Abstract
In earlier research, we identified a 43-kDa c-ErbAα1 protein (p43) in the mitochondrial matrix of rat liver. In the present work, binding experiments indicate that p43 displays an affinity for triiodothyronine (T3) similar to that of the T3 nuclear receptor. Using in organello import experiments, we found that p43 is targeted to the organelle by an unusual process similar to that previously reported for MTF1, a yeast mitochondrial transcription factor. DNA-binding experiments demonstrated that p43 specifically binds to four mitochondrial DNA sequences with a high similarity to nuclear T3 response elements (mt-T3REs). Using in organello transcription experiments, we observed that p43 increases the levels of both precursor and mature mitochondrial transcripts and the ratio of mRNA to rRNA in a T3-dependent manner. These events lead to stimulation of mitochondrial protein synthesis. In transient-transfection assays with reporter genes driven by the mitochondrial D loop or two mt-T3REs located in the D loop, p43 stimulated reporter gene activity only in the presence of T3. All these effects were abolished by deletion of the DNA-binding domain of p43. Finally, p43 overexpression in QM7 cells increased the levels of mitochondrial mRNAs, thus indicating that the in organello influence of p43 was physiologically relevant. These data reveal a novel hormonal pathway functioning within the mitochondrion, involving a truncated form of a nuclear receptor acting as a potent mitochondrial T3-dependent transcription factor.
The regulation of mitochondrial activity by thyroid hormone is well documented. Triiodothyronine (T3) increases the number of mitochondria (20, 24–25) and mitochondrial protein synthesis (33). This hormone is thus considered to be a major regulator of mammalian mitochondrial biogenesis (33). T3 also stimulates mitochondrial metabolism (46) and, in particular, oxidative phosphorylation (48, 50). Some of these effects could involve activation of mitochondrial genome transcription induced by T3 (10, 29, 34). However, the molecular nature of these influences remains highly controversial. In particular, the evidence of mitochondrial high-affinity T3 binding sites which has been provided (19, 23, 49) leads to the proposition of direct action of the hormone on the organelle. Unfortunately, conflicting data and the controversial identification of the ADP/ATP translocator as a T3 mitochondrial receptor has complicated interpretation of this hypothesis. In parallel, the observation that T3 could activate a nuclear gene encoding mt-TFA (18), a potent mitochondrial transcription factor (13, 14), suggests that almost all aspects of the regulation of mitochondrial activity by thyroid hormone could involve the nuclear pathway. However, a recent study by Enriquez et al. (11) demonstrated that T3 acts directly at mitochondrial level to regulate RNA synthesis in the organelle.
In line with this work, previous studies have shed new light on the action of T3 at the mitochondrial level. First, in addition to the 47-kDa full-length T3 nuclear receptor c-ErbAα1, Bigler and Eisenman (4) reported the occurrence of several smaller size cellular c-ErbAα proteins in chicken erythroid cells, some of which displayed an extranuclear location. Moreover, these authors demonstrated that these proteins are synthesized by the use of internal AUGs identified in the c-erbAα1 mRNA (5). Second, by a number of convergent experimental approaches, we established that a 43-kDa protein related to c-ErbAα1 is located in the mitochondrial matrix (54), i.e., via Western blots with highly purified rat liver mitochondrial extracts and two different antisera raised against c-ErbA, immunoprecipitation of a T3-binding 43-kDa mitochondrial protein with one of these antisera, and electron microscopy. In addition, by using DNA binding analysis, we presented evidence that this mitochondrial protein specifically bound to a natural or a synthetic T3 response element (T3RE). Finally, in CV1 cells, by using an expression vector driving the synthesis of a 43-kDa c-ErbAα1 protein from an internal AUG identified by Bigler et al. (5), we found that this protein was localized in the mitochondrion. Interestingly, a recent report of Andersson and Vennström (1) also pointed out that such a protein displays an extranuclear location. Moreover, overexpression of the protein in the same cells induced significant stimulation of mitochondrial activity (54). These data lead us to propose that this truncated c-ErbA protein we called p43 could be a mitochondrial T3 receptor involved in some aspects of the direct influence of the hormone on the organelle (54).
On the basis of these observations, in the present experiments we tested mitochondrial import of p43 and its transcriptional activity by using isolated rat liver mitochondria or living cells. We report here that p43 is a potent T3-dependent transcription factor of the mitochondrial genome, thus clearly demonstrating the existence of direct T3 regulation of mitochondrial gene expression.
MATERIALS AND METHODS
Construction of plasmids and reporter genes.
The mouse D loop was generated by PCR (5′-ACATCAAGAAGAAGGAGCTACTCCC [positions 15347 to 15372] and 3′-CGGT-CTATGGAGGTTTGCATGTG [positions 113 to 136]) from the pST41 plasmid encoding the complete mouse mitochondrial genome (3), kindly provided by D. Ricquier (Centre National de la Recherche Scientifique Meudon, France), and cloned into the pBluescript SK vector. The pD-loop-tk-CAT reporter gene was constructed by insertion of a 1.1-kb HindIII-BamHI fragment of the SK–D-loop into the HindIII-BamHI site of pBL-CAT8+. pDR2-tk-CAT and pRSV-tk-CAT reporter plasmids were constructed by insertion of an oligonucleotide sequence containing natural T3REs (DR2 oligonucleotide, 5′-AGCTTGTCAAGGCATGAAGGTCAGCACG-3′; RSV oligonucleotide, 5′-AGCTTGTCATGCCTTCCTCAACATAGCCGTCAAGGCATGAAGG-3′), found in the rat D loop, into the HindIII-BamHI sites of pBL-CAT. pSG5-FE6, pSG5-Δ1, and pSG5-Δ2 were constructed by insertion of the 1.5-, 1.1-, and 1.0-kb EcoRI fragments, respectively, encoding c-ErbAα1, p43, and p43-ΔDBD into the EcoRI site of pSG5. The inserts were generated by EcoRI digestion of the pF1, pF1Δmet1, and pF1Δmet2 plasmids, kindly provided by J. Bigler and R. N. Eisenman (5). pSG5-mtTFA was constructed by insertion of a 1.3-kb BamHI-BglII fragment of the pQE9-hmtTFA plasmid encoding h-mtTFA into the BamHI-BglII sites of pSG5.
Mitochondrial import.
Import experiments were performed according to the method of Komiya and Mihara (26) by using highly purified isolated rat liver mitochondria (54). Mitochondria were incubated for 45 min in the presence of 5% rabbit reticulocyte lysate containing [35S]methionine-labeled proteins (c-ErbAβ, c-ErbAβ0, c-ErbAα1, p43, p43-ΔDBD, mt-TFA, and pO-DHFR synthesized from the SG5-THRβ, SG5-THRβ0, SG5-FE6, SG5-Δ1, SG5-Δ2, SG5-h-mtTFA, and sp64-pO-DHFR plasmids, respectively, the latter kindly provided by G. C. Shore) (45). After import experiments, mitochondria were treated with proteinase K as previously described (26). Then, 10% of the amount of reticulocyte lysate added to mitochondria for the import experiments was loaded in the control lane. To assess the presence of p43 in the mitochondrial matrix, matrix extracts were prepared after p43 import studies by using an osmotic shock procedure as described by Goglia et al. (19). Purity of matrix extracts was assessed by measurement of cytochrome oxidase activity, which remained lower than the limit of detection of the assay. As indicated in the legend, mitoplasts were prepared by digitonin treatment as previously described by Darley-Usmar et al. (9), mitochondria and rabbit reticulocyte lysates were depleted for ATP and ADP by apyrase treatment, or the membrane potential was decreased by 1 μM fluoryl cyanide m-chlorophenylhydrazone (FCCP) as previously described (26).
Cytoimmunofluorescence.
Stable transfection of CV1 cells and cytoimmunofluorescence studies of p43 were performed as described by Wrutniak et al. (54). Staining was performed with a monoclonal antibody raised against a mitochondrial antigen (Anti-Mitok [Chemicom International, Inc.]; final dilution, 1/30) and with a polyclonal antibody raised against the c-ErbA carboxy-terminal domain (rabbit RHT II antiserum [54]), kindly provided by M. Dauça (Université de Nancy, France) (final dilution, 1/100).
Electromobility shift assays (EMSAs).
Gel retardation experiments were performed according to the method of Wrutniak et al. (54), with 32P-labeled oligonucleotide probes corresponding to the mitochondrial T3RE-like sequences identified on the Rattus norvegicus mitochondrial genome (16): DR0 (ACGTTAGGTCAAGGTGTAGCC; mitochondrial genome positions 743 to 764), Ipal7 (AGCGCGACCTATTTAAGAGTTCATATC; positions 2371 to 2397), DR2 (GTCAAGGCATGAAGGTCAGCAC; positions 15928 to 15949), and RSV1-TRE (TTGATGCCTTCCTCAACATAGCCGTCAAGGCATGAAG; positions 15904 to 15941). A perfect DR4 sequence (TCAGGTCACAGGAGGTCA) was used as control probe, and a sequence belonging to the myogenin promoter (AGCTTCTCTGTGATTTAATGCCAGCGCG) was used as an unrelated probe. A 0.7-μg portion of highly purified mitochondrial protein extracts prepared on heparin agarose columns, as described by Wrutniak et al. (54), was added to each lane. The presence of p43 in the binding complex was assessed by using a monoclonal antibody (LA038 [Quality Biotechnology]; final dilution, 1/10) raised against a specific sequence of the DNA-binding domain of c-ErbA (KSFFRRTIQKNLHPTYSC). A full-length c-ErbAα1 protein synthesized in a baculovirus system was used where indicated (kindly provided by J. Ghysdael, Orsay, France).
T3 binding assays.
p43 affinity for T3 (Ka) was measured in saturation experiments by using 125I-labeled T3 (3.3 mCi/μg; NEN Life Science Products) according to the method of Daadi et al. (8). Nonspecific binding was assessed in simultaneous assays in which a 1 μM concentration of cold T3 was added. Bound and free T3 were separated by using a Sephadex G-50 column.
Cell cultures and transcriptional activation assays.
Myoblasts of the QM7 cell line (2) were grown in Earle’s 199 medium supplemented with 0.2% tryptose phosphate broth, penicillin (100 IU/ml), and 10% T3-depleted fetal calf serum. Serum was T3 depleted according to the method of Samuels et al. (40). However, after hormonal depletion, significant free T3 and T4 levels were still detected by radioenzymology (3 and 15 pM, respectively, in the depleted serum against 6.8 and 86 pM in the control serum).
Transient-transfection assays were performed with QM7 cells by using expression plasmids and chloramphenicol acetyltransferase (CAT) reporter genes previously described, in addition to a Rous sarcoma virus (RSV)–β-galactosidase expression vector, according to the method of Cassar-Malek et al. (7). Where indicated, 10−8 M T3 was added to the culture medium. All transfections were normalized according to β-galactosidase activity, and results were expressed as a percentage of the control value. Data are presented as the means ± the standard errors of the means (SEM) of five separate experiments.
In vitro protein synthesis.
Proteins were synthesized by using the rabbit reticulocyte lysate transcription-translation TNT kit (Promega) according to the manufacturer’s instructions. For import experiments, reactions were performed by using [35S]methionine-labeled p43, p43-ΔDBD, c-ErbAα1, c-ErbAβ, c-ErbAβ0, and mt-TFA (pSG5-Δ1, pSG5-Δ2, pSG5-FE6, pSG5-THRβ, pSG5-THRβ0, pSG5-h-mtTFA, and psp64-pO-DHFR vectors, respectively).
In organello mitochondrial transcription and translation assays.
Transcription and translation assays were performed by using isolated rat liver mitochondria over a 60-min period at 37°C, as described by Ostronoff et al. (37), in the presence of 2% rabbit reticulocyte lysate (containing p43, p43-ΔDBD, or mt-TFA; unprogrammed lysate for control). Mitochondrial RNA was extracted twice at room temperature (37). Translation reactions were performed in the presence of [35S]methionine; newly synthesized mitochondrial proteins were precipitated with trichloroacetic acid (TCA) on a Whatman filter 41, washed five times with 5% TCA–0.1% methionine, twice with ethanol, and dried. Radioactivity was measured by liquid scintillation counting.
Mitochondrial DNA probes and Northern blot analysis.
DNA probes were constructed by specific digestion of the pST41 plasmid, which contains the mouse mitochondrial DNA, and purification of the specific insert by using the Qiaex II kit (Qiagen). Digestion by HincII gave a 1,494-pb insert encoding 12S and 16S rRNA. Specific DNA probes for the mouse mitochondrial cytochrome b (Cytb), cytochrome c oxidase subunit III (COX III), and cytochrome c oxidase subunit I (COX I) were generated by PCR from the pST41 plasmid with the following primers: Cytb (5′-CCTACCTGCCCCATCCAACAT and 3′-ATGGTTAGAGTCCTTAATAGC), COX III (5′-CAAACTCATGCATATCACATA and 3′-ATCTGCATTAGACTGAAAAGG), and COX I (5′-AATCGTTGATTATTCTCAACC and 3′-GTGTAAGCTCCTTGGTTGGAT). S26 ribosomal protein DNA probe was used as an invariant control as previously described (52). After in organello mitochondrial transcription studies, total mitochondrial RNA was analyzed by electrophoresis through 1.4% formaldehyde-agarose gels or through 1.4% agarose gels in the presence of deionized CH3HgOH (12) and then transferred to a nylon membrane. Membranes were prehybridized at 65°C for 30 min (0.5 M phosphate buffer, 1 mM EDTA, 7% sodium dodecyl sulfate [SDS], 1% bovine serum albumin [BSA]), and [32P]dCTP-labeled DNA probes were added. Hybridization was carried out at 65°C for 24 h. After hybridization, membranes were washed according to the following protocol: 2× SSC buffer (25 mM phosphate buffer, 0.25% SDS) (1× SSC is 150 mM NaCl plus 15 mM sodium citrate) for 5 min at 65°C, 0.6× SSC buffer (two times) for 20 min at 65°C, and 0.1× SSC buffer for 5 min at 65°C. Membranes were exposed to X-ray films and analyzed by densitometric scanning.
Western blot analysis.
A 100-μg portion of control or p43-overexpressing CV1 cells was analyzed by using RHTII antiserum raised against c-ErbA. Portions (100 μg) of control or p43-overexpressing QM7 cells were analyzed with rabbit antiserum raised against mt-TFA produced by injections of recombinant protein obtained by purification by Ni2+ affinity chromatography after E. coli transformation with pQE9–mt-TFA.
RESULTS
p43 displays a binding affinity for T3 that is similar to that of the c-ErbAα1 nuclear receptor.
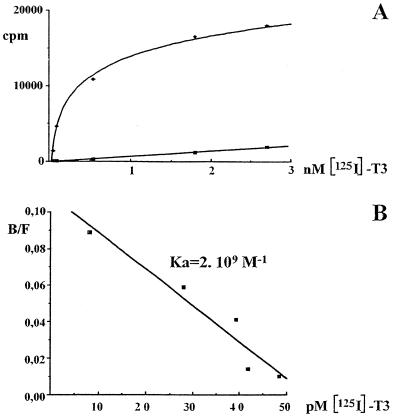
In order to define the p43 binding affinity for T3, we performed saturation experiments with [125I]T3. After Scatchard analysis, we found that p43 displayed a strong affinity (Ka = 2 · 109 M−1) for the hormone (Fig. 1), a finding similar to that previously recorded for the c-ErbAα1 nuclear receptor (Ka = 3 · 109 M−1 [42]). This result indicated that deletion of the amino-terminal domain of c-ErbAα1 did not alter its ability to bind T3.
FIG. 1.
Binding affinity of p43 for triiodothyronine. (A) Saturation experiments were performed with p43 synthesized in rabbit reticulocyte lysate and labeled with [125I]T3. (B) The affinity constant (Ka) was estimated by using a Scatchard linearization of saturation experiments (43). Data are the means ± the SEM of three separate experiments.
In contrast to full-length c-ErbA T3 nuclear receptors, p43 is imported into mitochondria.
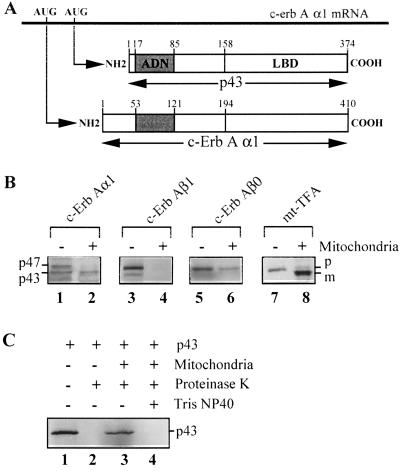
The mitochondrial import of p43 or of the full-length c-ErbAα1 and c-ErbAβ1 T3 receptors was studied by using purified isolated rat liver mitochondria and in vitro-synthesized 35S-labeled proteins in rabbit reticulocyte lysate. As a positive control, the simultaneous import of the mitochondrial transcription factor mt-TFA was observed. In addition, at the end of the experiment, the mitochondrial pellet was subjected to a proteinase K treatment in order to prevent any possible contamination of mitochondria by nonimported proteins (26).
As previously reported by Bigler et al. (5), translation with the pSG5-FE6 vector gave rise to two major c-ErbAα1 proteins with molecular masses of 47 kDa (the full-length T3 nuclear receptor) and 43 kDa (p43), as illustrated in Fig. 2A. Using reticulocyte lysates containing the two proteins, we detected only the shorter form in the organelle after import experiments (Fig. 2B, lane 2). Data obtained by using a 35S-labeled p43 protein synthesized from another vector, pSG5-Δ1, established the specific mitochondrial import of this protein as follows: (i) p43 is highly sensitive to proteinase K treatment (Fig. 2C, lane 2), and (ii) it was protected from proteolysis after the import experiment (Fig. 2C, lane 3). At this step, solubilization of mitochondria by Tris–NP-40 restored the sensitivity of p43 to proteinase K (Fig. 2C, lane 4). By contrast, we failed to detect any significant import of the c-ErbAβ1 form of the T3 nuclear receptor (Fig. 2B, lane 4), in agreement with our previous work where this protein was not detected in mitochondrial extracts (54). However, most nonmammalian vertebrates express a c-ErbAβ0 variant with a very short amino terminus, showing an important homology with p43 (15). To test the possibility that c-ErbAβ0 could enter into mitochondria, we performed import experiments with 35S-labeled TRβ0 protein synthesized from the pSG5-THRβ0 plasmid. As observed for p43, THRβ0 is imported into mitochondria (Fig. 2B, lane 6). Finally, as expected, mt-TFA was imported with the predicted protein cleavage (38), thus validating the experimental procedure (Fig. 2B, lane 8). All of these results demonstrated that, in contrast to the full-length c-ErbAα1 or the c-ErbAβ1 T3 receptors, the 43-kDa c-ErbAα1 protein is imported into the mitochondrial matrix.
FIG. 2.
Import of full-length thyroid hormone receptors (c-ErbAα1, c-ErbAβ1, and c-ErbAβ0) and mt-TFA into mitochondria. (A) p43 is synthesized through the use of an internal AUG of c-erbAα1 mRNA. (B) Import experiments (+) were performed with isolated rat liver mitochondria in the presence of 5% rabbit reticulocyte lysate containing 35S-labeled proteins and incubated at 30°C for 50 min. Import reactions were stopped by cooling on ice, and samples were treated with proteinase K to prevent contamination by nonimported proteins. Mitochondria were collected by centrifugation, and after SDS-polyacrylamide gel electrophoresis, the presence of labeled proteins was assessed by exposure to X-ray films. (C) Sensitivity of p43 to proteinase K before and after mitochondrial import. Lanes: 1, 10% of the amount of labeled proteins used in import experiments was loaded; lane 2, treatment of p43 with proteinase K; lane 3, p43 import experiment; lane 4, treatment with proteinase K after p43 import and solubilization of mitochondrial membranes by Tris–NP-40. −, Control lane (10% of the amount of labeled proteins used in import experiments was loaded; +, import experiment; p47, c-ErbAα1 nuclear receptor; p43, mitochondrial c-ErbAα1 protein; p, precursor protein; m, mature protein.
Typical steps involved in mitochondrial protein import have already been described: (i) binding of the protein to a mitochondrial outer membrane receptor; (ii) translocation through the outer and inner mitochondrial membranes, a process generally driven by the mitochondrial membrane potential; and (iii) unfolding to an active conformation involving ATP-dependent interactions with mitochondrial heat shock proteins. In addition, the targeting sequence of almost all mitochondrial protein imported is cleaved by matrix proteases; however, if this sequence is included in the mature protein sequence, protein import does not lead to a reduction in protein size (35).
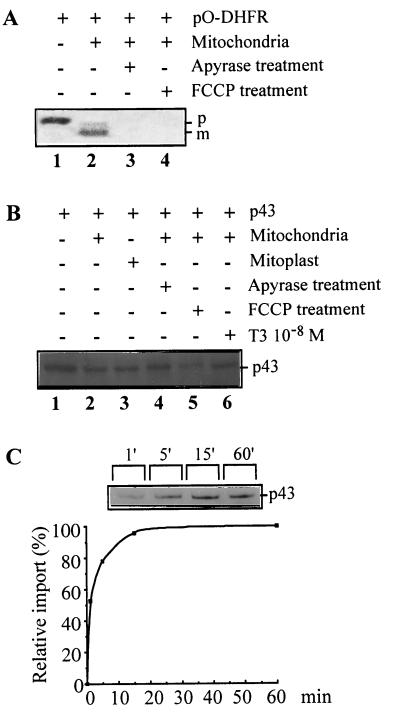
To assess the involvement of several of these steps in p43 mitochondrial import, we studied the influence of apyrase (used to deplete ATP and ADP stores) or FCCP (used to decrease the mitochondrial membrane potential). The efficiency of such treatment was validated in our experimental conditions by using pO-DHFR (a protein where the matrix targeting signal of mitochondrial preornithine carbamyl transferase was fused to dihydrofolate reductase [45]). As previously shown (45), pO-DHFR import occurred with the predicted protein cleavage (Fig. 3A, lane 2) and was strongly inhibited by apyrase (Fig. 3A, lane 3) or FCCP (Fig. 3A, lane 4). First, p43 size did not decrease after mitochondrial protein import (Fig. 3B, lane 2), thus ruling out the pre-sequence cleavage pathway for this protein. Moreover, removal of the outer membrane by treatment with digitonin before import did not significantly affect the amounts of p43 recovered in the matrix extract (Fig. 3B, lane 3), suggesting that interaction of p43 with the outer mitochondrial receptor is not a prerequisite for its translocation. In addition, reduction of the mitochondrial membrane potential by FCCP or depletion of mitochondrial ATP stores by apyrase did not affect p43 import (Fig. 3B, lanes 5 and 4). Finally, exogenous T3 addition did not influence p43 import (Fig. 3B, lane 6).
FIG. 3.
p43 import takes place through an unusual pathway. (A) pO-DHFR import. p, precursor protein; m, mature protein. (B) p43 import was studied as described in the legend to Fig. 2. After import and proteinase K treatment, matrix proteins were specifically extracted by osmotic shock and loaded in each well. A total of 10% of the amount of labeled p43 used in import experiments was loaded in the control lane (lane 1). Prior to import, when indicated, mitoplast and ATP-depleted mitochondria were obtained respectively after digitonin and apyrase treatments. Mitochondrial membrane potential was dissipated by the addition of FCCP prior to import experiments. (C) Time-related changes in the amount of labeled p43 imported into mitochondria and relative p43 import expressed as the percentage of the value recorded after 60 min of incubation. Mitochondrial proteins were collected after increasing times of incubation at 30°C.
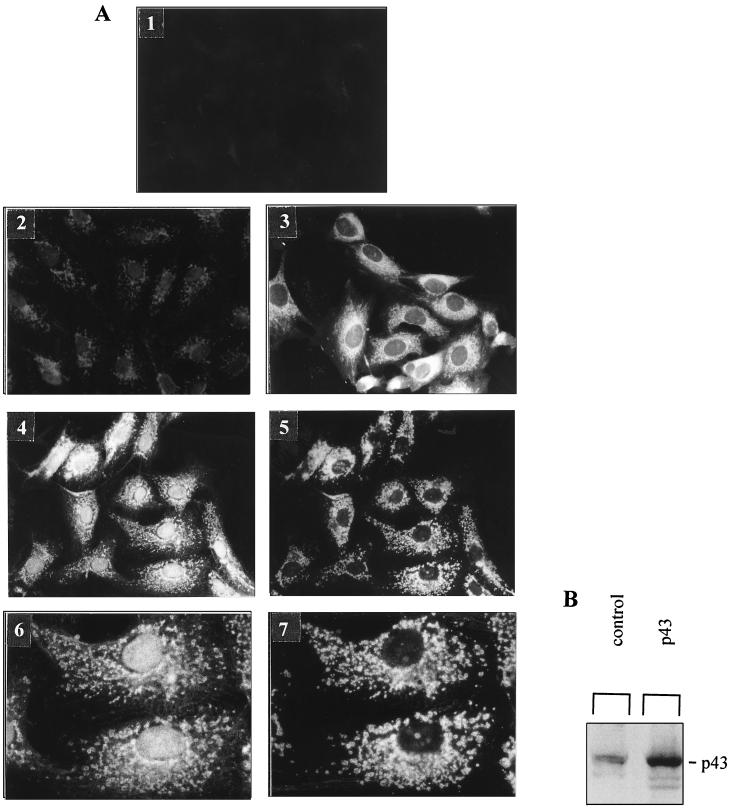
A study of the time course of p43 import demonstrated that the protein was rapidly imported (Fig. 3C). Significant import was detected within the first minute of the experiment; in addition, the maximal mitochondrial p43 level was recorded after 15 min of incubation and did not change thereafter. In agreement with these data, comparison of the total amount of p43 present in the reaction medium with the amount of imported p43 suggested the involvement of a saturable process. To test this hypothesis, we overexpressed p43 by stable transfection of simian CV1 cells, which do not express high levels of c-ErbA proteins. In cytoimmunofluorescence experiments that used the same procedure (54) no staining was observed with the c-ErbA preabsorbed RHTII antiserum in control cells tranfected with empty vector (Fig. 4A1). By contrast, with RHTII antiserum we found a slight labeling (Fig. 4A2). Moreover, as previously demonstrated when p43 is moderately expressed, the protein was predominantly localized in the mitochondrion (Fig. 4A3). However, when it was strongly overexpressed, the protein displayed both a mitochondrial and a nuclear location (Fig. 4A4). In the present experiments, the use of an antibody raised against a mitochondrial antigen (Fig. 4A5) confirmed the colocalization of p43 and mitochondria (Fig. 4A4 to 4A7). In addition, Western blot experiments were performed with control or p43 overexpressing CV1 cells in order to assess that the main protein overexpressed is 43 kDa (Fig. 4B). All of these data indicate that mitochondrial p43 import is a saturable process and that the protein is essentially imported into the nucleus after mitochondrial import saturation. Interestingly, in agreement with the nuclear location of a testis-specific mt-TFA isoform previously reported by Larsson et al. (27), we also observed that significant amounts of mt-TFA reached the nucleus after overexpression of this mitochondrial transcription factor (data not shown), thus suggesting that such a feature is not specific to p43.
FIG. 4.
p43 mitochondrial import occurs according to a saturable process. (A) Panels: 1, staining of CV1 cells with a c-ErbA-preabsorbed RHTII antiserum (antibody raised against c-ErbA; final dilution, 1/100); 2, staining of control CV1 cells transfected with empty vector; 3 to 7, staining of CV1 cells overexpressing p43 after stable transfection; 2 to 4, staining with rabbit RHTII antiserum (final dilution, 1/100); 3, mild p43 overexpression level leads to a major mitochondrial p43 location; 4, high p43 overexpression level leads to simultaneous mitochondrial and nuclear p43 locations; 5, staining with an antibody raised against a mitochondrial antigen (Anti-Mitok; final dilution, 1/30) (same microscopic field as in panel 4) (magnification, ×400); 6 and 7, magnification of microscopic fields 4 and 5 demonstrating colocalization of the two antigens. (B) High p43 overexpression in CV1 cells. Western blot experiments were performed with RHTII antiserum. Proteins (100 μg) were loaded into each lane.
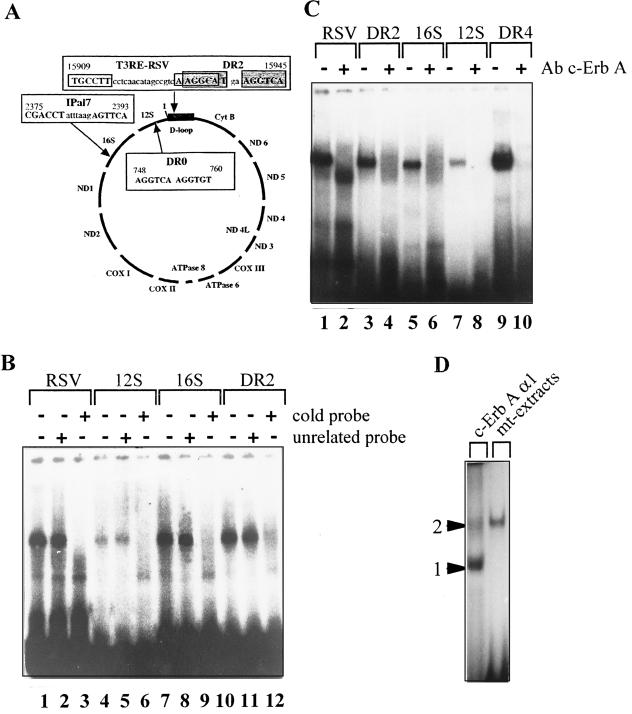
p43 binds to specific sequences of the mitochondrial genome.
The mitochondrial 43-kDa c-ErbAα1 protein is truncated by a number of amino acids upstream of the DNA-binding domain of the T3 nuclear receptor. In addition, we have previously demonstrated that p43 specifically binds to a synthetic palindromic and a natural nuclear T3RE sequence or to a DR2 sequence identified in the mitochondrial genome (54). This led us to search for other mitochondrial T3RE (mt-T3RE) sequences. In addition to the initial DR2 sequence, we found three other potential binding sequences with high similarity to other T3REs described to date (Fig. 5A): (i) a DR0 sequence (6) located in the gene encoding 12S rRNA; (ii) an Ipal7 (inverted palindrome [44]) located in the gene encoding 16S rRNA; and (iii) a perfect RSV-T3RE-like sequence differing from that previously published only by the number of nucleotides between the two half-sites (39). Interestingly, two of these sequences (DR2 and RSV-T3RE) are located in the D loop which contains the promoters used by mt-TFA to induce transcription of the mitochondrial genome.
FIG. 5.
p43 specifically binds to the four T3RE-like sequences recorded in the rat mitochondrial genome. (A) T3RE-like sequences recorded in the rat mitochondrial genome; the first and last nucleotides of the sequences are numbered according to the mitochondrial genome sequence reported by Bibb et al. (3). (B to D) EMSAs were performed with highly purified rat liver mitochondrial extracts and 32P-labeled nucleotidic probes. (B) When indicated, an excess of cold T3RE probe or unrelated probe was added to assess the specificity of binding (“+” = 200-fold molar excess). (C) When indicated, a monoclonal antibody raised against a specific sequence of the DNA-binding domain of c-ErbA (LA038; final dilution, 1/10) was incubated for 15 min with the mitochondrial extract prior to the addition of the labeled probe. The DR4 probe is used as a positive control. (D) Comparison of the binding patterns to a DR4 probe of the full-length c-ErbAα1 protein (synthesized in baculovirus) and the p43 mitochondrial complex. Bands: 1, c-ErbAα1 monomer; 2, c-ErbAα1 homodimer. The electrophoretic mobility of the mitochondrial complex is similar to that recorded for the c-ErbAα1 homodimer.
We performed EMSAs with highly purified mitochondrial extracts by using these mitochondrial sequences as probes (mt-T3REs). We found that a mitochondrial protein complex bound to all mt-T3REs, as well as to a DR4 sequence used as a positive control (Fig. 5B and C). The specificity of the binding was demonstrated by the strong inhibition of binding resulting from the addition of an excess of the corresponding cold probe (Fig. 5B, lanes 3, 6, 9, and 12) and by the inability of an unrelated DNA probe to compete for binding (Fig. 5B, lanes 2, 5, 8, and 11). Interestingly, whatever the T3RE probe used in the EMSAs, this mitochondrial complex displayed the same migration pattern, thus suggesting that a common complex binds to all T3REs. In addition, preincubation of mitochondrial extracts with an antibody raised against c-ErbA fully abrogated the binding of the mitochondrial complex to mt-T3REs, as well as to a DR4 T3RE (Fig. 5C, lanes 2, 4, 6, 8, and 10), whereas nonrelated antibodies (raised against ADP/ATP translocator or myosin heavy chain) did not (data not shown), demonstrating that p43 is a major component of this complex. Comparison of the respective binding patterns of this mitochondrial complex and an in vitro-synthesized full-length c-ErbAα1 protein (TRα) with a DR4-T3RE provided additional information. As expected, TRα monomer and dimer were easily detected; in addition, the mitochondrial complex displayed an electrophoretic mobility similar to that observed for the TRα homodimer (Fig. 5D).
All of these data indicating that p43 is a mitochondrial DNA-binding protein led us to test its possible transcriptional activity in mitochondria.
p43 is a T3-dependent transcription factor in the mitochondrial context.
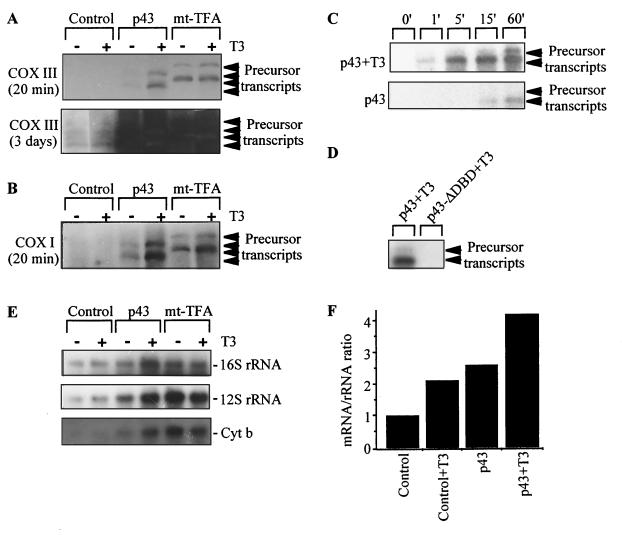
To examine more directly the ability of p43 to activate mitochondrial transcription, we performed in organello transcription experiments with isolated rat liver mitochondria and p43 or mt-TFA proteins synthesized in rabbit reticulocyte lysate.
By using a COX III probe, two precursor transcripts of >13 kb were detected on Northern blots after 20 min of film exposure in mitochondria incubated in the presence of p43 or mt-TFA (Fig. 6A). These transcripts were difficult to detect in mitochondria incubated in the presence of unprogrammed reticulocyte lysate, requiring a much longer film exposure (up to 3 days) to obtain evidence of their presence (Fig. 6A). These data indicate that, like mt-TFA, p43 induced a strong increase in the levels of mitochondrial precursor transcripts in the presence of exogenous T3. A reduced p43 influence was observed in the absence of exogenous T3, in agreement with the fact that mitochondria are a major compartment of T3 accumulation in the cell (32, 49). In addition, all of the precursor transcripts induced by p43 or mt-TFA were also detected in Northern blots performed with other mitochondrial probes (Cytb, ND 5, ND 4, ATPase 6/COX II, ND 6L, COX I, ND 2, or ND 1; Fig. 6B and data not shown). However, we failed to detect the precursor transcript population with the 12S and 16S rRNA probes. Interestingly, the size of precursor transcripts detected in control mitochondria after 3 days of film exposure corresponded to that obtained in the presence of p43 and mt-TFA (Fig. 6A). In addition, we found that in the presence of exogenous T3, p43 induced a strong rise in precursor transcript levels within 5 min of incubation; such a strong increase was not recorded in the absence of the hormone (Fig. 6C). Overall, these observations agree with our data from transient-transfection assays (see below) showing that p43 is a mitochondrial T3-dependent transcription factor. Finally, although p43-ΔDBD was imported in significant amounts into the mitochondrial matrix (data not shown), incubation of mitochondria in the presence of this protein did not induce any rise in the accumulation of precursor transcripts levels (Fig. 6D), thus establishing the absolute requirement of the p43 DNA-binding domain for transcriptional activity.
FIG. 6.
p43 induces a T3-dependent increase in mitochondrial transcripts levels. (A to E) In organello transcription experiments were performed as described by Ostronoff et al. (37) with purified rat liver mitochondria in the presence of 2% rabbit reticulocyte lysate (unprogrammed lysate for control mitochondria, p43, or mt-TFA) and incubated at 37°C for 60 min. When indicated, 10−8 M T3 was added to the incubation medium. Transcription experiments were stopped by cooling on ice. Mitochondria were collected by centrifugation, and mitochondrial RNA was extracted twice at room temperature. (A to D) After in organello transcription experiments, precursor transcripts were detected by Northern blotting with the indicated mitochondrial probes. Duration of film exposure is indicated under the probe. (A and B) Precursor transcript levels recorded after 60 min of in organello transcription. (C) Time-related changes in precursor transcript levels in the presence of p43 (±10−8 M T3). Transcription experiments were performed in the presence of cold p43 and incubated at 37°C for increasing times. (D) Transcription experiments were performed in the presence of cold p43 or p43-ΔDBD and then incubated at 37°C for 60 min with T3 (10−8 M). (E) Northern blot experiments were performed with agarose slab gels containing deionized CH3HgOH after in organello transcription studies. Mature transcripts are detected by hybridization with the indicated mitochondrial probes after 60 min of in organello transcription. (F) Influence of T3 and/or p43 on the value of the mitochondrial mRNA/rRNA ratio. The ratios were obtained after densitometric analysis of CytB and 12S plus 16S rRNA.
In further experiments, we studied the appearance of mature transcripts by using 16S and 12S rRNA and Cytb probes in Northern blot experiments. As expected, mt-TFA stimulated accumulation of these transcripts independently of the presence of exogenous T3 (31). A similar influence was observed for p43, and it was potentiated by exogenous T3 (Fig. 6E). Therefore, these data demonstrated that p43 and mt-TFA induced 16S and 12S rRNA transcription despite their not being detected in the precursor transcript population. As previously described (11), when the relative amount of mRNA was compared with the amount of rRNAs, T3 addition induced a twofold increase in the value of the mRNA/rRNA ratio relative to that of the control (Fig. 6F). Interestingly, we found that this hormonal influence was moderately potentiated by the presence of p43, increasing this ratio up to fourfold (Fig. 6F). These alterations in the mRNA/rRNA ratio suggest that this T3 receptor is involved in the hormone-induced shift of the relative expression of mitochondrial mRNA and rRNA.
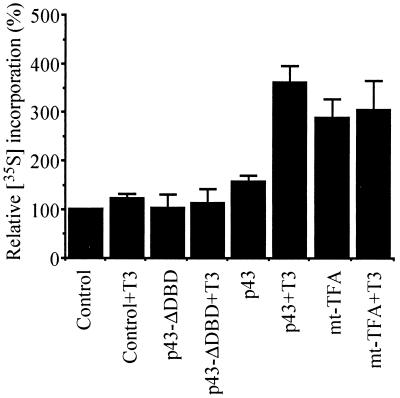
In addition, using the same experimental procedure, we tested the influence of p43 on mitochondrial protein translation by measuring [35S]methionine incorporation. Whereas mitochondrial translation was efficiently inhibited by chloramphenicol in these experiments (data not shown), incorporation of labeled methionine was stimulated threefold by mt-TFA (Fig. 7). Interestingly, in the presence of T3, p43 also enhanced this incorporation up to threefold compared to the level of incorporation of control mitochondria (Fig. 7). As expected from the previous results, p43-ΔDBD had no influence on methionine incorporation (Fig. 7).
FIG. 7.
p43 induces a T3-dependent increase in mitochondrial translation. In organello translation experiments were performed as described for Fig. 6 in the presence of [35S]methionine and incubated at 37°C for 120 min. Translation experiments were stopped by cooling on ice. Mitochondria were collected by centrifugation, and mitochondrial proteins were TCA precipitated on Whatman filter 41, washed five times with 5% TCA, and then washed twice with ethanol. Radioactivity was measured by liquid scintillation counting. [35S]methionine incorporation into mitochondrial proteins was expressed as a percentage of control values. Data are presented as the means ± the SEM of five separate experiments. When indicated, 10−8 M T3 was added to the incubation medium.
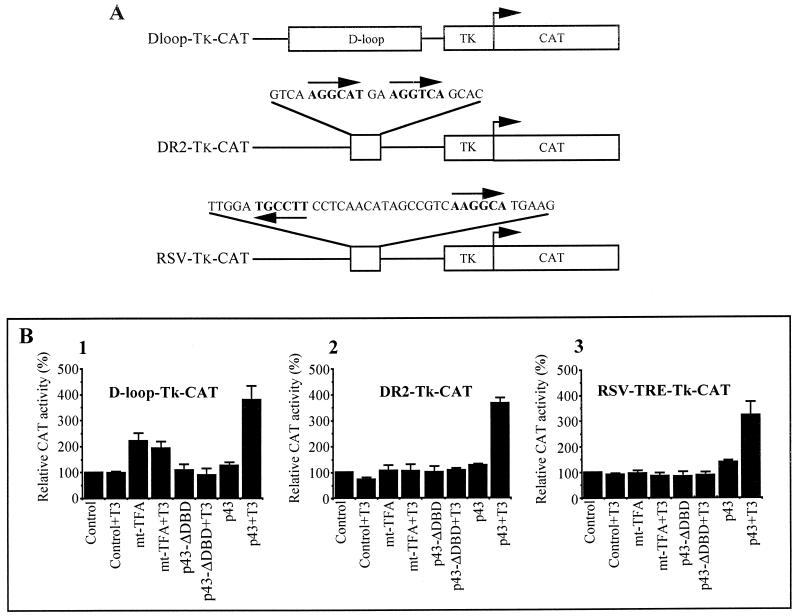
Transient-transfection experiments indicate that mt-T3REs located in the mitochondrial D loop are involved in p43 transcriptional activity.
Taking advantage of the fact that strong overexpression of p43 and mt-TFA results both in a mitochondrial and a nuclear location of the protein, we performed transient-transfection experiments with QM7 myoblasts to assess the involvement of the mt-T3RE located in the mitochondrial sequences in the transcriptional activity of p43. For this purpose, CAT reporter genes under the control of the mitochondrial D loop or the two sequences of mt-T3RE located in the D loop (RSV-T3RE and DR2) were constructed (Fig. 8A) and cotransfected with an expression vector for p43 or mt-TFA (pSG5–mt-TFA). Stimulatory activities of p43 and mt-TFA used as a positive control were simultaneously observed. In addition, a deleted p43 protein without a DNA-binding domain (p43-ΔDBD) was also expressed in these experiments to assess the direct involvement of p43 DNA binding in transcriptional activation.
FIG. 8.
p43 expression induces a T3-dependent stimulation of the activity of reporter genes driven by the D loop or mitochondrial T3REs. (A) Representation of the construction used in CAT assays. (B) Transient-transfection assays were performed with T3-deprived QM7 cells by using the expression vectors and CAT reporter genes indicated in the figure. All results were normalized according to the β-galactosidase activity and are expressed as percentages of the control value. Data are presented as the means ± the SEM of five separate experiments. When indicated, 10−8 M T3 was added to the culture medium.
When a reporter gene driven by the mitochondrial D loop was used, expression of mt-TFA induced a twofold stimulation of reporter gene activity (Fig. 8B1). Interestingly, p43 expression induced a fourfold induction of CAT activity dependent on the presence of T3. Moreover, in experiments with reporter genes under the control of the mt-DR2 or the mt-RSV-T3RE sequences, where mt-TFA was devoid of any transcriptional activity, p43 displayed activity similar to that recorded for the D-loop construct (Fig. 8B2 and 8B3). In all experiments, p43-ΔDBD failed to stimulate reporter gene activity, thus demonstrating that p43 binding to its response element is needed to induce gene reporter activation. Overall, these data suggest that p43 could be a T3-dependent transcription factor of the mitochondrial genome using response elements located in the D loop which differ from those used by mt-TFA.
p43 overexpression increases mitochondrial transcript levels in living cells.
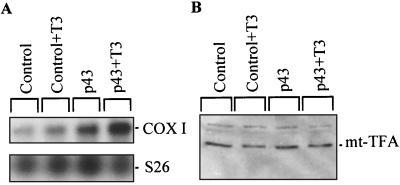
To test p43 mitochondrial transcriptional activity in living cells, we stably overexpressed p43 in QM7 myoblasts. In these QM7 cells cultured in a T3-depleted medium, Northern blot analysis with a COX I probe demonstrated that mitochondrial mRNA levels were enhanced by p43 overexpression in the presence of exogenous T3 (Fig. 9A). To exclude the possibility that this influence is mediated by stimulation of mt-TFA expression at the nuclear level, we performed Western blot experiments with cellular extracts and an antibody raised against mt-TFA (Fig. 9B). We found that p43 overexpression and/or T3 have no influence on mt-TFA protein expression.
FIG. 9.
Stable p43 overexpression in QM7 cells increases COX I mRNA levels without affecting mt-TFA expression. QM7 cells were cultured in a T3-depleted medium according to the method of Samuels et al. (40), and, when indicated, 10−8 M T3 was added to the incubation medium. (A) Northern blot analysis with total RNA extracted from the control or from p43-overexpressing QM7 cells demonstrated that p43 overexpression increases COX I mRNA levels in the presence of exogenous T3; amounts of RNA loaded in each lane were assessed by using S26 mRNA as a control. (B) p43 overexpression in QM7 cells and/or exogenous T3 do not influence mt-TFA protein. Western blot experiments were performed with an antiserum raised against mt-TFA. Proteins (100 μg) were loaded in each lane.
DISCUSSION
The influence of thyroid hormone at the mitochondrial level remains a complex and poorly understood field of research. Several kinds of action have been reported, obviously involving different pathways with regard to their respective latency periods (for a review, see reference 53). For instance, stimulation of oxidative phosphorylation occurs within minutes of thyroid hormone treatment (48, 50), whereas a significant enhancement of mitochondriogenesis is not recorded until after several days (20). Mitochondrial biogenesis is the result of complex mechanisms involving, in particular, a coordinated rise in the expression of nuclear genes encoding mitochondrial proteins and in the expression of the mitochondrial genome. This observation shows the importance of the regulation of mitochondrial transcription by T3.
Interestingly, numerous studies report that thyroid hormone stimulates mitochondrial genome expression (10, 17, 29, 34). This influence could be partly explained by the finding that T3 increases the steady-state levels of mt-TFA mRNA (18), encoding a mitochondrial transcription factor. However, this pathway involving T3 nuclear receptors does not provide a convincing explanation of the short-term stimulation of mitochondrial transcription induced by T3 in isolated mitochondria in the absence of nuclear influence (29). Therefore, one attractive possibility is that a T3-dependent transcription factor directly acts inside the organelle, a possibility supported by the recent data of Enriquez et al. (11).
The identification by our laboratory of a 43-kDa c-ErbAα1 protein located in the mitochondrial matrix displaying simultaneous DNA- and T3-binding activities (p43, 54) led to the hypothesis that p43 was similar to a truncated c-ErbAα1 protein previously characterized by Bigler et al. (5), synthesized by the use of an internal AUG of the c-erbAα1 messenger. Moreover, we found that stable overexpression of p43 in CV1 cells stimulated mitochondrial activity (54), suggesting that this mitochondrial T3-binding protein was directly involved in some aspects of the influence of the hormone on mitochondrial activity. This led us to study the mitochondrial import and function of p43.
In the present study, we first showed that the binding affinity of p43 for T3 is similar to that of the c-ErbAα1 nuclear receptor (42). This result indicated that the amino-terminal deletion occurring in p43 did not alter the T3-binding activity of the protein.
p43 is imported into mitochondria by an unusual pathway previously reported for a yeast mitochondrial transcription factor.
Import experiments with isolated mitochondria revealed several interesting points. First, the inability of the full-length T3 nuclear receptors c-ErbAα1 and β1 to be imported into mitochondria explains the previous failure to detect these proteins in the organelle (54). Second, the rapid import of p43 into the organelle suggests that the amino terminus of the nuclear receptor c-ErbAα1 efficiently abrogates the mitochondrial translocation of the protein. Finally, study of the time course of p43 import suggests that the protein is internalized into mitochondria according to a saturable process, in agreement with our cytoimmunofluorescence data indicating that p43 displays both a mitochondrial and a nuclear location only when highly overexpressed. As in our previous work (54), we failed to detect p43 in rat liver nuclei; p43 nuclear import probably does not occur under physiological conditions. However, these findings are in line with the study of Andersson and Vennström (1) indicating that, when overexpressed, a truncated c-ErbAα1 protein corresponding to p43 could display both a nuclear and an undefined cytoplasmic location. Moreover, comparison of c-ErbAα1 and p43 overexpression led these researchers to conclude that the T3 receptor amino terminus is needed to induce an exclusive nuclear location, in agreement with the present results. Interestingly, the mitochondrial import of TRβ0 observed in the present work is also in line with the results of Andersson and Vennström (1), indicating that, when overexpressed, the protein displays a nuclear and a cytoplasmic location. Therefore, these data suggest that p43 is probably not a unique receptor in mitochondria.
Another set of data demonstrates that p43 import is not affected by experimental alterations of several important factors normally involved in mitochondrial protein import, including interaction with the outer membrane receptor, membrane potential, or mitochondrial ATP stores. Several studies have underlined that the import of some mitochondrial proteins could be independent of one or more of the parameters studied. For instance, cytochrome c or cytochrome c oxidase subunit Va does not interact with the outer mitochondrial membrane receptor (30, 36, 51), whereas the import of apocytochrome c is also independent of the mitochondrial membrane potential (51). More interesting is the observation that import of the yeast mitochondrial transcription factor MTF1 (28) is similar to that recorded here for p43 (41). Therefore, although quite unusual, p43 mitochondrial import occurs according to a process already described. Finally, this import is not affected by hormone, indicating that T3 does not affect p43 activity by regulating its intramitochondrial level. However, import mechanisms of MTF1 or p43 are still unknown.
p43 specifically binds to mitochondrial DNA sequences related to T3REs.
We have previously shown that, in addition to a natural or a synthetic T3RE sequence, p43 specifically binds to a DR2 sequence located in the D loop of the mitochondrial genome (54). This led us to search for other T3RE-related sequences in the mitochondrial genome by using the following criteria: (i) the presence of two half-sites and (ii) the presence of no more than one base mutation in each half-site by comparison with the consensus sequence. These are more stringent criteria than those fulfilled by some natural nuclear T3REs. In addition to the DR2 motif, three other sequences fit these criteria.
In EMSA where the mt-DR2 and a DR4 sequence were used as positive controls, a mitochondrial protein complex specifically bound to all sequences. The abrogation of DNA binding by an antibody raised against c-ErbA demonstrated that p43 is a major component of the mt-T3RE-binding complex. In addition, the electrophoretic mobilities were similar with all probes, suggesting that the same complex binds to all T3REs. Finally, comparison of the respective electrophoretic mobilities of the mitochondrial complex and of the full-length c-ErbAα1 homodimer after DNA binding clearly suggests that p43 does not bind to mt-T3REs as a monomer. This result agrees with a recent study indicating that deletion of a sequence included in the c-ErbAα1 amino terminus strongly favors formation of homodimeric complexes (22). Moreover, this observation raised the question of the mitochondrial partner(s) involved in this binding. In Western blot studies, we failed to detect RXR isoforms in rat liver mitochondrial extracts (data not shown). Therefore, we conclude that the p43 mitochondrial complex does not include this major partner of T3 nuclear receptors.
p43 is a mitochondrial T3-dependent transcription factor.
These data led us to study the possible transcriptional activity of p43 mediated by binding to the previously identified mitochondrial DNA sequences. Using isolated rat liver mitochondria, we found that in the presence of exogenous T3 p43 induced a dramatic rise in the levels of mitochondrial precursor transcripts recorded after 60 min of incubation. In agreement with a previous study of T3 transcriptional activity in the organelle (29), these changes were detected after no more than 5 min of incubation. A weaker, more delayed rise in precursor transcript levels was also detected in the absence of exogenous T3, possibly as a result of the influence of T3 mitochondrial stores (32, 49). Overall, the extent and rapidity of the increase in precursor transcript levels rule out the possibility that changes in mitochondrial RNA stability are a major factor involved in the influence of p43 and T3. In addition, this conclusion is also supported by the observation that the half-lives of mitochondrial rRNA and mRNA are increased in hypothyroid rats (11).
Finally, the failure of p43-ΔDBD to stimulate precursor transcript accumulation indicates that p43 DNA binding is absolutely needed to induce transcriptional activation. Therefore, p43 fulfills all of the experimental criteria to be considered as a T3-dependent transcription factor of the mitochondrial genome: (i) binding to specific mitochondrial DNA sequences and (ii) T3-dependent transcriptional activity induced by binding to DNA. Interestingly, these data may also explain the earlier finding of a previously uncharacterized protein with a molecular mass of ca. 43 kDa in the mt-TFA-containing fraction that induces mitochondrial genome transcription (13).
In addition, Northern blot experiments indicated that the precursor transcripts induced by p43 contain all mitochondrial genes tested except for the 12S and 16S rRNAs. Such data suggest that these precursor transcripts detected in our experiments are partially processed. However, since p43 binds to a T3RE located in the DNA region coding for the 16 rRNA, the possibility that use of this response element induces transcription of a subunit of the mitochondrial genome, including all transcripts except the 12S and 16S rRNAs, cannot be excluded. Despite the lack of previous experimental evidence, this possibility could satisfactory explain the shift in the mRNA/rRNA ratio induced by T3 (reference 11 and the present study).
In parallel with the increase in precursor transcripts levels, mt-TFA, or p43 in the presence of exogenous T3, also induces a significant rise in the levels of mitochondrial mature transcripts. However, this influence was smaller than that on the accumulation of precursor transcripts, suggesting that cleavage of precursor transcripts is a limiting step for protein synthesis in isolated mitochondria. Interestingly, p43 induces a stimulation of mitochondrial protein synthesis to an extent similar to that recorded for mature transcripts. This observation suggests that, at least in these experimental conditions, mitochondrial transcription is a crucial target for regulators of mitochondrial activity.
Taking advantage of the nuclear location of mt-TFA or p43 after overexpression experiments, we used transfection assays to study the involvement in p43 transcriptional activity of the two mt-T3RE sequences located on the mitochondrial D loop. In these experiments, mt-TFA induced a significant activation of a D-loop reporter gene. However, mt-TFA activity was restricted to this reporter. By contrast, p43 stimulated all reporter genes driven by the D loop or mt-T3REs only in the presence of T3. Therefore, although transcriptional activity of both mt-TFA and p43 is mediated by the D loop, it appears that the response elements used by these two factors are not similar. In addition, these data demonstrate that the two mt-T3RE sequences located in the D loop support the mitochondrial transcriptional activity of p43.
Finally, stable overexpression of p43 in QM7 cells in the presence of exogenous T3 increased the levels of mitochondrial mRNAs, without involving an induction of mt-TFA, suggesting that direct stimulation of mitochondrial transcription by p43 is functional in living cells. Reduced stimulation of COX I mRNA by p43 was also recorded in the absence of exogenous T3. However, the remaining activity could be explained by the limited T3 levels measured in the culture medium after hormonal depletion and by the fact that mitochondria are a major compartment of T3 accumulation in the cell (32, 49). In agreement with this possibility, it has been shown that very low levels of T3 could significantly influence transcription with isolated mitochondria (11). However, reduced p43 T3-independent activity cannot be completely ruled out in the mitochondrial context.
Overall, in conjunction with in organello transcription assays, the data obtained in living cells clearly support the proposition that p43 is a potent mitochondrial transcription factor. However, while it provides an understanding of some aspects of T3 action on mitochondria, our work does not explain short-term hormonal influences, including the stimulation of oxidative phosphorylation, which occurs within minutes of T3 treatment and is insensitive to inhibitors of protein synthesis (48). Clearly, other pathways inducing activation of preexisting mitochondrial proteins are probably involved in such effects.
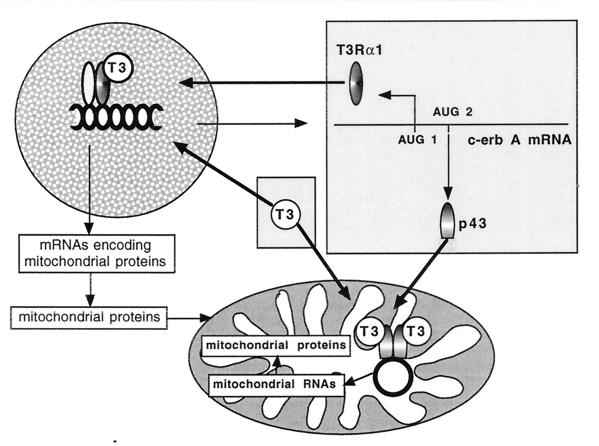
In conclusion, the present study emphasizes the mitochondria are a direct target of T3. Moreover, the presence of a c-ErbAα1-related protein in the organelle, and the additional observation that c-ErbAβ0 is also imported into mitochondria, suggest that other hormonal receptors belonging to the nuclear receptor subfamily could also be located in the organelle. Finally, the finding that the c-erbAα gene simultaneously encodes mitochondrial and nuclear T3 receptors reveals an interesting pathway involved in the coordination of the expression of (i) nuclear genes encoding mitochondrial proteins and (ii) the mitochondrial genome (Fig. 10). Such coordination is probably needed for the stimulation of mitochondrial biogenesis (25, 46), and our data provide a significant explanation for the major influence of T3 on mitochondrial activity.
FIG. 10.
The c-ErbA gene encodes a mitochondrial and a nuclear T3 receptor. With the nuclear receptor, T3 increases the expression of genes encoding the mitochondrial proteins. With the mitochondrial receptor, T3 stimulates mitochondrial genome expression. This mechanism could provide the basis for coordination of nuclear and mitochondrial transcription needed for stimulation of mitochondrial biogenesis.
ACKNOWLEDGMENTS
We thank J. Bigler and R. N. Eisenman for the gift of pF1, pF1Δmet1, and pF1Δmet2 plasmids; F. Flamant for the gift of pSG5-THRβ0 plasmid; G. C. Shore for the gift of psp64-pO-DHFR plasmid; D. Ricquier for the gift of pST41 plasmid; M. Dauça for the gift of RHT II antiserum; and J. Ghysdael for the gift of a baculovirus synthesized c-Erb A α1 protein.
This work was supported by grants from the Institut National de la Recherche Agronomique (INRA), the Association Française contre les Myopathies, the Association de Recherche contre le Cancer, and the Deutsche Forschungsgemeinschaft WI 889/3-2. François Casas, Pierrick Rochard, and Anne Rodier are recipients of fellowships from, respectively, INRA and the Direction Générale de l’Enseignement et de la Recherche, the Ministère de la Recherche et de l’Enseignement, and the Ligue Nationale contre le Cancer.
REFERENCES
- 1.Andersson M L, Vennström B. Chicken thyroid hormone receptor alpha requires the N-terminal amino acids for exclusive nuclear localization. FEBS Lett. 1997;416:291–296. doi: 10.1016/s0014-5793(97)01223-4. [DOI] [PubMed] [Google Scholar]
- 2.Antin P B, Ordhal C P. Isolation and characterization of an avian myogenic cell line. Dev Biol. 1991;143:111–121. doi: 10.1016/0012-1606(91)90058-b. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M J, Van Etten R A, Wright C T, Walberg M W, Clayton D A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 4.Bigler J, Einsenman R N. c-erbA encodes multiple proteins in chicken erythroid cells. Mol Cell Biol. 1988;8:4155–4161. doi: 10.1128/mcb.8.10.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigler J, Hokanson W, Eisenman R N. Thyroid hormone receptor transcriptional activity is potentially autoregulated by truncated forms of the receptor. Mol Cell Biol. 1992;12:2406–2417. doi: 10.1128/mcb.12.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnside J, Darling D S, Carr F E, Chin W W. Thyroid hormone regulation of the rat glycoprotein hormone alpha-subunit gene promoter activity. J Biol Chem. 1989;264:6886–6891. [PubMed] [Google Scholar]
- 7.Cassar-Malek I, Marchal S, Rochard P, Casas F, Wrutniak C, Samarut J, Cabello G. Induction of c-Erb A-AP-1 interactions and c-Erb A transcriptional activity in myoblasts by RXR. Consequences for muscle differentiation. J Biol Chem. 1996;271:11392–11399. doi: 10.1074/jbc.271.19.11392. [DOI] [PubMed] [Google Scholar]
- 8.Daadi M, Lenoir C, Dace A, Bonne J, Teboul M, Planells R, Torresani J. Nuclear factors specifically favor thyroid hormone binding to c-ErbAα1 protein (thyroid hormone receptor α) over-expressed in E. coli. FEBS Lett. 1995;358:137–141. doi: 10.1016/0014-5793(94)01410-3. [DOI] [PubMed] [Google Scholar]
- 9.Darley-Usmar V M, Rickwood D, Wilson M T. Mitochondria, a practical approach. Washington, D.C.: IRL Press, Ltd.; 1987. [Google Scholar]
- 10.De Leo T, Barletta A, Di Meo S, Goglia F, Liverini G, Bove G. Thyroid activity and hepatic mitochondrial population at various ages in the rat. Bull Soc Ital Biol Sper. 1976;52:1920–1925. [PubMed] [Google Scholar]
- 11.Enriquez J A, Fernandez-Silva P, Garrido-Perez N, Lopez-Perez M J, Perez-Martos A, Montoya J. Direct regulation of mitochondrial RNA synthesis by thyroid hormone. Mol Cell Biol. 1999;19:657–670. doi: 10.1128/mcb.19.1.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Silva P, Enriquez J A, Montoya J. A simple procedure for recovering the denaturing effect of methylmercury in agarose gel electrophoresis. BioTechniques. 1992;12:480–482. [PubMed] [Google Scholar]
- 13.Fisher R P, Clayton D A. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher R P, Lisowsky T, Parisi M A, Clayton D A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 15.Forrest D, Sjoberg M, Vennstrom B. Contrasting developmental and tissue-specific expression of alpha and beta thyroid hormone receptor genes. EMBO J. 1990;9:1519–1528. doi: 10.1002/j.1460-2075.1990.tb08270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 17.Gadaleta M N, Barletta A, Caldarazzo M, De Leo T, Saccone C. Triiodothyronine action on RNA synthesis in rat-liver mitochondria. Eur J Biochem. 1972;30:376–381. doi: 10.1111/j.1432-1033.1972.tb02108.x. [DOI] [PubMed] [Google Scholar]
- 18.Garstka H L, Fäcke M, Escribano J R, Wiesner R J. Stoichiometry of mitochondrial transcripts and regulation of gene expression by mitochondrial transcription factor A. Biochem Biophys Res Commun. 1994;200:619–626. doi: 10.1006/bbrc.1994.1493. [DOI] [PubMed] [Google Scholar]
- 19.Goglia F, Torresani J, Barletta A, Liverini G. In vitro binding of triiodothyronine to rat liver mitochondria. Pfleugers Arch Eur J Physiol. 1981;390:120–124. doi: 10.1007/BF00590193. [DOI] [PubMed] [Google Scholar]
- 20.Gross N J. Control of mitochondrial turnover under the influence of thyroid hormone. J Cell Biol. 1971;48:29–40. doi: 10.1083/jcb.48.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustafsson R, Tata J R, Lindberg J, Ernster L. The relationship between the structure and activity of rat skeletal muscle mitochondria after thyroidectomy and thyroid hormone treatment. J Cell Biol. 1965;26:555–578. doi: 10.1083/jcb.26.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadzic E, Habeos I, Raaka B M, Samuels H H. A novel multifunctional motif in the amino-terminal A/B domain of T3Ralpha modulates DNA binding and receptor dimerization. J Biol Chem. 1998;273:10270–10278. doi: 10.1074/jbc.273.17.10270. [DOI] [PubMed] [Google Scholar]
- 23.Hashizume K, Ichikawa K. Localization of 3,5,3′-l-triiodothyronine receptor in rat kidney mitochondrial membranes. Biochem Biophys Res Commun. 1982;106:920–926. doi: 10.1016/0006-291x(82)91798-3. [DOI] [PubMed] [Google Scholar]
- 24.Jakovilcic S, Swift H S, Gross N J, Rabinowitz R. Biochemical and stereological analysis of rat liver mitochondria in different thyroid states. J Cell Biol. 1978;77:887–901. doi: 10.1083/jcb.77.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadenbach B. Regulation of metabolic process in mitochondria. In: Targer J M, Papa S, Quagliariello E, Slater E C, editors. Biochemical aspects of the biogenesis of mitochondria. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1966. pp. 508–517. [Google Scholar]
- 26.Komiya T, Mihara K. Protein import into mammalian mitochondria: characterization of the intermediates along the import pathway of the precursor into the matrix. J Biol Chem. 1996;271:22105–22110. doi: 10.1074/jbc.271.36.22105. [DOI] [PubMed] [Google Scholar]
- 27.Larsson N G, Garman J D, Oldfords A, Barsh G S, Clayton D A. A single mouse gene encodes the mitochondrial transcription factor A and a testis-specific nuclear HMG-box protein. Nat Genet. 1996;13:296–302. doi: 10.1038/ng0796-296. [DOI] [PubMed] [Google Scholar]
- 28.Lisowsky T, Michaelis G. A nuclear gene essential for mitochondrial replication suppresses a defect of mitochondrial transcription in Saccharomyces cerevisiae. Mol Gen Genet. 1988;214:218–223. doi: 10.1007/BF00337714. [DOI] [PubMed] [Google Scholar]
- 29.Martino G, Covello C, De Giovanni R, Filipelli R, Pitrelli G. Direct in vitro action of thyroid hormones on mitochondrial RNA-polymerase. Mol Biol Rep. 1986;11:205–211. doi: 10.1007/BF00419598. [DOI] [PubMed] [Google Scholar]
- 30.Miller B R, Cumsky M G. An unusual mitochondrial import pathway for the precursor to yeast cytochrome c oxidase subunit Va. J Cell Biol. 1991;112:833–841. doi: 10.1083/jcb.112.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montoya J, Perez-Martos A, Garstka H L, Wiesner R J. Regulation of mitochondrial transcription by mitochondrial transcription factor A. Mol Cell Biochem. 1997;174:227–230. [PubMed] [Google Scholar]
- 32.Morel G, Ricard-Blum S, Ardail D. Kinetics of internalization and subcellular binding sites for T3 in mouse liver. Biol Cell. 1996;86:167–174. doi: 10.1016/0248-4900(96)84781-2. [DOI] [PubMed] [Google Scholar]
- 33.Mutvei A, Husman B, Andersson G, Nelson B D. Thyroid hormone and not growth hormone is the principal regulator of mammalian mitochondrial biogenesis. Acta Endocrinol. 1989;121:223–228. doi: 10.1530/acta.0.1210223. [DOI] [PubMed] [Google Scholar]
- 34.Mutvei A, Kuzela S, Nelson B D. Control of mitochondrial transcription by thyroid hormone. Eur J Biochem. 1989;180:235–240. doi: 10.1111/j.1432-1033.1989.tb14638.x. [DOI] [PubMed] [Google Scholar]
- 35.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson D W, Hergersberg C, Neupert W. Role of cytochrome c heme lyase in the import of cytochrome c into mitochondria. J Biol Chem. 1988;263:19034–19042. [PubMed] [Google Scholar]
- 37.Ostronoff L K, Izquierdo J M, Enriquez J A, Montoya J, Cuezva J M. Transient activation of mitochondrial translation regulates the expression of the mitochondrial genome during mammalian mitochondrial differentiation. Biochem J. 1996;316:183–191. doi: 10.1042/bj3160183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parisi M A, Clayton D A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 39.Saatcioglu F, Deng T, Karin M. A novel cis element mediating ligand-independent activation by c-ErbA: implications for hormonal regulation. Cell. 1993;75:1095–1105. doi: 10.1016/0092-8674(93)90319-l. [DOI] [PubMed] [Google Scholar]
- 40.Samuels H H, Stanley F, Casanova J. Depletion of l-3,5,3′-triiodothyronine and l-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology. 1979;105:80–85. doi: 10.1210/endo-105-1-80. [DOI] [PubMed] [Google Scholar]
- 41.Sanyal A, Getz G. Import of transcription factor MTF1 into the yeast mitochondria takes place through an unusual pathway. J Biol Chem. 1995;270:11970–11976. doi: 10.1074/jbc.270.20.11970. [DOI] [PubMed] [Google Scholar]
- 42.Sap J, Munoz A, Damm K, Goldberg Y, Ghysdael J, Leutz A, Beug H, Vennstrom B. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature. 1986;324:635–640. doi: 10.1038/324635a0. [DOI] [PubMed] [Google Scholar]
- 43.Scatchard G. The attractions of proteins for small molecules and ions. Ann N Y Acad Sci. 1949;51:660. [Google Scholar]
- 44.Schräder M, Muller K M, Nayeri S, Kahlen J P, Carlberg C. Vitamin D3-thyroid hormone receptor heterodimer polarity directs ligand sensitivity of transactivation. Nature. 1994;370:382–386. doi: 10.1038/370382a0. [DOI] [PubMed] [Google Scholar]
- 45.Skerjanc I S, Sheffield W P, Randall S K, Silvius J R, Shore G C. Import of precursor proteins into mitochondria: site of polypeptide unfolding. J Biol Chem. 1990;265:9444–9451. [PubMed] [Google Scholar]
- 46.Soboll S, Horst C, Hummerich H, Schumacher J P, Seitz H J. Mitochondrial metabolism in different thyroid states. Biochem J. 1992;281:171–173. doi: 10.1042/bj2810171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sterling K, Milch P O. Thyroid hormone binding by a component of mitochondrial membrane. Proc Natl Acad Sci USA. 1975;72:3225–3229. doi: 10.1073/pnas.72.8.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sterling K, Brenner M A, Sakurada T. Rapid effect of triiodothyronine on the mitochondrial pathway in rat liver in vivo. Science. 1980;210:340–342. doi: 10.1126/science.7423197. [DOI] [PubMed] [Google Scholar]
- 49.Sterling K, Campbell G A, Taliadouros G S, Nunez E A. Mitochondrial binding of triiodothyronine (T3). Demonstration by electron-microscopic radioautography of dispersed liver cells. Cell Tissue Res. 1984;236:321–325. doi: 10.1007/BF00214233. [DOI] [PubMed] [Google Scholar]
- 50.Sterling K, Milch P O, Brenner M A, Lazarus J H. Thyroid hormone action: the mitochondrial pathway. Science. 1977;197:996–999. doi: 10.1126/science.196334. [DOI] [PubMed] [Google Scholar]
- 51.Stuart R A, Nicholson D W, Neupert W. Early steps in mitochondrial protein import: receptor functions can be substituted by the membrane insertion activity of apocytochrome c. Cell. 1990;60:31–43. doi: 10.1016/0092-8674(90)90713-o. [DOI] [PubMed] [Google Scholar]
- 52.Vincent S, Marty L, Fort P. S26 ribosomal protein RNA: an invariant control for gene regulation experiments in eucaryotic cells and tissues. Nucleic Acids Res. 1993;21:1948–1953. doi: 10.1093/nar/21.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wrutniak C, Cabello G. La voie d’action mitochondriale de la triiodothyronine: mythe ou réalité? Med/Sci. 1996;12:475–484. [Google Scholar]
- 54.Wrutniak C, Cassar-Malek I, Marchal S, Rascle A, Heusser S, Keller J M, Fléchon J, Dauça M, Samarut J, Ghysdael J, Cabello G. A 43-kDa protein related to c-Erb A alpha 1 is located in the mitochondrial matrix of rat liver. J Biol Chem. 1995;270:16347–16354. doi: 10.1074/jbc.270.27.16347. [DOI] [PubMed] [Google Scholar]