic representation of the effect of N-(3-oxododecanoyl)-L-homoserine lactone (3O-C12-HSL) signaling molecule on the LasR system
Figure 7.
Representation of the effects of Pqs quorum sensing in virulence production
Quorum sensing inhibition as an anti-virulence strategy
QS is known to be an extremely important mechanism in the regulation of virulence factors as well as the formation of biofilms so it has become a potential target to minimize drug resistance during the treatment. Various in vivo studies showed that strains lacking in the expression of transcriptional regulators or auto-inducer (AI) biosynthetic pathway give rise to lower mortality of mice as compared with the animals treated with wild type of P. aeruginosa. Three different approaches can be considered while designing QSIs such as signal molecule inactivation, inhibition of AI syntheses, and interference with transcriptional regulators (156-158). Various studies have been conducted targeting QS inhibition as an anti-virulence strategy against various resistant pathogens including P. aeruginosa. For instance, a study (2015) shows that novel N,N-disubstituted biguanides were found to have QS inhibition activity against Chromobacterium violaceum (159). In another study by Singh S, et al. (2016), phenolic compounds from ginger rhizomes exhibited a QS inhibitory activity against C. violaceum and P. aeruginosa (160). Furthermore, in a study based on in silico docking, ADME, and toxicity, aryl glycoxamide derivatives were found to have substantial potential to develop as anti-virulent agents to inhibit QS in P. aeruginosa and E. coli (161). In another report, molecular docking studies were carried out for novel 1,8- Naphthyridine derivatives and showed moderate to good anti-bacterial activity tested against various strains such as P. aeruginosa, E. coli, Staphylococcus aureus, and Bacillus subtilis (162). Thus, these findings make a basis to consider the QS mechanism as a potential target for anti-virulence strategy.
Targeting quorum sensing proteins is a remedial solution to multi-drug resistant strains
Autoinducers involved in quorum sensing of P. aeruginosa
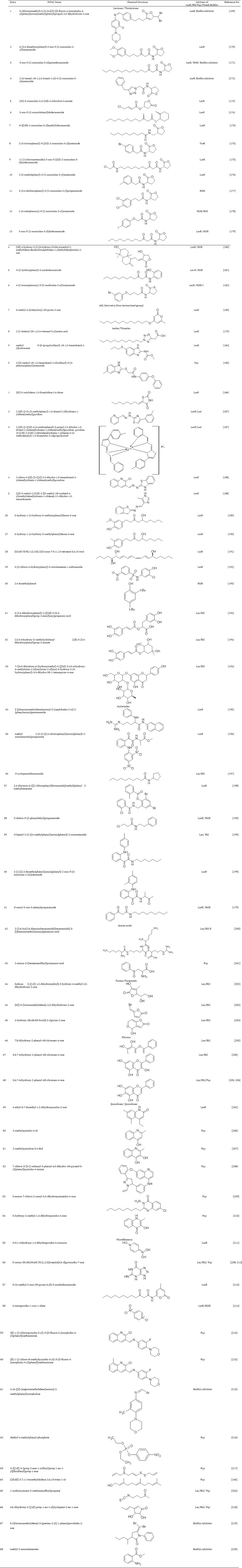
QS is a process in which both Gram-positive and Gram-negative bacteria monitor their species, modulate intra- and interspecies cell to cell communication, control expression of specific genes in response to fluctuation in cell population density and regulate diverse physiological functions by releasing chemical signaling molecules known as autoinducers (AI), example N-acyl-homoserine lactone (AHL). Table 1 (entry 1-9) shows a different kind of AI released during QS (163, 164).
Table 1.
Different analogs of Acyl Homoserine Lactone (AHL)
Chemical classes of compounds that inhibit quorum sensing in P. aeruginosa
P. aeruginosa contains an MvfR QS system. These systems can be targeted to attenuate the virulence of P. aeruginosa. Some research groups have found that the P. aeruginosa mutants lacking the las gene are a-virulent type and unable to cause pneumonia. Rhl mediated QS inhibition includes the rhlR encoded putative transcriptional activator, RhlR, and rhlI encoded putative AI synthase, RhlI. A second P. aeruginosa Auto-inducer (PAI-2), N-butyryl homoserine lactone, was shown to re-store rhamnolipids production in a P. aeruginosarhlI mutant and also require rhlI for its synthesis. Some compounds also have been found that can act as inhibitors of both Las and Rhl mediated QS and could be beneficial to curb the pathogenesis of P. aeruginosa (165). In contrast, the Pqs system is associated with QS through quinolone signaling molecules and can be targeted by QSIs to inhibit bacterial virulence (166, 167). In P. aeruginosa, both QS and biofilms are impacted by the surrounding environment representing these complex communities as a challenge (168). A variety of potent chemical compounds can be utilized to inhibit the process of QS and thereby reducing the QS-mediated biofilm formation in P. aeruginosa. In this quest, various reported synthetic compounds have been found which act on attenuating the P. aeruginosa virulence by targeting various QS mediated systems as represented in Table 2.
Table 2.
Chemical classes reported as quorum sensing inhibitors that act via different mechanisms against Pseudomonas aeruginosa
Concluding remarks
It is well-established that the pathogenic microbial strains possess an enhanced ability to adapt and develop a mechanism against the chemical compound that could impair its sustainability. The overuse of antibiotics increases the chances of development of resistant strains. This is especially true for the opportunistic pathogen P. aeruginosa and its inherent capability to transform into a multidrug-resistant phenotype. However, this pathogen can rapidly develop resistance to multiple classes of antibiotics during patient treatment. The chromosomal protein AmpG, the outer membrane porin OprD, and the multitude of efflux pumps are particularly responsible for this challenging therapeutic regime, and the discussion presented in this review highlights complex mechanisms and pathways by which P. aeruginosa regulates and/or co-regulates their expression. In the lack of a diminished antibiotic development pipeline towards antimicrobial therapeutics, we must look for novel strategies to combat the threat of antibacterial resistance. To solve this issue, an alternative strategy that involves the development of new active agents that are capable of targeting bacterial virulence besides its growth has to be devised. In this context, research for anti-QS has been largely explored during the last two decades to propose new alternatives to struggle against bacterial infection with limited selective pressure. The present study highlights the importance of QS in up-regulation of efflux pump genes for escaping from antibiotic attack. However, the scientific community has to admit the importance of QS in the development of bacterial resistance, and concealed pathways have to be explored for investigating the role of QSI in order to develop anti-QS therapeutics.
Acknowledgment
Authors are thankful to the Department of Pharmaceutical Sciences, Sam Higginbottom University of Agriculture Technology and Sciences (SHUATS), Prayagraj, India, for providing us the platform and infrastructure for preparing this manuscript.
Conflicts of Interest
The authors of this article ensure that there are no conflicts of interest.
References
- 1.Web Review of Todar’s Online Textbook of Bacteriology. The Good, the Bad, and the deadly. Sci Mag. 2004;304:1421–1632. [Google Scholar]
- 2.Kon K, Rai M. Antibiotic resistance mechanisms and new antimicrobial approaches. 1st ed. London, UK: Academic Press; 2016. [Google Scholar]
- 3.Gomez MI, Prince A. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr Opin Pharmacol. 2007;7:244–251. doi: 10.1016/j.coph.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Hill D, Rose B, Pajkos A, Robinson M, Bye P, Bell S, et al. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol. 2005;43:5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obritsch MD, Douglas NF, MacLaren R, Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–4610. doi: 10.1128/AAC.48.12.4606-4610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang CI, Kim SH, Kim HB, Park SW, Choe YJ, Oh MD, et al. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis. 2003;37:745–751. doi: 10.1086/377200. [DOI] [PubMed] [Google Scholar]
- 8.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore DM, Woodford N. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006;14:413–420. doi: 10.1016/j.tim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Hancock RE. Resistance mechanisms in Pseudomonas aeruginosa and other non-fermentative gram-negative bacteria. Clin Infect Dis. 1998;1:93–99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 11.Maura D, Ballok AE, Rahme LG. Considerations and caveats in anti-virulence drug development. Curr Opin Microbiol. 2016;33:41–46. doi: 10.1016/j.mib.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner S, Sommer R, Hinsberger S, Lu C, Hartmann RW, Empting M, et al. Novel strategies for the treatment of Pseudomonas aeruginosa infections. J Med Chem. 2016;59:5929–5969. doi: 10.1021/acs.jmedchem.5b01698. [DOI] [PubMed] [Google Scholar]
- 13.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: A new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 14.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs. Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 15.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 16.Yan S, Wu G. Can biofilm be reversed through quorum sensing in Pseudomonas aeruginosa. Front Microbiol. 2019;10:1582–1591. doi: 10.3389/fmicb.2019.01582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiteley M, Diggle SP, Greenberg EP. Bacterial quorum sensing: the progress and promise of an emerging research area. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariminik A, Majid BS, Kheirkhah B. Pseudomonas aeruginosa quorum sensing modulates immune responses: an updated review article. Immunol Lett. 2017;190:1–6. doi: 10.1016/j.imlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh R, Das A, Mallik S. Inhibition of quorum sensing in Pseudomonas aeruginosa: a review. Indian J Pharm Sci. 2019;81:797–806. [Google Scholar]
- 20.Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 2006;30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 21.Zeng L. Pseudomonas aeruginosa pathogenicity and antibiotic resistance. Doctor of Philosophy, A Dissertation. The Graduate School, University of Florida; 2004. [Google Scholar]
- 22.Antibiotic resistance threats in the United States, Centers for Disease Control and Prevention: Antibiotic Resistance . Threats in the United States 2013, Atlanta, GA: Centers for Disease Control and Prevention: [Google Scholar]
- 23.Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CY, Jerng JS, Chen KY, Lee LN, Yu CJ, Hsueh PR, et al. Pandrug-resistant Pseudomonas aeruginosa among hospitalised patients: clinical features, risk-factors and outcomes. Clin Microbiol Infect. 2006;12:63–68. doi: 10.1111/j.1469-0691.2005.01305.x. [DOI] [PubMed] [Google Scholar]
- 25.Wei Q, Ma LZ. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci. 2013;14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanwar J, Das S, Fatima Z, Hameed S. Multidrug resistance: an emerging crisis. Interdiscip Perspect Infect Dis. 2014;Article ID 541340:1–7. doi: 10.1155/2014/541340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayani M, Siadati S, Rajabnia R, Taher AA. Drug resistance of Pseudomonas aeruginosa and enterobacter cloacae Isolated from ICU, Babol, Northern Iran. Int J Mol Cell Med. 2013;2:204–209. [PMC free article] [PubMed] [Google Scholar]
- 28.Dash M, Padhi S, Narasimham MV, Pattnaik S. Antimicrobial resistance pattern of Pseudomonas aeruginosa isolated from various clinical samples in a tertiary care hospital, South Odisha, India. Saudi J Health Sci. 2014;3:15–19. [Google Scholar]
- 29.Antimicrobial resistance and healthcare-associated infections, Annual Epidemiological Reports. European Centre for Disease Prevention and Control. Annual epidemiological report 2014. Antimicrobial resistance and healthcare-associated infections. Stockholm. ECDC; 2015. [Google Scholar]
- 30.Kos VN, Deraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, et al. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother. 2015;59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali Z, Mumtaz N, Naz SA, Jabeen N, Shafique M. Multi-drug resistant pseudomonas aeruginosa: a threat of nosocomial infections in tertiary care hospitals. J Pak Med Assoc. 2015;65:12–16. [PubMed] [Google Scholar]
- 32.Chakraborty P, Mukherjee S. A Study on the prevalence and microbiological profile of nosocomial infections in the ICU of a tertiary care hospital in Eastern India. World Academy of Science, Engineering and Technology. Int J Curr Microbiol Appl Sci. 2016;5:920–925. [Google Scholar]
- 33.Khan MA, Faiz A. Antimicrobial resistance patterns of Pseudomonas aeruginosa in tertiary care hospitals of Makkah and Jeddah. Ann Saudi Med. 2016;36:23–28. doi: 10.5144/0256-4947.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Restrepo MI, Babu BL, Reyes LF, Chalmers JD, Soni NJ, Sibila O, et al. Burden and risk factors for Pseudomonas aeruginosa community-acquired pneumonia: a multinational point prevalence study of hospitalized patients. Eur Respir J. 2018;52:1701190–1701204. doi: 10.1183/13993003.01190-2017. [DOI] [PubMed] [Google Scholar]
- 35.Lila G, Mulliqi G, Raka L, Kurti A, Bajrami R, Azizi E. Molecular epidemiology of Pseudomonas aeruginosa in university clinical center of Kosovo. Infect Drug Resist. 2018;11:2039–2046. doi: 10.2147/IDR.S174940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barsic B, Tambic A, Santini M, Klinar I, Kutlesa M, Krajinovic V. Antibiotic resistance among nosocomial isolates in a Croatian intensive care unit--results of a twelve-year focal surveillance of nosocomial infections. J Chemother. 2004;16:273–281. doi: 10.1179/joc.2004.16.3.273. [DOI] [PubMed] [Google Scholar]
- 37.Benie CKD, Nathalie G, Adjehi D, Solange A, Ferniquekonan K, Desire K, et al. Prevalence and antibiotic resistance of Pseudomonas aeruginosa isolated from bovine meat, fresh fish and smoked fish. Arch Clin Microbiol. 2017;8:40–49. [Google Scholar]
- 38.Pragasam AK, Veeraraghavan B, Nalini E, Anandan S, Kaye KS. An update on antimicrobial resistance and the role of newer antimicrobial agents for Pseudomonas aeruginosa. Indian J Med Microbiol. 2018;36:303–316. doi: 10.4103/ijmm.IJMM_18_334. [DOI] [PubMed] [Google Scholar]
- 39.Gandra S, Mojica N, Klein EY, Ashok A, Nerurkar V, Kumari M, et al. Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008-2014. Int J Infect Dis. 2016;50:75–82. doi: 10.1016/j.ijid.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta R, Malik A, Rizvi M, Ahmed M. Presence of metallo-beta-lactamases (MBL), extended-spectrum beta-lactamase (ESBL) & AmpC positive non-fermenting gram-negative bacilli among intensive care unit patients with special reference to molecular detection of blaCTX-M&blaAmpC genes. Indian J Med Res. 2016;144:271275–271289. doi: 10.4103/0971-5916.195043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellappan K, Belgode Narasimha H, Kumar S. Coexistence of multidrug resistance mechanisms and virulence genes in carbapenem-resistant Pseudomonas aeruginosa strains from a tertiary care hospital in South India. J Glob Antimicrob Resist. 2018;12:37–43. doi: 10.1016/j.jgar.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 42.Dhaneria M, Jain S, Singh P, Mathur A, Lundborg CS, Pathak A. Incidence and determinants of health care-associated blood stream infection at a neonatal intensive care unit in Ujjain, India: A prospective cohort study. Dis. 2018;6:271–275. doi: 10.3390/diseases6010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wattal C, Raveendran R, Goel N, Oberoi JK, Rao BK. Ecology of blood stream infection and antibiotic resistance in intensive care unit at a tertiary care hospital in North India. Braz J Infect Dis. 2014;18:245–251. doi: 10.1016/j.bjid.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lila G, Mulliqi-Osmani G, Bajrami R, Kurti A, Azizi E, Raka L. The prevalence and resistance patterns of Pseudomonas aeruginosa in a tertiary care hospital in Kosovo. Infez Med. 2017;25:21–26. [PubMed] [Google Scholar]
- 45.Sala A, Di Ianni F, Pelizzone I, Bertocchi M, Santospirito D, Rogato F, et al. The prevalence of Pseudomonas aeruginosa and multidrug resistant Pseudomonas aeruginosa in healthy captive ophidian. Peer J. 2019;7:6706–6719. doi: 10.7717/peerj.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrios CC, Ciancotti-Oliver L, Bautista-Rentero D, Adan-Tomas C, Zanón-Viguer V. A new treatment choice against multi-drug resistant Pseudomonas aeruginosa: doripenem. J Bacteriol Parasitol. 2014;5:21–27. [Google Scholar]
- 47.BusiRizzi E, Schinina V, Bordi E, Buontempo G, Narciso P, Bibbolino C. HIV-related bronchopulmonary infection by Pseudomonas aeruginosa in the Haarat era: radiological findings. Acta Radiol. 2006;47:793–797. doi: 10.1080/02841850600827569. [DOI] [PubMed] [Google Scholar]
- 48.Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 49.Tredget EE, Shankowsky HA, Rennie R, Burrell RE, Logsetty S. Pseudomonas infections in the thermally injured patient. Burns. 2004;30:3–26. doi: 10.1016/j.burns.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Al-Wrafy F, Brzozowska E, Gorska S, Gamian A. pathogenic factors of Pseudomonas aeruginosa – the role of biofilm in pathogenicity and as a target for phage therapy. PostepyHig Med Dosw. 2016;70:78–91. doi: 10.5604/01.3001.0010.3792. [DOI] [PubMed] [Google Scholar]
- 51.Wagner VE, Filiatrault MJ, Picardo KF, Iglewski BH. Pseudomonas aeruginosa Virulence and Pathogenesis Issues. In: Cornelis P, editor. Pseudomonas Genomics and Molecular Biology. 1st ed. Norfolk, UK: Caister Academic Press; 2008. pp. 129–158. [Google Scholar]
- 52.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 53.Wiener-Kronish JP, Frank D, Sawa T. Mechanisms of lung epithelial cell injury by Pseudomonas aeruginosa. In: Wong HR, Shanley TP, editors , editors. Molecular and Cellular Biology of Critical Care Medicine Volume 1 Molecular Biology of Acute Lung Injury. Boston: Kluwer Academic Publishers; 2001. pp. 149–161. [Google Scholar]
- 54.Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit: A critical review. Genes Dis. 2019;6:109–119. doi: 10.1016/j.gendis.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kollef MH, Chastre J, Fagon JY, François B, Niederman MS, Rello J, et al. Global prospective epidemiologic and surveillance study of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit Care Med. 2014;42:2178–2187. doi: 10.1097/CCM.0000000000000510. [DOI] [PubMed] [Google Scholar]
- 56.Crouch BS, Wunderink RG, Jones CB, Leeper Jr KV. Ventilator associated pneumonia due to Pseudomonas aeruginosa. Chest. 1986;109:1019–1029. doi: 10.1378/chest.109.4.1019. [DOI] [PubMed] [Google Scholar]
- 57.Raineri E, Porcella L, Acquarolo A, Crema L, Albertario F, Candiani A. Ventilator-associated pneumonia caused by Pseudomonas aeruginosa in intensive care unit: epidemiology and risk factors. J Med Microbiol Diagn. 2014;3:185–193. [Google Scholar]
- 58.Rello J, Ausina V, Puzo C, Quintana E, Net A, Prats G. Risk factors for infection by Pseudomonas aeruginosa in patients with ventilator-associated pneumonia. Intensive Care Med. 1994;20:193–198. doi: 10.1007/BF01704699. [DOI] [PubMed] [Google Scholar]
- 59.Streeter K, Katouli M. Pseudomonas aeruginosa: a review of their pathogenesis and prevalence in clinical settings and the environment. Infect Epidemiol Med. 2016;2:25–32. [Google Scholar]
- 60.Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, et al. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J Paediatr. 2001;138:699–704. doi: 10.1067/mpd.2001.112897. [DOI] [PubMed] [Google Scholar]
- 61.Smith JJ, Travis SM, Greenberg EP, Welsh MJ. Cystic fibrosis airway epithelial fail to kill bacteria of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 62.Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:23–45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 63.Terheggen-Lagro SW, Rijkers GT, Van der Ent CK. The role of airway epithelium and blood neutrophils in the inflammatory response in cystic fibrosis. J Cyst Fibros. 2005;4:15–23. doi: 10.1016/j.jcf.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 64.Worlitzch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez-Solano L, Macia MD, Fajardo A, Oliver A, Martinez JL. Chronic Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Clin Infect Dis. 2008;47:1526–1533. doi: 10.1086/593186. [DOI] [PubMed] [Google Scholar]
- 66.Bhatia S, Singh S. Fungal Siderophores-From Mineral-Microbe Interactions to Anti-pathogenicity Springer Nature. Switzerland AG: 2021 . Inhibition of siderophores in blocking fungal infections; pp. 13–31. [Google Scholar]
- 67.Bhardwaj S, Singh S, Bhatia S. Fungal Siderophores - From Mineral-Microbe Interactions to Anti-pathogenicity by Springer Nature. Switzerland AG: 2021. Contrasting role of fungal siderophore in metal ion complex formation; pp. 99–117. [Google Scholar]
- 68.Cornelis P, Matthijs S. Pseudomonas Siderophores. In: Varma A, Chincholkar S, editors. Microbial Siderophores. Springer, Berlin, Heidelberg: 2007. [Google Scholar]
- 69.Peek ME, Bhatnagar A, McCarty NA, Zughaier SM. Pyoverdine, the major siderophore in Pseudomonas aeruginosa, Evades NGAL Recognition. Interdiscip Perspect Infect Dis. 2012:1–10. doi: 10.1155/2012/843509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Therrien C, Levesque RC. Molecular basis of antibiotic resistance and b-lactamase inhibition by mechanism-based inactivators: perspectives and future direction. FEMS Microbiol Rev. 2000;24:251–262. doi: 10.1111/j.1574-6976.2000.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 71.Vidaur L, Sirgo G, Rodriguez AH, Rello J. Clinical approach to the patient with suspected ventilator-associated pneumonia. Respir Care. 2005;50:965–974. [PubMed] [Google Scholar]
- 72.Bert F, Branger C, Lambert-Zechovsky N. Identification of PSE and OXA b-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J Antimicrob Chemother. 2000;50:11–18. doi: 10.1093/jac/dkf069. [DOI] [PubMed] [Google Scholar]
- 73.Mittal R, Aggarwal S, Sharma S, Chhibber S, Harjai K. Urinary tract infections caused by Pseudomonas aeruginosa: a minireview. J Infect Public Health. 2009;2:101–111. doi: 10.1016/j.jiph.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 75.Bass PF, Jarvis JA, Mitchell CK. Urinary tract infections. Prim Care. 2003;30:41–61. doi: 10.1016/s0095-4543(02)00057-x. [DOI] [PubMed] [Google Scholar]
- 76.Zulianello L, Canard C, Kohler T, Caille D, Lacroix JS, Meda P. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect Immun. 2006;74:3134–147. doi: 10.1128/IAI.01772-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lysczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 78.Ressner RA, Murray CK, Griffith ME, Rasnake MS, Hospenthal DR, Wolf SE. Outcomes of bacteremia in burn patients involved in combat operations overseas. J Am Coll Surg. 2008;206:439–444. doi: 10.1016/j.jamcollsurg.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 79.Estahbanati HK, Kashani PP, Ghanaatpisheh F. Frequency of Pseudomonas aeruginosa serotypes in burn wound infections and their resistance to antibiotics. Burns. 2002;28:637–641. doi: 10.1016/s0305-4179(02)00024-4. [DOI] [PubMed] [Google Scholar]
- 80.Jombo GT, Akpan S, Epoke J, DenenAkaa P, Odey F. Multi-drug resistant Pseudomonas aeruginosa infections complicating surgical wounds and the potential challenges in managing post-operative wound infections:,University of Calabar teaching hospital experience. Asian J Trop Med. 2010;3:479–482. [Google Scholar]
- 81.Hoiby N, Johnsen HK, Moser C, Song Z, Ciofu O, Kharazmi A. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 2001;3:23–35. doi: 10.1016/s1286-4579(00)01349-6. [DOI] [PubMed] [Google Scholar]
- 82.Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 83.Kuchma SL, Connoly JP, O’Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol. 2005;187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aerguinosa biofilm matrix. J Bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl: a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nagachandrikaa T, Kumarb U, Dumpatic S, Charyc S, Mandatharac PS, Rathi VM. Prevalence of contact lens related complications in a tertiary eye centre in India. Cont Lens Anterior Eye. 2011;34:266–268. doi: 10.1016/j.clae.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 88.Ramphal R, McNiece MT, Polack FM. Adherence of Pseudomonas aeruginosa to the injured cornea: a step in the pathogenesis of corneal infections. Ann Opthalmol. 1981;13:421–425. [PubMed] [Google Scholar]
- 89.Stern GA, Lubniewski A, Allen C. The interaction between Pseudomonas aeruginosa and the corneal epithelium. Arch Ophthalmol. 1985;103:1221–1225. doi: 10.1001/archopht.1985.01050080133033. [DOI] [PubMed] [Google Scholar]
- 90.Roberston DM, Petroll WM, Jester JV, Cavanagh HD. Current concepts: contact lens related Pseudomonas keratitis. Cont Lens Anterior Eye. 2007;30:94–107. doi: 10.1016/j.clae.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 91.Yeung KK, Forister JFY, Forister EF, Chung MY, Han S, Weissman BA. Compliance with soft contact lens replacement schedules and associated contact lens-related ocular complications: the UCLA contact lens study. Optometry. 2010;81:598–607. doi: 10.1016/j.optm.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 92.Wang MC, Liu CY, Shiao AS, Wang T. Ear problems in swimmers. J Chin Med Assoc. 2005;68:347–352. doi: 10.1016/S1726-4901(09)70174-1. [DOI] [PubMed] [Google Scholar]
- 93.Nussinovitch M, Rimon A, Volovitz B, Raveh E, Prais D, Amir J. Cottontip applicators as a leading cause of otitis externa. Int J Pediatr Otorhinolaryngol. 2004;73:1168–1172. doi: 10.1016/j.ijporl.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 94.Ninkovic G, Dullo V, Saunders NC. Microbiology of otitis externa in the secondary care in United Kingdom and antimicrobial sensitivity. Auris Nasus Larynx. 2008;35:480–484. doi: 10.1016/j.anl.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 95.Crnich CJ, Safdar N, Maki DG. The role of the intensive care unit environment in the pathogenesis and prevention of ventilator-associated pneumonia. Respir Care. 2005;50:813–836. [PubMed] [Google Scholar]
- 96.Ferrara AM. Potentially multidrug-resistant non-fermentative Gram-negative pathogens causing nosocomial pneumonia. Int J Antimicrob Agents. 2006;27:183–195. doi: 10.1016/j.ijantimicag.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 97.Shaw MJ. Ventilator-associated pneumonia. Curr Opin Pulm Med. 2005;11:236–241. doi: 10.1097/01.mcp.0000159834.05401.78. [DOI] [PubMed] [Google Scholar]
- 98.Chastre J, Fagon JY. Ventilator-associated pneumonia. Am J Respir Crit Care Med. 2002;165:867–903. doi: 10.1164/ajrccm.165.7.2105078. [DOI] [PubMed] [Google Scholar]
- 99.Ratjen F. Diagnosing and managing infection in CF. Paediatr Respir Rev. 2006;7:151–153. doi: 10.1016/j.prrv.2006.04.217. [DOI] [PubMed] [Google Scholar]
- 100.Nicotra MB, Rivera M, Dale AM, Shepherd R, Carter R. Clinical pathophysiologic and microbiologic characterization of bronchiectasis in an aging cohort. Chest. 1995;108:955–961. doi: 10.1378/chest.108.4.955. [DOI] [PubMed] [Google Scholar]
- 101.Porras-Gomez M, Vega-Baudrit J, Nunez-Corrales S. Overview of multidrug-resistant Pseudomonas aeruginosa and novel therapeutic approaches. J Biomater Nanobiotechnol. 2012;3:519–527. [Google Scholar]
- 102.Gellatly SL, Hancock RE. Pseudomonas aeruginosa : new insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 103.Todar, K. Kenneth Todar’s Online Textbook of Bacteriology. University of Wisconsin-Madison, Department of Bacteriology; 2009. pportunistic infections caused by Pseudomonas aeruginosa. [Google Scholar]
- 104.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2015;112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, et al. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Strateva T, Mitov I. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann Microbiol. 2011;61:717–732. [Google Scholar]
- 107.Galoway DR. Pseudomonas aeruginosa elastase and elasolysis revisited: recent developments. Mol Microbiol. 1991;5:2315–2321. doi: 10.1111/j.1365-2958.1991.tb02076.x. [DOI] [PubMed] [Google Scholar]
- 108.Mariencheck WI, Alcorn JF, Palmer SM, Wright JR. Pseudomonas aeruginosa elastase degrades surfactant proteins A and D. Am J Respir Cell Mol Biol. 2003;28:528–537. doi: 10.1165/rcmb.2002-0141OC. [DOI] [PubMed] [Google Scholar]
- 109.Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, van Strijp JA, et al. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J Immunol. 2012;188:386–393. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- 110.Hoegy F, Mislin GL, Schalk IJ. Pyoverdine and pyochelin measurements. Methods Mol Biol. 2014;1149:293–301. doi: 10.1007/978-1-4939-0473-0_24. [DOI] [PubMed] [Google Scholar]
- 111.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 112.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 113.Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, et al. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect. 2007;13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 115.Van Delden C. In: Virulence factors in Pseudomonas aeruginosa. Ramos J-L, editor. Pseudomonas. Vol. 2. New York: Kluwer Academic/Plenum Publishers; 2004 . [Google Scholar]
- 116.Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 117.Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4:481–511. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob Agents Chemother. 2007;51:4062–4070. doi: 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chang Q, Wu C, Lin C, Li P, Zhang K, Xu L, et al. The structure of ampG gene in Pseudomonas aeruginosa and its effect on drug resistance. Can J Infect Dis Med Microbiol. 2018;2018:1–8. doi: 10.1155/2018/7170416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sandoval-Motta S, Aldana M. Adaptive resistance to antibiotics in bacteria: a systems biology perspective. Wiley Interdiscip Rev Syst Biol Med. 2016;8:253–267. doi: 10.1002/wsbm.1335. [DOI] [PubMed] [Google Scholar]
- 121.Taylor PK, Yeung AT, Hancock RE. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol. 2014;191:121–130. doi: 10.1016/j.jbiotec.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 122.Ozkan S, Kaynak F, Kalkanci A, Abbasoglu U, Kustimur S. Slime production and proteinase activity of Candida species isolated from blood samples and the comparison of these activities with minimum inhibitory concentration values of antifungal agents. Mem Inst Oswaldo Cruz. 2005;100:319–323. doi: 10.1590/s0074-02762005000300019. [DOI] [PubMed] [Google Scholar]
- 123.Das T, Sehar S, Manefield M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep. 2013;5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- 124.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 126.Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers. Nat Rev Microbiol. 2013;11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 127.Lambert P. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95:22–26. [PMC free article] [PubMed] [Google Scholar]
- 128.Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 129.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare. Clin Infect Dis. 2002;34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 130.Fleming D, Rumbaugh KP. Approaches to dispersing medical biofilms. Microorganisms. 2017;5:15–31. doi: 10.3390/microorganisms5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 132.Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19:341–345. doi: 10.1016/j.cub.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 133.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Attinger C, Wolcott R. Clinically addressing biofilm in chronic wounds. Adv Wound Care. 2012;1:127–132. doi: 10.1089/wound.2011.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Romling U, Balsalbre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;271:541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- 136.Rogers SA, Huigens RW, Cavanagh J, Melander C. Synergistic effects between conventional antibiotics and 2-aminoimidazole-derived antibiofilm agents. Antimicrob Agents Chemother. 2010;54:2112–2118. doi: 10.1128/AAC.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lewis K. Persister Cells: Molecular mechanisms related to antibiotic tolerance. Handb Exp Pharmacol. 2012;211:121–133. doi: 10.1007/978-3-642-28951-4_8. [DOI] [PubMed] [Google Scholar]
- 138.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis. 2017;17:383–392. doi: 10.1016/S1473-3099(17)30316-X. [DOI] [PubMed] [Google Scholar]
- 140.Chang W, Li Y, Zhang L, Cheng A, Lou H. Retigeric acid B attenuates the virulence of Candida albicans via inhibiting adenylyl cyclase activity targeted by enhanced farnesol production. PLoS one. 2012;7:1–10. doi: 10.1371/journal.pone.0041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.West SA, Winzer K, Gardner A, Diggle SP. Quorum sensing and the confusion about diffusion. Trends Microbiol. 2012;20:586–594. doi: 10.1016/j.tim.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 143.Henke JM, Bassler BL. Bacterial social engagements. Trends Cell Biol. 2014;14:648–656. doi: 10.1016/j.tcb.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 144.Darch SE, West SA, Winzer K, Diggle SP. Density-dependent fitness benefits in quorum sensing bacterial populations. Proc Natl Acad Sci USA. 2012;109:8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.De Sordi L, Muhlschlegel FA. Quorum sensing and fungal bacterial interactions in Candida albicans: a communicative network regulating microbial coexistence and virulence. FEMS Yeast Res. 2009;9:990–999. doi: 10.1111/j.1567-1364.2009.00573.x. [DOI] [PubMed] [Google Scholar]
- 146.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seed PC, Passador L, Iglewski BH. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: An autoinduction regulatory hierarchy. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xiao G, Deziel E, He J, Lepine F, Lesic B, Castonguay MH, et al. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 152.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, et al. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol. 2013;9:339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 153.Christensen LD, Moser C, Jensen PO, Rasmussen TB, Christophersen L, Kjelleberg S, et al. Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiol. 2007;153:2312–2320. doi: 10.1099/mic.0.2007/006122-0. [DOI] [PubMed] [Google Scholar]
- 154.Hurley MN, Camara M, Smyth AR. Novel approaches to the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. Eur Respir J. 2012;40:1014–1023. doi: 10.1183/09031936.00042012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Smith RS, Iglewski BH P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 156.Soheili V, Tajani AS, Ghodsi R, Bazzaz BSF. Anti-PqsR compounds as next-generation antibacterial agents against Pseudomonas aeruginosa: A review. Eur J Med Chem. 2019;172:26–35. doi: 10.1016/j.ejmech.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 157.Jiang Q, Chen J, Yang C In Y, Yao K. Quorum sensing: a prospective herapeutic target for bacterial diseases. Biomed Res Int. 2019:1–16. doi: 10.1155/2019/2015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Remy B, Mion S, Plener L, Elias M, Chabriere E, Daude D. Interference in bacterial quorum sensing: a biopharmaceutical perspective. Front Pharmacol. 2018;9:1–17. doi: 10.3389/fphar.2018.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Singh S, Wanjari PJ, Bhatia S, Sonwane VC, Chakraborti AK, Bharatam PV. Design, synthesis, biological evaluation and toxicity studies of N, N-disubstituted biguanides as quorum sensing inhibitors. Med Chem Res. 2015;24:1974–1987. [Google Scholar]
- 160.Singh S, Bhatia S, Prasad SB. In silico identification of polyphenolic compounds from the grape fruit as quorum sensing inhibitors. J Chem Pharm Res. 2016;8:411–419. [Google Scholar]
- 161.Bhatia S, Singh S, Sivaiah K. In silico docking, ADME and toxicity studies of aryl glyoxamide derivatives as anti-virulence agents. Trends in Pharmaceuticals and Nanotechnology. 2019;1:1–12. [Google Scholar]
- 162.Sachdeva S, Bhatia S, Mittal A, Sinha M. Synthesis, evaluation and in silico studies of 1, 8-naphthyridine derivatives against antimicrobial activity. J App Pharm Sci. 2015;5:53–59. [Google Scholar]
- 163.Guan LL, Onuki H, Kamino K. Bacterial growth stimulation with exogenous siderophore and synthetic n-acyl homoserine lactone autoinducers under iron-limited and low-nutrient conditions. Appl Environ Microbiol. 2000;66:2797–2803. doi: 10.1128/aem.66.7.2797-2803.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 165.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cugini C, Calfee MW, Farrow JM 3rd, Morales DK, Pesci EC, Hogan DA. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- 167.Soheili V, Bazzaz BS, Abdollahpour N, Hadizadeh F. Investigation of Pseudomonas aeruginosa quorum-sensing signaling system for identifying multiple inhibitors using molecular docking and structural analysis methodology. Microb Pathog. 2015;89:73–78. doi: 10.1016/j.micpath.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 168.De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol . 2001;67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiol. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 170.Hodgkinson JT, Galloway WR, Wright M, Mati IK, Nicholson RL, Welch M, et al. Design, synthesis and biological evaluation of non-natural modulators of quorum sensing in Pseudomonas aeruginosa. Org Biomol Chem. 2012;10:6032–6044. doi: 10.1039/c2ob25198a. [DOI] [PubMed] [Google Scholar]
- 171.Kutty SK, Barraud N, Ho KK, Iskander GM, Griffith R, Rice SA, et al. Hybrids of acylated homoserine lactone and nitric oxide donors as inhibitors of quorum sensing and virulence factors in Pseudomonas aeruginosa. Org Biomol Chem. 2015;13:9850–9861. doi: 10.1039/c5ob01373a. [DOI] [PubMed] [Google Scholar]
- 172.Hansen MR, Jakobsen TH, Bang CG, Cohrt AE, Hansen CL, Clausen JW, et al. Triazole-containing N-acyl homoserine lactones targeting the quorum sensing system in Pseudomonas aeruginosa. Bioorg Med Chem. 2015;23:1638–1650. doi: 10.1016/j.bmc.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 173.Michael A, Welsh MA, Blackwell HE. Chemical probes of quorum sensing: from compound development to biological discovery. FEMS Microbiol Rev. 2016;40:774–794. doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Smith KM, Bu Y, Suga H. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem Biol. 2003;10:81–89. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 175.Champion , et al. IGE CH3 PEPTIDE VACCINE, United States Patent, US 9. 2016. 249,233 B2. [Google Scholar]
- 176.Gerdt JP, McInnis CE, Schell TL, Rossi FM, Blackwell HE. Mutational analysis of the quorum-sensing receptor LasR reveals interactions that govern activation and inhibition by non-lactone ligands. Chem Biol. 2014;21:1361–1369. doi: 10.1016/j.chembiol.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Eibergen NR, Moore JD, Mattmann ME, Blackwell HE. Potent and Selective Modulation of the RhlR Quorum Sensing Receptor by Using Non-native Ligands: An Emerging Target for Virulence Control in Pseudomonas aeruginosa. Chembiochem. 2015;16:2348–2356. doi: 10.1002/cbic.201500357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Moore JD, Gerdt JP, Eibergen NR, Blackwell HE. Active Efflux Influences the Potency of Quorum Sensing Inhibitors in Pseudomonas aeruginosa. ChemBioChem. 2014;15:435–442. doi: 10.1002/cbic.201300701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Muh U, Schuster M, Heim R, Singh A, Olson ER, Greenberg EP. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ma L, Liu X, Liang H, Che Y, Chen C, Dai H. Effects of 14-alpha-lipoyl andrographolide on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2012;56:6088–6094. doi: 10.1128/AAC.01119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Smith KM, Bu Y, Suga H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem Biol. 2003;10:563–571. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 182.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Park S, Kim HS, Ok K, Kim Y, Park HD, Byun Y. Design, synthesis and biological evaluation of 4-(alkyloxy)-6-methyl-2H-pyran-2-one derivatives as quorum sensing inhibitors. Bioorg Med Chem Lett. 2015;25:2913–2917. doi: 10.1016/j.bmcl.2015.05.054. [DOI] [PubMed] [Google Scholar]
- 184.Singh S, Bhatia S. In silico identification of albendazole as a quorum sensing inhibitor and its in vitro verification using CviR and LasBreceptors based assay systems. Bioimpacts. 2018;8:201–209. doi: 10.15171/bi.2018.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J, et al. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014;10:1–17. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lidor O, Al-Quntar A, Pesci EC, Steinberg D. Mechanistic analysis of a synthetic inhibitor of the Pseudomonas aeruginosa LasI quorum-sensing signal synthase. Sci Rep. 2015;5:16569. doi: 10.1038/srep16569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Borges A, Simoes M, Todorovic TR, Filipovic NR, Garcia-Sosa AT. Cobalt complex with thiazole-based ligand as new Pseudomonas aeruginosa quorum quencher, biofilm inhibitor and virulence attenuator. Mol. 2018;23:1385–1400. doi: 10.3390/molecules23061385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chourasiya SS, Kathuria D, Singh S, Sonawane VC, Chakraborti AK, Bharatam PV. Design, synthesis and biological evaluation of novel unsymmetrical azines as quorum sensing inhibitors. RSC Adv. 2015;5:80027–80038. [Google Scholar]
- 189.Snow Setzer M, Sharifi-Rad J, Setzer WN. The search for herbal antibiotics: an in silico investigation of antibacterial phytochemicals. J Antibiot. 2016;5:1–113. doi: 10.3390/antibiotics5030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kim H, Lee S, Byun Y, Park H. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci Rep. 2015;5:1–11. doi: 10.1038/srep08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu B, Capilato J, Pham MP, Walker J, Spur B, Rodriguez A, et al. Lipoxin A4 augments host defense in sepsis and reduces Pseudomonas aeruginosa virulence through quorum sensing inhibition. FASEB J. 2016;30:2400–2410. doi: 10.1096/fj.201500029R. [DOI] [PubMed] [Google Scholar]
- 192.Borlee BR, Geske GD, Blackwell HE, Handelsman J. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator LasR in Pseudomonas aeruginosa by high-throughput screening. Appl Environ Microbiol. 2010;76:8255–8258. doi: 10.1128/AEM.00499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pattnaik SS, Ranganathan SK, Ampasala DR, Syed A, Ameen F, Busi S. Attenuation of quorum sensing regulated virulence and biofilm development in Pseudomonas aeruginosa PAO1 by Diaporthephaseolorum SSP12. Microb Pathog. 2018;118:177–189. doi: 10.1016/j.micpath.2018.03.031. [DOI] [PubMed] [Google Scholar]
- 194.Rasamiravaka T, Labtani Q, Duez P, El Jaziri M. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int. 2015;2015:1–18. doi: 10.1155/2015/759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.El-Shaer S, Shaaban M, Barwa R, Hassan R. Control of quorum sensing and virulence factors of Pseudomonas aeruginosa using phenylalanine arginyl b-naphthylamide. J Med Microbiol. 2016;65:1194–1204. doi: 10.1099/jmm.0.000327. [DOI] [PubMed] [Google Scholar]
- 196.Nizalapur S, Kimyon O, Biswas NN, Gardner CR, Griffith R, Rice SA, et al. Design, synthesis and evaluation of N-aryl-glyoxamide derivatives as structurally novel bacterial quorum sensing. Org Biomol Chem. 2016;14:680–693. doi: 10.1039/c5ob01973g. [DOI] [PubMed] [Google Scholar]
- 197.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl Environ Microbiol. 2007;73:3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Muh U, Hare BJ, Duerkop BA, Schuster M, Hanzelka BL, Heim R, et al. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. PNAS. 2006;103:16948–16952. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nizalapur S, Ho KKK, KimyonÃn , Yee E, Berry T, Manefield M, et al. Synthesis and biological evaluation of N-naphthoyl-phenylglyoxamide-based small molecular antimicrobial peptide mimics as novel antimicrobial agents and biofilm inhibitors. Org Biomol Chem. 2016;14:3623–3637. doi: 10.1039/c6ob00298f. [DOI] [PubMed] [Google Scholar]
- 200.Issa R, Meikle ST, James SL, Cooper IR. Use of poly (ε-Lysine) dendrons: A strategy targeting bacterial quorum sensing and biofilm formation. J Conf Sci. 2014:1–8. [Google Scholar]
- 201.Kasper SH, Bonocora RP, Wade JT, Musah RA, Cady NC. Chemical inhibition of kynureninase reduces Pseudomonas aeruginosa quorum sensing and virulence factor expression. ACS Chem Biol. 2016;11:1106–1117. doi: 10.1021/acschembio.5b01082. [DOI] [PubMed] [Google Scholar]
- 202.El-Mowafy SA, Shaaban MI, Abd El Galil KH. Sodium ascorbate as a quorum sensing inhibitor of Pseudomonas aeruginosa. J Appl Microbiol. 2014;117:1388–1399. doi: 10.1111/jam.12631. [DOI] [PubMed] [Google Scholar]
- 203.Hentzer M, Eberl L, Nielsen J, Givskov M. Quorum sensing: a novel target for the treatment of biofilm infections. Bio Drugs. 2003;17:241–250. doi: 10.2165/00063030-200317040-00003. [DOI] [PubMed] [Google Scholar]
- 204.Jakobsen TH, Tolker-Nielsen T, Givskov M. Bacterial biofilm control by perturbation of bacterial signaling processes. Int J Mol Sci. 2017;18:1970–1997. doi: 10.3390/ijms18091970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Paczkowski JE, Mukherjee S, McCready AR, Cong JP, Aquino CJ, Kim H, et al. Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J Biol Chem. 2017;292:4064–4076. doi: 10.1074/jbc.M116.770552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Luo J, Dong B, Wang K, Cai S, Liu T, Cheng X, et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS One. 2017;12:176883–176915. doi: 10.1371/journal.pone.0176883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Morkunas B, Gal B, Galloway WRJD, Hodgkinson JT, Ibbeson BM, Tan YS, et al. Discovery of an inhibitor of the production of the Pseudomonas aeruginosa virulence factor pyocyanin in wild-type cells. Beilstein J Org Chem. 2016;12:1428–1433. doi: 10.3762/bjoc.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tan SYY, Chua SL, Chen Y, Rice SA, Kjelleberg S, Nielsen TE, et al. Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database. Antimicrob Agents Chemother. 2013;57:5629–5641. doi: 10.1128/AAC.00955-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Soukarieh F, Oton EV, Dubern JF, Gomes J, Halliday N, Crespo MDP, et al. In silico and in vitro-guided identification of inhibitors of alkylquinolone-dependent quorum sensing in Pseudomonas aeruginosa. Molecules. 2018;23:257–262. doi: 10.3390/molecules23020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Cámara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jha S K, Rashmi S, Shubhra , Singh HR. High throughput screening of quorum sensing inhibitors based lead molecules for Pseudomonas aeruginosa associated infections. Int J Pharm Clin Res. 2014;6:214–220. [Google Scholar]
- 212.Fong J, Zhang C, Yang R, Boo ZZ, Tan SK, Nielsen TE, et al. Combination therapy strategy of quorum quenching enzyme and quorum sensing inhibitor in suppressing multiple quorum sensing pathways of P. aeruginosa. Sci Rep. 2018;8:1155–1166. doi: 10.1038/s41598-018-19504-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Park S, Kim HS, Ok K, Kim Y, Park HD, Byun Y. Design, synthesis and biological evaluation of 4-(alkyloxy)-6-methyl-2H-pyran-2-one derivatives as quorum sensing inhibitors. Bioorg Med Chem Lett. 2015;25:2913–2917. doi: 10.1016/j.bmcl.2015.05.054. [DOI] [PubMed] [Google Scholar]
- 214.Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, Kote M, et al. screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol. 2005;187:1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sangshetti JN, Khan FAK, Patil RH, Marathe SD, Gade WN, Shinde DB. Biofilm inhibition of linezolid-like Schiff bases: Synthesis, biological activity, molecular docking and in silico ADME prediction. Bioorg Med Chem Lett. 2015;25:874–880. doi: 10.1016/j.bmcl.2014.12.063. [DOI] [PubMed] [Google Scholar]
- 216.Aybey A, Demirkan E. Inhibition of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa by human serum paraoxonase. J Med Microbiol. 2015;65:105–113. doi: 10.1099/jmm.0.000206. [DOI] [PubMed] [Google Scholar]
- 217.Jakobsen TH, Van-Gennip M, Phipps RK, Shanmugham MS, Christensen LD, Alhede M, et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother. 2012;56:2314–2325. doi: 10.1128/AAC.05919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kim B, Park JS, Choi HY, Yoon SS, Kim WG. Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa : A connection between quorum sensing and c-di-GMP. Sci Rep. 2018;8:8617–8630. doi: 10.1038/s41598-018-26974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Goh WK, Gardner CR, Chandra Sekhar KV, Biswas NN, Nizalapur S, Rice SA, et al. Synthesis, quorum sensing inhibition and docking studies of 1,5-dihydropyrrol-2-ones. Bioorg Med Chem. 2015;23:7366–7377. doi: 10.1016/j.bmc.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 220.Kim SK, Park HY, Lee JH. Anthranilate deteriorates the structure of Pseudomonas aeruginosa biofilms and antagonizes the biofilm-enhancing indole effect. Appl Environ Microbiol. 2015;81:2328–2338. doi: 10.1128/AEM.03551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]